Abstract
Cancer cells engage in a metabolic program to enhance biosynthesis and support cell proliferation. The regulatory properties of pyruvate kinase M2 (PKM2) influence altered glucose metabolism in cancer. PKM2 interaction with phosphotyrosine-containing proteins inhibits enzyme activity and increases availability of glycolytic metabolites to support cell proliferation. This suggests that high pyruvate kinase activity may suppress tumor growth. We show that expression of PKM1, the pyruvate kinase isoform with high constitutive activity, or exposure to published small molecule PKM2 activators inhibit growth of xenograft tumors. Structural studies reveal that small molecule activators bind PKM2 at the subunit interaction interface, a site distinct from that of the endogenous activator fructose-1,6-bisphosphate (FBP). However, unlike FBP, binding of activators to PKM2 promotes a constitutively active enzyme state that is resistant to inhibition by tyrosine-phosphorylated proteins. These data support the notion that small molecule activation of PKM2 can interfere with anabolic metabolism.
Cancer cells differ from many normal cells in the way they utilize extracellular nutrients, providing a strategy to interfere with tumor growth1,2. The increased cell proliferation that characterizes tumor growth imposes an enhanced need for biological building blocks to support production of new cells3. To provide for this increased biosynthetic demand, cancer cells exhibit higher uptake of nutrients such as glucose. In addition, the metabolic pathways of cancer cells are altered to allow production of macromolecules and withstand oxidative stress associated with tumorigenesis1,3-6.
Enhanced glucose uptake is a hallmark of several cancers and is exploited in the clinic as a diagnostic tool through PET imaging of the glucose analogue 18F-deoxyglucose (18FDG-PET)7. Moreover, in contrast to most normal tissues where much of the glucose is oxidized through the TCA cycle in mitochondria, cancer cells preferentially convert glucose to lactate3. The fate of glucose inside cells is influenced by the enzymatic properties of the specific glycolytic gene products expressed. Expression of the M2 isoform of pyruvate kinase (PKM2) can contribute to the characteristic glucose metabolism of tumors and replacement of PKM2 with its splice variant PKM1 cannot efficiently support biosynthesis and tumor growth8. Thus, pyruvate kinase regulates a step in glucose metabolism that can be critical for controlling cell proliferation.
Pyruvate kinase catalyzes the last step of glycolysis, transferring the phosphate from phosphoenolpyruvate (PEP) to adenosine diphosphate (ADP) to yield adenosine triphosphate (ATP) and pyruvate. In mammals, two genes encode a total of four pyruvate kinase isoforms. The Pkrl gene encodes the PKL and PKR isoforms, which are expressed in the liver and red blood cells respectively. Most tissues express either the PKM1 or PKM2 isoform encoded by the Pkm gene. PKM1 is found in many normal differentiated tissues whereas PKM2 is expressed in most proliferating cells including all cancer cell lines and tumors tested9. PKM1 and PKM2 are derived from alternative splicing of a Pkm gene transcript by mutual exclusion of a single conserved exon encoding 56 amino acids10-12. Despite very similar primary sequences, PKM1 and PKM2 have different catalytic and regulatory properties. PKM1 exhibits high constitutive enzymatic activity13. In contrast, PKM2 is less active, but can be allosterically activated by the upstream glycolytic metabolite fructose-1,6-bisphosphate (FBP)14. It has been hypothesized that FBP binding induces conformational changes that promote the association of the protein into homotetramers that comprise the most active form of the enzyme15,16.
Unlike other pyruvate kinase isoforms, PKM2 can interact with proteins harboring phosphorylated tyrosine residues leading to release of FBP which, in turn, reduces the activity of the enzyme17. Low PKM2 activity, in conjunction with increased glucose uptake, facilitates the flux of glucose carbons into anabolic pathways derived from glycolysis3,9,17,18. Also, PKM2, but not PKM1, can be inhibited by direct oxidation of cysteine 358 as an adaptive response to increased intracellular reactive oxygen species (ROS)19. Additionally, PKM2 expression in cancer cells is associated with enhanced phosphorylation of H11 on phosphoglycerate mutase 1 (PGAM1) by PEP20. This pathway provides an alternative route for pyruvate production while bypassing the generation of ATP via the pyruvate kinase step and thereby allows glycolysis to proceed at high rates21. Replacement of PKM2 with the constitutively active isoform PKM1 results in reduced lactate production, enhanced oxygen consumption, and a decrease in PGAM1 phosphorylation8,20. Furthermore, there appears to be selection for PKM2 expression for growth in vivo. However, it is also possible that PKM2 expression reflects selection against high pyruvate kinase activity and therefore against expression of PKM1, raising the possibility that activation of PKM2 may impede cancer cell proliferation by interfering with regulatory mechanisms critical for proliferative metabolism.
Recently, we identified small molecules that selectively activate PKM2 over other pyruvate kinase isoforms in vitro22,23. Here we show that synthetic PKM2 activators can increase PKM2 activity in cells to levels that are comparable to PKM1 expression. PKM2 activators bind to a pocket at the PKM2 subunit interface and thereby enhance association of PKM2 subunits into stable tetramers. Importantly, this mechanism of tetramer stabilization is refractory to inhibition by tyrosine-phosphorylated proteins and influences cell metabolism. Among the two classes of PKM2 activators described here, a member of the thieno[3,2-b]pyrrole[3,2-d]pyridazinones class, TEPP-46, has pharmacokinetic properties amenable to experiments in mice. Expression of PKM1 or continuous dosing of mice with TEPP-46, decreases development of human cancer cell xenografts, suggesting that increased pyruvate kinase activity can impair tumorigenesis.
RESULTS
Increased pyruvate kinase activity impairs tumor growth
We have previously demonstrated that replacement of PKM2 with PKM1 impairs the ability of H1299 human non-small cell lung cancer cells to form xenograft tumors in mice8. This observation may reflect selection for PKM2 expression in tumors. However, because the ability to decrease PKM2 activity correlates with increased cell proliferation17, it is also possible that high pyruvate kinase activity associated with the expression of the constitutively active PKM1 isoform can suppress tumor growth. To address whether PKM1 expression alone affects tumor formation in vivo, we engineered H1299 cells to stably express Flag-tagged PKM1 in the presence of endogenous PKM2 (henceforth referred to as H1299-PKM1 cells). Expression of Flag-PKM1 did not affect endogenous PKM2 levels (Supplementary Results, Supplementary Fig. 1a), but resulted in a 35±17% increase in total cellular pyruvate kinase activity (Fig. 1a). Both PKM1 and PKM2 can associate into tetramers24,25. To determine if PKM1 can associate with endogenous PKM2 we used an anti-Flag antibody to immunoprecipitate Flag-PKM1. SDS-PAGE of the immunoprecipitated protein followed by silver staining revealed the presence of stoichiometric amounts of Flag-PKM1 and endogenous co-precipitating PKM2 (Supplementary Fig. 1b). The identity of PKM2 was confirmed by mass spectrometry (Supplementary Fig. 1c). These data show that PKM1 can form heterocomplexes with endogenous PKM2, and that immunoprecipitation of exogenously expressed Flag-tagged pyruvate kinase can be used to assess formation of multimeric pyruvate kinase complexes in cells. Furthermore, these data suggest that expression of PKM1 in PKM2-expressing cells suffices to increase total pyruvate kinase activity.
Figure 1. PKM1 expression in cancer cells impairs xenograft tumor growth.

(a) H1299 human lung cancer cells were infected with a retrovirus to stably express Flag-PKM1 (referred to as H1299-PKM1 cells in the text) or empty vector (Parental) and after selection, cells were lysed and pyruvate kinase activity in the lysates was assayed. (b) Tumor formation over time of parental or H1299-PKM1 cells generated as in (a) and injected subcutaneously at equal numbers in nu/nu mice [p value calculated by logrank (Mantel-Cox) test]. (c) Final tumor weights from the experiment in (b). Mean tumor weights ± s.e.m. are shown and p value was calculated by unpaired Student’s t-test. (d) Expression of Flag-PKM1 and endogenous PKM2 in the cells used in (b) and (c) was determined by western blot with isoform-specific antibodies. Uncropped blots are shown in Supplementary Fig. 10. (e) Pyruvate kinase activity assays in lysates of the tumors shown in (d) (N=3, 1-way ANOVA and Tukey’s post-test).
To determine whether PKM1 expression with enhanced pyruvate kinase activity interferes with tumor growth, we compared the ability of H1299-PKM1 versus parental H1299 cells to form tumors in immunocompromised (nu/nu) mice. Tumors emerged in all sites where parental cells were injected after a median of 31.5 days (Fig. 1b). However, only 4 out of 10 sites injected with H1299-PKM1 cells gave rise to tumors. Notably, tumors derived from H1299-PKM1 cells were significantly smaller (Fig. 1c), occurred later and had reduced expression of Flag-PKM1 relative to the injected H1299-PKM1 cells (Fig. 1d and Supplementary Fig. 2). Furthermore, pyruvate kinase activity in tumor lysates was consistently lower than that seen in parental H1299 cells (Fig. 1e). As the same numbers of parental or PKM1-expressing cells were injected to initiate tumors, the low pyruvate kinase activity in all tumors suggests that the small tumors derived from H1299-PKM1 cells arose from a subset of cells that lost PKM1 expression. These data support the notion that decreased pyruvate kinase activity is associated with tumor growth, and suggest that high pyruvate kinase activity can suppress formation of cancer cell xenograft tumors in mice.
Small molecules can specifically activate PKM2 in cells
A recent screen identified two structurally distinct classes of small-molecule PKM2 activators22,33. A representative compound from each class was selected for further studies; the thieno[3,2-b]pyrrole[3,2-d]pyridazinone NCGC00186528 (TEPP-46; ML265, PubChem CID: 44246499) and the substituted N,N’-diarylsulfonamide NCGC00185916 (DASA-58; ML203, PubChem CID: 44543605) are both potent activators of recombinant PKM2 (TEPP-46: AC90=470 nM, AC50=92 nM; DASA-58: AC90=680 nM, AC50=38 nM; Fig. 2a) and are soluble in aqueous solution22,33. Furthermore, TEPP-46 and DASA-58 are selective for PKM2 as they do not activate recombinant PKM1 in vitro (Fig. 2b). To investigate whether these compounds are able to activate PKM2 selectively in cells, we engineered A549 cells to express Flag-PKM1 or Flag-PKM2 with concomitant knockdown of endogenous PKM2 (referred to as A549-PKM1/kd and A549-PKM2/kd, respectively) (Fig. 2c). We treated these cells with 40 μM DASA-58 and assayed pyruvate kinase activity in the corresponding cell lysates. Consistent with our results in H1299 cells, lysates from DMSO-treated A549-PKM1/kd cells had 233±27% more pyruvate kinase activity than A549-PKM2/kd cells. We observed no increase in pyruvate kinase activity following treatment of A549-PKM1/kd cells with DASA-58; however DASA-58 treatment resulted in a 248 ±21% increase of pyruvate kinase activity in A549-PKM2/kd cell lysates. These data suggest that DASA-58 can selectively activate PKM2 in cells.
Figure 2. TEPP-46 and DASA-58 isoform specificity in vitro and in cells.
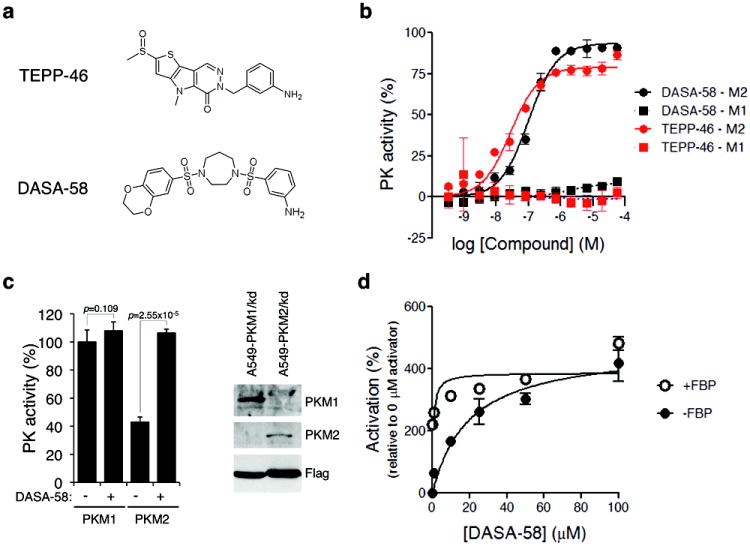
(a) Structures of the PKM2 activators TEPP-46 and DASA-58 used in this study (b) Purified recombinant human PKM1 or PKM2 expressed in bacteria were subjected to pyruvate kinase activity assays in the presence of increasing concentrations of TEPP-46 or DASA-58. (c) A549 cells were engineered to stably express Flag-PKM1 or Flag-PKM2 in the absence of endogenous PKM2 which was knocked down by shRNA. As the PKM1 and PKM2 cDNAs correspond to the mouse orthologues, their expression was resistant to knockdown8. Expression of Flag-PKM1 and Flag-PKM2 was confirmed by western blot with isoform-specific antibodies (right panel). These cell lines were then treated with 40 μM DASA-58 for 3 hours and the respective lysates were assayed for pyruvate kinase activity (N=3, Student’s t-test). Similar results were observed in H1299 cells (not shown). Uncropped blots are shown in Supplementary Fig. 10. (d) A549 cells were treated with the indicated doses of DASA-58 for 3 hours, lysed and assayed for pyruvate kinase activity in the presence or absence of 200 μM FBP.
In a manner analogous to FBP, both TEPP-46 and DASA-58 decrease the Km of PKM2 for PEP with no effect on the Km for ADP22,33, suggesting that TEPP-46 and DASA-58 activate PKM2 by a mechanism similar to that of the endogenous activator FBP. To determine whether FBP could further activate PKM2 in activator-treated cells, we incubated A549 cells with increasing concentrations (0-100 μM) of DASA-58 and assayed PKM2 activity in the corresponding lysates in the presence or absence of FBP. In the absence of FBP, DASA-58 activated PKM2 in a dose-dependent manner with an effective cellular half-activation concentration (EC50) of 19.6 μM (Fig. 2d). However, in the presence of high physiological levels of FBP (200 μM), we observed no significant additional increase in activity as a result of activator treatment. These data are consistent with the in vitro kinetic analysis suggesting that DASA-58 enhances PKM2 activity by a mechanism similar to FBP.
PKM2 activators stabilize subunit interactions
The most active form of PKM2 is as a tetramer, and subunit association is thought to be promoted by FBP. To test this hypothesis, we separated bacterially expressed recombinant PKM2 into monomers and tetramers by size exclusion chromatography (Supplementary Fig. 3a) and assayed the ability of FBP to increase PKM2 activity. PKM2 tetramers showed greater than 50-fold higher activity compared to PKM2 monomers (Supplementary Fig. 3b). Addition of FBP had minimal effects on PKM2 tetramer activity (consistent with bacterial FBP being trapped on the stable tetramers17), but FBP increased the activity of PKM2 monomers by approximately 70% (Supplementary Fig. 3c). The relatively modest activation of the monomers likely reflects the slow kinetics of assembling tetramers from monomers under dilute conditions. To determine if FBP activation of monomers is accompanied by changes in PKM2 subunit composition, we performed sucrose gradient ultracentrifugation on purified recombinant PKM2. Under these conditions, the majority of PKM2 is found to dissociate into monomers (Fig. 3a). Exposure of PKM2 to FBP throughout the experiment results in a shift of the protein into a tetrameric configuration that is comparable to that seen with the constitutively active PKM1 isoform (Supplementary Fig. 3d). To investigate if small-molecule activators also promote pyruvate kinase subunit association into tetramers, we incubated purified PKM2 with TEPP-46 or DASA-58. We observed only a partial shift of PKM2 into tetramers with both activators alone (Supplementary Fig. 3d). As only a fraction of bacterially expressed PKM2 is bound to FBP (Supplementary Fig. 3a-c) we reasoned that the activators may only stabilize FBP-bound PKM2. Consistent with this hypothesis, transient incubation of PKM2 with FBP prior to addition of TEPP-46 resulted in the activator fully stabilizing the PKM2 tetramer. Overall, these data suggest that unlike PKM2, PKM1 is a stable tetrameric enzyme, and both FBP and small-molecule activators increase PKM2 activity by promoting the tetrameric state.
Figure 3. Activators promote PKM2 tetramer formation and prevent inhibition by pTyr signaling.

(a) Sucrose gradient ultracentrifugation profiles of purified recombinant PKM2 and effects of FBP and TEPP-46 on PKM2 subunit stoichiometry. Recombinant PKM2 was transiently exposed to FBP prior to addition of TEPP-46. After centrifugation, fractions were collected, analyzed by SDS-PAGE and stained with Coomassie Blue. Relative protein amounts were calculated by band densitometry on a LiCOR Odyssey infrared imaging system. (b) A549 cells were treated with 100 μM pervanadate for 10 min. in the presence or absence of TEPP-46, lysed hypotonically, and were analyzed by size exclusion chromatography. Chromatographic fractions were then subjected to western blotting with a pyruvate kinase antibody to assess the stoichiometry of PKM2 subunit association under these conditions. Uncropped blots are shown in Supplementary Fig. 10. (c) Pyruvate kinase activity assays in A549 cells treated with pervanadate as in (b) in the presence of DMSO, 1 μM TEPP-46 or 1 μM DASA-58 (N=3, p=0.0044 by 2-way ANOVA).
To investigate whether PKM2 activators promote pyruvate kinase subunit association in cells, we generated A549 cells stably expressing Flag-PKM1 or Flag-PKM2 and treated them with DMSO or DASA-58. Following lysis and immunoprecipitation with Flag antibodies, we determined the relative amount of endogenous PKM2 that co-precipitated under the various conditions by western blot using an antibody that recognizes an epitope common to both pyruvate kinase isoforms. Flag-PKM1 immunoprecipitated equivalent amounts of endogenous PKM2 irrespective of activator treatment (Supplementary Fig. 3e). In contrast, DASA-58 treatment resulted in increased levels of endogenous PKM2 immunoprecipating with Flag-PKM2 when compared with DMSO-treated cells or cells treated with an inactive analog of DASA-5823. Similar results were obtained with TEPP-46 (Supplementary Fig. 3f). These data indicate that PKM2 activators can promote stable association of PKM2 subunits in cells.
Phosphotyrosine interaction with PKM2 downstream of growth factor signaling is critical for both cell proliferation and metabolic changes that promote anabolism17. Binding to phosphotyrosine decreases PKM2 activity by catalyzing release of FBP from the enzyme17,26. Pervanadate inhibits tyrosine phosphatases to acutely increase levels of tyrosine-phosphorylated proteins, and pervanadate treatment of cells results in inhibition of PKM2 but not inhibition of PKM1 activity17,20. To determine whether PKM2 activity inhibition caused by increased tyrosine phosphorylated proteins results in destabilization of PKM2 tetramers in cells, we treated cells with pervanadate and determined the stoichiometry of PKM2 subunit composition by size exclusion chromatography. In logarithmically growing A549 cells, approximately half of the PKM2 elutes as a tetramer while the other half dissociates to monomers during size exclusion chromatography (Fig. 3b). Under these conditions we do not detect a significant population of dimeric PKM2. Pervanadate treatment caused disappearance of PKM2 tetramers and the entire PKM2 population was detected as monomers in this assay. We then tested whether PKM2 activators influence regulation of PKM2 tetramerization by tyrosine phosphorylated proteins. In logarithmically growing cells treated with TEPP-46 all PKM2 was found as a tetramer (Fig. 3b). Moreover, PKM2 tetramers were preserved even after treatment of cells with pervanadate (Fig. 3b). Similar results were obtained with H1299 cells (Supplementary Fig. 3g).
To test whether PKM2 activators also affect PKM2 activity when levels of tyrosine-phosphorylated proteins are increased, we assayed PKM2 activity in lysates of cells treated with DMSO, TEPP-46 or DASA-58 followed by pervanadate treatment. Pre-treatment of cells with TEPP-46 or DASA-58 prevented pervanadate-induced inhibition of PKM2 activity (Fig. 3c). It is plausible that activator binding renders PKM2 resistant to an inhibitory modification induced by pervanadate treatment. However, a phosphotyrosine-containing peptide corresponding to the optimal PKM2 interaction motif17 can promote dissociation of PKM2 tetramers (Supplementary Fig. 3h), but does not inhibit the ability of TEPP-46 to activate recombinant PKM2 (Supplementary Fig. 3i). These results indicate that activators render PKM2 tetramers resistant to dissociation induced by phosphotyrosine signaling.
Overall, these data argue that PKM2 activators enhance PKM2 activity by promoting the stable (active) tetrameric form of PKM2. However, similar to PKM1 but unlike the endogenous activator FBP, small-molecule PKM2 activators promote the active form of the enzyme even in the presence of increased phosphotyrosine levels that would otherwise lower PKM2 activity.
Structural analysis of PKM2 activator mode of binding
Based on these biochemical studies, it is possible that these agents activate PKM2 by binding at the same site as FBP but fail to be released following interaction with phosphotyrosine. Alternatively, PKM2 activators may stabilize the tetramer in another way. To explore these possibilities, purified recombinant PKM2 was crystallized in the presence of TEPP-46 or DASA-58. Our refined model shows that one tetrameric PKM2 contains two activators and four FBP molecules (Fig. 4a, Supplementary Table 1 and Supplementary Fig. 4a). The four FBP molecules co-purified from the E. coli cells where PKM2 was produced and they were found to occupy all four of the FBP binding pockets of the PKM2 tetramer. In comparison, one activator was found in the interface (named A-A’) between the A domains of each dimer, approximately 35Å away from the FBP binding pocket (Fig. 4a and Supplementary Fig. 4a). The bound activator was completely buried within the A-A’ interface. The activator binding pocket was lined with equivalent sets of residues provided by each of the PKM2 molecules forming the A-A’ interface, where the activator was accommodated through polar and van der Waals interactions with pocket-lining residues (Fig. 4b and Supplementary Fig. 4b). Particularly in the case of TEPP-46, crystallographic evidence suggests that the binding orientation of the activator alternates based on a pseudo-twofold axis that is co-linear with both the nitrogen-methyl carbon bond of the N-methyl pyrrole moiety of TEPP-46 and the pseudo-twofold axis of the A-A’ interface. The extra space in the pocket was filled with solvent molecules or ions, which also mediate hydrogen bonds between the activator and the pocket-lining residues (Fig. 4b). These data show that TEPP-46 and DASA-58 bind PKM2 through a binding pocket distinct from that of FBP and that they stabilize a tetrameric conformational state.
Figure 4. Structural analysis of PKM2 activator mode of action.

(a) Interaction between tetrameric PKM2 and TEPP-46. The four PKM2 monomers are represented in cartoon mode with different colors, respectively. The bound FBP and the activator molecules are colored black and red, respectively, and shown as space-filling models. The interfaces between two monomers are indicated by dotted lines. (b) Interaction between TEPP-46 and surrounding residues. The bound activator is colored yellow and represented by ball and stick model. The residues involved in the interaction from two monomers are labeled and colored green and cyan, respectively. Hydrogen bonds are indicated by blue dotted lines with distance (Å). (c) DASA-58 stabilizes the interaction of Flag-PKM2(K305Q) with endogenous PKM2. Flag-PKM2(K305Q) stably expressed in A549 cells was immunoprecipitated from corresponding lysates and the levels of co-precipitating endogenous PKM2 were assessed by western blotting with a PKM2 antibody. Uncropped blots are shown in Supplementary Fig. 10.
FBP is thought to contribute to tetramer formation by stabilizing the C-C’ interface. Given the distinct location of the activator binding pocket, we investigated whether activator binding results in stabilization of subunit interaction along the A-A’ interface. A recent study showed that acetylation of K305 results in decreased PKM2 subunit association as another way to inactivate the enzyme27. The ε-amino group of K305 interacts via a salt bridge with E384 in the neighboring subunit along the A-A’ interface (Supplementary Fig. 4c). Acetylation may interfere with this interaction resulting in destabilization of the PKM2 tetramer. We mutated K305 to Q to mimic acetylation. Flag-PKM2(K305Q) failed to co-precipitate endogenous PKM2 (Fig. 4c). Given that the activator binds between two PKM2 subunits at the A-A’ interface where K305 and E384 reside, we asked if PKM2 activator can rescue the interaction between Flag-PKM2(K305Q) and endogenous PKM2. Indeed, treatment of cells with DASA-58 restored the ability of Flag-PKM2(K305Q) to co-precipitate endogenous PKM2 (Fig. 4c). These data further support a model where PKM2 activators function by binding at the A-A’ interface and stabilize the PKM2 tetramer, and suggest that the activators could circumvent in vivo mechanisms for inhibition of PKM2 activity.
PKM2 activators alter metabolism in cultured cells
Overall, our results suggest that PKM2 activators will mimic the regulatory properties of constitutively active PKM1, thereby promoting high PKM2 activity regardless of the known mechanisms cells use to decrease pyruvate kinase activity. Therefore, we next examined the effects of activator treatment on cellular proliferation. Similar to results observed when PKM2 is replaced with PKM18, under standard tissue culture conditions PKM2 activators had no significant effects on cell proliferation when tested across several lines (Fig. 5a and Supplementary Fig. 5). In contrast, when we assayed proliferation under hypoxic conditions (1% O2), PKM2 activator treatment resulted in a decreased rate of cell proliferation compared to DMSO-treated cells (Fig. 5a). Similarly, expression of PKM1 in the presence of endogenous PKM2 had no effect on cell proliferation in standard tissue culture conditions, but inhibited cell proliferation under hypoxia to a similar degree as treatment with activators. These observations are consistent with previous data showing that replacement of PKM2 with PKM1 also impairs cell proliferation under low oxygen8.
Figure 5. Metabolic effects of cell treatment with PKM2 activators.
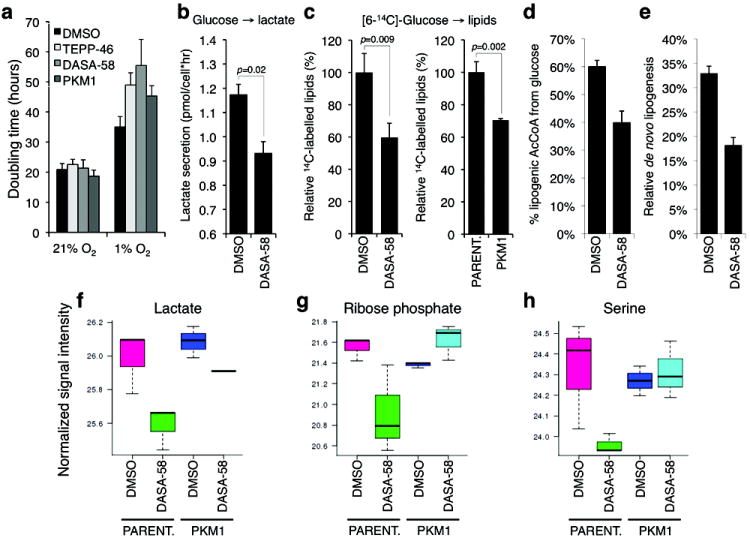
(a) Effects of TEPP-46, DASA-58 (both used at 30 μM), or PKM1 expression on the doubling time of H1299 cells under normoxia (21% O2) or hypoxia (1% O2). (b) Effects of DASA-58 on lactate production from glucose. Logarithmically growing H1299 cells were washed with Krebs buffer and incubated in Krebs buffer containing glucose in the presence of 50 μM DASA-58 (N=3, Student’s t-test). Produced lactate in the incubation medium was assayed after 20 min. as described in Methods. (c) Logarithmically growing H1299 cells (left), or parental H1299 and H1299-PKM1 cells (right) were incubated for 2 hours with 4 μCi/ml [6-14C]-glucose in the presence of DMSO or 30 μM DASA-58, cellular lipids were extracted and lipid-incorporated 14C was quantified by scintillation counting. (d) Contribution of [U-13C6]glucose to the lipogenic AcCoA pool, and fractional new synthesis of palmitate (e) were determined by isotopomer spectral analysis in A549 cells treated with DMSO or 100 μM DASA-58. Error bars indicate 95% confidence intervals. (f-h) Intracellular concentrations of lactate, ribose phosphate and serine in parental H1299 or H1299-PKM1 cells treated with DMSO or 25 μM of TEPP-46 for 36 hours were determined by targeted LC-MS/MS.
To test our hypothesis that PKM2 activators mimic a metabolic state found in PKM1-expressing cells, we interrogated the effects of activator treatment on cell metabolism. Replacement of PKM2 with PKM1 in cultured cells results in reduced lactate production and enhanced oxygen consumption8. Acute treatment of H1299 cells with DASA-58 also resulted in decreased lactate production (Fig. 5b). Unlike cells where PKM2 is replaced with PKM1, expression of PKM1 in the presence of endogenous PKM2 or activator treatment had no significant effect on oxygen consumption (Supplementary Fig. 6a). Furthermore, DASA-58 treatment did not affect glucose or glutamine consumption (Supplementary Fig. 6b-c), indicating that changes in uptake of major nutrients are unlikely to underlie the observed metabolic phenotypes. Inhibition of PKM2 activity mediated by phosphotyrosine signaling results in more efficient incorporation of glucose carbons into lipids17. To determine if PKM2 activator treatment inhibits glucose carbon incorporation into lipids, cells were incubated with [6-14C]-glucose in the presence of DMSO or DASA-58, cellular lipids were extracted, and 14C-labelled lipids were quantified by scintillation counting. DASA-58 treatment resulted in a significant decrease in glucose-derived carbon incorporation into lipids (Fig. 5c). Similarly, we observed a decrease in glucose carbon incorporation into lipids in H1299-PKM1 cells (Fig. 5c). We also used gas chromatography-mass spectrometry (GC-MS) to analyze metabolite extracts of cells incubated with 13C-labeled glucose. DASA-58 treatment resulted in diminished incorporation of glucose carbons into acetyl-CoA used for de novo palmitate synthesis (Fig. 5d and Supplementary Fig. 6d) and a decrease in overall de novo lipogenesis (Fig. 5e). TEPP-46 also induced a decrease in the intracellular levels of acetyl-coA, lactate, ribose phosphate and serine (Fig. 5f-h and Supplementary Fig. 6e). Ribose phosphate is a key intermediate for the biosynthesis of nucleotides via the pentose phosphate pathway, and serine, in addition to being incorporated directly into proteins can serve as a precursor for lipid head groups and glycine as well as provide carbon to the folate pool. Notably, these changes, along with overall changes in other intracellular metabolite concentrations (Supplementary Fig. 6f), were evident only in parental cells and not PKM1-expressing cells, indicating that the effects of the activator on cellular metabolism are specific to PKM2. No significant differences in the concentrations of lactate, ribose phosphate or serine were observed between PKM1-expressing and parental cells, most likely reflecting that, unlike transient activation of PKM2 by small molecules, selection for PKM1 expression in cells may lead to adaptive changes in metabolite levels to compensate for the effects of chronic pyruvate kinase activity elevation. Regardless, these data indicate that small-molecule activation of PKM2 can alter glucose metabolism and can decrease the intracellular concentrations of intermediates required for biosynthesis.
Low PKM2 activity is also associated with increased phosphorylation of PGAM1 on the catalytic histidine residue (His11)20. To determine if activator treatment decreases PGAM1 phosphorylation, we treated cells with DMSO or DASA-58, and analyzed the corresponding lysates by 2-D SDS-PAGE/isoelectric focusing to resolve PGAM1 species in cells20. Similar to PKM1-expressing cells20, we observed a decrease in PGAM1 phosphorylation as a result of activator treatment (Supplementary Fig. 7). Together, these data suggest that PKM2 activators induce a metabolic state in cells comparable to, but distinct from, that seen with PKM1 expression.
PKM2 activator inhibits xenograft tumor growth
PKM1 expression impairs the ability of cancer cells to grow in vivo as xenografts. To determine whether PKM2 activators also impede xenograft tumor growth, we determined which compound was suitable for experiments in mice. Based on in vitro absorption, distribution, metabolism and excretion (ADME) profiling studies, we predicted that TEPP-46 would have superior in vivo drug exposure compared to other analogs. To determine appropriate repeated drug doses for the mouse experiments, we performed single-dose pharmacokinetic and acute pharmacodynamic (PK-PD) studies with TEPP-46. Supplementary Fig. 8 shows mouse plasma drug concentration over time after a single intravenous, intraperitoneal or oral dose. TEPP-46 exhibits good oral bioavailability with relatively low clearance, long half-life, and good volume of distribution - parameters that predict for drug exposure in tumor tissues (Supplementary Table 2). Acute oral-dose of TEPP-46 at 150 mg/kg readily achieved maximal PKM2 activation measured in A549 xenograft tumors (Supplementary Fig. 9a). To ensure that drug treatment could be carried out safely for a multi-day dosing regimen, we also performed a 5-day repeat-dose tolerability study in mice and showed that 50 mg/kg twice daily was well-tolerated with no sign of weight loss (Supplementary Fig. 9b).
Because 50 mg/kg twice daily oral dosing of TEPP-46 was well tolerated and led to reasonable plasma levels in mice, we tested whether this dose of TEPP-46 could promote PKM2 tetramerization in xenograft tumors. We treated mice bearing H1299 xenograft tumors with vehicle or TEPP-46 and analyzed tumor lysates by size exclusion chromatography. In xenografts from vehicle-treated mice, very little PKM2 was found as tetramers (Fig. 6a). In contrast, tumors exposed to TEPP-46 harbored exclusively tetrameric PKM2. Metabolomic analysis revealed that, similar to the effects observed in cultured cells following treatment with TEPP-46, tumors derived from mice treated with TEPP-46 had lower concentrations of lactate, ribose phosphate and serine (Fig. 6b). We then tested whether these changes in PKM2 activity and tumor metabolism impact the ability of H1299 cells to form xenografts. H1299 cells were injected into immunocompromised (nu/nu) mice and the animals were randomly divided into two cohorts, one treated with vehicle and the other treated with 50 mg/kg of TEPP-46, dosed twice daily for the duration of the experiment. No apparent toxicity was observed in any mice, despite over 7 weeks of continuous drug exposure based on blood counts and serum chemistries (Supplementary Table 3) as well as histological examination of blood, bone marrow, liver, kidney, heart and the gastrointestinal tract. Tumors in activator-treated mice emerged with a delayed latency compared to tumors in vehicle-treated mice (Fig. 6c). In addition, the tumors from activator-treated mice were smaller than those arising in vehicle-treated animals (Fig. 6d). TEPP-46 was detectable in tumors from activator-treated mice suggesting that cells in the tumor were exposed to the drug.
Figure 6. PKM2 activators impair xenograft growth.

(a) Mice bearing H1299 xenograft tumors received bolus doses of TEPP-46 at 50 mg/kg 16 hours and 4 hours before sacrifice. Tumors were dissected and PKM2 complex stoichiometry in tumor lysates was determined by size exclusion chromatography. Uncropped blots are shown in Supplementary Fig. 10. (b) Concentrations of lactate, ribose phosphate and serine in H1299 xenograft tumors from mice treated with vehicle or TEPP-46 as in (a). (c) H1299 cells were injected subcutaneously into nu/nu mice which were subsequently randomly divided into two cohorts, one given vehicle and the other TEPP-46 at 50 mg/kg twice-daily throughout the duration of the experiment. Injection sites were monitored for tumor emergence [p value calculated by logrank (Mantel-Cox) test]. After 52 days, the tumors were dissected and final tumor weights were measured (d). Mean tumor weights ± s.e.m. are shown and p value was calculated by unpaired Student’s t-test.
These data demonstrate that cancer cells in xenograft tumors are exposed to TEPP-46 after several weeks of oral dosing, and that this can mimic PKM1 expression to impair growth of H1299 cells as xenograft tumors.
DISCUSSION
Cancer cells harbor genetic changes that allow them to increase nutrient uptake and alter metabolism to support anabolic processes, and interfering with this metabolic program is a strategy for cancer therapy1,2. Altered glucose metabolism in cancer cells is mediated in part by expression of PKM2, which has unique regulatory properties. Unlike its splice variant PKM1, which is found in many normal tissues, PKM2 is allosterically activated by FBP and can interact with tyrosine-phosphorylated proteins to release FBP and decrease enzyme activity. Thus, growth factor signaling promotes decreased PKM2 activity and availability of glycolytic metabolites for anabolic pathways that branch from glycolysis. This suggests that activation of PKM2 might oppose the effects of growth signaling and interfere with anabolic glucose metabolism.
Consistent with this hypothesis, our data show that high pyruvate kinase activity caused by PKM1 expression or small-molecule PKM2 activation impedes the ability of cancer cells to form tumors in mice. PKM1 can associate with endogenous PKM2 and form heterocomplexes that are likely insensitive to FBP regulation and thus exhibit higher activity. Recent publications have described non-enzymatic functions for PKM228-30. While these non-classical PKM2 activities may also play a role in tumor formation, our data suggest that the ability to decrease enzyme activity is also an important property of the enzyme that may drive PKM2 selection in tumors. Furthermore, these experiments provide evidence that elevated pyruvate kinase activity can be incompatible with efficient tumor growth.
Small-molecule PKM2 activators can mimic the enzymatic properties of PKM1 in PKM2-expressing cells and alter cell metabolism. Our studies focused on DASA-58 and TEPP-46, which are representative of two classes of PKM2-activating compounds22,23. These small molecules induce changes in the kinetic properties of PKM2 that are identical to those induced by the endogenous PKM2 activator FBP, suggesting that PKM2 could adopt a PKM1-like state in cells when FBP is high and/or in the absence of phosphotyrosine signaling. Surprisingly, structural analyses of the activators bound to PKM2 tetramers revealed a binding pocket at the interface of subunit interaction that is distinct from the site of FBP binding. Furthermore, unlike FBP, which stabilizes the C-C’ interface of the active tetramer, the small-molecule activators stabilize the A-A’ interface. Interestingly, FBP was also observed in the crystals containing small-molecule activators, and activators stabilize PKM2 tetramers more effectively in the presence of FBP. This raises the possibility that FBP binding is still required for small-molecule activation and that stabilization of the A-A’ interface inhibits FBP release. Nevertheless, these compounds enhance PKM2 enzymatic activity in a manner that renders PKM2 resistant to inhibition by phosphotyrosine binding, and interaction with phosphotyrosine is the only mechanism described to release FBP from PKM2. No natural ligands have been identified that bind PKM2 in the same site as the PKM2 activators, but given that multiple activator classes bind in the same pocket, it is possible that this represents a previously unknown site of pyruvate kinase regulation.
Our findings are consistent with PMK2 existing in a monomer-tetramer equilibrium that can be altered by the presence of FBP, TEPP-46 or DASA-58. Although this study agrees with previous reports demonstrating that the fully associated tetrameric form of PKM2 is the active form of the enzyme, dimeric PKM2 was not a predominant form of the enzyme observed in our assays. Previous studies have described PKM2 dimers following isoelectric focusing chromatography9. It is possible that the methods we used to assess PKM2 multimeric state favor complete tetramer dissociation to monomers. We suspect that much of the PKM2 in vivo exists in an equilibrium between loosely associated tetramers with low activity and tightly associated tetramers exhibiting high activity, and that this equilibrium is influenced by FBP levels and phosphotyrosine signaling or post-translational modifications that destabilize the loosely-associated (low activity) tetramer17,19,26,27.
PKM2 activators may impair tumor cell proliferation by interfering with anabolic metabolism. Activator treatment in vitro and in vivo results in decreased pools of ribose-phosphate and serine, which are key precursors for nucleotide, lipid and amino acid metabolism. However, no change in metabolite pools were seen when pyruvate kinase activity was elevated by chronic PKM1 expression in vitro, suggesting that the metabolic states elicited by these two treatments may not be equivalent. It is possible that this response to PKM1 expression may reflect adaptive events that can more effectively compensate for increased pyruvate kinase activity in vitro. PKM2 activator treatment also reduced incorporation of glucose carbons into lactate and lipids, thus interfering with the increased lactate production used by many tumors to establish a redox balance compatible with high glycolytic rates3. Lipids are essential components of new cells and reduced lipid production has been shown to inhibit proliferation31. Interestingly, the effects of PKM2 activation on proliferation in cell culture are only evident under hypoxic conditions, suggesting that glucose-dependent anabolic pathways may only be important for proliferation under some conditions. Oxidation-induced inhibition of PKM2 under hypoxia may support NADPH production via the oxidative branch of the pentose phosphate pathway to sustain antioxidant responses19. While the source of NADPH for biosynthetic processes, including lipid synthesis, remains a subject of active investigation, the ability of PKM2 to control cellular redox state may partly underlie the selective effects of PKM2 activation on proliferation under hypoxia.
While our data indicate that small-molecule activation of PKM2 can impede the proliferation of cancer cells in xenograft models, it remains to be determined whether such compounds will be similarly effective in autochthonous mouse tumor models or be efficacious as a cancer therapy in humans. PKM2 activators can also sensitize cells to oxidative stress-induced death raising the possibility that such compounds could enhance tumor killing in combination with drugs that increase cellular ROS6,19. Regardless, this study further supports the notion that pyruvate kinase acts as an important regulatory node in glycolysis to control glucose fate in cells and argues that high pyruvate kinase activity is not conducive to the anabolic metabolism necessary for tumor growth.
METHODS
All mouse studies were performed in accordance with institutional guidelines and approved by the MIT committee on animal care. Full experimental details are included in the Supplementary Methods.
Cell culture
293T and A549 cells were cultured in standard DMEM-based media, and H1299, T.T, SN12C, and SKMel28 cells were cultured in RPMI-based media. For all hypoxia experiments the media were supplemented with 20 mM HEPES. Cells expressing specific Flag-tagged isoforms of mouse pyruvate kinase M, or mutants thereof, in the absence of endogenous PKM2 were derived as described8. Cell doubling time was calculated by periodic measurement of cell mass accumulation over six days using crystal violet staining. Cell viability was assayed using CellTiter96® AQueous (Promega) or MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) assay (Promega) according to the manufacturer’s instructions.
Western blot
Cell or tissue lysates were analyzed by SDS-PAGE and western blot using standard protocols and the following primary antibodies: anti-pyruvate kinase (Abcam, ab6191), anti-PKM1 (Sigma, SAB4200094), anti-PKM2 (Cell Signaling, 4053), anti-FLAG (Sigma, F3165), and anti-actin (Abcam, ab1801). Where indicated Flag-agarose (Sigma-A2220) was used for immunoprecipitation of Flag-tagged proteins. Iso-electric focusing/SDS-PAGE two-dimensional Western blot analysis was performed as described previously20.
Xenograft experiments
H1299 cells with and without constitutive expression of mouse PKM1 were suspended in sterile PBS, and 5×106 cells were injected subcutaneously into nu/nu mice. Tumor growth was monitored, mice sacrificed after the time indicated, and tumors were harvested for analysis.
PKM2 activity
Pyruvate kinase activity was measured as described previously32. Where indicated, 100 μM pervanadate was added 10 minutes prior to cell lysis. For the phosphotyrosine peptide experiments, the amino acid sequences of the peptides were: GGAVDDDYAQFANGG (M2tide) and GGAVDDDpYAQFANGG (P-M2tide)17.
Sucrose-gradient ultracentrifugation
Recombinant PKM2 was incubated with 500 nM TEPP-46, 1 μM DASA-58, or 100 μM FBP for 30 minutes on ice before layering on a 10-40% sucrose gradient. For conditions involving FBP, 100 μM FBP was included in the sucrose gradient except where indicated that PKM2 was transiently exposed to FBP. Gradients were spun at 55,000 rpm for 10 hours using a Beckman TLS-55 rotor and fractions analyzed by SDS-PAGE or Coomassie blue staining. Coomassie blue staining intensity was quantified using IR fluorescence.
Size exclusion chromatography
Recombinant protein or ~2 mg of cellular protein was separated on a HiPrep 16/60 Sephacryl S-200 HR column (GE) in 50 mM sodium phosphate, 150 mM sodium chloride, pH 7.2. Fractions were analyzed by UV absorbance or SDS-PAGE and western blot as indicated.
Recombinant PKM2
Full-length human PKM2 was expressed as a His-tagged fusion protein in Escherichia coli strain BL21(DE3) and isolated by Ni-NTA affinity chromatography. For structural studies recombinant PKM2 was further purified by size exclusion chromatography. The final protein purity was confirmed by SDS-PAGE. Further details are in the Supplementary Methods.
Protein crystallization and structure determination
For co-crystallization, PKM2 was incubated overnight at room temperature in the presence of 5 mM activators (TEPP-46 or DASA-58) and crystallization trays were set up using the sitting-drop vapor diffusion method with droplets of protein solution (0.5 μl) and reservoir solution (0.5 μl). The best diffracting crystals were obtained from a reservoir solution containing 25% P3350, 0.2 M NH4OAc and 0.1 M Bis-Tris pH 6.5. Data collection was carried out at the Advanced Photon Source beamline 23ID-B. Data were reduced with HKL-200033 (DASA-58) or HKL-300034 (TEPP-46). Structures were solved by direct replacement with the isomorphous Protein Data Bank (PDB)35 entry 3GQY. Activator geometry restraints were obtained at the PRODRG36 server. Iterations of model rebuilding, refinement and geometry validation were performed with COOT37, REFMAC38 and MOLPROBITY39, respectively. Further details are in the Supplementary Methods.
Protein identification by LC-MS/MS
Pyruvate kinase immunoprecipitates were separated by SDS-PAGE, the band corresponding to pyruvate kinase by molecular weight excised and subjected to in-gel trypsin digestion, then analyzed by reversed-phase microcapillary/tandem mass spectrometry (LC/MS/MS) analysis. MS/MS spectra were searched against the concatenated target and decoy (reversed) Swiss-Prot protein database using Sequest [Proteomics Browser Software (PBS), Thermo Fisher Scientific]. Peptides passing a false discovery rate (FDR) threshold of 1% were accepted. Additional details are in the Supplementary Methods.
Metabolism measurements
Glucose-dependent lipid synthesis was performed as described previously31. Lactate levels were measured using YSI 7100 Select Biochemistry Analyzer (YSI Incorporated). Oxygen consumption rates were measured using a polarographic oxygen electrode40. Where indicated metabolites and lipid synthesis were measured using GC-MS. For determination of [U-13C6]glucose enrichment in lipogenic AcCoA, A549 cells were cultured for 3 days in the presence of tracer and relevant metabolites were extracted using methanol and chloroform. Fatty acid methyl esters were generated from lipid biomass by dissolving dried chloroform fractions in 50 μl of Methyl-8 reagent (Pierce) and incubating at 60°C for 1 hour. GC/MS analysis was performed using an Agilent 6890 GC equipped with a 30m DB-35MS capillary column connected to an Agilent 5975B MS operating under electron impact (EI) ionization at 70 eV. Mass isotopomer distributions (MIDs) were determined by integrating palmitate ion fragments in the m/z range of 270 to 286. Computational estimation of AcCoA enrichment and fractional new palmitate synthesis was accomplished as previously described41. For metabolite measurement by LC-MS/MS42, cells or snap-frozen xenograft tumor tissue were extracted with 4:1 v/v MeOH/H2O equilibrated at -80 °C, and the extracts were dried under nitrogen gas. Samples were re-suspended in water and analyzed using a 5500 QTRAP triple quadrupole mass spectrometer (AB/SCIEX) coupled to a Prominence UFLC system (Shimadzu) via selected reaction monitoring (SRM) of metabolites in both positive and negative ion mode. Peak areas from the total ion current (TIC) for each metabolite SRM transition were integrated using MultiQuant v2.0 software (AB/SCIEX). Integrated TIC areas corresponding to metabolite concentrations were imported into Metaboanalyst43 software for further analysis. Additional details for all metabolism measurements are provided in the Supplementary Methods.
ADME and PK/PD methods
Pharmacokinetic studies were performed in fasted male BALB/c mice. Following drug administration via the respective routes, plasma samples were collected at the indicated time points and TEPP-46 levels were analyzed by LC-MS/MS. Pharmacokinetics parameters (Cmax, Tmax, T½, AUC) were calculated using non-compartmental model with WinNonlin Ver 5.2 statistics software (Pharsight Corporation).
Supplementary Material
Acknowledgments
The Structural Genomics Consortium is a registered charity (Number 1097737) and receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knutand Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck and Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research, and the Wellcome Trust. Crystallography results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. The authors thank Paul Chang for experimental advice related to sucrose gradient ultracentrifugation and SAI Advantium Pharma Ltd. for help with pharmacokinetics studies. We also thank Mitali Kini for experimental help; and acknowledge Min Yuan and Susanne Breitkopf for help with mass spectrometry experiments. This work was supported by the Molecular Libraries Initiative of the National Institutes of Health Roadmap for Medical Research and the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health and by R03MH085679. This work was also funded by R01 GM56203 (L.C.C.). J.M.A. acknowledges funding from NIH 5P01CA120964 and NIH DF/HCC Cancer Center Support Grant 5P30CA006516. M.V.H. acknowledges additional funding support from the Smith Family Foundation, the Burroughs Wellcome Fund, the Damon Runyon Cancer Research Foundation, the Stern family, and the National Cancer Institute.
Footnotes
AUTHOR CONTRIBUTIONS
D.A., Y.Y. W.J.I. and M.V.H. designed and coordinated the study. M.B.B., C.J.T. L.C.C. H.-W.P. and L.D. advised on various aspects of the study. J.-K. J., M.B.B., M.S.; A.P.S., H.V., N.S., M.J.W., K.R.B., W.L., C.P.A., J.I., D.S.A., and C.J.T. designed and provided compounds. B.S.H., W.T., S.D. and H.-W.P. performed all structural studies; A.J. did additional structural analysis. H.Y., C.K., K.E.Y., K.K., F.G.S., S.J. and L.D. performed in vivo pharmacology and ADME studies. C.M.M., J.M.A, and G.S. did mass spectrometry. M.H.H. reviewed pathology. D.A., Y.Y., W.J.I., K.R.M., B.P.F., K.D.C., S.M., T.M.K., C.K., S.Y.L., Z.R.J., S.M.D., H.R.C, and M.V.H. all performed experiments. D.A. and M.V.H. wrote the paper with significant input from Y.Y. and W.J.I.
COMPETING FINANCIAL INTERESTS
L.C.C. is a founder, M.V.H. is a consultant, and A.J., H.Y., C.K., K.E.Y., K.K., F.G.S., S.J. L.D are employed by Agios Pharmaceuticals, a company seeking to target metabolic enzymes for cancer therapy.
References
- 1.Tennant DA, Duran RV, Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat Rev Cancer. 2010;10:267–77. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]
- 2.Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov. 2011;10:671–84. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 3.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 5.Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330:1340–4. doi: 10.1126/science.1193494. [DOI] [PubMed] [Google Scholar]
- 6.Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov. 2009;8:579–91. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 7.Weissleder R. Molecular imaging in cancer. Science. 2006;312:1168–71. doi: 10.1126/science.1125949. [DOI] [PubMed] [Google Scholar]
- 8.Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–3. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 9.Mazurek S. Pyruvate kinase type M2: a key regulator of the metabolic budget system in tumor cells. Int J Biochem Cell Biol. 2011;43:969–80. doi: 10.1016/j.biocel.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Noguchi T, Inoue H, Tanaka T. The M1- and M2-type isozymes of rat pyruvate kinase are produced from the same gene by alternative RNA splicing. J Biol Chem. 1986;261:13807–12. [PubMed] [Google Scholar]
- 11.Clower CV, et al. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci U S A. 2010;107:1894–9. doi: 10.1073/pnas.0914845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamada K, Noguchi T. Regulation of pyruvate kinase M gene expression. Biochem Biophys Res Commun. 1999;256:257–62. doi: 10.1006/bbrc.1999.0228. [DOI] [PubMed] [Google Scholar]
- 13.Ikeda Y, Tanaka T, Noguchi T. Conversion of non-allosteric pyruvate kinase isozyme into an allosteric enzyme by a single amino acid substitution. J Biol Chem. 1997;272:20495–501. doi: 10.1074/jbc.272.33.20495. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda Y, Noguchi T. Allosteric regulation of pyruvate kinase M2 isozyme involves a cysteine residue in the intersubunit contact. J Biol Chem. 1998;273:12227–33. doi: 10.1074/jbc.273.20.12227. [DOI] [PubMed] [Google Scholar]
- 15.Ashizawa K, Willingham MC, Liang CM, Cheng SY. In vivo regulation of monomer-tetramer conversion of pyruvate kinase subtype M2 by glucose is mediated via fructose 1,6-bisphosphate. J Biol Chem. 1991;266:16842–6. [PubMed] [Google Scholar]
- 16.Ashizawa K, McPhie P, Lin KH, Cheng SY. An in vitro novel mechanism of regulating the activity of pyruvate kinase M2 by thyroid hormone and fructose 1, 6-bisphosphate. Biochemistry. 1991;30:7105–11. doi: 10.1021/bi00243a010. [DOI] [PubMed] [Google Scholar]
- 17.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–6. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 18.Eigenbrodt E, Reinacher M, Scheefers-Borchel U, Scheefers H, Friis R. Double role for pyruvate kinase type M2 in the expansion of phosphometabolite pools found in tumor cells. Crit Rev Oncog. 1992;3:91–115. [PubMed] [Google Scholar]
- 19.Anastasiou D, et al. Inhibition of Pyruvate Kinase M2 by Reactive Oxygen Species Contributes to Antioxidant Responses. Science. 2011;334:1278–83. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vander Heiden MG, et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329:1492–9. doi: 10.1126/science.1188015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Locasale JW, Vander Heiden MG, Cantley LC. Rewiring of glycolysis in cancer cell metabolism. Cell Cycle. 2010;9:4253. doi: 10.4161/cc.9.21.13925. [DOI] [PubMed] [Google Scholar]
- 22.Jiang JK, et al. Evaluation of thieno[3,2-b]pyrrole[3,2-d]pyridazinones as activators of the tumor cell specific M2 isoform of pyruvate kinase. Bioorg Med Chem Lett. 2010;20:3387–93. doi: 10.1016/j.bmcl.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boxer MB, et al. Evaluation of substituted N,N’-diarylsulfonamides as activators of the tumor cell specific M2 isoform of pyruvate kinase. J Med Chem. 2010;53:1048–55. doi: 10.1021/jm901577g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikeda Y, Taniguchi N, Noguchi T. Dominant negative role of the glutamic acid residue conserved in the pyruvate kinase M(1) isozyme in the heterotropic allosteric effect involving fructose-1,6-bisphosphate. J Biol Chem. 2000;275:9150–6. doi: 10.1074/jbc.275.13.9150. [DOI] [PubMed] [Google Scholar]
- 25.Kato H, Fukuda T, Parkison C, McPhie P, Cheng SY. Cytosolic thyroid hormone-binding protein is a monomer of pyruvate kinase. Proc Natl Acad Sci U S A. 1989;86:7861–5. doi: 10.1073/pnas.86.20.7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hitosugi T, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal. 2009;2:ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lv L, et al. Acetylation Targets the M2 Isoform of Pyruvate Kinase for Degradation through Chaperone-Mediated Autophagy and Promotes Tumor Growth. Mol Cell. 2011;42:719–30. doi: 10.1016/j.molcel.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo W, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–44. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoshino A, Hirst JA, Fujii H. Regulation of cell proliferation by interleukin-3-induced nuclear translocation of pyruvate kinase. J Biol Chem. 2007;282:17706–11. doi: 10.1074/jbc.M700094200. [DOI] [PubMed] [Google Scholar]
- 30.Stetak A, et al. Nuclear translocation of the tumor marker pyruvate kinase M2 induces programmed cell death. Cancer Res. 2007;67:1602–8. doi: 10.1158/0008-5472.CAN-06-2870. [DOI] [PubMed] [Google Scholar]
- 31.Hatzivassiliou G, et al. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–21. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 32.Vander Heiden MG, et al. Identification of small molecule inhibitors of pyruvate kinase M2. Biochem Pharmacol. 2009;79:1118–24. doi: 10.1016/j.bcp.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otwinowski Z, Minor W. Methods in Enzymology. Vol. 276. Academic Press; 1997. Processing of X-ray diffraction data collected in oscillation mode; pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 34.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62:859–66. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 35.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–63. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 39.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–9. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schumacker PT, Chandel N, Agusti AG. Oxygen conformance of cellular respiration in hepatocytes. Am J Physiol. 1993;265:L395–402. doi: 10.1152/ajplung.1993.265.4.L395. [DOI] [PubMed] [Google Scholar]
- 41.Metallo CM, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2011;481:380–4. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc. 2012;7:872–81. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS. MetaboAnalyst 2.0--a comprehensive server for metabolomic data analysis. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


