Abstract
Adult stem cells undergo asymmetric cell division to self-renew and give rise to differentiated cells that comprise mature tissue1. Sister chromatids may be distinguished and segregated non-randomly in asymmetrically dividing stem cells2, although the underlying mechanism and the purpose it may serve remain elusive. We developed the CO-FISH (chromosome orientation fluorescence in situ hybridization) technique3 with single-chromosome resolution and show that sister chromatids of X and Y chromosomes, but not autosomes, are segregated non-randomly during asymmetric divisions of Drosophila male germline stem cells (GSCs). This provides the first direct evidence that two sister chromatids containing identical genetic information can be distinguished and segregated non-randomly during asymmetric stem cell divisions. We further show that the centrosome, SUN-KASH nuclear envelope proteins, and Dnmt2 are required for non-random sister chromatid segregation. Our data suggest that the information on X and Y chromosomes that enables non-random segregation is primed during gametogenesis in the parents. Moreover, we show that sister chromatid segregation is randomized in GSC overproliferation and dedifferentiated GSCs. We propose that non-random sister chromatid segregation may serve to transmit distinct information carried on two sister chromatids to the daughters of asymmetrically dividing stem cells.
The Drosophila male germline stem cell (GSC) system is an excellent model system for the study of asymmetric stem cell division. GSCs can be identified at single-cell resolution at the apical tip of the testis, where they attach to a cluster of somatic hub cells, a major component of the stem cell niche4. GSCs divide asymmetrically by orienting the mitotic spindle perpendicular to the hub5. We previously showed that the mother centrosome is inherited by the GSCs6.
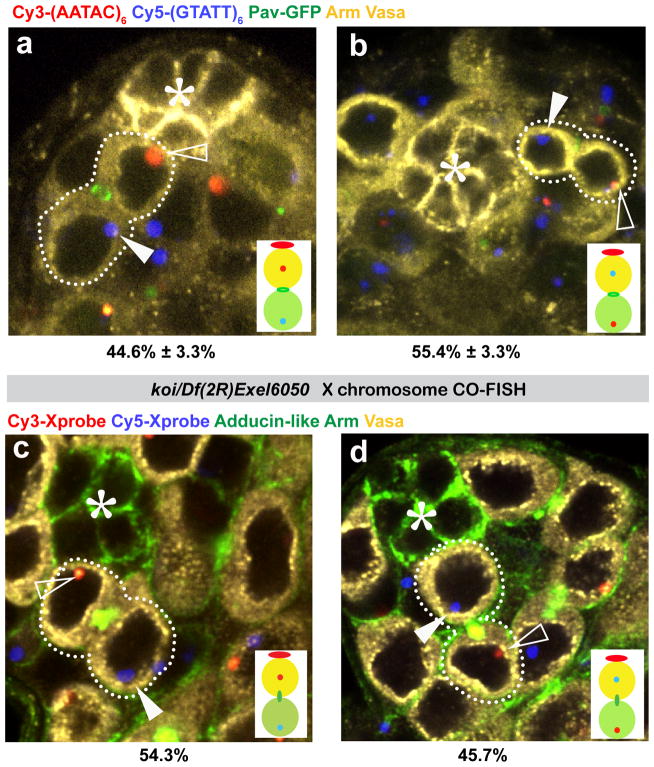
We adapted the CO-FISH (chromosome orientation fluorescence in situ hybridization) protocol, which allows strand-specific identification of sister chromatids3, combined with chromosome-specific probes7 (Figure 1a). Using this method, we identified the sister chromatids of each chromosome in GSCs and their differentiating daughter gonialblasts (GBs; Figure 1b, Figure S1). We found that sister chromatids of the Y chromosome are inherited with a strong bias during GSC division: In approximately 85% of cases, GSCs inherited the sister chromatid of the Y chromosome, whose template strand contains the (GTATT)6 satellite (and thus hybridizes to the Cy3-(AATAC)6 probe), and GBs inherited the sister chromatid whose template contains the (AATAC)6 sequence (and thus hybridizes to the Cy5-(GTATT)6 probe; Figure 1c, d). Using X chromosome-specific probes, we found that the X chromosome shows a similar bias (Figure 1e, f). Essentially the same results were obtained when the Cy5 probe for the X chromosome was replaced with a probe that is not complementary to the Cy3-labelled probe (Figure S2). Although both X and Y chromosomes show a similar bias in segregation (approximately 85:15), we found that the two chromosomes segregate independently of each other (Figure 1g, h, i) (see Methods for details).
Figure 1. Non-random segregation of Y and X chromosome strands during GSC divisions.
a) Chromosome-specific probes used in this study. b) Schematic diagram of the CO-FISH procedure. Cy3- and Cy5-labelled probes for the Y chromosome are shown as an example. Pavarotti-GFP27 (midbody/ring canal), ShAdd-Venus28 or anti-Add antibody (spectrosome) was used to identify GSC-GB pairs. Representative images of CO-FISH results using Y chromosome probes (c, d), X chromosome probes (e, f), and both X and Y probes (g–i). Expected segregation patterns based on co-segregation vs. random segregation are shown at the bottom of (g–i). In all figures the Cy5 signal is indicated by solid arrowheads and the Cy3 signal by open arrowheads. (*) Hub. N, number of GSC-GB pairs scored. Data are presented as mean ± standard deviation.
Two major scenarios can explain the observed bias of approximately 85:15. In the first scenario, approximately 85% of GSCs inherit the “red strand” (i.e. the sister chromatid containing the template strand that hybridizes to Cy3 probes) with near 100% accuracy, whereas approximately 15% of GSCs inherit the “blue strand” with near 100% accuracy. This would indicate that GSCs maintain particular strands of the X and Y chromosomes forever (“immortal strands”). In the second scenario, each GSC inherits the “red strand” with 85% probability and the “blue strand” with 15% probability at each division. In this case, GSCs do not retain immortal strands; instead, the “template strands” switch approximately once in every seven divisions (15%≅1/6.7). To distinguish between these possibilities, we conducted a long-pulse experiment where flies were continuously exposed to BrdU-containing medium (see Figure S3 for details). The results of this experiment clearly supported the second scenario.
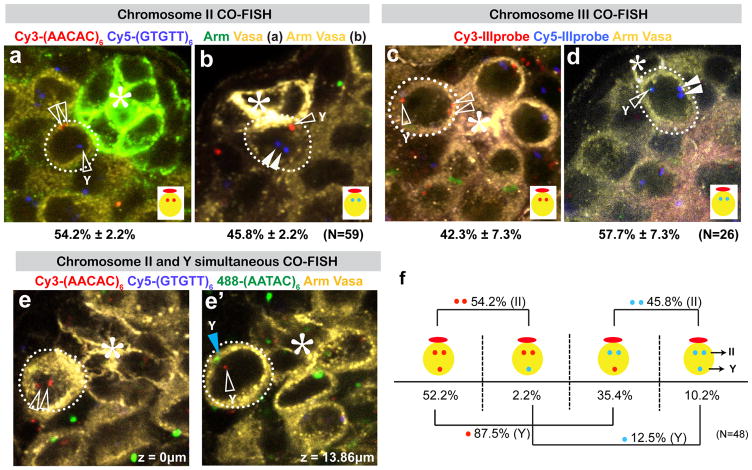
In contrast to X and Y chromosomes, we found that the autosomes (chromosomes II and III) do not show biased segregation (~50:50; Figure 2). Consistent with previous reports that homologous chromosomes are paired, even in non-meiotic cells in Drosophila8, we observed that two autosome signals corresponding to homologous chromosomes were always juxtaposed to each other (Figure 2a–d). In spite of the lack of biased segregation with regard to which strands are inherited by GSCs, cells always inherited two Cy3 signals or two Cy5 signals, the mechanism and significance of which remain elusive. It should be noted that the repeat sequences used as probes for chromosome II and III also exist on the Y chromosome9, yielding a third “lone” signal in addition to the paired autosome signals. The identity of the lone signal was confirmed by combining autosome probes and a Y chromosome probe, 488-(AATAC)6. The Y chromosome signal was always close to the lone signal (Figure 2e, e′). Importantly, the Y chromosome detected as a lone signal showed biased segregation, despite the fact that the paired autosome signals showed a random segregation pattern in the same set of samples (Figure 2f). This result further confirms our observation that sister chromatids of the Y chromosome are segregated non-randomly.
Figure 2. Autosomes are randomly segregated during GSC divisions.
Representative images of CO-FISH results using chromosome II probes (a, b), and chromosome III probes (c, d). Lone signals that correspond to the Y chromosome are marked with “Y”. N, number of GSCs scored. (*) Hub. e) A representative image showing that the lone signal of the (AACAC)6 probe (open arrowheads) is close to the (AATAC)6 signal (blue arrowhead). f) Summary of scoring results using chromosome II probes. Paired signals segregate randomly (Cy3-Cy3:Cy5-Cy5=54.4:45.6), whereas lone signals segregate non-randomly. (Cy3:Cy5=87.6:12.4). (AACAC)6 and (AATAC)6 sequences are on the same strand of the Drosophila Y chromosome.
Although many studies have reported biased sister chromatid segregation, the genes responsible for biased segregation have never been described. We found that centrosomin (cnn), a core component of the pericentriolar material10, SUN domain protein Koi11, and KASH domain protein Klar12 are required for biased sister chromatid segregation (Figure 3, Table S1). It is well established that the LINC (linker of nucleoskeleton and cytoskeleton) complex, composed of SUN- and KASH-domain proteins, tethers the nucleus to cytoskeletal components (such as microtubules, which in turn connect to the centrosome) via the nuclear envelope13. Thus, we speculate that specific sister chromatids are tethered to the mother centrosome of the GSC that is consistently located at the hub-GSC junction (see Figure 4e).
Figure 3. cnn, koi, and klar are required for non-random sister chromatid segregation.
a, b) Representative images of Y chromosome CO-FISH in cnn mutant. Open arrowheads indicate the Cy3-(AATAC)6 probe; closed arrowheads indicate the Cy5-(GTATT)6 probe; asterisk indicates hub.
c, d) Representative images of X chromosome CO-FISH in koi mutant. Open arrowheads indicate the Cy3-X probe; closed arrowheads indicate the Cy5-X probe; asterisk indicates hub.
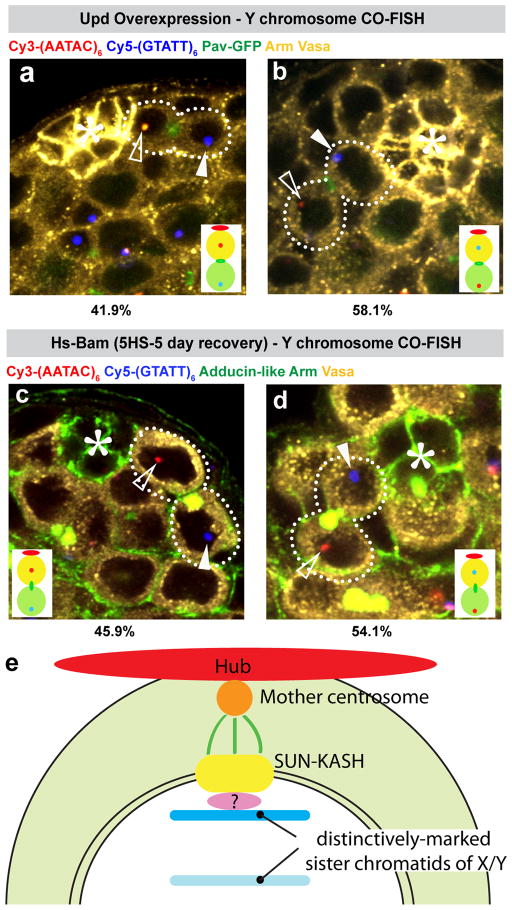
Figure 4. Non-random segregation of Y and X chromosomes is disrupted in upd-overexpressing testes and dedifferentiated stem cells.
a, b) Representative images of CO-FISH using the Y probe upon overexpression of Upd (nos-gal4>UAS-Upd). For this experiment we limited our analysis to GSCs juxtaposed to hub cells, because GSCs located away from the hub do not have a spatial reference point for assessment of the sister chromatid segregation pattern. N, number of GSC-GB pairs scored. (*) Hub. c, d) Representative images of CO-FISH using the Y probe in dedifferentiated GSCs. Differentiation was induced by heat shock treatment of hs-Bam flies followed by a 5-day recovery period29.
e) Model of non-random sister chromatid segregation (see text for details).
We further found that sister chromatid segregation of X and Y chromosomes was randomized in dnmt2 mutants (Table S2a and Figure S4). Although some studies indicated that Dnmt2 has DNA methyltransferase activity14,15, other studies showed that it functions as an RNA methyltransferase16 and that DNA methylation is barely detectable in the Drosophila genome17. Therefore, the mechanism by which Dnmt2 participates in non-random sister chromatid segregation remains elusive. However, our analysis, using various crossing schemes (crosses of homozygous mother/father with heterozygous father/mother), indicates that Dnmt2 confers heritable, DNA sequence-independent information on the X and Y chromosomes during gametogenesis in the parents, leading to non-random sister chromatid segregation of X and Y chromosomes in the GSCs of the progeny (Table S2b). For example, in GSCs from flies that are genetically heterozygous (dnmt2+/−), where the X chromosome is inherited from a mutant mother (dnmt2−/−) and the Y chromosome from a heterozygous father (dnmt2+/−), X chromosome segregation was randomized, whereas Y chromosome segregation remained non-random. These results suggest the striking possibility that the information that enables non-random sister chromatid segregation of X and Y chromosomes in adult stem cells is primed during gametogenesis in the parents, transmitted to the zygote on single X and Y chromosomes, and maintained through many cell divisions during embryogenesis and adult tissue homeostasis.
We found that sister chromatid segregation of X and Y chromosomes is randomized in GSC overproliferation induced by ectopic expression of Upd (Figure 4a, b, Table S3). Upd is a signalling ligand that is normally expressed exclusively in hub cells and activates the JAK-STAT pathway in GSCs and cyst stem cells to specify stem cell identity4. This finding suggests that non-random sister chromatid segregation is under the control of stem cell identity. However, it is unlikely that non-random sister chromatid segregation determines GSC identity, because the mutants defective in non-random segregation described above (cnn, koi, klar, dnmt2) do not exhibit GSC overproliferation or depletion.
We also found that sister chromatid segregation is randomized in dedifferentiated GSCs (Figure 4c–d, Table S3). Partially differentiated germ cells can revert back to GSC identity to replenish the stem cell pool18,19. Although these dedifferentiated GSCs are apparently functional because they can produce differentiating spermatogonia and reconstitute spermatogenesis18,20, they did not recover non-random sister chromatid segregation. This result may indicate that the information on X and Y chromosomes that allows non-random sister chromatid segregation is lost upon commitment to differentiation as a GB. Consistent with our earlier observation that dedifferentiation increases during aging20, we found that non-random sister chromatid segregation was compromised during aging (at day 30, 63:37 for the X chromosome [N=35] and 68:32 for the Y chromosome [N=28]).
This study provides the first evidence that adult stem cells can distinguish two sister chromatids and further, points to a model in which sister chromatids are distinctly recognized, leading to anchorage of particular strands to the mother centrosome through the SUN-KASH proteins (Figure 4e). Our data also indicate that non-random sister chromatid segregation does not necessarily mean that they are immortal21.
At present it is not clear why X and Y chromosomes segregate non-randomly. Considering the data presented in this study, we favour the possibility that certain epigenetic information is transmitted distinctively to GSCs and GBs. Indeed, X and Y chromosomes are subject to various forms of epigenetic regulation, such as dosage compensation22 and male-specific meiotic sex chromosome inactivation23. In addition, Stellate, a repetitive sequence that encodes a polypeptide known to reduce fertility, and Suppressor of Stellate [Su(Ste)], the piRNA that suppresses Stellate expression, are located on the X and Y chromosomes, respectively24,25. Intriguingly, we observed that Stellate is derepressed in mutants of cnn, dnmt2, koi, and klar (Figure S5), although determination of whether derepression of Stellate is due to a failure in non-random sister chromatid segregation awaits future investigation. Not surprisingly, we found that the mutants in which Stellate is derepressed show reduced fertility (Figure S6).
Recently, it was shown that old vs. new histones segregate asymmetrically during GSC divisions26. Our study demonstrates that GSCs do not segregate old (immortal) DNA strands. Thus, the relationship between biased sister chromatid segregation and histone segregation remains elusive. In summary, our study presents the first evidence of chromosome-specific non-random sister chromatid segregation in adult stem cells and provides mechanistic insights into how cells segregate sister chromatids non-randomly.
Methods
Fly husbandry
All fly stocks were raised on Bloomington Standard Media at 25°C unless otherwise noted. The following fly stocks were used: Ubi-Pavarotti-GFP, sh-adducin-Venus, cnnmfs3/CyO, cnnHK21/CyO, koiHRKO80.w, Df(2R)Exel6050/CyO, klar1, Df(3L)emc-E12, P(EP)Mt2G3429 (denoted dnmt2G3429 in the text), dnmt2Δ99, dnmt2149, Df(2L)ED775/CyO, hs-Bam, UAS-Upd/CyO, and nos-gal4. These stocks are described in FlyBase.
Combined immunofluorescence staining and CO-FISH
Newly eclosed adult flies (day 0) were fed food containing BrdU (950 μl 100% apple juice, 7 μg agar, and 50 μl 100 mg/ml BrdU solution in a 1:1 mixture of acetone and DMSO) for approximately 10 hours. After the feeding period, flies were transferred to regular fly food for approximately 10 hours. Because the average GSC cell cycle length is 12 hours, most GSCs undergo a single S phase followed by mitosis during our feeding procedure. GSCs that have undergone more or less than one S phase or mitosis were excluded from our analysis by limiting scoring to GSC-GB pairs that have complementary CO-FISH signals in the GSC and GB (i.e., red signal in one cell, blue signal in the other). All possible scenarios are explained in Figure S1. Samples were dissected in 1X PBS, fixed for 30–60 min with 4% formaldehyde in PBS, permeabilized for at least 1 hour in PBST (0.1% Triton X-100 in PBS) and incubated with primary antibodies overnight at 4°C. Samples were then washed with PBST (20 min, three times), incubated overnight at 4°C with Alexa Fluor-conjugated secondary antibodies (1:200; Molecular Probes), and washed again with PBST (20 min, three times). Samples were fixed for 10 min with 4% formaldehyde followed by three washes in PBST for 5 min each. Samples were then treated with RNase A (2 mg/ml in water) for 10 min at 37°C, washed with PBST for 5 min, and stained with 100 μl Hoechst 33258 (Sigma Aldrich) at 2 μg/ml for 15 min at room temperature. The samples were then rinsed with 2X SSC, transferred to a tray, and irradiated with ultraviolet light in a UV Stratalinker 1800 (calculated dose, 5400 J/m2). Nicked BrdU strands were digested with exonuclease III (New England Biolabs) at 3 U/μl in buffer supplied by the manufacturer (50 mM Tris-HCl, 5 mM MgCl2, and 5 mM dithiothreitol [DTT], pH 8.0) at 37°C for 10 min. Samples were rinsed once with PBST for 5 min and then fixed in 4% formaldehyde in PBS for 2 min and washed three times for 5 min each in PBST. To allow gradual transition into 50% formamide/2X SSC, samples were incubated sequentially for a minimum of 10 min each in 20% formamide/2X SSC, 40% formamide/2X SSC, and 50% formamide/2X SSC. The hybridization mixture consisted of 50% formamide, 2X SSC, 10% dextran sulfate, 0.5 μg/ml Cy3-labelled probe, and 0.5 μg/ml Cy-5-labeled probe. Fluorescence-labelled probes were obtained from Integrated DNA Technologies. The hybridization solution was added to the samples and hybridization was carried out at 37°C overnight. Using non-complementary pairs of probes for the X chromosome, we detected a similar bias in segregation pattern (Figure S2), excluding the possibility that annealing of complementary probes interferes with correct hybridization between the probes and the target sequences. Autosome probes were denatured in hybridization solution at 65°C for 3 min prior to hybridization. The samples were never heat-denatured. As a critical control, hub cells, which are predominantly quiescent and, thus, do not incorporate BrdU, did not show any CO-FISH signal (evident in all images).
Following hybridization, samples were washed once in 50% formamide/2X SSC, once in 25% formamide/2X SSC, and three times in 2X SSC. Samples were then mounted in VECTASHIELD (H-1200, Vector Laboratories) and images were recorded using a Leica TCS SP5 confocal microscope with a 63× oil immersion objective (NA=1.4) and processed using Adobe Photoshop software. The primary antibodies used were rabbit anti-Vasa (1:200; Santa Cruz Biotechnology), mouse anti-Adducin-like (1:20; developed by H. D. Lipshitz and obtained from the Developmental Studies Hybridoma Bank (DSHB), mouse anti-Armadillo (1:20; developed by Eric Wieschaus and obtained from DSHB), rabbit anti-Stellate (1:1000, a generous gift of Phillip Zamore30). The secondary antibodies used were Alexa Fluor 594- and 488-conjugated secondary antibodies (1:200; Molecular Probes).
CO-FISH with both X and Y probes
The X and Y probes were labelled such that GSCs retain the Cy3 signal in ~85% of cases. If segregation of X and Y chromosomes is correlated, the probability that a GSC inherits two Cy3 signals will be approximately 85%, and that of inheriting two Cy5 signals will be approximately 15%, whereas there will be few instances where a GSC inherits one Cy3 and one Cy5 signal. In contrast, if the X and Y chromosomes segregate asymmetrically independently of each other, the probability of GSCs inheriting two Cy3 signals will be 72% (85% × 85%), that of inheriting two Cy5 signals will be 2% (15% × 15%), and that of inheriting one Cy3 and one Cy5 signal will be 26% (85% × 15% × 2).
Supplementary Material
Acknowledgments
We thank Drs. Frank Lyko, Matthias Schaefer, Gunter Reuter, Phillip Zamore, Alexei Aravin, David Glover, Lynn Cooley, John Kim, Vladimir Gvozdev, Maria Pia Bozzetti, Bloomington Drosophila Stock Center and Vienna Drosophila RNAi Center for reagents and helpful information, and Yamashita laboratory members for discussions. This study was supported by the University of Michigan (Life Sciences Institute and Office of the Provost and Executive Vice President for Academic Affairs) (to YMY) and AHA (12PRE9630000) and NIH (1F31HD071727-01) (to SY). YMY is supported by the MacArthur Foundation.
Footnotes
Competing financial interest: The authors declare no competing financial interests.
Author contributions
SY conceived the project and developed the single chromosome CO-FISH protocol for Drosophila cells. SY and YMY designed and conducted experiments, interpreted the data, and wrote the manuscript.
References
- 1.Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. nature04956 [pii] [DOI] [PubMed] [Google Scholar]
- 2.Tajbakhsh S, Gonzalez C. Biased segregation of DNA and centrosomes: moving together or drifting apart? Nat Rev Mol Cell Biol. 2009;10:804–810. doi: 10.1038/nrm2784. nrm2784 [pii] [DOI] [PubMed] [Google Scholar]
- 3.Falconer E, et al. Identification of sister chromatids by DNA template strand sequences. Nature. 2010;463:93–97. doi: 10.1038/nature08644. nature08644 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fuller MT, Spradling AC. Male and female Drosophila germline stem cells: two versions of immortality. Science. 2007;316:402–404. doi: 10.1126/science.1140861. [DOI] [PubMed] [Google Scholar]
- 5.Yamashita YM, Jones DL, Fuller MT. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science. 2003;301:1547–1550. doi: 10.1126/science.1087795. 301/5639/1547 [pii] [DOI] [PubMed] [Google Scholar]
- 6.Yamashita YM, Mahowald AP, Perlin JR, Fuller MT. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 2007;315:518–521. doi: 10.1126/science.1134910. 315/5811/518 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dernburg AF. In: Drosophila Protocols. Sullivan W, Ashburner M, Hawley RS, editors. CSHL Press; 2000. [Google Scholar]
- 8.Fung JC, Marshall WF, Dernburg A, Agard DA, Sedat JW. Homologous chromosome pairing in Drosophila melanogaster proceeds through multiple independent initiations. J Cell Biol. 1998;141:5–20. doi: 10.1083/jcb.141.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makunin IV, et al. A novel simple satellite DNA is colocalized with the Stalker retrotransposon in Drosophila melanogaster heterochromatin. Mol Gen Genet. 1999;261:381–387. doi: 10.1007/s004380050979. [DOI] [PubMed] [Google Scholar]
- 10.Li K, Kaufman TC. The homeotic target gene centrosomin encodes an essential centrosomal component. Cell. 1996;85:585–596. doi: 10.1016/s0092-8674(00)81258-1. [DOI] [PubMed] [Google Scholar]
- 11.Kracklauer MP, Banks SM, Xie X, Wu Y, Fischer JA. Drosophila klaroid encodes a SUN domain protein required for Klarsicht localization to the nuclear envelope and nuclear migration in the eye. Fly (Austin) 2007;1:75–85. doi: 10.4161/fly.4254. [DOI] [PubMed] [Google Scholar]
- 12.Mosley-Bishop KL, Li Q, Patterson L, Fischer JA. Molecular analysis of the klarsicht gene and its role in nuclear migration within differentiating cells of the Drosophila eye. Curr Biol. 1999;9:1211–1220. doi: 10.1016/s0960-9822(99)80501-6. [DOI] [PubMed] [Google Scholar]
- 13.Razafsky D, Hodzic D. Bringing KASH under the SUN: the many faces of nucleo-cytoskeletal connections. J Cell Biol. 2009;186:461–472. doi: 10.1083/jcb.200906068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phalke S, et al. Retrotransposon silencing and telomere integrity in somatic cells of Drosophila depends on the cytosine-5 methyltransferase DNMT2. Nat Genet. 2009;41:696–702. doi: 10.1038/ng.360. [DOI] [PubMed] [Google Scholar]
- 15.Kunert N, Marhold J, Stanke J, Stach D, Lyko F. A Dnmt2-like protein mediates DNA methylation in Drosophila. Development (Cambridge, England) 2003;130:5083–5090. doi: 10.1242/dev.00716. dev.00716 [pii] [DOI] [PubMed] [Google Scholar]
- 16.Schaefer M, et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590–1595. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 18.Brawley C, Matunis E. Regeneration of male germline stem cells by spermatogonial dedifferentiation in vivo. Science. 2004;304:1331–1334. doi: 10.1126/science.1097676. [DOI] [PubMed] [Google Scholar]
- 19.Kai T, Spradling A. Differentiating germ cells can revert into functional stem cells in Drosophila melanogaster ovaries. Nature. 2004;428:564–569. doi: 10.1038/nature02436. [DOI] [PubMed] [Google Scholar]
- 20.Cheng J, et al. Centrosome misorientation reduces stem cell division during ageing. Nature. 2008;456:599–604. doi: 10.1038/nature07386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yadlapalli S, Cheng J, Yamashita YM. Drosophila male germline stem cells do not asymmetrically segregate chromosome strands. J Cell Sci. 2011;124:933–939. doi: 10.1242/jcs.079798. jcs.079798 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conrad T, Akhtar A. Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription. Nat Rev Genet. 2011;13:123–134. doi: 10.1038/nrg3124. [DOI] [PubMed] [Google Scholar]
- 23.Hense W, Baines JF, Parsch J. X chromosome inactivation during Drosophila spermatogenesis. PLoS Biol. 2007;5:e273. doi: 10.1371/journal.pbio.0050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aravin AA, et al. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- 25.Tulin AV, Kogan GL, Filipp D, Balakireva MD, Gvozdev VA. Heterochromatic Stellate gene cluster in Drosophila melanogaster: structure and molecular evolution. Genetics. 1997;146:253–262. doi: 10.1093/genetics/146.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tran V, Lim C, Xie J, Chen X. Asymmetric division of Drosophila male germline stem cell shows asymmetric histone distribution. Science. 2012;338:679–682. doi: 10.1126/science.1226028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minestrini G, Mathe E, Glover DM. Domains of the Pavarotti kinesin-like protein that direct its subcellular distribution: effects of mislocalisation on the tubulin and actin cytoskeleton during Drosophila oogenesis. J Cell Sci. 2002;115:725–736. doi: 10.1242/jcs.115.4.725. [DOI] [PubMed] [Google Scholar]
- 28.Petrella LN, Smith-Leiker T, Cooley L. The Ovhts polyprotein is cleaved to produce fusome and ring canal proteins required for Drosophila oogenesis. Development (Cambridge, England) 2007;134:703–712. doi: 10.1242/dev.02766. [DOI] [PubMed] [Google Scholar]
- 29.Sheng XR, Brawley CM, Matunis EL. Dedifferentiating spermatogonia outcompete somatic stem cells for niche occupancy in the Drosophila testis. Cell Stem Cell. 2009;5:191–203. doi: 10.1016/j.stem.2009.05.024. S1934-5909(09)00282-3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forstemann K, et al. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.