Abstract
Recent studies showed that Ten-eleven translocation (Tet) family dioxygenases can oxidize 5-methyl-2’-deoxycytidine (5-mdC) in DNA to yield the 5-hydroxymethyl, 5-formyl and 5-carboxyl derivatives of 2’-deoxycytidine (5-HmdC, 5-FodC and 5-CadC). 5-HmdC in DNA may be enzymatically deaminated to yield 5-hydroxymethyl-2’-deoxyuridine (5-HmdU). After their formation at CpG dinucleotide sites, these oxidized pyrimidine nucleosides, particularly 5-FodC, 5-CadC, and 5-HmdU, may be cleaved from DNA by thymine DNA glycosylase, and subsequent action of base-excision repair machinery restores unmethylated cytosine. These processes are proposed to be important in active DNA cytosine demethylation in mammals. Here we used a reversed-phase HPLC coupled with tandem mass spectrometry (LC-MS/MS/MS) method, along with the use of stable isotope-labeled standards, for accurate measurements of 5-HmdC, 5-FodC, 5-CadC and 5-HmdU in genomic DNA of cultured human cells and multiple mammalian tissues. We found that overexpression of the catalytic domain of human Tet1 led to marked increases in the levels of 5-HmdC, 5-FodC and 5-CadC, but only a modest increase in 5-HmdU, in genomic DNA of HEK293T cells. Moreover, 5-HmdC is present at a level that is approximately 2–3 and 3–4 orders of magnitude greater than 5-FodC and 5-CadC, respectively, and 35–400 times greater than 5-HmdU in the mouse brain and skin, and human brain. The robust analytical method built a solid foundation for dissecting the molecular mechanisms of active cytosine demethylation, for measuring these 5-mdC derivatives and assessing their involvement in epigenetic regulation in other organisms and for examining whether these 5-mdC derivatives can be used as biomarkers for human diseases.
INTRODUCTION
Eukaryotic chromatin possesses a wealth of information that is required for the development and growth of a multicellular organism. Such information is embedded both genetically in the DNA sequence itself and epigenetically via DNA methylation and posttranslational modifications of core histone proteins (1,2). In mammals, DNA methylation occurs at the C5 position of cytosine residues mainly at CpG dinucleotide sites (1). This methylation pattern is established during embryonic development by de novo DNA methyltransferases DNMT3a and DNMT3b, and maintained during cell division by maintenance DNA methyltransferase DNMT1 (1). The importance of cytosine methylation in mammalian development is manifested by the observations that mice deficient in DNMT3a die at 4 weeks of age (3), and mice lacking DNMT1 or DNMT3b are embryonically lethal (3,4).
Studies in the past decade showed that DNA methylation pattern in mammals can be lost through passive or active mechanisms (5,6). Elimination or inhibition of DNMT1 activity can lead to loss of cytosine methylation during cell division, a process known as passive cytosine DNA demethylation. Active cytosine DNA demethylation is the conversion of 5-methylcytosine (5-mC) to unmethylated cytosine in a process that is independent of DNA replication (5). Multiple lines of evidence support the existence of active cytosine demethylation in the entire genome or in specific genomic loci, and multiple mechanisms have been proposed for this process (6).
Recent discovery of Ten-eleven translocation (Tet) family dioxygenases, including Tet1, Tet2 and Tet3, in the oxidation of 5-mC sheds significant new light on active cytosine demethylation in mammals. Tet1 was first observed to catalyze the oxidation of 5-mC to 5-hydroxymethylcytosine (5-HmC) in DNA in vitro and in human cells (7), and all three members of mouse Tet family were later found to carry the same enzymatic activity (8). Aside from the generation of 5-HmC, Tet family proteins are also capable of oxidizing 5-mC to 5-formylcytosine (5-FoC) and 5-carboxylcytosine (5-CaC) (9,10). 5-FoC and 5-CaC in DNA can be readily excised by thymine DNA glycosylase (TDG) in vitro (9,11), and genetic depletion of TDG in mouse embryonic stem (ES) cells leads to substantial accumulation of 5-CaC (9). These results suggest that Tet-mediated oxidation of 5-mC to 5-FoC and 5-CaC, followed by the excision of the latter two by TDG and restoration of an unmethylated cytosine by base-excision repair (BER) machinery, constitutes a new pathway for active cytosine demethylation in mammals (Figure 1).
Figure 1.
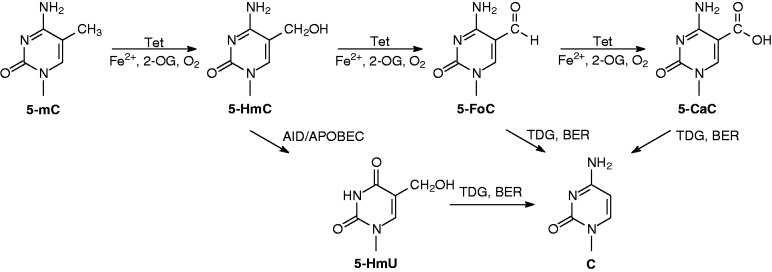
Proposed mechanisms for Tet-mediated oxidative demethylation of 5-methylcytosine in DNA. In the presence of Fe2+, 2-oxoglutarate (2-OG) and O2, Tet family proteins can catalyze the sequential oxidation of 5-mC in DNA to give 5-HmC, 5-FoC and 5-CaC. 5-CaC can then be recognized and cleaved by TDG, and subsequent action of BER machinery leads to the restoration of unmethylated cytosine. Alternatively, the Tet-produced 5-HmC can be deaminated by AID/APOBEC deaminases to give 5-HmU, which can be cleaved and replaced with unmethylated cytosine by TDG and BER machinery.
Apart from the direct removal of Tet-produced oxidation products (i.e. 5-FoC and 5-CaC) by TDG, it was also proposed that 5-HmC in DNA can be deaminated, by AID (activation-induced deaminase)/APOBEC (apolipoprotein B mRNA-editing enzyme complex) families of cytidine deaminases, to yield 5-hydroxymethyluracil (5-HmU) (12,13). While 5-HmC in DNA is a poor substrate for TDG, 5-HmU, when paired with a guanine, can be readily excised by DNA glycosylases (e.g. TDG) (11,12,14). Thus, the oxidation of 5-mC to 5-HmC by Tet proteins, deamination of the latter nucleobase by AID/APOBECs and TDG-induced BER of the resulting 5-HmU may also give rise to active cytosine demethylation in mammals (Figure 1) (12,13). The involvement of this sequential oxidation-deamination mechanism in active cytosine demethylation was recently challenged by the apparent lack of significant biochemical activity of recombinant AID or APOBEC toward 5-HmC deamination in vitro or in cultured cells because of failure in detecting 5-HmU (15). Nevertheless, it remains possible that such deamination may occur in specific cellular context(s).
Liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) method has played an important role in the identification of the Tet-induced oxidation products of 5-mC (7,9,10). The method, along with the use of external standards, has also been used for assessing the levels of 5-HmC, 5-FoC and 5-CaC in cells or tissues (10,15). However, we reason that the use of stable isotope-labeled internal standards will offer unambiguous identification and more accurate measurements of levels of intermediates that are proposed to be involved in active cytosine demethylation, which, along with genetic manipulation, may provide important insights into the mechanisms of active cytosine demethylation.
Aberrant epigenetic regulation of gene expression has long been known to be associated with cancer development (16). Along this line, Tet1 is a fusion partner of the mixed lineage leukemia (MLL) gene in acute myeloid leukemia (17,18). Lower levels of 5-HmC have been observed in myeloid leukemia and a number of solid tumors (including carcinomas of the breast, liver, lung, pancreas and prostate), as shown by immunohistochemistry or dot blot analysis (19,20), and in lung and brain tumors, as revealed by both immunohistochemistry and LC-MS/MS analysis (21). Thus, a robust analytical method for the accurate quantification of the aforementioned 5-mC oxidation intermediates may also result in the discovery of biomarkers for the diagnosis of cancer and perhaps other human diseases.
Herein, we developed an LC-MS/MS/MS coupled with the stable isotope-dilution method for the sensitive and accurate quantification of 5-hydroxymethyl-2’-deoxycytidine (5-HmdC), 5-formyl-2’-deoxycytidine (5-FodC), 5-carboxyl-2’-deoxycytidine (5-CadC) and 5-hydroxymethyl-2’-deoxyuridine (5-HmdU) in genomic DNA of cultured human cells and multiple mammalian tissues.
MATERIALS AND METHODS
Materials
All chemicals and enzymes, unless otherwise specified, were from Sigma-Aldrich (St. Louis, MO). [1,3-15N2-2’-D]-5-hydroxymethyl-2’-deoxycytidine ([1,3-15N2-2’-D]-5-HmdC), [1,3-15N2-2’-D]-5-hydroxymethyl-2’-deoxyuridine ([1,3-15N2-2’-D]-5-HmdU) and [1,3-15N2-2’-D]-5-formyl-2’-deoxycytidine ([1,3-15N2-2’-D]-5-FodC) were synthesized previously (22,23). [4-amino-1,3-15N3]-2’-deoxycytidine ([4-amino-1,3-15N3]-dC) was purchased from Cambridge Isotope Laboratories (Andover, MA). Proteinase K was obtained from New England Biolabs (Ipswich, WA). The HEK293T human embryonic kidney cells, HeLa human cervical cancer cells, WM-266-4 human melanoma cells and cell culture reagents were purchased from ATCC (Manassas, VA, USA). Expression vectors for the catalytic domain of human Tet1 (amino acids 1418–2136) and its corresponding mutant Tet1m (H1672Y/D1674A) were previously described (12) and were deposited to Addgene (#39454 and #39455). The DNA of mouse ES cells, which were derived from mouse blastocysts, was provided by Prof. Gerd P. Pfeifer (City of Hope).
Synthesis and characterization of [4-amino-1,3-15N3]-5-Carboxyl-2’-deoxycytidine ([4-amino-1,3-15N3]-5-CadC)
The title compound was prepared at a microscale according to previously published methods for the synthesis of the corresponding unlabeled compounds with some modifications (24). [4-amino-1,3-15N3]-dC (3.0 mg, 13.0 μmol) was dissolved in dimethylformamide (100 µl) and allowed to stir at room temperature (rt) for 10 min. Slight excess of iodine (2.13 mg), followed by m-chloroperbenzoic acid (2.97 mg), was added and allowed to stir at rt for 1.5 h. The reaction mixture was then dried by Speed-vac. The sample was reconstituted with water and filtered. The resulting [4-amino-1,3-15N3]-5-iodo-2’-deoxycytidine ([4-amino-1,3-15N3]-5-IdC) was purified by HPLC.
The purified [4-amino-1,3-15N3]-5-IdC was dissolved in methanol (500 µl), and CO was bubbled through the solution for 30 min, to which solution were subsequently added triphenylphosphine (1.37 mg), triethylamine (2 µl) and tris(dibenzylideneacetone)dipalladium(0) (0.81 mg). The reaction was stirred at 50°C under CO atmosphere overnight. The mixture was then cooled to rt and the solid removed by filtration. The resulting [4-amino-1,3-15N3]-5-methoxycarbonyl-2′-deoxycytidine was readily converted to the desired [4-amino-1,3-15N3]-5-CadC by treating with 0.1 M K2CO3 in MeOH/H2O at 40°C overnight. The latter compound was isolated from the reaction mixture by HPLC. The identity of [4-amino-1,3-15N3]-5-CadC was confirmed by LC-MS/MS analysis and by high-resolution MS analysis on an ESI-TOF instrument. The latter measurement gave m/z 273.0636 for the [M – H]− ion, whose calculated m/z was 273.0641.
Cell culture, transfection and DNA extraction
HEK293T cells were cultured in Dulbecco's Modified Eagle Medium (ATCC). HeLa human cervical cancer cells and WM-266-4 human melanoma cells were cultured in Eagle's Minimum Essential Medium (ATCC). All cells were incubated at 37°C in 5% CO2 atmosphere. The culture medium was supplemented with 10% fetal bovine serum and 100 IU/ml penicillin. The HEK293T cells were seeded in six-well plates at 50–60% confluence and, 24 h later, the cells were transfected with 1.5 μg Tet1 or Tet1m expression plasmids using Lipofectamine 2000 (Invitrogen). The cells were harvested for DNA extraction 48 h after plasmid transfection.
Genomic DNA was extracted from the cells and mouse brain tissues using a high-salt method, following published procedures (25). With this method, we could typically obtain ∼30 µg of DNA from 5 million cells or 100 µg of DNA from 100 mg of tissues. The whole brain tissues were dissected from 8-week-old male mice of C57/B6 strain, snap-frozen in liquid nitrogen and stored in −80°C freezer before DNA extraction. Genomic DNA samples from human brain (cerebellum, Supplementary Table S1 gives the patient information) and mouse skin tissues were previously isolated (26,27). The DNA pellet was redissolved in doubly distilled water and its concentration measured by UV absorbance at 260 nm.
Enzymatic digestion
For the enzymatic digestion of DNA, nuclease P1 (0.1 U/μg DNA), phosphodiesterase 2 (0.00025 U/μg DNA) and erythro-9-(2-hydroxy-3-nonyl)adenine (1 mM in final concentration) were added to 30–80 μg of cellular or tissue DNA in a solution containing 30 mM sodium acetate (pH 5.6) and 1.0 mM zinc acetate. The digestion mixture was incubated at 37°C for 48 h and continued for another 2 h after alkaline phosphatase (0.05 U/μg DNA), phosphodiesterase 1 (0.0005 U/μg DNA) and Tris–HCl (pH 8.9, final concentration 50 mM) were added. To the mixture were then added 30 fmol of [1,3-15N2-2’-D]-5-FodC, 25 fmol of [4-amino-1,3-15N3]-5-CadC, and 2 pmol of [1,3-15N2-2’-D]-5-HmdU. The enzymes were removed by chloroform extraction. The aqueous layer was dried, reconstituted in doubly distilled water and subjected to off-line HPLC for the enrichment of target nucleosides.
For LC-MS/MS/MS quantification of 5-HmdC, 50 fmol of [1,3-15N2-2’-D]-5-HmdC was added to the enzymatic digestion mixture of 10–50 ng genomic DNA. The samples were subsequently extracted with chloroform to remove the enzymes and the aqueous layer subjected to LC-MS/MS/MS analysis.
HPLC
HPLC purification of [4-amino-1,3-15N3]-5-IdC was performed on an Agilent 1100 series HPLC system equipped with a binary pump and a UV detector. A 4.6 × 250 mm Apollo C18 column (5 μm in particle size and 300 Å in pore size, Grace Inc., Deerfield, IL) was used and the wavelength was set at 260 nm. A solution of 10 mM ammonium formate (pH 6.3, solution A) and a mixture of 10 mM ammonium formate and acetonitrile (70:30, v/v, solution B) were used as mobile phases. The flow rate was 0.50 ml/min, and a gradient of 5 min 0–20% B, 40 min 20–55% B and 5 min 55–98% B was used. The purification of [4-amino-1,3-15N3]-5-CadC was conducted under the same conditions except that a 4.6 × 250 mm Alltima C18 column (5 μm in particle size and 300 Å in pore size, Grace Inc., Deerfield, IL) with a gradient of 10 min 0–2% B, 15 min 2–15% B and 5 min 15–98% B was used.
The enrichment of 5-FodC, 5-CadC and 5-HmdU from the enzymatic digestion products of DNA was carried out on a Beckman HPLC system with pump module 125 and a UV detector (module 126). A 4.6 × 250 mm Aeris Widepore C18 column (3.6 μm in particle size, Phenomenex, Torrance, CA) was used. An isocratic elution at a flow rate of 0.8 ml/min and with a solution of 10 mM ammonium formate (pH 8.5) as mobile phase was used. A typical HPLC trace is depicted in Supplementary Figure S1 in the Supporting Information. From the enrichment HPLC trace, we often observed a low level (<5%) of RNA contamination for the DNA samples, and the DNA amount was corrected based on the ratio of peak areas found for guanosine (rG) to 2’-deoxyguanosine (dG) in the HPLC trace (Supplementary Figure S1). The HPLC fractions eluting at 3.0–4.0, 9.5–10.5 and 26.0–27.5 min were collected for 5-CadC, 5-HmdU and 5-FodC, respectively. The collected fractions were dried, redissolved in water and injected for LC-MS/MS/MS analysis.
During the above enrichment, 5-methyl-2’-deoxycytidine (5-mdC) could also be resolved from other nucleosides. The levels of 5-mdC, expressed as percentage of dG, were quantified based on the peak areas of 5-mdC and dG found in the chromatogram with the consideration of the extinction coefficients of the two nucleosides at 260 nm (5020 and 11 700 L mol−1 cm−1 for 5-mdC and dG, respectively) (28,29).
Quantification with LC-MS/MS/MS
The LC-MS/MS/MS experiments were conducted using a 0.5 × 250 mm Zorbax SB-C18 column (5 μm in particle size, Agilent Technologies, Santa Clara, CA) and an Agilent 1200 capillary HPLC pump. The flow rate was 8.0 μl/min. The effluent from the LC column was directed to an LTQ linear ion-trap mass spectrometer (Thermo Fisher Scientific), which was set up for monitoring the labeled and unlabeled 5-HmdC, 5-FodC and 5-CadC in the positive-ion mode, and the labeled and unlabeled 5-HmdU in the negative-ion mode. A solution of 0.1% (v/v) formic acid in water (solution A) and a solution of 0.1% (v/v) formic acid in methanol (solution B) were used as mobile phases for 5-HmdC, 5-FodC and 5-CadC analyses. A solution of 2 mM ammonium formate in water (solution A) and a solution of 2 mM ammonium formate in methanol (solution B) were used as mobile phases for 5-HmdU analysis. A gradient of 5 min 0–20% B and 25 min 20–70% B was used for the separation of all the modified nucleosides. The temperature for the ion transport tube was maintained at 275 and 300°C in the positive- and negative-ion modes, respectively. The sheath gas flow rate was 15 arbitrary units, and no auxiliary gas was used. The electrospray, capillary and tube lens voltages for the positive-ion mode analyses were 5 kV, 14 V and 20 V, respectively, and these values for the negative-ion mode analyses were 4.5 kV, −10 V and −100 V, respectively. The sensitivities for detecting all these modified nucleosides were optimized by varying two working parameters of the LTQ mass spectrometer, i.e. normalized collision energy and activation Q (Supplementary Table S2).
The numbers of moles of 5-HmdC, 5-FodC, 5-CadC and 5-HmdU in the nucleoside mixtures were calculated from the area ratios of peaks found in the selected-ion chromatograms for the analytes over their corresponding isotope-labeled standards, the amounts of the labeled standards added and the equations derived from the calibration curves. The final data, expressed in terms of numbers of modifications per 106 nucleosides (or per 103 5-mdC), were calculated by dividing the number of moles of the modified nucleosides with the number of moles of total nucleosides (or 5-mdC) in the digested DNA (Supplementary Table S3).
RESULTS
Quantification of 5-HmdC, 5-FodC, 5-CadC and 5-HmdU in DNA of HEK293T cells with Tet1 overexpression and in DNA of cultured human cancer cells
We set out to develop an LC-MS/MS/MS combined with stable isotope-dilution method for the accurate assessment of levels of 5-HmdC, 5-FodC, 5-CadC and 5-HmdU in genomic DNA of cultured cells and tissues. We first examined how overexpression of Tet1 affects the levels of the four modified pyrimidine nucleosides in genomic DNA of cultured human cells. We expressed the catalytic domain of Tet1 or its corresponding inactive mutant in HEK293T cells, isolated the genomic DNA from the cells and digested the DNA to nucleosides with a cocktail of four enzymes (See ‘Materials and Methods’ section). Three of the aforementioned pyrimidine nucleosides, 5-HmdU, 5-FodC and 5-CadC, were subsequently enriched from the digestion mixture with reversed-phase HPLC, and the enriched fractions were subjected to LC-MS3 analyses using an LTQ linear ion trap mass spectrometer. As displayed in Supplementary Figure S1, the three modified pyrimidine nucleosides can be readily resolved from each other and from the four canonical nucleosides by reversed-phase HPLC with isocratic elution. Owing to the high level of 5-HmdC in cellular and tissue DNA, the digestion mixture of much lower amount of genomic DNA (i.e. 10 ng of tissue DNA or 10–50 ng of cellular DNA) was directly subjected for LC-MS3 analysis of 5-HmdC.
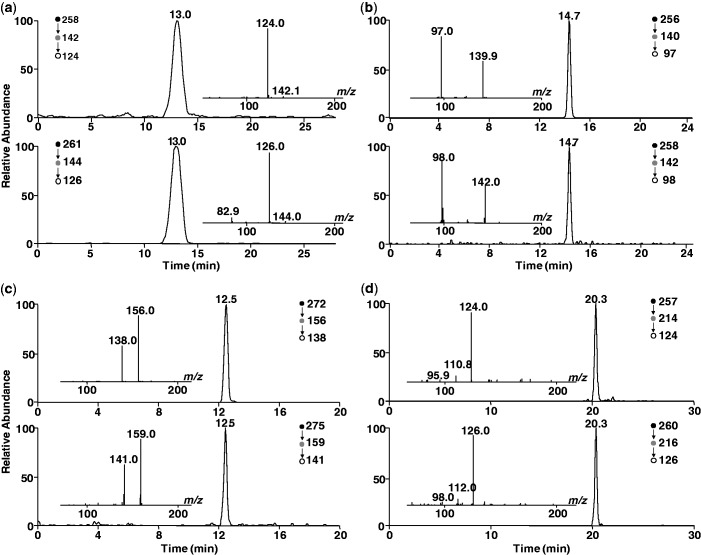
The fragmentation of the protonated ions of unlabeled and labeled 5-HmdC and 5-FodC was previously described (23). Briefly, upon collisional activation, the [M + H]+ ions of unlabeled 5-HmdC and 5-FodC can readily eliminate a 2-deoxyribose moiety to yield the protonated ions of the nucleobase portion (m/z 142 and 140 for 5-HmdC and 5-FodC, respectively) as the dominant fragment ions in MS/MS. Further collisional activation of the ions of m/z 142 and 140 leads to facile loss of H2O and HNCO moieties to yield fragment ions of m/z 124 and 97 in MS3 for 5-HmdC and 5-FodC, respectively (Figure 2a and b, inset). Collisional activation of the protonated ion of 5-CadC again resulted in facile formation of the protonated ion of 5-CaC (m/z 156) in MS/MS (spectrum not shown), the latter fragment can lose a H2O molecule to give the ion of m/z 138 in MS3 (Figure 2c, inset). As we found previously, 5-HmdU exhibits better sensitivity when analyzed in the negative-ion mode (22). The deprotonated 5-HmdU loses readily an HNCO moiety to yield an ion of m/z 214 in MS/MS, and further collisional activation of the latter leads to the loss of part of the 2-deoxyribose component to produce the ion of m/z 124 in MS3 (Figure 2d, inset). The proposed structures for the major fragment ions observed in MS/MS and MS3 are displayed in Supplementary Figure S2.
Figure 2.
Representative LC-MS/MS/MS results for the quantification of 5-HmdC (a), 5-FodC (b), 5-CadC (c) and 5-HmdU (d) in HEK293T cells expressing the catalytic domain of Tet1. Shown are the selected-ion chromatograms for monitoring the indicated transitions for the analytes (top trace) and the isotope-labeled standards (bottom trace), and the inset gives the corresponding MS/MS/MS for the analytes and internal standards.
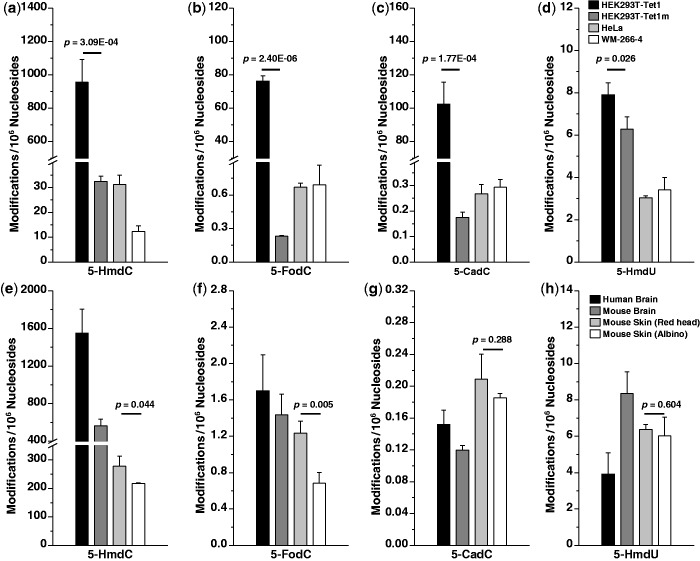
The nearly identical elution time and similar MS3 spectra for the analytes and their stable isotope-labeled counterparts allowed for the unambiguous identification and reliable quantification of the four pyrimidine nucleosides in the digestion mixture of DNA isolated from the HEK293T cells [Figure 2; the calibration curves for 5-HmdC, 5-FodC and 5-CadC are depicted in Supplementary Figure S3, and the calibration curve for 5-HmdU was shown previously (30)]. In this vein, because the levels of 5-HmdC, 5-FodC and 5-CadC cover a wide range in different samples, calibration curves in two concentration ranges were used for determining accurately the levels of these mdC modifications. Our quantification results revealed that the levels of 5-HmdC, 5-FodC, 5-CadC and 5-HmdU in HEK293T cells overexpressing the catalytically active form of Tet1 were 960, 76, 102 and 7.9 modifications per 106 nucleosides, respectively (Figure 3a–d and Supplementary Table S3). The levels of the four nucleosides were 32.5, 0.23, 0.18 and 6.3 modifications per 106 nucleosides, respectively, in DNA of HEK293T cells overexpressing the catalytically inactive form of Tet1 (Figure 3a–d and Supplementary Table S3). The catalytic activity of Tet1 led to markedly higher levels (by ∼29.5-, 330- and 570-fold for 5-HmdC, 5-FodC and 5-CadC, respectively) of the three 5-mdC oxidation products. Thus, we confirmed the previous findings that 5-HmdC, 5-FodC and 5-CadC can be induced from Tet1-mediated oxidation of 5-mdC in genomic DNA (7–10). Our results also provided, for the first time, unambiguous evidence to support that, in contrast to the substantial elevation of the levels of the three 5-mdC oxidation products described above, Tet1 catalytic activity alone only gave rise to a modest (by ∼25%) increase in the level of 5-HmdU in genomic DNA of cultured cells (Figure 3d).
Figure 3.
Quantification results for the levels of 5-HmdC (a), 5-FodC (b), 5-CadC (c) and 5-HmdU (d) in HEK293T cells expressing the active or inactive forms of Tet1 (n = 3) and in HeLa and WM-266-4 cells (n = 3). Quantification results for the levels of 5-HmdC (e), 5-FodC (f), 5-CadC (g) and 5-HmdU (h) in human brain tissues (black bar, n = 4), mouse brain tissues (dark gray bar, n = 3), and skin tissues of redhead (light gray bar, n = 3) and albino (open bar, n = 3) mice. The data represent the mean and standard deviation of the measurement results. The P values were calculated using unpaired two-tailed t-test.
We next assessed whether these oxidation products of 5-mdC are present in genomic DNA of cultured human cancer cells. Our results showed that, in genomic DNA of HeLa cells, the levels of 5-HmdC, 5-FodC, 5-CadC and 5-HmdU were 31.2, 0.67, 0.27 and 3.0 per 106 nucleosides, respectively (Figure 3a–d). In addition, these four nucleosides were present in genomic DNA of WM-266-4 human melanoma cells at the levels of 12.2, 0.69, 0.29 and 3.4 modifications per 106 nucleosides, respectively (Figure 3a–d). Thus, the levels of these modifications are similar as what were found for HEK293T cells expressing catalytically inactive form of Tet1, except that the level of 5-HmdC in WM-266-4 cells is ∼2.5-fold lower than that in HEK293T or HeLa cells.
We further measured the levels of 5-HmdC, 5-FodC and 5-CadC in DNA of mouse ES cells (Supplementary Table S3). For comparison with previous LC-MS/MS measurement results obtained from the use of the external standards (10), we converted the data to the number of modified nucleosides per 106 dC. Our results showed that the levels of 5-HmdC, 5-FodC and 5-CadC are 680 ± 50, 14.5 ± 2.9 and 3.4 ± 0.3 modifications per 106 dC, respectively. Our data for 5-FodC and 5-CadC are largely consistent with those reported previously (10), where the levels of the two nucleosides from duplicate measurements were 17.9/19.8 and 3.6/3.1 modifications per 106 dC, respectively. However, the level of 5-HmdC from the previous measurement, i.e. 1120/1380 modifications per 106 dC (10), was ∼2-fold higher than what we measured here.
Quantification of 5-HmdC, 5-FodC, 5-CadC and 5-HmdU in genomic DNA from human and mouse brain
We next quantified the levels of the modified nucleosides in human and mouse brain tissues. In human brain (cerebellum) DNA, the average levels of 5-HmdC, 5-FodC and 5-CadC are 1550, 1.7 and 0.15 modifications per 106 nucleosides, respectively, with the three nucleosides exhibiting a ratio of ∼10 000:11:1 (Figure 3e–g and Supplementary Table S3). Given these brain tissues were from four individuals at different ages (Supplementary Table S1), the relatively small standard deviations associated with measurement results suggested a lack of substantial age-dependent effect on the levels of these modifications in human brain. In mouse brain genomic DNA, the contents of 5-HmdC, 5-FodC and 5-CadC display a ratio of ∼4700:12:1, at levels of 560, 1.4 and 0.12 modifications per 106 nucleosides, respectively (Figure 3e–g and Supplementary Table S3). The levels of 5-HmdU in DNA from human and mouse brain were 3.9 and 8.3 modifications per 106 nucleosides, respectively, which are drastically (by ∼400- and 70-fold, respectively) lower than those of 5-HmdC (Figure 3h), but higher than 5-FodC (by 2.3- and 5.9-fold, respectively) and markedly higher than 5-CadC (by 26- and 69-fold, respectively).
Quantification of 5-HmdC, 5-FodC, 5-CadC and 5-HmdU in DNA from mouse skin tissues
Mitra et al. (27) recently studied genomic consequences of functional deficiency in the melanocortin 1 receptor gene (Mc1re/e), the ‘redhead-fairskin’ gene in numerous species including mouse and man. The redhair-fairskin Mc1r allele results in elevated production of pheomelanin (red pigment), and elevations in endogenous reactive oxygen species (ROS) were suggested to contribute to UV-independent generation of melanoma in ‘redhair-fairskin’ (redhead) mice (27). We reasoned that the increased generation of ROS may also result in a rise in levels of 5-mdC modifications within skin tissues of redhead mice as compared with albino mice, the latter of which carry a dysfunctional tyrosinase gene (Tyrc/c) that abolishes all melanin (including pheomelanin) biosynthesis. Thus, we also analyzed the levels of the four pyrimidine nucleosides in DNA isolated from the skin tissues of these two strains of mice (Mc1re/e versus Mc1re/e;Tyrc/c). Our results showed that indeed the skin of redhead mice contains significantly higher levels of 5-HmdC (280 versus 217 per 106 nucleosides) and 5-FodC (1.2 versus 0.7 per 106 nucleosides) as compared with skin from Mc1re/e albino mice (Figure 3e and f, representative LC-MS/MS/MS results are shown in Supplementary Figure S4). The levels of 5-CadC (0.21 versus 0.19 per 106 nucleosides) and 5-HmdU (6.4 versus 6.0 per 106 nucleosides, Figure 3g and h) in these two strains of mice are, however, similar.
DISCUSSION
Our LC-MS/MS/MS coupled with stable isotope-dilution method facilitates the accurate and reproducible detection of 5-HmdC, 5-FodC, 5-CadC and 5-HmdU in DNA isolated from cultured human cells and multiple mammalian tissues. In this context, it is worth emphasizing the advantages of the isotope-dilution method for the measurements of these mdC oxidation products. In this method, we added the stable isotope-labeled standards to the nucleoside mixture; thus, the analytes and the corresponding internal standards are enriched by HPLC and analyzed by LC-MS/MS/MS under identical conditions. Any variation in experimental conditions during the HPLC enrichment and LC-MS/MS/MS analysis will not influence the measurement of the molar ratios of the analytes over their corresponding internal standards. Therefore, the determination of the levels of the analytes will not be affected by alterations in these experimental conditions. Additionally, the co-elution of the analyte and its isotope-labeled standard, along with the similar fragmentation pattern of the analyte and internal standard, offers unequivocal chemical specificity for analyte identification.
Relative to the previously reported LC-MS/MS method with the use of external standards, our method also displays superior sensitivity and reproducibility. The limits of detection for 5-HmdC, 5-FodC, 5-CadC (10) and 5-HmdU were reported to be 2.5 fmol, 5 fmol, 10 fmol and 2–3 modifications per 105 dC, resepectively (15). With our method, the limits of quantitation for 5-HmdC, 5-FodC, 5-CadC and 5-HmdU were 0.056, 0.098, 0.14 and 80 fmol, respectively (Supplementary Table S2). Along this line, with the use of external standards, 5-CadC was not detectable in DNA from HEK293T cells or most mouse organs (10), and 5-HmdU could not be detected in DNA isolated from HEK293T cells (15). We, however, were able to quantify these two modified nucleosides in all cell lines and mammalian tissues tested. Moreover, while reasonably good reproducibility was obtained for the levels of the abundant 5-HmdC in cellular or tissue DNA with the use of the previous method, relatively large variations were observed for the levels of 5-FodC between the two replicate measurements of various mouse tissue DNA samples (10). In contrast, excellent reproducibilities were achieved with our isotope-dilution method (Figure 3 and Supplementary Table S3).
Both endogenous ROS and Tet-mediated oxidation may contribute to the generation of the 5-mdC oxidation products in vivo. Whereas no in vitro experiments have been performed to assess the frequencies of ROS-induced formation of 5-CadC, a previous study showed that the Fenton-type reagent, Cu(II)/H2O2/ascorbate, induced the generation of 5-HmdC at a rate that is ∼1.5 times higher than that of 5-FodC (31). Here we observed that overexpression of the catalytic domain of Tet1 in HEK293T cells led to drastic increases in the levels of 5-HmdC, 5-FodC and 5-CadC, which is accompanied with a modest increase of 5-HmdU. In addition, we found that 5-HmdC is present at a level that is ∼2–3 orders of magnitude greater than 5-FodC, and 3–4 orders of magnitude greater than 5-CadC, in brain tissues of humans and mice, and skin tissues of mice (Figure 3). We also observed that 5-HmdC exists at a level that is ∼35–70 times greater than 5-HmdU in skin and brain tissues of mice, and ∼400 times greater in human brain tissues. To our knowledge, this is the first direct quantitative comparison between the levels of multiple oxidized 5-mdC derivatives in mammalian tissues. The difference in the levels of 5-HmdU and 5-HmdC in DNA is remarkable viewing that mouse and human DNA carries at least 20 times more dT than mdC. While relatively high level of 5-HmdC in human brain tissues has been documented (32), this is the first demonstration that this modified nucleoside is also present in mouse skin at levels that are only a few times lower than that in mouse brain. Previous biochemical studies support that human TDG exhibits high cleavage rates for 5-FodC, 5-HmdU, 5-CadC, but extremely low cleavage rate for 5-HmdC at CpG dinucleotide site (11,14,33). Therefore, the much higher endogenous levels of 5-HmdC than the other three oxidized pyrimidine nucleosides could be attributed, at least in part, to the poor removal of 5-HmdC from the genome. Considering the recent notion that 5-HmdC, apart from being an intermediate for active cytosine demethylation, may itself serve as an epigenetic mark (34), accumulation of 5-HmdC in skin tissues may also alter the epigenetic regulation of gene expression.
In summary, we developed a sensitive LC-MS/MS coupled with stable isotope-dilution method for the quantification of 5-HmdC, 5-FodC, 5-CadC and 5-HmdU in cellular and tissue DNA. Our results provided the first direct comparison of the levels of these four modified nucleosides in same mammalian tissue samples. It can be envisaged that the robust analytical method, when combined with manipulations of genes involved in Tet-mediated active cytosine demethylation pathway, will provide important insights into the roles of various protein factors in active DNA cytosine demethylation. Additionally, the method developed here should be generally applicable for the detection of these 5-mdC modification products in other organisms, which will provide important knowledge for understanding the involvement of Tet family enzymes and their orthologs in epigenetic regulation of gene expression in these organisms. Moreover, considering that aberrant levels of 5-hmdC has been found in a number of different cancers, the analytical method reported here should also be useful for examining whether 5-HmdC and other 5-mdC modifications discussed in this article can be used as biomarkers for cancer and perhaps other human diseases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3 and Supplementary Figures 1–5.
ACKNOWLEDGEMENTS
The authors want to thank Prof. Gerd P. Pfeifer for providing mouse ES cell DNA. Human tissue was obtained from the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland, Baltimore, MD, contract HHSN275200900011C, Ref. No. N01-HD-9-0011.
FUNDING
National Institutes of Health (NIH) [DK082779 to Y.W., NS047344 and ES021957 to H.S. and AR043369 to D.E.F.]; Maryland Stem Cell Research Fund postdoctoral fellowship (to Y.S and Y.Z). Funding for open access charge: NIH [DK082779].
Conflict of interest statement. None declared.
REFERENCES
- 1.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 4.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 5.Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat. Rev. Mol. Cell. Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J. Biol. Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bennett MT, Rodgers MT, Hebert AS, Ruslander LE, Eisele L, Drohat AC. Specificity of human thymine DNA glycosylase depends on N-glycosidic bond stability. J. Am. Chem. Soc. 2006;128:12510–12519. doi: 10.1021/ja0634829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nabel CS, Jia H, Ye Y, Shen L, Goldschmidt HL, Stivers JT, Zhang Y, Kohli RM. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat. Chem. Biol. 2012;8:751–758. doi: 10.1038/nchembio.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lund AH, van Lohuizen M. Epigenetics and cancer. Genes Dev. 2004;18:2315–2335. doi: 10.1101/gad.1232504. [DOI] [PubMed] [Google Scholar]
- 17.Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23) Cancer Res. 2002;62:4075–4080. [PubMed] [Google Scholar]
- 18.Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23) Leukemia. 2003;17:637–641. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- 19.Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu J, Xu ZD, Zhu HG, Ling ZQ, Ye D, et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene. 2013;32:663–669. doi: 10.1038/onc.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin SG, Jiang Y, Qiu R, Rauch TA, Wang Y, Schackert G, Krex D, Lu Q, Pfeifer GP. 5-Hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res. 2011;71:7360–7365. doi: 10.1158/0008-5472.CAN-11-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong H, Cao H, Wang Y, Wang Y. Identification and quantification of a guanine-thymine intrastrand cross-link lesion induced by Cu(II)/H2O2/ascorbate. Chem. Res. Toxicol. 2006;19:614–621. doi: 10.1021/tx060025x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao H, Wang Y. Collisionally activated dissociation of protonated 2'-deoxycytidine, 2'-deoxyuridine, and their oxidatively damaged derivatives. J. Am. Soc. Mass Spectrom. 2006;17:1335–1341. doi: 10.1016/j.jasms.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 24.Dai Q, He C. Syntheses of 5-formyl- and 5-carboxyl-dC containing DNA oligos as potential oxidation products of 5-hydroxymethylcytosine in DNA. Org. Lett. 2011;13:3446–3449. doi: 10.1021/ol201189n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Clauson CL, Robbins PD, Niedernhofer LJ, Wang Y. The oxidative DNA lesions 8,5′-cyclopurines accumulate with aging in a tissue-specific manner. Aging. Cell. 2012;11:714–716. doi: 10.1111/j.1474-9726.2012.00828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Cao H, You C, Yuan B, Bahde R, Gupta S, Nishigori C, Niedernhofer LJ, Brooks PJ, Wang Y. Endogenous formation and repair of oxidatively induced G[8-5 m]T intrastrand cross-link lesion. Nucleic Acids Res. 2012;40:7368–7374. doi: 10.1093/nar/gks357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitra D, Luo X, Morgan A, Wang J, Hoang MP, Lo J, Guerrero CR, Lennerz JK, Mihm MC, Wargo JA, et al. An ultraviolet-radiation-independent pathway to melanoma carcinogenesis in the red hair/fair skin background. Nature. 2012;491:449–453. doi: 10.1038/nature11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan B, Zhang J, Wang H, Xiong L, Cai Q, Wang T, Jacobsen S, Pradhan S, Wang Y. 6-Thioguanine reactivates epigenetically silenced genes in acute lymphoblastic leukemia cells by facilitating proteasome-mediated degradation of DNMT1. Cancer Res. 2011;71:1904–1911. doi: 10.1158/0008-5472.CAN-10-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, Yuan B, Zhang F, Xiong L, Wu J, Pradhan S, Wang Y. Cyclophosphamide perturbs cytosine methylation in Jurkat-T cells through LSD1-mediated stabilization of DNMT1 protein. Chem. Res. Toxicol. 2011;24:2040–2043. doi: 10.1021/tx2003849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, Yuan B, Guerrero C, Bahde R, Gupta S, Wang Y. Quantification of oxidative DNA lesions in tissues of Long-Evans Cinnamon rats by capillary high-performance liquid chromatography-tandem mass spectrometry coupled with stable isotope-dilution method. Anal. Chem. 2011;83:2201–2209. doi: 10.1021/ac103099s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao H, Wang Y. Quantification of oxidative single-base and intrastrand cross-link lesions in unmethylated and CpG-methylated DNA induced by Fenton-type reagents. Nucleic Acids Res. 2007;35:4833–4844. doi: 10.1093/nar/gkm497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L, Lu X, Lu J, Liang H, Dai Q, Xu GL, Luo C, Jiang H, He C. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat. Chem. Biol. 2012;8:328–330. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Rando OJ, Fazzio TG. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.