Abstract
Drug resistance remains a vexing problem in the treatment of cancer patients. While many studies have focused on cell autonomous mechanisms of drug resistance, we hypothesized that the tumor microenvironment may confer innate resistance to therapy. Here we developed a co-culture system to systematically assay the ability of 23 stromal cell types to influence the innate resistance of 45 cancer cell lines to 35 anti-cancer drugs. We found that stroma-mediated resistance is surprisingly common – particularly to targeted agents. We further characterized the stroma-mediated resistance of BRAF-mutant melanoma to RAF inhibition because most of these patients exhibit some degree of innate resistance1-4. Proteomic analysis showed that stromal secretion of the growth factor hepatocyte growth factor (HGF) resulted in activation of the HGF receptor MET, reactivation of the MAPK and PI3K/AKT pathways, and immediate resistance to RAF inhibition. Immunohistochemistry confirmed stromal HGF expression in patients with BRAF-mutant melanoma and a statistically significant correlation between stromal HGF expression and innate resistance to treatment. Dual inhibition of RAF and MET resulted in reversal of drug resistance, suggesting RAF/MET combination therapy as a potential therapeutic strategy for BRAF-mutant melanoma. A similar resistance mechanism was uncovered in a subset of BRAF-mutant colorectal and glioblastoma cell lines. More generally, these studies indicate that the systematic dissection of tumor-microenvironment interactions may reveal important mechanisms underlying drug resistance.
Oncoprotein-targeted drugs hold enormous promise for the future of cancer treatment. However, complete clinical responses are rare, suggesting that mechanisms exist to render a substantial portion of tumor cells resistant to treatment. For example, melanomas harboring the BRAF V600E mutation show a dramatic response to RAF inhibitors, but responses are almost always partial, and tumors often recur within 6 months1-4.
We hypothesized that innate drug resistance might be caused at least in part by factors secreted by the tumor microenvironment. While growth and metastasis-promoting effects of the microenvironment have been well documented5,6, a role in drug resistance has only been partially explored7-11. To test the hypothesis that stromal cells might confer innate resistance to cancer cells, we developed a co-culture system whereby GFP-labeled tumor cells are co-cultured with stromal cells, and the ability of the stromal cells to modulate drug sensitivity is measured by monitoring GFP levels over time (Supplementary Fig. 1). Forty-five GFP-labeled human cancer cell lines were cultured either alone or in combination with a panel of up to 23 human stromal cell lines in the presence of increasing doses of 35 widely used anti-cancer drugs (Supplementary Tables 1 and 2).
Our analysis of cancer cell-stromal cell-drug interactions (Supplementary Tables 3 and 4) yielded a striking result – anti-cancer drugs capable of killing tumor cells when cultured alone, frequently are rendered ineffective when tumor cells are cultured in the presence of stroma (Figure 1a). For example, certain dermal fibroblasts were able to confer complete resistance of colorectal and pancreatic cancer cell lines to the cytotoxic agent gemcitabine (Figure 1b and Supplementary Fig. 2). Different stromal cells conferred resistance to BRAF-mutant melanoma cell lines treated with RAF inhibitors, and ERBB2 over-expressing breast cancer cell lines treated with ERBB2 inhibitors (Figure 1c, 1d and Supplementary Fig. 3 and 4). The stroma-mediated resistance phenomenon was particularly pronounced with targeted agents compared with conventional cytotoxic chemotherapy (P < 0.001; Supplementary Table 2). Overall, of the 23 targeted agents in the panel, 15 (65%) showed evidence of microenvironment-mediated resistance (Supplementary Table 2 and Supplementary methods).
Figure 1. Effect of stromal cells on chemoresistance of cancer cell lines.
a, 45 GFP labeled cancer cell lines were treated with 35 drugs either alone or in co-culture with a panel of up to 23 stromal cell lines and primary cells. The drug effect was calculated by normalizing the number of cells (GFP) after 7 days of treatment to the number of cells (GFP) in DMSO control wells. X-axis represents drug effect in the absence of stromal cells while Y-axis represents drug effect in the presence of stromal cells. The Y axis was also normalized to the effect that each stromal cell type has on cancer cell proliferation when no drug is present (in order to distinguish true rescue from stromal effects on proliferation). The middle diagonal line represents the expected result when stromal cells do not confer resistance. Upper and lower diagonal lines represent one standard deviation from the mid-diagonal line. b, Hierarchical clustering of stromal cells according to their ability to rescue colorectal (CRC) and pancreatic cancer cell lines from 0.1uM gemcitabine. c, Hierarchical clustering of stromal cells according to their ability to rescue melanoma cancer cell lines with V600E BRAF mutation from 2uM PLX4720. d, Hierarchical clustering of stromal cells according to their ability to rescue HER2 amplified breast cancer cell lines from 2uM lapatinib. See Supplementary Figures 1 to 3 for details.
We next explored the mechanism of stroma-mediated innate resistance to the RAF inhibitor PLX4720 (an analog of which, vemurafenib, was recently FDA-approved for the treatment of BRAF-mutant melanoma). In a recent phase 3 clinical trial, 48% of BRAF mutant melanoma patients treated with vemurafenib had a confirmed response, and only 0.9% of patients had a complete response, indicating a high rate of innate resistance2. We tested 18 stromal cell lines for their ability to confer resistance of 7 BRAF V600E melanoma cell lines to PLX4720. Of these, 6 fibroblast lines conferred resistance (Figure 1c and Supplementary Fig. 3).
To determine if the rescue effect was mediated by direct fibroblast-tumor contact or by the secretion of soluble factors, we tested the ability of fibroblast-conditioned growth media to recapitulate the resistance effect. Fibroblast-conditioned media rescued BRAF-mutant melanoma cells from PLX4720, indicating that the rescue was due to a factor secreted by the fibroblasts (Figure 2a). To identify the rescuing secreted factor, we performed an antibody array-based analysis of 567 secreted factors (Supplementary Tables 5 and 6), comparing the conditioned media from the 6 rescuing to 12 non-rescuing stromal cells. The factor best correlated with PLX4720 resistance was hepatocyte growth factor (HGF), a well-characterized growth factor whose secretion by mesenchymal cells induces activation of the MET receptor tyrosine kinase (Figure 2b and Supplementary Fig. 5 and 6). While MET has been reported to be overexpressed12,13 and contribute to the progression of melanoma12, it has not been previously implicated in RAF-inhibitor resistance. A potential role of MET activation in the development of resistance to the EGFR inhibitor gefitinib in non-small cell lung cancer, however, has been recently reported10,14.
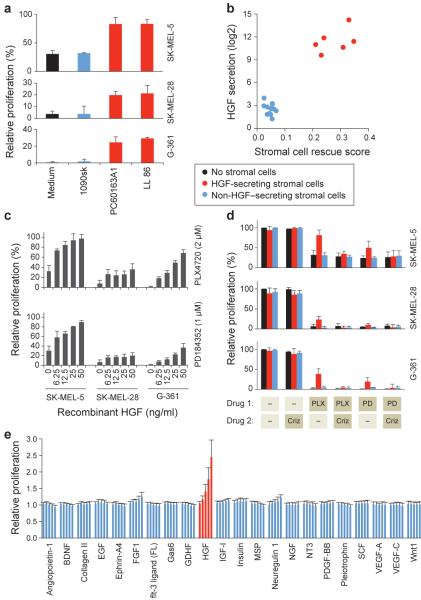
Figure 2. HGF rescues melanoma cancer cell lines from RAF and MEK inhibitors.
a, 3 melanoma cell lines were co-cultured with conditioned media from three fibroblast cell lines or with fresh media and treated with 2uM PLX4720. Proliferation was quantified after 7 days and compared to non-treated cells. Bars represent standard error between replicates (n = 3). b, the HGF secretion level of 18 stromal cell lines measured by a protein cytokine array (Supplementary Table 5) is plotted vs. the ability of each stromal cell line to rescue BRAF V600E melanoma cell lines from PLX4720 (Supplementary Fig. 3). c, Effect of HGF (6.25-50ng/ml) on proliferation of melanoma cell lines under PLX4720 or PD184352 treatment. Bars represent standard error between replicates (n = 3). d, Drug resistance manifests only in the presence of HGF-secreting stromal cells, and is reversed by MET inhibitor. Melanoma cell lines were co-cultured with nine stromal cell lines, representing HGF secreting and non-secreting stromal cells and treated with PLX4720 (2uM) or PD184352 (1um) with or without 0.2uM crizotinib. Proliferation was quantified after 7 days and normalized to non-treated cells. Results were averaged across 4 stromal cell lines that secrete HGF and 5 that do not. Non-averaged results are presented in Supplementary Fig. 11. Bars represent standard error between replicates (n = 3). e, 22 cytokines were added to 6 melanoma cell lines that were then treated with 2uM PLX4720 or 1uM PD184352 or DMSO control. Proliferation quantified after 7 days and normalized to No-Cytokine. Results shown are averaged for all cell lines and both drugs. Bars represent standard error between replicates (n = 3).
We next tested HGF expression by immunohistochemistry in 34 BRAF V600E melanoma patient-derived biopsies taken just prior to treatment with BRAF inhibitor (or a combination of BRAF and MEK inhibitors). HGF was detected in the tumor-associated stromal cells in 23/34 patients (68%) (Fig. 3a, 3b and Supplementary Table 7), and phospho-MET immunofluorescence studies similarly documented MET phosphorylation (activation) in patient samples (Supplementary Fig. 7).
Figure 3. HGF is present in the stromal cells of melanoma and correlates with poor response to therapy.
a, Pre-treatment melanoma section from patient # 32 was analyzed for HGF expression by immunohistochemistry (IHC). Black arrow: normal epidermis. Red arrow: tumor cells. Blue arrow: HGF-expressing stroma (brown staining). Low magnification image shown on the left (scale bar - 200μm) while high magnification image shown on the right (scale bar - 50μm). b, Melanoma sections from patient # 23 analyzed for HGF expression by IHC. On treatment biopsy was obtained 2 weeks after the initiation of treatment with the BRAF inhibitor vemurafenib (PLX4032) and one month after the pre-treatment biopsy was obtained. Third biopsy was obtained 12 months after the initiation of treatment while the patient was progressing under treatment. Low magnification images are shown on top (scale bar - 100μm) while high magnification images are shown on the bottom (scale bar - 50μm). c, Maximal response to treatment of BRAF V600E melanoma patients with or without stromal HGF as measured by IHC. Patients with stromal HGF had a significantly poorer response to treatment compared to those lacking expression (*P < 0.05 by two-sample t-test assuming equal variance). Median values for each group are depicted above the median line.
Our in vitro studies predict that the presence of stromal HGF should be associated with innate resistance. Indeed, patients with stromal HGF had a significantly poorer response to treatment compared to those lacking expression (P < 0.05; Fig 3c). Interestingly, only one of the 34 patients had a durable complete response (14 months and continuing), and this patient lacked HGF expression (Supplementary Table 7). On-treatment biopsies taken 2 weeks after treatment initiation were also available from 10 patients, and for 5 of those (50%), stromal HGF expression was found to be increased compared to pre-treatment (Fig. 3 and Supplementary Table 7). Whether this increase is attributable to recruitment of HGF-secreting fibroblasts to the tumor or up-regulation of HGF in existing fibroblasts remains to be determined. Of note, both normal skin and benign nevi exhibited stromal HGF expression (Supplementary Fig. 8). Our results thus support the clinical relevance of HGF-mediated resistance to BRAF inhibitors. Importantly, Settleman and colleagues similarly observed an association between plasma HGF levels and response to BRAF inhibitor treatment (Wilson et al, manuscript submitted15).
To establish HGF as the cause of drug resistance, and not simply a biomarker of it, we tested the ability of recombinant HGF to induce resistance, as well as the ability of HGF-neutralizing antibody or the MET-inhibitory small-molecule crizotinib to block fibroblast-induced PLX4720 resistance. These experiments indicated that HGF is both necessary and sufficient for conferring the resistance phenotype (Figure 2c, 2d and Supplementary Fig. 9,10 and 11). Consistent with this observation, the extent to which different BRAF-mutant melanoma cell lines (n=20) could be rescued by HGF was highly correlated with their level of MET expression (Supplementary Fig. 12).
While our stromal cell profiling studies pointed to the HGF/MET axis as the most relevant in mediating PLX4720 resistance, it is conceivable that other ligands of receptor tyrosine kinases (RTK) might similarly confer resistance. To test this possibility, we collected 22 well-characterized RTK ligands and tested their ability to rescue BRAF-mutant melanoma cells from either PLX4720 or the MEK inhibitor PD184352. Surprisingly, despite many RTKs being expressed and activated by their cognate ligands, HGF was the only ligand that conferred substantial resistance to RAF or MEK inhibition (Figure 2e, Supplementary Fig. 13, 14 and 15 and Supplementary Table 8).
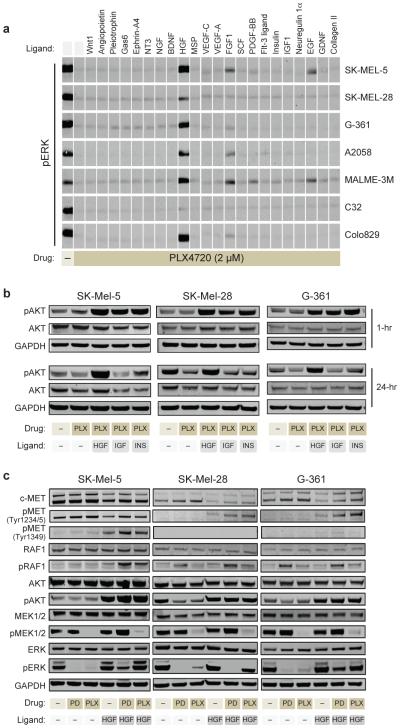
We next sought to clarify the precise mechanism by which HGF/MET is uniquely capable of inducing primary resistance to PLX4720. MET is known to activate both the MAP kinase (MEK/ERK) and the PI-3-kinase (PI3K/AKT) pathways (Supplementary Fig. 16), and both pathways have been suspected to be involved in acquired resistance to BRAF inhibitors16-18. We used Western blotting to assess ERK and AKT activation status in a panel of 7 BRAF-mutant melanoma cell lines treated with BRAF inhibitor together with various RTK ligands. HGF treatment led to sustained activation of both ERK and AKT, whereas such dual activation was not seen with any of the other RTK ligands in any of the melanoma lines (Figure 4a and Supplementary Fig. 17). We note that while EGF, FGF-1 and PDGF-BB reactivated ERK in most cell lines, phospho-ERK levels were modest compared to cells treated with HGF. Moreover, these ligands failed to activate AKT. Similarly, insulin or IGF-1 treatment led to a transient increase in phospho-AKT, but did not activate ERK (Figure 4b and Supplementary Fig. 18). HGF was thus unique in its ability to induce sustained activation of both ERK and AKT (Figure 4c and Supplementary Fig. 19 and 20). Importantly, we found that HGF-mediated activation of ERK was most profound under BRAF inhibition compared to MEK inhibition (Figure 4c and Supplementary Fig. 19). This may be best explained by the fact that in the presence of BRAF inhibitors, MET can reactivate MEK through RAF1 (CRAF), thus bypassing BRAF, which is not possible under conditions of direct MEK inhibition (Figure 4c, Supplementary Fig. 16).
Figure 4. Characterizing the molecular mechanism of HGF-induced primary resistance.
a, Activation of ERK by cytokines. Levels of phosphorylated ERK (T202/Y204) were assayed by immunoblotting 1 hour after treatment with media (-) or with 22 cytokines in the presence of PLX4720 or DMSO (DM) control. b, The activation of AKT by HGF, IGF-1 (IGF), and Insulin (INS). Levels of phosphorylated AKT (S473) were assayed 1 hour and 24 hours after treatment with HGF, IGF-1, or insulin in the presence of PLX4720 (2uM). c, Effect of HGF (25ng/ml) on melanoma cell lines treated with 2uM PLX4720 or 1uM PD184352. MAPK and PI3K/AKT pathways activation was assessed after 24 hours of treatment by immunoblot analysis of pRAF1, pMEK, pERK, pAKT and pMET.
Our model thus predicts that both the MAPK pathway and the PI3K/AKT pathway contribute to the primary resistance induced by HGF-secreting stromal cells. In agreement with this model, we have found that i) HGF-induced resistance is greater under BRAF inhibition compared to MEK inhibition (Figure 2d), ii) combination BRAF- and MEK-inhibitor treatment is not sufficient to eliminate HGF-induced resistance, as this combination does not silence AKT (Supplementary Fig. 21), and iii) combination treatment with MEK and AKT inhibitors suppresses the majority of HGF-induced drug resistance (Supplementary Fig. 21).
Our discovery of HGF-mediated innate resistance to BRAF inhibitors should be distinguished from recent reports proposing dysregulation of IGF, PDGF, COT, BRAF or MEK as BRAF-inhibitor resistance mechanisms18-22. In these reports, the emergence of late, acquired drug resistance was studied (e.g. following exposure to drug for many months), whereas we find that HGF-secreting stromal cells confer immediate, innate resistance to BRAF inhibitors. For example, the p61BRAF(V600E) splice variant that was recently shown to confer resistance to RAF inhibitors21 was never seen in tumors prior to RAF inhibitor treatment, implicating this splice variant as a mechanism for acquired rather than innate resistance. Whether HGF has a role in acquired resistance as well remains to be determined.
Activation of the EGF receptor was recently shown to drive the resistance of some BRAF V600E colorectal cancer cell lines to RAF inhibition23,24. In order to explore a possible role for MET activation in BRAF-mutant non-melanoma cancers, we tested 7 non-melanoma BRAF-mutant cell lines (5 colorectal and 2 glioblastoma), and found that all 7 had evidence of phospho-MET expression (Supplementary Fig. 22). Although stromal HGF expression is less common in colorectal cancer compared to melanoma (Supplementary Fig. 8a), MET overexpression and HGF autocrine secretion have been documented in colorectal cancer25-27. We indeed identified two HGF-secreting, BRAF-mutant non-melanoma cell lines (one colorectal (RKO) and one glioblastoma (KG-1-C); Supplementary Fig. 6), and in these cell lines, combined RAF and MET (but not EGFR) inhibition resulted in a clear synergistic effect (Supplementary Fig. 22 and 23). Synergy between BRAF and MET inhibitors was more variable among non-HGF-secreting BRAF-mutant cell lines (Supplementary Fig. 22). As predicted by our proposed mechanism of resistance, mono-therapy with BRAF or MEK inhibitors had no effect on pAKT and caused little inhibition of pERK in HGF-secreting cell lines. However, dual inhibition of BRAF and MET resulted in significant inhibition of both pERK and pAKT (Supplementary Fig. 24). The extent to which autocrine or microenvironment-mediated MET activation in non-melanoma BRAF-mutant tumors explains their failure to respond to BRAF inhibition deserves further investigation.
The findings reported here have potentially immediate clinical implications. Several small-molecule or antibody inhibitors of HGF/MET are in clinical development or have been FDA-approved for other indications. Given the tolerability of those agents and the similar tolerability of BRAF inhibitors, combination clinical trials in BRAF-mutant melanoma, colorectal cancer, and possibly other tumor types should be considered.
Lastly, we note that the stroma-derived, HGF-mediated RAF inhibitor resistance mechanism detailed here was but one of many such stroma-mediated drug resistance interactions uncovered in our initial screen (Figure 1a). Our findings point to the microenvironment as an important, yet under-studied source of cancer drug resistance. Moreover, the results suggest that such resistance mechanisms can be uncovered through the systematic dissection of tumor-microenvironment interactions. Future studies should therefore aim to identify such resistance mechanisms for all drugs currently in development, potentially leading to mechanism-based combination regimens such as the BRAF-MET combination proposed here.
METHODS SUMMARY
Stromal mediated chemoresistance co-culture screen
On day 0 stromal cells (1700 cells in 20ul/well) were plated in 384-clear bottom plates (Corning #3712), together with GFP-labeled cancer cells (1700 cells/20ul). Cells were treated on day 1 with 10 uL of 5X drug using the Cybi-Well Vario 384/25 (CyBio). On day 4, the media in all wells was replaced with fresh media and fresh drug was added to all wells containing melanoma cell lines (all other cancers were treated on day 1 only). GFP was read on Days 1, 4, and 7 by SpectraMax M5e microplate reader (Molecular Devices). Fluorescence microscope with high throughput screening (HTS) capabilities (Zeiss Axio observer Z1) was used to document bright field and GFP images on day 7. All screens were done in quadruplicate. See Supplementary information for full methods description.
Methods
1. Stromal mediated chemoresistance co-culture screen
On day 0 stromal cells (1700 cells in 20ul/well) were plated in 384-clear bottom plates (Corning #3712), together with GFP-labeled cancer cells (1700 cells/20ul). Cells were treated on day 1 with 10 uL of 5X drug using the Cybi-Well Vario 384/25 (CyBio). On day 4, the media in all wells was replaced with fresh media and fresh drug was added to all wells containing melanoma cell lines (all other cancers were treated on day 1 only). GFP was read on Days 1, 4, and 7 by SpectraMax M5e microplate reader (Molecular Devices). Fluorescence microscope with high throughput screening (HTS) capabilities (Zeiss Axio observer Z1) was used to document bright field and GFP images on day 7. All screens were done in quadruplicate.
2. Cell lines and reagents
The sources of all used cell lines are listed in Supplementary table 1. All cells were in maintained in DMEM (Invitrogen – 10569-010) with 10% FBS and 1x Pen Strep Glutamine (Invitrogen - 15140-122). Cancer cell lines were lentivirally transduced using pLex_TRC206 plasmid. The sources of all used drugs are listed in Supplementary table 2. Antibodies to MET (#3148), pMET (#3077, 3133), pRAF1 (#9427), pERK (#4370), AKT (#2920), pAKT (#4060), MEK1/2 (#4694), pMEK1/2 (#9154), and GAPDH (#2118) were purchased from Cell Signaling. Antibody to Raf1 (#ab656) was purchased from Abcam. Antibody to ERK (#sc-135900) was purchased from Santa Cruz. Anti-rabbit (#926-32211) and anti-mouse (#926-32220) secondary antibodies purchased from Licor. The following cytokines were purchased from R&D systems: Angiopoietin-1 (923-AN-025), BDNF (248-BD-005), EGF (236-EG-200), Ephrin-A4 (369-EA-200), FGF1 (231-BC-025), flt-3 ligand (308-FK-025), Gas6 (885-GS-050), GDNF (212-GD-010), IGF-1 (291-G1-050), MSP (352-MS-010), neuregulin 1 alpha (5898-NR-050), NGF (256-GF-100), NT3 (267-N3-005), PDGF-BB (220-BB-010), Pleiotrophin (252-PL-050), VEGF-A (293-VE-010), VEGF-C (2179-VC-025). HGF (228-10702-2) was purchased from Raybiotech. Insulin (I9278) was purchased from Sigma. Stem Cell Factor (569600-10UG) was purchased from EMD. Type II collagen (ab7534) and Wnt1 (ab84080) were purchased from Abcam. Skin tissue microarrays (TMA) of Normal skin, Nevi and Melanomas were purchased from US Biomax (#ME1004a and #ME803a). Colorectal cancer TMAs were prepared as previously described31.
3. Clinical samples
Patients with metastatic melanoma containing BRAF V600E mutation (confirmed by genotyping) were enrolled on clinical trials for treatment with a BRAF inhibitor or combined BRAF + MEK inhibitors (Supplementary Table 7) and were consented for tissue acquisition per IRB-approved protocol. Tumor biopsies were performed pre-treatment (day 0), at 10-14 days on treatment, and at time of progression. Formalin-fixed tissue was analyzed to confirm that viable tumor was present via hematoxylin and eosin (H&E) staining. Tumor responses were determined by the investigators according to the Response Evaluation Criteria in Solid Tumors (RECIST)
4. Analysis of Co-culture screen data
The GFP readings from each well on day 7 were background subtracted by the readings in the same wells on day 1, and quadruplicates were averaged. The drug effect for each cancer cell line in the presence or absence of stromal cells was calculated by normalizing the number of cells (GFP) after 7 days of treatment to the number of cells (GFP) in DMSO control wells. The drug effect in the presence of stromal cells was further normalized to the effect that each stromal cell type has on cancer cell proliferation when no drug is present (See “Without Stroma” and “With Stroma” columns in Supplementary table 3). “Rescue score” was calculated by subtracting the “Without stroma” drug effect from the “With stroma” drug effect.
5. Analysis of Co-culture screen data
The GFP readings from each well on day 1 were subtracted from the readings in the same wells on day 7. Quadruplicates were averaged. The effect of each drug on the proliferation of each cancer cell line was calculated by normalizing the GFP reading of the cancer cell line when treated with a drug to the GFP reading of the same cancer cell line when treated with DMSO control (“No stroma” column in Supplementary table 3). The proliferation of the cancer cell under a drug when co-cultured with a stroma cell line was normalized first to the proliferation of the same cancer cell line when grown without drug or stromal cell line and then normalized again to the effect that the specific stromal cell line has on the proliferation of the cancer cell line (“With Stroma” column in Supplementary table 3). “Rescue score” was calculated by subtracting the “No stroma” drug effect from the “With stroma” drug effect.
6. Assigning “rescue by stroma” score to all screened drugs (Supplementary table 2)
Only cases in which a drug slowed the proliferation of a cancer cell line to <30% were analyzed (Supplementary table 4). Rescue was counted as positive if the rescue score was > 0.3. Drugs that were rescued by stromal cells in at least 3 different cancer cell lines representing >40% of all screened cancer cell lines for this drug got the maximal score: “++”. Drugs that were rescued by stromal cells in 3 cancer cell lines that represent only 20-40% of screened cancer cell lines or that were rescued in only 1 or 2 cell lines that represent more than 40% of screened cancer cell lines were scored: “+”.
7. Antibody arrays
Soluble proteins in the media of the stromal cell-lines were measured using RayBio Human cytokine array G4000 (#AAH-CYT-G4000-8) and RayBio Biotin Label-based Human Antibody Array (#AAH-BLG-1-4), according to recommended protocols. These arrays can detect 274 and 507 proteins, respectively. Stromal cells were plated 3 days before the experiment in DMEM containing 10% FBS and were 75-90% confluent when media was collected and filtered. Media with 10% FBS was also hybridized to the arrays and used later for normalization. 10 Technical and Biological replicates were done – both showing a very high correlation (Correlation coefficient > 0.9) (Data not shown). Hybridization was done overnight in 4°C. All slides were scanned using Axon’s GenePix 4000B scanner and analyzed using GenePix Pro 6.0. The F532 Median - B532 score was used and averaged across triplicates on each array. Results were then normalized using internal controls and values of cytokines in clear media + 10% FBS were subtracted. All results are available in Supplementary tables 5 and 6.
8. Stromal averaged Melanoma rescue scores
The averaged melanoma rescue effect of each stromal cell line was calculated by averaging the rescue scores of this cell line (Supp. Table 3) across all melanoma cell lines and all PLX4720 concentrations. Only instances in which the treatment caused a drop of proliferation below 0.3 when no stromal cells are present were included in this calculation.
9. The effect of pre-conditioned media (PCM)
PCM was prepared by filtering media from 80-90% confluent 15cm plates that were plated 3 days earlier and then diluting it 1:1 with fresh media. Experiments were performed according to the previously described co-culture experiment protocol except for the following changes: 1. On day 0, 384-well plates were seeded with 20ul/well of PCM instead of 20ul of stromal cells. 2. On day 1, the media from all wells was changed to fresh PCM. 3. On day 4, media was changed to fresh PCM instead of fresh media before re-treating the cells.
10. Hierarchical clustering
Unsupervised Hierarchical clustering of stromal cell lines according to their ability to rescue melanoma cancer cell lines from 2uM of PLX4720 (Supp. Table 3) was done using GENE-E (http://www.broadinstitute.org/cancer/software/GENE-E/). Euclidean distance metric was used.
11. HGF ELISA
Cells were plated 3 days before the experiment in DMEM containing 10% FBS and were 75-90% confluent when media was collected and filtered. HGF ELISA was performed using RayBio Human HGF ELISA kit (#ELH-HGF-001) according to the kit’s instructions. The media was diluted 1:1 with diluents B before it was added to the assay microplate. For the standard HGF curve we used the same HGF that was used for all other experiments (#228-10702-2) and not the HGF that comes with the kit. Absorbance at 450nm was read using Spectramax M5e (Molecular Devices).
12. Neutralizing HGF by Anti-HGF antibodies
Co-culture experiments were performed as described above except for the addition of Neutralizing anti-HGF antibodies (R&D Systems #MAB294) on days 0 and on day 4 after the media was changed.
13. Western blot analysis and quantification
Cells were plated a day before treatment in a 6-well plate at 5×105 cells/well, and were treated the next day. At the designated time points, cells were lysed with lysis buffer containing 50mM Tris (pH 7.4), 150mM NaCl, 2mM EDTA, 1% NP-40, 1mg/mL NaF, a one pellet per 10ml each of Roche PhosStop phosphatase inhibitor (04906837001) and Roche Complete Mini protease inhibitor (Roche). Protein concentrations were determined by BioRad DC Protein Assay Kit II. Samples were mixed with 4x protein sample loading buffer (Li-Cor #928-40004) and NuPage sample reducing agent (Invitrogen #NP0009), and run on a 4-12% Bis-Tris gel (NuPage #WG1402BOX) at 120V. Membranes were transferred using Program 4 on the iBlot Gel Transfer Device (Invitrogen #IB1001). Western blotting was performed with standard methods, with immunoblotting performed according to antibody manufacturer specifications. Near-infrared (NIR) fluorescence was detected with the Odyssey Infrared Imaging System (Licor), and signal intensity was quantified with Odyssey Application Software (Licor). All quantifications were first normalized to background intensity, and then to GAPDH loading control.
14. High throughput westerns
High throughput western blot experiments (Fig 4a and Supp. Figure 17) were performed as described above, except for the following changes: 1) Cells were seeded in a 96-well plate at 5×104 cells/well, 2) Samples were mixed with E-Page 4x loading buffer (Invitrogen #EPBUF-01) and run on 6% E-PAGE 96-well gels (Invitrogen #EP09606). For the transfer Program 3 of the iBlot Gel Transfer Device was used.
15. Rescue of melanoma cell lines by cytokines
One day prior to treatment, cancer cells were seeded in black, 384-well plates (Corning #3712) at a concentration of 2500 cells/well. On day 1, all 22 ligands were added at 5 different concentrations to 6 melanoma cell lines treated with PLX4720, PD184352, or DMSO control. On day 4, media was changed to fresh and cells were retreated with drugs and cytokines. GFP was read of the plates on Days 1, 4, and 7 by SpectraMax M5e microplate reader (Molecular Devices).
16. Tyrosine kinase phosphorylation profiling
Luminex immunosandwich assays were performed as previously described32 with the following modifications: Antibodies were conjugated to Luminex MagPlex microspheres (Luminex). Assays were carried out in 384-well ThermoMatrix square bottom plates (Thermo) in conjunction with a 96-well (CyBio) and a 384-well (BioMek) liquid handler. The data was acquired with a FlexMap 3D instrument (Luminex) according to the manufacturer’s instructions. The raw data was normalized by subtracting sample and antibody backgrounds.
17. Immunohistochemistry
Deparaffinized tissue sections were treated with Antigen Retrieval Citra Solution (Biogenex Laboratories, #HK086-9K) in microwave for 15 min. Tissue sections were then incubated with Peroxidase Blocking Reagent (15 min; DAKO #S2001) and Protein Block (15 min; DAKO #X0909). Primary antibody against HGF (R&D #AB-294-NA; 0.75 μg/mL), MET (Invitrogen #187366; 4 μg/mL), pERK (Cell Signaling #4376; 1:200 dilution) or pAKT (Cell Signaling #4060; 1:50 dilution) was applied, and slides were incubated for 16 h at 4 degrees. For HGF, sections were then incubated with rabbit anti-goat antibody (Vector #BA-5000) for 30 min. Signals were visualized using EnVision™+/HRP, Rabbit (for HGF; DAKO, #K4003) or Mouse (for MET; DAKO, #K4001) or SignalStain® Boost IHC Detection Reagent (for pERK and pAKT; Cell Signaling #8114), diaminobenzidine (DAKO #K3468) and hematoxylin counterstain. To detect MET expression in melanoma, VECTOR VIP Peroxidase Substrate Kit (Vector #SK-4600) was used instead of diaminobenzidine. All Immunostained slides were scored by a pathologist (T.M.) blinded to the clinical outcome data.
18. Immunofluorescence
Fresh frozen tissue sections were stained using Cell Signaling Technology’s general protocol. Sections were blocked using PBS with 5% normal goat serum (Cell Signaling # 5425S) and 0.3% Triton-X for 1 hour. Primary antibody against p-Met (Y1234/1235) (Cell signaling #3077S;1:100 dilution) diluted in PBS with 1% BSA, and 0.3% Triton-X was applied, and slides were incubated for 16 h at 4 degrees. Sections were then washed with PBS and incubated with goat anti-rabbit IgG conjugated to Dylight 488 (ThermoFisher Scientific, #35552; 1:500 dilution) for 1 hour. Slides were mounted with Prolong anti-fade reagent with DAPI (Life Technologies, CA, #P36935). Experiments were done in parallel with SignalSlide Phospho-Met (Tyr1234/1235) IHC control slide (Cell Signaling, #8118) for proper staining. Images were captured using a Nikon Eclipse 80i fluorescence microscope.
19. Calculating excess over Bliss
The Bliss independence model predicts the combined response C for two single compounds with response A and B according to the relationship C = A + B - A * B, where A is the fractional inhibition of compound A at the particular concentration and B is the fractional inhibition of compound B at the particular concentration. According to this model, the excess above the predicted Bliss independence represents the synergistic effect of the combination treatment33.
Supplementary Material
Acknowledgements
We thank all members of the Golub lab for helpful discussion. We thank Cory M. Johannessen, Ithai Rabinowitch, Noam Shoresh and Alon Goren for critical reading of the manuscript, and Leslie Gaffney for expert assistance with graphics. This work was supported by the Howard Hughes Medical Institute, NCI grants P50CA093683 and U54CA112962 to TRG, and a Melanoma Research Alliance Team Science Award.
Footnotes
Supplementary information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions. R.S. and T.R.G. conceived and designed the experiments. R.S. performed the primary cancer-stroma-drugs screen with help from K.S., M.B.R. and A.D. R.S. performed the protein arrays. R.S., K.S., M.B.R., A.D. and M.M.M. Performed secondary screens, westerns and ELISA. J.D. performed tyrosine kinase phosphorylation profiling. Clincal samples and clinical data were collected by J.A.W., K.T.F., D.T.F., P.B.C., D.B.S., A.R. and R.S.L. Immunohistochemistry was performed and analyzed by S.O., T.M. and Z.R.Q. Immunofluorescence was performed by J.A.W. and Z.A.C. R.S. and T.R.G. Produced the text and figures, including supplementary information. K.S. helped produce some of the text and figures. All authors discussed results and contributed to the final manuscript.
Author Information. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Bollag G, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. doi:10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chapman PB, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. doi:10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flaherty KT, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. doi:10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sosman JA, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. doi:10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. doi:10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. doi:10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 7.McMillin DW, et al. Tumor cell-specific bioluminescence platform to identify stroma-induced changes to anticancer drug activity. Nat Med. 2010;16:483–489. doi: 10.1038/nm.2112. doi:10.1038/nm.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shekhar MP, Santner S, Carolin KA, Tait L. Direct involvement of breast tumor fibroblasts in the modulation of tamoxifen sensitivity. Am J Pathol. 2007;170:1546–1560. doi: 10.2353/ajpath.2007.061004. doi:10.2353/ajpath.2007.061004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teicher BA, et al. Tumor resistance to alkylating agents conferred by mechanisms operative only in vivo. Science. 1990;247:1457–1461. doi: 10.1126/science.247.4949.1457. [DOI] [PubMed] [Google Scholar]
- 10.Wang W, et al. Crosstalk to stromal fibroblasts induces resistance of lung cancer to epidermal growth factor receptor tyrosine kinase inhibitors. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15:6630–6638. doi: 10.1158/1078-0432.CCR-09-1001. doi:10.1158/1078-0432.CCR-09-1001. [DOI] [PubMed] [Google Scholar]
- 11.Williams RT, Roussel MF, Sherr CJ. Arf gene loss enhances oncogenicity and limits imatinib response in mouse models of Bcr-Abl-induced acute lymphoblastic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6688–6693. doi: 10.1073/pnas.0602030103. doi:10.1073/pnas.0602030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puri N, et al. c-Met is a potentially new therapeutic target for treatment of human melanoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13:2246–2253. doi: 10.1158/1078-0432.CCR-06-0776. doi:10.1158/1078-0432.CCR-06-0776. [DOI] [PubMed] [Google Scholar]
- 13.Lee YJ, et al. Expression of the c-Met Proteins in Malignant Skin Cancers. Ann Dermatol. 2011;23:33–38. doi: 10.5021/ad.2011.23.1.33. doi:10.5021/ad.2011.23.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. doi:10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 15.Wilson T, et al. Widespread potential for growth factor-driven resistance to anti-cancer kinase inhibitors. Nature. 2012 doi: 10.1038/nature11249. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang CC, et al. MEK-independent survival of B-RAFV600E melanoma cells selected for resistance to apoptosis induced by the RAF inhibitor PLX4720. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17:721–730. doi: 10.1158/1078-0432.CCR-10-2225. doi:10.1158/1078-0432.CCR-10-2225. [DOI] [PubMed] [Google Scholar]
- 17.Shao Y, Aplin AE. Akt3-mediated resistance to apoptosis in B-RAF-targeted melanoma cells. Cancer research. 2010;70:6670–6681. doi: 10.1158/0008-5472.CAN-09-4471. doi:10.1158/0008-5472.CAN-09-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villanueva J, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1 R/PI3K. Cancer cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. doi:10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johannessen CM, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. doi:10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nazarian R, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. doi:10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poulikakos PI, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. doi:10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, et al. Identification of the MEK1(F129L) activating mutation as a potential mechanism of acquired resistance to MEK inhibition in human cancers carrying the B-RafV600E mutation. Cancer research. 2011;71:5535–5545. doi: 10.1158/0008-5472.CAN-10-4351. doi:10.1158/0008-5472.CAN-10-4351. [DOI] [PubMed] [Google Scholar]
- 23.Corcoran RB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2012;2:227–235. doi: 10.1158/2159-8290.CD-11-0341. doi:10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prahallad A, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012 doi: 10.1038/nature10868. doi:10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 25.Kammula US, et al. Molecular co-expression of the c-Met oncogene and hepatocyte growth factor in primary colon cancer predicts tumor stage and clinical outcome. Cancer Lett. 2007;248:219–228. doi: 10.1016/j.canlet.2006.07.007. doi:10.1016/j.canlet.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 26.Di Renzo MF, et al. Overexpression and amplification of the met/HGF receptor gene during the progression of colorectal cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 1995;1:147–154. [PubMed] [Google Scholar]
- 27.Liu C, Park M, Tsao MS. Overexpression of c-met proto-oncogene but not epidermal growth factor receptor or c-erbB-2 in primary human colorectal carcinomas. Oncogene. 1992;7:181–185. [PubMed] [Google Scholar]
- 28.Chan AT, Ogino S, Fuchs CS. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. N Engl J Med. 2007;356:2131–2142. doi: 10.1056/NEJMoa067208. doi:10.1056/NEJMoa067208. [DOI] [PubMed] [Google Scholar]
- 29.Du J, et al. Bead-based profiling of tyrosine kinase phosphorylation identifies SRC as a potential target for glioblastoma therapy. Nature biotechnology. 2009;27:77–83. doi: 10.1038/nbt.1513. doi:10.1038/nbt.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keith CT, Borisy AA, Stockwell BR. Multicomponent therapeutics for networked systems. Nat Rev Drug Discov. 2005;4:71–78. doi: 10.1038/nrd1609. doi:10.1038/nrd1609. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.