Abstract
The intestinal microbiota have critical roles in immune system and metabolic homeostasis, but they must be tolerated by the host to avoid inflammatory responses that can damage the epithelial barrier separating the host from the luminal contents1-6. Breakdown of this regulation and the resulting inappropriate immune response to commensals are thought to lead to the development of inflammatory bowel diseases (IBDs) such as Crohn's disease and ulcerative colitis7. We hypothesized that the intestinal immune system is instructed by the microbiota to limit responses to luminal antigens. We demonstrate that, at steady state, the microbiota inhibit the transport of both commensal and pathogenic bacteria from the lumen to a key immune inductive site, the mesenteric lymph node (MLN). However, in the absence of Myd88 or under conditions of antibiotic-induced dysbiosis, non-invasive bacteria trafficked to the MLN in a CCR7-dependent manner and induced both T cell responses and IgA production. Trafficking was carried out by CX3CR1hi mononuclear phagocytes, an intestinal cell population previously reported to be non-migratory8. These findings define a central role for commensals in regulating the migration to the MLN of CX3CR1hi mononuclear phagocytes endowed with the ability to capture luminal bacteria, thereby compartmentalizing the intestinal immune response to avoid inflammation.
How the intestinal immune system discriminates between commensal and pathogenic microorganisms remains an enigma. Commensals display the same immunostimulatory molecules as pathogenic bacteria and have been shown to trigger inflammation and disease if they penetrate the intestinal epithelial barrier. Host sensing of commensals has been shown to be important for the proper development and functionality of the immune system, as germ-free mice have an altered immune system organization and reduced cellularity, especially in the small intestinal lamina propria, compared to mice harboring a complex microbiota (reviewed in REF9). This feature of host immune system dependence on the microbiota led us to examine whether commensal bacteria have a role in modulating mucosal immune responses to specific microbes. To this end, we investigated the consequence of antibiotic-mediated depletion of the intestinal microbiota5 (Figure S1a) on the host response to a non-invasive strain of Salmonella enterica serovar Typhimurium. We utilized this strain because, while it has limited CD18-dependent access to the blood and spleen, it cannot cross the epithelium overlying intestinal lymphoid tissues, and hence does not reach the Peyer's patches10,11. We were thus able to investigate a potential role for commensals in regulating access of the non-invasive Salmonella to the lamina propria-draining lymphatics that traffic luminal contents to the MLN, a site of immune system induction.
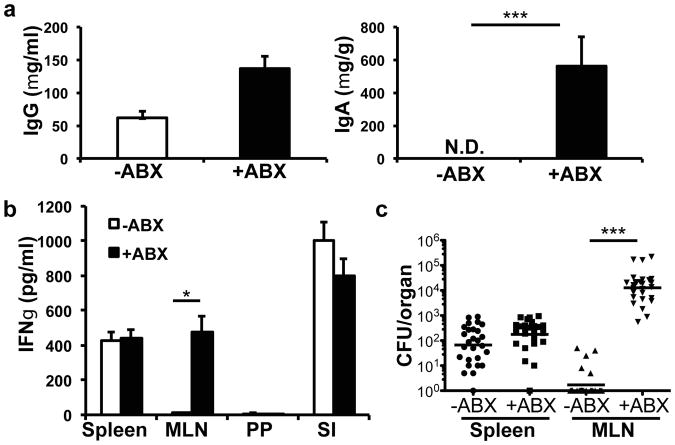
Oral infection with non-invasive Salmonella normally induces an IgG but not an IgA response10 (Figure 1a). Unexpectedly, after treatment with antibiotics, infected mice mounted a strong Salmonella-specific fecal IgA response in addition to an IgG response (Figure 1a). In untreated mice, Salmonella-specific T cells were limited to the spleen and small intestinal lamina propria, but antibiotic-treated mice additionally had antigen-specific T cells in the MLN (Figures 1b, S1b). This was a Salmonella-specific response, as there was no difference in IFNγ-producing T cells following anti-CD3 stimulation and there was little IFNγ production from MLN T cells isolated from mice that did not receive antigenic stimulation or that were Salmonella-naive (Figure S1b and data not shown). This indicates that immune responses specific for non-invasive Salmonella are only observed in the lamina propria-draining MLN if commensal bacteria are depleted before infection.
Figure 1. Induction of immune response against non-invasive Salmonella after antibiotic treatment.
Mice were left untreated or antibiotic treated for four weeks. (a) Mice were orally infected with non-invasive, non-pathogenic Salmonella (invA/aroA) and Salmonella-specific IgG in the blood and IgA in the feces were measured by ELISA. Bars represent the average from 5 mice from one of 3 independent experiments. ***P<0.001, unpaired t-test. Error bars represent S.E.M. (b) T cells from spleen (SP), mesenteric lymph node (MLN), and small intestine lamina propria (SI) of animals infected with non-invasive Salmonella were cultured with irradiated splenocytes and boiled Salmonella antigen and IFNγ was measured by ELISA. Bars represent three mice per treatment group and are representative of two independent experiments. *P<0.05, unpaired t-test. Error bars represent S.E.M. (c) Bacterial titers in the spleen and MLN were determined for mice infected with non-invasive Salmonella (invA). Data points represent organs from a single mouse; data were pooled from six experiments. ***P<0.0001, Mann-Whitney Test. Error bars represent the geometric mean.
We hypothesized that commensals could influence the amount of antigen (in this case, Salmonella) that reaches the MLN. In untreated mice, infection with both pathogenic and non-pathogenic strains of non-invasive Salmonella resulted in bacteria reaching the spleen, but very few bacteria were observed in the MLN (Figure 1c and S1c). Importantly, after antibiotic treatment, there were high titers of the bacteria in the MLN, as previously reported12, in addition to the spleen (Figure 1c and S1c). As expected, no bacteria were found in the Peyer's patches in either condition (data not shown). The increase in bacteria reaching the MLN was unlikely the result of increased epithelial permeability after treatment with antibiotics, since we did not observe an increase in bacteria reaching the spleen. This was confirmed by using a direct test for intestinal permeability with FITC-dextran gavage (Figure S1d). In addition, the increased Salmonella in the MLN was not a consequence of impaired colonization resistance upon depletion of the microbiota, as there was equivalent expansion of Salmonella in the presence of any individual antibiotic, yet only vancomycin or ampicillin treatment allowed for its trafficking to the MLN and induction of the bacterium-specific IgA (Fig. S2).
Our results raised the question of whether commensal bacteria could gain access to the MLN in the same way as non-invasive Salmonella, allowing for the induction of mucosal-specific immune responses. Antibiotic-treated and untreated mice were infected by oral gavage with a non-invasive, non-pathogenic bacterium, E. coli K-12. In the control mice, the bacteria could not be found in the MLN, nor was there induction of E. coli-specific IgA (Figure S3a and S3b). In contrast, in antibiotic-treated mice E. coli were detected in the MLN and there was induction of a specific IgA response (Figure S3a and S3b). These results indicate that the microbiota function to limit the colonization of the MLN by both commensal and pathogenic bacteria, thereby constraining the induction of intestinal immune responses against both.
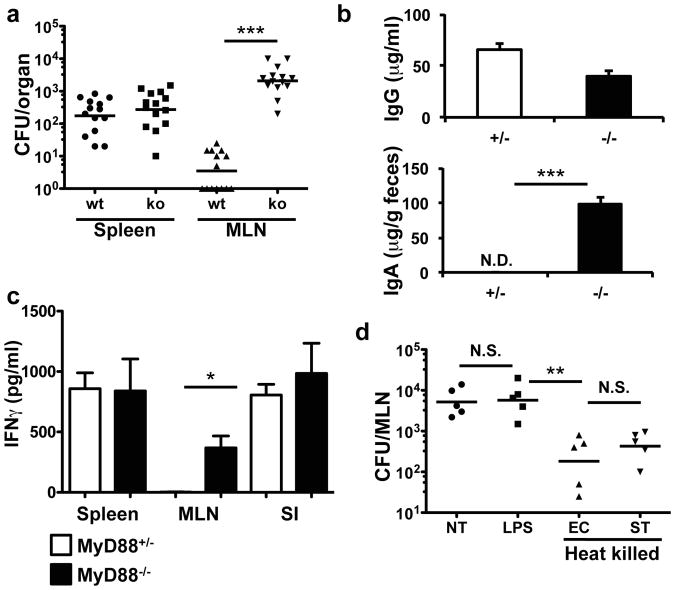
To investigate whether a specific microbial recognition system is involved in microbiota-dependent restraint of Salmonella trafficking to the MLN, we examined mice bearing mutations in diverse signaling pathways. As observed in antibiotic-treated mice, Myd88-/- mice infected with non-invasive Salmonella had increased bacterial counts in the MLN compared to Myd88+/- littermate controls (Figure 2a). Consistent with the observations in commensal-depleted animals, there was also induction of Salmonella-specific IgA and antigen-induced IFNγ-producing T cells in the MLN in Myd88-/- animals but not in littermate controls (Figures 2b and 2c). In contrast, there was no increase in bacteria reaching the MLN or induction of IgA in NOD2- or NALP3-deficient animals, suggesting that MyD88 specifically relays a microbial signal via a Toll-like receptor (Figure S4a). This indicates that detection of commensals in a MyD88-dependent manner functions to reduce bacterial colonization of the MLN and thereby reduce intestinal immune responses.
Figure 2. MyD88-dependent signals limit non-invasive Salmonella entry in the MLN.
(a) Myd88-/- or heterozygous littermates were infected with noninvasive Salmonella (invA) and bacterial titers were determined in the spleen and MLN. Data points represent organs from a single mouse. Data were pooled from three independent experiments. ***P<0.0001, Mann-Whitney Test. Error bars represent the geometric mean. (b) Myd88-/- or heterozygous littermates were infected with non-invasive, non-pathogenic Salmonella and blood and feces were analyzed for Salmonella-specific IgG and IgA. Bars represent data from three mice per genotype. Data are representative of three independent experiments. ***P<0.001, unpaired t-test. Error bars represent S.E.M. (c) IFNγ production by spleen, MLN and small intestinal lamina propria (SI) T cells from Salmonella-infected mice of the indicated genotype. Analysis was as in Figure 1b. Bars represent three animals per genotype. Data are representative of two independent experiments. *P<0.05, unpaired t-test. Error bars represent S.E.M. (d) Antibiotic-treated B6 mice were left untreated (NT) or gavaged twice with LPS or heat killed E. coli (EC) or Salmonella (ST). 24h later the mice were infected orally with non-invasive Salmonella. Bacterial titers in the MLN were determined two days later. Each point represents individual mice from one of two independent experiments. **P<0.002, one-way ANOVA. Error bars represent the geometric mean.
We next sought to identify microbial products that could prevent trafficking of non-invasive Salmonella to the MLN in antibiotic-treated mice. Re-colonizing antibiotic-treated mice with the cecal contents of unmanipulated mice reduced the amount of Salmonella that reached the MLN (Figure S4b), as did short pretreatment with cecal contents (two days before infection with non-invasive Salmonella) or with heat-killed E. coli or Salmonella (Figure 2d). These results indicate that complete re-colonization was not required to abrogate trafficking of the bacteria. Pre-feeding mice with heat killed bacteria before infection did not reduce the load of Salmonella in the intestine, even though the amount of bacteria reaching the MLN was reduced (Figures 2d and S4c). This indicates that recognition of bacterial products can function to reduce colonization of internal organs with a non-invasive bacterium without affecting the intestinal load of that bacterium. As expected, pre-treating MyD88-deficient animals with heat-killed Salmonella had no effect (Figure S4d). Pretreating mice with LPS, bacterial DNA, or zymosan (Figure 2d and data not shown) also failed to limit MLN titers of Salmonella, despite the ability of LPS to induce up-regulation of the anti-microbial protein RegIIIγ (Figure S4e). Thus, commensal-dependent intestinal epithelial responses may not be sufficient to prevent trafficking of non-invasive Salmonella to the MLN.
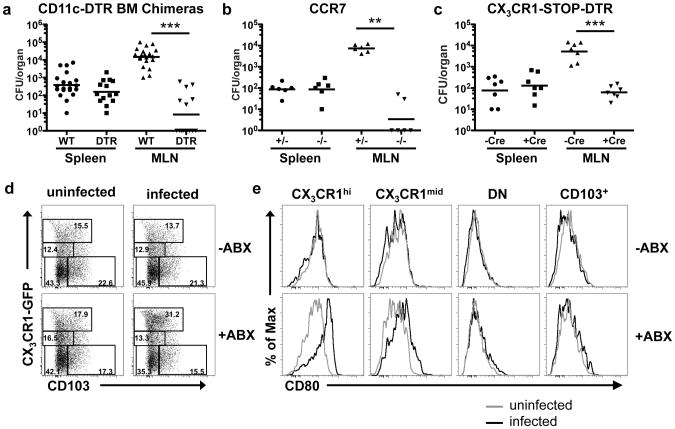
Antigens and pathogens that can penetrate the mucus layer and cross the intestinal epithelium to enter the lamina propria are thought to be trafficked by dendritic cells to the MLN, where they can prime immune responses13,14. This conclusion was based on experiments with invasive bacteria that colonized Peyer's patches14,15. Although lamina propria CD11c+ cells were also shown to be required for non-invasive Salmonella colonization of the lamina propria and the MLN, it was concluded that the DC were required only for bacterial translocation across the epithelium12,16. We wished to determine whether non-invasive bacteria, that cannot colonize Peyer's patches, would additionally be transported to the MLN by lamina propria DCs. Our observation that Salmonella trafficked to the MLN only after dysbiosis induced by antibiotics, in the absence of barrier disruption, suggested a specific mechanism that restricts access of bacteria to intestinal immune priming sites. We therefore examined whether DCs or other lamina propria myeloid cells were responsible for transporting Salmonella to the MLN and initiating immune responses to it. We first utilized CD11c-DTR mice, in which injection of diphtheria toxin (DT) results in selective depletion of CD11c+ cells17,18. CD11c-DTR bone marrow chimeric mice were treated with antibiotics and then injected with DT to deplete DCs (Figure S5) before oral infection with non-invasive Salmonella. There was no decrease in splenic titers in DT-treated versus untreated mice (Figure 3a) arguing against a role for DCs in bacterial entry to the tissue. However, bacterial titers in the MLN were significantly reduced following ablation of CD11c+ cells (Figure 3a). Thus, in microbiota-depleted mice, CD11c+ cells are at least partially responsible for transporting non-invasive Salmonella to the MLN. As DC migration to the MLN requires their expression of CCR719,20, we treated CCR7+/+ or CCR7-/- mice with antibiotics to deplete commensal bacteria before infecting orally with non-invasive Salmonella. There was a substantial reduction in Salmonella in the MLN of commensal-depleted CCR7-/- mice compared to wild-type mice (Figure 3b), indicating that this chemokine receptor is required for transport of the bacteria to the MLN.
Figure 3. Colonization of MLN by non-invasive Salmonella requires CCR7-dependent trafficking of CX3CR1hicells.
(a-c) Antibiotic-treated mice of the indicated genotype and littermate controls were orally infected with non-invasive Salmonella, and bacterial titers in spleen and MLN were determined. For (a) and (c), animals were treated with diphtheria toxin for two consecutive days before infection. For (a) ***P<0.0001, for (b) **P<0.005, for (c) ***P=0.0006, all Mann-Whitney Test. Error bars represent the geometric mean. (d) Analysis of dendritic cell subsets in MLN of untreated or antibiotic-treated Cx3cr1gfp/+ mice that were mock infected or infected with non-invasive Salmonella. MLN cells were isolated at 48h, gated on the MHCII+CD11c+ population, and analyzed for expression of CX3CR1 and CD103. Percentages are shown in each gate. Absolute numbers are shown in Figure S9a. (e) Expression of CD80 on intestinal myeloid cell subsets. Cells were gated on the indicated cell populations as shown in (d). For data in (a-c), points represents individual mice pooled from independent experiments. Panels in (d) represent individual mice from one of five independent experiments.
The CD11c+ mononuclear phagocytes within the lamina propria have been subdivided into two major populations, CX3CR1+ and CD103+ cells (Figure S6a and reviewed in Ref. 21). CX3CR1+ cells can be further divided into cells expressing high and intermediate levels of the chemokine receptor. CX3CR1hi cells, which also express CD14 and low levels of CD103 (Figure S6a and Ref. 16,21), have been described to have both DC- and macrophage-like characteristics. They differentiate from a monocyte precursor15,16,22, are thought to be non-migratory in response to in vivo TLR activation8, and are poorly immunostimulatory in vitro. CX3CR1low cells, which also differentiate from monocyte precursors, have been described to have DC-like characteristics22. CD103+ cells, which do not express CX3CR1, are thought to be conventional myeloid DCs as they develop from a classical DC precursor15,16, express CCR7, can activate naïve T cells in vitro, and migrate to the MLN upon TLR stimulation8,15,23. In contrast with previous reports that found CCR7 only expressed on CD103+ cells8,15, we observed upregulation of CCR7 on both CX3CR1+ and CD103+ cells after in vitro stimulation with LPS (Figure S6b), suggesting that both have migratory potential.
To assess whether intestinal CD103+ or CX3CR1+ cells were involved in transporting Salmonella to the MLN in the absence of commensals, we next examined mice in which a loxP-flanked stop cassette upstream of the DTR coding region was knocked into the Cx3cr1 locus (Figure S7a). Expression of CD11c-Cre excises the stop cassette, allowing for DTR expression and selective depletion of CD11c+CX3CR1hi cells upon administration of DT, with no effect on CD11c+CX3CR1low or CD11c+CD103+ cells (Figure S7b and c). Because monocytes have been reported to take up wildtype Salmonella24, we determined that they were not depleted in the DT-treated animals, but were actually increased in number (Figure S8). When mice were orally infected with non-invasive Salmonella following DT-mediated depletion of the CX3CR1hi mononuclear cells, there was a marked reduction in the number of bacteria reaching the MLN (Figure 3c). This result, which was unexpected in light of recent studies showing that only the CD103+ cells migrate from the lamina propria to the MLN after TLR activation, suggested that CX3CR1hi cells are responsible for trafficking of the non-invasive bacteria to the MLN upon disturbance of the microbiota.
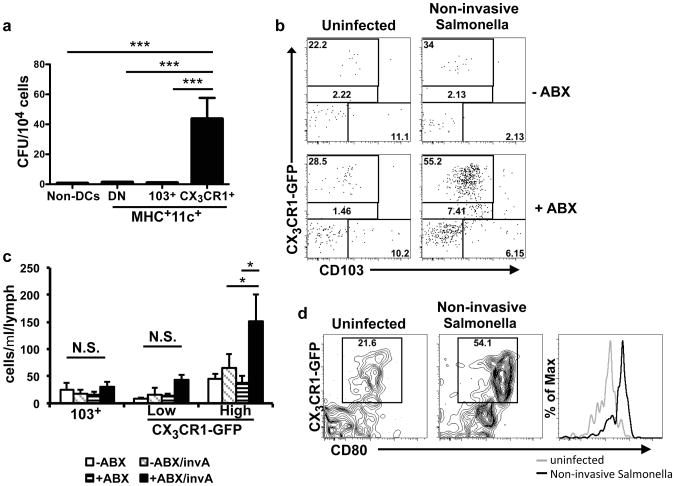
To further evaluate the hypothesis that CX3CR1hi cells are responsible for trafficking of non-invasive Salmonella to the MLN, we orally infected antibiotic-treated CX3CR1-GFP mice25 with the bacterium and analyzed the cell populations in the MLN. There was an increase in DCs of all subsets after antibiotic treatment (Figure S9a), but after infection with non-invasive Salmonella there was further increase only of CX3CR1hi cells (Figure 3d and S9a), and this was not observed if mice were pretreated with heat-killed Salmonella before infection (Figure S9b), consistent with the limited number of bacteria reaching the MLN in these animals (Figure 2d). Further, the co-stimulatory molecule CD80 was upregulated on CX3CR1hi cells, but not CD103+ DC, after infection of antibiotic-treated mice with non-invasive Salmonella (Figure 3e), implicating the CX3CR1hi cells in priming the anti-Salmonella T cell response. To confirm that CX3CR1+ cells had taken up non-invasive Salmonella, we sorted CX3CR1+ and CD103+ cells from the MLN of infected antibiotic-treated mice (Figure S9c) and assayed for cells containing colony-forming units. Most of the bacteria were recovered from the CX3CR1+ cells rather than the CD103+ cells, and no bacteria could be detected from cells derived from untreated infected mice (Figure 4a). These results suggest that commensal-derived signals prevent CX3CR1+ cells from ferrying bacteria to the MLN, although we cannot rule out additional mechanisms, e.g. reduced killing of internalized bacteria under these conditions.
Figure 4. CD103-CX3CR1+cells migrate into afferent lymphatics of antibiotic-treated animals.
(a) CX3CR1+ or CD103+ cells (gated on the MHCII+CD11c+ population, as in Figure 3d) from the MLN were isolated from antibiotic-treated, non-invasive Salmonella-infected mice. The numbers of bacteria per 104 cells were determined by plating on LB-strep plates. No bacteria were observed from cells isolated from uninfected or infected but antibiotic-untreated mice. Bars represent pooled data from 15 individual mice from 4 independent experiments. ***P<0.0001, one-way ANOVA with Bonferroni correction. Error bars represent S.E.M. (b) – (d) Untreated or antibiotic-treated Cx3cr1gfp/+ were left uninfected or were infected with non-invasive Salmonella. At 48h, cells in the intestinal lymph were isolated and analyzed by flow cytometry. (b) Cells were gated on the MHCII+CD11c+ population and analyzed for expression of CX3CR1-GFP and CD103. Data are representative of three independent experiments. (c) Quantitation of MHCII+CD11c+CX3CR1-GFP+ cells in the afferent lymph of the indicated mice. **P<0.01, one-way ANOVA with Bonferroni correction. Error bars represent S.E.M. (d) CD80 expression on CX3CR1-GFP+ cells from lymph of antibiotic-treated mice with and without Salmonella infection.
To examine more directly whether CX3CR1+ cells migrate from the small intestine to the MLN, we assayed the afferent intestinal lymph in antibiotic-treated or untreated CX3CR1-GFP mice after oral infection with non-invasive Salmonella. In uninfected or non-antibiotic treated animals, very few cells were detected and, as expected, TLR activation with R848 resulted in the preferential release of CD103+ DC into the lymph (Figure S9d). Unlike previous reports8, we observed CX3CR1+ cells in the afferent lymph of un-manipulated animals, with a higher proportion of CX3CR1hi than CX3CR1low cells (Figures 4b, 4c and S9d). The number and proportion of these cells in the afferent lymph was markedly higher after infection of antibiotic-treated mice with non-invasive Salmonella (Figures 4b and 4c). Additionally, in antibiotic-treated mice infected with non-invasive Salmonella there was up-regulation of CD80 in lymph-derived CX3CR1+ cells (Figure 4d).
Our results indicate the existence of a novel CX3CR1hi cell-mediated pathway for antigen access from the intestinal lumen and for immune priming. Inhibition of this pathway through a MyD88-dependent mechanism may contribute to maintenance of immunological tolerance towards commensal microbes. There is thus another role for microbiota-initiated signaling in the host: limiting immune priming against intestinal antigens by inhibiting trafficking of a lamina propria cell population and phagocytosed bacteria to the MLN. We speculate that disruption of this pathway for inhibition of antigen trafficking may be an important defense strategy upon infection with invasive microbes, but could also contribute to inflammatory bowel disease (IBD) in humans. IBD is thought to be an inappropriate immune response against commensal microorganisms, and it has been reported that patients with IBD have changes in their microbiota26. Our results suggest that this could lead to increased intestinal priming and in increased intestinal inflammation, which can itself cause dysbiosis27,28, thus amplifying the pathogenic process. Further, in a susceptible individual, reductions in certain classes of bacteria might be sufficient to trigger this increased trafficking. In conditions of infection, the dysbiosis that results from intestinal inflammation28,29 may be beneficial, as it may allow for induction of immune responses against pathogens and not commensals. Further work will be needed to determine if a particular microbe or microbial signal is required for regulation of CX3CR1+ cell migration. Therapeutic modulation of these cells may attenuate intestinal inflammation or enhance priming for mucosal vaccines.
Methods
Mice
C57BL/6 mice were purchased from Taconic Farms or Jackson Laboratories. Ccr7 and Nod2 mutant mice were purchased from Jackson Laboratories. CX3CR1-GFP and CD11c-DTR mice were previously described17,25. Myd88 and Nalp3 mutant mice were from Ruslan Medzhitov and Jurg Tschopp, respectively. For inducible CX3CR1-DTR mice, we introduced a loxP-floxed stop cassette followed by DTR into the Cx3cr1 locus. Mice were subsequently crossed to CD11c-Cre mice (Jackson Laboratories) allowing deletion of the stop cassette in CD11c+ cells and induction of DTR in CX3CR1+ cells. All animal experiments were performed in accordance with approved protocols for the NYU Institutional Animal Care and Usage Committee.
Depletion of gut commensal microbiota
Animals were provided ampicillin (A; 1 g/L; Sigma), vancomycin (V; 500 mg/L; Amresco), neomycin sulfate (N; 1 g/L; Sigma), and metronidazole (M; 1 g/L; Sigma Labs) in drinking water for four weeks as described5. Mice were switched to water without antibiotics for two days before infecting them as below.
Infection with S. Typhimurium and E. coli
Mice were infected orally with 1×108 Salmonella ΔinvA cfu or 2×108 ΔAroA/ΔinvA cfu in 100μl of PBS. For titers, organs were homogenized two days after infection, diluted in phosphate-buffered saline (PBS) and plated on LB agar with streptomycin (50μg/ml). Colony counts were expressed as cfu per organ. For T cell analysis, mice were analyzed 10 days after infection. For antibody titers, mice were infected on day 0 and day 15. Blood and feces were collected 2 weeks after the second infection. Mice were infected with 1×109 E. coli K12 (ATCC #29425) and analyzed for bacterial titers or induction of antibodies as above. A description of Salmonella strains can be found in Table S1.
Measurement of T cell and antibody responses
Analyses of serum and fecal proteins for Salmonella-specific IgG and IgA by enzyme-linked immunosorbent assay (ELISA) were performed as described10. Concentrations were determined by interpolating from a standard curve generated by plating IgG or IgA. For T cell responses, single-cell suspensions from the indicated organ were first negatively depleted with anti-B220 and -CD8 magnetic microbeads (Miltenyi Biotec). The depleted fraction was stained and cell sorting was performed on an Aria flow cytometer (BD Biosciences) to obtain memory/effector TCR+CD4+CD44+CD62L-T cells (>99% purity). These cells were plated at 1×105 cells/well with 5×105 irradiated splenocytes and 1μg of heat killed Salmonella. At 72h, IFNγ in the supernatant was measured by ELISA (BD Biosciences).
Generation of CD11c-DTR bone marrow chimeras
6 week old Ly5.1 congenic mice (Jackson Labs) were lethally irradiated and reconstituted with bone marrow from wild-type or CD11 c-DTR transgenic mice (Ly5.2). After 8-10 weeks, recipients were examined for immune reconstitution by analyzing blood for Ly5.2+ hematopoietic cells before treating with antibiotics.
Cell isolation
Small intestinal lamina propria cells were isolated as previously described30. Spleen and MLN cells were isolated by injecting the organ with complete RPMI containing 100 U ml−1 type VIII collagenase (Sigma) and 150 μg ml−1 DNaseI (Sigma). The organ was disassociated using frosted glass slides and the tissue was incubated at 37°C for 45 minutes.
Lymph collection
Afferent lymph was isolated as described8. Mice were gavaged with 200μl corn oil 1 hour before isolating lymph to visualize the lymphatics. Afferent lymphatics were identified by anatomical location and confirmed by cellular composition as afferent lymph contains a majority of MHCII+ cells whereas efferent lymph contains a majority of T cells.
Antibodies, cell staining, and flow cytometry
Flow cytometric analysis was performed on a LSR II (BD Biosciences) and analyzed using FlowJo software (Tree Star Inc.). Antibodies were from BD Pharmingen or eBiosciences. DAPI was used to exclude dead cells.
In vivo cytokine analysis
Mice were infected with 2×108 ΔAroA/ΔinvA cfu. 9d later mice were injected with Brefeldin A (Sigma) as described31. 4h later, mice were injected with 50 μg boiled Salmonella. Spleen and MLN T cells were isolated 2h later. During cell isolation and extracellular staining, all solutions contained Brefeldin (GolgiPlug, BD Biosciences). Cells were stained for cytokines as per the manufactures instructions (BD Bioscience).
RNA extraction and real-time RT-PCR
The terminal ileum was disrupted in Trizol (Ambion) using a TissueLyserII (Qiagen). Realtime PCR for RegIIIγ was performed as described32.
Intestinal permeability
Intestinal permeability was assessed by P.O. administration of FITC-conjugated dextran (Sigma-Aldrich) at 400 mg/kg as described33. Serum FITC-dextran concentrations were determined using an Envision 2104 Multiplate Reader (Perkin Elmer).
Quantitation of fecal bacterial DNA by 16S rRNA gene amplification
Fecal pellets were weighed and resuspended in 200mM Tris pH 8/20mM NaCl/20mM EDTA with 3% SDS. Bacteria was lysed by mechanical disruption with silica beads using a Retsch TissueLyser. DNA was extracted using phenol-chloroform. qPCR was performed on a Lightcycler 480 (Roche Applied Science) using SYBR green (Qiagen). 16S rRNA was amplified using forward primer 5′-ACTCCTACGGGAGGCAGCAGT-3′ and reverse primer 5′-ATTACCGCGGCTGCTGGC-3′. A standard curve was generated using limiting dilutions of bacterial DNA to allow conversion of 16S rRNA amplification to ng of bacterial DNA. The data were normalized to fecal pellet weight and expressed as ng bacterial DNA/mg of fecal pellet.
Sorting Salmonella infected cells
Cx3cr1gfp/+ mice were treated with antibiotics and infected with non-invasive Salmonella. Two days later MLN cells were isolated and stained. Cell sorting was performed on an Aria cytometer (BD Biosciences). TCRP-CD19-MHCII+CD11c+ cells were sorted into CX3CR1+, CD103+ or CX3CR1-CD103- cell populations. CD19+ or TCRβ+ cells were plated as non-DCs. Sorted cells were plated on LB-Strep plates.
Heat-killed Salmonella and E. coli
Salmonella or E. coli were heated to 85° for 60 minutes. Bacteria were plated and allowed to grow overnight to confirm killing. Mice were administered 1×1010 heat killed bacteria by P.O. daily for two days before infecting with non-invasive Salmonella.
Statistical analysis
One-way analysis of variance (ANOVA) with Bonferroni's posttest or unpaired t-test was performed using a 95% confidence interval. All analyses were performed using GraphPad Prism version 4.0. Differences were considered to be significant at P values of <0.05.
Supplementary Material
Acknowledgments
We thank H. Yue and J. Hall for reading the manuscript and members of the Littman laboratory for their suggestions. We thank the NYU Histology Core which is supported in part by grant 5P30CA016087-32 from the National Cancer Institute. Supported by the American Cancer Society and NIH T32 CA009161 (G.E.D.), NIH T32 DK083256-02 (R.S.L.), Human Frontier Science Program Long-Term Fellowship (B.B.), NIH R01AI085166 (S.R.S.) and the Howard Hughes Medical Institute (D.R.L.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions: G.E.D. designed and performed the experiments. G.E.D. and D.R.L planned experiments and wrote the manuscript with input from the coauthors. R.S.L., J.X.Z., S.R.S., B.B., C.G., and A.C., helped plan and perform experiments.
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Macpherson AJ, Uhr T. Compartmentalization of the mucosal immune responses to commensal intestinal bacteria. Ann N Y Acad Sci. 2004;1029:36–43. doi: 10.1196/annals.1309.005. [DOI] [PubMed] [Google Scholar]
- 2.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 3.Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 4.Hooper LV, et al. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 5.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Balfour Sartor R. Bacteria in Crohn's disease: mechanisms of inflammation and therapeutic implications. J Clin Gastroenterol. 2007;41(1):S37–43. doi: 10.1097/MCG.0b013e31802db364. [DOI] [PubMed] [Google Scholar]
- 8.Schulz O, et al. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J Exp Med. 2009;206:3101–3114. doi: 10.1084/jem.20091925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 10.Vazquez-Torres A, et al. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature. 1999;401:804–808. doi: 10.1038/44593. [DOI] [PubMed] [Google Scholar]
- 11.Griffin AJ, McSorley SJ. Development of protective immunity to Salmonella, a mucosal pathogen with a systemic agenda. Mucosal Immunol. 2011;4:371–382. doi: 10.1038/mi.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hapfelmeier S, et al. Microbe sampling by mucosal dendritic cells is a discrete, MyD88-independent step in DeltainvG S. Typhimurium colitis. The Journal of experimental medicine. 2008;205:437–450. doi: 10.1084/jem.20070633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mowat AM. Anatomical basis of tolerance and immunity to intestinal antigens. Nat Rev Immunol. 2003;3:331–341. doi: 10.1038/nri1057. [DOI] [PubMed] [Google Scholar]
- 14.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 15.Bogunovic M, et al. Origin of the lamina propria dendritic cell network. Immunity. 2009;31:513–525. doi: 10.1016/j.immuni.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Varol C, et al. Intestinal lamina propria dendritic cell subsets have different origin and functions. Immunity. 2009;31:502–512. doi: 10.1016/j.immuni.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 17.Jung S, et al. In vivo depletion of CD11c(+) dendritic cells abrogates priming of CD8(+) T cells by exogenous cell-associated antigens. Immunity. 2002;17:211–220. doi: 10.1016/s1074-7613(02)00365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vallon-Eberhard A, Landsman L, Yogev N, Verrier B, Jung S. Transepithelial pathogen uptake into the small intestinal lamina propria. J Immunol. 2006;176:2465–2469. doi: 10.4049/jimmunol.176.4.2465. [DOI] [PubMed] [Google Scholar]
- 19.Jang MH, et al. CCR7 is critically important for migration of dendritic cells in intestinal lamina propria to mesenteric lymph nodes. J Immunol. 2006;176:803–810. doi: 10.4049/jimmunol.176.2.803. [DOI] [PubMed] [Google Scholar]
- 20.Worbs T, et al. Oral tolerance originates in the intestinal immune system and relies on antigen carriage by dendritic cells. J Exp Med. 2006;203:519–527. doi: 10.1084/jem.20052016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varol C, Zigmond E, Jung S. Securing the immune tightrope: mononuclear phagocytes in the intestinal lamina propria. Nat Rev Immunol. 2010;10:415–426. doi: 10.1038/nri2778. [DOI] [PubMed] [Google Scholar]
- 22.Rivollier A, He J, Kole A, Valatas V, Kelsall BL. Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. The Journal of experimental medicine. 2012;209:139–155. doi: 10.1084/jem.20101387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansson-Lindbom B, et al. Functional specialization of gut CD103+ dendritic cells in the regulation of tissue-selective T cell homing. J Exp Med. 2005;202:1063–1073. doi: 10.1084/jem.20051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rydstrom A, Wick MJ. Monocyte recruitment, activation, and function in the gut-associated lymphoid tissue during oral Salmonella infection. Journal of immunology. 2007;178:5789–5801. doi: 10.4049/jimmunol.178.9.5789. [DOI] [PubMed] [Google Scholar]
- 25.Jung S, et al. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol. 2000;20:4106–4114. doi: 10.1128/mcb.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thiennimitr P, Winter SE, Baumler AJ. Salmonella, the host and its microbiota. Curr Opin Microbiol. 2012;15:108–114. doi: 10.1016/j.mib.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lupp C, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 30.Valdez Y, et al. Nramp1 expression by dendritic cells modulates inflammatory responses during Salmonella Typhimurium infection. Cellular microbiology. 2008;10:1646–1661. doi: 10.1111/j.1462-5822.2008.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu F, Whitton JL. Cutting edge: re-evaluating the in vivo cytokine responses of CD8+ T cells during primary and secondary viral infections. Journal of immunology. 2005;174:5936–5940. doi: 10.4049/jimmunol.174.10.5936. [DOI] [PubMed] [Google Scholar]
- 32.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Napolitano LM, Koruda MJ, Meyer AA, Baker CC. The impact of femur fracture with associated soft tissue injury on immune function and intestinal permeability. Shock. 1996;5:202–207. doi: 10.1097/00024382-199603000-00006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.