Abstract
Objective
To highlight the contribution of the gut microbiota to the modulation of host metabolism by dietary inulin-type fructans (ITF prebiotics) in obese women.
Methods
A double blind, placebo controlled, intervention study was performed with 30 obese women treated with ITF prebiotics (inulin/oligofructose 50/50 mix; n=15) or placebo (maltodextrin; n=15) for 3 months (16 g/day). Blood, faeces and urine sampling, oral glucose tolerance test, homeostasis model assessment and impedancemetry were performed before and after treatment. The gut microbial composition in faeces was analysed by phylogenetic microarray and qPCR analysis of 16S rDNA. Plasma and urine metabolic profiles were analysed by 1H-NMR spectroscopy.
Results
Treatment with ITF prebiotics, but not the placebo, led to an increase in Bifidobacterium and Faecalibacterium prausnitzii; both bacteria negatively correlated with serum lipopolysaccharide levels. ITF prebiotics also decreased Bacteroides intestinalis, Bacteroides vulgatus and Propionibacterium, an effect associated with a slight decrease in fat mass and with plasma lactate and phosphatidylcholine levels. No clear treatment clustering could be detected for gut microbial analysis or plasma and urine metabolomic profile analyses. However, ITF prebiotics led to subtle changes in the gut microbiota that may importantly impact on several key metabolites implicated in obesity and/or diabetes.
Conclusions
ITF prebiotics selectively changed the gut microbiota composition in obese women, leading to modest changes in host metabolism, as suggested by the correlation between some bacterial species and metabolic endotoxaemia or metabolomic signatures.
Keywords: Obesity, Prebiotic, Nutrition, Intestinal Bacteria
Significance of this study.
What is already known on this subject?
Dysbiosis is associated with obesity and related metabolic disorders.
Dietary inulin-type fructans (ITF) selectively change the gut microbiota composition and improve host physiological functions in obese rodents.
The beneficial effect of ITF (prebiotic effect) in non-obese humans is often linked to a promotion of bifidobacteria in the colon.
What are the new findings?
ITF prebiotics induce a huge bifidogenic effect but also modify numerous other bacteria in obese women.
The changes in gut microbiota induced by ITF prebiotics are correlated with bacterial-related metabolites (phosphatidylcholine, lactate, hippurate) and with serum lipopolysaccharide levels, despite a lack of significant effect on body weight.
As inferred from the Human Intestinal Tract Chip (HITChip) analysis (phylogenetic microarray based on 16S rRNA gene sequences for the study of the human gastrointestinal microbiota) and the metabolomic signatures, the prebiotic approach selectively changes microbiota-related host functions.
How might it impact on clinical practice in the foreseeable future?
ITF prebiotics might be used to support the dietary advice to control obesity and related metabolic disorders including diabetes, and cardiovascular or liver disease. Our results indicate bacterial types or functions that could constitute novel therapeutic targets in this major public health issue.
Introduction
Obesity represents a major public health problem, as it leads to metabolic disorders involved in the development of type 2 diabetes, fatty liver disease or cardiovascular disease.1 The gut microbiota is now considered a key ‘endogenous’ organ contributing to the modulation of host physiology and constitutes a plausible target in the control of obesity and related metabolic disorders.2 3 Indeed, several studies in rodents (ie, germ-free vs conventionally raised mice) suggest that the gut microbiota promotes energy storage by changing host gene expression.4–6 Moreover, we have proposed that metabolic endotoxaemia (ie, increased blood lipopolysaccharide (LPS) levels) triggers low-grade inflammation occurring with overfeeding, obesity and diabetes,7 and thus contributes to these metabolic disorders.8–11 In humans, obesity is characterised by dysbiosis. Initial studies have shown that changes in the proportion and/or level of predominant bacterial phyla (a decrease in Bacteroidetes and/or an increase in Firmicutes) occur in obese individuals and may be reversed by dieting and weight loss,4 12–14 but these changes remain a matter of debate.15 16 Specific changes also occur at the genus level, such as lower bifidobacteria in obese versus lean and diabetic versus non-diabetic individuals.15 17
The concept of prebiotic is based on the possibility of favourably modulating the gut microbiota composition to maintain and/or promote health, using nutrients such as non-digestible, fermentable carbohydrates.18 19 Among these, inulin-type fructans (ITF) have been proposed as modulators of microbial ecology and host physiology in animals and humans.3 20 In diet-induced or genetic models of obesity in rodents, ITF were shown to decrease body weight gain and fat mass accumulation, improve glucose tolerance and insulin resistance, decrease inflammation and improve gut barrier function.9 21–23 Moreover, in these animal models, ITF modulated gut microbiota composition leading mainly to a bifidogenic effect.23 24 Recently, an extensive analysis of the gut microbiota composition after feeding obese mice short-chain ITF revealed that prebiotic treatment decreased Firmicutes and increased Bacteroidetes, and changed more than 100 taxa of bacteria. This was associated with beneficial effects on host metabolism, such as an improvement in glucose and lipid metabolism and a reduction of plasma LPS.25
Some of these effects have also been observed in humans. We, and others, have demonstrated that in healthy humans, short-chain ITF promoted satiety,26 27 increased breath-hydrogen excretion (a marker of gut microbiota fermentation) and modulated gut peptides regulating food intake.27 28 In overweight and obese adults, short-chain ITF supplementation promoted weight loss, improved glucose regulation and modulated the production of peptides implicated in energy intake.29 The aim of our exploratory study was to evaluate the impact of ITF prebiotics on the gut microbial ecosystem in obese women to highlight the contribution of gut microbial changes in the modulation of host metabolism on nutritional intervention. A metabolomic analysis in urine and plasma was also performed in parallel to evaluate the potential interest of microbial-related metabolites as novel biomarkers of prebiotic intervention.
Methods
Subjects
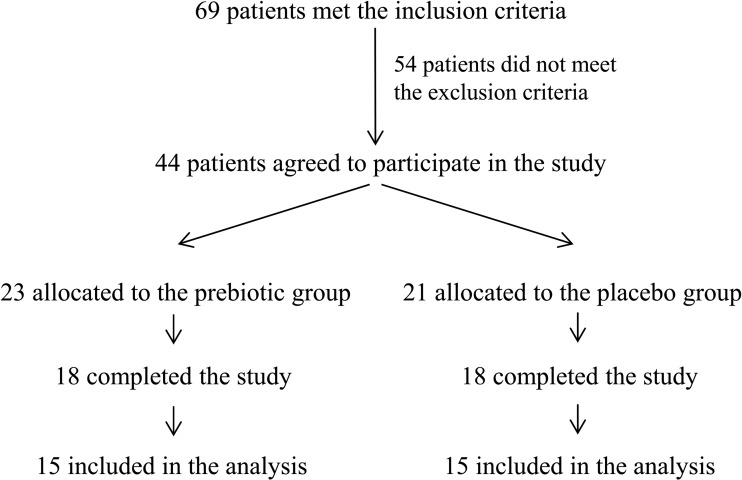
Forty-four obese women (body mass index (BMI) >30 kg/m2) between the ages of 18 and 65 years were voluntarily recruited from the Cliniques Universitaires Saint Luc, Brussels, Belgium between March 2008 and May 2011. Eight patients failed to complete the study for the following reasons: pregnancy (one patient), loss of phone contact during follow-up (one patient), gastro-oesophageal reflux (two patients), personal reasons (two patients) and absence of weight loss (two patients). Of the 36 patients who completed the study, three patients per group were excluded from the analysis because of antibiotic treatment during the study and inadequate or missing faecal sampling. Finally, 15 patients per group were included in the analysis (figure 1). The subjects were assigned to the ITF or placebo group following a systematic allocation method. The exclusion criteria were: presence of gastrointestinal disorders or a history of chronic physical/mental disease, use of antidiabetic drugs or weight-loss treatments, recent consumption (<6 weeks) of antibiotics or treatments that influence gut microbiota composition, chronic consumption of antacids or anti-inflammatory drugs, pregnancy, unusual diets (vegetarians, vegans and high consumption of dietary supplements), consumption of pre/probiotics or fibre supplements, excessive alcohol consumption (>30 glasses of alcoholic beverages/week), intensive physical training, a history of bariatric surgery, and gastro-oesophageal reflux treated with high doses of proton pump inhibitors. The Commission d'Ethique Biomédicale Hospitalo-facultaire from the Université catholique de Louvain (Brussels, Belgium) provided ethical approval for this study and written informed consent was obtained from each participant. The trial was registered at clinicaltrials.gov as NCT00616057.
Figure 1.
Trial profile. Of the 44 enrolled patients, eight patients failed to complete the study for the following reasons: pregnancy (one patient), loss of contact during follow-up (one patient), gastro-oesophageal reflux (two patients), personal reasons (two patients) and absence of weight loss (two patients). Of the 36 patients who completed the study, three patients per group were excluded from the analysis because of antibiotic treatment during the study and inadequate or missing faecal sampling.
Dietary intervention
The subjects were assigned to receive either a daily supplement of 16 g (8 g twice a day) of ITF (Synergy 1, namely, inulin/oligofructose 50/50 mix) or placebo (maltodextrin) for 3 months (both products were kindly provided by Orafti, Oreye, Belgium). Maltodextrin has a taste and appearance similar to Synergy 1 and has been used as a placebo for ITF in previous studies.26–29 The patients were asked to take a half dose during the first week of treatment to promote adaptation to the fibre and reduce gastrointestinal symptoms. Both the placebo and ITF were provided in identical opaque packages (named A or B and distributed as follows: patient 1/A, patient 2/B, patient 3/A, patient 4/B,…). The powder was added in warm drinks such as coffee, tea or hot chocolate or in dairy products. The subjects and the researchers were blinded to the treatment.
Trial protocol
After verifying that the patients met the inclusion criteria and obtaining consent, the patients were submitted to an anamnesis and received dietary advice for weight loss. At the beginning (T0) and the end of the treatment (T3 months), blood, faeces and urine were sampled. Blood was collected in EDTA tubes and a portion was sent directly to the hospital laboratory to measure the following classical biological parameters: glycaemia, insulinaemia, HbA1c, aspartate transaminase, alanine transaminase, alkaline phosphatase, C-reactive protein (CRP), total cholesterol, high-density and low-density lipoproteins (HDL and LDL, respectively) cholesterol and triglycerides. The remainder of the blood was centrifuged at 4200 g for 10 min at 4°C and the plasma was frozen at −80°C. Adiponectinaemia was determined using the RayBio ELISA kit (RayBiotech, Norcross, Georgia, USA). Faeces and urine samples were frozen at −20°C. Patients were also submitted to an oral glucose tolerance test (OGTT) (75 g glucose in drinking solution and measurement of glycaemia and insulinaemia 2 h after ingestion) and to a homeostasis model assessment (HOMA) test. HOMA estimates insulin sensitivity from the fasting glucose and insulin concentrations.30 Finally, weight and body composition were determined by impedancemetry (Tanita BC-418MA, Tanita UK, West Drayton, UK). The BMI was calculated and waist and hip circumferences were measured. A telephone follow-up was performed three times during the study (after 2 weeks, 1 month and 2 months of treatment) to verify compliance and enquire about possible side effects.
The primary endpoint of this study was to assess the prebiotic effect of ITF on gut microbiota composition in obese women. We focused on the bifidogenic effect because several studies with ITF in healthy humans had highlighted a significant increase in bifidobacteria following the prebiotic treatment.24 31 Patient enrolment was stopped when the primary endpoint, namely a statistically significant increase in faecal bifidobacteria quantified by qPCR in the ITF group, was effective (ie, 15 patients/group). The effect of ITF on clinical outcome and metabolic profiling was then evaluated as a secondary aim.
Gut microbiota analysis
Faecal samples were collected and stored at −20°C until further processing. The DNA was isolated as previously described,32 using the repeated bead beating procedure with a modified protocol for the QiAamp Stool DNA Mini Kit (Qiagen, Hilden, Germany). Faecal microbiota composition was assessed using qPCR (see online supplemental methods) and the Human Intestinal Tract Chip (HITChip), a phylogenetic microarray.33 The HITChip contains over 4800 probes based on 16S rRNA gene sequences of over 1100 intestinal bacterial phylotypes and identifies both variation and relative quantity of the human intestinal tract communities.34 Hybridisations were performed with samples labelled in duplicate with Cy3 and Cy5 dyes. Slides data were extracted from the microarray images using Agilent Feature Extraction software, V.10.7.3.1 (http://www.agilent.com). Array normalisation was performed as previously described,34 35 with a set of R-based scripts (http://r-project.org) in combination with a designed relational database which runs under the MySQL database management system (http://www.mysql.com). This procedure was applied on both dyes for each sample, and duplicates with a Pearson correlation over 0.98 were considered for further analysis. Ward's minimum variance method was used for the construction of hierarchical clusters of the total microbiota probe profiles, while the distance matrix between the samples was based on Pearson correlations.
Metabolic profiling by 1H NMR spectroscopy
400 µl of the urine samples was mixed with 200 μl of phosphate buffer (0.2M, pH 7.4) made in D2O containing 0.01% deuterated 3-(trimethysilyl)-1-propanesulfonic acid and 3 mM sodium azide. 200 µl of the plasma samples was mixed with 400 μl of D2O. Samples were centrifuged 10 min at 4°C (8000 g) and transferred to 5 mm tubes for nuclear magnetic resonance (NMR) analysis.
All 1H NMR spectra were acquired on a Bruker Avance 700 MHz spectrometer (Bruker Analytische, Rheinstetten, Germany) operating at 700.19 MHz and equipped with a 5 mm CryoProbe from the same manufacturer. See online supplemental methods for details.
Serum LPS measurement
LPS concentrations were measured by using Endosafe-MCS (Charles River Laboratories, Lyon, France) based on the Limulus Amoebocyte Lysate kinetic chromogenic methodology, as previously described.36 Samples were diluted from 1/30 to 1/100.
Statistical analyses
Raw data are expressed as mean±SD. Treatment effect was assessed based on differential values obtained by subtracting the value at T0 from the value at T3 months for each patient. Differential values are expressed as mean±95% CI. As most of the parameters had an abnormal distribution (assessed using a Shapiro–Wilk test), the Mann–Whitney test was used to compare differential values or both groups at T0. Correlations were analysed by using Spearman's correlation in GraphPad Prism (V.5.00 for Windows, GraphPad Software, San Diego, California, USA). The level of significance was set at p<0.05.
For the HITChip analysis, the Wilcoxon signed-rank test corrected for false discovery rate,37 was applied to evaluate the significant differences of individual genus-level groups in the microbiota of both groups after 3 months. For comprehensive multivariate statistical analyses, Canoco software for Windows 4.5 was used.38 Redundancy analysis was used to assess correlations between the microbial groups detected by the HITChip analysis in the function of sample characteristics. This redundancy analysis is a multivariate analysis that shows the distribution of the samples in a three-dimensional space based on species variables and environmental variables. The log transformed hybridisation signals of 131 genus-level phylogenetic groups targeted by the HITChip were used as species variables. As environmental variables, we included age, BMI, gender, antibiotic use, sample collection time and sample source (placebo or prebiotic group at T0 and T3 months). The Monte Carlo Permutation Procedure as implemented in the Canoco software package was used to assess statistical significance of the variation in large datasets.
Statistical analyses for metabolic profiling are described in the online supplemental methods.
Results
As described in the Methods section, 15 patients per group were included in the analysis (except for the HITChip analysis, in which we excluded one outlier sample in the treated group). Minor side effects were reported at the beginning of the treatment: six patients in the placebo group and 14 patients in the treated group had slight bloating, flatulence and/or abdominal pain. Those symptoms disappeared within a few days in most patients.
The baseline characteristics of the patients are reported in table 1. There were no differences between the two groups at the beginning of the study.
Table 1.
Descriptive characteristics at the beginning of treatment for all subjects included in the data analysis; n=15 for each group
| Placebo (n=15) | Prebiotic (n=15) | |
|---|---|---|
| Age (years) | 48±8 | 47±9 |
| Body weight (kg) | 97.5±15.8 | 99.1±16.3 |
| Body mass index (kg/m2) | 35.6±4.3 | 36.1±4.1 |
| Fat mass (%) | 46.1±3.9 | 45.4±3.5 |
| Waist circumference (cm) | 114.3±11.8 | 115.6±11.9 |
| HbA1c (%) | 5.8±0.5 | 5.6±0.5 |
| Fasting glycaemia (mg/dl) | 99±15 | 97±18 |
Results are given as mean±SD.
Analysis of the gut microbiota composition
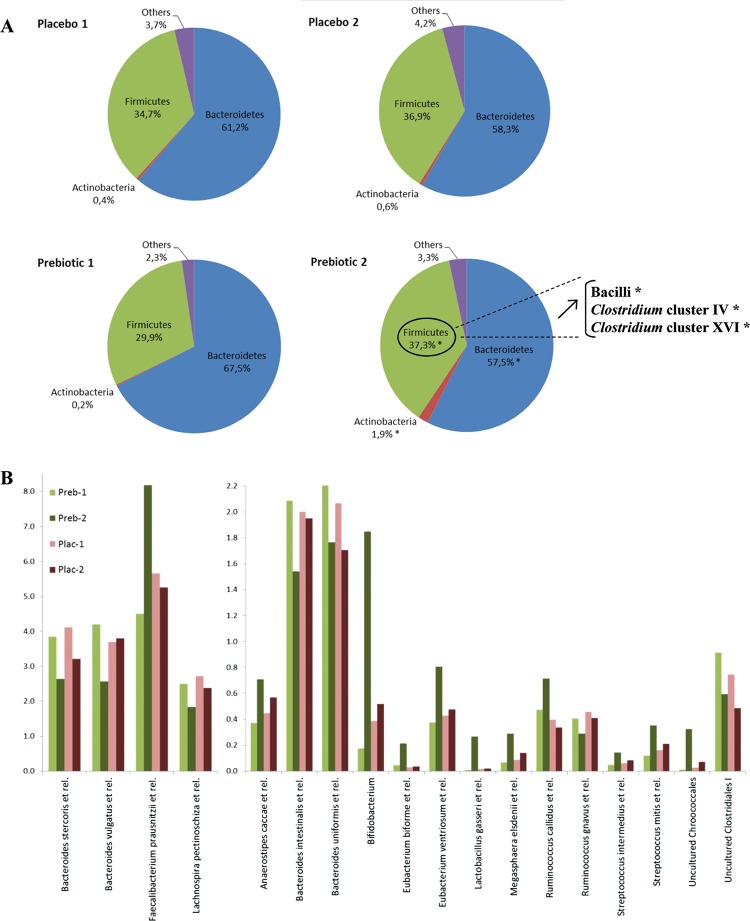
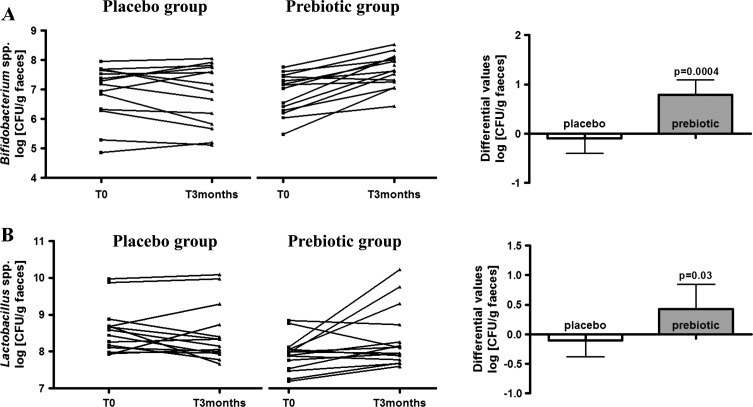
The HITChip analysis revealed an important modulation of the gut microbiota composition in patients treated with ITF prebiotics for 3 months. These changes occurred at the phylum level (subdivided for the Firmicutes over classes and Clostridium clusters) and the genus-like taxonomic level with phylotypes with >90% 16S rRNA gene sequence similarity (see online table S1). ITF prebiotics induced a significant increase in Firmicutes and Actinobacteria and a decrease in Bacteroidetes (figure 2A). Changes in Firmicutes were due to a significant increase of three groups belonging to this phylum, namely the bacilli, and Clostridium clusters IV and XVI (the bacterial groups belonging to these three classes are described in online table S2). At the genus-like taxonomic level (figure 2B), an increase in Bifidobacterium and Faecalibacterium prausnitzii and a decrease in Bacteroides intestinalis and B vulgatus occurred after ITF prebiotic treatment but not after placebo treatment. The important increase in bifidobacteria following prebiotic treatment was confirmed by qPCR (figure 3A). The HITChip analysis also revealed an important change for Lactobacillus gasseri (a 34.4-fold change), occurring mainly in three patients belonging to the prebiotic group. Lactobacilli analysis by qPCR highlighted a significant effect of the prebiotic treatment on the genus Lactobacillus spp. with an increase in the prebiotic group and no change in the placebo group (figure 3B).
Figure 2.
Human Intestinal Tract Chip analysis. (A) Relative contribution (mean percentage of total detected bacteria at the phylum level) of the major phyla in both groups (placebo and treated): (1) before; and (2) after treatment. *p<0.05 according to the Wilcoxon signed rank test (T0 vs T3 months in the prebiotic group). Exact p values are given in online supplemental table S1. (B) Relative contribution (mean percentage of total detected bacteria at the genus-like taxonomic level) of the species/genus of bacteria in both groups before and after treatment (Preb-1: prebiotic group, T0; Preb-2: prebiotic group, T3 months; Plac-1: placebo group, T0; Plac-2: placebo group, T3 months).
Figure 3.
Gut microbiota analysis by quantitative PCR. (A) Bifidobacterium spp.; (B) Lactobacillus spp. Left: individual levels in log (CFU/g faeces) for each patient of the placebo and prebiotic groups before (T0) and after (T3 months) treatment. Right: differential values (T3 months–T0) in log (CFU/g faeces). Results are given as mean±95% CI. p Values according the Mann-Whitney test (placebo vs prebiotic) to assess treatment effect.
A hierarchical clustering of the HITChip data did not reveal any clustering between the patients of each group but confirmed the important inter-individual variations with clusters observed between samples coming from the same patient (see online figure S1).
To assess for differences in microbiota composition of both groups at T0 and T3 months, a multivariate statistics approach was used. A redundancy analysis showed that, although the different study groups did not change significantly in composition, the samples belonging to the prebiotic group at 3 months positively correlated with Actinobacteria (including the Bifidobacterium group) and negatively correlated with Bacteroidetes and Proteobacteria (see online figure S2).
Anthropometric and blood parameters
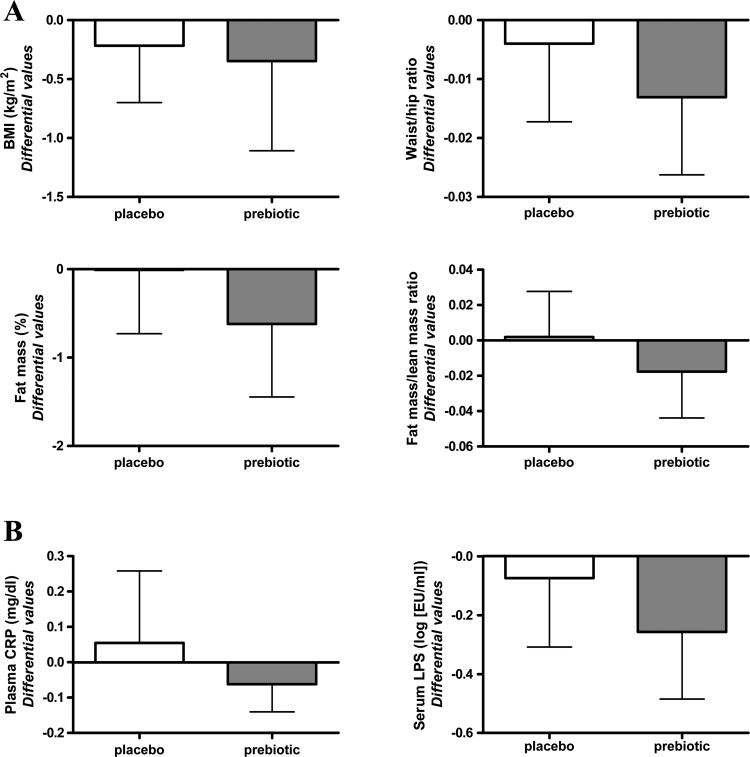
After 3 months of treatment, ITF prebiotics had no significant impact on BMI and waist/hip ratio but tended to decrease fat mass even if the differential values were not significantly different between both groups (figure 4A).
Figure 4.
(A) Anthropometric characteristics. (B) Plasma C-reactive protein (CRP) and serum lipopolysaccharide (LPS) of patients in both groups (placebo and treated) before (T0) and after (T3 months) treatment. Differential values (T3 months–T0) are given as mean±95% CI. Statistical analysis performed on transformed values (log) for LPS. BMI, body mass index.
The prebiotic treatment did not significantly modify HbA1c, fasting glycaemia and insulinaemia, post-OGTT insulinaemia, HOMA index or adiponectinaemia. However, we observed a significant difference in the post-OGTT glycaemia, with an increase occurring in the placebo group and a slight decrease observed in the treated group. The prebiotic treatment had no significant effect on cholesterol (total, HDL or LDL) and triglycerides (see online table S3).
Finally, treatment effect on plasma CRP was not significant. Metabolic endotoxaemia was decreased in both groups but to a higher extent in the prebiotic group, even if the treatment effect did not reach significance (figure 4B).
Correlations between gut bacteria and biological parameters
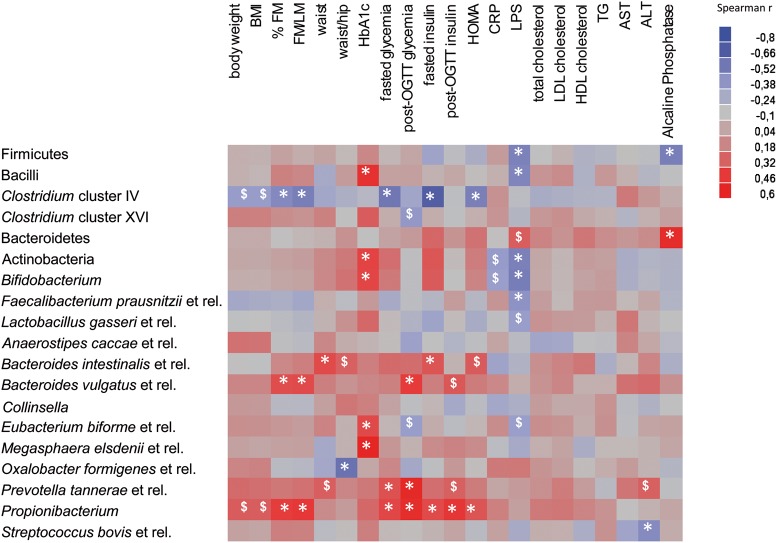
A Spearman correlation analysis was performed to evaluate the potential link between significant changes in gut microbiota composition induced by prebiotics and host metabolism (figure 5). LPS changes significantly and negatively correlated with several phyla and species of bacteria that were increased by the prebiotics, namely Firmicutes (bacilli appear to be responsible for this correlation), Actinobacteria, Bifidobacterium and Faecalibacterium prausnitzii. Changes in Clostridium cluster IV group (which was also increased by ITF) negatively correlated with anthropometric parameters and with the fasting glycaemia, insulinaemia and HOMA index. In contrast, changes in Propionibacterium, Bacteroides intestinalis and Bacteroides vulgatus, all three being significantly decreased by prebiotic treatment, positively correlated with changes in body composition and glucose homeostasis.
Figure 5.
Heat map of the Spearman r correlations between the gut bacteria significantly modified by the prebiotic treatment and anthropometric/biological parameters. Correlations were performed on differential values (T3 months–T0) for each patient in both groups (placebo and treated). *p<0.05; $p<0.1 following the Spearman correlation. ALT, alanine transaminase; AST, aspartate transaminase; BMI, body mass index; CRP, C-reactive protein; FM, fat mass; HDL, high-density lipoprotein; HOMA, homeostasis model assessment; LDL, low-density lipoprotein; LM, lean mass; LPS, lipopolysaccharides; OGTT, oral glucose tolerance test; TG, triglycerides.
Metabolic profiling by 1H NMR spectroscopy
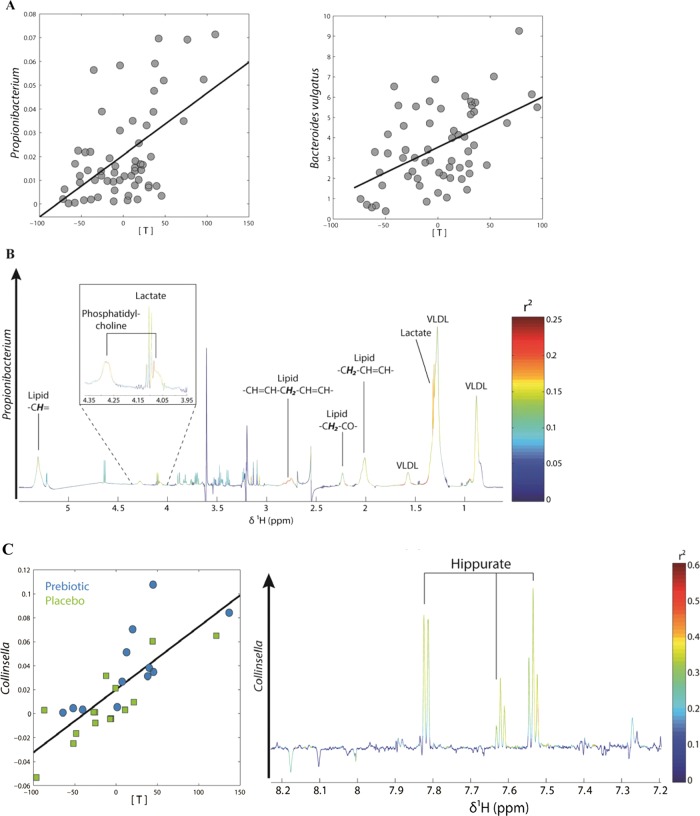
Overall, the multivariate statistical analysis of urine and plasma NMR spectra did not allow separating the metabolic profiles observed in the placebo and treated groups at either time point (data not shown). Nevertheless, in plasma, we observed a significant, positive correlation between the partial least square (PLS) scores of the NMR spectra and Propionibacterium and Bacteroides vulgatus (figure 6A). The discriminant metabolites explaining this correlation were lactate and phosphatidylcholine (PC), meaning that these two metabolites were increased in patients where Propionibacterium and Bacteroides vulgatus were more abundant (figure 6B shows the PLS loading plot for Propionibacterium).
Figure 6.
(A) Partial least square (PLS) regression analysis between plasma metabolic profiles and gut levels of Propionibacterium and Bacteroides vulgatus: score plots showing the correlation between bacterial levels (relative contribution: y axis) and PLS scores (x axis). (B) PLS loading plot for Propionibacterium showing the contribution of plasma lactate and phosphatidylcholine to the model. (C) PLS regression analysis between changes in urinary metabolic profiles and changes in the gut levels of Collinsella performed on differential values (T3 months–T0). Left: PLS scores (x axis) are plotted against Collinsella levels (relative contribution: y axis). Right: PLS loading plot showing the contribution of hippurate to the model. Details of model parameters are described in the online supplemental methods section.
The PLS scores of the urine NMR spectra significantly correlated with the waist/hip ratio and with the post-OGTT insulin (see online figure S3). The PLS loadings of these two models showed that creatinine and hippurate negatively correlated with post-OGTT insulin, whereas creatinine alone correlated with the waist/hip ratio. Finally, changes in the levels of Collinsella, a bacterial genus significantly increased by prebiotic treatment, significantly correlated with the PLS scores of urinary metabolic profiles (figure 6C). Hippurate was associated with these changes in Collinsella as depicted by the PLS loadings. The PLS parameters of the listed models are given in the online supplemental methods.
Discussion
Over the past few years, interest has grown regarding the potential role of the gut microbiota in the occurrence of obesity and related diseases. Nutritional approaches are considered potential tools to modulate the gut microbiota with a concomitant impact on health.2 3 We show, for the first time, that a 3-month treatment with ITF prebiotics led to a selective modulation of the gut microbiota composition in obese women. At the phylum level, the ITF treatment increased Firmicutes and Actinobacteria and decreased Bacteroidetes. This result was intriguing because other studies observed an increased Firmicutes and/or a decreased Bacteroidetes count in obese versus lean individuals. These changes could be inverted after a diet-induced weight loss and were division-wide with no specific subgroup being preferentially decreased or increased.4 13 39 In our study, increased levels of Firmicutes following prebiotic treatment were due to increases in bacilli, and Clostridium clusters IV and XVI. Interestingly, changes in bacilli were negatively associated with changes in LPS, whereas changes in Clostridium cluster IV negatively correlated with changes in fat mass and fasting glucose homeostasis. This observation suggests a positive impact of the prebiotics on these specific Firmicutes. Consistent with this, Schwiertz et al recently showed a higher level of the Clostridium leptum group, Faecalibacterium prausnitzii being the major bacterium of this group, in lean patients compared to obese individuals.15
In addition, other studies have shown similar levels of Bacteroidetes in lean and obese individuals,16 or even an increased proportion of this phylum in overweight and obese patients,15 thus showing that the potential change of Bacteroidetes in obesity is controversial. Moreover, Wu et al recently studied the impact of diet on gut microbiome using diet inventories and 16S rDNA sequencing to characterise faecal samples from 98 individuals.40 They observed that bacterial phyla were related to diet, with Bacteroidetes being positively associated with fat and negatively associated with fibre, and Firmicutes showing an opposite correlation. Interestingly, these associations were in accord with the results obtained with ITF prebiotics, which are fermentable dietary fibres.
Among the prebiotic-induced specific changes at the genus-like taxonomic level, significant increases in Bifidobacterium and Faecalibacterium prausnitzii were of particular interest. Bifidobacteria have been shown to decrease in obesity and diabetes,7 15 17 41 and to increase on prebiotic treatment, leading to improved host metabolism in rodents and healthy humans.23 24 31 42 43 F prausnitzii is depleted in inflammatory bowel disease (IBD) and Crohn's disease patients and exhibits anti-inflammatory effects in vitro (cellular models) and in vivo (TNBS-induced colitis in mice).44–46 It is also decreased in obese diabetic patients compared to non-diabetic individuals (lean and obese) and tends to increase after bariatric surgery. Furthermore, it is negatively correlated with inflammatory markers in obese patients both before and after bariatric surgery.47 In healthy humans, ITF prebiotics significantly increase the levels of F prausnitzii.31 In our study, prebiotic-induced changes in Bifidobacterium and F prausnitzii negatively correlated with changes in LPS, a pro-inflammatory component. This result is of particular interest in view of the central role played by metabolic endotoxaemia in the process of low-grade inflammation in obesity.7 8 23
At the beginning of the treatment, there was no statistical difference in anthropometric and blood parameters between both groups of patients. Although ITF prebiotics showed promising results on gut microbiota composition in obese women, they did not significantly impact BMI, lipid or glucose homeostasis but they tended to decrease fat mass. We cannot exclude that tendencies observed for the clinical parameters could have become significant with a higher number of patients.
Nevertheless, the metabolic profiling of urine and plasma revealed interesting correlations between metabolites implicated in obesity and some gut bacteria. Levels of Propionibacterium and Bacteroides vulgatus, both decreased by prebiotic treatment, positively correlated with plasma levels of PC and lactate. Propionibacterium is a bacterial genus with probiotic properties, mainly isolated from dairy products.48 A Mouse Intestinal Tract Chip analysis has revealed a significant increase of this genus in db/db mice compared to lean mice (Cani PD, personal communication). Bacteroides vulgatus has been found to constitute part of the core gut microbiota in healthy humans and is generally considered beneficial.49–51 However, it has also been associated with a higher genetic risk of developing coeliac disease and some specific strains of B vulgatus are capable of promoting colitis,52 53 thus suggesting a controversial impact on health. Dumas et al have demonstrated that mice developing insulin resistance and non-alcoholic fatty liver disease have lower levels of PC, leading to an impairment of very low density lipoprotein secretion and, thereby, hepatic triglycerides accumulation.54 Nevertheless, other studies have shown that PC levels are higher in obesity and decrease following diet-induced weight loss or bariatric surgery.55 56 Moreover, PC molecules are increased in conventionally raised (CONV-R) animals compared to germ-free mice, which have lower adiposity and hepatic triglycerides than their CONV-R counterparts.57 Disturbances in whole body lactate turnover appear to contribute to the development of insulin resistance. Elevated plasma levels of lactate have been reported in both obesity and type 2 diabetes.58–60
Thus, low levels of plasma PC and lactate, which were observed in patients with low levels of Propionibacterium and Bacteroides vulgatus in the prebiotic group, appeared to be associated with lower adiposity. Moreover, changes in these bacteria positively correlated with changes in fat mass and glucose homeostasis. This correlation was in accord with the higher levels of Propionibacterium in diabetic db/db mice, thus confirming their potential negative role on host metabolism, at least in pathological situations linked to glucose homeostasis disturbances (Cani PD, personal communication).
Finally, increased levels of Collinsella, a genus belonging to Actinobacteria and significantly augmented with prebiotics, correlated with higher urinary levels of hippurate. Collinsella aerofaciens has been associated with a low risk of colon cancer,61 and patients with IBD present lower gut levels of this genus than do control individuals.62 Hippurate is a gut-derived metabolite commonly associated with a ‘healthy phenotype’. It is decreased in diabetes and obesity and increases following bariatric surgery. Moreover, it is one of the main discriminant metabolites explaining the difference in urine metabolic profiles between lean and obese or diabetic individuals.63–65 Thus, increased levels of Collinsella and urinary hippurate could be considered as a beneficial effect associated with ITF fermentation.
In conclusion, the prebiotic intervention was not sufficient to induce significant clustering in the overall metabolomic or gut microbiota profiles between the two groups of patients. However, we discovered subtle changes in the gut microbiota which correlated with changes in fat mass, serum LPS levels and metabolism (hippurate, lactate and PC). Thus, by modelling symbiotic biology systems (eg, the gut microbiota–host interactions) with prebiotics in obese women, we identified potential supersystem interactions associated with the occurrence of obesity and related diseases. Accordingly, our study may be considered as a prerequisite to better define the target population and patient characteristics for further larger intervention studies.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgments
We wish to thank the patients and investigators who participated in this study. PDC is a Research Associate and LBB a Research Fellow of the FRS-FNRS (Fond de la Recherche Scientifique), Belgium.
Footnotes
Contributors: EMD: protocol and trial design, patient selection, random allocation sequence, patient-related study procedures, performance of the experiments, data analysis and interpretation, manuscript writing. PDC: protocol and trial design, patient selection, performance of the experiments, data analysis and interpretation. SPC, SF, PGBP, AMN and LBB: performance of the experiments, data analysis and interpretation. WMdV and GRG: data analysis and interpretation. JPT: responsibility for clinical trial and patient care, protocol and trial design, patient selection. NMD: principal investigator, protocol and trial design, patient selection, random allocation sequence, data analysis and interpretation, manuscript writing. All of the authors have read, revised and approved the final manuscript.
Funding: This project was supported by a FNRS grant (no. 1.5.095.09F).
Competing interests: None.
Patient consent: Obtained.
Ethics approval: Commission d'Ethique Biomédicale Hospitalo-facultaire, Université catholique de Louvain.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Hotamisligil GS. Inflammation and metabolic disorders. Nature 2006;444:860–7 [DOI] [PubMed] [Google Scholar]
- 2.Delzenne NM, Cani PD. Interaction between obesity and the gut microbiota: relevance in nutrition. Annu Rev Nutr 2011;31:15–31 [DOI] [PubMed] [Google Scholar]
- 3.Delzenne NM, Neyrinck AM, Cani PD. Modulation of the gut microbiota by nutrients with prebiotic properties: consequences for host health in the context of obesity and metabolic syndrome. Microb Cell Fact 2011;10(Suppl 1):S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006;444:1027–31 [DOI] [PubMed] [Google Scholar]
- 5.Turnbaugh PJ, Ridaura VK, Faith JJ, et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 2009;1:6ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Claus SP, Ellero SL, Berger B, et al. Colonization-induced host-gut microbial metabolic interaction. MBio 2011;2:e00271–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cani PD, Amar J, Iglesias MA, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007;56:1761–2 [DOI] [PubMed] [Google Scholar]
- 8.Cani PD, Bibiloni R, Knauf C, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 2008;57:1470–81 [DOI] [PubMed] [Google Scholar]
- 9.Cani PD, Possemiers S, Van de Wiele T, et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut 2009;58:1091–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muccioli GG, Naslain D, Backhed F, et al. The endocannabinoid system links gut microbiota to adipogenesis. Mol Syst Biol 2010;6:392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cani PD, Delzenne NM. Interplay between obesity and associated metabolic disorders: new insights into the gut microbiota. Curr Opin Pharmacol 2009;9:737–43 [DOI] [PubMed] [Google Scholar]
- 12.Nadal I, Santacruz A, Marcos A, et al. Shifts in clostridia, bacteroides and immunoglobulin-coating fecal bacteria associated with weight loss in obese adolescents. Int J Obes (Lond) 2009;33:758–67 [DOI] [PubMed] [Google Scholar]
- 13.Ley RE, Backhed F, Turnbaugh P, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 2005;102:11070–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature 2009;457:480–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwiertz A, Taras D, Schafer K, et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring) 2010;18:190–5 [DOI] [PubMed] [Google Scholar]
- 16.Duncan SH, Lobley GE, Holtrop G, et al. Human colonic microbiota associated with diet, obesity and weight loss. Int J Obes (Lond) 2008;32:1720–4 [DOI] [PubMed] [Google Scholar]
- 17.Wu X, Ma C, Han L, et al. Molecular characterisation of the faecal microbiota in patients with type II diabetes. Curr Microbiol 2010;61:69–78 [DOI] [PubMed] [Google Scholar]
- 18.Gibson GR, Roberfroid MB. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr 1995;125:1401–12 [DOI] [PubMed] [Google Scholar]
- 19.Roberfroid M, Gibson GR, Hoyles L, et al. Prebiotic effects: metabolic and health benefits. Br J Nutr 2010;104(Suppl 2):S1–63 [DOI] [PubMed] [Google Scholar]
- 20.Delzenne NM, Neyrinck AM, Backhed F, et al. Targeting gut microbiota in obesity: effects of prebiotics and probiotics. Nat Rev Endocrinol 2011;7:639–46 [DOI] [PubMed] [Google Scholar]
- 21.Cani PD, Neyrinck AM, Maton N, et al. Oligofructose promotes satiety in rats fed a high-fat diet: involvement of glucagon-like Peptide-1. Obes Res 2005;13:1000–7 [DOI] [PubMed] [Google Scholar]
- 22.Cani PD, Knauf C, Iglesias MA, et al. Improvement of glucose tolerance and hepatic insulin sensitivity by oligofructose requires a functional glucagon-like peptide 1 receptor. Diabetes 2006;55:1484–90 [DOI] [PubMed] [Google Scholar]
- 23.Cani PD, Neyrinck AM, Fava F, et al. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia 2007;50:2374–83 [DOI] [PubMed] [Google Scholar]
- 24.Meyer D, Stasse-Wolthuis M. The bifidogenic effect of inulin and oligofructose and its consequences for gut health. Eur J Clin Nutr 2009;63:1277–89 [DOI] [PubMed] [Google Scholar]
- 25.Everard A, Lazarevic V, Derrien M, et al. Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes 2011;60:2775–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cani PD, Joly E, Horsmans Y, et al. Oligofructose promotes satiety in healthy human: a pilot study. Eur J Clin Nutr 2006;60:567–72 [DOI] [PubMed] [Google Scholar]
- 27.Verhoef SP, Meyer D, Westerterp KR. Effects of oligofructose on appetite profile, glucagon-like peptide 1 and peptide YY3-36 concentrations and energy intake. Br J Nutr 2011;106:1757–62 [DOI] [PubMed] [Google Scholar]
- 28.Cani PD, Lecourt E, Dewulf EM, et al. Gut microbiota fermentation of prebiotics increases satietogenic and incretin gut peptide production with consequences for appetite sensation and glucose response after a meal. Am J Clin Nutr 2009;90:1236–43 [DOI] [PubMed] [Google Scholar]
- 29.Parnell JA, Reimer RA. Weight loss during oligofructose supplementation is associated with decreased ghrelin and increased peptide YY in overweight and obese adults. Am J Clin Nutr 2009;89:1751–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matthews DR, Hosker JP, Rudenski AS, et al. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985;28:412–19 [DOI] [PubMed] [Google Scholar]
- 31.Ramirez-Farias C, Slezak K, Fuller Z, et al. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr 2009;101:541–50 [DOI] [PubMed] [Google Scholar]
- 32.Salonen A, Nikkila J, Jalanka-Tuovinen J, et al. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J Microbiol Methods 2010;81:127–34 [DOI] [PubMed] [Google Scholar]
- 33.Zoetendal EG, Rajilic-Stojanovic M, de Vos WM. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 2008;57:1605–15 [DOI] [PubMed] [Google Scholar]
- 34.Rajilic-Stojanovic M, Heilig HG, Molenaar D, et al. Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ Microbiol 2009;11:1736–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jalanka-Tuovinen J, Salonen A, Nikkila J, et al. Intestinal microbiota in healthy adults: temporal analysis reveals individual and common core and relation to intestinal symptoms. PLoS One 2011;6:e23035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Everard A, Geurts L, Van RM, et al. Tetrahydro iso-alpha acids from hops improve glucose homeostasis and reduce body weight gain and metabolic endotoxemia in high-fat diet-fed mice. PLoS One 2012;7:e33858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B Methodological 1995;57:289–300 [Google Scholar]
- 38.Leps J, Smilauer P. Multivariate analysis of ecological data using CANOCO. Cambridge: Cambridge University Press, 2003 [Google Scholar]
- 39.Ley RE, Turnbaugh PJ, Klein S, et al. Microbial ecology: human gut microbes associated with obesity. Nature 2006;444:1022–3 [DOI] [PubMed] [Google Scholar]
- 40.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011;334:105–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalliomaki M, Collado MC, Salminen S, et al. Early differences in fecal microbiota composition in children may predict overweight. Am J Clin Nutr 2008;87:534–8 [DOI] [PubMed] [Google Scholar]
- 42.Bouhnik Y, Raskine L, Champion K, et al. Prolonged administration of low-dose inulin stimulates the growth of bifidobacteria in humans. Nutr Res 2007;27:187–93 [Google Scholar]
- 43.Kolida S, Meyer D, Gibson GR. A double-blind placebo-controlled study to establish the bifidogenic dose of inulin in healthy humans. Eur J Clin Nutr 2007;61:1189–95 [DOI] [PubMed] [Google Scholar]
- 44.Martinez-Medina M, Aldeguer X, Gonzalez-Huix F, et al. Abnormal microbiota composition in the ileocolonic mucosa of Crohn's disease patients as revealed by polymerase chain reaction-denaturing gradient gel electrophoresis. Inflamm Bowel Dis 2006;12:1136–45 [DOI] [PubMed] [Google Scholar]
- 45.Frank DN, St Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA 2007;104:13780–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA 2008;105:16731–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Furet JP, Kong LC, Tap J, et al. Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes 2010;59:3049–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cousin FJ, Mater DDG, Foligne B, et al. Dairy propionibacteria as human probiotics: A review of recent evidence. Dairy Sci Technol 2011;91:1–26 [Google Scholar]
- 49.Tap J, Mondot S, Levenez F, et al. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol 2009;11:2574–84 [DOI] [PubMed] [Google Scholar]
- 50.Noor SO, Ridgway K, Scovell L, et al. Ulcerative colitis and irritable bowel patients exhibit distinct abnormalities of the gut microbiota. BMC Gastroenterol 2010;10:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cuiv PO, Klaassens ES, Durkin AS, et al. Draft genome sequence of Bacteroides vulgatus PC510, a strain isolated from human feces. J Bacteriol 2011;193:4025–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanchez E, De PG, Capilla A, et al. Influence of environmental and genetic factors linked to celiac disease risk on infant gut colonization by Bacteroides species. Appl Environ Microbiol 2011;77:5316–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rath HC, Wilson KH, Sartor RB. Differential induction of colitis and gastritis in HLA-B27 transgenic rats selectively colonized with Bacteroides vulgatus or Escherichia coli. Infect Immun 1999;67:2969–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dumas ME, Barton RH, Toye A, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci USA 2006;103:12511–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oberbach A, Bluher M, Wirth H, et al. Combined proteomic and metabolomic profiling of serum reveals association of the complement system with obesity and identifies novel markers of body fat mass changes. J Proteome Res 2011;10:4769–88 [DOI] [PubMed] [Google Scholar]
- 56.Schwab U, Seppanen-Laakso T, Yetukuri L, et al. Triacylglycerol fatty acid composition in diet-induced weight loss in subjects with abnormal glucose metabolism–the GENOBIN study. PLoS One 2008;3:e2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Velagapudi VR, Hezaveh R, Reigstad CS, et al. The gut microbiota modulates host energy and lipid metabolism in mice. J Lipid Res 2010;51:1101–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lovejoy J, Newby FD, Gebhart SS, et al. Insulin resistance in obesity is associated with elevated basal lactate levels and diminished lactate appearance following intravenous glucose and insulin. Metabolism 1992;41:22–7 [DOI] [PubMed] [Google Scholar]
- 59.Qvisth V, Hagstrom-Toft E, Moberg E, et al. Lactate release from adipose tissue and skeletal muscle in vivo: defective insulin regulation in insulin-resistant obese women. Am J Physiol Endocrinol Metab 2007;292:E709–14 [DOI] [PubMed] [Google Scholar]
- 60.Reaven GM, Hollenbeck C, Jeng CY, et al. Measurement of plasma glucose, free fatty acid, lactate, and insulin for 24 h in patients with NIDDM. Diabetes 1988;37:1020–4 [DOI] [PubMed] [Google Scholar]
- 61.Moore WE, Moore LH. Intestinal floras of populations that have a high risk of colon cancer. Appl Environ Microbiol 1995;61:3202–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kassinen A, Krogius-Kurikka L, Makivuokko H, et al. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology 2007;133:24–33 [DOI] [PubMed] [Google Scholar]
- 63.Salek RM, Maguire ML, Bentley E, et al. A metabolomic comparison of urinary changes in type 2 diabetes in mouse, rat, and human. Physiol Genomics 2007;29:99–108 [DOI] [PubMed] [Google Scholar]
- 64.Waldram A, Holmes E, Wang Y, et al. Top-down systems biology modeling of host metabotype-microbiome associations in obese rodents. J Proteome Res 2009;8:2361–75 [DOI] [PubMed] [Google Scholar]
- 65.Calvani R, Miccheli A, Capuani G, et al. Gut microbiome-derived metabolites characterize a peculiar obese urinary metabotype. Int J Obes (Lond) 2010;34:1095–8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.