Abstract
Large-scale covariance of cortical thickness or volume in distributed brain regions has been consistently reported by human neuroimaging studies. The mechanism of this population covariance of regional cortical anatomy has been hypothetically related to synchronized maturational changes in anatomically connected neuronal populations. Brain regions that grow together, i.e., increase or decrease in volume at the same rate over the course of years in the same individual, are thus expected to demonstrate strong structural covariance or anatomical connectivity across individuals. To test this prediction, we used a structural MRI dataset on healthy young people (N = 108; aged 9–22 years at enrollment), comprising 3–6 longitudinal scans on each participant over 6–12 years of follow-up. At each of 360 regional nodes, and for each participant, we estimated the following: (1) the cortical thickness in the median scan and (2) the linear rate of change in cortical thickness over years of serial scanning. We constructed structural and maturational association matrices and networks from these measurements. Both structural and maturational networks shared similar global and nodal topological properties, as well as mesoscopic features including a modular community structure, a relatively small number of highly connected hub regions, and a bias toward short distance connections. Using resting-state functional magnetic resonance imaging data on a subset of the sample (N = 32), we also demonstrated that functional connectivity and network organization was somewhat predictable by structural/maturational networks but demonstrated a stronger bias toward short distance connections and greater topological segregation. Brain structural covariance networks are likely to reflect synchronized developmental change in distributed cortical regions.
Introduction
The human brain network or connectome is being explored with rapidly expanding arrays of techniques that increasingly include multiple brain imaging modalities. Thus, understanding inter-relationships between brain connectivity networks as derived from different imaging modalities has emerged as a central challenge. Most empirical studies of cross-modal integration have focused on diffusion imaging (DTI and DSI) and functional magnetic resonance imaging (fMRI) (Bullmore and Sporns, 2009), demonstrating striking convergence but also important differences between networks of white matter connections and functional coactivation within the brain (Damoiseaux and Greicius, 2009; Honey et al., 2009).
Complementary to fMRI and DTI-based connectomics, population (intersubject) covariance in brain anatomy represents another source of information about inter-regional anatomical associations. The existence of statistically robust and anatomically plausible correlations between the individually variable thickness or volume of pairs of brain regions, each measured once in each of multiple individuals, has been recognized for over a decade (Rockel et al., 1980; White et al., 1997; Wright et al., 1999; Lerch et al., 2006). Structural covariance networks are highly heritable (Schmitt et al., 2008) and show systematic differences with age and disease status (He et al., 2008; Seeley et al., 2009; Bernhardt et al., 2011; Chen et al., 2011).
It has been proposed that structural covariance of cortical thickness between two brain regions reflects their synchronized maturational change, perhaps mediated by axonal connections forming and reforming over the course of development (Mechelli et al., 2005; Lerch et al., 2006). Thus early and reciprocal axonal connectivity between cortical regions is expected to have a mutually trophic effect on regional growth in an individual brain, leading to covariance of regional volumes at a population level. There is some evidence for such developmental models of structural covariance but they have not yet been directly and comprehensively tested (Wright et al., 1999; Lerch et al., 2006; Zielinski et al., 2010; Raznahan et al., 2011b).
Structural MRI networks demonstrate economical, small-world and modular properties qualitatively similar to those reported for functional brain networks (He et al., 2007; Bassett et al., 2008; Chen et al., 2008). Pairs of regions that are functionally connected may also demonstrate strong structural covariance (Seeley et al., 2009; Kelly et al., 2012; Zhou et al., 2012) and highly correlated rates of anatomical change over adolescence (Raznahan et al., 2011b). However, the relationship between structural or maturational networks and functional networks has not yet been systematically explored.
The current study used a structural MRI dataset of healthy young people (N = 108), scanned longitudinally on at least three occasions over 6 years, to estimate cortical thickness and maturational change (linear increase or decrease of cortical thickness) at each of 360 regional nodes. We compared brain networks based on structural covariance and maturational change measurements, testing the hypothesis that structural covariance is related to synchronized maturational change between distributed cortical regions (Fig. 1). Using resting state fMRI in a subset of the sample (N = 32) to measure the vascular signal in response to intrinsic neuronal activity, we quantified convergence between structural/maturational and functional networks.
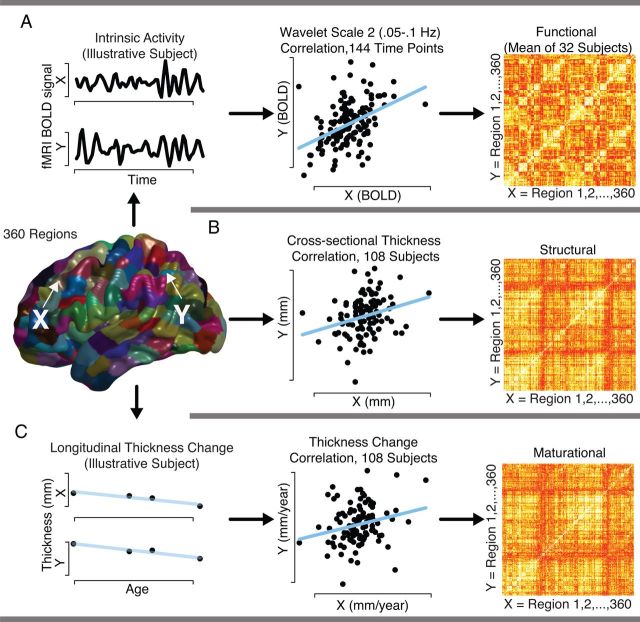
Figure 1.
Schematic illustration of the estimation of the three types of brain connectivity, between the same 360 cortical regions: functional connectivity, structural covariance, and synchronized maturational change. A, For functional connectivity, wavelet correlations of intrinsic activity at 0.05–0.1 Hz were calculated between every pair of regions and averaged across subjects. B, For structural covariance, each region's cortical thickness was estimated using the median scan in terms of age of acquisition for each subject, and pairwise correlations across subjects were calculated after regressing out linear effects of age and gender. C, For synchronized maturational change, each region's slope of maturation with age was calculated via linear regression for each subject, using all scans acquired in the age range 9–22, and pairwise correlations in the rate of maturation were calculated across subjects.
Materials and Methods
All subjects were recruited for the National Institute of Mental Health (NIMH) study of human brain development. To rule out psychiatric diagnosis, a structured diagnostic interview was performed at every visit. Subjects returned every 2 years for follow-up scans. All subjects who had three or more usable MRI scans between the ages of 9 and 22 years were included in this study, because of prior evidence that the cortex matures linearly with age during this period (Raznahan et al., 2011a). This resulted in 108 total subjects (67 male; 41 females) with a total of 376 scans. Participant characteristics are summarized in Table 1. Informed consent was obtained from subjects > 18 years old or from parents of younger subjects, and an additional written assent was obtained from subjects < 18 years old. This study used the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health (NIH), Bethesda, MD (http://biowulf.nih.gov). This study was approved by the NIMH institutional review board.
Table 1.
Demographic information about the study population
| Number of subjects | Number of scans | Age | IQ | Handedness | Gender | |
|---|---|---|---|---|---|---|
| Maturational coupling analysis | 108 | 376 | Range = 9.0–22.8 | Range = 87–141 | R = 99 | M = 67 |
| Mean = 15.2 | Mean = 115.3 | L = 5 | F = 41 | |||
| SD = 3.4 | SD = 11.8 | M = 4 | ||||
| Structural covariance analysis | 108 | 108 | Range = 11.1–20.0 | Range = 87–141 | R = 99 | M = 67 |
| Mean = 14.8 | Mean = 115.3 | L = 5 | F = 41 | |||
| SD = 2.3 | SD = 11.8 | M = 4 | ||||
| Functional connectivity analysis | 32 | 32 | Range = 15.21–33.7 | Range = 87–139 | R = 31 | M = 12 |
| Mean = 22.2 | Mean = 115.3 | L = 1 | F = 20 | |||
| SD = 4.8 | SD = 12.4 |
Image acquisition.
All scanning was performed on a 1.5 T General Electric Signa MRI scanner located at the NIH Clinical Center (Bethesda, MD). The structural scan consisted of a T1-weighted fast spoiled gradient echo sequence: TE 5 ms; TR 24 ms; flip angle 45 degrees; matrix 256 × 256 × 124; field of view 24 cm. Resting-state fMRI was acquired on a subset of the same subjects included in the structural network analyses (see Table 1). The fMRI scan consisted of two sequential 3 min echo-planar imaging (EPI) scans acquired while subjects were lying quietly in the scanner with eyes closed: TR 2.3 s; TE 40 ms; voxel 3.75 × 3.75 × 5 mm; matrix size 64 × 64; field of view 240 × 240 mm; 27 interleaved slices. These two sequential 3 min scans were concatenated, providing 6 min of total scan time for each subject. A recent study reported that estimates of correlation strength between regions stabilize within 5–6 min of resting-state data acquisition (Van Dijk et al., 2010). In addition, our analyses of functional connectivity were performed on the group average of 32 subjects, which should further increase signal to noise. Nonetheless the relatively short acquisition time is a potential weakness of our study, and indeed many other neuroimaging studies that include young subjects who cannot tolerate longer scanning sessions, as longer durations could yield more precise estimates of functional connectivity.
Structural image processing.
The Montreal Neurological Institute's (MNI) CIVET pipeline was used to estimate cortical thickness. Inner and outer cortical surfaces were modeled using triangular meshes generated with ∼40,000 vertices per hemisphere, by a constrained Laplacian algorithm (CLASP) (Kim et al., 2005). These meshes were downsampled into 360 regions using a modified N-cut algorithm to generate spatially contiguous, approximately equally sized regions (Craddock et al., 2012). For each subject, each region's rate of change (thickness/year) was estimated using linear regression. These rates were correlated across subjects, yielding a 360 × 360 matrix of regional synchronized maturational change.
The median scan in terms of age, for each subject, was also used for a cross-sectional structural covariance network analysis. For this analysis, regional estimates of thickness were estimated as above. The residuals from a linear model that included age and gender as independent variables were correlated across subjects to yield a 360 × 360 matrix of structural covariance. In a supplemental analysis, we also used regression to control the cross-sectional cortical thickness values across subjects for the slope of maturational change within each subject.
Functional image processing.
fMRI imaging processing used standard methods, combining AFNI (Cox, 1996) and FSL (Jenkinson and Smith, 2001), as described previously. For each EPI scan, the first four volumes were discarded from the time series, and the data were despiked to limit the impact of statistical outliers on signal intensity. A series of six degrees of freedom rigid body transformations were performed to correct for small amounts of motion occurring in between fMRI volumes. The anatomical scans were segmented into three tissue types, with a probability threshold of 0.8 to identify white matter and CSF. Using a 6 degrees of freedom transformation from each functional scan to that subject's structural scan, and a 12 degrees of freedom transformation from each structural scan to the template, all scans were coregistered to the MNI adult brain template image. A linear model was fit to each voxel's time series including as independent variables the average CSF signal, the average white matter signal, and the six parameters from motion correction, with the residuals from this model used for all further analysis.
The 360 regions defined along the cortical surface of the CIVET mesh for the structural analysis were projected into volume space for use in the functional network analysis. Cortical gray matter was initially defined by combining all of the regions in FSL's Harvard-Oxford probabilistic cortical atlas thresholded at 25%. This gray matter template was then subparcellated with a nearest neighbor approach, using the MNI coordinates of the CIVET vertices. Every voxel in the volumetric gray matter template was assigned to the same brain region as the nearest vertex, to yield 360 regions in volume space for fMRI analysis corresponding to the regions of the cortical surface used for structural MRI analysis. The average fMRI time series was extracted for each of these regions. In a supplemental analysis to test for partial volume-related artifacts, the mean regional signals were also recalculated using a weighted average approach, where each voxel's contribution was weighted by its gray matter density as determined by the anatomical segmentation of the high resolution structural MRI scan.
Recent attention has focused on the possibility for systematic bias in fMRI scans resulting from in-scanner motion (Power et al., 2012; Satterthwaite et al., 2012; Van Dijk et al., 2012), and its impact on fMRI studies of pediatric populations who tend to move more than adults. To limit the impact of motion artifact on these time series, we implemented a scrubbing procedure as part of fMRI preprocessing. An estimate of motion at each time point was calculated as the framewise displacement (FD), using the three translational and three rotational displacements from rigid body motion correction procedure described above:
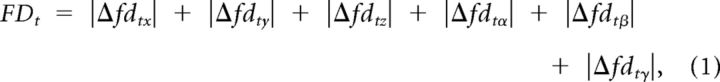 |
where Δfdtx was the displacement in the x direction between frame t − 1 frame t. Rotational displacements were converted from degrees to millimeters by calculating displacement on the surface of a sphere of radius 50 mm. Any frame t with FDt > 0.3 mm was removed from the estimates of biregional fMRI connectivity (Carp, 2012; Power et al., 2012). This scrubbing procedure is similar to that described by Power et al. (2012), but, following Carp (2012), linear interpolations of the deleted frames were used during the wavelet decomposition to prevent contaminated signal from spreading to nearby frames during the bandpass filter. As the optimal procedures for removing motion artifact are still an ongoing area of research, and it is unclear exactly how different methods impact downstream analyses, we also tested our main hypotheses on unscrubbed data. There was no material difference in the results obtained from scrubbed versus unscrubbed data.
To limit the impact of physiological confounds such as respiration, we regressed out the CSF and white matter mean signal from the data and subsequently performed a wavelet decomposition to the scale 2 frequency band (0.05–0.11 Hz) using the maximal overlap discrete wavelet transform with a Daubechies 4 wavelet. Sequential scans were then concatenated, resulting in a single series of wavelet coefficients. The wavelet correlation, −1 ≤ r ≤ 1, was averaged across subjects for every pair of regions to yield a 360 × 360 matrix of functional connectivity. Although considerable physiological noise can remain in fMRI signals even after regressing out CSF and white matter signals (Weissenbacher et al., 2009), due to the nonstationarity and probable scale invariance (“1/f ” characteristics) of noise in fMRI time series, wavelets may be preferable to more commonly used bandpass filters in terms of removing low-frequency noise from the data (Bullmore et al., 2004).
Graph analysis.
Graph analysis was conducted in R (www.r-project.org/) using the package igraph (Csardi and Nepusz, 2006) as well as in-house functions. The association matrices—whether derived from synchronized maturational change, structural covariance, or functional connectivity—were thresholded to make graph models. In these graphs, the nodes are the brain regions included in the graph, and the edges are the included connections. Sparse networks—with relatively few edges representing relatively strong functional, maturational, or structural connections—were constructed using a minimum spanning tree followed by global thresholding (Alexander-Bloch et al., 2010) using positive associations only. Networks were constructed for a range of connection densities, from 1 to 30% at 1% intervals, where connection density refers to the percentage of all possible edges included, such that at 100% there would be edges from each node to every other node. Summary statistics were calculated at 10%, and sparse networks at 2% connection density were used for visual representation. All networks were compared with random networks with the same number of nodes and edges, as well as to random networks with the same degree distributions (Viger and Latapy, 2005). The following, previously described, graph theoretical properties were calculated for all networks.
The minimum path length, Li,j, between nodes i and j is defined as the lowest number of edges that must be traversed to go from i to j. A node's average path length L(i,G) is the average length of the shortest paths between i and all other nodes. The global efficiency, E, of a graph G is as follows:
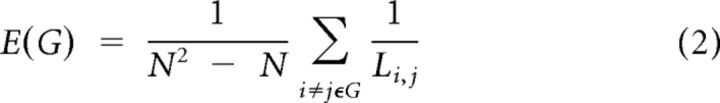 |
where Li,j is the minimum path length between i and j. Global efficiency can be interpreted as the capacity for integrative information transfer across the network (Latora and Marchiori, 2001).
The clustering coefficient, C, of a node i in a graph G is the ratio of triangles, δv, to connected triples, τv, or the proportion of i's neighbors that are themselves directly connected by a single edge. This measure quantifies the cliquishness of the network in the vicinity of the node. At the level of the whole graph, the clustering coefficient is as follows:
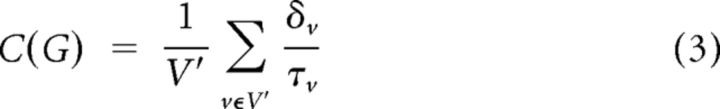 |
where V′ is the set of nodes with degree (number of edges) greater than 2 (Schank and Wagner, 2005).
The modularity, Q, of a graph G is a function of the proportion of G's edges that fall within modules (topological communities of varying size) (Newman and Girvan, 2004). This can be defined as follows:
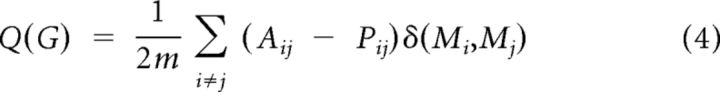 |
where m is the total number of edges; Aij = 1 if an edge links i and j and 0 otherwise; δ (Mi, Mj) = 1 if i and j are in the same module and 0 otherwise; and Pij is the probability that a random graph with the same degree distribution as G has an edge between i and j, as follows:
 |
where ki is i's degree. Nodes should be assigned to modules to yield the largest possible Q, which is approximated using a simulated annealing algorithm (Reichardt and Bornholdt, 2006). For the analysis of community structure, nodes were assigned to their most stable modules based on 100 runs of the simulated annealing algorithm (Alexander-Bloch et al., 2012).
The connection length or distance dij between two nodes i and j was simply estimated as the Euclidean distance (“as the crow flies”) between the centroids of the two graphically connected brain regions, , where x, y, and z are the coordinates of the centroid of each region in MNI space. The mean connection distance or wiring cost, D(G), of a graph G was the average distance of all of its edges as follows:
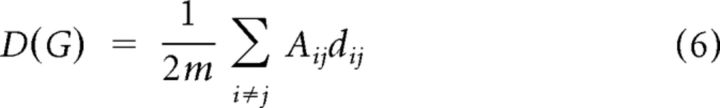 |
where Aij = 1 if an edge links i and j and 0 otherwise. This value was normalized to range from 0 to 1 by dividing by the maximum possible mean connection distance of a graph with the same number of edges.
The impact of age and in-scanner motion on structural-functional convergence.
The follow-up fMRI scan acquired on some of the subjects occurred outside of the 9–22 age range included in the longitudinal analysis, resulting in a difference in age between the fMRI population and structural/maturational population, with 12 of the 32 subjects > 22 years old. Given prior work demonstrating that there are differences in functional connectivity between children, adolescents, and adults (Fair et al., 2007, 2008, 2009; Supekar et al., 2010), it was important to assess the impact of this age difference on structural–functional convergence. We therefore assessed the structural–functional and maturational–functional convergence separately, using only the subjects whose fMRI scans were acquired at the same time as the structural scans used in the structural analysis (N = 16; age range 15.2–22.68) and those whose fMRI scans were acquired as adults at a later date (N = 16; age range 24.2–33.8). Additionally, we assessed the impact of the motion scrubbing procedure on the measures of structural–functional convergence, by repeating the fMRI analysis without performing motion scrubbing and recalculating the edgewise correlation between average functional connectivity and structural covariance and synchronized maturational change.
The choice of anatomically defined brain regions.
In a supplemental analysis, we also constructed structural, maturational, and functional networks using 74 cortical regions of the Automatic Anatomical Labeling (AAL) atlas, which incorporates a priori knowledge about functional neuroanatomical boundaries. This atlas has been used in numerous fMRI studies since it was originally reported (Tzourio-Mazoyer et al., 2002), and it has been previously used in studies of cortical thickness covariance across subjects (Gong et al., 2009; Khundrakpan et al., 2012). This atlas is less fine-grained and less uniform than our custom atlas of 360 cortical regions.
Results
Network-level similarity
Brain networks derived from cross-sectional structural covariance, synchronized maturational change, and functional connectivity demonstrated convergent, nonrandom network configurations. Compared with random networks with the same number of nodes and edges, the brain networks were all highly clustered and modular (Fig. 2A,B). Most of the edges in the sparsely thresholded networks tended to be physically short-distance connections (see Fig. 4B), and the degree distributions were skewed toward having a small number of relatively high-degree nodes (Fig. 4C).
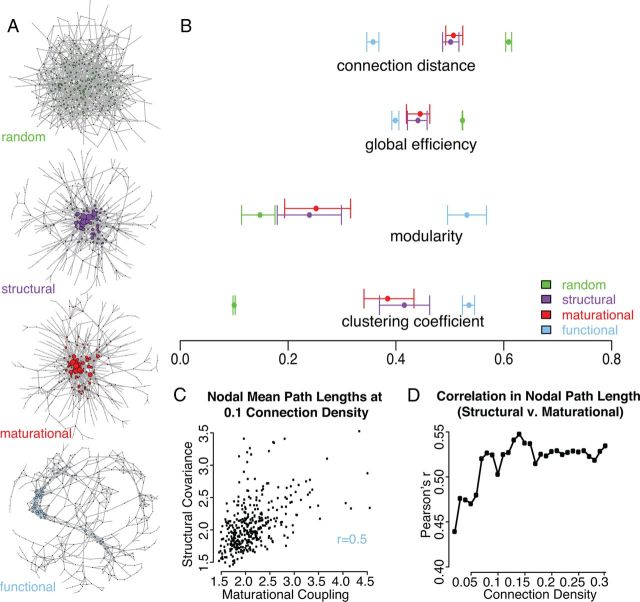
Figure 2.
Topological characteristics of multimodal brain networks constructed from the strongest connections in each modality. A, Topological layouts using the same force-directed algorithm (Csardi and Nepusz, 2006) for all networks thresholded at a sparse 2% connection density, as well as a random graph with the same number of nodes and edges. B, Global topological properties of the networks at 10% connection density, with bootstrapped 95% confidence intervals generated by resampling 5000 times with replacement across subjects. C, Nodal average path length for structural versus maturational networks at 10% connection density. D, Correlation coefficient of nodal average path length for structural versus maturational networks, across a range of connection densities.
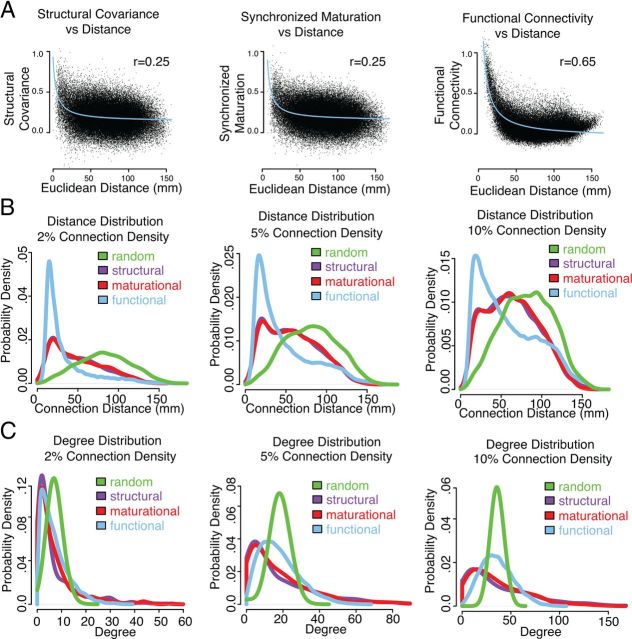
Figure 4.
Network profiles of edge connection distance and nodal degree distributions. A, Across all pairs of brain regions, connection distance is inversely proportional to structural covariance, synchronized maturational change, and functional connectivity. B, At three different connection densities, connection distance probability distributions for structural, maturational, functional, and benchmark random networks. C, At three different connection densities, nodal degree probability distributions for structural, maturational, functional, and benchmark random networks.
Further convergence was evident between structural, maturational, and functional networks at the level of nodal properties. The average path length was correlated across connection densities, for the maturational and structural networks (Fig. 2C,D; r > 0.4; p < 0.00001), indicating that the same central nodes tend to have the shortest paths to other nodes in both modalities. The correlation with the functional networks was substantially lower but still significant over a wide range of connection densities (r > 0.3, p < 0.001). The average anatomical distance of connections was also correlated across nodes, for all of the networks (r > 0.3, p < 0.001).
Despite all three networks showing highly nonrandom organization, and several convergent nodal properties, structural and maturational networks could be distinguished from functional networks in a number of ways. At the global level functional networks were relatively more clustered and modular, showing enhanced segregative properties. In contrast the structural and maturational networks demonstrate greater global efficiency and degree distributions that are skewed to a greater extent toward a small number of relatively high-degree nodes (Figs. 2, 4). The nodal-level corollary of these differences was a lack of correlation between nodal clustering coefficients seen in structural and maturational networks as compared with functional networks, reflecting the fact that fMRI networks were more topologically clustered and spatially localized.
Edge-level similarity
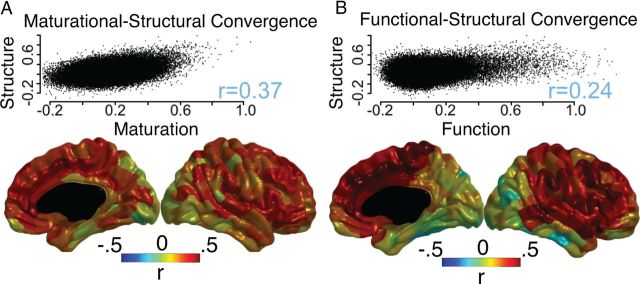
Across the 64,620 possible edges between the 360 brain regions, there is a strong similarity between maturational and structural measures of connectivity (Pearson's r = 0.37, Mantel permutation test p ≪ 0.001; Fig. 3). The strength of this relationship varies across the cortex, with the strongest convergence occurring in cingulate, frontal medial, and inferior frontal areas (Fig. 3). On average growth rates were negative during the observed age range for the vast majority of regions (308 out of 360), indicating synchronized losses in cortical thickness. The correlation between structural and maturation networks was not substantially altered by regressing out the maturational rate of change for each subject from the cross-sectional measures of cortical thickness, and then constructing the structural covariance networks (original r = 0.366, recalculated r = 0.369). This indicates that the convergence between structural and maturational covariance is not a statistical artifact but rather evidence for a substantive link between coordinated maturational change and inter-regional covariation of cortical thickness.
Figure 3.
Association between structural covariance, maturational coupling, and functional connectivity. A, The correlation between structural covariance and maturational coupling, across all pairs of brain regions (scatterplot) and for each region separately. B, The correlation between structural covariance and functional connectivity across all pairs of brain regions (scatterplot) and for each region separately.
There is also a predictive relationship between structural and functional connectivity metrics, across all edges (r = 0.24, p ≪ 0.001; Fig. 3). When including only the strongest 10% of edges in terms of structural covariance, the correlation across this subset of edges increases to r = 0.35. In contrast, including only the strongest 10% of edges in terms of functional connectivity only marginally increases the correlation (r = 0.27). The amount of multimodal convergence is regionally heterogeneous, and is highest in cingulate, superior temporal, and prefrontal regions (Fig. 3). A supplemental analysis that weighted each voxel's contribution to the regional fMRI signal by its gray matter density found the same edge-level convergence between structural and functional networks, suggesting that this convergence was not due to partial volume-related artifacts.
All of the connectivity measures have a strong negative relationship with the physical distance between regions, as approximated by the Euclidean distance between regional centroids, such that connectivity strength tends to decrease as the distance between regions increases (Fig. 4A). All three types of connectivity were inversely proportional to distance, with C(G) ∼ 1/D(G): structural networks, r = 0.25; maturational networks, r = 0.25; functional networks, r = 0.65; Mantel permutation test p ≪ 0.001 in all cases. Inverse proportionality was a better fit than a linear relationship where C(G) ∼ D(G) (structural networks, r = −0.23; maturational networks, r = −0.23; functional networks, r = − 0.36), or an inverse-square law where C(G) ∼ 1/D(G)2 (structural networks, r = 0.21; maturational networks, r = 0.20; functional networks, r = 0.61).
The influence of functional community structure
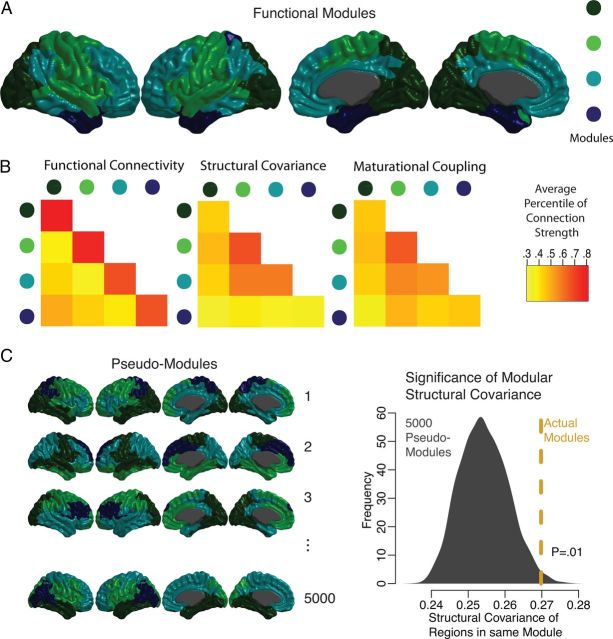
Synchronized maturational change and structural covariance were greater for regions in the same functional modules than for regions in different functional modules, indicating that covariation in regional cortical anatomy and maturation across subjects coincided with cortical modular functional organization (Fig. 5). Within this broad convergence, heterogeneity was evident in the level of functional–structural convergence between modules. In particular, a module that comprised almost the entire occipital lobe demonstrated relatively low intramodular structural covariance, as did a medial temporal module that also included the temporal pole.
Figure 5.
Structural covariance and maturational coupling of functional modular communities. A, Population-level functional communities based on 100 runs of a simulated annealing algorithm to maximize network modularity, at 10% connection density. B, The average functional connectivity, structural covariance, and maturational coupling, within and between the functional communities. C, The structural covariance within functional communities tested using a permutation procedure. Five thousand pseudo-modules were generated having the same number of spatially contiguous clusters as the actual modules and similar symmetry about the midline.
Since strong maturational and structural covariance within functional modules could be explicable by the spatial contiguity and contralateral homology of functional modules, rather than any more fundamental cross-modal convergence, we examined whether these factors alone explained the relatively high within-module structural covariance. We generated pseudo-modules that were the same size as the actual functional modules, contained the same number of spatially contiguous anatomical clusters, and were symmetric about the midline. The actual within-module structural covariance was greater than would be expected for random modules, even when controlling for these important factors (Fig. 5). This result was replicated across connection densities from 10–20%, at 1% intervals, using functional modules derived from both weighted and unweighted networks (p < 0.02 in all cases).
The impact of age and in-scanner motion
To test whether alterations in the functional connectome with age impacted on our measures of functional–structural convergence, we split the study population into a younger and an older group of subjects and tested the relationship between functional, structural, and maturational connectivity separately. The correlations across all edges were similar in the younger group (N = 16; age range 15.2–22.68; structural–functional correlation, r = 0.23; maturational–functional correlation, r = 0.19) and the older group (age range 24.2–33.8; functional–structural correlation, r = 0.22; maturational–functional correlation, r = 0.21). This indicates that the overall convergence is not being driven preferentially by one of these age groups.
Although previous studies have demonstrated a relationship between age and in-scanner motion, such that younger subjects tend to have more scans contaminated by motion artifact (Satterthwaite et al., 2012), in our sample there was no evidence for a correlation between age and the number of high motion frames > 0.3 mm (r = −0.066, p = 0.7). Similarly, when the subjects were divided into the 50% oldest and the 50% youngest subjects, there was no evidence for a difference in the number of “clean” frames that did not contain high amounts of motion (younger group, mean = 137 frames; older group, mean = 141 frames; Welch's t(18) = 0.75, p = 0.46).
To assess the impact of in-scanner motion on the measures of structural–functional convergence, we repeated the fMRI analysis without performing motion scrubbing and recalculated the correlation across all edges between functional, structural, and maturational connectivity. The results without scrubbing (structural–functional correlation, r = 0.23; maturational–functional correlation, r = 0.20) were comparable to the results with scrubbing (structural–functional correlation, r = 0.24; maturational–functional correlation, r = 0.21). The stability of these correlations indicates that the structural–functional convergence is unlikely to be due either to motion artifact or to alterations in the correlational structure induced by the scrubbing procedure.
The choice of anatomically defined brain regions
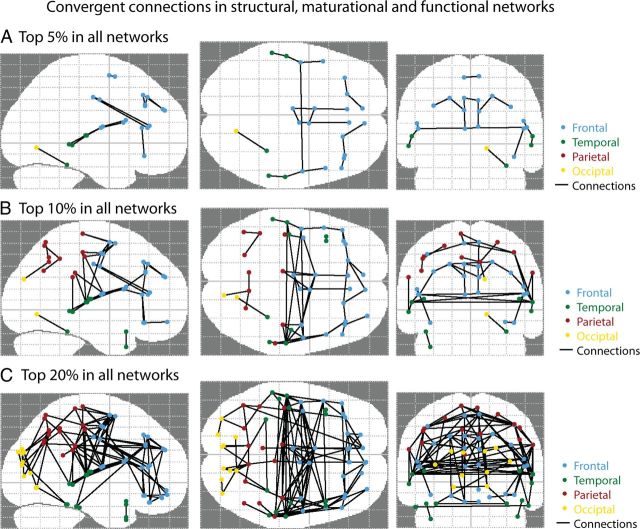
The convergence across structural, maturational, and functional networks was robust to replacing the 360 cortical regions used in the original analysis with 74 anatomically-predefined cortical regions of the AAL atlas. In fact using the AAL regions resulted in stronger convergence between structural and maturational networks (r = 0.6) and structural and functional networks (r = 0.37), possibly because these regions were larger on average resulting in greater signal to noise. This re-analysis also allowed greater specificity as to the anatomical labels of the regions with highest multimodal convergence. The “multimodal network backbone” comprised a set of inter-regional connections that were among the strongest in functional, maturational, and also structural networks. The consistently interconnected regions included medial and inferior frontal cortex, sensorimotor cortex (supplementary motor area and precentral gyrus), posterior parietal cortex (supramarginal gyrus and precuneus), and superior temporal cortex bilaterally (Fig. 6).
Figure 6.
Pairs of brain regions that are strongly connected in all three of the networks under study: structural, maturational, and functional. The conjunction map of correlations that are among the strongest 5% (A), 10% (B), and 20% (C) in all three networks. All of the correlations shown are significant in each of the three networks using an FDR-adjusted p < 0.0001 (Benjamani et al., 2006). Visualization was performed in MATLAB using the Network Based Statistics Toolbox (Zalesky et al., 2010). Anatomical labels were defined using the AAL atlas (Tzourio-Mazoyer et al., 2002).
Discussion
By applying graph-theory approaches to three different facets of connectivity between brain regions we show that structural, maturational, and functional brain networks all possess nonrandom economical small-world and modular organization, with amplified local connectivity in functional networks. In addition, patterns of cross-sectional structural covariance recapitulate patterns of coordinated structural maturation and, to a lesser extent, coordinated functional activation, in a regionally heterogeneous manner that is related to brain functional community structure.
Conserved global organization
Nonrandom brain configuration has previously been revealed by network analysis of diverse brain properties including those derived from histology, structural and fMRI, magnetoencephalography, and electroencephalography (Bullmore and Sporns, 2009, 2012) suggesting consistent organizational principles. Evolutionary pressure may exist to maximize communication efficiency while minimizing connection cost, with this tension resulting in cost-efficient, “small-world” networks having both low cost and high efficiency. The physical necessity to embed this network in anatomical space results in a characteristic balance between short- and long-distance connections that constrain the brain's topological characteristics (Kaiser and Hilgetag, 2006; Bassett et al., 2010; Vértes et al., 2012; Alexander-Bloch et al., 2013). Our data reveal that similar organizational principles also operate on maturational networks, shaping neurodevelopment across adolescence.
Structural–maturational convergence
Our results strongly support the hypothesis that correlated anatomical structure between brain regions results from similarities in maturational trajectories. Not only are these networks topologically extremely similar, but also maturational and structural connectivity are highly correlated across brain regions. These findings allow for the reinterpretation of the pre-existing literature describing patterns of brain structural covariance across human populations, in terms of both normal brain anatomy and clinical abnormalities. Since the first MRI studies demonstrating population covariance in brain regional anatomy, it has been suggested that structural covariance could be mediated by coordinated neurodevelopment (Wright et al., 1999; Zielinski et al., 2010); regions that are connected to each other may grow and shrink at the same rate, leading to inter-regional correlations of cortical thickness. In addition, longitudinal imaging studies have established that different brain systems demonstrate specific maturational trajectories during childhood and adolescence (Gogtay et al., 2004; Shaw et al., 2008; Raznahan et al., 2011a). Our results explicitly address and confirm this explanatory link. As thickness decreases across the cortex during adolescence, with synaptic pruning leading to reduced neuropil volume, it is probable that synchronized pruning drives population covariance in cortical thickness.
Local functional convergence
That synchronized maturational change likely underlies cross-sectional covariance in cortical thickness raises subsequent questions about the relationships of these measures with functional connectivity. Our findings advance previous arguments in favor of the functional relevance of structural covariance (Andrews et al., 1997; Seeley et al., 2009), in several novel directions. First, by carrying out the first brain-wide test of structural–functional convergence, querying all possible pairs of brain regions rather than a priori regions of interest, we demonstrate a systematic association between the functional connectivity and the structural covariance between pairs of brain regions. Second, we specify how the strength of this convergence varies across the brain: relatively high in prefrontal, cingulate, motor, and perisylvian areas; diminished in the medial temporal and occipital lobes. Third, our graph-theoretical approach reveals that structural covariance reflects the mesoscopic functional community structure of the brain. Regions in the same fMRI module show higher structural covariance than regions in different modules, even when we control for the spatial contiguity of modular structure. As in the “edge-level” analysis of convergence, we also find regional differences in modular coincidence, such that functional modules that include canonical regions of the default-mode network, in addition to primary motor and perisylvian regions, show relatively high structural covariance compared with the rest of the brain. Although we cannot be certain of the behavioral or cognitive significance of our neuroanatomical findings, the greatest multimodal convergence included regions implicated in sensorimotor, language/auditory processing, and working memory systems (Fig. 6).
Cellular mechanisms and interpretations
Although formally testing mechanisms underlying the cross-modal convergences and divergences we describe would require experimental approaches difficult to implement in humans, a number of mechanisms can be considered speculatively. At the cellular level, functional coupling has a bidirectional relationship with the strength of axonal connections. Synaptogenesis in neuronal systems is known to occur in the presence of synchronous firing (Katz and Shatz, 1996; Bi and Poo, 1999), and experience and learning have been shown to induce structural alterations in gray matter (Draganski et al., 2004; Ilg et al., 2008). For some regions within the default-mode network, functional connectivity has been shown to precede white matter connectivity developmentally (Supekar et al., 2010; Zielinski et al., 2010). Axonal connectivity could, in turn, explain coordinated growth between connected brain regions, for example, due to synchronous exposure to trophic factors (Ferrer et al., 1995; Pezawas et al., 2004) and/or mechanical tension (Van Essen, 1997). Strong population covariance has been repeatedly observed between brain regions that are known to be densely axonally interconnected, such as frontoparietal (Wright et al., 1999; Lerch et al., 2006) and frontotemporal systems (Woodruff et al., 1997; Bullmore et al., 1998). It is therefore possible that shared functional demand drives maturational coupling, which in turn drives structural covariance, although our data are also consistent with other, less straightforwardly causal relationships between these networks of brain regions.
Consistent with prior work comparing functional and white matter (diffusion imaging) networks (Koch et al., 2002; Greicius et al., 2009; Honey et al., 2009; Adachi et al., 2012), structural covariance does not display a simple one-to-one equivalence with functional connectivity, but rather a broadly correlated relationship that becomes anatomically and topologically complex in its details. Local divergences could reflect regional variation in the extent to which functional coupling is underpinned by direct white matter connections or in the developmental stability of functional connectivity (Fair et al., 2008, 2009; Zielinski et al., 2010). Thus, convergence between structural/maturational and functional networks may be strongest where functional connectivity is developmentally sustained and mediated by direct white matter connectivity. Explicitly addressing how white matter and functional connectivity may interact to influence cortical maturation awaits the existence of large longitudinal datasets including both diffusion and functional imaging components. Finally, partially attenuated and anatomically heterogeneous cross-modal convergence could relate to noisiness of MRI data. A potential weakness of this study is the lack of in-scanner respiratory and cardiac data. Future work could assess the impact of physiological noise on structural/functional convergence (Glover et al., 2000; He and Liu, 2012). The relatively lower cross-modal convergence in occipital pole and medial temporal cortex could result from lower signal to noise in image acquisition, or because maturational coordination in these regions is completed in early childhood and therefore not observable in our cohort. An extra source of error exists in the estimation of regional rates of growth from a limited number of longitudinal scans.
The role of anatomical distance
The relationship between measures of brain connectivity and the physical distance between brain regions is both a fascinating area of ongoing research and a potential experimental confound. In-scanner motion, cardiac and respiratory signal, and blurring due to spatial registration and interpolation can bias neuroimaging data in favor of the discovery of short-distance connections. On the other hand, a host of experimental results confirm that there is a genuinely biological, inverse relationship between anatomical distance and connectivity, indicating that physical distance represents a metabolic cost that constraints and enables network topological characteristics (Honey et al., 2009; Kaiser and Varier, 2011; Vértes et al., 2012; Alexander-Bloch et al., 2013). Our results confirm the strong inverse relationship between connectivity and distance in structural networks and functional networks, and demonstrate a similar relationship for the first time in maturational networks.
Anatomical proximity is more predictive of functional connectivity than either maturational or structural covariance. This stronger relationship with distance in turn predicts the relatively higher clustering and modularity observed in the functional networks (Vértes et al., 2012; Alexander-Bloch et al., 2013). The connection between distance and functional activity could result from the preponderance of short-range lateral connections in the cortex, combined with spatial autocorrelation of cortico-thalamo-cortical and more complex network-level effects (Honey et al., 2009; Adachi et al., 2012). It is also possible that the vascular system mediating the fMRI signal has a higher extent of spatial autocorrelation than neuronal activity itself. Finally, the lower voxel resolution in fMRI compared with structural MRI increases spatial smoothing at short distances when the images are interpolated into the same space.
Conclusions
Using one of the largest extant longitudinal datasets of typical brain maturation in humans, we show that network organizational principles that have been previously shown to operate on structural and functional networks also constrain maturational networks, shaping neurodevelopment across adolescence. Furthermore, coordinated maturational trajectories likely underlie cross-sectional structural covariance in thickness and other anatomical properties across healthy individuals. These networks also recapitulate intrinsic brain activity in an anatomically heterogeneous fashion that respects the organization of functional subcommunities in the brain. A three-way synergy may exist between wiring, firing, and developmental trajectories that is visible at the spatial resolution of MRI.
Footnotes
This study was funded through the National Institute of Mental Health, the National Institutes of Health Intramural Research Program, and the National Institutes of Health–Oxford-Cambridge Scholarship Program (A.A.B.). The Behavioural and Clinical Neuroscience Institute is supported by the Medical Research Council (UK) and the Wellcome Trust. We wish to thank Francois Lalonde and Liv Clasen for help with data collection and analysis, Budhachandra Khundrakpam for assistance with the AAL atlas, and also the participants who took part in this study.
References
- Adachi Y, Osada T, Sporns O, Watanabe T, Matsui T, Miyamoto K, Miyashita Y. Functional connectivity between anatomically unconnected areas is shaped by collective network-level effects in the macaque cortex. Cereb Cortex. 2012;22:1586–1592. doi: 10.1093/cercor/bhr234. [DOI] [PubMed] [Google Scholar]
- Alexander-Bloch AF, Gogtay N, Meunier D, Birn R, Clasen L, Lalonde F, Lenroot R, Giedd J, Bullmore ET. Disrupted modularity and local connectivity of brain functional networks in childhood-onset schizophrenia. Front Syst Neurosci. 2010;4:147. doi: 10.3389/fnsys.2010.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander-Bloch AF, Vértes PE, Stidd R, Lalonde F, Clasen L, Rapoport J, Giedd J, Bullmore ET, Gogtay N. The anatomical distance of functional connections predicts brain network topology in health and schizophrenia. Cereb Cortex. 2013;23:127–138. doi: 10.1093/cercor/bhr388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander-Bloch A, Lambiotte R, Roberts B, Giedd J, Gogtay N, Bullmore E. The discovery of population differences in network community structure: new methods and applications to brain functional networks in schizophrenia. Neuroimage. 2012;59:3889–3900. doi: 10.1016/j.neuroimage.2011.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews TJ, Halpern SD, Purves D. Correlated size variations in human visual cortex, lateral geniculate nucleus, and optic tract. J Neurosci. 1997;17:2859–2868. doi: 10.1523/JNEUROSCI.17-08-02859.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Bullmore E, Verchinski BA, Mattay VS, Weinberger DR, Meyer-Lindenberg A. Hierarchical organization of human cortical networks in health and schizophrenia. J Neurosci. 2008;28:9239–9248. doi: 10.1523/JNEUROSCI.1929-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Greenfield DL, Meyer-Lindenberg A, Weinberger DR, Moore SW, Bullmore ET. Efficient physical embedding of topologically complex information processing networks in brains and computer circuits. PLoS Comput Biol. 2010;6:e1000748. doi: 10.1371/journal.pcbi.1000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Krieger AM, Yekutieli D. Adaptive linear step-up procedures that control the false discovery rate. Biometrika. 2006;93:491–507. [Google Scholar]
- Bernhardt BC, Chen Z, He Y, Evans AC, Bernasconi N. Graph-theoretical analysis reveals disrupted small-world organization of cortical thickness correlation networks in temporal lobe epilepsy. Cereb Cortex. 2011;21:2147–2157. doi: 10.1093/cercor/bhq291. [DOI] [PubMed] [Google Scholar]
- Bi G, Poo M. Distributed synaptic modification in neural networks induced by patterned stimulation. Nature. 1999;401:792–796. doi: 10.1038/44573. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. The economy of brain network organization. Nat Rev Neurosci. 2012;13:336–349. doi: 10.1038/nrn3214. [DOI] [PubMed] [Google Scholar]
- Bullmore ET, Woodruff PW, Wright IC, Rabe-Hesketh S, Howard RJ, Shuriquie N, Murray RM. Does dysplasia cause anatomical dysconnectivity in schizophrenia? Schizophr Res. 1998;30:127–135. doi: 10.1016/s0920-9964(97)00141-2. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Fadili J, Maxim V, Sendur L, Whitcher B, Suckling J, Brammer M, Breakspear M. Wavelets and functional magnetic resonance imaging of the human brain. Neuroimage. 2004;23(Suppl 1):S234–S249. doi: 10.1016/j.neuroimage.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Carp J. Optimizing the order of operations for movement scrubbing: comment on Power et al. Neuroimage. 2012 doi: 10.1016/j.neuroimage.2011.12.061. Advanced online publication. Retrieved Jan. 15, 2013. [DOI] [PubMed] [Google Scholar]
- Chen ZJ, He Y, Rosa-Neto P, Germann J, Evans AC. Revealing modular architecture of human brain structural networks by using cortical thickness from MRI. Cereb Cortex. 2008;18:2374–2381. doi: 10.1093/cercor/bhn003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, He Y, Rosa-Neto P, Gong G, Evans AC. Age-related alterations in the modular organization of structural cortical network by using cortical thickness from MRI. Neuroimage. 2011;56:235–245. doi: 10.1016/j.neuroimage.2011.01.010. [DOI] [PubMed] [Google Scholar]
- Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29:162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- Craddock RC, James GA, Holtzheimer PE, 3rd, Hu XP, Mayberg HS. A whole brain fMRI atlas generated via spatially constrained spectral clustering. Hum Brain Mapp. 2012;33:1914–1928. doi: 10.1002/hbm.21333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal Complex Systems. 2006;18:1695. [Google Scholar]
- Damoiseaux JS, Greicius MD. Greater than the sum of its parts: a review of studies combining structural connectivity and resting-state functional connectivity. Brain Struct Funct. 2009;213:525–533. doi: 10.1007/s00429-009-0208-6. [DOI] [PubMed] [Google Scholar]
- Draganski B, Gaser C, Busch V, Schuierer G, Bogdahn U, May A. Neuroplasticity: changes in grey matter induced by training. Nature. 2004;427:311–312. doi: 10.1038/427311a. [DOI] [PubMed] [Google Scholar]
- Fair DA, Dosenbach NU, Church JA, Cohen AL, Brahmbhatt S, Miezin FM, Barch DM, Raichle ME, Petersen SE, Schlaggar BL. Development of distinct control networks through segregation and integration. Proc Natl Acad Sci U S A. 2007;104:13507–13512. [Google Scholar]
- Fair DA, Cohen AL, Dosenbach NU, Church JA, Miezin FM, Barch DM, Raichle ME, Petersen SE, Schlaggar BL. The maturing architecture of the brain's default network. Proc Natl Acad Sci U S A. 2008;105:4028–4032. doi: 10.1073/pnas.0800376105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair DA, Cohen AL, Power JD, Dosenbach NU, Church JA, Miezin FM, Schlaggar BL, Petersen SE. Functional brain networks develop from a “local to distributed” organization. PLoS Comput Biol. 2009;5:e1000381. doi: 10.1371/journal.pcbi.1000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer I, Blanco R, Carulla M, Condom M, Alcántara S, Olivé M, Planas A. Transforming growth factor-alpha immunoreactivity in the developing and adult brain. Neuroscience. 1995;66:189–199. doi: 10.1016/0306-4522(94)00584-r. [DOI] [PubMed] [Google Scholar]
- Glover GH, Li TQ, Ress D. Image-based method for retrospective correction of physiological motion effects in fMRI: RETROICOR. Magn Reson Med. 2000;44:162–167. doi: 10.1002/1522-2594(200007)44:1<162::aid-mrm23>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Gogtay N, Giedd JN, Lusk L, Hayashi KM, Greenstein D, Vaituzis AC, Nugent TF, 3rd, Herman DH, Clasen LS, Toga AW, Rapoport JL, Thompson PM. Dynamic mapping of human cortical development during childhood through early adulthood. Proc Natl Acad Sci U S A. 2004;101:8174–8179. doi: 10.1073/pnas.0402680101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong G, Rosa-Neto P, Carbonell F, Chen ZJ, He Y, Evans AC. Age- and gender-related differences in the cortical anatomical network. J Neurosci. 2009;29:15684–15693. doi: 10.1523/JNEUROSCI.2308-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Supekar K, Menon V, Dougherty RF. Resting-state functional connectivity reflects structural connectivity in the default mode network. Cereb Cortex. 2009;19:72–78. doi: 10.1093/cercor/bhn059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Liu TT. A geometric view of global signal confounds in resting-state functional MRI. Neuroimage. 2012;59:2339–2348. doi: 10.1016/j.neuroimage.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Chen ZJ, Evans AC. Small-world anatomical networks in the human brain revealed by cortical thickness from MRI. Cereb Cortex. 2007;17:2407–2419. doi: 10.1093/cercor/bhl149. [DOI] [PubMed] [Google Scholar]
- He Y, Chen Z, Evans A. Structural insights into aberrant topological patterns of large-scale cortical networks in Alzheimer's disease. J Neurosci. 2008;28:4756–4766. doi: 10.1523/JNEUROSCI.0141-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey CJ, Sporns O, Cammoun L, Gigandet X, Thiran JP, Meuli R, Hagmann P. Predicting human resting-state functional connectivity from structural connectivity. Proc Natl Acad Sci U S A. 2009;106:2035–2040. doi: 10.1073/pnas.0811168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilg R, Wohlschläger AM, Gaser C, Liebau Y, Dauner R, Wöller A, Zimmer C, Zihl J, Mühlau M. Gray matter increase induced by practice correlates with task-specific activation: a combined functional and morphometric magnetic resonance imaging study. J Neurosci. 2008;28:4210–4215. doi: 10.1523/JNEUROSCI.5722-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Med Image Anal. 2001;5:143–156. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- Kaiser M, Hilgetag CC. Nonoptimal component placement, but short processing paths, due to long-distance projections in neural systems. PLoS Comput Biol. 2006;2:e95. doi: 10.1371/journal.pcbi.0020095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser M, Varier S. Evolution and development of brain networks: from Caenorhabditis elegans to Homo sapiens. Network. 2011;22:143–147. doi: 10.3109/0954898X.2011.638968. [DOI] [PubMed] [Google Scholar]
- Katz LC, Shatz CJ. Synaptic activity and the construction of cortical circuits. Science. 1996;274:1133–1138. doi: 10.1126/science.274.5290.1133. [DOI] [PubMed] [Google Scholar]
- Kelly C, Toro R, Di Martino A, Cox CL, Bellec P, Castellanos FX, Milham MP. A convergent functional architecture of the insula emerges across imaging modalities. Neuroimage. 2012;61:1129–1142. doi: 10.1016/j.neuroimage.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khundrakpam BS, Reid A, Brauer J, Carbonell F, Lewis J, Ameis S, Karama S, Lee J, Chen Z, Das S, Evans AC Brain Development Cooperative Group. Developmental changes in organization of structural brain networks. Cereb Cortex. 2012 doi: 10.1093/cercor/bhs187. Advance online publication. Retrieved Jan. 15, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Singh V, Lee JK, Lerch J, Ad-Dab'bagh Y, MacDonald D, Lee JM, Kim SI, Evans AC. Automated 3-D extraction and evaluation of the inner and outer cortical surfaces using a Laplacian map and partial volume effect classification. Neuroimage. 2005;27:210–221. doi: 10.1016/j.neuroimage.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Koch MA, Norris DG, Hund-Georgiadis M. An investigation of functional and anatomical connectivity using magnetic resonance imaging. Neuroimage. 2002;16:241–250. doi: 10.1006/nimg.2001.1052. [DOI] [PubMed] [Google Scholar]
- Latora V, Marchiori M. Efficient behavior of small-world networks. Phys Rev Lett. 2001;87:198701. doi: 10.1103/PhysRevLett.87.198701. [DOI] [PubMed] [Google Scholar]
- Lerch JP, Worsley K, Shaw WP, Greenstein DK, Lenroot RK, Giedd J, Evans AC. Mapping anatomical correlations across cerebral cortex (MACACC) using cortical thickness from MRI. Neuroimage. 2006;31:993–1003. doi: 10.1016/j.neuroimage.2006.01.042. [DOI] [PubMed] [Google Scholar]
- Mechelli A, Friston KJ, Frackowiak RS, Price CJ. Structural covariance in the human cortex. J Neurosci. 2005;25:8303–8310. doi: 10.1523/JNEUROSCI.0357-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman ME, Girvan M. Finding and evaluating community structure in networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2004;69 doi: 10.1103/PhysRevE.69.026113. 026113. [DOI] [PubMed] [Google Scholar]
- Pezawas L, Verchinski BA, Mattay VS, Callicott JH, Kolachana BS, Straub RE, Egan MF, Meyer-Lindenberg A, Weinberger DR. The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. J Neurosci. 2004;24:10099–10102. doi: 10.1523/JNEUROSCI.2680-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raznahan A, Lerch JP, Lee N, Greenstein D, Wallace GL, Stockman M, Clasen L, Shaw PW, Giedd JN. Patterns of coordinated anatomical change in human cortical development: a longitudinal neuroimaging study of maturational coupling. Neuron. 2011a;72:873–884. doi: 10.1016/j.neuron.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raznahan A, Shaw P, Lalonde F, Stockman M, Wallace GL, Greenstein D, Clasen L, Gogtay N, Giedd JN. How does your cortex grow? J Neurosci. 2011b;31:7174–7177. doi: 10.1523/JNEUROSCI.0054-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt J, Bornholdt S. Statistical mechanics of community detection. Phys Rev E Stat Nonlin Soft Matter Phys. 2006;74 doi: 10.1103/PhysRevE.74.016110. 016110. [DOI] [PubMed] [Google Scholar]
- Rockel AJ, Hiorns RW, Powell TP. The basic uniformity in structure of the neocortex. Brain. 1980;103:221–244. doi: 10.1093/brain/103.2.221. [DOI] [PubMed] [Google Scholar]
- Satterthwaite TD, Wolf DH, Loughead J, Ruparel K, Elliott MA, Hakonarson H, Gur RC, Gur RE. impact of in-scanner head motion on multiple measures of functional connectivity: relevance for studies of neurodevelopment in youth. Neuroimage. 2012;60:623–632. doi: 10.1016/j.neuroimage.2011.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schank T, Wagner D. Approximating clustering-coefficient and transitivity. J Graph Algorithms and Applications. 2005;9:265–275. [Google Scholar]
- Schmitt JE, Lenroot RK, Wallace GL, Ordaz S, Taylor KN, Kabani N, Greenstein D, Lerch JP, Kendler KS, Neale MC, Giedd JN. Identification of genetically mediated cortical networks: a multivariate study of pediatric twins and siblings. Cereb Cortex. 2008;18:1737–1747. doi: 10.1093/cercor/bhm211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD. Neurodegenerative diseases target large-scale human brain networks. Neuron. 2009;62:42–52. doi: 10.1016/j.neuron.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P, Kabani NJ, Lerch JP, Eckstrand K, Lenroot R, Gogtay N, Greenstein D, Clasen L, Evans A, Rapoport JL, Giedd JN, Wise SP. Neurodevelopmental trajectories of the human cerebral cortex. J Neurosci. 2008;28:3586–3594. doi: 10.1523/JNEUROSCI.5309-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supekar K, Uddin LQ, Prater K, Amin H, Greicius MD, Menon V. Development of functional and structural connectivity within the default mode network in young children. Neuroimage. 2010;52:290–301. doi: 10.1016/j.neuroimage.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Van Dijk KR, Hedden T, Venkataraman A, Evans KC, Lazar SW, Buckner RL. Intrinsic functional connectivity as a tool for human connectomics: theory, properties, and optimization. J Neurophysiol. 2010;103:297–321. doi: 10.1152/jn.00783.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk KR, Sabuncu MR, Buckner RL. The influence of head motion on intrinsic functional connectivity MRI. Neuroimage. 2012;59:431–438. doi: 10.1016/j.neuroimage.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC. A tension-based theory of morphogenesis and compact wiring in the central nervous system. Nature. 1997;385:313–318. doi: 10.1038/385313a0. [DOI] [PubMed] [Google Scholar]
- Vértes PE, Alexander-Bloch AF, Gogtay N, Giedd JN, Rapoport JL, Bullmore ET. Simple models of human brain functional networks. Proc Natl Acad Sci U S A. 2012;109:5868–5873. doi: 10.1073/pnas.1111738109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viger F, Latapy M. Efficient and simple generation of random simple connected graphs with prescribed degree sequence. In: Wang L, editor. Computing and combinatorics: lecture notes in computer science. Vol 3595. Berlin: Springer; 2005. [Google Scholar]
- Weissenbacher A, Kasess C, Gerstl F, Lanzenberger R, Moser E, Windischberger C. Correlations and anticorrelations in resting-state functional connectivity MRI: a quantitative comparison of preprocessing strategies. Neuroimage. 2009;47:1408–1416. doi: 10.1016/j.neuroimage.2009.05.005. [DOI] [PubMed] [Google Scholar]
- White LE, Andrews TJ, Hulette C, Richards A, Groelle M, Paydarfar J, Purves D. Structure of the human sensorimotor system. II: lateral symmetry. Cereb Cortex. 1997;7:31–47. doi: 10.1093/cercor/7.1.31. [DOI] [PubMed] [Google Scholar]
- Woodruff PW, Wright IC, Shuriquie N, Russouw H, Rushe T, Howard RJ, Graves M, Bullmore ET, Murray RM. Structural brain abnormalities in male schizophrenics reflect fronto-temporal dissociation. Psychol Med. 1997;27:1257–1266. doi: 10.1017/s0033291797005229. [DOI] [PubMed] [Google Scholar]
- Wright IC, Sharma T, Ellison ZR, McGuire PK, Friston KJ, Brammer MJ, Murray RM, Bullmore ET. Supra-regional brain systems and the neuropathology of schizophrenia. Cereb Cortex. 1999;9:366–378. doi: 10.1093/cercor/9.4.366. [DOI] [PubMed] [Google Scholar]
- Zalesky A, Fornito A, Bullmore ET. Network-based statistic: identifying differences in brain networks. Neuroimage. 2010;53:1197–1207. doi: 10.1016/j.neuroimage.2010.06.041. [DOI] [PubMed] [Google Scholar]
- Zhou J, Gennatas ED, Kramer JH, Miller BL, Seeley WW. Predicting regional neurodegeneration from the healthy brain functional connectome. Neuron. 2012;73:1216–1227. doi: 10.1016/j.neuron.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski BA, Gennatas ED, Zhou J, Seeley WW. Network-level structural covariance in the developing brain. Proc Natl Acad Sci U S A. 2010;107:18191–18196. doi: 10.1073/pnas.1003109107. [DOI] [PMC free article] [PubMed] [Google Scholar]