Abstract
Mycobacterium tuberculosis infection alters macrophage gene expression and macrophage response to interferon gamma (IFNγ), a critical host defense cytokine. However, regulation of these changes is poorly understood. We report discordance of changes in nascent transcript and total nuclear RNA abundance for the transcription factors STAT1 and IRF1, together with lack of effect on their RNA half-lives, in human THP-1 cells infected with M. tuberculosis and stimulated with IFNγ. The results indicate that negative post-initiation regulation of mRNA biogenesis limits the expression of these factors, which mediate host defense against M. tuberculosis through the cellular response to IFNγ. Consistent with the results for STAT1 and IRF1, transcriptome analysis reveals down-regulation of post-initiation mRNA biogenesis processes and pathways by infection, with and without IFNγ stimulation. Clinical relevance for regulation of post-initiation mRNA biogenesis is demonstrated by studies of donor samples showing that post-initiation mRNA biogenesis pathways are repressed in latent tuberculosis infection compared to cured disease and in active tuberculosis compared to ongoing treatment or to latent tuberculosis. For active disease and latent infection donors from two populations (London, UK, and The Gambia), each analyzed using a different platform, pathway-related gene expression differences were highly correlated, demonstrating substantial specificity in the effect. Collectively, the molecular and bioinformatic analyses point toward down-regulation of post-initiation mRNA biogenesis pathways as a means by which M. tuberculosis infection limits expression of immunologically essential transcription factors. Thus, negative regulation of post-initiation mRNA biogenesis may constrain the macrophage response to infection and overall host defense against tuberculosis.
Keywords: macrophage, nuclear RNA, transcriptome
Introduction
Morbidity and mortality from tuberculosis are extremely high, with about 8 million new cases of active disease per year and about 2 million deaths (1); in the absence of effective treatment, mortality is about 50% (2). Increased failure of anti-tuberculous chemotherapy (3, 4) with the emergence of multi-drug and extensively-drug resistant strains of Mycobacterium tuberculosis has added urgency to the goal of developing effective vaccines and immunotherapies. Macrophages are the immune cells predominantly targeted by M. tuberculosis. Bacterial replication occurs in macrophages at two points in the immunological life cycle of tuberculosis: during the innate immune response to infection (before adaptive immunity forces a transition to latent infection), and during reactivation (when adaptive immunity fails to maintain latent infection) (reviewed in 5). Determining effects of M. tuberculosis infection on host macrophage gene expression and relating those effects to differences in gene expression between individuals who maintain latent tuberculosis infection (LTBI)4 and those who develop pulmonary tuberculosis (PTB) should facilitate efforts to apply host immune pressure against tuberculosis.
The interaction between macrophages infected with M. tuberculosis and the host immune mediator IFNγ is a major determinant of host response to M. tuberculosis (6). The host transcription factors Signal Transducer and Activator of Transcription 1 (STAT1) and IFN Regulatory Factor 1 (IRF1) are essential mediators of the response to IFNγ and of host defense against M. tuberculosis (for example, (7–10). In humans, mutations in STAT1 confer susceptibility to normally non-pathogenic mycobacterial infections (11, 12); in mice, a deficiency of STAT1 or IRF1 abolishes immune control of M. tuberculosis growth, which leads to a fatal fulminant infection rather than a chronic illness with slow disease progression (13, 14). The consequences of deficiencies in these transcription factors emphasize that their regulation is essential for an effective host response to M. tuberculosis. Moreover, both are induced by M. tuberculosis infection and by IFNγ stimulation (15–21). IFNγ induction of STAT1 and IRF1 and M. tuberculosis induction of IRF1 are attributable at least in part to increased transcription. However, little is known about whether mechanisms other than regulation of transcription initiation control their expression, or any other transcriptome changes, with or without IFNγ stimulation in cells infected with M. tuberculosis.
Post-initiation steps in mRNA biogenesis (5' end capping, elongation, splicing, 3' end cleavage and polyadenylation) allow regulation of gene expression in addition to control of transcription initiation (22–25). Alternative splicing and polyadenylation can determine tissue-specific or signal-mediated levels of transcript isoform expression (reviewed in 26, 27). For example, in patients with chronic granulomatous disease, increased levels of functional NADPH (phagocyte) oxidase as a response to IFNγ therapy result from mutations that alter CYBB gene exon usage (28, 29). In other examples, stimulation of toll-like receptors by bacteria-derived ligands alters transcripts through effects on alternative splicing and polyadenylation (30–36). With M. tuberculosis infection, alternatively spliced transcripts of IL12Rb are produced (37). Even without alternative mRNA processing, changing the rate of a single processing event can control gene expression level, as demonstrated for glucocorticoid-mediated repression of gonadotropin-releasing hormone expression through inhibition of pre-mRNA splicing (38). Thus, post-initiation regulation of mRNA biogenesis might be an important host response to M. tuberculosis infection.
In the present work we characterized expression of genes responsive to M. tuberculosis infection and IFNγ stimulation and analyzed transcriptome data to better understand the basis for their regulated expression. Data from in vitro infection of THP-1 cells indicated that negative post-initiation regulation of mRNA biogenesis, superimposed on IFNγ-stimulated activation of transcription, limits increases in STAT1 and IRF1 gene expression. Analysis of transcriptome data demonstrated that down-regulation of post-initiation mRNA biogenesis pathways occurs with in vitro infection and distinguishes individuals who develop PTB from those who maintain LTBI.
Materials and Methods
Cell growth and infections
All manipulations with viable M. tuberculosis were performed under biosafety level 3 containment. M. tuberculosis TN913, a prevalent, drug-sensitive clinical isolate of the C strain from the 1990–1994 New York City tuberculosis outbreak (39), was obtained from the Public Health Research Institute Tuberculosis Center. The human monocytic cell line THP-1 was obtained from ATCC. The bacteria and cells were maintained, and THP-1 cells were differentiated and infected, as previously described (17). Differentiated THP-1 cells model human alveolar macrophages, as judged by a variety of criteria (17, 20, 40, 41). Three days post-infection, infected cells and parallel cultures of uninfected cells were left untreated or were treated for two hours with IFNγ (Peprotech, Rocky Hill, NJ) at 1 ng/ml. In some experiments, actinomycin D (Calbiochem, La Jolla, CA) was then added to a concentration of 10 µg/ml, and cells were harvested at various times thereafter to determine transcript half-life. The titer of the inoculum on the day of infection and the presence of intracellular bacteria on the day of harvest were confirmed by plating to determine CFU.
Cell fractionation and recovery of RNA
All steps were carried out at 0–4° C. Cells were scraped from flasks and collected by centrifugation at 400 × g for 5 min. To isolate total cellular RNA, cell pellets were extracted immediately. Alternatively, nuclear and cytoplasmic fractions were prepared as previously described (20). Cytoplasmic RNA was extracted and the nuclei were suspended in nuclear run-on buffer containing 0.1% NP-40 and lacking nucleotide triphosphates. Nuclei then were used in the nuclear run-on assay (below) or were further purified by sedimentation through a cushion of 60% glycerol in the same buffer (42) before extraction of RNA. RNA was recovered using TRI-Reagent (Molecular Resource Center) as recommended by the vendor.
Nuclear Run-on Assay
Nascent transcript abundance was determined as previously described (21). Signal from hybridization to pGem1 provided a negative control for specificity and for background correction. Signal from hybridization to human GAPDH provided a positive control and an internal standard for normalization. Probes for GAPDH, IRF1, ISG15, and STAT1 were described previously (15, 16, 20, 43). All genes are denoted by Human Genome Organization official gene symbols. Data are from 6 independent experiments. Quantification of nascent transcript abundance is shown as the average of fold induction relative to expression in uninfected, unstimulated cells. Fold induction is used to allow comparison both among genes and among experimental conditions. Error bars for each perturbation (M. tuberculosis infection, IFNγ stimulation, or both) represent ± SEM. Statistical significance is taken as p < 0.05 based on a two-tailed Student's t test.
Quantitative RT-PCR
Reverse transcription reactions, quantitative PCR reactions using molecular beacons for amplicon detection, and calculation of target abundance were performed as previously described for IRF1. The results were normalized to the level of GAPDH exon 9 for the respective samples (21). Mock reverse transcription samples for each RNA preparation demonstrated negligible DNA contamination. The specificity of assays for introns and exon junctions was confirmed with genomic DNA and cDNA templates. Data are from 4 to 6 independent experiments; in some experiments, not all genes were assayed. RNA half-life was calculated as the average of values determined from exponential decay curves for individual experiments (n = 2–4). Error bars represent ± SEM (for n > 2) or the range of values (for n = 2). Statistical significance for data sets with n ≥ 3 was taken as p < 0.05 based on a two-tailed Student's t test.
Transcriptome analysis
Gene expression profiles for THP-1 cells were determined for four replicate experiments that each included all four experimental conditions (control, M. tuberculosis infection, IFNγ stimulation, infection and stimulation) using Affymetrix Human Genome U133A 2.0 Gene Chips, following the vendor's protocols for cRNA preparation, labeling, hybridization, and scanning. Quantification of probe expression levels was based on the "preferred methods" of Choe et al. (44). This analysis included MAS5 for background correction, global scale to 500 for probe-level normalization, PM-MM for hybridization specificity correction, RMA’s median polish for gene expression summary calculation, GAPDH expression for gene expression normalization, and Loess normalization for intensity skew correction. The use of GAPDH as an internal standard for expression level normalization was empirically validated based on 60 different segments of the GAPDH gene. The signal varied among the probes, but was comparable under all conditions for each one (data not shown). These data are publicly available (GEO accession no. GSE17477 http://www.ncbi.nlm.nih.gov/geo/ ).
For GO annotation analysis of gene expression, we first identified regulated genes based on statistically significant differential expression in comparisons of gene expression between each perturbation and uninfected, unstimulated cells. A dynamically thresholded t-test (Cyber-T) (45) was used to identify differentially expressed genes (p < 0.05 or p < 0.01). A more stringent threshold for differential expression was set using a permutation-based implementation to calculate false discovery rate (q < 0.1). Fisher's exact test was used to assess the statistical significance (p < 0.05) for the proportion of regulated genes having particular combinations of annotations taken from the GO database (http://www.geneontology.org/) (46) among down-regulated genes in comparison to the overall proportion of genes having those annotations among those probed.
The transcriptomes of samples from clinical studies were obtained from Gene Expression Omnibus (GEO, GSE19491 (47), GSE19439 (48), and GSE11199 (49)). The first two studies included comparison of whole blood from LTBI and PTB donors. The third study included comparison of monocyte-derived macrophages from LTBI and cured PTB donors. The publishing authors’ normalized expression values were used.
For pathway analysis of data from in vitro infection and from the clinical studies, the significance of gene sets defined by Reactome pathway annotations (www.reactome.org) (50, 51) was evaluated using the Coincident Extreme Ranks in Numerical Observations (CERNO) method (52). The CERNO test uses the rank order of significance for differential expression of the genes in a pathway to determine significance for the pathway annotation without a threshold on expression differences for individual genes. A nested CERNO testing routine was applied (53). Benjamini-Hochberg false discovery rates (FDR) (54) were calculated for the entire collection of gene sets, but for each sample comparison separately. Eighteen post-initiation mRNA biogenesis Reactome gene sets were identified with at least three representative genes on the Affymetrix Human Genome U133A 2.0 and the Agilent-014850 Whole Human Genome Microarray. A one-sided Student's t-test was performed to test whether individual genes were down-regulated. Statistical analysis methods not otherwise cited were performed in the R programming environment (55).
Results
Differential induction of nascent transcripts and total nuclear RNA for both STAT1 and IRF1
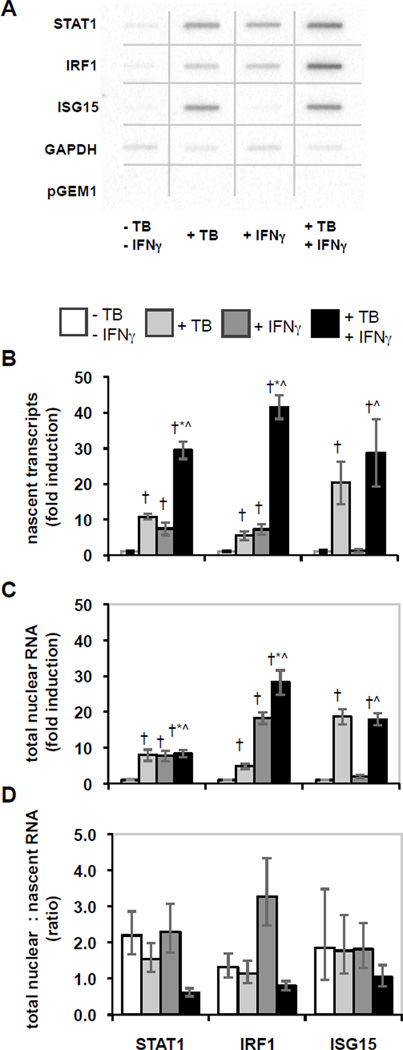
Comparing induction at different points in the course of gene expression will reveal the presence of regulated steps. To set a baseline for identifying post-initiation regulation, we first measured the levels of nascent transcripts for STAT1 and IRF1, two central regulators of response to M. tuberculosis infection and to IFNγ stimulation. M. tuberculosis infection and IFNγ stimulation each induced expression of the two genes (Fig. 1 A and B). When infected cells were also stimulated with IFNγ, the increases in nascent transcripts for STAT1 and IRF1 were about two- to three-fold greater than the sum of the responses to each perturbation alone. Thus, a synergistic response occurs. To test for specificity in the effects of M. tuberculosis infection on the response to IFNγ, we measured transcription of ISG15, since it is induced by M. tuberculosis, but not by IFNγ (15, 20). In contrast to STAT1 and IRF1, the level of ISG15 nascent transcripts was similar with and without IFNγ stimulation of infected cells (Fig. 1 A and B). These data demonstrate specific synergistic induction of STAT1 and IRF1 nascent transcripts with IFNγ stimulation of M. tuberculosis-infected cells.
Figure 1.
Effects of M. tuberculosis infection and IFNγ stimulation on nascent and total nuclear RNA. THP-1 cells were differentiated, infected with M. tuberculosis and/or stimulated with IFNγ, and then nascent transcripts were measured using the nuclear run-on assay, or total nuclear RNA was extracted and quantified by qRT-PCR. A) The hybridization results for nuclear run-on assays were imaged and quantified using a phosphorimager. The figure is a composite of noncontiguous portions from a single phosphorimager exposure that included the membranes for all four conditions in a representative experiment. B) Nascent transcript measurement is shown as average fold induction ± SEM for 6 replicate experiments. Statistically significant differences (p < 0.05) are indicated compared to control (†), compared to M. tuberculosis (*), and compared to IFNγ (^). C) Total nuclear RNA abundance is shown as average fold induction ± SEM for 4–6 replicate experiments as for panel B. D) The ratios of the abundance of total nuclear RNA relative to the level of nascent transcripts were calculated from the averages for each. Error bars represent ± SEM.
If regulation were only at the level of transcription initiation, the synergistic induction of nascent transcripts for STAT1 and IRF1 would lead to comparable increases in the nuclear and cytoplasmic pools of the corresponding mRNAs. This was the case with M. tuberculosis infection alone, since the levels of STAT1, IRF1, and ISG15 increased in total nuclear RNA as much as in nascent transcripts (compare Fig. 1B to 1C; the ratio is shown in Fig. 1D). IFNγ-mediated induction of STAT1 in total nuclear RNA and in nascent transcripts was also similar. However, induction of IRF1 in total nuclear RNA was 2.5-fold greater than in nascent transcripts, indicating that induction of nascent transcripts and additional positive regulation determine the expression of IRF1 in response to IFNγ stimulation. In sharp contrast, when cells were infected with M. tuberculosis and stimulated with IFNγ, the ratio of the total nuclear RNA to nascent transcript for both STAT1 and IRF1 was one-fourth the ratio in uninfected cells stimulated with IFNγ (Fig. 1B, 1C, 1D). ISG15 showed less effect. These results suggest that expression of STAT1 and IRF1 is subject to gene-specific negative regulation that limits their induction by IFNγ in infected cells. Comparing induction of polyA+ mRNA to nascent transcripts also revealed the limit on expression of STAT1 and IRF1 in M. tuberculosis-infected cells stimulated with IFNγ (data not shown). The reduced induction of STAT1 and IRF1 associated with IFNγ stimulation of infected cells was specific for M. tuberculosis, since it was not observed in cells infected with the non-pathogenic M. bovis BCG (Fig. S1). Thus, in cells infected with M. tuberculosis, regulation in addition to initiation of transcription controls IFNγ-stimulated STAT1 and IRF1 expression.
Post-initiation regulation of response to M. tuberculosis and IFNγ
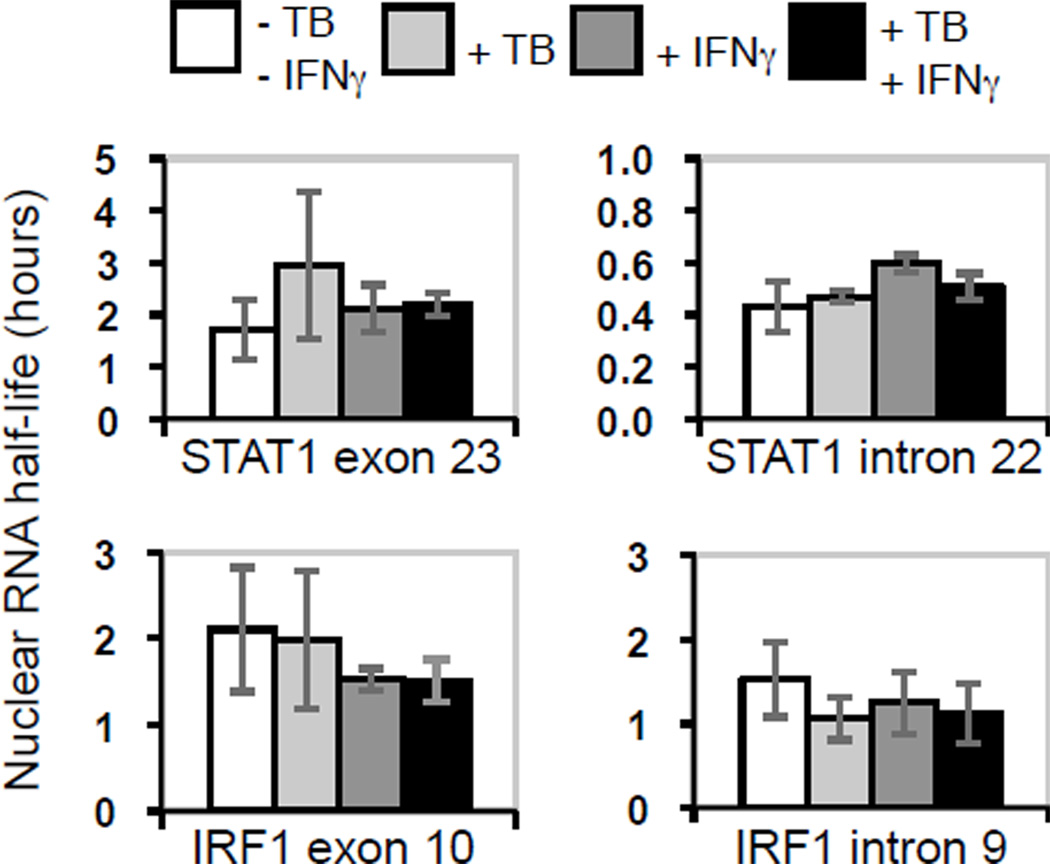
The differentials between induction in total nuclear RNA and in nascent transcripts for both STAT1 and IRF1 seen with IFNγ stimulation of M. tuberculosis-infected cells could result from increased RNA turnover, from negative post-initiation regulation of mRNA biogenesis, or from both. We first addressed the turnover hypothesis by measuring the half-life of exon and intron sequences for STAT1 and IRF1 in total nuclear RNA. We found no significant difference in turnover under any condition (Fig. 2), suggesting that neither degradation nor nuclear export were affected. We next determined that the induction of poly-A+ nuclear RNA was proportional to the induction of total nuclear RNA for both STAT1 and IRF1 (Fig. S2), suggesting that cleavage and polyadenylation was not regulated for either gene. Moreover, the half-lives of the total cellular poly-A+ RNAs were comparable in uninfected and infected cells with and without IFNγ stimulation for each of the two genes (Fig. S3), indicating that the overall turnover of those transcripts is also not regulated in response to these perturbations. Since regulation of neither turnover nor final maturation explains differences between induction of nascent transcripts and total nuclear RNA, we interpret the data as indicating that STAT1 and IRF1 expression is limited at a post-initiation step in mRNA biogenesis, i.e., before a mature mRNA transcript is produced.
Figure 2.
Effects of M. tuberculosis infection and IFNγ stimulation on STAT1 and IRF1 exon and intron half-life in total nuclear RNA. STAT1 and IRF1 transcripts were assayed and the half-life of each target region was calculated. The averages are shown with the range or SEM (see Materials and Methods).
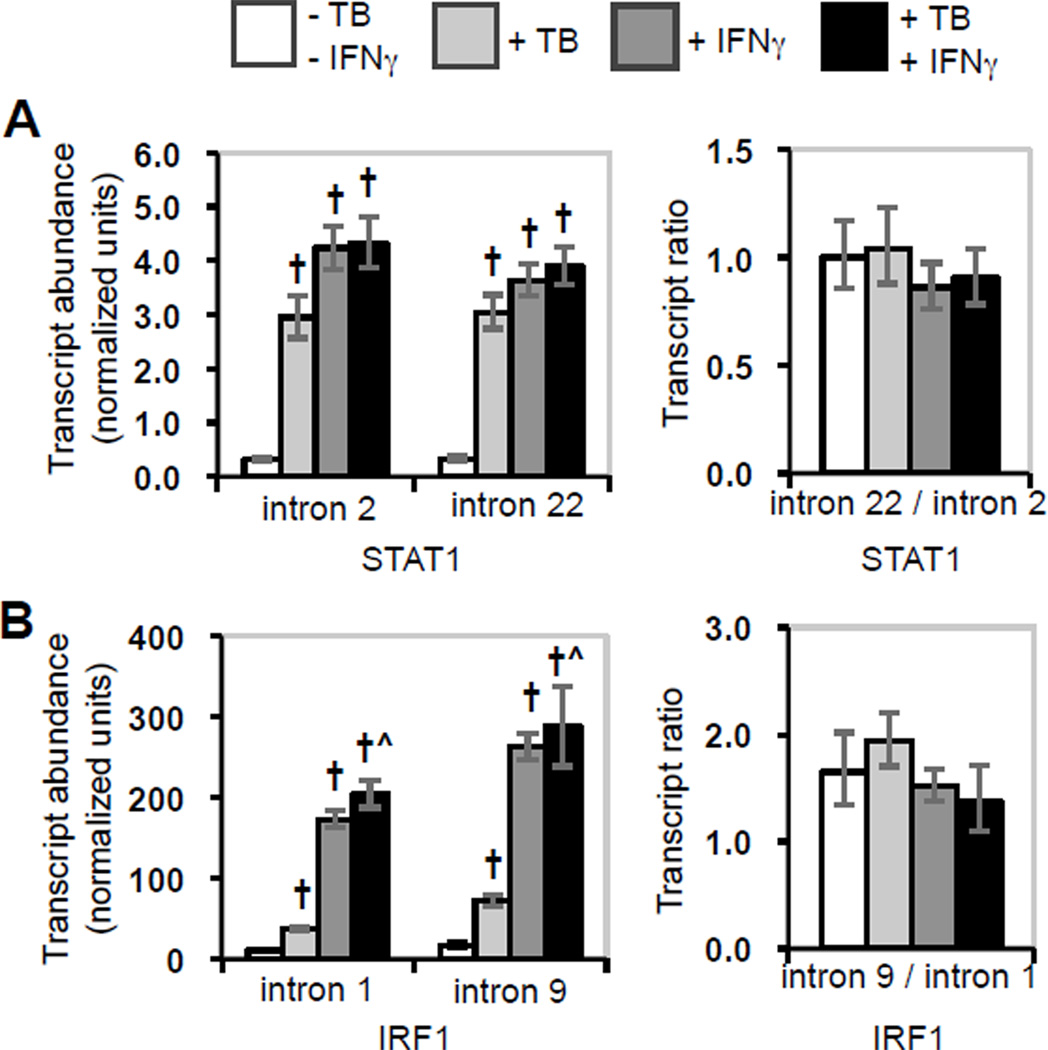
Control of transcript elongation is one mechanism for post-initiation regulation of mRNA biogenesis. Therefore, we considered whether the observed limited induction of STAT1 and IRF1 in M. tuberculosis-infected cells stimulated with IFNγ was caused by down-regulation of elongation. When we assayed introns throughout the STAT1 and in IRF1 transcripts, each target exhibited the induction caused by M. tuberculosis infection, IFNγ stimulation, or both, that was shown by the exon measurements (compare Fig. 1C to Fig. 3, left panel). Little or no change occurred in the ratio of STAT1 intron 2 to intron 22 or in the ratio of IRF1 intron 1 to intron 9 under any condition (Fig. 3, right panel). Thus, regulated elongation contributes to neither positive post-initiation regulation of IRF1 by IFNγ nor negative post-initiation regulation of STAT1 and IRF1 in cells infected with M. tuberculosis and stimulated with IFNγ.
Figure 3.
Effects of M. tuberculosis infection and IFNγ stimulation on STAT1 and IRF1 transcript elongation. Intron sequences in total nuclear STAT1 and IRF1 transcripts were assayed. The average fold-induction of each target region is shown. Error bars represent ± SEM for each average and for the ratios. A) STAT1 intron 2 and intron 22. B) IRF1 intron 1 and intron 9. Statistically significant differences are indicated as described in the legend for Fig. 1.
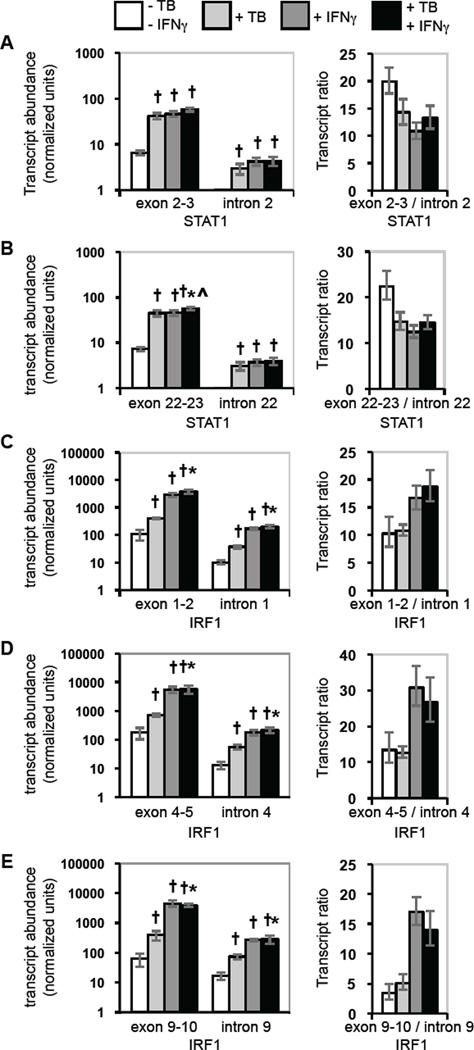
To assess the possibility that regulated splicing accounts for the observed differences between induction of nascent and total nuclear RNA for STAT1 and for IRF1, we measured the abundance of exon-exon junctions and of the respective introns for both transcripts. Because the ratio of spliced to unspliced transcripts reflects the rate of each splice, a change in gene expression due to a regulated splice would alter the ratio. For STAT1, the ratios of the spliced exon 2:3 junction compared to intron 2 and of the spliced exon 22:23 junction compared to intron 22 were similar in uninfected, unstimulated cells, and they decreased similarly with M. tuberculosis infection, IFNγ stimulation, and stimulation of infected cells (Fig. 4A and 4B). In contrast, analysis of IRF1 splices showed that the ratios between the spliced exon 1:2, 4:5 and 9:10 junctions and the respective introns differed from each other in uninfected, unstimulated cells (Fig. 4C–4E). This result suggests that individual splices occur at different rates. The characteristic ratio of each splice was changed little by M. tuberculosis infection alone, but each ratio increased in uninfected and infected cells stimulated with IFNγ. The increase in the level of spliced relative to unspliced transcripts could contribute to the up-regulation of IRF1 expression by IFNγ stimulation of uninfected and infected cells. However, the data do not support a role for regulated splicing in the negative post-initiation regulation that limits induction of STAT1 and IRF1 in infected cells stimulated with IFNγ.
Figure 4.
Effects of M. tuberculosis infection and IFNγ stimulation on spliced and unspliced STAT1 and IRF1 transcripts. Exon junction and intron sequences in total nuclear STAT1 and IRF1 transcripts were assayed. The average fold-induction of each target region is shown. Error bars represent ± SEM for each average and for the ratios. A) STAT1 exon 2–3 junction and intron 2. B) STAT1 exon 22–23 junction and intron 22. C) IRF1 exon 1–2 junction and intron 1. D) IRF1 exon 4–5 junction and intron 4. E) IRF1 exon 9–10 junction and intron 10. Statistically significant differences are indicated as described in the legend for Fig. 1.
Regulated expression of genes that mediate post-initiation mRNA biogenesis
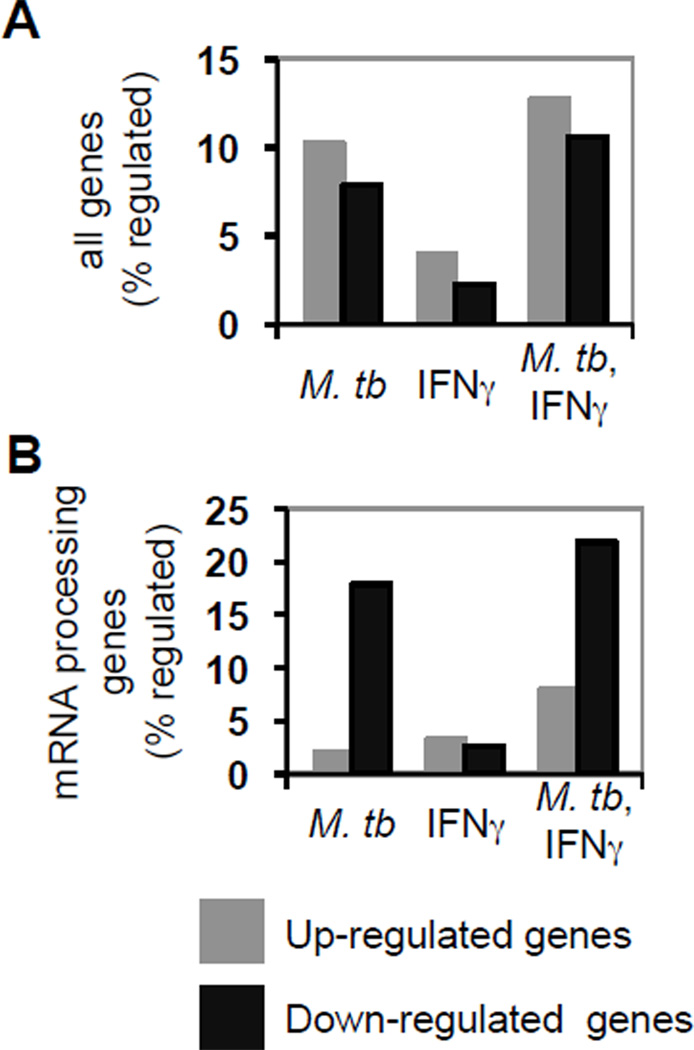
The finding that induction of STAT1 and IRF1 is limited by negative post-initiation regulation in M. tuberculosis-infected cells stimulated with IFNγ suggests that the processes and pathways involved may be down-regulated. To test this possibility, we analyzed gene expression profiles (GEO accession no. GSE17477) for differentiated THP-1 cells that were uninfected or infected with M. tuberculosis with and without IFNγ stimulation. We found that among all regulated genes the number induced was much greater than the number repressed under all conditions tested (Fig. 5A). The same result was obtained in IFNγ-treated cells with genes annotated for mRNA processing in Gene Ontology (GO) (46) (Fig 5B). In contrast, more of those genes were repressed than induced in cells that were infected with M. tuberculosis with and without IFNγ stimulation (Fig. 5B). Thus, the effect of M. tuberculosis infection was different for expression of genes annotated for mRNA processing than for the overall transcriptome. Moreover, mRNA processing genes were over-represented among all the down-regulated genes when various stringency criteria were used for defining regulated genes (see Experimental Procedures). Among the 14 mRNA processing genes deemed regulated under the more stringent criteria, four were responsive to infection alone, while the other 10 were down-regulated only in cells that were infected and stimulated with IFNγ (Table I). Thus, the transcriptome analysis demonstrated a statistically significant down-regulation of mRNA processing genes consistent with limited induction of STAT1 and IRF1 expression due to post-initiation down-regulation when cells infected with M. tuberculosis are stimulated with IFNγ.
Figure 5.
M. tuberculosis infection and IFNγ stimulation of infected cells preferentially down-regulates genes annotated for mRNA processing. The percentage of A) all genes probed and B) genes having a Gene Ontology annotation for mRNA processing or a descendant that were up or down regulated is shown for THP-1 cells that were infected with M. tuberculosis, stimulated with IFNγ, and infected then stimulated, as indicated. Among down-regulated genes defined by a two-sided Cyber-T p value < 0.05 for cells infected with M. tuberculosis or infected with M. tuberculosis then stimulated with IFNγ, each compared to uninfected cells, genes annotated for mRNA processing or a descendant were over-represented (Fisher's exact test p < 0.0001).
Table I.
mRNA processing genes down-regulated by M. tuberculosis-infection with or without IFNγ stimulation
| Gene Symbola | Regulationb | Biological Processc |
|---|---|---|
| HNRPA0 | M. tb ± IFNγ | nuclear mRNA splicing, via spliceosome |
| LSM5 | M. tb ± IFNγ | spliceosome assembly |
| NUDT21 | M. tb ± IFNγ | mRNA 3' end formation |
| SFRS11 | M. tb ± IFNγ | mRNA splicing |
| SNRNP25 | M. tb + IFNγ | mRNA splicing |
| CPSF1 | M. tb + IFNγ | mRNA splicing, mRNA 3' end formation |
| GEMIN4 | M. tb + IFNγ | spliceosome assembly |
| LSM4 | M. tb + IFNγ | spliceosome assembly |
| NHP2L1 | M. tb + IFNγ | spliceosome assembly |
| RBM8A | M. tb + IFNγ | exon junction complex formation |
| SF1 | M. tb + IFNγ | pre-mRNA 3' splice site recognition, spliceosome assembly |
| SFRS14 | M. tb + IFNγ | mRNA splicing |
| SNRPN | M. tb + IFNγ | snRNP assembly |
| TARDBP | M. tb + IFNγ | alternative mRNA splicing |
Human Genome Organization Gene Nomenclature Committee official symbol
- M. tb ± IFNγ (down-regulated by infection with or without IFNγ stimulation)
- M. tb + IFNγ: (down-regulated only by infection and IFNγ stimulation)
biological process annotations from Gene Ontology
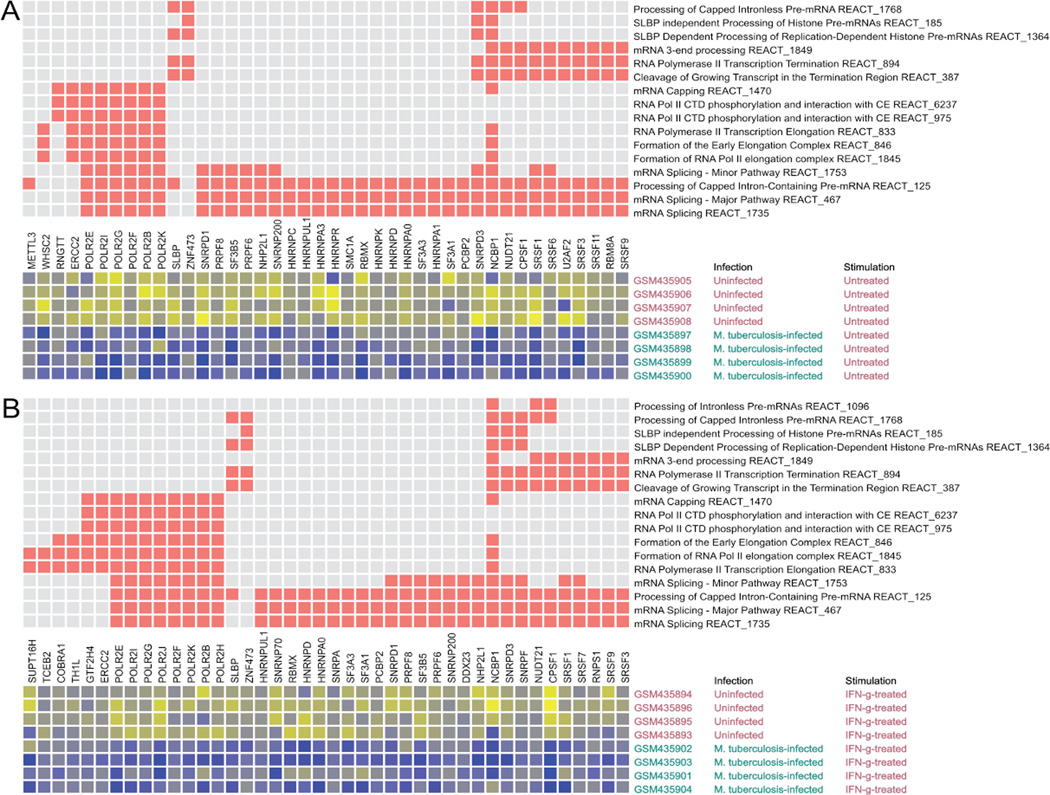
Analogous results were obtained when the transcriptome results were analyzed using a different annotation knowledge-base focused on pathways (Reactome) (http://www.reactome.org) (50, 51) and a different statistical approach (CERNO, see Experimental Procedures) (52, 53) (Fig. 6 and Fig. S4). Of 18 Reactome pathways pertaining to post-initiation mRNA biogenesis, 16 were significantly down-regulated in infected cells (Fig. 6a and Fig. S4a). Significant down-regulation occurred for 17 of these pathways in cells that were infected and stimulated with IFNγ compared to cells that were only IFNγ-stimulated (Fig. 6b and Fig S4b). The analyses of transcriptome data based on annotations for process and for pathways, taken together with the results for expression of STAT1 and IRF1, identify negative post-initiation regulation of mRNA biogenesis as a factor in an essential immunological response to IFNγ stimulation of M. tuberculosis-infected cells.
Figure 6.
Post-initiation mRNA biogenesis Reactome pathways and genes are down-regulated by M. tuberculosis infection of THP-1 cells. Eighteen pathways were tested. The pathways shown are significantly different for the indicated comparisons (A and B) based on a false discovery rate (FDR) criterion of 0.05 for the CERNO test results. The pathway tests were based on the rank order of differential expression for all genes in a pathway (among all transcript measurements of all named genes targeted by the gene expression platform). The top matrix depicts the membership of genes (columns) in pathways (rows) with red squares. The rows and columns of the top matrix are sorted to bring together similar pathway membership patterns. Genes shown are members of one or more pathways that exhibited differential expression for the indicated comparison (one-sided Student's T-test to assess down-regulation, p < 0.05 unless otherwise noted). The lower matrix is a heatmap of expression for each of the significantly regulated genes in each of the samples that were compared. The heatmap shows gradations from higher to lower expression as yellow to blue. The decrease in expression (blue) is evident for infected cells. Gene symbols are shown with the NCBI Gene IDs. A) 16 pathways were down-regulated when comparing infected THP-1 cells to uninfected cells. B) 17 pathways were down-regulated when comparing THP-1 cells infected with M. tuberculosis and stimulated with IFNγ to uninfected, IFN-γ-stimulated THP-1 cells.
Pulmonary tuberculosis is associated with down-regulation of post-initiation mRNA biogenesis pathways
To investigate the clinical relevance of the above results from in vitro infection of macrophage-like THP1 cells, we analyzed transcriptome data obtained from clinical samples. In two studies comparing whole blood from LTBI and PTB donors (47, 48), the analysis revealed that PTB was associated with significant down-regulation of post-initiation mRNA biogenesis pathways (Fig. S4c and S4d). Moreover, PTB donors also exhibited this down-regulation in longitudinal comparisons to gene expression data from samples collected 2 and 12 months after initiation of anti-tuberculous treatment (Fig. S4e and S4f). In a study comparing monocyte-derived macrophages from LTBI donors and cured PTB donors (49), significant down-regulation for 10 of the 18 post-initiation mRNA biogenesis pathways was associated with LTBI (Fig. S4g). Thus, the negative post-initiation regulation of mRNA biogenesis described above as an effect of in vitro infection is also identified as a difference associated with infection/disease status in vivo.
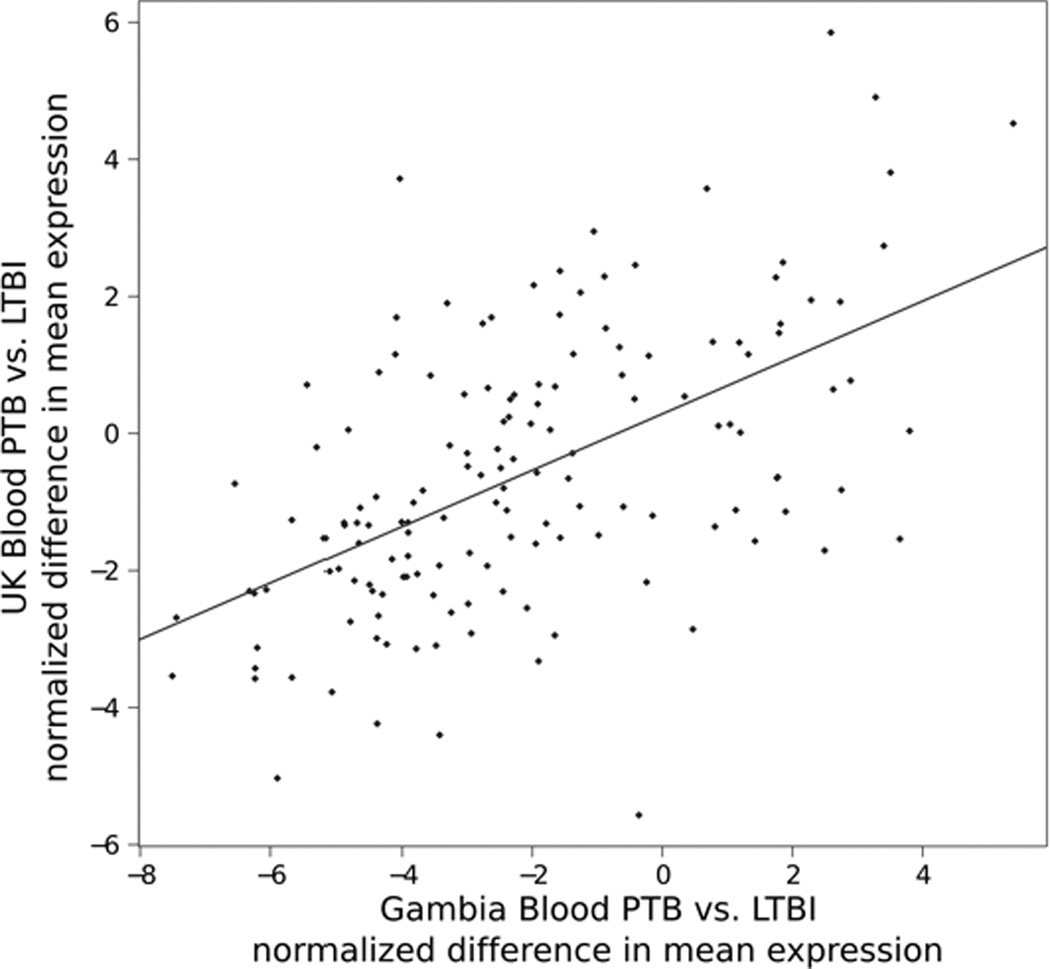
We next considered the relation between expression of individual genes and the disease-stage-specific effects on post-initiation mRNA biogenesis pathways. Even though the data from the two studies of PTB and LTBI donors come from different patients in different geographic regions measured on completely different gene expression platforms (Affymetrix versus Agilent), differential expression of individual transcripts was significantly similar in the two studies (Fig 7). The significance of the correlation provides evidence that PTB is associated with specific changes in the expression of particular genes that mediate post-initiation mRNA biogenesis. Moreover, querying for mRNA post-initiation biogenesis genes differentially expressed at a nominal p < 0.05 level revealed nine genes common across sample comparisons in four independent studies involving three distinct biological systems (THP-1 cells infected in vitro, two studies of whole blood comparing LTBI and PTB donors from different regions using different platforms, and monocyte-derived macrophages from LTBI and cured donors): RNPS1, HNRNPA0, HNRNPUL1, COBRA1, GTF2H4, CPSF1, NHP2L1, SNRNP70, TCEB1. The existence of a core set of regulated genes and the correlation for differential gene expression in data from completely independent studies of PTB and LTBI donors provide strong support for the clinical relevance of decreased gene expression related to post-initiation mRNA biogenesis.
Figure 7.
Two comparisons of PTB to LBTI establish specificity in the differential expression of post-initiation mRNA biogenesis genes. Transcript measurements from two independent studies in two geographic regions that implemented different gene expression microarray platforms were compared at the level of differential expression. Differential expression was measured as the difference in mean expression between PTB and LTBI. For the data from each study, the difference in mean expression was normalized to the respective standard error in the estimate of the difference to provide a T-statistic for each probe or probe set. Positive values indicate higher expression in PTB disease relative to LTBI. T-statistics calculated for differential expression of individual genes in each study demonstrated correlation of the direction and magnitude of differential expression of the same genes measured in the two studies. The linear regression trend (red line) is highly significant (p < 1e-12). The significance provides evidence that the pathway level effect arises from particular changes in expression of specific differentially regulated genes.
Discussion
When M. tuberculosis infects macrophages, complex changes occur in host gene expression. Infection increases host production of IFNγ, which then alters macrophage gene expression. Infection also affects the macrophage response to IFNγ. To understand the consequences for regulation of host cell gene expression, we characterized the induction of STAT1 and IRF1, transcription factors that are key to the cellular IFNγ response and to host defense against the pathogen, and we analyzed transcriptome data obtained from in vitro and in vivo infection. These studies revealed negative post-initiation regulation of mRNA biogenesis as a novel way in which M. tuberculosis alters macrophage gene expression. With IFNγ stimulation of cells infected with M. tuberculosis, but not with IFNγ stimulation alone or with infection alone, less induction of STAT1 and IRF1 transcripts occurred in total nuclear RNA than in nascent RNA, and no difference occurred in transcript turnover. These results strongly suggest that negative post-initiation regulation in vitro limits the expression of these genes induced by IFNγ stimulation of infected cells. In agreement with the molecular data interpretation, analysis of biological process and pathway annotations identified significant effects on genes annotated for post-initiation mRNA biogenesis among genes down-regulated by M. tuberculosis infection with or without IFNγ stimulation in vitro. Moreover, analysis of publicly available transcriptome data from clinical studies identified reduced expression of post-initiation mRNA biogenesis pathways with i) PTB compared to LTBI, ii) PTB compared to treated PTB, and iii) LTBI compared to cured PTB. These data demonstrate that M. tuberculosis infection licenses novel responses to IFNγ that limit induction of STAT1 and IRF1 through negative post-initiation regulation. This limit to induction constitutes a new constraint on IFNγ-induced gene expression in macrophages. Moreover, the limit on induction of STAT1 and IRF1, and perhaps other genes, may be associated with clinically relevant host response to M. tuberculosis, particularly PTB, inasmuch as post-initiation mRNA biogenesis pathways are down-regulated in PTB compared to LTBI or to treated PTB, and in LTBI compared to cured PTB.
The results from both in vitro infection and clinical samples help elucidate the consequences of M. tuberculosis infection for post-initiation regulation of mRNA biogenesis. For in vitro infection, the comparison of different transcript pools for STAT1, IRF1 and several other immune response genes (e.g. CCL2, CXCL10, FCGR1, and MX1; Y.Q. and R.P., unpublished) provided evidence that negative post-initiation regulation does not alter expression uniformly because these genes exhibited various ratios for induction in total nuclear RNA relative to nascent transcripts. Moreover, the transcriptome analyses indicated the involvement of a specific subset of significantly down-regulated genes associated with post-initiation mRNA biogenesis. Nine genes were found common to the in vitro and in vivo results, and the two comparisons of PTB to LTBI transcriptomes from blood demonstrated similar extents of differential expression for individual transcripts. Thus, the consequences of M. tuberculosis infection for post-initiation regulation of mRNA biogenesis are gene-specific.
The negative post-initiation regulation that limits STAT1 and IRF1 expression is not merely an inhibition of a positive response, for at least two reasons. First, IFNγ stimulation caused an increase in the ratio of spliced to unspliced IRF1 nuclear RNA that was similar in uninfected and infected cells, indicating that positive post-initiation regulation of IRF1 by IFNγ still occurs in cells infected with M. tuberculosis. The positive effect may be related to the presence of exonic splicing enhancer sequences in the IRF1 transcript (J.C. and R.P., unpublished), strongly suggesting that IFNγ mediates post-translational regulation of SR family proteins that act at such sites. Second, IFNγ stimulation did not cause positive post-initiation regulation of STAT1 expression, yet STAT1 is subject to the negative regulation. Taken together, the results reveal the simultaneous occurrence of a positive post-initiation response to IFNγ and a distinct negative post-initiation response for these two genes in cells that are also infected with M. tuberculosis.
The case reported above for STAT1 and IRF1 is the first example for the immune response in which down-regulation of genes in post-initiation mRNA biogenesis pathways is associated with negative post-initiation regulation of target gene expression. A parallel occurs in the neuroendocrine system, where a glucocorticoid-mediated decrease in expression of the mRNA processing factor NOVA1 decreases the splicing of gonadotropin releasing hormone pre-mRNA to reduce the level of the mature mRNA in the hypothalamus (38). The negative post-initiation regulation identified by our study is layered over synergistic transcriptional activation of STAT1 and IRF1 expression as a response to M. tuberculosis infection and IFNγ stimulation. Limiting the consequences of synergistic transcriptional activation can account for the comparable induction of IRF1 mRNA in uninfected and M. tuberculosis-infected cells stimulated with IFNγ (21, 56). Many studies have examined changes in macrophage gene expression caused by M. tuberculosis and other mycobacteria (17, 19, 20, 56–64), among others). Effects of infection were described for RNA stability, total RNA abundance, nascent transcript levels, promoter activity, and epigenetic modifications, but changes in nascent and total transcript abundance were not compared. In the present work, comparison of target gene expression at different levels, such as total nuclear RNA relative to nascent transcripts, or spliced relative to unspliced, allowed detection of post-initiation regulation.
This report opens new avenues to investigate the ability of M. tuberculosis to persist or grow despite ongoing macrophage response to IFNγ. Future studies will further define the mechanisms that affect host defense by limiting expression of STAT1 and IRF1. For example, detailed characterization of RNA polymerase II distribution on the STAT1 and IRF1 genes may discover local fluctuations in elongation that were not discovered by comparing the abundance of one upstream and one downstream intron in total nuclear RNA for each transcript. Regulation of 3' transcript cleavage and polyadenylation also needs to be addressed. While characterizing mechanisms for the newly discovered negative regulation that limits macrophage response to IFNγ, it will be important to determine whether manipulating mRNA processing can restore an unconstrained macrophage response to IFNγ and thereby impede M. tuberculosis infection. Moreover, the discovery that PTB down-regulates post-initiation mRNA biogenesis pathways indicates a need to better understand how that effect influences the course of infection.
Supplementary Material
Acknowledgements
We thank Karl Drlica for critically reading the manuscript; Anthony Galante, Olga Tkachenko, and Antony Canova for technical assistance; Yoshihiko Hoshino for assistance with nuclear run-on assays; and Barry Kreiswirth for providing M. tuberculosis TN913.
Footnotes
Support was provided by the National Institutes of Health (Grant Nos. AI37877 and HL68517 to R.P. and HL106788 to M.G. and R.P.) and by the Foundation of UMDNJ (to R.P.).
Abbreviations: IFN regulatory factor (IRF), IFNα/β-stimulated gene (ISG), latent tuberculosis infection (LTBI), pulmonary tuberculosis (PTB)
References
- 1.Corbett EL, Watt CJ, Walker N, Maher D, Williams BG, Raviglione MC, Dye C. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch. Intern. Med. 2003;163:1009–1021. doi: 10.1001/archinte.163.9.1009. [DOI] [PubMed] [Google Scholar]
- 2.Enarson DA, Chiang C-Y, Murray, F J. Global Epoidemiology of Tuberculosis. In: Rom WN, Garay SM, editors. Tuberculosis. second ed. New York: Lippincott Williams & Wilkins; 2004. pp. 13–29. [Google Scholar]
- 3.Kim HR, Hwang SS, Kim HJ, Lee SM, Yoo CG, Kim YW, Han SK, Shim YS, Yim JJ. Impact of extensive drug resistance on treatment outcomes in non-HIV-infected patients with multidrug-resistant tuberculosis. Clin. Infect. Dis. 2007;45:1290–1295. doi: 10.1086/522537. [DOI] [PubMed] [Google Scholar]
- 4.Sotgiu G, Ferrara G, Matteelli A, Richardson MD, Centis R, Ruesch-Gerdes S, Toungoussova O, Zellweger JP, Spanevello A, Cirillo D, Lange C, Migliori GB. Epidemiology and clinical management of XDR-TB: a systematic review by TBNET. Eur. Respir. J. 2009;33:871–881. doi: 10.1183/09031936.00168008. [DOI] [PubMed] [Google Scholar]
- 5.Ernst JD. The immunological life cycle of tuberculosis. Nat Rev Immunol. 2012;12:581–591. doi: 10.1038/nri3259. [DOI] [PubMed] [Google Scholar]
- 6.Chan J, Flynn J. The immunological aspects of latency in tuberculosis. Clin. Immunol. 2004;110:2–12. doi: 10.1016/s1521-6616(03)00210-9. [DOI] [PubMed] [Google Scholar]
- 7.Durbin JE, Hackenmiller R, Simon MC, Levy DE. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 8.Meraz MA, White JM, Sheehan KCF, Bach EA, Rodig SJ, Dighe AS, Kaplan DH, Riley JK, Greenlund AC, Campbell D, Carver-Moore K, DuBois RN, Clark R, Aguet M, Schreiber RD. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 9.Kamijo R, Harada H, Matsuyama T, Bosland M, Gerecitano J, Shapiro D, Le J, Koh SI, Kimura T, Green SJ, Mak TW, Taniguchi T, Vilcek J. Requirement for transcription factor IRF-1 in NO synthase induction in macrophages. Science. 1994;263:1612–1615. doi: 10.1126/science.7510419. [DOI] [PubMed] [Google Scholar]
- 10.Martin E, Nathan Q-W, Xie C. Role of interferon regulatory factor 1 in induction of nitric oxide synthase. J. Exp. Med. 1994;180:977–984. doi: 10.1084/jem.180.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapgier A, Wynn RF, Jouanguy E, Filipe-Santos O, Zhang S, Feinberg J, Hawkins K, Casanova JL, Arkwright PD. Human complete Stat-1 deficiency is associated with defective type I and II IFN responses in vitro but immunity to some low virulence viruses in vivo. J. Immunol. 2006;176:5078–5083. doi: 10.4049/jimmunol.176.8.5078. [DOI] [PubMed] [Google Scholar]
- 12.Dupuis S, Jouanguy E, Al-Hajjar S, Fieschi C, Al-Mohsen IZ, Al-Jumaah S, Yang K, Chapgier A, Eidenschenk C, Eid P, Al Ghonaium A, Tufenkeji H, Frayha H, Al-Gazlan S, Al-Rayes H, Schreiber RD, Gresser I, Casanova JL. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat. Genet. 2003;33:388–391. doi: 10.1038/ng1097. [DOI] [PubMed] [Google Scholar]
- 13.Sugawara I, Yamada H, Mizuno S. STAT1 knockout mice are highly susceptible to pulmonary mycobacterial infection. Tohoku J. Exp. Med. 2004;202:41–50. doi: 10.1620/tjem.202.41. [DOI] [PubMed] [Google Scholar]
- 14.Yamada H, Mizuno S, Sugawara I. Interferon regulatory factor 1 in mycobacterial infection. Microbiol. Immunol. 2002;46:751–760. doi: 10.1111/j.1348-0421.2002.tb02760.x. [DOI] [PubMed] [Google Scholar]
- 15.Pine R, Decker T, Kessler DS, Levy DE, Darnell JE., Jr Purification and cloning of interferon-stimulated gene factor 2 (ISGF2): ISGF2 (IRF-1) can bind to the promoters of both beta interferon- and interferon-stimulated genes but is not a primary transcriptional activator of either. Mol. Cell. Biol. 1990;10:2448–2457. doi: 10.1128/mcb.10.6.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pine R, Canova A, Schindler C. Tyrosine phosphorylated p91 binds to a single element in the ISGF2/IRF-1 promoter to mediate induction by IFN α and IFN γ, and is likely to autoregulate the p91 gene. EMBO J. 1994;13:158–167. doi: 10.1002/j.1460-2075.1994.tb06245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiden M, Tanaka N, Qiao Y, Zhao BY, Honda Y, Nakata K, Canova A, Levy DE, Rom WN, Pine R. Differentiation of Monocytes to Macrophages Switches the Mycobacterium tuberculosis Effect on HIV-1 Replication from Stimulation to Inhibition: Modulation of Interferon Response and CCAAT/Enhancer Binding Protein β Expression. J. Immunol. 2000;165:2028–2039. doi: 10.4049/jimmunol.165.4.2028. [DOI] [PubMed] [Google Scholar]
- 18.Ting LM, Kim AC, Cattamanchi A, Ernst JD. Mycobacterium tuberculosis inhibits IFN-gamma transcriptional responses without inhibiting activation of STAT1. J. Immunol. 1999;163:3898–3906. [PubMed] [Google Scholar]
- 19.Qiao Y, Prabhakar S, Coccia EM, Weiden M, Canova A, Giacomini E, Pine R. Host defense responses to infection by Mycobacterium tuberculosis. Induction of IRF-1 and a serine protease inhibitor. J. Biol. Chem. 2002;277:22377–22385. doi: 10.1074/jbc.M202965200. [DOI] [PubMed] [Google Scholar]
- 20.Prabhakar S, Qiao Y, Hoshino Y, Weiden M, Canova A, Giacomini E, Coccia E, Pine R. Inhibition of Response to Alpha Interferon by Mycobacterium tuberculosis . Infect. Immun. 2003;71:2487–2497. doi: 10.1128/IAI.71.5.2487-2497.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiao Y, Prabhakar S, Canova A, Hoshino Y, Weiden M, Pine R. Posttranscriptional inhibition of gene expression by Mycobacterium tuberculosis offsets transcriptional synergism with IFN-gamma and posttranscriptional up-regulation by IFN-gamma. J. Immunol. 2004;172:2935–2943. doi: 10.4049/jimmunol.172.5.2935. [DOI] [PubMed] [Google Scholar]
- 22.Topisirovic I, Svitkin YV, Sonenberg N, Shatkin AJ. Cap and cap-binding proteins in the control of gene expression. Wiley interdisciplinary reviews. RNA. 2011;2:277–298. doi: 10.1002/wrna.52. [DOI] [PubMed] [Google Scholar]
- 23.Weake VM, Workman JL. Inducible gene expression: diverse regulatory mechanisms. Nature reviews. 2010;11:426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- 24.de Nadal E, Ammerer G, Posas F. Controlling gene expression in response to stress. Nature reviews. 2011;12:833–845. doi: 10.1038/nrg3055. [DOI] [PubMed] [Google Scholar]
- 25.Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 26.Shin C, Manley JL. Cell signalling and the control of pre-mRNA splicing. Nat Rev Mol Cell Biol. 2004;5:727–738. doi: 10.1038/nrm1467. [DOI] [PubMed] [Google Scholar]
- 27.Lutz CS. Alternative polyadenylation: a twist on mRNA 3' end formation. ACS chemical biology. 2008;3:609–617. doi: 10.1021/cb800138w. [DOI] [PubMed] [Google Scholar]
- 28.Condino-Neto A, Newburger PE. Interferon-gamma improves splicing efficiency of CYBB gene transcripts in an interferon-responsive variant of chronic granulomatous disease due to a splice site consensus region mutation. Blood. 2000;95:3548–3554. [PubMed] [Google Scholar]
- 29.Ishibashi F, Mizukami T, Kanegasaki s, Motoda L, Kakinuma R, Endo F, Nunoi H. Improved superoxide-generating ability by interferon g due to splicing pattern change of transcripts in neutrophils from patients with a splice site mutation in CYBB gene. Blood. 2001;98:436–441. doi: 10.1182/blood.v98.2.436. [DOI] [PubMed] [Google Scholar]
- 30.Leeman JR, Gilmore TD. Alternative splicing in the NF-kappaB signaling pathway. Gene. 2008;423:97–107. doi: 10.1016/j.gene.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaresova I, Rozkova D, Spisek R, Janda A, Brazova J, Sediva A. Kinetics of Toll-like receptor-4 splice variants expression in lipopolysaccharide-stimulated antigen presenting cells of healthy donors and patients with cystic fibrosis. Microbes Infect. 2007;9:1359–1367. doi: 10.1016/j.micinf.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 32.Leung E, Hong J, Fraser A, Krissansen GW. Splicing of NOD2 (CARD15) RNA transcripts. Mol. Immunol. 2007;44:284–294. doi: 10.1016/j.molimm.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi K, Hernandez LD, Galan JE, Janeway CA, Jr, Medzhitov R, Flavell RA. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell. 2002;110:191–202. doi: 10.1016/s0092-8674(02)00827-9. [DOI] [PubMed] [Google Scholar]
- 34.Janssens S, Burns K, Tschopp J, Beyaert R. Regulation of interleukin-1- and lipopolysaccharide-induced NF-kappaB activation by alternative splicing of MyD88. Curr. Biol. 2002;12:467–471. doi: 10.1016/s0960-9822(02)00712-1. [DOI] [PubMed] [Google Scholar]
- 35.Hardy MP, O'Neill LA. The murine IRAK2 gene encodes four alternatively spliced isoforms, two of which are inhibitory. J. Biol. Chem. 2004;279:27699–27708. doi: 10.1074/jbc.M403068200. [DOI] [PubMed] [Google Scholar]
- 36.Shell SA, Hesse C, Morris SM, Jr, Milcarek C. Elevated levels of the 64-kDa cleavage stimulatory factor (CstF-64) in lipopolysaccharide-stimulated macrophages influence gene expression and induce alternative poly(A) site selection. J. Biol. Chem. 2005;280:39950–39961. doi: 10.1074/jbc.M508848200. [DOI] [PubMed] [Google Scholar]
- 37.Robinson RT, Khader SA, Martino CA, Fountain JJ, Teixeira-Coelho M, Pearl JE, Smiley ST, Winslow GM, Woodland DL, Walter MJ, Conejo-Garcia JR, Gubler U, Cooper AM. Mycobacterium tuberculosis infection induces il12rb1 splicing to generate a novel IL-12Rbeta1 isoform that enhances DC migration. J. Exp. Med. 2010;207:591–605. doi: 10.1084/jem.20091085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park E, Lee MS, Baik SM, Cho EB, Son GH, Seong JY, Lee KH, Kim K. Nova-1 mediates glucocorticoid-induced inhibition of pre-mRNA splicing of gonadotropin-releasing hormone transcripts. J. Biol. Chem. 2009;284:12792–12800. doi: 10.1074/jbc.M807386200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friedman CR, Quinn GC, Kreiswirth BN, Perlman DC, Salomon N, Schluger N, Lutfey M, Berger J, Poltoratskaia N, Riley LW. Widespread dissemination of a drug-susceptible strain of Mycobacterium tuberculosis . J. Infect. Dis. 1997;176:478–484. doi: 10.1086/514067. [DOI] [PubMed] [Google Scholar]
- 40.Riendeau CJ, Kornfeld H. THP-1 cell apoptosis in response to Mycobacterial infection. Infect. Immun. 2003;71:254–259. doi: 10.1128/IAI.71.1.254-259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sly LM, Lopez M, Nauseef WM, Reiner NE. 1alpha,25-Dihydroxyvitamin D3-induced monocyte antimycobacterial activity is regulated by phosphatidylinositol 3-kinase and mediated by the NADPH-dependent phagocyte oxidase. J. Biol. Chem. 2001;276:35482–35493. doi: 10.1074/jbc.M102876200. [DOI] [PubMed] [Google Scholar]
- 42.Boulikas T. At least 60 ADP-ribosylated variant histones are present in nuclei from dimethylsulfate-treated and untreated cells. EMBO J. 1988;7:57–67. doi: 10.1002/j.1460-2075.1988.tb02783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pine R, Levy DE, Reich N, Darnell JE., Jr Transcriptional stimulation by CaPO4-DNA precipitates. Nucleic Acids Res. 1988;16:1371–1378. doi: 10.1093/nar/16.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choe SE, Boutros M, Michelson AM, Church GM, Halfon MS. Preferred analysis methods for Affymetrix GeneChips revealed by a wholly defined control dataset. Genome Biol. 2005;6:R16. doi: 10.1186/gb-2005-6-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t -test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 46.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T, Wilkinson KA, Banchereau R, Skinner J, Wilkinson RJ, Quinn C, Blankenship D, Dhawan R, Cush JJ, Mejias A, Ramilo O, Kon OM, Pascual V, Banchereau J, Chaussabel D, O'Garra A. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maertzdorf J, Ota M, Repsilber D, Mollenkopf HJ, Weiner J, Hill PC, Kaufmann SH. Functional correlations of pathogenesis-driven gene expression signatures in tuberculosis. PloS one. 2011;6:e26938. doi: 10.1371/journal.pone.0026938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thuong NT, Dunstan SJ, Chau TT, Thorsson V, Simmons CP, Quyen NT, Thwaites GE, Thi Ngoc Lan N, Hibberd M, Teo YY, Seielstad M, Aderem A, Farrar JJ, Hawn TR. Identification of tuberculosis susceptibility genes with human macrophage gene expression profiles. PLoS Pathog. 2008;4:e1000229. doi: 10.1371/journal.ppat.1000229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Joshi-Tope G, Gillespie M, Vastrik I, D'Eustachio P, Schmidt E, de Bono B, Jassal B, Gopinath GR, Wu GR, Matthews L, Lewis S, Birney E, Stein L. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Croft D, O'Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, Jupe S, Kalatskaya I, Mahajan S, May B, Ndegwa N, Schmidt E, Shamovsky V, Yung C, Birney E, Hermjakob H, D'Eustachio P, Stein L. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamaguchi KD, Ruderman DL, Croze E, Wagner TC, Velichko S, Reder AT, Salamon H. IFN-beta-regulated genes show abnormal expression in therapy-naive relapsing-remitting MS mononuclear cells: gene expression analysis employing all reported protein-protein interactions. Journal of Neuroimmunology. 2008;195:116–120. doi: 10.1016/j.jneuroim.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 53.Croze E, Yamaguchi KD, Knappertz V, Reder AT, Salamon H. Interferon-beta-1b-induced short- and long-term signatures of treatment activity in multiple sclerosis. The pharmacogenomics journal. 2012 doi: 10.1038/tpj.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 55.R Development Core Team. A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2010. [Google Scholar]
- 56.Kincaid EZ, Ernst JD. Mycobacterium tuberculosis exerts gene-selective inhibition of transcriptional responses to IFN-gamma without inhibiting STAT1 function. J. Immunol. 2003;171:2042–2049. doi: 10.4049/jimmunol.171.4.2042. [DOI] [PubMed] [Google Scholar]
- 57.Ehrt S, Schnappinger D, Bekiranov S, Drenkow J, Shi S, Gingeras TR, Gaasterland T, Schoolnik G, Nathan C. Reprogramming of the macrophage transcriptome in response to interferon- gamma and Mycobacterium tuberculosis: signaling roles of nitric oxide synthase-2 and phagocyte oxidase. J. Exp. Med. 2001;194:1123–1140. doi: 10.1084/jem.194.8.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gehring AJ, Rojas RE, Canaday DH, Lakey DL, Harding CV, Boom WH. The Mycobacterium tuberculosis 19-kilodalton lipoprotein inhibits gamma interferon-regulated HLA-DR and Fc gamma R1 on human macrophages through Toll-like receptor 2. Infect. Immun. 2003;71:4487–4497. doi: 10.1128/IAI.71.8.4487-4497.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alvarez GR, Zwilling BS, Lafuse WP. Mycobacterium avium inhibition of IFN-gamma signaling in mouse macrophages: Toll-like receptor 2 stimulation increases expression of dominant-negative STAT1 beta by mRNA stabilization. J. Immunol. 2003;171:6766–6773. doi: 10.4049/jimmunol.171.12.6766. [DOI] [PubMed] [Google Scholar]
- 60.Shi S, Nathan C, Schnappinger D, Drenkow J, Fuortes M, Block E, Ding A, Gingeras TR, Schoolnik G, Akira S, Takeda K, Ehrt S. MyD88 primes macrophages for full-scale activation by interferon-gamma yet mediates few responses to Mycobacterium tuberculosis . J. Exp. Med. 2003;198:987–997. doi: 10.1084/jem.20030603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pai RK, Pennini ME, Tobian AA, Canaday DH, Boom WH, Harding CV. Prolonged toll-like receptor signaling by Mycobacterium tuberculosis and its 19-kilodalton lipoprotein inhibits gamma interferon-induced regulation of selected genes in macrophages. Infect. Immun. 2004;72:6603–6614. doi: 10.1128/IAI.72.11.6603-6614.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Y, Curry HM, Zwilling BS, Lafuse WP. Mycobacteria inhibition of IFN-gamma induced HLA-DR gene expression by up-regulating histone deacetylation at the promoter region in human THP-1 monocytic cells. J. Immunol. 2005;174:5687–5694. doi: 10.4049/jimmunol.174.9.5687. [DOI] [PubMed] [Google Scholar]
- 63.Sow FB, Alvarez GR, Gross RP, Satoskar AR, Schlesinger LS, Zwilling BS, Lafuse WP. Role of STAT1, NF-κB, and C/EBPβ in the macrophage transcriptional regulation of hepcidin by mycobacterial infection and IFN-γ. J. Leukoc. Biol. 2009;86:1247–1258. doi: 10.1189/jlb.1208719. [DOI] [PubMed] [Google Scholar]
- 64.Benson SA, Ernst JD. TLR2-dependent inhibition of macrophage responses to IFN-gamma is mediated by distinct, gene-specific mechanisms. PloS one. 2009;4:e6329. doi: 10.1371/journal.pone.0006329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.