Abstract
In the developing peripheral nervous system, axon-derived signals stimulate Schwann cells to undergo a global genetic reprogramming involving the cessation of cellular division and the upregulation of myelin genes. How such a comprehensive change in gene transcription is regulated is poorly understood. Here we report that BRG1/SMARCA4, the central helicase of the mammalian SWI/SNF-related chromatin remodeling complex, is required for Schwann cells to differentiate and form myelin, both in vitro and in vivo, in the mouse. BRG1 was highly activated in Schwann cells at early stages of myelination, and loss of the enzyme inhibited their differentiation and completely prevented myelin formation. Furthermore, we identify NF-κB as a key transcription factor that associates with the BRG1 complex in response to neuregulin 1 type III. During myelination, BRG1 was activated through the formation of a complex with NF-κB, and both proteins bound to the promoter region of Sox10, an inducer of myelination. These findings delineate a novel mechanism whereby axonal signals promote myelination through the remodeling of chromatin structure.
Introduction
During peripheral nervous system development, axonal signals stimulate Schwann cell precursors to vastly extend their cell membrane, wrap around an axonal segment, and upregulate a wide array of specialized proteins. Ultimately, myelin is formed, which allows for saltatory conduction and provides trophic support to axons (Edgar and Garbern, 2004). The striking morphological and physiological changes that the Schwann cell must undergo during differentiation require equally extensive changes in gene expression. Previous research on Schwann cell gene regulation has focused largely on the role of individual promyelinating transcription factors (Svaren and Meijer, 2008); however, recent studies have begun to reveal a number of epigenetic mechanisms for controlling gene transcription, including chromatin modification. Densely packed chromatin inhibits the binding of proteins such as transcription factors to DNA, thereby repressing gene expression. Chromatin compaction can be altered by histone-modifying enzymes, which post-translationally modify the protein tails protruding from histones. These modifications alter DNA interaction with histones and serve to recruit chromatin-associated factors and transcription regulators (Li et al., 2007; Bannister and Kouzarides, 2011). Such chromatin modifications were recently shown to play an essential role in Schwann cell differentiation through the deletion of histone deacetylases 1 and 2 (Chen et al., 2011; Jacob et al., 2011).
Another mechanism by which chromatin structure can alter gene transcription is through ATPase-dependent remodelers that physically separate DNA–histone interactions to slide and reposition nucleosomes, allowing previously repressed sequences of DNA to become available to transcriptional regulators (de la Serna et al., 2006; Tang et al., 2010). One of the best characterized chromatin remodelers is the SWI/SNF family, which is conserved from yeast to humans. These complexes are defined by a central ATPase subunit and, in mammals, the subfamily containing the highly homologous BRG1 or BRM ATPases has been implicated in the development of various tissues. Previous studies have documented an essential role for BRG1 in the differentiation of neurons (Wu et al., 2007), T-cells (Zhao et al., 1998; Gebuhr et al., 2003), and muscle (de la Serna et al., 2001). Therefore, we hypothesized that a BRG1 complex may be involved in Schwann cell differentiation.
As BRM and BRG1 have no intrinsic sequence specificity, they must be targeted to DNA through interaction with sequence-specific DNA binding proteins; for example, in differentiating muscle the BRG1 complex is recruited to chromatin through interaction with the transcription factors MyoD and MEF2 (Ohkawa et al., 2006). These findings suggest that differentiation-specific transcription factors interact with chromatin remodeling complexes to regulate gene transcription through reorganization of chromatin.
Schwann cells require the coordinated activity of several key transcription factors to initiate differentiation into a myelinating phenotype. NF-κB is one example of such a promyelinating transcription factor (Nickols et al., 2003; Yoon et al., 2008). It is a dimer formed by five different subunits and, in Schwann cells, the p65/RelA subunit complexes with p50 and regulates the expression of Oct6 (Yoon et al., 2008), a POU domain transcription factor required for proper timing of myelination (Bermingham et al., 1996; Ghazvini et al., 2002), and Sox10 (Chen et al., 2011), a transcription factor required for Schwann cell specification (Kuhlbrodt et al., 1998; Britsch et al., 2001) and differentiation (Peirano et al., 2000; Ghislain and Charnay, 2006; LeBlanc et al., 2007; Schreiner et al., 2007; Finzsch et al., 2010).
In this study we report that the chromatin remodeling factor BRG1 is essential for myelin formation by Schwann cells in the peripheral nervous system and delineates a molecular pathway by which axonal signals effect Schwann cell differentiation through the activation of an NF-κB – BRG1 chromatin remodeling complex.
Materials and Methods
Antibodies and reagents.
Antibodies recognizing BRG1, p65, BRM, Sox10, Sox2, Krox20, and Oct6 were purchased from Santa Cruz Biotechnology. Antibodies raised against Tuj1, MBP, and Krox20 were from Covance, while antibodies against α-tubulin were from Calbiochem. Oct6 antibodies used for Western blotting and supershift were a gift of Dies Meijer (Erasmus University Medical Center, Rotterdam, The Netherlands). Rabbit IgG was purchased from Jackson ImmunoResearch Laboratories. Caspase3 was from Cell Signaling Technology. Ki67 was from Thermo Scientific. Dibutyryl-cAMP (dbcAMP) was purchased from Biomol, and SN50 was from Enzo Life Sciences. SMARCA4/BRG1 siRNA and control siRNA was obtained from Dharmacon.
Cell culture.
Cos7 and HEK 293 cells were maintained in DMEM with 10% fetal bovine serum (FBS) (Sigma) and 1% penicillin/streptomycin (Invitrogen ). Rat Schwann cells were isolated from sciatic nerves of 4- to 5-day-old Sprague Dawley rats and purified as described previously by Yoon et al. (2008). Passaged rat Schwann cells were grown with 2 μm forskolin in DMEM with 10% FBS (Sigma). All experiments using animals were approved by the Animal Care and Use Committee at Vanderbilt University (Nashville, TN).
SiRNA transfection.
Freshly isolated Rat Schwann cells were transfected with siRNA to SMARCA4/BRG1 (Dharmacon) or nontargeting siRNA using DharmaFECT transfection reagent (Dharmacon) according to the manufacturer's directions.
DRG/Schwann cell cocultures.
Myelinating dorsal root ganglion (DRG)/Schwann cell cocultures were established using DRGs isolated from embryonic day (E)15 rats as described (Nickols et al., 2003; Yoon et al., 2008) and plated in Ultraculture media (BioWhittiker) supplemented with 10% FBS (Hyclone), 2 mm l-glutamine (Invitrogen ), and 50 ng/ml nerve growth factor (NGF) (Harlan Laboratories) at a density of 80,000 cells/2.2 cm2 on collagen-coated coverslips. Myelination was induced 5 days later by adding 50 μg/ml ascorbic acid in growth media. Growth media and ascorbic acid were replaced every 2 days. SN50 peptide inhibitor of NF-κB was added at 50 μg/ml for 48 h.
Membrane purification.
Cos7 or HEK293 cells were transfected with NRG 1 type III constructs (Wang et al., 2001) using Lipofectamine (Invitrogen) according to the manufacturer's directions. After 2 days, transfected and untransfected cells were rinsed in PBS and then scraped into homogenization buffer (20 mm HEPES, pH8.0, 1.5 mm MgCl2, and 1 mm EGTA), incubated on ice for 10 min, and then homogenized in a Dounce homogenizer. Homogenates were centrifuged at 1000 × g for 10 min at 4°C, and the supernatants were collected and centrifuged at 35,000 × g for 1 h at 4°C. The membrane pellets were resuspended in PBS and immediately added to cells. Aliquots of purified membrane were used for protein assays and Western blotting.
Immunoprecipitation and Western blotting.
For immunoprecipitation experiments, confluent rat Schwann cells were cultured in DMEM plus 10% FBS for 2 days then in a 1:1 mixture of DMEM and Ham's F-12 with N2 supplement (Invitrogen) for another 2 days. Cells were treated for 1 h with membrane fragments and then lysed in lysis buffer containing 20 mm Tris-HCl, pH 7.5, 137 mm NaCl, 1% Nonidet P-40, 10% glycerol, 1 mm MgCl2, 1 mm EGTA, 1 mm Na3VO4, 20 mm β-glycerol phosphate, 1 mm phenylmethylsulfonyl fluoride (PMSF), and a protease inhibitor cocktail (Roche). Protein concentrations were then determined by Bradford assay (Bio-Rad). Cell lysates (0.5 to 1 mg of protein) were incubated with 2 μg of either p65 or BRG1 antibodies (Santa Cruz Biotechnology) overnight at 4°C and then immunoprecipitated with protein G agarose beads (Zymed). The immunoprecipitates were separated by SDS-PAGE and Western blotted with antibodies to p65 (1:1000) or BRG1 (1:1000; Santa Cruz Biotechnology).
In some cases lysates of rat Schwann cells as well as rat sciatic nerves were subjected to SDS-PAGE and Western blotting without immunoprecipitation. The membranes were blocked in Tris-buffered saline, pH 7.4, with 0.1% Tween (TBST) containing 3% bovine serum albumin and treated with primary antibody overnight at 4°C. Secondary antibodies conjugated to horseradish peroxidase were added, and proteins were visualized by chemiluminescence.
Electrophoretic mobility shift assay.
NF-κB was analyzed by gel-shift assay with a 32P-labeled, double-stranded NF-κB consensus oligonucleotide (Promega) as described previously (Nickols et al., 2003). Two oligonucleotides containing the Krox20 consensus site, 5′-GGGAGTCAGTCTTGCGTGGGCCTTAGTCAGTCGGG- 3′ and 5′-CCCGACTGACTAAGGCCCACGCAAGACTGACTCCC-3′ (Warner et al., 1999), were synthesized, annealed, and 32P labeled. The probe for Oct6 was purchased from Promega. Supershift assays were performed by adding 1–2 μg of specific antibody to the electrophoretic mobility shift assay (EMSA) reaction for 15 min at RT.
ATPase assays.
ATPase assays were carried out as described (Bultman et al., 2005) with modifications. Samples were lysed in RIPA buffer containing 100 mm NaCl, 20 mm Tris, pH 7.6, 0.2% deoxycholate, 0.2% Triton X-100, 0.2% NP-40, 1 mm DTT, 2 mm β-mercaptoethanol, and a protease inhibitor tablet (Roche), sonicated, and clarified by centrifugation. Protein concentration of lysates was quantified by Bradford assay. Lysates were incubated with 2 μg of BRG1-specific antibodies overnight at 4°C and then with protein G agarose beads (Zymed). The precipitates were washed with wash buffer containing 20 mm HEPES, pH 7.6, 10% glycerol, 12.5 mm MgCl2, 0.1 mm EDTA, pH 8.0, 0.2% NP-40, 0.1 mm DTT, and 0.6 m KOAc and then resuspended in assay buffer containing 10 mm Tris, pH 7.5, 50 mm NaCl, 5 mm MgCl2, 20% glycerol in the presence of 1 mg/ml BSA, 0.5 mm PMSF, 1 mm DTT, 0.5 mm ATP, 20 nm plasmid DNA, and 1 μCi [32P]ATP. The reaction was carried out at 37°C for 1 h and stopped with the addition of 1 μl 0.5 μm EDTA per 20 μl reaction volume. Immediately, 1 μl reaction was spotted on a polyethyleneimine cellulose TLC plate and separated using 1 m formic acid and 0.5 m LiCl. The ratio of inorganic phosphate to ATP was quantified using Scion image software.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation (ChIP) assays were performed as described by Chen et al. (2011) with slight alterations. Postnatal day (P)5 mouse sciatic nerves were dissected and minced in 1% formaldehyde for 25 min at room temperature (RT), rinsed with PBS, and resuspended in 150 mm NaCl, 10% glycerol, 50 mm Tris (pH 8.0). Nerves were homogenized with Triton X-100 added to a final concentration of 0.3% and then sonicated for 30 × 10 s pulses with a 50 s incubation on ice between pulses. DNA concentrations were measured and samples containing 20–50 μg DNA were diluted with lysis buffer to 1 ml total volume and incubated with 2 μg antibodies overnight at 4°C. Dynabeads (Dynal Biotech) were added for 2 h at 4°C and washed with a low salt buffer containing 0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris, pH 8.0, and 150 mm NaCl and then with high salt buffer containing 0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris, pH8.0, 500 mm NaCl. Beads were then washed five times with 0.25 m LiCl, 1% NP-40, 1% deoxycholic acid, 1 mm EDTA, 10 mm Tris, pH 8, and once with TE buffer. The beads were treated with 1% SDS, 0.1% NaHCO3, and 200 mm NaCl for 10 min at 65°C, proteinase K (0.1 μg/μl) was added, and samples were incubated for 2 h at 42°C and then overnight at 65°C. DNA was purified using the PCR cleanup kit from Qiagen and subjected to PCR using primers and the protocol described previously by Chen et al. (2011). IκB primers were designed to encompass the six NF-κB binding sites in the rat NFKBIA gene (forward: 5′CCAAAGTCGTCGGTGGGAA3′; reverse: 5′GGTGGCGAGTTCAGACTGT3′). The relative signal intensity was quantified using Scion Image Quant Software and normalized to the IgG control for each experiment.
Mouse breeding.
For Schwann cell lineage-specific ablation, the mice carrying the Brg1 loxP allele (Brg1F/F) were bred with Dhh-Cre strains (Sumi-Ichinose et al., 1997). Both strains were in a 129Sv/B6 mixed background. Brg1F/+;Dhh-Cre mice were then crossed with Brg1F/F mice to generate conditional knockout (cko) (Brg1F/F;Dhh-Cre) and heterozygous control (Brg1F/+;Dhh-Cre) mice. Animals of either sex were analyzed. Genotypes were determined by PCR on genomic DNA. Due to the difficulty in movement exhibited by the CKO mice, moistened food chow was provided in the bottom of the cage. Animal use was approved by the Institutional Animal Care and Use Committee of the University of Texas Southwestern Medical Center at Dallas, TX.
Electron microscopy.
Mice were anesthetized and perfused with fix solution containing 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 m cacodylate buffer. Sciatic nerves were dissected, rinsed, and postfixed overnight at 4°C and processed for electron microscopy.
Immunocytochemistry.
Sciatic nerves from mice, at the indicated age, were isolated, fixed with 4% paraformaldehyde overnight, protected with 15% sucrose, and cryosectioned into 10 μm sections. The sections were then incubated with blocking solution (5% normal goat serum/0.3% Triton X-100/PBS). Primary antibodies were diluted in the blocking solution and incubated overnight at 4°C. The primary antibodies were used in various combinations such as anti-Sox10 goat antiserum (1:200), anti-Oct6 goat antiserum (1:200), anti-Krox20 rabbit antiserum (1:200), anti-Ki67 rabbit antiserum (1:200), and anti-caspase3 rabbit antiserum (1:200). After rinsing in PBS, the secondary antibodies conjugated to Cy2 or Cy3 immunofluorescent dyes were diluted in the blocking solution and added to slides. Topro3 (Invitrogen) or 4′,6-diamidino-2-phenylindole (DAPI) was used to label the nuclei. All images were acquired using a Zeiss LSM 510 confocal microscope.
Rat sciatic nerves from P5 pups were similarly fixed and cryoprotected. Following sectioning, the tissue was blocked in 1.5% FBS, 1.5% BSA, and 0.5% Triton X-100 with 0.03 m glycine. Brg1 antibody (Santa Cruz Biotechnology) diluted in PBS containing 1.5% FBS and 1.5% BSA at 1:200 was applied, followed by PBS washing and incubation with Alexa Fluor 488 donkey anti-mouse IgG (Invitrogen) at 1:1000. The section was then washed with PBS and incubated with goat anti-Sox10 (Santa Cruz Biotechnology) at 1:50 followed by Alexa Fluor 594 donkey anti-goat IgG, Invitrogen) at 1:1000. Coverslips were mounted using Vectastain reagent (Vector Laboratories with DAPI to stain nuclei.
Statistical analysis.
Statistical significance was determined by a one-way ANOVA followed by a Tukey's multiple-comparison test when comparing three or more conditions or by a two-tailed Student's t test when comparing two conditions. In all cases, p < 0.05 was considered to be statistically significant.
Results
Active BRG1 is expressed during early stages of peripheral myelination
Schwann cells undergo extensive changes in gene expression to differentiate into a myelinating phenotype; therefore, we hypothesized that ATP-dependent chromatin remodeling was required for axon-induced Schwann cell differentiation. We first investigated BRG1 expression and activity in Schwann cells in vivo in the myelinating rat sciatic nerve. Peripheral myelination occurs during the first 14 days after birth in rodents, and we found BRG1 highly expressed during the first postnatal week but undetectable in the adult, fully myelinated sciatic nerve (Fig. 1A). Localization of BRG1 to Schwann cells was confirmed by colabeling with Sox10 in postnatal day 5 sciatic nerves (Fig. 1B). When bound to DNA, the ATPase activity of BRG1 is increased; therefore, to determine whether the enzyme was active in the developing sciatic nerve, we used an in vitro ATPase assay and found that BRG1 was most active in early stages of myelination close to birth, but the activity decreased as myelination progressed (Fig. 1C,D). These results suggested that BRG1 is remodeling chromatin during myelination and may have a role in Schwann cell differentiation.
Figure 1.
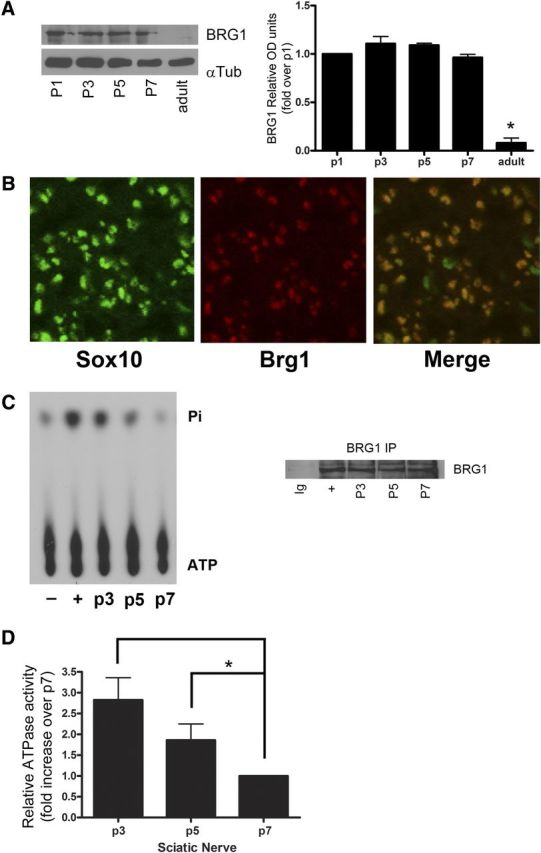
BRG1 is expressed and activated in myelinating rat sciatic nerves. A, Sciatic nerves from rats at postnatal days 1, 3, 5, 7, and adult (∼3–4 months of age) were analyzed by Western blotting for BRG1 and α-tubulin as a loading control. The level of expression was quantified, and the graph indicates BRG1 relative units as compared to postnatal day 1. To determine statistical significance a one-way ANOVA was performed followed by Tukey's post hoc test. B, P5 sciatic nerves were fixed, cryosectioned, and immunostained with antibodies to BRG1 and Sox10. C, BRG1 ATPase activity was measured in P3, P5, and P7 rat sciatic nerve lysates by immunoprecipitation (IP) of BRG1 (right), followed by an in vitro ATPase assay using radiolabeled ATP. The released Pi (inorganic phosphate) was separated from ATP using thin layer chromatography. Positive (+) and negative (−) controls were performed with lysate from E15 rat embryos (Bultman et al., 2005) with or without DNA added to the reaction mixture, respectively. D, The intensity of the signal was quantified by densitometry. Statistical significance was performed by a one-way ANOVA followed by a Tukey's post hoc test (n = 3).
BRG1 is required for Schwann cell differentiation in vitro
BRG1-mediated chromatin remodeling is essential for differentiation in several cell types (Zhao et al., 1998; de la Serna et al., 2001; Gebuhr et al., 2003; Wu et al., 2007); therefore, we investigated whether BRG1 is necessary for the differentiation of Schwann cells into a myelinating phenotype. BRG1 expression was silenced by siRNA knockdown in cultured Schwann cells, which were then stimulated to undergo differentiation with dibutyryl cAMP. To assess differentiation, we measured the level of Oct6, which is upregulated during myelination and in response to cAMP. In cells lacking BRG1, there was a significant inhibition of Oct6 induction relative to a control siRNA (Fig. 2A,B). These results indicated that BRG1 is indeed involved in regulating the differentiation of Schwann cells and suggested that it may have a role in myelin formation.
Figure 2.
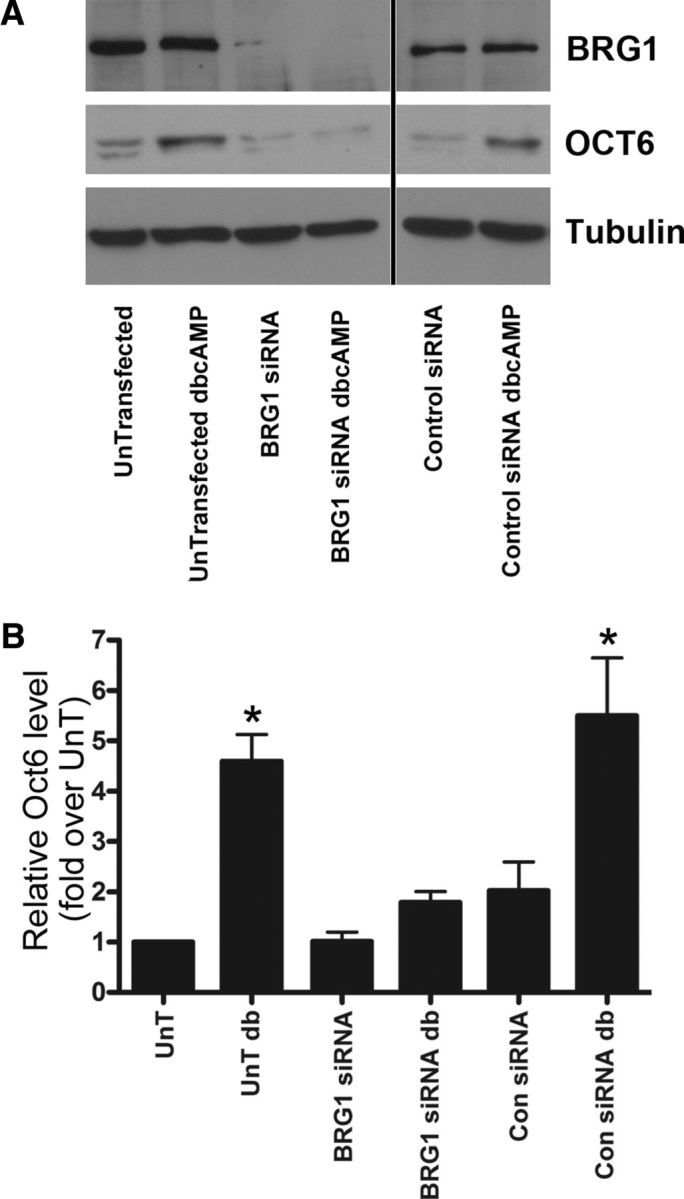
BRG1 is required for Schwann cell differentiation in response to elevated cAMP. A, Primary rat Schwann cells were transfected with siRNA to BRG1 or control siRNA overnight and then stimulated with 500 μm dbCAMP for 72 h. Cells were lysed and analyzed by Western blotting for BRG1, OCT6, and α-tubulin. (The image shown is from the same blot, but the order of the lanes was altered for the figure, as indicated by the line.) B, Quantitative analysis of A was performed using Scion image quant, and the results were normalized to the untransfected signal (UnT) in each experiment. A one-way ANOVA was used to calculate significance, followed by a Tukey's post hoc test (n = 4). Con, Control; db, dibutyryl.
Conditional deletion of BRG1 results in a dramatic deficit in myelin
To further evaluate the requirement for BRG1 in Schwann cell development, we bred mice homozygous for a floxed BRG1 allele (Olave et al., 2002) with mice expressing Cre recombinase under the desert hedgehog (Dhh) promoter, which is expressed almost exclusively in Schwann cell precursors starting around E12.5 (Joseph et al., 2004). Brg1flox/flox; Dhh-Cre (hereafter referred to as cko or conditional knockout) mice were viable at birth but appeared to have severe motor defects such as severe tremors, ataxia, and hindlimb weakness. By P18 the conditional knockout mice exhibited severe weakness and difficulty in movement, and by 3 weeks of age most had died despite access to moistened food (Fig. 3A). The sciatic nerves of BRG1 cko mice were notably thinner and transparent as compared to that of wild-type (wt) animals at P11, suggesting a severe deficiency in myelin (Fig. 3B). There were no defects detected in either control heterozygous mutants (Brg1flox/+;Dhh-Cre) or Dhh-Cre or the Brg1flox/flox mice. Morphological analysis of these nerves by electron microscopy (EM) revealed virtually a complete absence of myelin (Fig. 3c). In cross-sections from the nerves of postnatal day 7 and 18 pups there were no myelin profiles detected. In addition, although a few Schwann cells were able to sort the axons, many of the Schwann cells in the cko mice were associated with large axon bundles, especially at P18, indicating a failure to segregate axons and form a 1:1 relationship (Fig. 3C). This result suggests that the Schwann cells lacking BRG1 are blocked at an early stage in differentiation, before radial sorting.
Figure 3.
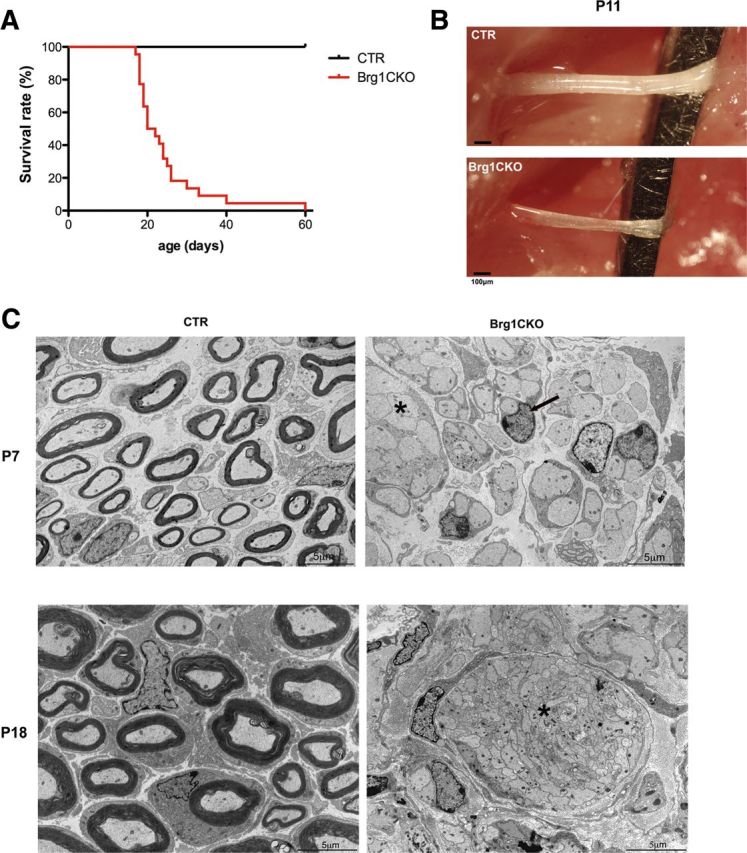
BRG1 cko mice display severe hypomyelination. A, A Kaplan–Meier survival curve indicating the survival of wild-type (CTR, control) or Brg1F/F;Dhh-Cre (Brg1CKO) mice (n = 31). B, Image of wild-type (CTR) and BRG conditional knockout (Brg1CKO) sciatic nerves at p11. C, Electron micrographs of cross sections of control, Brg1F/+;Dhh-Cre (CTR), and Brg1F/F;Dhh-Cre (Brg1CKO) nerves at P7 (top) and P18 (bottom). The asterisks indicate atypical bundles of axons, and the arrow points out a single Schwann cell surrounding an unmyelinated axon. Note the complete lack of myelin profiles in the cko nerves. Magnification, 4200×.
Conditional deletion of BRG1 results in the loss in the expression of promyelinating transcription factors
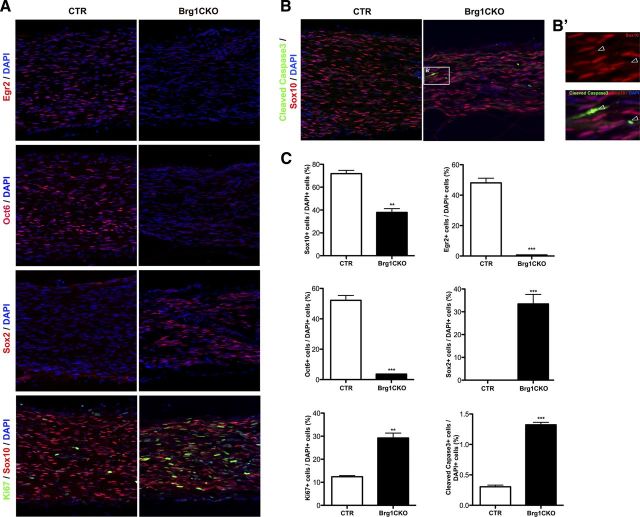
To further establish that deletion of BRG1 prevented Schwann cell differentiation, postnatal day 7 sciatic nerves were analyzed for the expression of the promyelinating transcription factors Oct6, EGR2/Krox20, and Sox10. In the sciatic nerve of mutant mice, Oct6+ promyelinating and EGR2/Krox20+ mature Schwann cells are barely detectable; however, immature Schwann cells (Sox2+) persist (Fig. 4A). This data indicated that the lack of BRG1 was preventing immature Schwann cells from differentiating into promyelinating cells. We also detected a significant reduction in Sox10+ cells (Fig. 4A,C), which is interesting since Sox10 is turned on as neural crest commit to a glial lineage and continues to be expressed in both myelinating and nonmyelinating cells (Kuhlbrodt et al., 1998). Therefore, a reduction in Sox10 expression is not indicative of the cell stalling at a particular stage of differentiation, but may reflect an ability of BRG1 to more directly regulate Sox10 expression.
Figure 4.
Conditional deletion of BRG1 results in the loss in the expression of promyelinating transcription factors. A, Control, Brg1F/+;Dhh-Cre, (CTR, control), and Brg1F/F;Dhh-Cre (BRG1CKO) sciatic nerves from P7 were fixed, cryosectioned, and immunostained for EGR2/Krox20, Oct6, or Sox2 (red) or Sox10 (red) and Ki67 (green). The nuclei were labeled with Topro3 (blue). B, Cryosections were immunostained with antibodies to the cleaved form of caspase 3 (green) and Sox10 (red), and the nuclei were labeled with Topro3 (blue). The boxed area is depicted enlarged on the right (B′), where a couple of Sox10+ cells with activated caspase 3 are indicated by the arrows. (Note that some of the caspase positive cells were weakly Sox10 positive, possibly due to degradation of Sox10 during apoptosis.) C, The fraction of Topro3-labeled cells that were positive for each of the indicated markers was quantified, and a Student's t test was used to calculate significance. Mean ± SEM is shown (n = 3; *p < 0.05).
Although there was no obvious loss of Schwann cells in the EM images, the myelin defect in the BRG1 ckos could be also be caused by cell death or a failure of the precursors to proliferate. However, in nerves from mice lacking BRG1 there was actually a slight increase in proliferation coupled with a modest increase in apoptosis (Fig. 4A–C). These data suggest that in the absence of BRG1 there is a slight increase in proliferating precursors, but the excess cells ultimately undergo cell death. Together, these results reveal an essential role for the BRG1 chromatin remodeling factor in the differentiation of Schwann cell precursors to a myelinating phenotype.
BRG1 and NF-κB form a DNA binding complex in myelinating cells
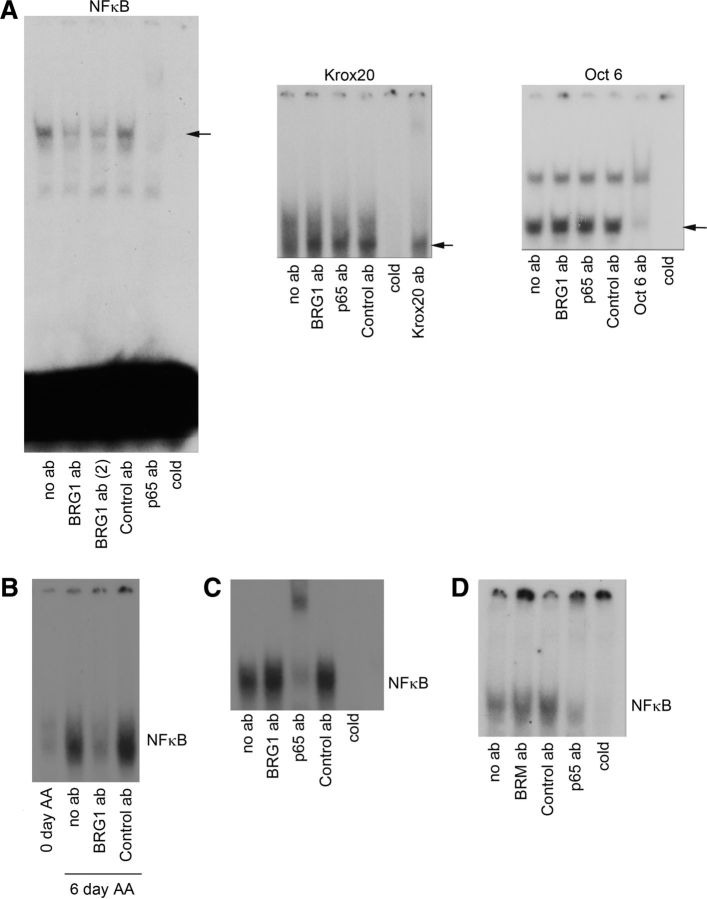
Chromatin remodeling enzymes typically associate with DNA through interaction with specific transcription factors; therefore, we investigated a potential association between BRG1 and a number of promyelinating transcription factors using electrophoretic mobility shift assays or EMSAs. Using extracts from early postnatal sciatic nerves and DNA probes specific for Oct6, EGR2/Krox20, or NF-κB, we detected DNA binding by all three factors as expected (Fig. 5A). The potential presence of BRG1 in the DNA-protein complex was assessed by addition of an antibody to the ATPase, which would result in a supershift or diminished signal due to reduced stability of the protein-DNA complex. The anti-BRG1 had no effect on the Oct6 or EGR2/Krox20 DNA binding; however, it markedly reduced the NF-κB signal, indicating its presence in the complex (Fig. 5A). To confirm this result, we tested another, independent antibody to BRG1 and observed a similar result (Fig. 5A). An antibody to the p65 subunit of NF-κB served as a positive control since this subunit is involved in the complex activated during myelination (Nickols et al., 2003).
Figure 5.
BRG1 and NF-κB form a DNA binding complex in myelinating cells. A, P2 or P5 mouse sciatic nerve lysates were analyzed for protein binding to DNA using a consensus sequence for NF-κB in an EMSA. To identify the proteins in the complex bound to the NF-κB consensus sites, specific antibodies (ab) to BRG1 (two different antibodies were used, as indicated), p65 or a control antibody, were added to the binding reaction. The complex and unbound DNA were then separated by gel electrophoresis. Excess unlabeled NF-κB consensus DNA (cold) was added as a control (n = 3). Note the reduction in the binding complex caused by either BRG1 antibody and the antibody to p65. Similar EMSAs were performed on nerve lysates using consensus sequences for EGR2/Krox20 and Oct6, as indicated. B, EMSA analysis using an NF-κB probe was performed on lysates from DRG/Schwann cell cocultures either before stimulation with ascorbic acid (AA) to induce myelination or after 6 days of AA treatment in the presence or absence of antibodies to BRG1 or control (n = 3). C, HEK 293 cells were treated with TNFα for 1 h to stimulate NF-κB and then lysed, and the lysates were subjected to an EMSA using an NF-κB consensus sequence. In contrast to experiments in myelinating cells, addition of antibodies to BRG1 had no effect on the protein–DNA complex. D, P2 rat sciatic nerve lysate was analyzed by EMSA using a NF-κB probe. Supershift analysis was used to identify DNA-bound proteins. Addition of antibodies to BRM or control had no effect on protein-DNA binding. Unlabeled probe (cold) was added as a control.
To further explore the formation of a BRG1-NF-κB complex during myelination, we cocultured dorsal root ganglia neurons and Schwann cells. NF-κB is not activated in these cultures until ascorbic acid is added to stimulate myelination (Nickols et al., 2003). After 6 days in ascorbic acid, NF-κB binding to DNA was detected in an EMSA assay (Fig. 5B). Similar to the extracts from neonatal sciatic nerves, addition of an antibody to BRG1 disrupted NF-κB binding to DNA, indicating that these proteins are in a complex.
To determine whether BRG1 generally associates with NF-κB under conditions that lead to its activation, or if the association occurs only under certain conditions, such as during Schwann cell differentiation, HEK 293 cells were treated with TNFα, which stimulates NF-κB binding. However, addition of the BRG1 antibody did not diminish the complex induced by the cytokine (Fig. 5C), indicating that the NF-κB–BRG1 interaction is unique to specific conditions, including Schwann cell differentiation. In addition, we assessed whether BRM, the BRG1 homolog, also associates with NF-κB in Schwann cells, since this ATPase is expressed in these glial cells (data not shown). However, no BRM–NF-κB complex was detected (Fig. 5D), suggesting that this chromatin remodeler does not interact with NF-κB during myelination.
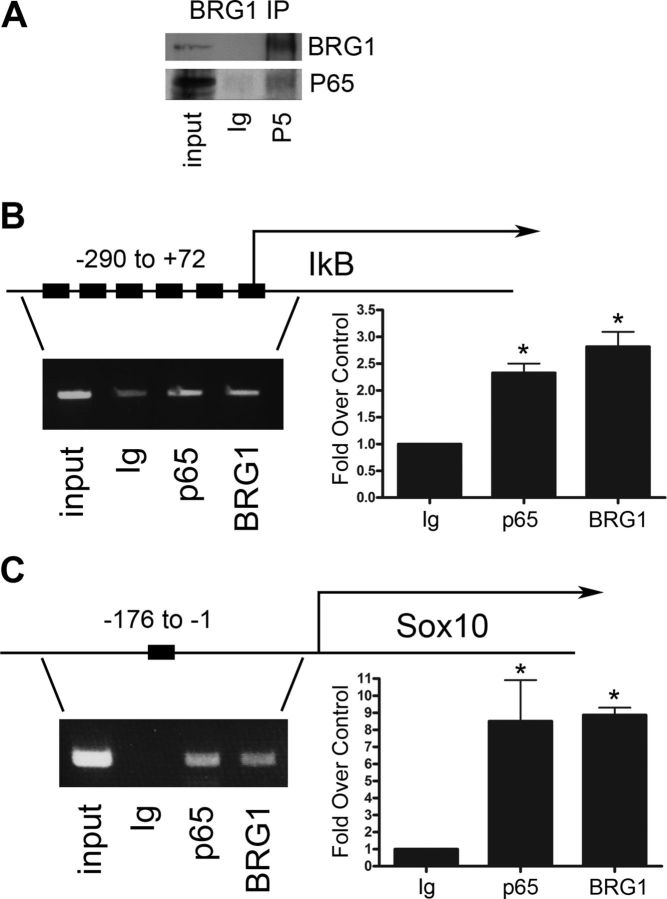
Since our EMSA data indicated that BRG1 participates in a DNA binding complex with NF-κB, we next tested whether an interaction between BRG1 and the p65 subunit of NF-κB could be detected during myelination by coimmunoprecipitation. Using postnatal day 5 sciatic nerves, the time when BRG1 ATPase was activated (Fig. 1C), we detected an interaction between these proteins (Fig. 6A), confirming that BRG1 and NF-κB form a complex during myelin formation.
Figure 6.
BRG1 and NF-κB complex at NF-κB-DNA binding sites in myelinating cells. A, BRG1 was immunoprecipitated from P5 rat sciatic nerve. Proteins immunoprecipitated by BRG1 antibodies or control IgG were analyzed by Western blotting using antibodies for either the p65 subunit of NF-κB or BRG1. B, The binding of the p65 subunit of NF-κB to the region 5′ of the IκBα gene was analyzed by ChIP. The DNA locus is diagramed with solid boxes indicating the known NF-κB binding sites. The nerves were fixed, lysed, sonicated, and immunoprecipitated with antibodies to BRG1, p65, or a nonspecific IgG. The cross-linking was then reversed, the proteins were digested with proteinase K, and the remaining DNA fragments were subjected to PCR with primers flanking the NF-κB sites in the IκBα promoter region. A sample of the PCR is shown. Quantification, displayed as fold over Ig is shown on the right (n = 3). C, ChIP assays were performed on P5 mouse sciatic nerve using PCR primers to the NF-κB site in the Sox10 promoter region (Chen et al., 2011), (n = 3). The NF-κB binding site at −1 to −176 of the Sox10 gene is the solid box in the diagram. A sample PCR is shown, and the quantification relative to the control IgG antibody is shown on the right (n = 3). A one-way ANOVA was used to calculate significance, followed by Tukey's post hoc test. The mean ± SEM is shown (*p < 0.05).
Knowing that BRG1 is necessary for myelin formation and that it can form an active DNA-binding complex with NF-κB, we next hypothesized that BRG1 is localized to known NF-κB target genes. One of the best characterized targets of NF-κB is its own inhibitor, IκBα (Sun et al., 1993); therefore, we tested for the presence of NF-κB and BRG1 at the IκBα promoter during myelination by ChIP using extracts of P5 sciatic nerves. We detected binding of both BRG1 and the p65 subunit of NF-κB 5′ of the IκBα gene, where there are six known NF-κB binding sites (Fig. 6B). In addition, we assessed the Sox10 promoter, since we recently identified this gene as a target of NF-κB during Schwann cell differentiation (Chen et al., 2011), and we found that Sox10 expression was significantly reduced in the nerves of the BRG1 cko mice (Fig. 4). We were able to ChIP both the p65 subunit of NF-κB and BRG1 to the same region in the Sox10 promoter (Fig. 6C). These results suggest that BRG1 and NF-κB form a functional complex on genes involved in myelination.
Neuregulin 1 type III induces the association of BRG1 with NF-κB and increases BRG1 ATPase activity
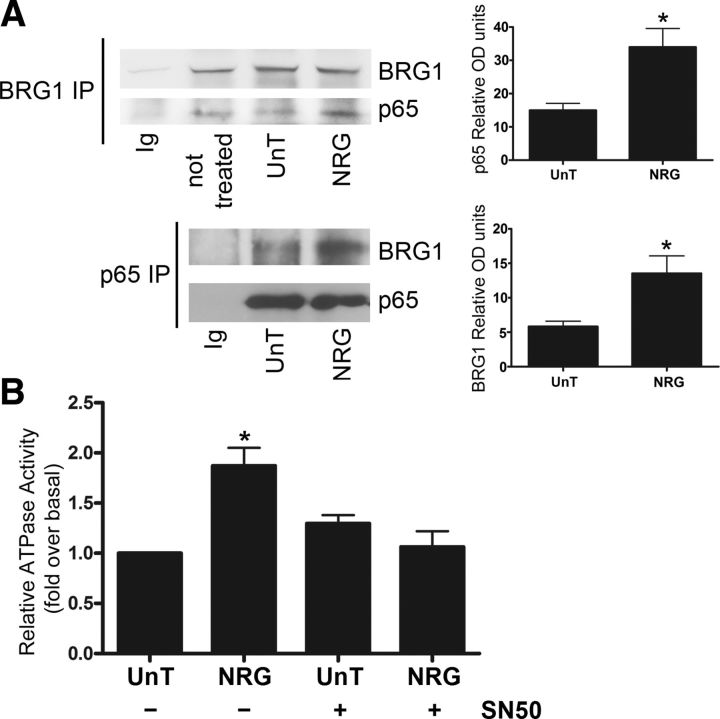
We previously reported that NF-κB binding to DNA is stimulated by neuregulin (NRG1) type III (Limpert and Carter, 2010), a transmembrane ligand expressed by axons that is essential for peripheral myelin formation (Taveggia et al., 2005). Therefore, we hypothesized that NRG1 type III could promote the interaction between NF-κB and BRG1. To test this hypothesis, primary rat Schwann cells were treated with membranes isolated from untransfected cells or those transfected with NRG1 type III. After treatment, either BRG1 or p65 was immunoprecipitated and the precipitates were probed for both BRG1 and p65. We discovered that neuregulin significantly increased the p65 and BRG1 interaction (Fig. 7A), indicating that that this axonal signal triggers the formation of an NF-κB-BRG1 chromatin remodeling complex.
Figure 7.
Neuregulin I (NRG1) type III stimulates the association of BRG1 with NF-κB and elevates BRG1 ATPase activity. A, Isolated rat Schwann cells were left untreated or treated with either untransfected cell membranes (UnT) or cell membranes isolated from HEK 293 cells transfected with NRG1 type III (NRG). Following 1 h of membrane treatment, the Schwann cells were rinsed, lysed, and subjected to immunoprecipitation with BRG1 or p65 antibodies or with an IgG control (Ig). Immunoprecipitates were analyzed by Western blotting using anti-BRG1 or anti-p65. Quantification of the Western blots is shown to the right. A two-tailed Student's t test was used to calculate significance. The mean ± SEM is shown (n = 3; *p < 0.05). B, Rat Schwann cells were treated for 1 h with cell membranes from untransfected HEK 293 cells (UnT) or cells transfected with NRG1 type III (NRG). Some Schwann cells were pretreated with SN50 (50 μg/ml) to inhibit NF-κB activity. The Schwann cells were then lysed, BRG1 protein was immunoprecipitated, and the precipitates were subjected to in vitro ATPase assays. The amount of released Pi (inorganic phosphate) was quantified and normalized to the ATPase activity of the Schwann cells that were treated with untransfected cell membranes. A one-way ANOVA was used to calculate significance, followed by Tukey's post hoc test. The mean ± SEM is shown (n = 4; *p < 0.05).
BRG1 hydrolyzes ATP to shuffle nucleosomes and remodel chromatin upon being recruited to DNA (Hargreaves and Crabtree, 2011); therefore, we hypothesized that an axonal signal that induced a BRG1-NF-κB complex would also stimulate its enzymatic activity. To test this, Schwann cells were treated with membranes expressing NRG1 type III, and ATP hydrolysis by BRG1 was measured. Neuregulin induced approximately a twofold increase in the ATPase activity of BRG1 relative to the control, indicating activation of the chromatin remodeling complex by this ligand (Fig. 7B). To determine whether NF-κB DNA binding was required for the activity of BRG,1 we pretreated the Schwann cells with the NF-κB peptide inhibitor SN50 and analyzed BRG1 activation following the addition of NRG1 type III-expressing membranes. The inhibition of NF-κB led to a reduction in NRG1 stimulated BRG1 activity (Fig. 7B). These results demonstrate that neuregulin activates the ATPase of BRG1 in Schwann cells in an NF-κB-dependent manner.
NF-κB is required for BRG1 activation in myelinating cells
To determine whether NF-κB binding to DNA, independent of upstream activators, is sufficient to stimulate BRG1, the p65 subunit of NF-κB was overexpressed in Schwann cells to induce DNA binding (Yoon et al., 2008), and BRG1 ATPase activity was analyzed. Rat Schwann cells overexpressing p65 displayed a 2.24 ± 0.49-fold increase in BRG1 activity as compared to cells transfected with a control plasmid (data not shown), indicating that p65 not only interacts with BRG1 but also affects its ability to remodel chromatin.
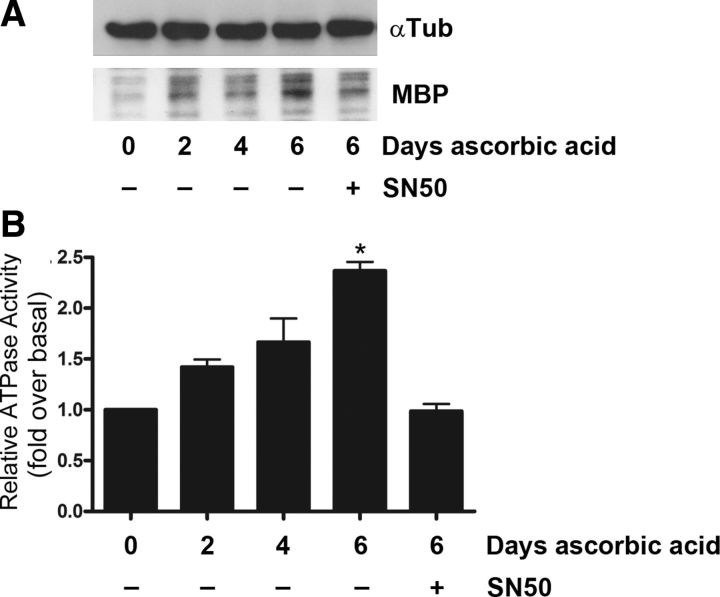
We further hypothesized that BRG1 activity during myelin formation would require the activation of NF-κB. To test this hypothesis, we used an in vitro myelination assay where Schwann cells were cultured with DRG neurons and myelination was induced by the addition of ascorbic acid. BRG1 activity increased following 6 days of ascorbic acid treatment (Fig. 8B). Previous analysis of NF-κB activation in this culture system demonstrated similar kinetics for the increase in NF-κB activity (Nickols et al., 2003). Inhibition of NF-κB resulted in decreased myelin formation (Fig. 8A), as reported previously (Nickols et al., 2003), and importantly, a significant reduction in BRG1 activity (Fig. 8B). These results suggest that NF-κB is required for the activation of BRG1 in myelinating cells.
Figure 8.
NF-κB is required for BRG1 activation in myelinating cells. A, DRG/Schwann cell cocultures were stimulated for 6 days with ascorbic acid to induce myelination. Lysates were collected every two days. The NF-κB inhibitor SN50 (50 μg/ml) was added to some cultures for 48 h before cell lysis at the 6 day time point. Cell lysates were Western blotted for myelin basic protein (MBP) and tubulin (αTub). B, Lysates from myelinating DRG/Schwann cells were collected every 2 days. In some cultures, SN50 (50 μg/ml) was added for 48 h before cell lysis at 6 days. The cell lysates were immunoprecipitated with BRG1 antibodies, subjected to ATPase assays, and the amount of released Pi (inorganic phosphate) was quantified and normalized to the activity at day 0 before stimulating myelination by ascorbic acid addition. A one-way ANOVA used to calculate significance, followed by Tukey's post hoc test. The mean ± SEM is shown (n = 3; *p < 0.05).
Discussion
Cellular differentiation requires the complex orchestration of a wide variety of genes, often activated in response to common signaling pathways. Cellular specificity in the expression profile is gained through the use of selective transcription factors as well as epigenetic mechanisms; of particular relevance here, the regulation of chromatin compaction. Transcription factors are unable to bind to promoters and facilitate the transcription of genes that are occluded by nucleosomes. Two mechanisms for modifying the interaction of DNA with histone proteins have been described. First, histone modifying enzymes, such as histone deacetylases (HDACs) and histone acetyltransferases (HATs), can alter the strength of the association between histones and DNA. Second, chromatin remodeling enzymes can use ATP hydrolysis to reposition nucleosomes. In this study, we demonstrated that chromatin remodeling is essential for myelination through the identification of BRG1 as a required factor for Schwann cell differentiation. Furthermore, we elucidated a mechanism by which the axonal signal neuregulin 1 type III stimulates BRG1 activation through an interaction with the transcription factor NF-κB. This study is the first to identify a pathway from extrinsic signals to chromatin remodeling required for the differentiation of myelinating Schwann cells.
Previous studies revealed a key role for HDACs in myelinating cells. Inhibition of HDAC activity in progenitor cells of the CNS blocked the differentiation of these cells into oligodendrocytes, the myelin-forming cells of the CNS (Marin-Husstege et al., 2002). Similarly, conditional ablation of HDAC 1 and 2 in Schwann cells inhibited Schwann cell development and myelin formation (Chen et al., 2011; Jacob et al., 2011). The Schwann cells in the double-null animals appeared to be arrested at an early stage before axonal segregation and upregulation of Oct6, an early marker of the myelinating lineage; however, there is some controversy as to whether Oct6 expression is increased, suggesting the cells may arrest a bit later, at the promyelinating stage (Chen et al., 2011; Jacob et al., 2011). In the BRG1-null nerves we observed a similar phenotype, with Schwann cells failing to upregulate Oct6 (Fig. 4A) and sort axons (Fig. 3), indicating that the cells remained at the precursor stage, before commitment to the myelinating lineage. Interestingly, an additional role for HDAC 1 and 2 in Schwann cell differentiation, beyond modifying histones, has been shown. Both HDACs were able to associate with and deacetylate the p65 subunit of NF-κB, which was necessary for its recruitment to the Sox10 promoter and the activation of transcription (Chen et al., 2011). Taken together with our results, these findings suggest that NF-κB may be a nodal point for multiple levels of epigenetic regulation, including histone modification and chromatin compaction.
Unlike HDAC 1/2 cko mice, which did not exhibit any change in Schwann cell proliferation (Chen et al., 2011), BRG1 cko mice displayed an increase in Ki67 labeling. These data are consistent with a role for BRG1 in cell cycle arrest. Cellular differentiation is comprised of two components, withdrawal from the cell cycle and expression of lineage-specific genes. Previous studies demonstrated that BRG1 has the capacity to promote both events. In addition to promoting differentiation in some cell types (Zhao et al., 1998; de la Serna et al., 2001; Gebuhr et al., 2003; Wu et al., 2007), BRG1 was shown to suppress the expression of the cell cycle proteins cyclin A, D1, and E (Giacinti and Giordano, 2006) and was required for the induction of the Cip/Kip family of Cdk inhibitors in cell lines, leading to growth arrest (Hendricks et al., 2004). During Schwann cell differentiation early glial precursor cells proliferate, and as the cells begin expressing genes associated with the Schwann cell lineage and myelin formation, including Oct6 and the lipid antigen O4, they become promyelinating. Cell division peaks in the promyelinating stage, when Notch expression reaches its maximum (Woodhoo et al., 2009); but then the cells begin upregulating genes such as the cyclin-dependent kinase inhibitor p27 (Tikoo et al., 2000; Parkinson et al., 2004), leading to withdrawal from the cell cycle. The cells also radially sort axons and establish a 1:1 relationship. Eventually, the Schwann cells fully transition out of the cell cycle as they initiate myelination, begin wrapping axons, and express early myelin-specific genes such as P0 (Stewart et al., 1993). The increase in proliferating cells and the failure to segregate axons, along with reduced expression of the promyelinating gene Oct6 in the Brg1−/− nerves, are all consistent with this chromatin remodeler being important for cell cycle withdrawal, in addition to regulating the expression of genes required for transition to the promyelinating stage. The excess in proliferating precursor cells in the Brg1−/− nerves appears to be eliminated through apoptosis, as there was a corresponding increase in cells with activated Caspase 3 at this stage.
Chromatin remodeling through BRG1 complexing with lineage specifying transcription factors has been demonstrated to occur during the differentiation of several cell types. For example, during myogenesis BRG1 binds the transcription factors MyoD and MEF2, which target the chromatin remodeling complex to early genes essential in skeletal muscle differentiation, such as myogenin (de la Serna et al., 2001; de la Serna et al., 2005; Ohkawa et al., 2006). Ironically, these transcription factors not only target the BRG1 complex to DNA sequences, but they also are dependent on BRG1 for DNA binding, since in the absence of BRG1, MyoD, and MEF2 were unable to bind to the myogenin promoter (de la Serna et al., 2005). Neuronal differentiation also requires cooperation between neuron-specific transcription factors and chromatin remodeling enzymes. In neuronal precursors, BRG1 is associated with the NRSF/ REST inhibitor of neuronal gene expression; however; during differentiation this association is lost and the Ca2+ responsive coactivator, CREST, is recruited to the BRG1-containing chromatin remodeling complex (Wu et al., 2007). These findings suggest that BRG1 may be targeted to chromatin by association with cell lineage-specific transcription factors, thereby facilitating the chromatin remodeling that is required for the differentiation of multiple cell types.
In differentiating Schwann cells, we propose a model whereby the myelination program is initiated by axonal signals, including membrane-bound NRG1 type III, which stimulate an interaction between the transcription factor NF-κB and the chromatin remodeling ATPase BRG1. This interaction elevates the activity of BRG1, potentially by bringing it in contact with DNA binding sites where it facilitates chromatin rearrangement. In this study, we have identified the promyelinating transcription factor NF-κB, specifically the p65 subunit, as an interacting partner of BRG1. This interaction was found at p65 DNA binding sites, in particular, at the Sox10 promoter, which was previously identified as an NF-κB target gene (Chen et al., 2011) and is required for myelin formation (Finzsch et al., 2010) and maintenance (Bremer et al., 2011). We also observed a reduction in Sox10 immunostaining in the Brg1−/− nerves, consistent with BRG1 regulating the expression of this gene (Fig. 4A,C). However, Sox10 expression is not completely ablated in concordance with its role as a Schwann cell lineage marker that is expressed in Schwann cell precursors, immature (Sox2+) and promyelinating Schwann cells (Oct6+), and even persists in the myelinating (Egr2/Krox20+) and nonmyelinating Schwann cells of the adult PNS. Due to the multiple essential roles of this transcription factor in Schwann cell development, its expression is likely to be controlled by multiple transcriptional mechanisms in vivo.
The interaction between BRG1 and NF-κB was stimulated by the axonal ligand neuregulin 1 type III, which initiates Schwann cell differentiation and activates both NF-κB (Limpert and Carter, 2010) as well as BRG1 ATPase activity (Fig. 7B). Recently, NRG1 has been identified as the upstream activator of several transcription factors required for myelination, including YY1 (He et al., 2010) and NFAT (Kao et al., 2009). It will be interesting in future studies to determine the sequential interaction among YY1, NFAT, and NF-κB and whether they all associate with chromatin remodeling complexes.
While this manuscript was under review, Weider et al. (2012) reported an identical phenotype in mice with BRG1 conditionally deleted in Schwann cells. Interestingly, these authors identified an association between BRG1 and Sox10. Hence, there may be a positive feedback loop with a BRG1-NF-κB complex upregulating Sox10, which also interacts with BRG1 to regulate Schwann cell differentiation.
Footnotes
This work was supported by National Institutes of Health Grants NS048249 and NS064278 (to B.D.C.), NS072427 and NS075243 (to Q.R.L.), and NS039472 (to S.O.Y.), and Muscular Dystrophy Association Development Grant (MDA4023) (to A.S.L.).
The authors declare no competing financial interests.
References
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham JR, Jr, Scherer SS, O'Connell S, Arroyo E, Kalla KA, Powell FL, Rosenfeld MG. Tst-1/Oct-6/SCIP regulates a unique step in peripheral myelination and is required for normal respiration. Genes Dev. 1996;10:1751–1762. doi: 10.1101/gad.10.14.1751. [DOI] [PubMed] [Google Scholar]
- Bremer M, Fröb F, Kichko T, Reeh P, Tamm ER, Suter U, Wegner M. Sox10 is required for Schwann-cell homeostasis and myelin maintenance in the adult peripheral nerve. Glia. 2011;59:1022–1032. doi: 10.1002/glia.21173. [DOI] [PubMed] [Google Scholar]
- Britsch S, Goerich DE, Riethmacher D, Peirano RI, Rossner M, Nave KA, Birchmeier C, Wegner M. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 2001;15:66–78. doi: 10.1101/gad.186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman SJ, Gebuhr TC, Magnuson T. A Brg1 mutation that uncouples ATPase activity from chromatin remodeling reveals an essential role for SWI/SNF-related complexes in β-globin expression and erythroid development. Genes Dev. 2005;19:2849–2861. doi: 10.1101/gad.1364105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wang H, Yoon SO, Xu X, Hottiger MO, Svaren J, Nave KA, Kim HA, Olson EN, Lu QR. HDAC-mediated deacetylation of NF-kappaB is critical for Schwann cell myelination. Nat Neurosci. 2011;14:437–441. doi: 10.1038/nn.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna IL, Carlson KA, Imbalzano AN. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat Genet. 2001;27:187–190. doi: 10.1038/84826. [DOI] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Berkes CA, Bergstrom DA, Dacwag CS, Tapscott SJ, Imbalzano AN. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol Cell Biol. 2005;25:3997–4009. doi: 10.1128/MCB.25.10.3997-4009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat Rev Genet. 2006;7:461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- Edgar JM, Garbern J. The myelinated axon is dependent on the myelinating cell for support and maintenance: molecules involved. J Neurosci Res. 2004;76:593–598. doi: 10.1002/jnr.20063. [DOI] [PubMed] [Google Scholar]
- Finzsch M, Schreiner S, Kichko T, Reeh P, Tamm ER, Bösl MR, Meijer D, Wegner M. Sox10 is required for Schwann cell identity and progression beyond the immature Schwann cell stage. J Cell Biol. 2010;189:701–712. doi: 10.1083/jcb.200912142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebuhr TC, Kovalev GI, Bultman S, Godfrey V, Su L, Magnuson T. The role of Brg1, a catalytic subunit of mammalian chromatin-remodeling complexes, in T cell development. J Exp Med. 2003;198:1937–1949. doi: 10.1084/jem.20030714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazvini M, Mandemakers W, Jaegle M, Piirsoo M, Driegen S, Koutsourakis M, Smit X, Grosveld F, Meijer D. A cell type-specific allele of the POU gene Oct-6 reveals Schwann cell autonomous function in nerve development and regeneration. EMBO J. 2002;21:4612–4620. doi: 10.1093/emboj/cdf475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain J, Charnay P. Control of myelination in Schwann cells: a Krox20 cis-regulatory element integrates Oct6, Brn2 and Sox10 activities. EMBO Rep. 2006;7:52–58. doi: 10.1038/sj.embor.7400573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacinti C, Giordano A. RB and cell cycle progression. Oncogene. 2006;25:5220–5227. doi: 10.1038/sj.onc.1209615. [DOI] [PubMed] [Google Scholar]
- Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Kim JY, Dupree J, Tewari A, Melendez-Vasquez C, Svaren J, Casaccia P. Yy1 as a molecular link between neuregulin and transcriptional modulation of peripheral myelination. Nat Neurosci. 2010;13:1472–1480. doi: 10.1038/nn.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks KB, Shanahan F, Lees E. Role for BRG1 in cell cycle control and tumor suppression. Mol Cell Biol. 2004;24:362–376. doi: 10.1128/MCB.24.1.362-376.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob C, Christen CN, Pereira JA, Somandin C, Baggiolini A, Lötscher P, Ozçelik M, Tricaud N, Meijer D, Yamaguchi T, Matthias P, Suter U. HDAC1 and HDAC2 control the transcriptional program of myelination and the survival of Schwann cells. Nat Neurosci. 2011;14:429–436. doi: 10.1038/nn.2762. [DOI] [PubMed] [Google Scholar]
- Joseph NM, Mukouyama YS, Mosher JT, Jaegle M, Crone SA, Dormand EL, Lee KF, Meijer D, Anderson DJ, Morrison SJ. Neural crest stem cells undergo multilineage differentiation in developing peripheral nerves to generate endoneurial fibroblasts in addition to Schwann cells. Development. 2004;131:5599–5612. doi: 10.1242/dev.01429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao SC, Wu H, Xie J, Chang CP, Ranish JA, Graef IA, Crabtree GR. Calcineurin/NFAT signaling is required for neuregulin-regulated Schwann cell differentiation. Science. 2009;323:651–654. doi: 10.1126/science.1166562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M. Sox10, a novel transcriptional modulator in glial cells. J Neurosci. 1998;18:237–250. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc SE, Ward RM, Svaren J. Neuropathy-associated Egr2 mutants disrupt cooperative activation of myelin protein zero by Egr2 and Sox10. Mol Cell Biol. 2007;27:3521–3529. doi: 10.1128/MCB.01689-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Limpert AS, Carter BD. Axonal neuregulin 1 type III activates NF-kappaB in Schwann cells during myelin formation. J Biol Chem. 2010;285:16614–16622. doi: 10.1074/jbc.M109.098780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Husstege M, Muggironi M, Liu A, Casaccia-Bonnefil P. Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J Neurosci. 2002;22:10333–10345. doi: 10.1523/JNEUROSCI.22-23-10333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickols JC, Valentine W, Kanwal S, Carter BD. Activation of the transcription factor NF-kappaB in Schwann cells is required for peripheral myelin formation. Nat Neurosci. 2003;6:161–167. doi: 10.1038/nn995. [DOI] [PubMed] [Google Scholar]
- Ohkawa Y, Marfella CG, Imbalzano AN. Skeletal muscle specification by myogenin and Mef2D via the SWI/SNF ATPase Brg1. EMBO J. 2006;25:490–501. doi: 10.1038/sj.emboj.7600943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olave I, Wang W, Xue Y, Kuo A, Crabtree GR. Identification of a polymorphic, neuron-specific chromatin remodeling complex. Genes Dev. 2002;16:2509–2517. doi: 10.1101/gad.992102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson DB, Bhaskaran A, Droggiti A, Dickinson S, D'Antonio M, Mirsky R, Jessen KR. Krox-20 inhibits Jun-NH2-terminal kinase/c-Jun to control Schwann cell proliferation and death. J Cell Biol. 2004;164:385–394. doi: 10.1083/jcb.200307132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirano RI, Goerich DE, Riethmacher D, Wegner M. Protein zero gene expression is regulated by the glial transcription factor Sox10. Mol Cell Biol. 2000;20:3198–3209. doi: 10.1128/mcb.20.9.3198-3209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner S, Cossais F, Fischer K, Scholz S, Bösl MR, Holtmann B, Sendtner M, Wegner M. Hypomorphic Sox10 alleles reveal novel protein functions and unravel developmental differences in glial lineages. Development. 2007;134:3271–3281. doi: 10.1242/dev.003350. [DOI] [PubMed] [Google Scholar]
- Stewart HJ, Morgan L, Jessen KR, Mirsky R. Changes in DNA synthesis rate in the Schwann cell lineage in vivo are correlated with the precursor–Schwann cell transition and myelination. Eur J Neurosci. 1993;5:1136–1144. doi: 10.1111/j.1460-9568.1993.tb00968.x. [DOI] [PubMed] [Google Scholar]
- Sumi-Ichinose C, Ichinose H, Metzger D, Chambon P. SNF2beta-BRG1 is essential for the viability of F9 murine embryonal carcinoma cells. Mol Cell Biol. 1997;17:5976–5986. doi: 10.1128/mcb.17.10.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SC, Ganchi PA, Ballard DW, Greene WC. NF-kappa B controls expression of inhibitor I kappa B alpha: evidence for an inducible autoregulatory pathway. Science. 1993;259:1912–1915. doi: 10.1126/science.8096091. [DOI] [PubMed] [Google Scholar]
- Svaren J, Meijer D. The molecular machinery of myelin gene transcription in Schwann cells. Glia. 2008;56:1541–1551. doi: 10.1002/glia.20767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Nogales E, Ciferri C. Structure and function of SWI/SNF chromatin remodeling complexes and mechanistic implications for transcription. Prog Biophys Mol Biol. 2010;102:122–128. doi: 10.1016/j.pbiomolbio.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taveggia C, Zanazzi G, Petrylak A, Yano H, Rosenbluth J, Einheber S, Xu X, Esper RM, Loeb JA, Shrager P, Chao MV, Falls DL, Role L, Salzer JL. Neuregulin-1 type III determines the ensheathment fate of axons. Neuron. 2005;47:681–694. doi: 10.1016/j.neuron.2005.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikoo R, Zanazzi G, Shiffman D, Salzer J, Chao MV. Cell cycle control of Schwann cell proliferation: role of cyclin-dependent kinase-2. J Neurosci. 2000;20:4627–4634. doi: 10.1523/JNEUROSCI.20-12-04627.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JY, Miller SJ, Falls DL. The N-terminal region of neuregulin isoforms determines the accumulation of cell surface and released neuregulin ectodomain. J Biol Chem. 2001;276:2841–2851. doi: 10.1074/jbc.M005700200. [DOI] [PubMed] [Google Scholar]
- Warner LE, Svaren J, Milbrandt J, Lupski JR. Functional consequences of mutations in the early growth response 2 gene (EGR2) correlate with severity of human myelinopathies. Hum Mol Genet. 1999;8:1245–1251. doi: 10.1093/hmg/8.7.1245. [DOI] [PubMed] [Google Scholar]
- Weider M, Kuspert M, Bischof M, Vogl MR, Hornig J, Loy K, Kosian T, Muller J, Hillgartner S, Tamm ER, Metzger D, Wegner M. Chromatin-remodeling factor Brg1 is required for Schwann cell differentiation and myelination. Dev Cell. 2012;23:193–201. doi: 10.1016/j.devcel.2012.05.017. [DOI] [PubMed] [Google Scholar]
- Woodhoo A, Alonso MB, Droggiti A, Turmaine M, D'Antonio M, Parkinson DB, Wilton DK, Al-Shawi R, Simons P, Shen J, Guillemot F, Radtke F, Meijer D, Feltri ML, Wrabetz L, Mirsky R, Jessen KR. Notch controls embryonic Schwann cell differentiation, postnatal myelination and adult plasticity. Nat Neurosci. 2009;12:839–847. doi: 10.1038/nn.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Olave IA, Qiu Z, Ghosh A, Graef IA, Crabtree GR. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- Yoon C, Korade Z, Carter BD. Protein kinase A-induced phosphorylation of the p65 subunit of nuclear factor-kappaB promotes Schwann cell differentiation into a myelinating phenotype. J Neurosci. 2008;28:3738–3746. doi: 10.1523/JNEUROSCI.4439-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]