Abstract
Trypanosomes are protozoan parasites that cycle between a mammalian host (bloodstream form) and an insect host, the Tsetse fly (procyclic stage). In trypanosomes, all mRNAs are trans-spliced as part of their maturation. Genome-wide analysis of trans-splicing indicates the existence of alternative trans-splicing, but little is known regarding RNA-binding proteins that participate in such regulation. In this study, we performed functional analysis of the Trypanosoma brucei heterogeneous nuclear ribonucleoproteins (hnRNP) F/H homologue, a protein known to regulate alternative splicing in metazoa. The hnRNP F/H is highly expressed in the bloodstream form of the parasite, but is also functional in the procyclic form. Transcriptome analyses of RNAi-silenced cells were used to deduce the RNA motif recognized by this protein. A purine rich motif, AAGAA, was enriched in both the regulatory regions flanking the 3′ splice site and poly (A) sites of the regulated genes. The motif was further validated using mini-genes carrying wild-type and mutated sequences in the 3′ and 5′ UTRs, demonstrating the role of hnRNP F/H in mRNA stability and splicing. Biochemical studies confirmed the binding of the protein to this proposed site. The differential expression of the protein and its inverse effects on mRNA level in the two lifecycle stages demonstrate the role of hnRNP F/H in developmental regulation.
INTRODUCTION
Trypanosomes are parasitic protozoa causing infamous diseases such as African sleeping sickness (Trypanosoma brucei), Leishmaniasis and Chagas’ disease or American trypanosomiasis (Trypanosoma cruzi). In addition, this family is one of the best model systems to study the role of posttranscriptional regulation in gene expression. These organisms lack conventional polymerase II promoters for protein coding genes. Although histone modification was recently shown to play a role in gene expression of T. brucei (1), most gene expression regulation is posttranscriptional. The genes are transcribed as polycistronic mRNAs that are processed by the concerted action of trans-splicing and polyadenylation. These processes are coupled, and perturbation of splicing signals affects the polyadenylation of the upstream gene (2–5). In trans-splicing, a common spliced leader (SL) is added to all mRNAs, donated by a small RNA, the SL RNA (6–8). Several recent studies have shed light on the contribution of trans-splicing and polyadenylation to global gene expression and identified alternative processing of transcripts at either their 5′ end, or, more commonly, at their 3′ end (9,10). Despite these recent studies, the most robust mechanism shown so far to regulate the trypanosome transcriptome is mRNA stability (11,12). Recently, it was demonstrated that basal splicing factors such as U2AF35, U2AF65 and SF1 regulate the stability of mature mRNAs (13).
Despite extensive studies on factors that affect mRNA stability and preferential translation, little is known regarding factors that regulate trans-splicing (12). Several RNA-binding proteins (RBP) that were shown to participate in splicing regulation in metazoa also exist in trypanosomes. Among these are heterogeneous nuclear ribonucleoproteins (hnRNP) and proteins carrying a serine-arginine motif (SR) (6–8). Three SR proteins were described in trypanosomes: TSR1, TSR1IP and TRRM1 (14–16). However, their exact role in trans-splicing is not currently known. Polypyrimidine tract binding proteins (PTB) or hnRNP I homologues were shown to be required for trans-splicing of mRNAs carrying a C-rich polypyrimidine tract (17). The PTB proteins were also shown to regulate mRNA stability (17,18).
The hnRNP proteins are modular proteins that generally consist of multiple domains connected by linker regions that vary in length. The most ubiquitous domain of these proteins is the RNA recognition motif (RRM), which is composed of two motifs, ribonucleoprotein domains RNP-1 and RNP-2, through which the protein associates with the RNA (19,20). The hnRNP proteins undergo nucleocytoplasmic shuttling. In metazoa, hnRNP proteins are known to participate in almost every step of gene expression, including transcription, capping, splicing and polyadenylation, in addition to transport, translation and degradation (19,20).
One of the most interesting and highly selective hnRNP protein families is hnRNP F/H, which includes hnRNP F and several spliced variants of hnRNP H. In mammals, these proteins appear to bind specifically to the poly (G) tracts (21). The RNA-binding domain of these proteins differs from those of most hnRNPs; as the residues that contact RNA in the RRM are not conserved, this class of RRM was named quasi RRM (qRRM) (22,23). The hnRNP F and hnRNP H are highly similar in sequence and structure (24); nevertheless, they antagonize each other in regulation of polyadenylation of mRNAs and have different binding specificities for gene regulatory elements (25). Moreover, hnRNP F is localized in the cytoplasm, whereas hnRNPH1 and H2 (also known as hnRNPH and hnRNPH', respectively) are nuclear (22).
The hnRNP F/H proteins are known for their role in regulation of alternative splicing (21,25–27). Although these proteins are in most cases inhibitors of alternative splicing, they can also function as activators (28–32). The hnRNP F also regulates polyadenylation site choice, by blocking recruitment of a cleavage stimulation polyadenylation factor (25). Recently, a binding site consensus sequence of hnRNP F and H1 was determined using cross-linking immunoprecipitation assay (CLIP): GU rich for F, and GA rich for H1 (33).
In this study, we identified the T. brucei hnRNP F/H homologue based on its domain architecture and the similarity of its RRM domains to qRRM domains of the mammalian hnRNP F. In addition, we defined its cellular localization and preferred binding motif and showed that it is highly expressed in the bloodstream form (BSF) of the parasite. Transcriptome analysis by microarray of cells silenced for the factor in the two lifecycle stages of the parasite demonstrated that a subset of the affected genes is inversely regulated at the two stages. Using two independent motif search approaches, we identified enrichment of purine rich sequences within the upregulated genes in both lifecycle stages. We found significant enrichment of the AAGAA motif in regulatory regions flanking the splice site and the poly (A) site, suggesting that the trypanosome protein is similar to hnRNP H in its binding preference. The predicted binding motif of the trypanosome protein was further confirmed using mini-genes and by ultraviolet (UV)-induced cross-linking.
Taken together, our findings suggest that the T. brucei hnRNP F/H homologue regulates both mRNA stability and splicing. This is the first trypanosome factor shown to be involved not only in splicing and mRNA stability but also in differential stage-specific regulation of gene expression.
MATERIALS AND METHODS
The oligonucleotides used in this study are listed in Supplementary Material S1.
Cell growth and transfection
Procyclic forms of T. brucei strain 29–13, which carries integrated genes for T7 polymerase and the tetracycline repressor (34), were grown and transfected as previously described (35). The BSF of T. brucei strain 427, cell line 1313–514 (a gift from C. Clayton, ZMBH, Heidelberg, Germany) (36) were cultivated at 37°C under 5% CO2 in HMI-9 medium (37). Transfections were performed as previously described (38,39).
Construction of RNAi constructs
The stem-loop construct for silencing of hnRNP F/H in PCF was generated using primers listed in Supplementary Material S1, as described (34). The constructs expressing dsRNA were linearized with EcoRV. The expression of dsRNA was induced using 8 µg/ml tetracycline.
The construct to silence hnRNP F/H in the BSF was prepared by cloning a polymerase chain reaction (PCR) product generated using oligonucleotides listed in Supplementary Material S1, to the p2T7TA-177 vector, as described (38–39).
Preparation of nuclear and cytoplasmic extracts
Trypanosoma brucei procyclics (108) were harvested and washed with phosphate buffered saline (PBS). The cell pellet was resuspended in hypotonic buffer [10 mM HEPES (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol and 5 µg/ml leupeptin]. Next, the cells were broken by 20 strokes in a Dounce homogenizer in the presence of 0.1% Nonidet P-40, and the extract was loaded on the top of 3 ml of sucrose cushion (0.8 M sucrose, 0.5 M MgCl2). The nuclei were collected at 8000g for 10 min. The pellet and the cytoplasmic fractions were analysed by western blotting.
Microarray analysis
Total RNA was isolated from uninduced cells and from silenced cells after 2.5 days of induction and then labelled using the Ambion Amino Allyl MessageAmp II aRNA kit (Ambion). DNA microarrays were obtained through NIAID’s Pathogen Functional Genomics Resource Center (managed and funded by the Division of Microbiology and Infectious Diseases, NIAID, NIH, DHHS, and operated by the J. Craig Venter Institute), hybridized using the Gene Expression hybridization kit (Agilent Technologies) and processed as previously described in detail (17). The data from all arrays were first subjected to Normexp-Background correction (40) and Loess within array normalization (41) using the Bioconductor Limma package (42). The rest of the analysis was performed by Partek® Genomics SuiteTM software, version 6.6 (© 2012 Partek Inc., St. Louis, MO, USA). Normalized data from two to four biological replicates were analysed to identify genes whose expression was up- or downregulated by an arbitrary cutoff of at least 1.5-fold and had P-value < 0.05 in all replicates when testing for differential expression (t-test). Heat maps were generated using Euclidean distance as a similarity measure.
Motif search
All gene sequences were derived from T. brucei genome version 4 (ftp://ftp.sanger.ac.uk/pub/databases/T.brucei_sequences/T.brucei_genome_v4/). For binding site analysis, 300 nt downstream of the 3′ splice site and 300 nt upstream to the poly (A) were used. The 3′ splice site and poly (A) sites were defined based on Kolev et al. (9). In the case of alternative poly (A) sites, the most downstream site was chosen.
Sequence motifs were detected using two complementary approaches: The first was the SFmap web server for predicting binding sites of protein motifs (43). SFmap is based on a weighted rank scoring approach, which computes similarity scores for a given regulatory motif based on information derived from its sequence environment (44). SFmap was used using ‘exact match’ mode. Further, we counted the number of hits per given motif within each sequence and used enrichment analyses. Enrichment of a given motif in the upregulated versus the downregulated genes was evaluated using the Mann–Whitney test; the threshold for statistical significance was defined using Bonferroni correction for multiple testing. The de novo motif search algorithm, DRIMUST (45,46), was independently applied to search for enrichment of motifs in the regions flanking the splice site and poly (A) sites of the unregulated genes. For the DRIMUST analysis, the genes were ranked according to the fold change observed in the microarray experiment. The full ranked list was provided as an input for the algorithm, which searched the entire motif space to detect enriched motif at the top of the list, where the top is data driven.
Northern and primer extension analyses
Primer extension was performed as previously described (47–49). The extension products were analysed on 6% acrylamide denaturing gels. Primers are listed in Supplementary Material S1. For northern analysis, total RNA was extracted, separated on agarose-formaldehyde gel and analysed using a DNA probe that was prepared by random labeling (35).
mRNA stability analysis
Uninduced cells and cells 2.5 days after induction (1.5 × 109 cells) were concentrated and resuspended into 25 ml of SDM-79 medium. Cells were aliquotted into five batches and incubated at 27°C for 30 min. Cells were pre-treated with 2 µg/ml sinefungin (Sigma) for 10 min, and then with 30 µg/ml Actinomycin D (Sigma). The RNA was subjected to northern analysis as previously described (17). Each experiment was repeated three times. The RNA level was normalized to 1 at t = 0, and the decay was fitted to Exp[-R*t]. R was calculated by regressing –ln [(normalized RNA level)] on t (in min), using ordinary least squares without the intercept term. The half life was then calculated as ln(2)/R.
Preparation of hnRNP F/H antibody
The T. brucei hnRNP F/H gene was amplified by PCR using primers listed in Supplementary Material S1. The amplified fragments were cloned into the pHIS expression vector (Novagen). To raise antibodies, 400 µg of the protein was emulsified with an equal volume of complete adjuvant (Difco). The emulsions were injected subcutaneously to female New Zealand white rabbits. The first injection was followed by additional two injections of 200 µg of protein at 2-week intervals. Sera were collected and examined for reactivity by immunofluorescence and western analysis.
Construction of the mini genes
The regulatory elements present in pNS21b, carrying a luciferase reporter gene, were used as described (17,50).
To fuse the AATP11-3′UTR regulatory sequences to the luciferase reporter in pNS21b, different sized fragments (1186, 975, 365 and 109 nt) were generated by PCR using primers listed in Supplementary Material S1 and were cloned into the BamHI and XhoI sites, replacing the pre-existing 3′UTR of pNS21b (17). The ‘AAGAA’ binding motif was mutated by site-directed mutagenesis using PCR, with primers carrying the mutation, and 5′and 3′ primers as specified in Supplementary Material S1.
To fuse the ‘multidrug resistance protein A (Tb927.8.2160)’- 5′UTR regulatory sequences to the luciferase reporter in pNS21b, the ‘multidrug resistance protein A (Tb927.8.2160)’- 5′ regulatory elements (400 nt) were amplified using primers specified in Supplementary Material S1. The sequence between PstI and BglII sites of pNS21b were replaced by the sequence from the Tb927.8.2160 gene. The ‘AAGAA’ binding motifs were mutated by site-directed mutagenesis using PCR, with primers carrying the mutation, and 5′ and 3′ primers specified in Supplementary Material S1.
Western blot analysis
Whole cell lysates (107 cells) were fractionated by SDS–PAGE, transferred to PROTRAN membranes (Whatman) and reacted with the anti hnRNP F/H antibodies described earlier in the text (diluted 1:4000). The bound antibodies were detected with goat anti-rabbit immunoglobulin G coupled to horseradish peroxidase and were visualized by ECL (Amersham Biosciences).
Immunofluorescence assay
Cells were washed with PBS, mounted on poly-L-lysine-coated slides, fixed in 8% formaldehyde and immunofluorescence was performed as described (51), using anti hnRNP F/H antisera. The cells were visualized by Nikon eclipse 90i microscope with Retiga 2000R (QIMAGING) camera.
RNase protection assay
The anti-sense RNA probe was transcribed in vitro by T7 polymerase (Ambion Megascript T7) using a PCR product encoding for the gene and carrying the T7 promoter. Total RNA (30 µg) was mixed with 105 cpm of gel-purified RNA probe and concentrated by ethanol precipitation. RNase protection was performed essentially as described (52). The protected fragments were precipitated with ethanol in the presence of sodium acetate and analysed on a 6% polyacrylamide, 7 M urea denaturing gel.
Quantitative real-time PCR
Real-time PCR was performed in a two-step reaction. First, cDNA was prepared from total RNA (1 µg) derived from uninduced cells (−Tet) or cells after 2.5 days of silencing (+Tet), using random primer and the RevertAid™ First Strand cDNA synthesis kit (Fermentas) following the manufacturer’s instructions. Next, real-time PCR was performed using 1 µl of cDNA (diluted 1: 100), 1 µM primers and Absolute Blue QPCR SYBR® Green ROX mix (Thermo Scientific). Quantitative RT-PCR was performed on a Chromo4 Real-Time PCR detection system (Bio-Rad), as follows: 95° for 2 min, followed by 40 cycles of 95° for 30 s, 60° for 30 s and 72° for 10 s. A concentration curve of amplified product, purified using the QIAquick PCR purification kit (Qiagen), was determined using the Opticon Monitor3 software supplied with the Opticon4 apparatus. The concentration curve was used to determine the amount of PCR product present in each sample (53–54).
In vitro cross-linking of hnRNP F/H to RNA substrates
Pre-mRNA was produced by in vitro transcription with T7 RNA polymerase (Promega) using a template constructed by PCR carrying the T7 promoter from primers listed in Supplementary Table S1. Whole cell extract was prepared from PCF as previously described (55). BSF extract was prepared by lysis in hypotonic buffer using 0.1% NP-40 (35), then proteins were extracted at 400 mM KCl, and the extract was diluted to 100 mM KCl. Gel purified RNA (10 000–50 000 cpm ∼2–10 ng) was incubated for 20 min on ice in the presence of (20 mM Tris HCl at pH 7.7, 5 mM MgCl2, 1 mM EDTA, 100 mM KCl) and extract (0.5–15 µg of protein per reaction). The reaction was then UV cross-linked at 254 nm (120 mJ/cm2) using a Bio-Link cross-linker (Vilber Lurant). Samples were treated with 20 µg of RNaseA for 30 min at 37°C. Proteins were analysed on a 10% SDS–PAGE and detected by autoradiography. For competition experiments with unlabelled RNA, wild-type and mutant transcripts were in vitro transcribed using the T7 Polymerase (Promega).
RESULTS
A T. brucei hnRNP F/H homologue
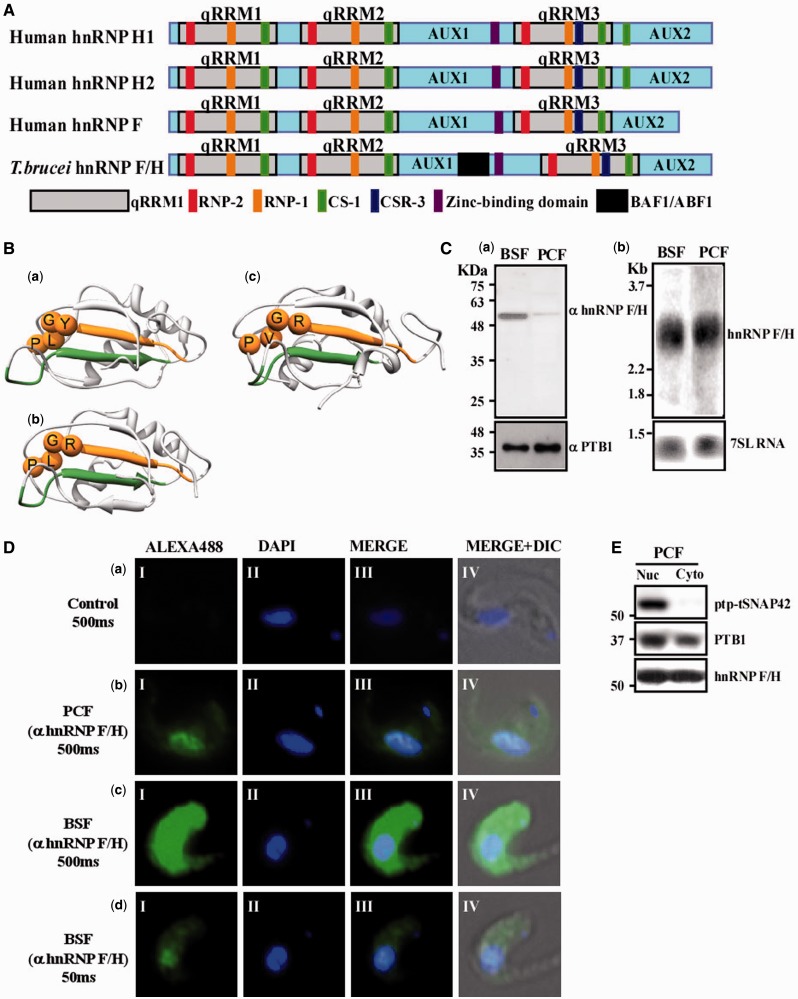
The Tb927.2.3880 protein was previously annotated by us as the homologue of mammalian hnRNP F/H (7). To assess the similarity of Tb927.2.3880 to hnRNP F/H, the sequence of the T. brucei protein was compared with the human hnRNP F, H1 and H2 proteins and showed a similar domain structure to hnRNP F (Figure 1A). Further, we conducted Multiple Sequence Alignment (Supplementary Figure S2) of the T. brucei, human and mouse hnRNP F/H proteins, demonstrating a high sequence similarity of the T. brucei qRRM domains to the qRRMs of the human hnRNP F and hnRNP H domains. Consistently also, the auxiliary domains (AUX1 and 2) (58) and the cold sensitive domain (CS) (Supplementary Material S2) (59) of the T. brucei protein show high sequence similarity with the counterpart domains of hnRNP F and hnRNP H proteins. A characteristic feature of hnRNP F/H is the deviation of its RRM, which is termed qRRM, from the classical RRM_1 domain found in other members of the hnRNP family (21–23). Inspecting the sequence of RNP1 and RNP2 in the qRRM revealed that the T. brucei protein carries the consensus RNP-2 [R/Y]G[L/V]P sequence of the qRRM (Supplementary Material S2). Another conserved domain characteristic of hnRNP F/H is the zinc-binding domain (CHHC motif-pfam08080) at the C-terminal end of AUX1. As shown in Figure 1A, the T. brucei protein also contains this domain, further supporting its close homology to the hnRNP F/H family. However, although human hnRNP H contains two CS-1 domains at the C-terminal, the second one located at the beginning of AUX2, the human hnRNP F contains a single domain, like the T. brucei protein, suggesting that the T. brucei protein might be a common ancestor of the human hnRNP F and hnRNP H.
Figure 1.
Identification and localization of the hnRNP F/H homologue in trypanosomes. (A) Schematic comparison of human hnRNP H1, hnRNP H2 and hnRNP F, with Tb hnRNP F/H. Each protein contains three qRRM domains (light grey boxes) together with two auxiliary domains (AUX1 and AUX2, light blue boxes). The positions of the RNP-1 (brown), RNP-2 (red), CS-1 (green) CSR-3 (dark blue), BAF1/ABF1 (black) and zinc-binding domain (purple) consensus sequences are indicated (22). (B) The predicted 3D structure of the RRM of T.brucei hnRNP F/H proteins. The structures of (a) first RRM, (b) second RRM and (c) third RRM were predicted using MODELLER (56). The RNP domains are coloured in deep green and orange for RNP-1 and RNP-2, respectively. The Cα atoms of characteristic residues of the RNP-2 of the qRRM [R/Y]G[L/V]P are shown as spheres. Sequence identity of the targets to the template are 30, 37 and 37% for the first, second and third RRMs, respectively. Graphics were prepared and analyses performed using the UCSF Chimera package (57). (C) (a) hnRNP F/H is differentially expressed in two stages of the parasite lifecycle. Whole cell extracts (107 cells) from both parasite stages (BSF and PCF) were separated on 10% SDS–PAGE and subjected to western analysis using anti-hnRNP F/H antibodies (diluted 1:4000). Molecular mass markers are indicated. The level of PTB1 was used as a control for equal loading. (b) Northern analysis for the hnRNP F/H in two stages of the parasites. RNA was prepared from PCF and BSF. Total RNA (20 µg) was subjected to northern analysis with a T7 transcribed antisense RNA probe specific to the hnRNP F/H genes. 7SL RNA, used to control for equal loading, is indicated. (D) Immunofluorescence of hnRNP F/H in two stages of the parasites (PCF and BSF). Cells were fixed with 8% paraformaldehyde for 20 min, incubated with anti-hnRNP F/H antibody as described in ‘Materials and Methods’ section, and detected by a Alexa488-conjugated secondary antibody. In control, cells were incubated with PBS lacking anti-hnRNP F/H antibody. (Panel I) fluorescence of the hnRNP F/H protein; (panel II) nuclei stained with DAPI; (panel III) merge of panels I and II and; (panel IV) DIC merge with panel III. The exposure time is indicated. (E) Cellular localization of the hnRNP F/H in PCF. Nuclear and cytoplasmic extracts were prepared from the PCF cells expressing TAP-PTP-tSNAP42. Proteins were prepared from the same cell equivalent and were separated on a 10% SDS–PAGE and subjected to western analysis with antibodies to PTB1 (1:10 000) (17) and anti-hnRNP F/H (1: 4000, this study).
To further test the hypothesis that the Tb927.2.3880 protein is an hnRNP F/H orthologue, we used HHpred, which is a powerful homology detection algorithm. HHpred relies on a Hidden Markov Model comparison with search for homology (60). HHpred was used on each of the predicted qRRM domains of the T. brucei protein. The automatic search against all domains in the Protein Data Bank (PDB) selected the second qRRM of the human hnRNP F as the closest homologue of each of the T. brucei domains with high sequence identity of 30, 37 and 37% to the first, second and third qRRMs of the T. brucei protein, respectively. The high homology to qRRMs of the human hnRNP F supports our contention that the T. brucei protein is the hnRNP F homologue. A structural model for each of the T. brucei qRRMs based on the NMR-based structure of the second qRRM domain of human hnRNP F (PDB ID 2hgm) is shown in Figure 1B (a–c). As expected from the high sequence identities between the predicted T. brucei qRRMs and the human hnRNP F qRRMs, the 3D models generated by the comparative modelling program MODELLER (56) were of high quality, with Z-scores values well within the range of Z-scores of experimentally solved structures in PDB (61). The high sequence identity and similar domain composition to the human hnRNP F and hnRNP H isoforms, as well as the high quality of the model we obtained of the T. brucei RRM regions based on the human hnRNP F qRRM, further support our contention that the T. brucei protein is an hnRNP F/H homologue. Nevertheless, based on evolutionary conservation alone, we could not distinguish whether the protein is a homologue of H or F.
Interestingly, analysis of the T. brucei protein domains using CD-search (62) revealed partial resemblance to a protein domain present in BAF1/ABF1, which is a chromatin-associated factor in yeast, known to remodel chromatin and to affect gene expression by modifying local chromatin architecture (63); this similarity suggests that T. brucei hnRNP F/H may interact with chromatin remodelling factors.
hnRNP F/H is more abundant in the BSF and is localized both to the nucleus and the cytoplasm
As first steps in deciphering the function of the hnRNP F/H, we examined its expression level and localization in the cell. Antibodies were prepared as described in ‘Materials and Methods’ section, and their specificity was examined using whole cell extracts from both stages of the parasites. The results, presented in Figure 1C-a, demonstrate the specificity of the recognition and the differential expression of the protein, which was ∼100-fold more abundant in the BSF. To examine the molecular mechanism of the differential expression of the protein in the two life stages, the mRNA levels were determined by northern analysis (Figure 1C-b) and indicated no major difference in the steady-state level of the mRNA, suggesting that the differential expression of the protein must be regulated at the level of translation or protein stability. Next, the localization of the protein was examined in the BSF and PCF parasites by immunofluorescence. The results (Figure 1D) indicate the location of the protein in nuclear speckles, similar to the localization observed for T. brucei PTB (17). However, the proteins can also be seen in the cytoplasm, but not in speckles. The difference in the amount of the protein in the two stages is reflected by the exposure time used to obtain the images. To confirm the presence of hnRNPF/H in the cytoplasm, cellular fractionation was performed using a cell line expressing the SL RNA transcription factor PTP-tagged tSNAP42. The results demonstrate that although tSNAP42 is found only in the nucleus, both PTB1 and hnRNP F/H are also found in the cytoplasmic fraction (Figure 1E).
Silencing of hnRNP F/H in BSF and PCF differentially affect the parasite transcriptome
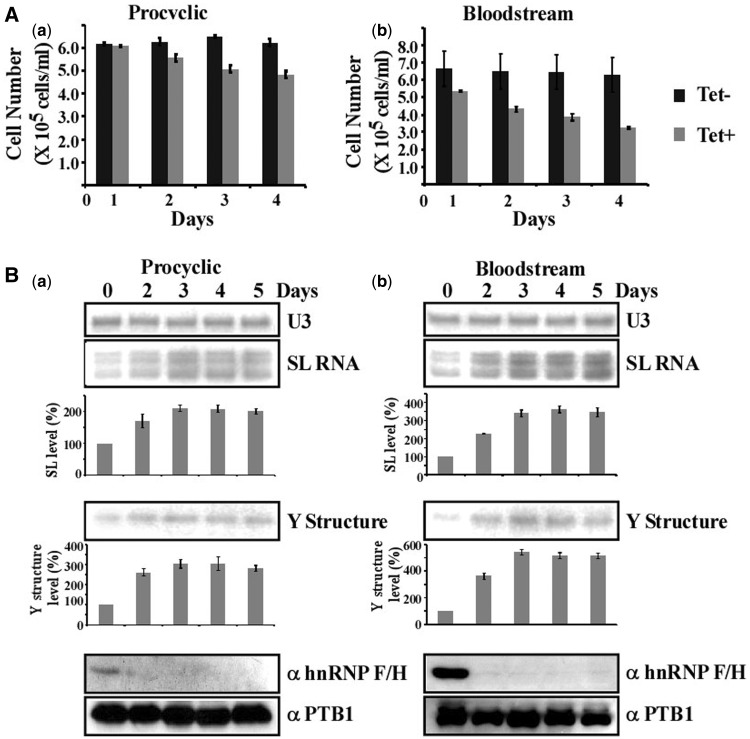
To examine the role of hnRNP F/H in regulating gene expression in the parasite, and because of the vast difference in expression of the protein in the two stages, gene knock-down by RNAi was conducted in each of the two lifecycle stages. Silencing in BSF was carried out by expression of dsRNA from a T7 opposing promoter construct (36) and in the procyclic stage by expression of a stem-loop construct (34). Growth of the silenced cells was compared with uninduced cells, and minor growth inhibition was observed in the PCF, and more profound growth inhibition was observed in BSF (Figure 2A). The greater effect on growth in BSF cells was not due to differential silencing because the level of the protein was reduced by >90% on the second day of silencing in both BSF and PCF silenced cells (Figure 2B).
Figure 2.
Silencing of hnRNP F/H in BSF and PCF stages, and its effects on the level of trans-splicing. (A) Growth pattern of T.brucei cells silenced for hnRNP F/H at the BSF and PCF stage. Growth of uninduced cells was compared with growth after tetracycline addition. Both uninduced and induced cultures were diluted daily to 105 cells per ml. The experiments were repeated three times; each bar corresponds to the average, with the standard deviation also indicated. (B) hnRNP F/H silencing affects trans-splicing in both the BSF and PCF stage. Cells expressing hnRNP F/H silencing construct were silenced for the number of days indicated. Protein (from 107 cells) was extracted from the silenced cells at the time points indicated, separated on a 10% SDS–PAGE and subjected to western analysis with anti-hnRNP F/H antibody, prepared as described in ‘Materials and Methods’ section. Reactivity with PTB1 antibodies was used as a control for equal loading. Total RNA (10 µg) from the same cells was subjected to primer extension with an oligonucleotide complementary to the intron region of the SL RNA (Supplementary Material S1). Primer extension of U3 was used to determine the amount of RNA in the samples. The products were separated on a 6% denaturing acrylamide gel. Quantitative analysis shows the percentage increase in the level of SL RNA and ‘Y structure’ intermediate’, as determined by ImageJ densitometry of three independent experiments. Standard deviation is indicated by error bars. The levels of SL RNA and Y structure intermediate are given as percentage increase with respect to the amount present at day 0 and were normalized to the level of U3 snoRNA. (a) PCF, (b) BSF.
Silencing of splicing factors that have a global effect on splicing (e.g. U2AF35, SF1, LSm, PRP31, PRP43 and various snRNP proteins) lead to an increase in the level of SL RNA (5–10-fold) accompanied by either a decrease or an increase in the Y structure splicing intermediate (13,64–68). The silencing of hnRNP F/H had only a minor effect on splicing in PCF, whereas a more profound effect was found in the BSF-silenced cells (Figure 2B). However, the splicing defect was not as severe as reported for basal splicing factors (13,64–68).
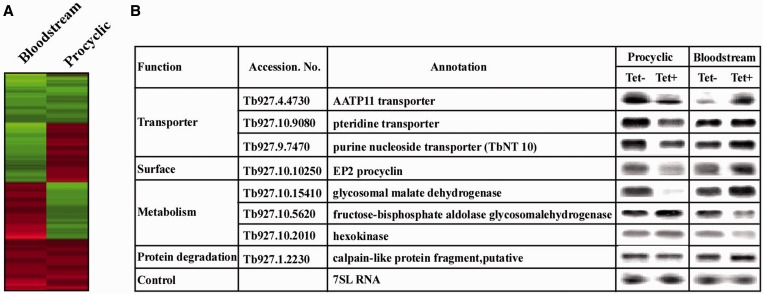
As the results presented in Figure 2 suggest that depletion of hnRNP F/H does not affect the splicing of all mRNAs, the effect of the depletion on the transcriptome was examined by microarray using RNA from PCF and BSF cells that were silenced for 2.5 days. The data were used to identify genes whose expression appeared to be significantly (P < 0.05) up- or downregulated, with an arbitrary cutoff of 1.5-fold for both BSF and PCF. A list of such genes is given in Supplementary Table S3. The transcriptome changes indicate that 1048 genes were upregulated and 1033 were downregulated in BSF, and only 422 and 425 were upregulated or downregulated, respectively, in PCF. This is expected, as the protein is more highly expressed in BSF compared with PCF. To visualize the globally observed change in gene expression in the hnRNP F/H-silenced cells, a heat map was constructed for the total collection of genes in both life stages (Figure 3A). Interestingly, from 239 genes regulated by hnRNP F/H at both stages, 159 genes were inversely regulated (upregulated in one stage and downregulated in the other) with a significant enrichment (P-value 5.9 × 10−7, Fisher’s exact test).
Figure 3.
Silencing of hnRNP F/H in the BSF and PCF stages differentially affects the parasite transcriptome. (A) Heat map of the total collection of genes that are expressed between PCF and BSF stages of the parasite on hnRNP F/H silencing. All transcripts were presented for the analysis. Each column represents the average of two to four biological replicates. The diagram represents the differential expression or fold change according to the following colour scale: red, upregulated genes; green, downregulated genes. (B) Northern analysis for the differentially expressed genes. RNA was prepared from uninduced cells (−Tet) and cells after 2.5 days of induction (+Tet) for hnRNP F/H silencing in PCF and BSF. Total RNA (20 µg) was subjected to northern analysis with T7 transcribed antisense RNA probes specific to the genes. The transcript identity, and the 7SL RNA that was used to control for equal loading, are indicated.
To verify the differential effect of hnRNP F/H regulation in the two lifecycle stages, RNA was prepared from the BSF- and PCF-silenced cells and subjected to northern analysis of the inversely regulated genes. The results (Figure 3B) support inverse effects on the steady-state levels of mRNA as a result of silencing in the two stages. These results support the notion that hnRNP F/H contributes to differential regulation of gene expression in the two life stages either by controlling mRNA stability, trans-splicing or both.
Interestingly, from the 239 genes regulated by hnRNP F/H at both stages, 37 were reported to change expression levels during the natural transition between the two stages (69), which is significantly higher than expected by random (22 expected, P-value = 0.0013, Fisher’s exact test). Recently, the RBP-10 was shown to be expressed exclusively in the BSF form and to control stage-specific gene expression (70). Indeed, of the 239 genes that were regulated in the two stages by hnRNP F/H, 25 are also controlled by RBP10 (16 expected, P-value = 0.018, Fisher’s exact test) (Supplementary Material S4).
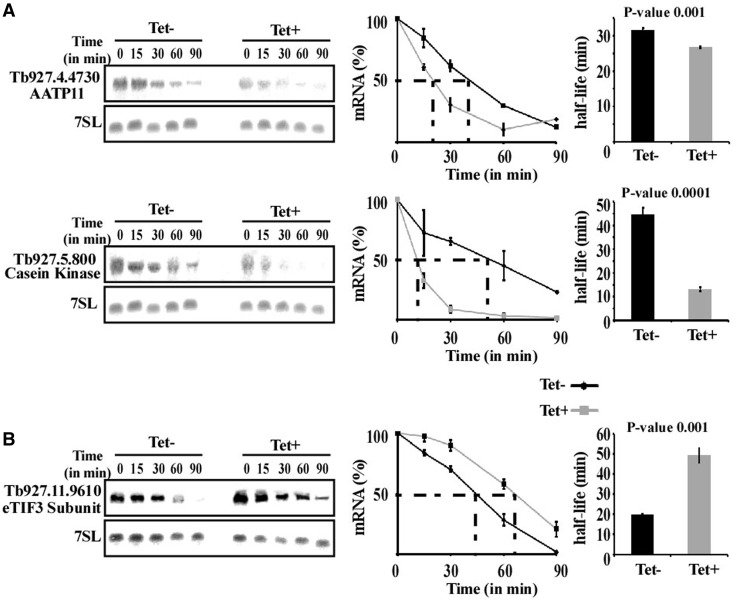
To examine whether up- and downregulation of the transcripts stem from changes in mRNA stability, the half-lives of regulated mRNAs were compared between silenced and uninduced cells in PCF. To measure half-lives, cells were treated with sinefungin and Actinomycin D to completely inactivate mRNA production (71). Northern analysis was performed, mRNA levels were measured by densitometry, the values were normalized using the level of 7SL RNA and the decay of the mRNAs in uninduced cells was compared with the decay in silenced cells. Notably, one of the transcripts examined in this experiment is AATP11, whose level is inversely regulated in the two lifecycle stages. For most mRNAs, half-lives were significantly different between un-induced and silenced cells [P < 0.01 (t-test)], suggesting that changes in the level of mRNAs are correlated with changes in mRNA stability (Figure 4).
Figure 4.
Changes in stability of mRNAs following silencing of hnRNP F/H. Uninduced and hnRNP F/H-silenced cells (2.5 days after induction) were treated with sinefungin (2 µg/ml) and, after 10 min, with Actinomycin D (30 µg/ml). RNA was prepared at the time points indicated above the lanes, separated on a 1.2% agarose-formaldehyde gel, and subjected to northern analysis with the indicated gene-specific probes. 7SL RNA was used to control for equal loading. (A) Half-life of downregulated transcripts. (B) Half-life of an upregulated transcript. The hybridization signals were measured by densitometry. The decay curves are shown with the blots, and the half-life (as obtained by linear interpolation) is indicated by the dashed lines. The decay in the absence of induction (–Tet) is indicated by black lines, and following induction (+Tet) by grey lines. The experiments were repeated three times; each data point corresponds to the average, with the standard deviation also indicated. The half-life was also calculated by fitting the normalized RNA levels to an exponential decay. The half-lives (averaged over the three experiments) are shown as bars with standard deviations, along with P-values (t-test) for the difference between the half-lives in uninduced compared with silenced cells.
Predicting the putative binding site of the hnRNP F/H homologue
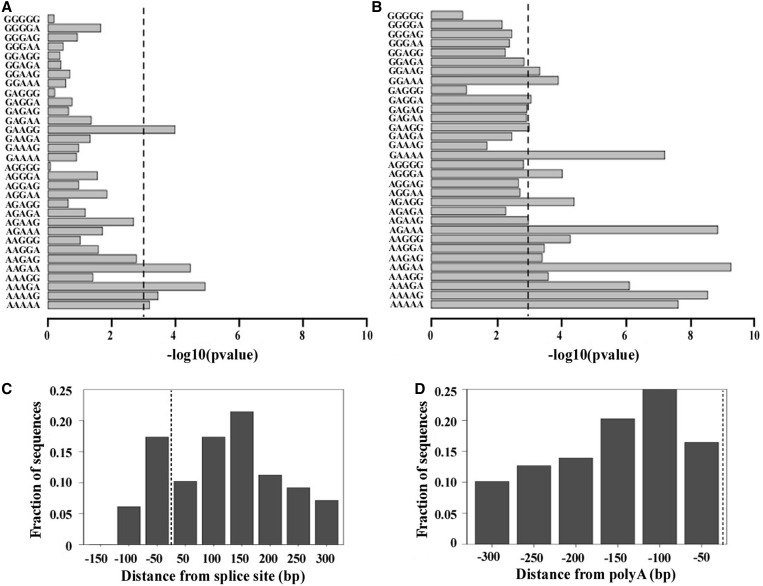
To decipher the potential binding site of the trypanosome protein, and compare it with the binding sites of the protein in mammalian cells, we concentrated on the microarray data from the BSF stage, which showed a much more profound effect following protein silencing owing to the fact that the protein is highly expressed and controls the expression of a greater number of transcripts at this stage. To this end, the sequences of the 5′ and 3′ sequences flanking the coding regions of the regulated transcripts were derived from GeneDB (see ‘Materials and Methods’ section) including 139 and 136 sequences from the 5′and 3′ UTRs of the upregulated transcripts (>1.5 fold change, P < 0.05) for which 300 nts were available, respectively. In addition, we extracted an equal number of sequences from the most down-regulated transcripts. Further SFmap was used to search for the presence of known splicing factor binding motifs in the 5′ and 3′ UTR regions (43,44). Next, we compared the results of SFmap detected in the upregulated sequences versus the downregulated sequences and searched for motifs, which were enriched in corresponding regions of the 5′ and 3′ UTRs of the upregulated genes. Interestingly, although we did not observe enrichment of the experimentally verified human hnRNP F motifs (characterized by G triplets flanked by pyrimidines) at either the 5′or at the 3′UTR regions, we nevertheless noticed a significant enrichment of a purine-rich motif, AAGAA, with significant P-values of 3.4 × 10−5 and 5.2 × 10−10 for 5′UTR and 3′UTR regions, respectively. Interestingly, the enriched motif resembles the human hnRNP H1 purine rich motif, recently identified from a high throughput CLIP experiment (33). We further searched for the presence of other purine rich motifs (independently testing all possible purine pentamers) and found that in general, purine rich motifs tended to be significantly more abundant in the UTRs of the upregulated genes. Among the enriched motifs, which were mostly stretches of adenines with singly embedded guanines, the motif AAGAA was the most significant motif when considering both the 5′ and 3′ UTR regions, with P-values of 3.4 × 10−5 and 5.2 × 10−10, respectively, using the Mann–Whitney test (Figure 5A and B).
Figure 5.
(A) Enrichment of purine rich motifs at the 5′UTR region. Enrichment of purine rich pentamers at the 5′region (300 nt downstream the splice site) of upregulated sequences (showing greater than 1.5-fold change in the RNA expression in the mutants versus the wild-type (WT) in the BSF) compared with the motifs found in the downregulated sequences (an equal number of downregulated sequences from the bottom of the ranked list). Purine rich motifs were detected using the SFmap algorithm (43). Enrichment was calculated using the Mann–Whitney test, –log10 of the P-value for the enrichment of each motif is demonstrated by the length of the bar. Dashed line represents the cutoff for significance following Bonferroni correction (P = 0.001). (B) Enrichment of purine rich motifs at the 3′UTR region. Enrichment of purine rich motifs at the 3′UTR region (300 nt upstream to the polyA site) of upregulated sequences (showing greater than 1.5-fold change in the RNA expression in the silenced cells versus the WT in the BSF) compared with the motifs found in the downregulated sequences (equal number of downregulated sequences from the bottom of the ranked list). Purine rich motifs were detected using the SFmap algorithm (43). Enrichment was calculated using the Mann–Whitney test, –log10 of the P-value for the enrichment of each motif is demonstrated by the length of the bar. Dashed line represents the cutoff for significance following Bonferroni correction (P = 0.001). (C) Distribution of the AAGAA motif in the upregulated sequences at the 5′UTR. Distribution of the detected motifs in the upregulated transcripts (showing greater than 1.5-fold change in the RNA expression in the silenced cells versus the uninduced BSF) at the 5′UTR region (150 nts upstream and 300 nts downstream to the splice site). The AAGAA motifs are concentrated around the splice site (dashed line). (D) Distribution of the AAGAA motif in the upregulated sequences at the 3′UTR. Distribution of the detected motifs in the upregulated sequences (showing greater than 1.5-fold change in induced relative to uninduced cells) at the 3′ UTR region.
To verify that the results were unbiased by the approach used, we also used a pure de novo ranked motif search algorithm based on the minimal hyper-geometric distribution (45,46). The advantage of the DRIMUST algorithm (http://drimust.technion.ac.il/) is that it searches for statistical enrichment of all possible motifs in the top of a ranked list and does not require prior definition of a threshold. When running DRIMUST on the entire list of 5′ and 3′ UTR regions, which we ranked based on fold-change, the AAGAA motif was clearly enriched in the most upregulated genes, with a P-value of 3 × 10−12 and 5 × 10−19, for the 5′ and 3′ UTR regions, respectively. Notably, although the AAGAA motif was enriched in the upregulated genes both in the 5′ and 3′ UTR regions, the motif was also present at a relatively high frequency in the downregulated genes. Nevertheless, when focusing on the upregulated transcripts (>1.5-fold change, P < 0.05) versus the equal number of downregulated ones, we noticed that the number of AAGAA motifs within the UTR regions was significantly higher in the upregulated versus the downregulated transcripts, with an average of 1.6 and 1.7 motifs per UTR compared with 0.6 and 0.3, for the 5′ and 3′ UTR regions, respectively, and up to 11 copies per UTR. The P-values for the Mann–Whitney test comparing the number of motifs per UTR in the up- versus downregulated genes was 1.2 × 10−6 and 2.2 × 10−16, for the 5′ and 3′ UTR regions, respectively. We further analysed the distribution of the AAGAA motif along the UTRs of the upregulated genes (Figure 5C and D). A higher density of the motif was found around the 3′ splice sites, between 50 nt upstream of the site to 150 nt downstream. In the 3′ UTR, the AAGAA motif was concentrated between 100 and 150 nt upstream of the poly (A) site.
The hnRNP F/H homologue binds to the 3′ UTR of AATP11 and regulates its level at the PCF stage
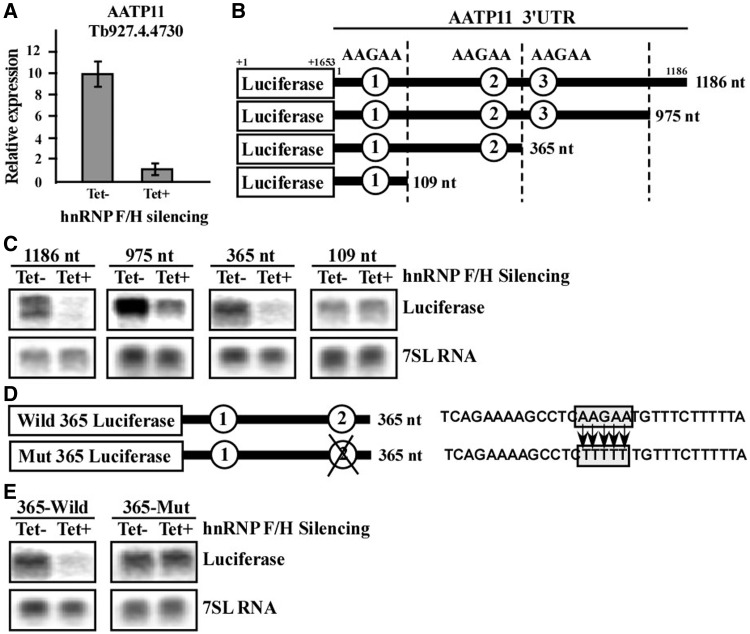
To validate the binding site suggested by the enrichment analyses, we chose to examine the role of hnRNP F/H in the regulation of AATP11. Previous studies suggest that AATP11 is highly expressed in the procyclic stage and is regulated via mRNA stability, dictated by sequences present in the 3′ UTR (72). We previously reported that PTB1 controls the stability of this mRNA, and that its splicing depends on PTB1 via a polypyrimidine tract that is C rich and contains a PTB binding site (17). The downregulation of AATP11 during hnRNP F/H silencing in PCF was verified by real-time PCR (Figure 6A). To map the domain responding to hnRNP F/H, we inspected the 3′ UTR region of 1186 nt for the presence of the putative binding site, AAGAA. The presence of AAGAA sites is schematically presented in Figure 6B. To investigate which of these sites serves as the binding site for hnRNP F/H, the 3′ UTR was cloned into the pNS21b vector (17,50), which is carrying a luciferase reporter gene (‘Materials and Methods’ section), and three deletions were prepared (Figure 6B). The constructs were transfected into PCF cells carrying the hnRNP F/H silencing construct. The level of lucifease mRNA was examined by northern analysis before and after silencing. The results suggest that site 2 is responsible for this regulation, as the regulation was lost in the transcript carrying 109 nt of the 3′ UTR but not in the one carying 365 nt (Figure 6C). To verify that the regulation is due to the AAGAA motif, the site was mutated to TTTTT (Figure 6D), and transgenic cells were generated with this construct, as described earlier in the text. Introducing this, mutation abolished the regulation, as no change in the luciferase transcript was observed on silencing of hnRNP F/H, compared with the downregulation observed with the wild-type domain (Figure 6E). These data suggest that AAGAA is most probably the binding site of hnRNP F/H protein, as changing only this sequence was sufficient to eliminate the regulation mediated by the hnRNP F/H protein. However, the ultimate proof for the sequence being the binding site of the hnRNP F/H awaits the genome-wide iCLIP (individual-nucleotide resolution UV Cross-Linking and ImmunoPrecipitation) analysis.
Figure 6.
hnRNP F/H binds to the 3′ UTR of AATP11 and regulates its level at the PCF stage. (A) Quantitative real-time PCR analysis of the AATP11 transcript on RNAi silencing of hnRNP F/H in the PC stage. cDNA was prepared from total RNA (1 µg) derived from uninduced cells (−Tet) or cells after 2.5 days of silencing (+Tet), as described in ‘Materials and Methods’ section. Real-time PCR was performed as described in ‘Materials and Methods’ section using cDNA (diluted 1:100), and concentration curves were used to determine the amount of PCR product amplified in uninduced cells (−Tet) or cells after 2.5 days of silencing (+Tet). The results shown are the average of three independent experiments. (B) Schematic representation of the mini-genes containing different sized 3′UTRs of AATP11 cloned downstream to the luciferase gene. Four different sized (1186, 975, 365 and 109 nt) fragments of 3′UTR of AATP11 were amplified as described in ‘Materials and Methods’ section and cloned downstream to the luciferase gene in the pNS21b expression vector. The constructs were then transfected into a cell line silenced for hnRNP F/H using RNAi. The sizes of the 3′UTRs and AAGAA hnRNP F/H-binding motifs are indicated. (C) Expression of the mini-gene transcripts in cells expressing the hnRNP F/H silencing construct. RNA was prepared from transgenic parasites expressing the luciferase AATP11 minigenes shown in panel B. Expression was monitored in hnRNP F/H cells after 2.5 days of silencing. Total RNA (30 μg) was separated on a 1.2% agarose, 2.2 M formaldehyde gel. The RNA was blotted and hybridized with a randomly labelled probe specific for the luciferase gene. The 7SL RNA was used as a control for equal loading. (D) Schematic representation of the luciferase minigenes carrying the 365 nt long 3′UTR of the AATP11 model gene with two AAGAA motifs. The ‘wild 365 luciferase’ minigene consists of a 365 nt long 3′UTR of AATP11 cloned downstream to the luciferase gene. The ‘mut 365 luciferase’ minigene carries 3′UTR of AATP11 with a second AAGAA mutated motif. The sequence of the domain carrying the mutations is boxed, and the base substitutions used to generate the mutation are depicted. (E) Expression of the wild and mutated minigenes. RNA was prepared from the transgenic parasites expressing the above luciferase minigenes as described in (D), and northern analysis was performed as described in (C).
HnRNP F/H serves a trans-splicing repressor
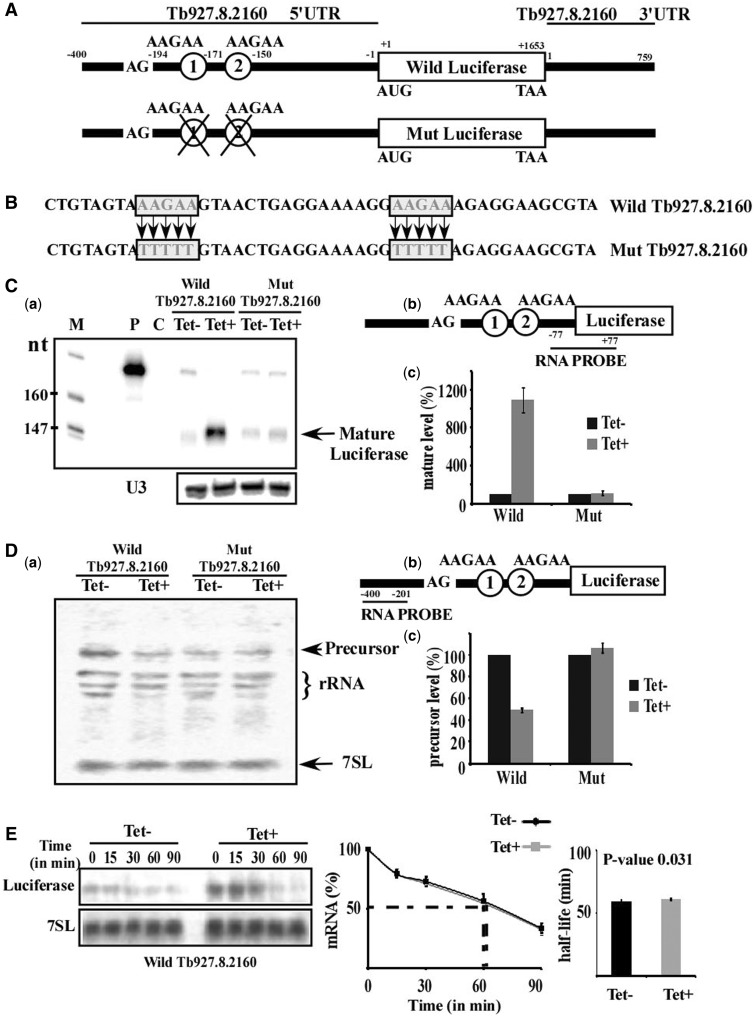
As hnRNP F/H proteins are known to function in alternative splicing and because of our interest in examining the role of trypanosome proteins in this process, the effect of hnRNP F/H on splicing was examined. We were encouraged by the finding that the putative binding sites of the protein were centered around the 3′ splice site, and at distances (−50 to +150), which are potentially relevant for controlling splicing (Figure 5C). We searched the regulated transcripts for a gene that carries the AAGAA site downstream of the 3′ splice site and found Tb927.8.2160, a gene encoding a multidrug resistance protein. The 400 nt long 5′-flanking sequence of this multidrug resistance protein, including a 194 nt long 5′UTR with two copies of the putative binding site AAGAA (Figure 7A), was cloned upstream to the luciferase gene in pNS21b (17,50). The vector therefore contained a luciferase gene whose expression depends on the 5′ flanking sequences of the multidrug resistance protein. To verify that the putative AAGAA binding site is the site that governs the splicing regulation of the multidrug resistance protein by hnRNP F/H, the AAGAA motifs were mutated to TTTTT (Figure 7A and B). Transgenic parasites carrying the reporter gene as well as the stem-loop construct to silence hnRNP F/H were generated. The effect of silencing on expression was examined by RNase protection assay using a probe that consists of region [−77 to −1, relative to the AUG] of the 5′UTR of the multidrug resistance protein fused with region of the luciferase gene [+1 to +77, relative to the AUG of luciferase sequence], as shown in Figure 7C-b. RNA was prepared from uninduced cells and cells after 2.5 days of induction. The results demonstrate elevation of mature transcript levels following silencing. However, when the two potential binding sites were mutated, the transcript was no longer elevated upon silencing (Figure 7C-a). The statistic of three experiments is presented (Figure 7C-c). The increase in the level of the mature transcripts may stem from either changes in stability, as the mutated sequence is found in the 5′ UTR of the transcripts and/or from the role of this factor as a splicing repressor. To examine these two possibilities, the level of the pre-mRNA was evaluated using a probe specific for pre-mRNA sequences. The probe (Figure 7D-b) exclusively detects the transgenic transcripts, as no signal was found in the parental strain RNA because the expression of the transgene is driven by the strong EP promoter and the expression of the authentic gene is much weaker. The results (Figure 7D-a) demonstate that under silencing, the level of the precursor decreased. However, almost no change was observed in the pre-mRNA carrying mutations in the AAGAA motifs, suggesting that hnRNP F/H serves as a splicing repressor; following hnRNP F/H depletion the repression is relieved, and more pre-mRNA is processed to mature mRNA (statistic of three experiments is given in Figure 7D-c). However, hnRNP F/H may also exert its regulation at the level of mRNA stability. To examine this possibility, the half life of the wild-type transcript was measured in un-induced cells and after 2.5 days of silencing. The results (Figure 7E) demonstrate no significant change in the half-life of the mRNA before and after silencing (P = 0.031), suggesting that the effect on the level of mRNA during silencing is not as a result of the role of the protein in regualting mRNA stability but because of its role as a splicing repressor. The function of hnRNP F/H as a repressor was demonstated here for a single transcript. Genome-wide mapping of the protein binding should indicate the extent by which the protein serves as a repressor based on its binding in the vicinity to the 3′splice site.
Figure 7.
HnRNP F/H serves as a trans-splicing repressor; validation of AAGAA as a binding motif of hnRNP F/H in 5′UTR. (A) Schematic representation of the luciferase minigene carrying the 5′ and 3′ UTR of the multidrug resistance protein A (Tb927.8.2160). The ‘wild luciferase’ minigene consists of the 5′UTR of the gene and ‘mut luciferase’ carries mutation as indicated. Both luciferase minigenes carry 759 nt long 3′UTR of Tb927.8.2160. Genomic coordinates are with respect to ATG (5′ UTR) and stop codon for 3′UTR. (B) Base substitutions used to generate the mutation in 5′UTR. The sequence of the domain carrying the mutations is boxed, and the base substitutions used to generate the mutation are depicted. (C) Role of hnRNP F/H in trans-splicing. (a) RNase protection of the luciferase fused transcript carrying the wild-type and mutated 5′UTRs. Expression was monitored in hnRNP F/H cells after 2.5 days of silencing by RNase protection assay. The protected fragments were separated on a 6% acrylamide–7 M urea gel. P indicates probe; C, control (no RNA was added to the RNase protection assay). Primer extension of U3 was used as control. (b) Schematic representation of the probe used for RNase protection assay in (a). Genomic coordinates of the probe are shown (−77 to +77, with respect to luciferase ATG to give a protected fragment of 154 nt). (c) Quantitative analysis of the mature luciferase fused transcript in (a). Quantitative analysis shows the percentage increase (with respect to amount in Tet- cells) in the level of mature fused-luciferase transcript (indicated in figure as mature)”, based on three independent experiments. The results were normalized to the level of U3 snoRNA. (D) (a) Northern analysis to detect the pre-mRNA of Tb927.8.2160-luciferase fused transcript of wild-type and mutated 5′UTRs. Northern analysis was with an RNA probe specific to the pre-mRNA. The level of 7SL RNA was used as a control for the amount of RNA. (b) Schematic representation of the probe used for northern analysis in (a). Genomic coordinates of the probe are shown (−400 to −201). (c) Quantitative analysis of the precursor luciferase fused transcript in (a). Quantitative analysis shows the percentage decrease/increase in the level of pre-fused transcript (indicated in figure as precursor), based on three independent experiments. The levels of pre-fused -luciferase transcripts are given as percentage increase with respect to the amount present in Tet-, and were normalized to the level of U3 snoRNA. (E) mRNA stability assay. Uninduced and hnRNP F/H silenced cells carrying wild-type fused-luciferase transcript (2.5 days after induction) essentially as described in Figure 4. RNA was prepared, separated on a 1.2% agarose-formaldehyde gel and subjected to northern analysis with the luciferase RNA probes. The 7SL RNA was used to control for equal loading. The half-life (as obtained by linear interpolation) is indicated by the dashed lines. The decay in the absence of induction (–Tet) is in black lines, and after induction (+Tet) is by grey lines (based on three experiments); each data point corresponds to the average, and standard deviations are indicated. The half-life was calculated as described in Figure 4.
HnRNP F/H binds directly to the substrates it regulates
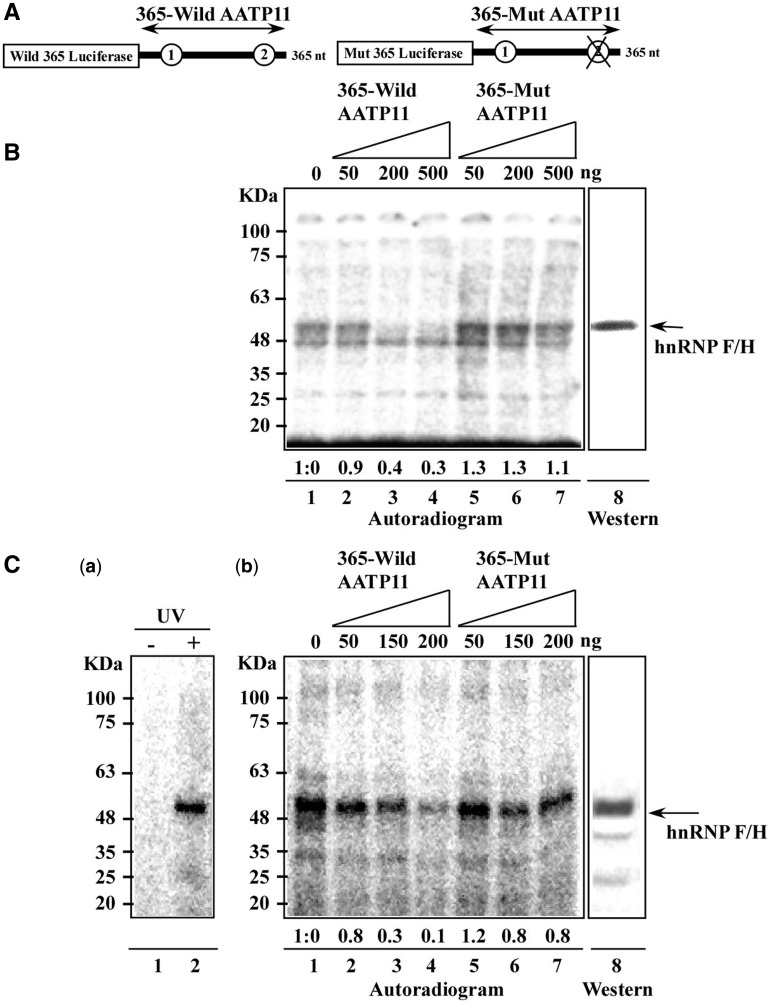
To gain further support for the direct binding of hnRNP F/H to the AAGAA site, binding was assessed by the UV cross-linking approach, which enables the detection of proteins that become cross-linked to radioactively labelled RNA. The pre-mRNA of AATP-11 and its corresponding mutant (Figure 6D and 8A) were used for this analysis. Whole cell extracts from PCF and BSF were incubated with a 365 nt radioactively labelled RNA, carrying the 3′ UTR of the transcript in the presence of elevated concentrations of either ‘cold’ wild-type or mutant transcripts (Figure 8B and C). The position of the hnRNP F/H in the gel was verified by western blotting (Figure 8B and C-b). The results indicate specific reduction in the cross-linking to the hnRNP F/H protein when unlabelled mRNA was added (reduction of ∼60% on addition of 200 ng excess unlabelled RNA), but not when the unlabelled mutated transcript was used, which lacked the AAGAA-binding sites (no reduction on addition of 200 ng excess unlablled mutant RNA). Another binding protein present in the PCF (just below the hnRNP F/H) was also bound to the substrate, but this binding was not competed by the relevant substrate, and this protein is most probably PCF-specific, as it was not detected in the BSF extract (Figure 8B). The same cross-linking results were obtained when BSF extracts were used (Figure 8C-b). Ten times less protein was used from BSF compared with PCF, which is in accordance with the higher level of the protein in BSF. Moreover, the same cross-linked protein was detected when an 5′ labelled RNA oligonucleotide carrying two binding sites was used for the cross-linking (Figure 8C-a).
Figure 8.
HnRNP F/H protein is cross-linked to a transcript carrying the AAGAA site and to any RNA oligonucleotide coding for the two putative binding sites. (A) Schematic representation of the probes used for cross-linking. (B) Cross-linking using PCF extract. Cross-linking was performed as described in ‘Materials and Methods’ section, using whole cell extracts (15 µg per reaction) of PCF extract and in the presence of (lanes 1–4) increasing amounts of unlabelled 365-wild AATP11 transcripts (0, 50, 200 and 500 ng, respectively). Lanes 5–7 show increasing amounts of non-radioactive 365-mut AATP11 transcripts carrying the AAGAA mutation (50, 200 and 500 ng, respectively). Lane 8, a portion of the gel was subjected to western analysis with anti hnRNP F/H antibodies. The size of the protein marker is indicated. (C) Cross-linking using BSF extract. Extract (0.7 µg per lane) was cross-linked to the same substrates as in (B). (a) oligonucleotide (5′-AAGAAAAGAA-3′) (40 000 cpm) was end labelled at the 5′ end with [γ-32P]-ATP and was incubated with extracts in the absence (lane 1) or after UV irradiation (lane 2), (b). BSF extract (0.7 µg per lane) was cross-linked to (lanes 1–4) increasing amounts of unlabelled 365-wild AATP11 transcripts (0, 50, 150, and 200 ng, respectively). Lanes 5–7 show increasing amounts of non-radioactive 365-mut AATP11 transcripts carrying the AAGAA mutation (50, 150 and 200 ng, respectively). Lane 8, a portion of the gel was subjected to western analysis with anti hnRNP F/H antibodies.
DISCUSSION
In this study, we characterized a trypanosome homologue of the mammalian hnRNP F/H proteins. Although the trypanosome protein RRM domains show higher sequence identity to the human hnRNP F qRRMs, its proposed RNA binding motif identified in this study (AAGAA) resembles the binding site of the human hnRNP H, which was recently determined from high-throughput binding experiments (33). Based on these data, we suggest that the trypanosome protein is an hnRNP F/H homologue. Interestingly, the stretches of Adenines flanking a single Guanine, specifically the AAGAA motif, was highly enriched in genes that were upregulated on hnRNP F/H depletion in BSF, most probably because these transcripts tend to carry multiple binding sites that can control their stability, splicing, or both. Most striking and novel was the finding that a significant number of the regulated genes were inversely affected by hnRNP F/H in the two lifecycle stages, suggesting the role of this factor in differential regulation of gene expression of the parasite when cycling between its two hosts. The stronger effect on splicing in the BSF is most probably because more substrates are regulated by hnRNP F/H at this stage. We cannot, however, rule out the possibility that the robust effect is a consequence of a secondary effect owing to perturbation of a factor(s) that acts as a master regulator.
Genome-wide studies mapping of SL addition sites suggested extensive alternative splicing changes throughout the lifecycle of the parasite (73). Alternative splicing was also shown recently to control protein localization, enabling the generation of two isoforms of tRNA-synthetase, a mitochondrial and cytoplasmic enzyme (74). Trans-splicing must therefore be a regulated process to generate this rich repertoire of alternative spliced forms that are developmentally regulated. However, little is known about factors that can participate in such regulation. Early studies from our group suggested that PTB proteins are involved in trans-splicing of a distinct subset of transcripts having a C rich polypyrimidine tract (17). Our current results suggest that hnRNP F/H might be a good candidate for mediating stage-specific splicing regulation. The protein is differentially regulated, highly expressed in the BSF and affects the level of a large number of genes at this stage. The protein recognition site, AAGAA, is located around the 3′ splice site (mostly 50 nt upstream to 150 nt downstream) in most substrates (Figure 5C). Such sites may serve as exonic or intronic enhancer or silencers. The one example provided in this study (Figure 7) supports the role of the protein as a splicing repressor. However, the extent of regulation on splicing awaits the iCLIP mapping of the protein in sites located in the vicinity to AG splice site, suggested in this study by the bioinformatic analysis.
The data presented here suggest that hnRNP F/H participates in differential gene expression in both life stages. There are only a few RBPs that were shown to affect differential regulation during cycling between the hosts. One such protein is TbZFP3, which acts as an anti-repressor to stabilize EP1 procyclin and to promote its translation (75). More recently, it was shown that the same protein regulates mRNA stability of transcripts enriched in the stumpy from of the parasite (76). ALBA3/4 proteins are present throughout trypanosome development in the Tsetse fly, with the striking exception of the transition stages, when the parasite is found in the proventiculus region of the fly, again demonstrating the involvement of an RBP in trypanosome developmental regulation. These proteins do not affect mRNA stability, but rather regulate translation (77).
Another protein that was shown to govern developmental gene expression is RBP10 (70). RBP10 does not bind mRNAs directly, but its tethering to a reporter mRNA inhibits translation and reduces to half the abundance of bound mRNA. It was suggested that this factor may affect the expression of regulatory proteins that are specific to the procyclic form (70). Most recently, overexpression of a single RBP (TbRBP6) in PCF was reported to induce transformation of the parasites to infective metacyclic forms expressing the variant surface glycoprotein. The mechanism that induces this remarkable phenotype is currently unknown (78). As opposed to most of the factors aforementioned, hnRNP F/H is unique because it is the first protein that demonstrates dual function in T. brucei, involved in both splicing and mRNA stability, and regulates the differential expression of genes in both lifecycle stages, in some cases, even in opposite directions.
Interestingly, there is a significant overlap between the genes shown to be regulated in the two lifecycle stages of the parasite (69) and the genes regulated by hnRNP F/H (Supplementary Material S4). A large number of the stage-specific regulated genes by RBP10 are also regulated by hnRNP F/H (Supplementary Material S4), suggesting that the differential regulation is governed by the cooperative action of several factors, which are required to orchestrate the differentiation programming. Elimination or overproduction of such factor(s) can change the balance and induce or suppress stage-specific gene expression. Sometimes, as in the case of TbRBP6, a single factor is sufficient to induce the switch from PCF to metacyclic trypanosomes (78).
The effect of hnRNP F/H on gene regulation might be even more complex than demonstrated in this study. At present, most of the differential stage-specific gene regulation in trypanosomes is attributed to the coordinate function of RBPs (12). However, the process might be also governed by chromatin remodelling. Although evidence was provided for regulation of gene expression by chromatin remodelling, there is no report to date demonstrating changes in chromatin modifications between the two lifecycle stages of the parasite (1). Recent studies have shown that splicing is connected to chromatin remodelling, and proteins like PTB were shown to orchestrate such cross-talk in mammals (79). Stage-specific trans-splicing might be governed by chromatin remodelling, and the T. brucei PTB as well as hnRNP F/H may participate in such cross-talk. Indeed, hnRNP F/H includes a domain present in BAF1/ABF1, which is a chromatin-associated factor in yeast. This intriguing observation should lead to experiments that search for changes in chromatin modifications under hnRNP F/H silencing and finding whether any of the chromatin modifiers associate with hnRNP F/H.
Both trypanosome hnRNP proteins: hnRNP F/H (this study) and PTB (hnRNP I) (17), affect the transcriptome at two levels, splicing and mRNA stability. In mammals, it was found that PTB binds 10-fold more strongly to intron sequences than to exons, and that the binding to exons is always near the splice sites, suggesting a dominant role in splicing regulation (80). In contrast to mammals, the T. brucei PTBs were shown to directly affect not only splicing but also mRNA stability. Thus, hnRNP proteins in trypanosomes have acquired also a role in mRNA stability regulation (17). The binding of PTB as well as hnRNP F/H to the 3′ UTR of genes regulates mRNA stability; if this binding already takes place in the nucleus, it may affect the splicing of the downstream gene as well. Indeed, proteins of the hnRNP F/H family were shown to affect polyadenylation in mammals (25). As polyadenylation and trans-splicing are coupled in trypanosomes, we can envision a scenario whereby the binding of hnRNP F/H upstream to the poly (A) site, as suggested by the bioinformatics analysis (Figure 5), could interfere with the cross-talk between the splicing and the polyadenylation machineries and thus affect the trans-splicing of the downstream gene. In addition, binding of the hnRNP F/H around the 3′ splice site may not only affect splicing but also affect mRNA stability. Recently, it was demonstrated that alternative-spliced forms of T. brucei tRNA synthase are regulated at the mRNA stability via specific sequences on the 5′ UTR (74). Thus, alternative splicing does not only create different proteins but also generates different mRNAs, which differ in their stability. Genome-wide mapping of both PTB- and hnRNP F/H-binding sites should shed light on the distribution of these proteins on mature and pre-mRNA sequences and help define the precise contribution of these factors to splicing and stability.
This study describes the role of a central RBP that regulates stage-specific gene expression. We showed that hnRNP F/H controls gene expression (often inversely) in the two lifecycle stages, possibly by interacting with a different RBP at each stage. We are only at the earliest stages of a full understanding of the regulatory circuits exerted by this essential multifunctional factor, which may be involved in regulating splicing, polyadenylation and mRNA stability and might also orchestrate the interactions with stage-specific chromatin remodelling events.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Materials 1–4.
FUNDING
Deutsche Forschungsgemeinschaft via Deutsche-Israelische Projektkooperation and the Israel Science Foundation as well as the I-core grant 41/11. (to S.M. in part). S.M. holds the David and Inez Myers Chair in RNA silencing of diseases. S.C. thanks the Human Frontier Science Program for financial support. Funding for open access charge: The I-CORE Program of the Planning and Budgeting Committee and The Israel Science Foundation [grant No 41/11].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Inbal Paz for her help with SFmap and DRIMust.
REFERENCES
- 1.Siegel TN, Hekstra DR, Kemp LE, Figueiredo LM, Lowell JE, Fenyo D, Wang X, Dewell S, Cross GA. Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev. 2009;23:1063–1076. doi: 10.1101/gad.1790409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hug M, Hotz HR, Hartmann C, Clayton C. Hierarchies of RNA-processing signals in a trypanosome surface antigen mRNA precursor. Mol. Cell. Biol. 1994;14:7428–7435. doi: 10.1128/mcb.14.11.7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LeBowitz JH, Smith HQ, Rusche L, Beverley SM. Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev. 1993;7:996–1007. doi: 10.1101/gad.7.6.996. [DOI] [PubMed] [Google Scholar]
- 4.Matthews KR, Tschudi C, Ullu E. A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes. Genes Dev. 1994;8:491–501. doi: 10.1101/gad.8.4.491. [DOI] [PubMed] [Google Scholar]
- 5.Vassella E, Braun R, Roditi I. Control of polyadenylation and alternative splicing of transcripts from adjacent genes in a procyclin expression site: a dual role for polypyrimidine tracts in trypanosomes? Nucleic Acids Res. 1994;22:1359–1364. doi: 10.1093/nar/22.8.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gunzl A. The pre-mRNA splicing machinery of trypanosomes: complex or simplified? Eukaryot. Cell. 2010;9:1159–1170. doi: 10.1128/EC.00113-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang XH, Haritan A, Uliel S, Michaeli S. trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot. Cell. 2003;2:830–840. doi: 10.1128/EC.2.5.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michaeli S. Trans-splicing in trypanosomes: machinery and its impact on the parasite transcriptome. Future Microbiol. 2011;6:459–474. doi: 10.2217/fmb.11.20. [DOI] [PubMed] [Google Scholar]
- 9.Kolev NG, Franklin JB, Carmi S, Shi H, Michaeli S, Tschudi C. The transcriptome of the human pathogen Trypanosoma brucei at single-nucleotide resolution. PLoS Pathog. 2010;6:e1001090. doi: 10.1371/journal.ppat.1001090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siegel TN, Hekstra DR, Wang X, Dewell S, Cross GA. Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res. 2010;38:4946–4957. doi: 10.1093/nar/gkq237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clayton C, Shapira M. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol. Biochem. Parasitol. 2007;156:93–101. doi: 10.1016/j.molbiopara.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 12.Kramer S, Carrington M. Trans-acting proteins regulating mRNA maturation, stability and translation in trypanosomatids. Trends Parasitol. 2011;27:23–30. doi: 10.1016/j.pt.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta SK, Carmi S, Waldman Ben-Asher H, Tkacz ID, Naboishchikov I, Michaeli S. Basal splicing factors regulate the stability of mature mRNAs in trypanosomes. J. Biol. Chem. 2013;288:4991–5006. doi: 10.1074/jbc.M112.416578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ismaili N, Perez-Morga D, Walsh P, Cadogan M, Pays A, Tebabi P, Pays E. Characterization of a Trypanosoma brucei SR domain-containing protein bearing homology to cis-spliceosomal U1 70 kDa proteins. Mol. Biochem. Parasitol. 2000;106:109–120. doi: 10.1016/s0166-6851(99)00205-4. [DOI] [PubMed] [Google Scholar]
- 15.Ismaili N, Perez-Morga D, Walsh P, Mayeda A, Pays A, Tebabi P, Krainer AR, Pays E. Characterization of a SR protein from Trypanosoma brucei with homology to RNA-binding cis-splicing proteins. Mol. Biochem. Parasitol. 1999;102:103–115. doi: 10.1016/s0166-6851(99)00091-2. [DOI] [PubMed] [Google Scholar]
- 16.Manger ID, Boothroyd JC. Identification of a nuclear protein in Trypanosoma brucei with homology to RNA-binding proteins from cis-splicing systems. Mol. Biochem. Parasitol. 1998;97:1–11. doi: 10.1016/s0166-6851(98)00118-2. [DOI] [PubMed] [Google Scholar]
- 17.Stern MZ, Gupta SK, Salmon-Divon M, Haham T, Barda O, Levi S, Wachtel C, Nilsen TW, Michaeli S. Multiple roles for polypyrimidine tract binding (PTB) proteins in trypanosome RNA metabolism. RNA. 2009;15:648–665. doi: 10.1261/rna.1230209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Estevez AM. The RNA-binding protein TbDRBD3 regulates the stability of a specific subset of mRNAs in trypanosomes. Nucleic Acids Res. 2008;36:4573–4586. doi: 10.1093/nar/gkn406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han SP, Tang YH, Smith R. Functional diversity of the hnRNPs: past, present and perspectives. Biochem. J. 2010;430:379–392. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- 20.Pinol-Roma S, Choi YD, Matunis MJ, Dreyfuss G. Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev. 1988;2:215–227. doi: 10.1101/gad.2.2.215. [DOI] [PubMed] [Google Scholar]
- 21.Matunis MJ, Xing J, Dreyfuss G. The hnRNP F protein: unique primary structure, nucleic acid-binding properties, and subcellular localization. Nucleic Acids Res. 1994;22:1059–1067. doi: 10.1093/nar/22.6.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Honore B, Vorum H, Baandrup U. hnRNPs H, H' and F behave differently with respect to posttranslational cleavage and subcellular localization. FEBS Lett. 1999;456:274–280. doi: 10.1016/s0014-5793(99)00911-4. [DOI] [PubMed] [Google Scholar]
- 23.Honore B, Rasmussen HH, Vorum H, Dejgaard K, Liu X, Gromov P, Madsen P, Gesser B, Tommerup N, Celis JE. Heterogeneous nuclear ribonucleoproteins H, H', and F are members of a ubiquitously expressed subfamily of related but distinct proteins encoded by genes mapping to different chromosomes. J. Biol. Chem. 1995;270:28780–28789. doi: 10.1074/jbc.270.48.28780. [DOI] [PubMed] [Google Scholar]
- 24.Honore B, Baandrup U, Vorum H. Heterogeneous nuclear ribonucleoproteins F and H/H' show differential expression in normal and selected cancer tissues. Exp. Cell Res. 2004;294:199–209. doi: 10.1016/j.yexcr.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 25.Veraldi KL, Arhin GK, Martincic K, Chung-Ganster LH, Wilusz J, Milcarek C. hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol. Cell. Biol. 2001;21:1228–1238. doi: 10.1128/MCB.21.4.1228-1238.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caputi M, Zahler AM. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H'/F/2H9 family. J. Biol. Chem. 2001;276:43850–43859. doi: 10.1074/jbc.M102861200. [DOI] [PubMed] [Google Scholar]
- 27.Krecic AM, Swanson MS. hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 1999;11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- 28.Caputi M, Zahler AM. SR proteins and hnRNP H regulate the splicing of the HIV-1 tev-specific exon 6D. EMBO J. 2002;21:845–855. doi: 10.1093/emboj/21.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen CD, Kobayashi R, Helfman DM. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat beta-tropomyosin gene. Genes Dev. 1999;13:593–606. doi: 10.1101/gad.13.5.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chou MY, Rooke N, Turck CW, Black DL. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol. Cell. Biol. 1999;19:69–77. doi: 10.1128/mcb.19.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garneau D, Revil T, Fisette JF, Chabot B. Heterogeneous nuclear ribonucleoprotein F/H proteins modulate the alternative splicing of the apoptotic mediator Bcl-x. J. Biol. Chem. 2005;280:22641–22650. doi: 10.1074/jbc.M501070200. [DOI] [PubMed] [Google Scholar]
- 32.Min H, Chan RC, Black DL. The generally expressed hnRNP F is involved in a neural-specific pre-mRNA splicing event. Genes Dev. 1995;9:2659–2671. doi: 10.1101/gad.9.21.2659. [DOI] [PubMed] [Google Scholar]
- 33.Huelga SC, Vu AQ, Arnold JD, Liang TY, Liu PP, Yan BY, Donohue JP, Shiue L, Hoon S, Brenner S, et al. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep. 2012;1:167–178. doi: 10.1016/j.celrep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z, Morris JC, Drew ME, Englund PT. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 2000;275:40174–40179. doi: 10.1074/jbc.M008405200. [DOI] [PubMed] [Google Scholar]
- 35.Mandelboim M, Barth S, Biton M, Liang XH, Michaeli S. Silencing of Sm proteins in Trypanosoma brucei by RNA interference captured a novel cytoplasmic intermediate in spliced leader RNA biogenesis. J. Biol. Chem. 2003;278:51469–51478. doi: 10.1074/jbc.M308997200. [DOI] [PubMed] [Google Scholar]
- 36.Haile S, Estevez AM, Clayton C. A role for the exosome in the in vivo degradation of unstable mRNAs. RNA. 2003;9:1491–1501. doi: 10.1261/rna.5940703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirumi H, Hirumi K. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 1989;75:985–989. [PubMed] [Google Scholar]
- 38.Comini MA, Guerrero SA, Haile S, Menge U, Lunsdorf H, Flohe L. Validation of Trypanosoma brucei trypanothione synthetase as drug target. Free Radic. Biol. Med. 2004;36:1289–1302. doi: 10.1016/j.freeradbiomed.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 39.Goldshmidt H, Matas D, Kabi A, Carmi S, Hope R, Michaeli S. Persistent ER stress induces the spliced leader RNA silencing pathway (SLS), leading to programmed cell death in Trypanosoma brucei. PLoS Pathog. 2010;6:e1000731. doi: 10.1371/journal.ppat.1000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, Smyth GK. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23:2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- 41.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 42.Wettenhall JM, Smyth GK. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20:3705–3706. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- 43.Paz I, Akerman M, Dror I, Kosti I, Mandel-Gutfreund Y. SFmap: a web server for motif analysis and prediction of splicing factor binding sites. Nucleic Acids Res. 2010;38:W281–W285. doi: 10.1093/nar/gkq444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akerman M, David-Eden H, Pinter RY, Mandel-Gutfreund Y. A computational approach for genome-wide mapping of splicing factor binding sites. Genome Biol. 2009;10:R30. doi: 10.1186/gb-2009-10-3-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eden E, Lipson D, Yogev S, Yakhini Z. Discovering motifs in ranked lists of DNA sequences. PLoS Comput. Biol. 2007;3:e39. doi: 10.1371/journal.pcbi.0030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leibovich L, Yakhini Z. Efficient motif search in ranked lists and applications to variable gap motifs. Nucleic Acids Res. 2012;40:5832–5847. doi: 10.1093/nar/gks206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hury A, Goldshmidt H, Tkacz ID, Michaeli S. Trypanosome spliced-leader-associated RNA (SLA1) localization and implications for spliced-leader RNA biogenesis. Eukaryot. Cell. 2009;8:56–68. doi: 10.1128/EC.00322-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang XH, Liu L, Michaeli S. Identification of the first trypanosome H/ACA RNA that guides pseudouridine formation on rRNA. J. Biol. Chem. 2001;276:40313–40318. doi: 10.1074/jbc.M104488200. [DOI] [PubMed] [Google Scholar]
- 49.Xu Y, Liu L, Lopez-Estrano C, Michaeli S. Expression studies on clustered trypanosomatid box C/D small nucleolar RNAs. J. Biol. Chem. 2001;276:14289–14298. doi: 10.1074/jbc.M007007200. [DOI] [PubMed] [Google Scholar]
- 50.Siegel TN, Tan KS, Cross GA. Systematic study of sequence motifs for RNA trans splicing in Trypanosoma brucei. Mol. Cell. Biol. 2005;25:9586–9594. doi: 10.1128/MCB.25.21.9586-9594.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lustig Y, Sheiner L, Vagima Y, Goldshmidt H, Das A, Bellofatto V, Michaeli S. Spliced-leader RNA silencing: a novel stress-induced mechanism in Trypanosoma brucei. EMBO Rep. 2007;8:408–413. doi: 10.1038/sj.embor.7400930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ben-Shlomo H, Levitan A, Beja O, Michaeli S. The trypanosomatid Leptomonas collosoma 7SL RNA gene. Analysis of elements controlling its expression. Nucleic Acids Res. 1997;25:4977–4984. doi: 10.1093/nar/25.24.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lustig Y, Wachtel C, Safro M, Liu L, Michaeli S. ‘RNA walk' a novel approach to study RNA-RNA interactions between a small RNA and its target. Nucleic Acids Res. 2010;38:e5. doi: 10.1093/nar/gkp872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shaked H, Wachtel C, Tulinski P, Yahia NH, Barda O, Darzynkiewicz E, Nilsen TW, Michaeli S. Establishment of an in vitro trans-splicing system in Trypanosoma brucei that requires endogenous spliced leader RNA. Nucleic Acids Res. 2010;38:e114. doi: 10.1093/nar/gkq065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gunzl A, Tschudi C, Nakaar V, Ullu E. Accurate transcription of the Trypanosoma brucei U2 small nuclear RNA gene in a homologous extract. J. Biol. Chem. 1995;270:17287–17291. doi: 10.1074/jbc.270.29.17287. [DOI] [PubMed] [Google Scholar]
- 56.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics. 2006 doi: 10.1002/0471250953.bi0506s15. Chapter 5, Unit 5.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 58.Bandziulis RJ, Swanson MS, Dreyfuss G. RNA-binding proteins as developmental regulators. Genes Dev. 1989;3:431–437. doi: 10.1101/gad.3.4.431. [DOI] [PubMed] [Google Scholar]
- 59.Bagga PS, Arhin GK, Wilusz J. DSEF-1 is a member of the hnRNP H family of RNA-binding proteins and stimulates pre-mRNA cleavage and polyadenylation in vitro. Nucleic Acids Res. 1998;26:5343–5350. doi: 10.1093/nar/26.23.5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miyake T, Loch CM, Li R. Identification of a multifunctional domain in autonomously replicating sequence-binding factor 1 required for transcriptional activation, DNA replication, and gene silencing. Mol. Cell. Biol. 2002;22:505–516. doi: 10.1128/MCB.22.2.505-516.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vazquez MP, Mualem D, Bercovich N, Stern MZ, Nyambega B, Barda O, Nasiga D, Gupta SK, Michaeli S, Levin MJ. Functional characterization and protein-protein interactions of trypanosome splicing factors U2AF35, U2AF65 and SF1. Mol. Biochem. Parasitol. 2009;164:137–146. doi: 10.1016/j.molbiopara.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 65.Tkacz ID, Gupta SK, Volkov V, Romano M, Haham T, Tulinski P, Lebenthal I, Michaeli S. Analysis of spliceosomal proteins in Trypanosomatids reveals novel functions in mRNA processing. J. Biol. Chem. 2010;285:27982–27999. doi: 10.1074/jbc.M109.095349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tkacz ID, Cohen S, Salmon-Divon M, Michaeli S. Identification of the heptameric Lsm complex that binds U6 snRNA in Trypanosoma brucei. Mol. Biochem. Parasitol. 2008;160:22–31. doi: 10.1016/j.molbiopara.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 67.Luz Ambrosio D, Lee JH, Panigrahi AK, Nguyen TN, Cicarelli RM, Gunzl A. Spliceosomal proteomics in Trypanosoma brucei reveal new RNA splicing factors. Eukaryot. Cell. 2009;8:990–1000. doi: 10.1128/EC.00075-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liang XH, Liu Q, Liu L, Tschudi C, Michaeli S. Analysis of spliceosomal complexes in Trypanosoma brucei and silencing of two splicing factors Prp31 and Prp43. Mol. Biochem. Parasitol. 2006;145:29–39. doi: 10.1016/j.molbiopara.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 69.Queiroz R, Benz C, Fellenberg K, Hoheisel JD, Clayton C. Transcriptome analysis of differentiating trypanosomes reveals the existence of multiple post-transcriptional regulons. BMC Genomics. 2009;10:495. doi: 10.1186/1471-2164-10-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wurst M, Seliger B, Jha BA, Klein C, Queiroz R, Clayton C. Expression of the RNA recognition motif protein RBP10 promotes a bloodstream-form transcript pattern in Trypanosoma brucei. Mol. Microbiol. 2012;83:1048–1063. doi: 10.1111/j.1365-2958.2012.07988.x. [DOI] [PubMed] [Google Scholar]
- 71.Colasante C, Robles A, Li CH, Schwede A, Benz C, Voncken F, Guilbride DL, Clayton C. Regulated expression of glycosomal phosphoglycerate kinase in Trypanosoma brucei. Mol. Biochem. Parasitol. 2007;151:193–204. doi: 10.1016/j.molbiopara.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 72.Robles A, Clayton C. Regulation of an amino acid transporter mRNA in Trypanosoma brucei. Mol. Biochem. Parasitol. 2008;157:102–106. doi: 10.1016/j.molbiopara.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 73.Nilsson D, Gunasekera K, Mani J, Osteras M, Farinelli L, Baerlocher L, Roditi I, Ochsenreiter T. Spliced leader trapping reveals widespread alternative splicing patterns in the highly dynamic transcriptome of Trypanosoma brucei. PLoS Pathog. 2010;6:e1001037. doi: 10.1371/journal.ppat.1001037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rettig J, Wang Y, Schneider A, Ochsenreiter T. Dual targeting of isoleucyl-tRNA synthetase in Trypanosoma brucei is mediated through alternative trans-splicing. Nucleic Acids Res. 2012;40:1299–1306. doi: 10.1093/nar/gkr794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Walrad P, Paterou A, Acosta-Serrano A, Matthews KR. Differential trypanosome surface coat regulation by a CCCH protein that co-associates with procyclin mRNA cis-elements. PLoS Pathog. 2009;5:e1000317. doi: 10.1371/journal.ppat.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walrad PB, Capewell P, Fenn K, Matthews KR. The post-transcriptional trans-acting regulator, TbZFP3, co-ordinates transmission-stage enriched mRNAs in Trypanosoma brucei. Nucleic Acids Res. 2012;40:2869–2883. doi: 10.1093/nar/gkr1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mani J, Guttinger A, Schimanski B, Heller M, Acosta-Serrano A, Pescher P, Spath G, Roditi I. Alba-domain proteins of Trypanosoma brucei are cytoplasmic RNA-binding proteins that interact with the translation machinery. PLoS One. 2011;6:e22463. doi: 10.1371/journal.pone.0022463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kolev NG, Ramey-Butler K, Cross GA, Ullu E, Tschudi C. Developmental progression to infectivity in Trypanosoma brucei triggered by an RNA-binding protein. Science. 2012;338:1352–1353. doi: 10.1126/science.1229641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol. Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.