Abstract
Neuronal DEG/ENaC (degenerin and epithelial Na+ channel) Na+ channels have been implicated in touch sensation. For example, MEC-4 is expressed in touch neurons in Caenorhabditis elegans and mediates gentle-touch response. Similarly, homologous mammalian ASIC2 and ASIC3 are expressed in sensory neurons and produce touch phenotypes when knocked out in mice. Here, we show that novel DEG/ENaC subunits DELM-1 and DELM-2 (degenerin-like channel mechanosensory linked-1 and degenerin-like channel mechanosensory linked-2) are expressed in glia associated with touch neurons in C. elegans and that their knock-out causes defects in mechanosensory behaviors related to nose touch and foraging, which are mediated by OLQ and IL1 sensory neurons. Cell-specific rescue supports that DELM-1 and DELM-2 are required cell-autonomously in glia to orchestrate mechanosensory behaviors. Electron microscopy reveals that in delm-1 knock-outs, OLQ and IL1 sensory neurons and associated glia are structurally normal. Furthermore, we show that knock-out of DELM-1 and DELM-2 does not disrupt the expression or cellular localization of TRPA-1, a TRP channel needed in OLQ and IL1 neurons for touch behaviors. Rather, rescue of the delm-1 nose-touch-insensitive phenotype by expression of a K+ channel in socket glia and of a cationic channel in OLQ neurons suggests that DELM channels set basal neuronal excitability. Together, our data show that DELM-1 and DELM-2 are expressed in glia associated with touch neurons where they are not needed for neuronal structural integrity or cellular distribution of neuronal sensory channels, but rather for their function.
Introduction
In 1991, our understanding of the molecular basis of touch sensation made a major leap forward with the cloning of the Caenorhabditis elegans ion channel subunit MEC-4 (Driscoll and Chalfie, 1991). MEC-4 and the mammalian epithelial Na+ channel ENaC (Canessa et al., 1993), were the first two members of an ion channel class now known as the DEG/ENaCs (degenerins and epithelial Na+ channels) (Bianchi and Driscoll, 2002). MEC-4 is expressed in body-touch neurons in C. elegans and forms a channel complex with the homologous subunit MEC-10 and accessory subunits MEC-2 and MEC-6 (the MEC channel) (Driscoll and Chalfie, 1991; Chalfie, 1993; Huang and Chalfie, 1994; Huang et al., 1995; Chelur et al., 2002; Goodman et al., 2002). The MEC channel is closed at rest and gated by mechanical forces (O'Hagan et al., 2005). The cloning of MEC-4 opened the door for the molecular identification of mammalian homologs of the ASIC (acid-sensing ion channel) subfamily that subsequent knock-out studies have implicated in touch sensation in mammals (Price et al., 2000, 2001; Kang et al., 2012). All the DEG/ENaC channels implicated to date in touch sensation are expressed in sensory neurons (Chalfie, 1993; Adams et al., 1998; Price et al., 2000, 2001; Chatzigeorgiou et al., 2010b; Zhong et al., 2010; Geffeney et al., 2011).
In mammals, three of the four types of mechanosensors (the Meissner's corpuscles, the Pacinian corpuscles, and the Merkel receptors) are composed of nerve endings and accessory cells. Recent work has implicated accessory cells of mechanosensors in touch sensation. For example, Maricich and colleagues using a mouse model in which Merkel cells do not develop, showed that Merkel cells are required for the proper encoding of Merkel receptor responses (Maricich et al., 2009) and Pawson and colleagues showed that GABA, released by the lamellar cells in Pacinian corpuscles, inhibits glutamatergic excitation during the static portion of sustained pressure (Pawson et al., 2009). However, the genes expressed in accessory cells that contribute to touch sensation are still largely unknown.
In C. elegans, 18 of the 30 postulated mechanosensory neurons (Goodman, 2006) have accessory glial sheath and socket cells (Altun and Hall, 2010). Previous studies have shown that glial cells are important for the function of the sensory neurons that they ensheath (Bacaj et al., 2008b; Wang et al., 2008, 2012). For example, Bacaj and colleagues showed that the activity of the polymodal sensory neuron ASH requires the function of associated glial sheath cells (Bacaj et al., 2008b). Among the mechanosensory neurons whose dendrites are ensheathed by glia are the OLQ and IL1 neurons. OLQ and IL1 neurons have been implicated in nose-touch avoidance and foraging suppression upon anterior body touch (Kaplan and Horvitz, 1993; Hart et al., 1995; Kindt et al., 2007).
We show here that DEG/ENaC channel subunits DELM-1 and DELM-2 (degenerin-like channel mechanosensory linked-1 and degenerin-like channel mechanosensory linked-2) are expressed in OLQ and IL1 glial socket cells and are needed cell-autonomously for nose-touch and foraging-suppression behaviors. DELM-1 and DELM-2 are not needed for cellular structural integrity or subcellular localization of neuronal sensory channels. Rather our data support that, by setting the level of neuronal excitability, DELM-1 and DELM-2 are required for the function of associated touch neurons. Our results demonstrate that DEG/ENaC channels, thought to be needed solely in sensory neurons to orchestrate touch sensation, are also required in glia.
Materials and Methods
C. elegans strains and growth.
Nematode strains were maintained at 20°C on standard nematode growth medium seeded with Escherichia coli strain OP50 (Brenner, 1974). Wild-type animals were N2 Bristol. All animals used in this study were hermaphrodites. Males were used for crosses only. Germline transformation was performed as described previously (Mello et al., 1991). Mutants were outcrossed three times before phenotypic analysis, crossing with other strains, and generation of transgenic animals. Double and triple mutants were generated by standard genetic crosses. Mutations were followed through the crosses by PCR.
Molecular biology.
We constructed Pdelm-1::RFP by swapping vap-1 promoter in the previously described Pvap-1::RFP construct (Wang et al., 2008) with the promoter of DELM-1 (2 kb upstream of the start codon). Pdelm-1::GFP and Pdelm-2::GFP were generated by subcloning the promoter regions of DELM-1 (2 kb upstream of the start codon) and DELM-2 (1.4 kb upstream of the start codon) into the pPD95.75 vector in frame with GFP. To generate Pitx-1::GFP, we replaced the promoter of DELM-2 in the Pdelm-2::GFP with the promoter of the ITX-1 gene (1.8 kb upstream of the start codon). To construct Pdelm-1::DELM-1, we amplified the DELM-1 genomic sequence by PCR and subcloned it into Pdelm-1::RFP vector downstream of delm-1 promoter. To generate Pdelm-2::DELM-2, DELM-2 genomic sequence was amplified by PCR and subcloned into Pdelm-2::GFP vector downstream of delm-2 promoter. To target expression of DELM-1, DELM-2, and ACD-1 in OLQ and IL1 socket glia, we subcloned DELM-1, DELM-2, and ACD-1 (Wang et al., 2008) cDNA sequences into the Pitx-1::GFP vector downstream of the promoter of the ITX-1 gene. DELM-1 and DELM-2 cDNA were cloned by RT-PCR using the one-step RT-PCR kit Titanium (Clontech) and gene-specific primers designed according to the DELM-1-predicted and DELM-2-predicted cDNA sequences available on WormBase (www.wormbase.org). Total RNA from a C. elegans mixed-age population was used as template. All PCR products were initially cloned into pCR-2.1-TOPO vector for sequence verification and amplification. To drive expression of DELM-1 in sensory neurons and amphid socket glia, we replaced itx-1 promoter, functional in OLQ and IL1 socket glia, with the ocr-4 promoter [∼4.8 kb from the start codon, OLQ neurons (Tobin et al., 2002)], the egl-46 promoter [∼3 kb from the start codon, FLP and PVD neurons (Yu et al., 2003)], the dat-1 promoter [∼0.8 kb from the start codon, CEP, ADE, and PDE neurons (Nass et al., 2001)], the sra-6 promoter [∼3 kb from the start codon, ASH neurons (Troemel et al., 1995)], and the itr-1 promoter [∼2.3 kb between the end of exon 1 and the end of exon 2, amphid socket glia (Gower et al., 2001)]. The promoter sequences used for the construction of these rescue constructs were according to Chatzigeorgiou and colleagues (Chatzigeorgiou et al., 2010a). We constructed Pocr-4::GCaMP1.0 for the calcium imaging experiments by swapping str-2 promoter in the Pstr-2::GCaMP1.0 construct (Chalasani et al., 2007) with the promoter of ocr-4 (∼4.8 kb upstream of the start codon). To target expression of IRK-2 in the OLQ and IL1 socket glia, we subcloned IRK-2 cDNA sequence into pPD95.75 under the control of the itx-1 promoter. To drive expression of mosquito TRPA1 (AgTRPA1) in the OLQ neurons, we replaced GCaMP1.0 in the Pocr-4::GCaMP1.0 construct with AgTRPA1 (Kang et al., 2011).
C. elegans strains.
C. elegans strains used were as follows: RB1177 delm-1(ok1226), RB1523 delm-2(ok1822), RB1052 trpa-1(ok999), CX10 osm-9(ky10), Ljex114 TRPA1::GFP. We created the following arrays in this work: blcEx33 [Pdelm-2::GFP], blcEx43 [Pdelm-1::GFP], blcEx38 [Pdelm-1::RFP; Pdelm-2::GFP], blcEx60 [Pdelm-1::RFP; Pitx-1::GFP], blcEx79 [Pdelm-2::IRK-2;Pdelm-2::GFP], blcEx89 [Pocr-4::GCaMP1.0;rol-6]. Strains made were as follows: BLC100 delm-2(ok1822) I;delm-1(ok1226) IV, BLC 166 delm-2(ok1822) I;delm-1(ok1226) IV;trpa-1(ok999) IV, BLC113 delm-1(ok1226) IV;blcEx53 [Pdelm-1::DELM-1; Pdelm-2::GFP], BLC152 delm-1(ok1226) IV;blcEx70 [Pitx-1::cDELM-1; Pdelm-2::GFP], BLC98 delm-2(ok1822) I;blcEx51 [Pdelm-2::DELM-2; Pdelm-2::GFP], BLC156 delm-2(ok1822) I;blcEx72 [Pitx-1::cDELM-2; Pdelm-2::GFP], BLC160 delm-1(ok1226) IV;blcEx51 [Pdelm-2::DELM-2; Pdelm-2::GFP], BLC161 delm-2(ok1822) I;blcEx53 [Pdelm-1::DELM-1; Pdelm-2::GFP], BLC185 delm-1(ok1226) IV; blcEx85 [Pegl-46::cDELM-1; Pdelm-2::GFP], BLC191 delm-1(ok1226) IV; blcEx86 [Pitr-1::cDELM-1; Pdelm-2::GFP], BLC192 delm-1(ok1226) IV; blcEx87 [Pdat-1::cDELM-1; Pdelm-2::GFP], BLC193 delm-1(ok1226) IV; blcEx88 [Pocr-4::cDELM-1; Pdelm-2::GFP], BLC219 delm-1(ok1226) IV; blcEx99 [Psra-6::cDELM-1; Pdelm-2::GFP], BLC240 trpa-1(ok999) IV; blcEx53[Pdelm-1::DELM-1; Pdelm-2::GFP], BLC173 delm-1(ok1226) IV; blcEx79 [Pdelm-2::IRK-2;Pdelm-2::GFP], BLC207 delm-1(ok1226) IV; blcEx89 [Pocr-4::GCaMP1.0;rol-6], BLC226 delm-2(ok1822) I;delm-1(ok1226) IV; blcEx79 [Pdelm-2::IRK-2;Pdelm-2::GFP], BLC237 delm-1(ok1226) IV; blcEx100[Pitx-1::ACD-1; Pdelm-2::GFP;unc-122], BLC240 trpa-1(ok999) IV; blcEx53 [Pdelm-1::DELM-1; Pdelm-2::GFP], BLC247 delm-1(ok1226) IV; blcEx102 [Pocr-4::AgTRPA1;Pdelm-2::GFP], BLC249 delm-1(ok1226) IV;TRPA-1::GFP, BLC250 delm-2(ok1882) I;TRPA-1::GFP, BLC252 trpa-1(ok999) IV; blcEx102 [Pocr-4::AgTRPA1;Pdelm-2::GFP], BLC256 trpa-1(ok999) IV;blcEx79 [Pdelm-2::IRK-2;Pdelm-2::GFP],BLC257 delm-1(ok1226) IV;blcEx89 [Pocr-4::GCaMP1.0;rol-6]; blcEx70 [Pitx-1::cDELM-1; Pdelm-2::GFP], BLC258 trpa-1(ok999) IV; blcEx89 [Pocr-4::GCaMP1.0;rol-6]. DELM-1 and DELM-2 indicate genomic DNA. cDELM-1 and cDELM-2 indicate cDNA.
In the delm-1(ok1226) mutant, a ∼2 kb fragment from the nucleotide 103 to nucleotide 2093 of the genomic sequence is missing. In the delm-2(ok1822) mutant, a 1kb fragment, including 568 bp before the start codon up to part of the fourth exon, is deleted. Since in both strains the coding regions are lacking multiple exons (see Fig. 2A), the translation of the full-length proteins is compromised. To verify the homozygosis of delm-1(ok1226) in the crosses, we used the following primers: 5′-ATGAATTCGCCTCCTATATCCCCTTATCATGTTG-3′, 5′-CTGATCCTCCCAAGAACATTCCCATATTTCCAC-3′, and 5′-ATGAATCAGTGTGGATGCACTCCGG-3′. The size of the amplified delm-1(ok1226) and wild-type bands are ∼400 and ∼600 bp respectively. To verify the homozygosis of delm-2(ok1822) in crosses, we used the following primers: 5′-GGTGTTCACGGGCGTGAG-3′, 5′-CCGTAGAACGGGAGACCCAG-3′, and 5′-ACAATTGCCAATTCGAGCTGCC-3′. The size of the amplified delm-2(ok1822) and wild-type bands are ∼430 and ∼620 bp respectively. Note that we did not detect the WT bands in delm-1 and delm-2 knock-out strains, suggesting that duplication and insertion elsewhere in the genome of DELM-1 and DELM-2 wild-type sequences is highly unlikely in these strains.
Figure 2.
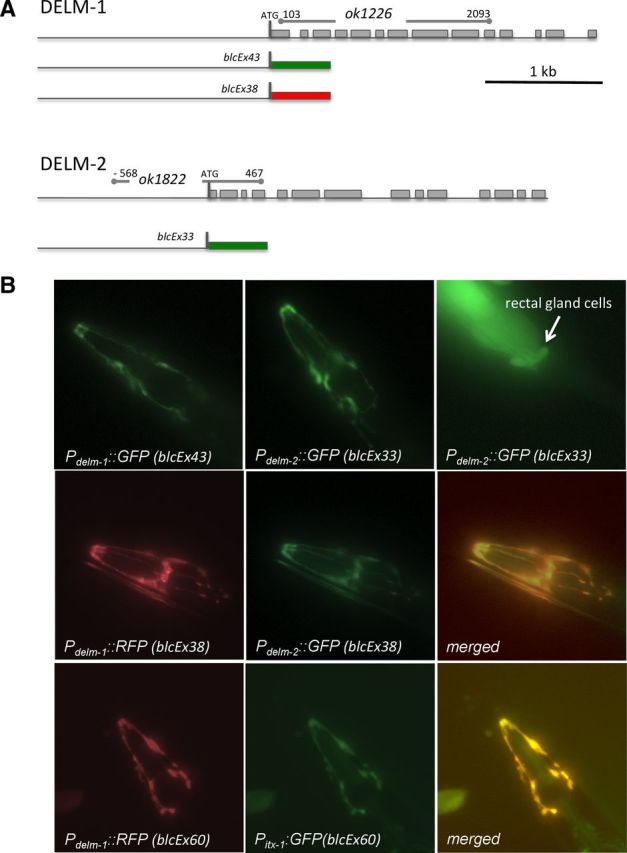
DELM-1 and DELM-2 expression patterns. A, Schematic representation of DELM-1 and DELM-2 gene structures, of the location of the deleted region in the knock-outs, and of the fluorescent protein constructs used. Lines and boxes indicate predicted untranslated and translated regions respectively. Green and red boxes designate GFP and RFP respectively. B, Fluorescent micrographs of transgenic C. elegans expressing GFP and RFP under the control of delm-1, delm-2, and itx-1 promoters as indicated. In the second and third rows on the far right, two merged images show overlapping of GFP and RFP signals supporting the idea that DELM-1 and DELM-2 are expressed in OLQ and IL1 glial socket cells (Haklai-Topper et al., 2011). Except for weak fluorescent signal in rectal gland cells associated with expression of Pdelm-2::GFP (upper far right), we could not detect expression of the fluorescent proteins under the control of delm-1 or delm-2 promoter in any other cell type.
Behavioral assays.
All behavioral assays were undertaken blind to the genotypes following previously described procedures. Briefly, for nose-touch assays, an eyelash hair was laid on the surface of freshly seeded plate in front of a forward-moving animal. The animal's response was recorded as “reversal” if the animal backed up and as “head withdrawal” if the animal moved the head away from the eyelash hair (Kindt et al., 2007). Data are displayed as responses given by each animal over the total of five trials (at least 15 s interval). Means were obtained by averaging these ratios obtained from different animals. Ten animals per strain were tested in each experiment. For the foraging suppression assay, forward-moving animals were touched with an eyelash hair on the anterior part of the body. The percentage of animals that suppressed foraging when touched was recorded in each strain. Thirty animals were assayed per strain in each experiment (Kindt et al., 2007). For the octanol avoidance assays, we dipped an eyelash hair glued on a toothpick in 30% octanol and placed it in front of a forward-moving animal on a plate without food. We recorded the time it took for the animal to respond to the odor by reversing direction (Troemel et al., 1995). For the SDS-avoidance assay, a drop of 0.1% SDS solution was delivered ∼1 mm anterior to a forward-moving animal on an unseeded plate (Bargmann et al., 1990; Hilliard et al., 2002). We scored the response as positive if the animal moved backward upon encountering the drop. Twenty animals were assayed per genotype and the percentage of animals responding to the drop was then calculated. For the osmotic-avoidance assay, a 1 cm diameter ring of 8 m glycerol was made on an unseeded plate (Hart et al., 1999). Twenty animals were then transferred inside the ring and allowed to crawl for 8 min. The animals that remained inside the ring were then counted and expressed as ratio of retained animals.
Fluorescence microscopy.
Fluorescent photographs were taken using a Leica microscope equipped with GFP and rhodamine filters and a Spot RTslider camera (Diagnostic Instruments) with Spot32 acquisition software. For TRPA-1::GFP quantification, photographs were taken using the same exposure time (200 ms, an exposure time that did not cause saturation of the camera detector) and quantifications were performed in ImageJ.
Transmission electron microscopy.
Previously described fixation methods were used (Perens and Shaham, 2005). Ultrathin serial sections (60 nm) were cut using a Reichert Ultra-Cut-E ultramicrotome and collected on Pioloform-coated single-slot copper grids. Electron microscopy (EM) images for every other section were acquired using an FEI Tecnai G2 Spirit BioTwin transmission electron microscope operating at 80 kV with a Gatan 4000 × 4000-pixel digital camera.
Calcium imaging.
For nose-touch stimulation and calcium imaging, we used previously described procedures (Kindt et al., 2007). Animals were glued on 2% agarose pads prepared using extracellular saline (145 mm NaCl, 5 mm KCl, 1 mm CaCl2, 5 mm MgCl2, 20 mm d-glucose, 10 mm HEPES, pH 7.2) using surgical glue (Gluture) and immediately immersed in the extracellular saline plus 2 mm serotonin. Serotonin was added to the extracellular saline because it was reported to enhance nose-touch avoidance response (Hilliard et al., 2005; Kindt et al., 2007). Animals were then transferred to a chamber mounted on a Nikon Eclipse E600FN microscope equipped with GFP filters. The nose-touch stimulator was a glass capillary (outer diameter, 1.5 mm; inner diameter, 0.83 mm) pulled by a micropipette puller. The tip of the glass capillary was flamed to ∼10 μm. The position and the movement of the stimulator were controlled by a Physik Instrumente M-111.1 DG microtranslation stage with C-863.10 Mercury DC-motor controller. The tip of the pipette was placed perpendicularly to the animal's nose at a distance of 150 μm and then moved forward to indent the animal nose 8 μm. The pipette was held in place for 2 s and then moved back.
Images were acquired at 5 Hz using TILLvisION v3.3 software and CCD camera Imago (T.I.L.L. Photonics). Typical exposure time was 50–80 ms. The image stacks were then analyzed using ImageJ. Before fluorescence intensity determination, images were corrected for movement using ImageJ TurboReg. The fluorescence intensity of the region of interest was output to a data log file. Data were analyzed and plotted using Origin.
Xenopus laevis oocytes.
DELM-1 and DELM-2 complementary RNAs (cRNAs) were synthesized using T7 mMESSAGE mMACHINE kit. cRNAs were then purified and run on denaturating agarose gels for size and integrity verification. cRNA quantification was performed spectroscopically. Stage V–VI oocytes were selected among multistaged oocytes dissected by 2 h collagenase treatment (2 mg/ml in Ca2+-free OR2 solution) from Xenopus laevis ovaries. Oocytes were injected with 5 ng/oocyte of cRNA and incubated in OR2 medium, which consists of 82.5 mm NaCl, 2.5 mm KCl, 1 mm CaCl2, 1 mm MgCl2, 1 mm Na2HPO4, 0.5 g/l polyvinyl pyrolidone, and 5 mm HEPES, pH 7.2, supplemented with penicillin and streptomycin (0.1 mg/ml) and 2 mm Na-pyruvate at 20°C for 2–3 d before recordings. Currents were measured using a two-electrode voltage-clamp amplifier (GeneClamp 500B; Molecular Devices) at room temperature. Electrodes (0.2– 0.5 MΩ) were filled with 3 m KCl, and oocytes were perfused with a solution containing (in mm) the following: 100 NaCl, 2 KCl, 1 CaCl2, 2 MgCl2, and 10 HEPES, pH 7.2. For ion selectivity experiments, we substituted 100 mm NaCl with 100 mm LiCl, 100 mm KCl, and 100 mm N-methyl-d-glucamine (NMDG), respectively. The oocyte membrane was clamped at −30 mV and stepped from −160 to +60 mV.
Single-channel recordings were obtained from Xenopus oocytes after manual removal of the vitelline membrane. Oocytes were perfused and electrodes (1–2 MΩ) were filled with a solution containing (in mm) 100 NaCl, 2 KCl, 1 CaCl2, 2 MgCl2, and 10 HEPES, pH 7.2. Currents were recorded using a patch-clamp amplifier (Axopatch 200B; Molecular Devices) at room temperature. Oocytes were held at 0 mV and stepped from −160 to + 100 mV. We used the pCLAMP suite of programs (Molecular Devices) for data acquisition and analysis. Currents were filtered at 200 Hz and sampled at 1 kHz.
Statistics.
Data were compared by ANOVA with post hoc Bonferroni correction or by t test (*p ≤ 0.05; **p ≤ 0.01).
Results
F23B2.3 and C24G7.1 genes encode two DEG/ENaC channel subunits
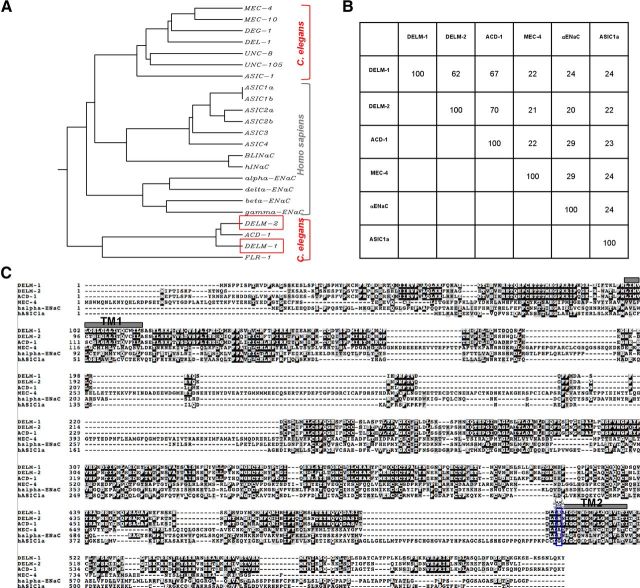
Previous work from our laboratory demonstrated that the DEG/ENaC channel ACD-1 is expressed in C. elegans glial amphid sheath cells (Wang et al., 2008). Knock-out of ACD-1 in combination with mutations in the neuronal channels TAX-2 and DEG-1 impairs chemotaxis to tastants and odors, and avoidance of acidic solutions, respectively (Wang et al., 2008, 2012). In this work we sought to identify C. elegans DEG/ENaC subunits with similarity with ACD-1. There are 28 predicted genes in C. elegans that encode DEG/ENaC subunits (Goodman and Schwarz, 2003). We focused on F23B2.3 and C24G7.1, renamed DELM-1 and DELM-2, which share high similarity with ACD-1 (67% identity, 78% similarity; and 70% identity, 83% similarity, respectively) (Fig. 1A–C). DELM-1 and DELM-2 share lower homology with C. elegans mechanosensitive channel subunit MEC-4 and mammalian α ENaC and ASIC1a subunits (Fig. 1B,C).
Figure 1.
The C. elegans DEG/ENaC channels DELM-1 and DELM-2 share similarity with glial channel ACD-1. A, Dendrogram showing similarity between known C. elegans and human DEG/ENaC channel subunits. B, Percentage of identity between DELM-1, DELM-2 and C. elegans ACD-1 and MEC-4, and human α ENaC and ASIC1a. C, DELM-1 and DELM-2 protein sequences and alignment with ACD-1, MEC-4, α ENaC, and ASIC1a. Identical and similar amino acids are in black and gray backgrounds respectively. Gray boxes TM1 and TM2 indicate the transmembrane domains. The blue box and skull indicate the residue that, when mutated to a bulky amino acid, induces hyperactivation of some of the neuronal DEG/ENaCs (Driscoll and Chalfie, 1991; Champigny et al., 1998; Darboux et al., 1998; García-Añoveros et al., 1998; Brown et al., 2007).
DELM-1 and DELM-2 are expressed in C. elegans glia associated with touch neurons
To establish DELM-1 and DELM-2 expression patterns, we generated transgenic animals expressing fluorescent proteins under the control of the promoters of DELM-1 and DELM-2 (Fig. 2A). We first generated transgenic animals expressing Pdelm-1::GFP and Pdelm-2::GFP separately. We found that the GFP signal was localized in cells in the head of C. elegans that appeared to be strikingly similar (Fig. 2B, first row). To test whether these were the same cells, we generated transgenic animals coexpressing Pdelm-1::RFP and Pdelm-2::GFP. Colocalization of RFP and GFP signals confirmed that indeed DELM-1 and DELM-2 promoters are functional in the same cells (Fig. 2B, second row). Based on the position and shape of these cells, we hypothesized that they might be OLQ and IL1 glial socket cells, which ensheath the dendrites of OLQ and IL1 sensory neurons. We thus generated transgenic animals expressing Pdelm-1::RFP and GFP under the control of the promoter of the ITX-1 gene, expressed in OLQ and IL1 glial socket cells (Haklai-Topper et al., 2011). The overlapping of GFP and RFP signals supported that DELM-1 and DELM-2 are expressed in OLQ and IL1 glial socket cells (Fig. 2B, third row).
Knock-out of DELM-1 and DELM-2 causes touch defects
OLQ and IL1 sensory neurons are implicated in nose-touch avoidance response and suppression of foraging upon anterior body touch (Kaplan and Horvitz, 1993; Hart et al., 1995; Alkema et al., 2005; Kindt et al., 2007). To determine whether DELM-1 and DELM-2 are needed for the function of OLQ and IL1 sensory neurons, we acquired delm-1 and delm-2 knock-out mutants (delm-1(ok1226) and delm-2(ok1822); Fig. 2A) and subjected them to nose-touch and foraging-suppression assays. In the delm-1(ok1226) mutant a ∼2 kb fragment from nucleotide 103 to nucleotide 2093 of the genomic sequence is missing. This results in the deletion of ¾ of the coding region (from amino acid 24 to amino acid 447, which includes part of the N terminus, the first transmembrane domain, and almost the entire extracellular domain). In the delm-2(ok1822) mutant, a 1 kb fragment, including 568 bp before the start codon up to part of the fourth exon, is deleted, causing the introduction of a premature stop codon at nucleotide 492. Since in both strains the coding regions are either lacking multiple exons (delm-1) (Fig. 2A) or contain premature stop (delm-2), the translation of the full-length proteins is compromised.
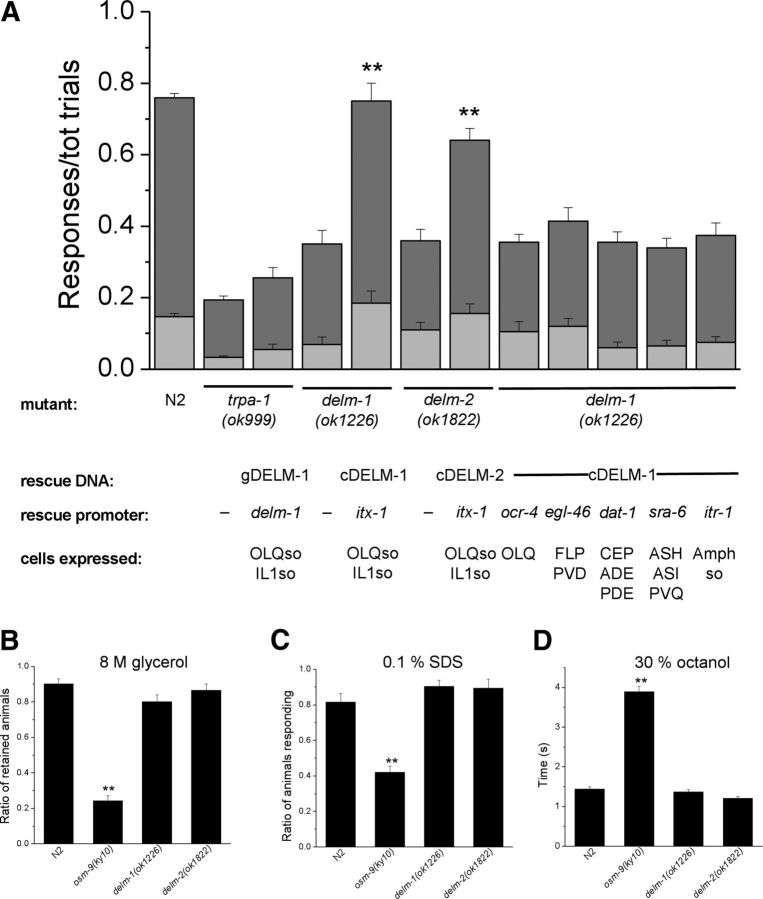
We found that delm-1 and delm-2 mutants were defective in nose-touch response (Fig. 3A). Interestingly, the nose-touch sensitivity of delm-1 and delm-2 mutants was comparable to that of trpa-1 mutants, although the ratio of positive responses was statistically higher. TRPA-1 is a TRP channel expressed in OLQ and IL1 neurons and needed for nose-touch responses and foraging-suppression behavior (Kindt et al., 2007). The nose-touch defective phenotype of delm-1 and delm-2 mutants was substantially rescued by expression of DELM-1 and DELM-2 genomic sequences in delm-1 and delm-2 mutants (Fig. 3A). These data support the idea that the nose-touch defects of delm-1 and delm-2 mutants are caused by DELM-1 and DELM-2 molecular lesions and not by other underlying mutations in these knock-out strains.
Figure 3.
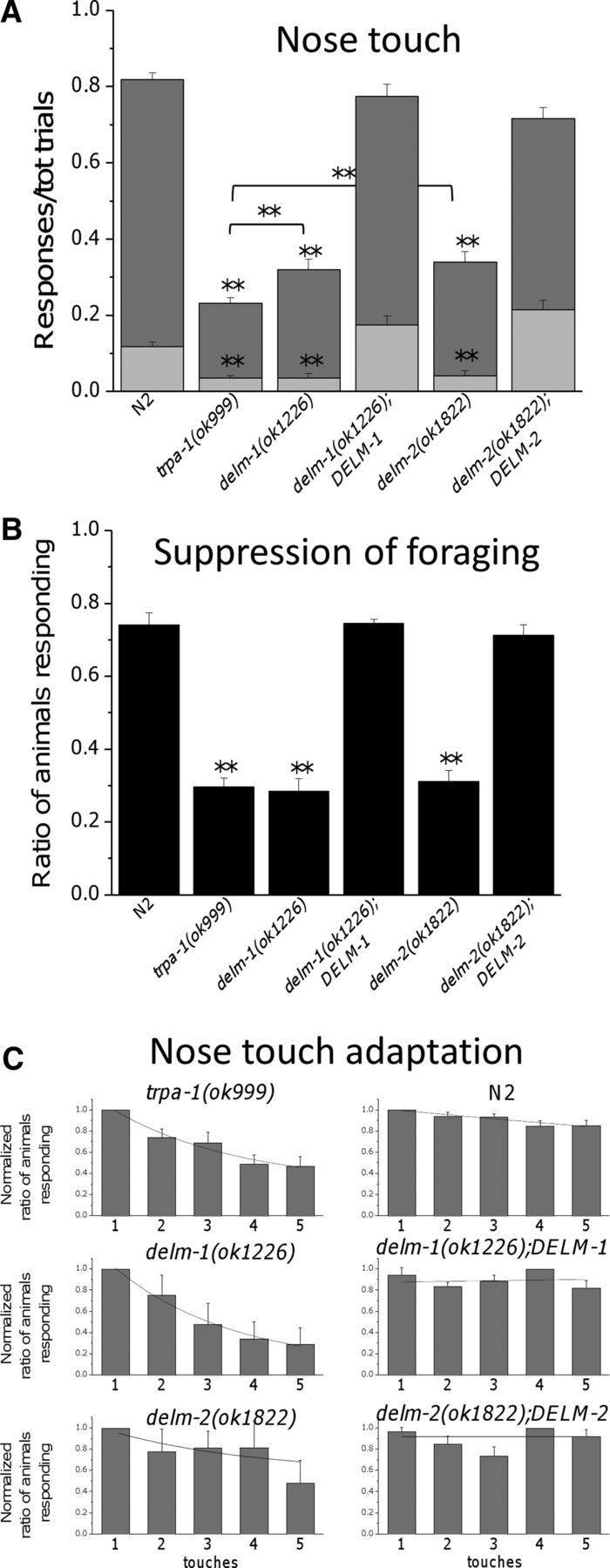
Knock-out of DELM-1 and DELM-2 reduces C. elegans nose-touch sensitivity and foraging suppression. A, Ratios of reversals (dark-gray bars) and head withdrawals (light-gray bars) in wild type (N2, 0.701 ± 0.016 and 0.117 ± 0.012), delm-1(ok1226) (0.284 ± 0.026 and 0.036 ± 0.010), delm-2(ok1822) (0.298 ± 0.027 and 0.040 ± 0.013), delm-1;DELM-1 (0.600 ± 0.031 and 0.174 ± 0.022) and delm-2;DELM-2 (0.501 ± 0.028 and 0.214 ± 0.024) C. elegans upon nose-touch stimulation. trpa-1(ok999) mutant was used as a control (0.196 ± 0.015 and 0.007). Number of animals assayed was 160, 157, 50, 70, 59, and 69. B, delm-1 and delm-2 mutant C. elegans do not suppress head oscillations associated with foraging when moving backward in response to anterior touch. Ratios of animals responding were as follows: 0.740 ± 0.032 for N2 (wild type), 0.296 ± 0.023 for trpa-1, 0.283 ± 0.034 for delm-1, 0.744 ± 0.011 for delm-1;DELM-1, 0.311 ± 0.029 for delm-2, and 0.711 ± 0.029 for delm-2;DELM-2. Number of assays was 9, 9, 4, 3, 3, and 3, with 30 animals tested in each assay. Data are expressed as means ± SE, **p < 0.01 (ANOVA), statistically significant difference with wild type or between two indicated strains. C, The ratio of animals responding to each of the five consecutive touches (30 s interval) was calculated for each experiment, averaged, and then normalized. Number of assays was 16 for trpa-1(ok999), 5 for delm-1(ok1226), 6 for delm-2(ok1822), 15 for N2 (wild type), 7 for delm-1(ok1226);DELM-1, and 7 for delm-2(ok1822);DELM-2 with 10 animals tested in each assay. Data are means ± SE and are fitted by exponential decay. The ratio of animals that responded to the first touch, before normalization, was as follows: 0.38 ± 0.06 ** for trpa-1(ok999), 0.6 ± 0.08 ** for delm-1(ok1226), 0.51 ± 0.13 ** for delm-2(ok1822), 0.89 ± 0.04 for N2 (wild type), 0.83 ± 0.07 for delm-1(ok1226);DELM-1, and 0.78 ± 0.04 for delm-2(ok1822);DELM-2. Data were fitted by single exponential decay. Tau values were 2.75 for trpa-1(ok999), 2.55 for delm-1(ok1226), 3.13 for delm-2(ok1822), 26.7 for N2 (wild type), and 20.1 for delm-1(ok1226);DELM-1. Fit for delm-2(ok1822);DELM-2 with single exponential was not optimal and yielded unusually large tau values (>25,000). Nevertheless, data are consistent with slower adaptation in this transgenic strain compared with delm-2(ok1822). The average normalized ratio of animals that responded to the fifth touch was 0.468 ± 0.09 ** for trpa-1(ok999), 0.292 ± 0.015 ** for delm-1(ok1226), 0.480 ± 0.21 for delm-2(ok1822), 0.852 ± 0.05 for N2 (wild type) *, 0.823 ± 0.07 for delm-1(ok1226);DELM-1, and 0.921 ± 0.06 for delm-2(ok1822);DELM-2. *p ≤ 0.05; **p ≤ 0.01; by ANOVA. Statistics were by comparison with the first touch.
We also found that delm-1 and delm-2 mutants, in contrast with wild-type animals, did not exhibit suppression of foraging when touched on the anterior body (Fig. 3B). The touch suppression of foraging defect was also rescued by expression of DELM-1 and DELM-2 genomic sequences in delm-1 and delm-2 mutants (Fig. 3B). We conclude that knock-out of the DEG/ENaC channels DELM-1 and DELM-2 reduces C. elegans nose-touch sensitivity and foraging suppression upon anterior body touch, mediated by OLQ and IL1 sensory neurons (Kaplan and Horvitz, 1993; Hart et al., 1995; Alkema et al., 2005; Kindt et al., 2007). These data suggest that DELM-1 and DELM-2 may function in glia to support the activity of associated sensory neurons.
Adaptation properties of delm-1 and delm-2 mutants
The residual nose-touch sensitivity of trpa-1 mutants displays pronounced adaptation (Kindt et al., 2007). Similarly, we found that the ratio of animals responding to nose touch decreased with consecutive touches more in delm-1 and delm-2 mutants than it did in wild-type animals (Fig. 3C) (Kindt et al., 2007). This phenotype was rescued by expression of DELM-1 and DELM-2 genomic sequences in delm-1 and delm-2 mutants. These results suggest that (1) nose-touch sensitivity is mediated by at least two distinct mechanisms (or neurons or both) with distinct adaptation properties (one slower and dependent on TRPA-1 and DELM channels, and one faster independent of TRPA-1 and DELM channels) and that (2) knock-out of DELM-1 and DELM-2 compromises the same mechanism (or neurons) that is compromised by knock-out of TRPA-1. These data again support the idea that that DELM-1 and DELM-2 are required for the function of OLQ and IL1 neurons, where TRPA-1 is expressed.
Knock-out of DELM-1 compromises OLQ neuronal response to touch similarly to knock-out of TRPA-1
Our behavioral data suggested that the activity of OLQ and IL1 sensory neurons in response to nose touch is compromised by knock-out of DELM-1 and DELM-2. To test this directly, we used the calcium sensor GCaMP to monitor in vivo responses of OLQ neurons to nose touch. We found that while calcium transients induced by two consecutive touches in OLQ neurons of wild-type animals were of similar amplitude, the calcium transient induced by the second touch was smaller in delm-1 knock-outs, similarly to what was seen in OLQ neurons of trpa-1 knock-outs (Fig. 4A–F,I) (Kindt et al., 2007). Importantly, the calcium transients in response to two consecutive touches were again of similar amplitude in delm-1 animals in which DELM-1 was expressed in socket cells using the glial-specific promoter itx-1 (Fig. 4G–I). At this point, it is not clear why only the Ca2+ transient upon second touch is reduced in trpa-1 and delm-1 mutants, while behavioral defects are apparent at first touch (Fig. 3A). However, this was observed for trpa-1 by Kindt and colleagues as well (Kindt et al., 2007). In addressing the inconsistency between the behavioral and the cellular phenotypes, Kindt and colleagues suggested that the activity of IL1 neurons may be also compromised in trpa-1 knock-outs, thus contributing to the more severe behavioral phenotype. However, no suitable Ca2+ sensor line is available to test this. Alternatively, it is possible that the mechanical stimulation used in imaging experiments is harsher than what the animals experience when bumping into an eyelash hair. Unfortunately, we were unable to test lower mechanical stimulations because they generated Ca2+ transients too small to be reliably analyzed. Moreover, we note that there are precedents for partial losses of neural response having strong effects on behavior (Hilliard et al., 2005). We also note that in our imaging experiments, the second touch stimulation was given 5 min after the first one. This protocol by Kindt and colleagues was used here to determine whether knock-out of delm-1 reproduced the cellular phenotype produced by knock-out of trpa-1 (Kindt et al., 2007). While future experiments are needed to study Ca2+ changes upon temporally closer and gentler mechanical stimulations, our results support that knock-out of DELM-1 in glia compromises OLQ neuronal response to nose touch, similarly to knock-out of TRPA-1.
Figure 4.
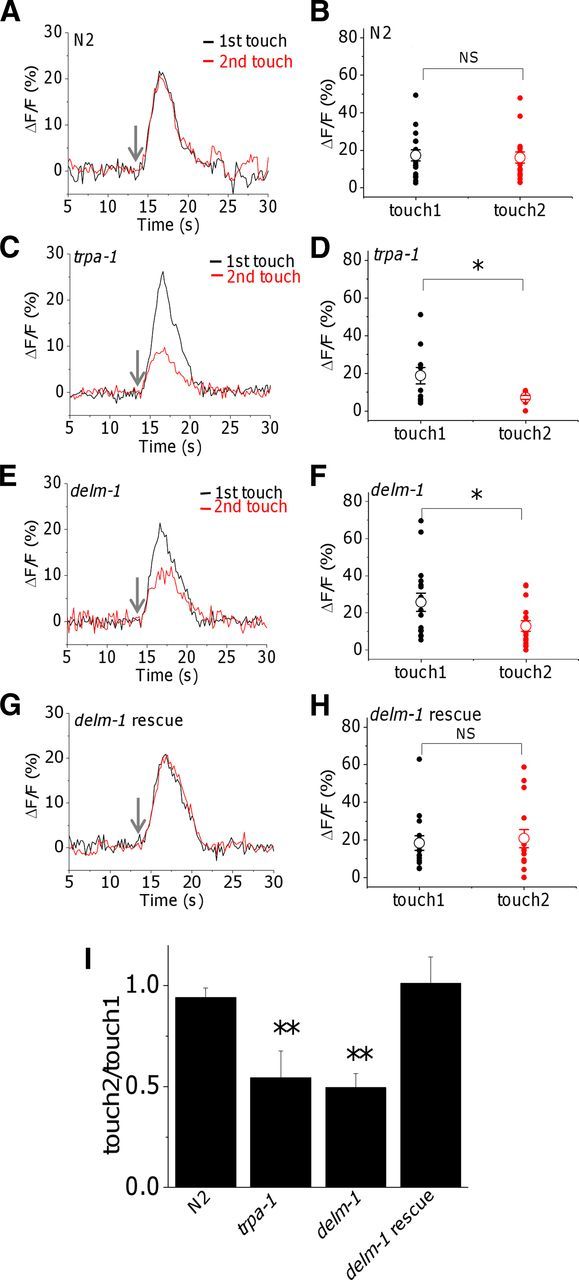
Effect of knock-out of delm-1 on touch-induced calcium transients in OLQ sensory neurons. A, C, E, G, Representative Ca2+ transients evoked in OLQ neurons of wild-type animals and mutants as indicated, following two consecutive touches to the nose (5 min interval). The arrows point to when the touch was delivered. B, D, F, H, Quantification of the Ca2+ transients generated by the first and second touches. Individual values and averages are shown by filled and open symbols respectively. The average fluorescence changes (ΔF/F) were as follows: 17.2 ± 3 for the first touch and 16 ± 3 for the second touch (n = 17), in N2 (wild type); 18.7 ± 4.4 for the first touch and 7.1 ± 1 for the second touch (n = 11), in trpa-1; 25.8 ± 4.9 for the first touch and 12.9 ± 2.9 for the second touch (n = 16), in delm-1; and 18.3 ± 3.8 for the first touch and 20.7 ± 4.8 for the second touch (n = 15), in delm-1 rescue (delm-1;Pitx-1::cDELM-1). Note that there is no statistical difference for calcium transients induced by the first touch between WT and all the mutants. I, Averages of the ratios of calcium transients generated by the second and first touches. *p ≤ 0.05 by t test; NS, not significantly different.
DELM-1 and DELM-2 channels function cell-autonomously in glia to orchestrate nose-touch behavior
Although our fluorescent protein reporters indicated that DELM-1 and DELM-2 are expressed in the OLQ and IL1 glial socket cells (Fig. 2B), we wondered whether DELM-1 and DELM-2 in fact act in these cells to orchestrate touch behaviors. To determine this, we assayed whether nose-touch defects of delm-1 and delm-2 mutants were rescued by expression of DELM-1 and DELM-2 cDNA sequences, under the control of the itx-1 promoter, functional in OLQ and IL1 glial socket cells (Haklai-Topper et al., 2011). We observed that the nose-touch defects were significantly rescued in these transgenic strains (Fig. 5A). To establish whether rescue of nose-touch defects could be achieved by expression of DELM-1 in nose-touch neurons (OLQ, FLP, CEP, and ASH sensory neurons (Kaplan and Horvitz, 1993; Kindt et al., 2007; Chatzigeorgiou and Schafer, 2011), we generated rescue strains in which DELM-1 cDNA sequence was under the control of promoters that function in these neurons. Thus we used the promoter of the ocr-4 gene to drive expression of DELM-1 cDNA in OLQ neurons (Tobin et al., 2002), the promoter of the egl-46 gene for expression in FLP neurons (and PVD) (Yu et al., 2003), the promoter of dat-1 for expression in CEP (and ADE and PDE) (Nass et al., 2001), and the promoter of sra-6 for expression in ASH neurons (Troemel et al., 1995). However, we found no rescue (Fig. 5A). We wanted to further test the function of polymodal ASH neurons, which previous studies showed to heavily contribute to nose touch (Kaplan and Horvitz, 1993). We found that delm-1 and delm-2 mutants displayed normal ASH-mediated behaviors, while osm-9 mutants as expected did not (Colbert and Bargmann, 1997) (Fig. 5B–D). These data support the idea that DELM-1 does not function in any of the nose-touch neurons to orchestrate nose-touch sensitivity.
Figure 5.
DELM-1 and DELM-2 are required cell-autonomously in glia for nose-touch behavior. A, Ratios of reversals (dark gray) and head withdrawals (light gray) in mutants and transgenic strains as indicated. Ratios of reversals and head withdrawals were as follow: N2 (wild-type), 0.617 ± 0.012 and 0.146 ± 0.008; trpa-1, 0.160 ± 0.010 and 0.038 ± 0.005; trpa-1;DELM-1, 0.200 ± 0.029 and 0.056 ± 0.014; delm-1, 0.280 ± 0.039 and 0.070 ± 0.019; delm-1;Pitx-1::cDELM-1, 0.565 ± 0.049 and 0.185 ± 0.033; delm-2, 0.250 ± 0.031 and 0.110 ± 0.021; delm-2;Pitx-1::cDELM-2, 0.485 ± 0.032 and 0.156 ± 0.026; delm-1;Pocr-4::cDELM-1, 0.250 ± 0.022 and 0.105 ± 0.027; delm-1;Pegl-46::cDELM-1, 0.295 ± 0.036 and 0.120 ± 0.022; delm-1;Pdat-1::cDELM-1, 0.295 ± 0.029 and 0.060 ± 0.016; delm-1;Psra-6::cDELM-1, 0.275 ± 0.026 and 0.065 ± 0.016; and delm-1;Pitr-1::cDELM-1, 0.300 ± 0.034 and 0.075 ± 0.016. Number of animals assayed was 299, 259, 50, 40, 70, 40, 40, 40, 40, 40, 40, and 80. B–D, delm-1 and delm-2 mutants are repelled by 8 m glycerol (B, ratios of retained animals was 0.902 ± 0.027 for N2, 0.243 ± 0.028 for osm-9, 0.801 ± 0.037 for delm-1, and 0.864 ± 0.037 for delm-2), by 0.1% SDS (C, ratios of animals responding was 0.816 ± 0.049 for N2, 0.420 ± 0.033 for osm-9, 0.904 ± 0.034 for delm-1, and 0.894 ± 0.051 for delm-2), and by 30% octanol (D, time to response was 1.438 ± 0.057 s for N2, 3.890 ± 0.141 s for osm-9, 1.366 ± 0.057 s for delm-1, and 1.209 ± 0.044 s for delm-2) to the same degree as wild-type animals. osm-9(ky10), which abolishes all ASH-mediated sensory responses (Colbert et al., 1997), serves as a control. Number of assays were 14, 14, 22, and 7 for N2 (wild type), osm-9(ky10), delm-1(ok1226), and delm-2(ok1822) with 20 animals per strain used in each assay for 8 m glycerol, 5 per strain with 20 animals tested in each assay for 0.1% SDS, and 105, 100, 101, and 105 animals tested for 30% octanol. Data are means ± SE. **p < 0.01 (ANOVA).
We next wondered whether expression of DELM-1 in glial amphid socket cells would rescue delm-1 nose-touch defects. Amphid socket cells are a pair of glial cells located in the head of C. elegans that wrap around the dendrites of eight pairs of sensory neurons (Altun and Hall, 2010), including the dendrites of ASH neurons (Kaplan and Horvitz, 1993). We found no rescue in delm-1;Pitr-1::cDELM-1 transgenic animals (Fig. 5A), suggesting that DELM-1 does not function in amphid socket cells.
Previous work has shown that TRPA-1 acts in OLQ sensory neurons to mediate nose-touch sensitivity (Kindt et al., 2007). Our data suggest that DELM-1 and DELM-2 function cell-autonomously in associated glial socket cells to allow or mediate nose-touch sensitivity. Since TRPA-1 and DELM-1 appear to be expressed in two different cell types, we hypothesized that overexpression of DELM-1 in glial socket cells would not rescue the nose-touch defects of trpa-1 mutants. Indeed, we found that nose-touch defects were still present in trpa-1;DELM-1 animals (Fig. 5A). These results again support the idea that TRPA-1 and DELM-1 act in sensory neurons and glia respectively and are both needed to orchestrate nose-touch sensitivity.
No structural defects or mislocalization of TRPA-1 in delm mutants
In C. elegans, amphid sheath cells are required for the elongation of the dendrites of sensory neurons and for the establishment and remodeling of their receptive sensory endings, the cilia (Bacaj et al., 2008a; Yoshimura et al., 2008; Procko et al., 2011). We thus wondered whether DELM-1 and DELM-2 were needed for neuronal structural development or integrity. EM analysis though revealed that OLQ and IL1 sensory neurons and associated glia had normal morphologies in delm-1 mutants (Fig. 6A–D, wild-type sections not shown).
Figure 6.
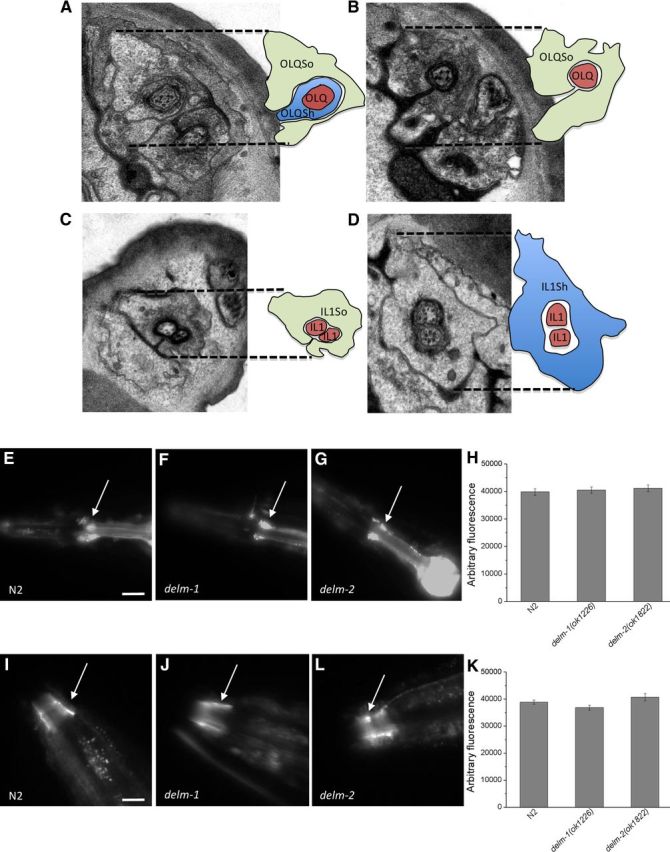
Ultrastructure of OLQ and IL1 neurons and glia, and TRPA-1::GFP expression and localization in delm mutants. A–D, EM images show normal structures of OLQ (A, B) and IL1 (C, D) touch neurons and associated socket glia (So) and sheath glia (Sh) in delm-1 mutants. Each image has a schematic representation on the right. E–G, Fluorescent micrographs of transgenic C. elegans expressing TRPA-1::GFP. A representative image is shown for wild type (N2), delm-1, and delm-2 mutants. Arrows point to OLQ and IL1 neurons. Scale bar, 100 μm. H, Quantification of the GFP signal in TRPA-1::GFP-expressing neurons. Number of animals analyzed was 10, 10, and 9 for N2 (wild type), delm-1, and delm-2 mutants respectively. I, J, Representative photographs of TRPA-1::GFP localization at the cilia of sensory neurons (arrows) in wild type (N2), delm-1, and delm-2 mutants as indicated. Scale bar, 40 μm. K, Quantification of the level of GFP signal at the cilia of sensory neurons. Number of animals analyzed was 16, 13, and 11 for N2 (wild type), delm-1, and delm-2 mutants respectively.
We next analyzed TRPA-1::GFP expression and cilia localization in delm-1 and delm-2 mutants. We found no significant difference compared with wild type (Fig. 6E–K). Note that RNAi of DELM-2 was shown to cause axon-guidance defects in DD/VD motor neuron in C. elegans (Schmitz et al., 2007). We did not notice abnormal axons in OLQ or IL1 neurons of delm-2;TRPA-1::GFP animals, and locomotion of delm-2 knock-out animals is normal. These data, combined with the known caveats of RNAi experiments using a feeding library (i.e., false-positives; Kamath et al., 2003), suggest that axon-guidance defects, if they occur at all in delm-2 knock-out animals, are mild. Nevertheless, the cell-specific rescue experiments still support the idea that DELM-2 functions cell-autonomously in OLQ and IL1 socket glia to control nose-touch behavior. In conclusion, our data support the idea that knock-out of DELM-1 and DELM-2 affects the function, rather than the ultrastructure, of touch neurons or the subcellular localization of neuronal TRPA-1.
DELM-1 shares functional features with glial channel ACD-1
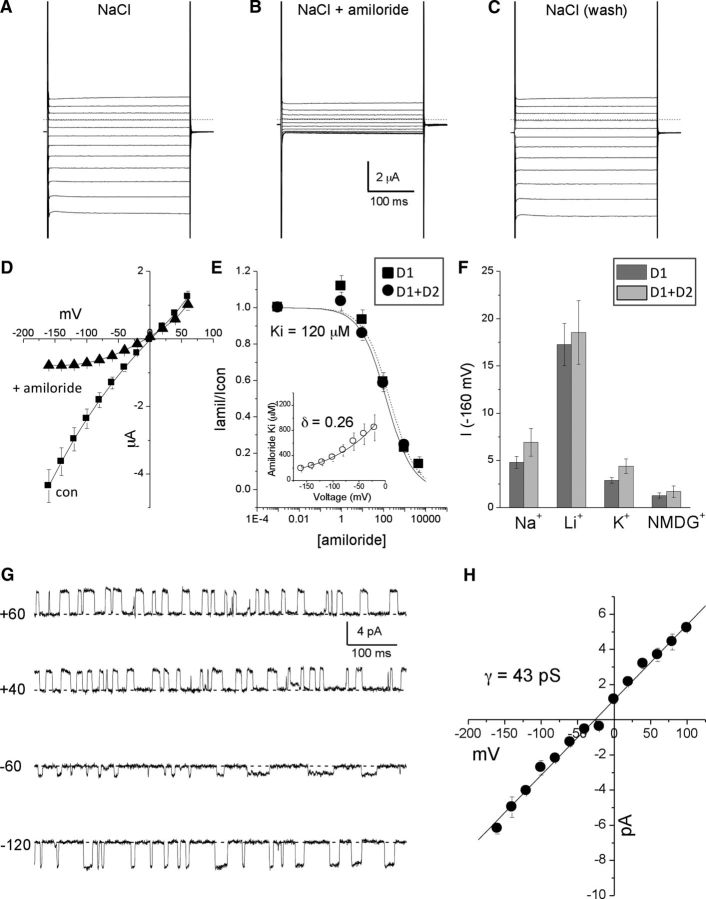
Glial DEG/ENaC channel ACD-1 produces large amiloride-sensitive currents when expressed in Xenopus oocytes (Wang et al., 2008). We found that DELM-1 produced similar currents (Fig. 7A–D). The concentration of amiloride that inhibited 50% of the current (Ki) was 120 μm (at −160 mV, Fig. 7E). Amiloride block was voltage dependent (δ = 0.26, Fig. 7E, inset), suggesting that amiloride interacted within the channel pore. DELM-1 was permeable to Li+ and Na+, was permeable to a lesser degree to K+, and had a small permeability to NMDG+ (PLi ≫ PNa > Pk > PNMDG) (Fig. 7F). When we analyzed single-channel properties, we found that DELM-1 is open ∼50% of the time and has a single-channel conductance of 43 pS (Fig. 7G,H). To conclude, these data show that DELM-1 shares properties with previously characterized members of the DEG/ENaC family, including glial ACD-1 (Brown et al., 2007; Wang et al., 2008).
Figure 7.
Electrophysiological properties of DELM-1 channel expressed in Xenopus oocytes. A, Example of DELM-1 currents elicited by voltage steps from −160 to +60 mV from a holding potential at −30 mV. A physiological NaCl solution was used. B, The same oocyte was perfused with a physiological NaCl solution plus 500 μm amiloride. C, Washout. D, DELM-1 current–voltage relationships in control (squares, n = 8) and the presence of 500 μm amiloride (triangles, n = 8). The relatively low reversal potential of the current reflected the phenomenon of Na+ overload (Goodman et al., 2002; Bianchi et al., 2004; Wang et al., 2008). E, Amiloride dose–response curves for DELM-1 (n = 7, squares) and DELM-1 plus DELM-2 (n = 6, circles). Data are fitted by sigmoid curves. The Kis were 120 and 190 μm for DELM-1 and DELM-1 plus DELM-2 currents, respectively. Insert shows voltage dependence of amiloride blockade for DELM-1. Data were fitted using a Woodhull model (Woodhull, 1973) (δ = 0.26, n = 7). F, Ionic selectivity for DELM-1 (dark gray) and DELM-1 plus DELM-2 currents (light gray). Currents were recorded at −160 mV (n = 6 for DELM-1; n = 5 for DELM-1 plus DELM-2). G, Example of DELM-1 single-channel currents at the indicated voltages. H, DELM-1 single-channel current–voltage relationships (n = 3). Data are means ± SE.
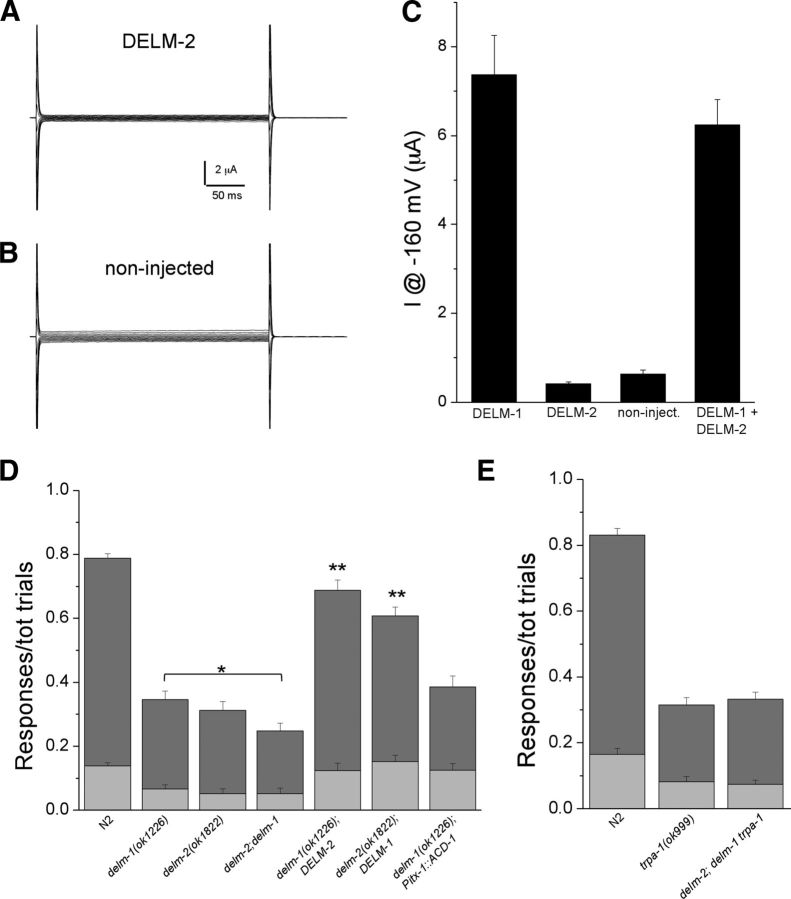
DELM-1 and DELM-2 can function independently
When we expressed DELM-2 in the Xenopus oocytes, we did not detect any current (Fig. 8A,B). Some DEG/ENaC subunits are not functional on their own but can modulate the function of other DEG/ENaCs (Huang and Chalfie, 1994; McDonald et al., 1995; Lingueglia et al., 1997). We thus tested whether DELM-2 modulated DELM-1 properties. We found no significant difference in current amplitude (Fig. 8C), amiloride sensitivity (Fig. 7E), and ion selectivity (Fig. 7F) between DELM-1 plus DELM-2 compared with DELM-1 alone. The DELM-2 cDNA used for Xenopus oocytes studies is the same that rescued nose-touch sensitivity in vivo (Fig. 5A). This suggests that DELM-2 is not functional in Xenopus oocytes, or that it is activated under conditions that were not tested, or that it modulates features of DELM-1 that we did not study.
Figure 8.
DELM-1 and DELM-2 can function independently in vivo. A, B, Examples of currents in a Xenopus oocyte injected with DELM-2 cRNA (A) and in a noninjected oocyte (B). Currents are elicited by voltage steps from −160 to +60 mV in 20 mV increments from a holding potential of −30 mV. C, Currents measured at −160 mV in oocytes expressing DELM-1 (n = 33, 7.36 ± 0.88 μA), DELM-2 (n = 8, 0.41 ± 0.04 μA), in noninjected oocytes (n = 7, 0.62 ± 0.09 μA), and DELM-1 plus DELM-2 (n = 33, 6.23 ± 0.57 μA). D, Ratios of reversals (dark-gray bar) and head withdrawals (light-gray bar) of N2 (wild type, 0.650 ± 0.014 and 0.138 ± 0.010), delm-1 (0.280 ± 0.026 and 0.066 ± 0.013), delm-2 (0.260 ± 0.028 and 0.052 ± 0.016), delm-2;delm-1 double mutants (0.196 ± 0.024 and 0.052 ± 0.016), delm-1 mutants overexpressing DELM-2 (0.564 ± 0.032 and 0.124 ± 0.022), delm-2 mutants overexpressing DELM-1 (0.456 ± 0.027 and 0.152 ± 0.020), and delm-1 mutants expressing ACD-1 in socket cells (0.260 ± 0.035 and 0.125 ± 0.021) upon nose-touch stimulation. Number of animals tested was 240, 60, 50, 50, 50, 50, and 40. E, Same as in D for N2 (0.665 ± 0.02 and 0.165 ± 0.018), trpa-1 (0.232 ± 0.022 and 0.082 ± 0.014) and delm-2;delm-1 trpa-1 (0.257 ± 0.020 and 0.075 ± 0.012) mutants. Number of animals tested was 80 each. Data are means ± SE.
Bearing in mind the caveats of heterologous expression systems, we turned again to in vivo studies. First, we generated delm-2;delm-1 double mutants. We found that the ratio of responses to nose touch of delm-2;delm-1 double mutants was statistically smaller than those of delm-1 single mutants (Fig. 8D). This result indicated that DELM-2 has some residual activity in the absence of DELM-1, suggesting that DELM-1 and DELM-2 may function independently of each other. To test this, we overexpressed DELM-2 in delm-1, and overexpressed DELM-1 in delm-2 mutants and indeed found that DELMs could rescue each other in reciprocal knock-outs (Fig. 8D). These data show that DELM-1 and DELM-2 can function independently of each other and that perhaps a certain number of functional DELM channels are needed in glial socket cells for normal nose-touch sensitivity. Interestingly though, expression of homologous glial channel ACD-1 in socket glia did not rescue delm-1 knock-out touch defects (Fig. 8D). This result suggests either that specific still unknown functional features of DELM channels are needed in socket glia, or that socket glia do not express accessory or regulatory proteins needed for ACD-1 function in vivo (though not in Xenopus oocytes). Future experiments should help distinguish between these possibilities. We also analyzed delm-2;delm-1 trpa-1 triple mutants and found that their touch deficit was not statistically different from that of trpa-1 mutants (Fig. 8E). This result confirms once more that neuronal TRPA-1 acts downstream of DELM-1 and DELM-2 in glia.
Rescue of delm-1 knock-out nose-touch defects by depolarization of sensory neurons and enhanced excretion of K+ from glia
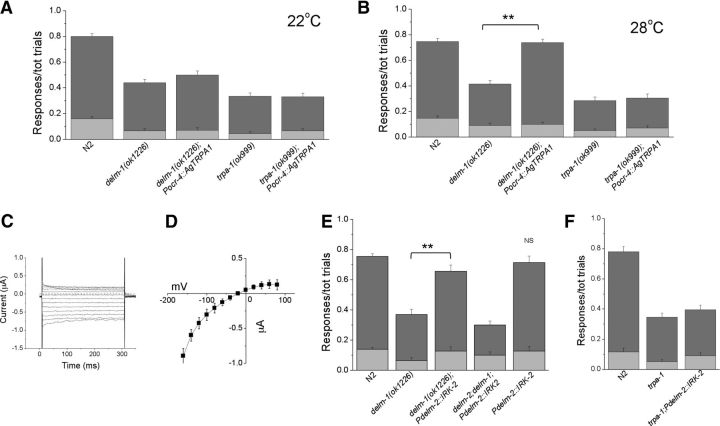
How may DELM-1 and DELM-2 be needed for the activity of OLQ and IL1 sensory neurons? In our previous work, we showed that homologous glial channel ACD-1 controls sensory neurons function by setting their basal excitability or calcium level (Wang et al., 2012). Given the level of similarity between ACD-1 and DELM channels and the fact that they are all expressed in glia, we hypothesized a similar mechanism. To test this hypothesis, we expressed the temperature-sensitive cationic TRP channel from Anopheles gambiae TPRA1 (AgTRPA1) in OLQ neurons of delm-1 knock-out. AgTRPA1 is expressed in distal antennal sensory structures of the malaria mosquito and it functions as a heat detector to guide navigation toward the prey (Wang et al., 2009). AgTRPA1 is closed at temperatures of <25°C and is robustly activated at temperatures of >28°C (Wang et al., 2009; Kang et al., 2011). Note that despite the name similarity, contrarily to C. elegans TRPA-1, AgTRPA1 is not thought to be activated by mechanical forces. We reasoned that if we could switch AgTRPA1 on and off by changing the temperature, we could directly test whether depolarization and/or increase in intracellular calcium of the sensory neurons rescued delm-1 knock-out touch defects. We found that, while at 22°C, delm-1;Pocr-4::AgTRPA1 animals had nose-touch defects similar to those of delm-1 knock-out animals (Fig. 9A), at 28°C, when AgTRPA1 is open, nose-touch defects in these animals were substantially rescued (Fig. 9B). Consistent with the idea that C. elegans TRPA-1 is part of the mechanotransducing machinery in OLQ and IL1 neurons and that AgTRPA1 is not mechanosensitive (Kindt et al., 2007), expression of AgTRPA1 in trpa-1 knock-out animals did not rescue nose-touch defects (Fig. 9A,B). In summary, our experiments using AgTRPA1 support the idea that DELM channels regulate OLQ and IL1 function by increasing their basal excitability and/or calcium level, a mechanism that we have previously described for homologous glial channel ACD-1 (Wang et al., 2012).
Figure 9.
Effect of expression of cationic channels in OLQ neurons and K+ channels in socket glia on delm-1 knock-out nose-touch defects. A, Ratios of reversals (dark-gray bar) and head withdrawals (light-gray bar) assayed at 22°C for N2 (wild type, 0.640 ± 0.022 and 0.160 ± 0.017), delm-1 (0.375 ± 0.026 and 0.065 ± 0.016), delm-1 expressing mosquito TRPA1 (AgTRPA1) in OLQ neurons (delm-1(ok1226);Pocr-4::AgTRPA1, 0.430 ± 0.030 and 0.070 ± 0.019), trpa-1 knock-out (0.290 ± 0.025 and 0.045 ± 0.013), and trpa-1 knock-out expressing AgTRPA1 in OLQ neurons (trpa-1(ok999);Pocr-4::AgTRPA1, 0.265 ± 0.029 and 0.065 ± 0.016). n = 80, 40, 40, 40, and 40. B, Same as in A except that behavioral assays were conducted at 28°C. N2 (wild type, 0.600 ± 0.024 and 0.147 ± 0.017), delm-1 (0.325 ± 0.028 and 0.090 ± 0.020), delm-1 expressing mosquito TRPA1 (AgTRPA1) in OLQ neurons (delm-1(ok1226);Pocr-4::AgTRPA1, 0.640 ± 0.026 and 0.100 ± 0.017), trpa-1 knock-out (0.235 ± 0.028 and 0.050 ± 0.013) and trpa-1 knock-out expressing AgTRPA1 in OLQ neurons (trpa-1(ok999);Pocr-4::AgTRPA1, 0.235 ± 0.032 and 0.070 ± 0.018) (n = 80, 40, 40, 40, and 40). C, Example of ionic currents recorded in an oocyte injected with IRK-2 cRNA and perfused with a solution containing 100 mm KCl. Currents were elicited by voltage steps from −160 to + 80 mV in 20 mV increments. The dashed line is the zero current level. IRK-2 currents were obtained by subtracting the BaCl2 resistant current from the control currents. BaCl2 is a blocker of inward rectifier K+ channels and was used at the concentration of 1 mm. D, Average IRK-2 current–voltage relationship shows that this channel conducts outward current (n = 12). E, Ratios of reversals (dark-gray bar) and head withdrawals (light-gray bar) of N2 (wild type), delm-1, delm-1 expressing IRK-2 in socket glia, delm-2;delm-1 double mutant expressing IRK-2 in socket glia, and wild type expressing IRK-2 in socket glia (n = 160, 40, 50, and 30, respectively). F, Same as in E for N2 (wild type), trpa-1 knock-out, and trpa-1 knock-out expressing IRK-2 in socket glia (n = 40 for each). Data are means ± SE. **p < 0.01 (ANOVA); NS, nonsignificant.
Extracellular K+ concentration is well known to influence neuronal excitability by setting the neurons' resting potential. Thus, to further test our hypothesis that DELM channels in glia establish the basal level of excitability of OLQ and IL1 neurons, we expressed a weak inward rectifier K+ channel in OLQ and IL1 glial socket cells. Weak inward rectifier K+ channels are expected to increase excretion of K+ (Ho et al., 1993), thus increasing the concentration of K+ in the extracellular space. We characterized C. elegans IRK-2 as a weak inward rectifier K+ by heterologous expression in Xenopus oocytes (Fig. 9C,D). We then expressed it in OLQ and IL1 socket glia of delm-1 knock-out animals to determine whether it rescued nose-touch defects. We found that it did (Fig. 9E). Interestingly, expression of IRK-2 in socket cells of wild-type animals has no effect on nose-touch sensitivity (Fig. 9E). We interpret these results to suggest that this intervention is not likely to push basal neuronal excitability outside the optimal range. We also found no rescue by expressing IRK-2 in delm-2;delm-1 double-mutant animals, suggesting that a minimum number of DEG/ENaCs is needed in glia to drive K+ excretion (Fig. 9E) (Gray et al., 2005). Importantly, expression of IRK-2 in socket glia does not rescue trpa-1 knock-out nose-touch defects (Fig. 9F), again underscoring the idea that TRPA-1 acts downstream of DELM-1 and DELM-2 to orchestrate nose-touch sensitivity. To conclude, our experiments with weak inward rectifier K+ channel IRK-2 further support the idea that DELM channels in glia are involved in a molecular mechanism that sets the basal excitability of associated sensory neurons.
Discussion
We have shown here that Na+ channel subunits of the DEG/ENaC family are expressed in glia associated with touch neurons in C. elegans, where they function cell-autonomously to orchestrate touch sensation. The C. elegans channel subunit MEC-4 and all the other homologous channels of the DEG/ENaC family across species that have been involved to date in touch sensation are expressed in sensory neurons (Chalfie, 1993; Adams et al., 1998; Price et al., 2000, 2001; Chatzigeorgiou et al., 2010b; Zhong et al., 2010; Geffeney et al., 2011). However, recently published data showing that ASIC1a, ASIC2, and ASIC3 triple knock-out mice have enhanced touch sensitivity instead of being touch insensitive, as was predicted, have cast doubts on the direct involvement of neuronal DEG/ENaC channels in touch (Kang et al., 2012). Our work prompts consideration of non-neuronal DEG/ENaC channels for their involvement in touch sensation and suggests that cell-specific knock-out of mammalian DEG/ENaCs may be necessary to discern the role of neuronal versus glial channels.
Work from other laboratories has indeed shown that DEG/ENaC subunits are expressed in glia associated with mechanosensors in mammals. Hitomi and colleagues showed that β ENaC is expressed on the plasma membrane of Schwann cells associated with Ruffini endings of the rat incisor, while Calavia and colleagues demonstrated the expression of ASIC2 in the inner lamellae of human Pacinian corpuscles, which are thought to be of Schwann cell origin (Hitomi et al., 2009; Calavia et al., 2010).
Interestingly though, not all mechanosensory neurons have associated cells. For example, in C. elegans the touch neurons that express MEC-4 are not associated with glia and in mammals hair follicle endings and most Ruffini endings do not have associated cells. It is possible that associated cells of certain types of mechanosensors provide an additional layer of modulation, as suggested by Pawson and colleagues (Pawson et al., 2009). The activity of glial DEG/ENaCs may be modulated by pH (Bianchi and Driscoll, 2002), extracellular Ca2+ (de Weille and Bassilana, 2001; Babini et al., 2002; Immke and McCleskey, 2003; Bianchi et al., 2004), or FMRF-type peptides (Askwith et al., 2000), resulting in changes in touch neurons' output.
While our experiments show that DELM channels in glia are needed to establish basal neuronal excitability, the molecular mechanism in which DELM channels are involved is still not clear. However, an appealing model can be suggested. DEG/ENaC channels expressed in kidney and lung epithelia are known to establish a favorable driving force for K+ excretion through inward rectifier K+ channels (Gray et al., 2005). Importantly, epithelial DEG/ENaCs can function in this capacity because they are open at baseline, like DELM-1 (Fig. 7). Thus, it is possible that DELM-1 and DELM-2 control touch neuron excitability by allowing excretion of K+ in the microenvironment between glia and neurons. Neuronal excitability is well known to depend on the concentration of extracellular K+. However, more attention has been dedicated so far on understanding the molecular mechanisms underlying K+ clearing from the extracellular space in the nervous system, important for maintaining normal excitability during high neuronal activity (Kofuji and Newman, 2004). If DELM channels favor K+ excretion, which will be established by future experiments, then this would be a novel mechanism, may be operational in other parts of the nervous system of C. elegans and other organisms, thereby extracellular K+ is increased above the basal level to increase neuronal excitability.
Our results also support the findings by Chatzigeorgiou and colleagues (Chatzigeorgiou and Schafer, 2011). Contrary to what laser ablation studies had originally suggested (Kaplan and Horvitz, 1993), OLQ sensory neurons play a major role in nose-touch sensitivity. Chatzigeorgiou and colleagues showed that OLQ (and CEP) neurons through RIH interneuron-mediated electrical synapses facilitate the activity of FLP neurons when they are active and inhibit their activity when they are inactive. Moreover, OLQ and CEP neurons reduce the threshold for touch sensitivity of FLP neurons (Chatzigeorgiou and Schafer, 2011). The picture emerging from these studies is quite complex. In C. elegans, nose-touch neurons are interconnected with each other to allow tuning of each other's properties. In addition, associated glia introduce another level of modulation. Such a system is likely designed to allow refinement of touch sensation based on external and internal stimuli. We propose that similar mechanisms may exist in the somatosensory or nociceptive pathways of the nervous system of higher organisms.
Footnotes
This work was supported by American Cancer Society Grant RGS-09-043-01-DDC and National Institutes of Health (NIH) Grant RO1NS070969 to L.B., and by NIH Grants R01NS073121, R01HD052677, and R01NS064273 to S.S. R.S. was an exchange graduate student from the Department of Biotechnology and Molecular Sciences, University of Insubria, Italy.
We thank the C. elegans Genetics Center and William R. Schafer for providing nematode strains. We thank Niels Ringstad for the IRK-2 cDNA, Paul A. Garrity for the AgTRPA1 cDNA, and Sreekanth Chalasani for providing the Pstr-2::GCaMP1.0 vector. We also thank Kenneth Muller for granting the use of his fluorescent microscopes; Gerhard Dahl, Peter H. Larsson, Michael Kim, and Marios Chatzigeorgiou for suggestions; and Jeff Grant for critical reading of the manuscript.
The authors declare no competing financial interests.
References
- Adams CM, Anderson MG, Motto DG, Price MP, Johnson WA, Welsh MJ. Ripped pocket and pickpocket, novel Drosophila DEG/ENaC subunits expressed in early development and in mechanosensory neurons. J Cell Biol. 1998;140:143–152. doi: 10.1083/jcb.140.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkema MJ, Hunter-Ensor M, Ringstad N, Horvitz HR. Tyramine functions independently of octopamine in the Caenorhabditis elegans nervous system. Neuron. 2005;46:247–260. doi: 10.1016/j.neuron.2005.02.024. [DOI] [PubMed] [Google Scholar]
- Altun ZF, Hall DH. Nervous system, neuronal support cells. WormAtlas. 2010 [Google Scholar]
- Askwith CC, Cheng C, Ikuma M, Benson C, Price MP, Welsh MJ. Neuropeptide FF and FMRFamide potentiate acid-evoked currents from sensory neurons and proton-gated DEG/ENaC channels. Neuron. 2000;26:133–141. doi: 10.1016/s0896-6273(00)81144-7. [DOI] [PubMed] [Google Scholar]
- Babini E, Paukert M, Geisler HS, Grunder S. Alternative splicing and interaction with di- and polyvalent cations control the dynamic range of acid-sensing ion channel 1 (ASIC1) J Biol Chem. 2002;277:41597–41603. doi: 10.1074/jbc.M205877200. [DOI] [PubMed] [Google Scholar]
- Bacaj T, Lu Y, Shaham S. The conserved proteins CHE-12 and DYF-11 are required for sensory cilium function in Caenorhabditis elegans. Genetics. 2008a;178:989–1002. doi: 10.1534/genetics.107.082453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacaj T, Tevlin M, Lu Y, Shaham S. Glia are essential for sensory organ function in C. elegans. Science. 2008b;322:744–747. doi: 10.1126/science.1163074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI, Thomas JH, Horvitz HR. Chemosensory cell function in the behavior and development of Caenorhabditis elegans. Cold Spring Harb Symp Quant Biol. 1990;55:529–538. doi: 10.1101/sqb.1990.055.01.051. [DOI] [PubMed] [Google Scholar]
- Bianchi L, Driscoll M. Protons at the gate: DEG/ENaC ion channels help us feel and remember. Neuron. 2002;34:337–340. doi: 10.1016/s0896-6273(02)00687-6. [DOI] [PubMed] [Google Scholar]
- Bianchi L, Gerstbrein B, Frøkjaer-Jensen C, Royal DC, Mukherjee G, Royal MA, Xue J, Schafer WR, Driscoll M. The neurotoxic MEC-4(d) DEG/ENaC sodium channel conducts calcium: implications for necrosis initiation. Nat Neurosci. 2004;7:1337–1344. doi: 10.1038/nn1347. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AL, Fernandez-Illescas SM, Liao Z, Goodman MB. Gain-of-function mutations in the MEC-4 DEG/ENaC sensory mechanotransduction channel alter gating and drug blockade. J Gen Physiol. 2007;129:161–173. doi: 10.1085/jgp.200609672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calavia MG, Montaño JA, García-Suárez O, Feito J, Guervós MA, German à A, Del Valle M, Pérez-Piñera P, Cobo J, Vega JA. Differential localization of acid-sensing ion channels 1 and 2 in human cutaneus pacinian corpuscles. Cell Mol Neurobiol. 2010;30:841–848. doi: 10.1007/s10571-010-9511-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canessa CM, Horisberger JD, Rossier BC. Epithelial sodium channel related to proteins involved in neurodegeneration. Nature. 1993;361:467–470. doi: 10.1038/361467a0. [DOI] [PubMed] [Google Scholar]
- Chalasani SH, Chronis N, Tsunozaki M, Gray JM, Ramot D, Goodman MB, Bargmann CI. Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans. Nature. 2007;450:63–70. doi: 10.1038/nature06292. [DOI] [PubMed] [Google Scholar]
- Chalfie M. Touch receptor development and function in Caenorhabditis elegans. J Neurobiol. 1993;24:1433–1441. doi: 10.1002/neu.480241013. [DOI] [PubMed] [Google Scholar]
- Champigny G, Voilley N, Waldmann R, Lazdunski M. Mutations causing neurodegeneration in Caenorhabditis elegans drastically alter the pH sensitivity and inactivation of the mammalian H+-gated Na+ channel MDEG1. J Biol Chem. 1998;273:15418–15422. doi: 10.1074/jbc.273.25.15418. [DOI] [PubMed] [Google Scholar]
- Chatzigeorgiou M, Schafer WR. Lateral facilitation between primary mechanosensory neurons controls nose touch perception in C. elegans. Neuron. 2011;70:299–309. doi: 10.1016/j.neuron.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzigeorgiou M, Grundy L, Kindt KS, Lee WH, Driscoll M, Schafer WR. Spatial asymmetry in the mechanosensory phenotypes of the C. elegans DEG/ENaC gene mec-10. J Neurophysiol. 2010a;104:3334–3344. doi: 10.1152/jn.00330.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzigeorgiou M, Yoo S, Watson JD, Lee WH, Spencer WC, Kindt KS, Hwang SW, Miller DM, 3rd, Treinin M, Driscoll M, Schafer WR. Specific roles for DEG/ENaC and TRP channels in touch and thermosensation in C. elegans nociceptors. Nat Neurosci. 2010b;13:861–868. doi: 10.1038/nn.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelur DS, Ernstrom GG, Goodman MB, Yao CA, Chen L, O'Hagan R, Chalfie M. The mechanosensory protein MEC-6 is a subunit of the C. elegans touch-cell degenerin channel. Nature. 2002;420:669–673. doi: 10.1038/nature01205. [DOI] [PubMed] [Google Scholar]
- Colbert HA, Bargmann CI. Environmental signals modulate olfactory acuity, discrimination, and memory in Caenorhabditis elegans. Learn Mem. 1997;4:179–191. doi: 10.1101/lm.4.2.179. [DOI] [PubMed] [Google Scholar]
- Colbert HA, Smith TL, Bargmann CI. OSM-9, a novel protein with structural similarity to channels, is required for olfaction, mechanosensation, and olfactory adaptation in Caenorhabditis elegans. J Neurosci. 1997;17:8259–8269. doi: 10.1523/JNEUROSCI.17-21-08259.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darboux I, Lingueglia E, Champigny G, Coscoy S, Barbry P, Lazdunski M. dGNaC1, a gonad-specific amiloride-sensitive Na+ channel. J Biol Chem. 1998;273:9424–9429. doi: 10.1074/jbc.273.16.9424. [DOI] [PubMed] [Google Scholar]
- de Weille J, Bassilana F. Dependence of the acid-sensitive ion channel, ASIC1a, on extracellular Ca(2+) ions. Brain Res. 2001;900:277–281. doi: 10.1016/s0006-8993(01)02345-9. [DOI] [PubMed] [Google Scholar]
- Driscoll M, Chalfie M. The mec-4 gene is a member of a family of Caenorhabditis elegans genes that can mutate to induce neuronal degeneration. Nature. 1991;349:588–593. doi: 10.1038/349588a0. [DOI] [PubMed] [Google Scholar]
- García-Añoveros J, García JA, Liu JD, Corey DP. The nematode degenerin UNC-105 forms ion channels that are activated by degeneration- or hypercontraction-causing mutations. Neuron. 1998;20:1231–1241. doi: 10.1016/s0896-6273(00)80503-6. [DOI] [PubMed] [Google Scholar]
- Geffeney SL, Cueva JG, Glauser DA, Doll JC, Lee TH, Montoya M, Karania S, Garakani AM, Pruitt BL, Goodman MB. DEG/ENaC but not TRP channels are the major mechanoelectrical transduction channels in a C. elegans nociceptor. Neuron. 2011;71:845–857. doi: 10.1016/j.neuron.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MB. Jorgensen EM, Kaplan JM, editors. Mechanosensation. WormBook. 2006:1–14. doi: 10.1895/wormbook.1.62.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MB, Schwarz EM. Transducing touch in Caenorhabditis elegans. Annu Rev Physiol. 2003;65:429–452. doi: 10.1146/annurev.physiol.65.092101.142659. [DOI] [PubMed] [Google Scholar]
- Goodman MB, Ernstrom GG, Chelur DS, O'Hagan R, Yao CA, Chalfie M. MEC-2 regulates C. elegans DEG/ENaC channels needed for mechanosensation. Nature. 2002;415:1039–1042. doi: 10.1038/4151039a. [DOI] [PubMed] [Google Scholar]
- Gower NJ, Temple GR, Schein JE, Marra M, Walker DS, Baylis HA. Dissection of the promoter region of the inositol 1,4,5-trisphosphate receptor gene, itr-1, in C. elegans: a molecular basis for cell-specific expression of IP3R isoforms. J Mol Biol. 2001;306:145–157. doi: 10.1006/jmbi.2000.4388. [DOI] [PubMed] [Google Scholar]
- Gray DA, Frindt G, Palmer LG. Quantification of K+ secretion through apical low-conductance K channels in the CCD. Am J Physiol Renal Physiol. 2005;289:F117–F126. doi: 10.1152/ajprenal.00471.2004. [DOI] [PubMed] [Google Scholar]
- Haklai-Topper L, Soutschek J, Sabanay H, Scheel J, Hobert O, Peles E. The neurexin superfamily of Caenorhabditis elegans. Gene Expr Patterns. 2011;11:144–150. doi: 10.1016/j.gep.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AC, Sims S, Kaplan JM. Synaptic code for sensory modalities revealed by C. elegans GLR-1 glutamate receptor. Nature. 1995;378:82–85. doi: 10.1038/378082a0. [DOI] [PubMed] [Google Scholar]
- Hart AC, Kass J, Shapiro JE, Kaplan JM. Distinct signaling pathways mediate touch and osmosensory responses in a polymodal sensory neuron. J Neurosci. 1999;19:1952–1958. doi: 10.1523/JNEUROSCI.19-06-01952.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard MA, Bargmann CI, Bazzicalupo P. C. elegans responds to chemical repellents by integrating sensory inputs from the head and the tail. Curr Biol. 2002;12:730–734. doi: 10.1016/s0960-9822(02)00813-8. [DOI] [PubMed] [Google Scholar]
- Hilliard MA, Apicella AJ, Kerr R, Suzuki H, Bazzicalupo P, Schafer WR. In vivo imaging of C. elegans ASH neurons: cellular response and adaptation to chemical repellents. EMBO J. 2005;24:63–72. doi: 10.1038/sj.emboj.7600493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitomi Y, Suzuki A, Kawano Y, Nozawa-Inoue K, Inoue M, Maeda T. Immunohistochemical detection of ENaCbeta in the terminal Schwann cells associated with the periodontal Ruffini endings of the rat incisor. Biomed Res. 2009;30:113–119. doi: 10.2220/biomedres.30.113. [DOI] [PubMed] [Google Scholar]
- Ho K, Nichols CG, Lederer WJ, Lytton J, Vassilev PM, Kanazirska MV, Hebert SC. Cloning and expression of an inwardly rectifying ATP-regulated potassium channel. Nature. 1993;362:31–38. doi: 10.1038/362031a0. [DOI] [PubMed] [Google Scholar]
- Huang M, Chalfie M. Gene interactions affecting mechanosensory transduction in Caenorhabditis elegans. Nature. 1994;367:467–470. doi: 10.1038/367467a0. [DOI] [PubMed] [Google Scholar]
- Huang M, Gu G, Ferguson EL, Chalfie M. A stomatin-like protein necessary for mechanosensation in C. elegans. Nature. 1995;378:292–295. doi: 10.1038/378292a0. [DOI] [PubMed] [Google Scholar]
- Immke DC, McCleskey EW. Protons open acid-sensing ion channels by catalyzing relief of Ca2+ blockade. Neuron. 2003;37:75–84. doi: 10.1016/s0896-6273(02)01130-3. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kang K, Panzano VC, Chang EC, Ni L, Dainis AM, Jenkins AM, Regna K, Muskavitch MA, Garrity PA. Modulation of TRPA1 thermal sensitivity enables sensory discrimination in Drosophila. Nature. 2011;481:76–80. doi: 10.1038/nature10715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Jang JH, Price MP, Gautam M, Benson CJ, Gong H, Welsh MJ, Brennan TJ. Simultaneous disruption of mouse ASIC1a, ASIC2 and ASIC3 genes enhances cutaneous mechanosensitivity. PLoS One. 2012;7:e35225. doi: 10.1371/journal.pone.0035225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan JM, Horvitz HR. A dual mechanosensory and chemosensory neuron in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1993;90:2227–2231. doi: 10.1073/pnas.90.6.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindt KS, Viswanath V, Macpherson L, Quast K, Hu H, Patapoutian A, Schafer WR. Caenorhabditis elegans TRPA-1 functions in mechanosensation. Nat Neurosci. 2007;10:568–577. doi: 10.1038/nn1886. [DOI] [PubMed] [Google Scholar]
- Kofuji P, Newman EA. Potassium buffering in the central nervous system. Neuroscience. 2004;129:1045–1056. doi: 10.1016/j.neuroscience.2004.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingueglia E, de Weille JR, Bassilana F, Heurteaux C, Sakai H, Waldmann R, Lazdunski M. A modulatory subunit of acid sensing ion channels in brain and dorsal root ganglion cells. J Biol Chem. 1997;272:29778–29783. doi: 10.1074/jbc.272.47.29778. [DOI] [PubMed] [Google Scholar]
- Maricich SM, Wellnitz SA, Nelson AM, Lesniak DR, Gerling GJ, Lumpkin EA, Zoghbi HY. Merkel cells are essential for light-touch responses. Science. 2009;324:1580–1582. doi: 10.1126/science.1172890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald FJ, Price MP, Snyder PM, Welsh MJ. Cloning and expression of the beta- and gamma-subunits of the human epithelial sodium channel. Am J Physiol. 1995;268:C1157–1163. doi: 10.1152/ajpcell.1995.268.5.C1157. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nass R, Miller DM, Blakely RD. C. elegans: a novel pharmacogenetic model to study Parkinson's disease. Parkinsonism Relat Disord. 2001;7:185–191. doi: 10.1016/s1353-8020(00)00056-0. [DOI] [PubMed] [Google Scholar]
- O'Hagan R, Chalfie M, Goodman MB. The MEC-4 DEG/ENaC channel of Caenorhabditis elegans touch receptor neurons transduces mechanical signals. Nat Neurosci. 2005;8:43–50. doi: 10.1038/nn1362. [DOI] [PubMed] [Google Scholar]
- Pawson L, Prestia LT, Mahoney GK, Güçl ü B, Cox PJ, Pack AK. GABAergic/glutamatergic-glial/neuronal interaction contributes to rapid adaptation in pacinian corpuscles. J Neurosci. 2009;29:2695–2705. doi: 10.1523/JNEUROSCI.5974-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perens EA, Shaham S. C. elegans daf-6 encodes a patched-related protein required for lumen formation. Dev Cell. 2005;8:893–906. doi: 10.1016/j.devcel.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Price MP, Lewin GR, McIlwrath SL, Cheng C, Xie J, Heppenstall PA, Stucky CL, Mannsfeldt AG, Brennan TJ, Drummond HA, Qiao J, Benson CJ, Tarr DE, Hrstka RF, Yang B, Williamson RA, Welsh MJ. The mammalian sodium channel BNC1 is required for normal touch sensation. Nature. 2000;407:1007–1011. doi: 10.1038/35039512. [DOI] [PubMed] [Google Scholar]
- Price MP, McIlwrath SL, Xie J, Cheng C, Qiao J, Tarr DE, Sluka KA, Brennan TJ, Lewin GR, Welsh MJ. The DRASIC cation channel contributes to the detection of cutaneous touch and acid stimuli in mice. Neuron. 2001;32:1071–1083. doi: 10.1016/s0896-6273(01)00547-5. [DOI] [PubMed] [Google Scholar]
- Procko C, Lu Y, Shaham S. Glia delimit shape changes of sensory neuron receptive endings in C. elegans. Development. 2011;138:1371–1381. doi: 10.1242/dev.058305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz C, Kinge P, Hutter H. Axon guidance genes identified in a large-scale RNAi screen using the RNAi-hypersensitive Caenorhabditis elegans strain nre-1(hd20) lin-15b(hd126) Proc Natl Acad Sci U S A. 2007;104:834–839. doi: 10.1073/pnas.0510527104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin D, Madsen D, Kahn-Kirby A, Peckol E, Moulder G, Barstead R, Maricq A, Bargmann C. Combinatorial expression of TRPV channel proteins defines their sensory functions and subcellular localization in C. elegans neurons. Neuron. 2002;35:307–318. doi: 10.1016/s0896-6273(02)00757-2. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Chou JH, Dwyer ND, Colbert HA, Bargmann CI. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell. 1995;83:207–218. doi: 10.1016/0092-8674(95)90162-0. [DOI] [PubMed] [Google Scholar]
- Wang G, Qiu YT, Lu T, Kwon HW, Pitts RJ, Van Loon JJ, Takken W, Zwiebel LJ. Anopheles gambiae TRPA1 is a heat-activated channel expressed in thermosensitive sensilla of female antennae. Eur J Neurosci. 2009;30:967–974. doi: 10.1111/j.1460-9568.2009.06901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Apicella A, Jr, Lee SK, Ezcurra M, Slone RD, Goldmit M, Schafer WR, Shaham S, Driscoll M, Bianchi L. A glial DEG/ENaC channel functions with neuronal channel DEG-1 to mediate specific sensory functions in C. elegans. EMBO J. 2008;27:2388–2399. doi: 10.1038/emboj.2008.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, D'Urso G, Bianchi L. Knockout of glial channel ACD-1 exacerbates sensory deficits in a C. elegans mutant by regulating calcium levels of sensory neurons. J Neurophysiol. 2012;107:148–158. doi: 10.1152/jn.00299.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhull AM. Ionic blockage of sodium channels in nerve. J Gen Physiol. 1973;61:687–708. doi: 10.1085/jgp.61.6.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S, Murray JI, Lu Y, Waterston RH, Shaham S. mls-2 and vab-3 Control glia development, hlh-17/Olig expression and glia-dependent neurite extension in C. elegans. Development. 2008;135:2263–2275. doi: 10.1242/dev.019547. [DOI] [PubMed] [Google Scholar]
- Yu H, Pretot RF, Burglin TR, Sternberg PW. Distinct roles of transcription factors EGL-46 and DAF-19 in specifying the functionality of a polycystin-expressing sensory neuron necessary for C. elegans male vulva location behavior. Development. 2003;130:5217–5227. doi: 10.1242/dev.00678. [DOI] [PubMed] [Google Scholar]
- Zhong L, Hwang RY, Tracey WD. Pickpocket is a DEG/ENaC protein required for mechanical nociception in Drosophila larvae. Curr Biol. 2010;20:429–434. doi: 10.1016/j.cub.2009.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]