Abstract
Heparan sulfates (HSs) have potential therapeutic value as anti-inflammatory and antimetastasis drugs, in addition to their current use as anticoagulants. Recent advances in chemoenzymatic synthesis of HS provide a way to conveniently produce homogenous HS with different biological properties. Crystal structures of sulfotransferases involved in this process are providing atomic detail of their substrate binding clefts and interactions with their HS substrates. In theory, the flexibility of this method can be increased by modifying the specificities of the sulfotransferases based on the structures, thereby producing a new array of products.
Introduction
Heparan sulfate (HS) is an abundant molecule present on the mammalian cell surface and plays a key role in a wide range of physiological functions, including regulation of embryonic development and of inflammatory responses [1]. The position and density of the sulfo groups and iduronic acid (IdoA) units dictate the function of these polysaccharides [2]. The HS sulfotransferases are a class of enzyme that transfer a sulfo group from the sulfo donor, 3′-phosphoadenosine 5′-phosphosulfate, to the hydroxyl (–OH) and amino (–NH2) positions of the saccharide residues present in polysaccharides (reviewed in Ref. [3]). Four different types of HS sulfotransferases have been discovered, including N-deacetylase/N-sulfotransferases (NDSTs), 2-O-sulfotransferases (2OSTs), 6-O-sulfotransferases (6OSTs), and 3-O-sulfotransferases (3OSTs), and each subtype has regioselectivity toward a specific position of the saccharide unit (Supplementary Figure 1). All HS sulfotransferases act on the polysaccharide backbone with a repeating disaccharide unit of glucuronic acid (GlcA) or IdoA and glucosamine to form the highly sulfated HS polysaccharide. In the past few decades, gene knockout experiments in mice and lower organisms have revealed the physiological significance of HS sulfotransferases [4]. However, the overall mechanism controlling HS biosynthesis remains enigmatic. Understanding how sulfotransferases specifically recognize their substrates is advancing our knowledge of this biosynthesis process.
The availability of crystal structures of HS sulfotransferases, especially those of ternary complexes containing oligosaccharide substrates, has elevated the understanding of the substrate specificities of HS sulfotransferases to a new level. This review aims to summarize recent advances in HS synthesis with crystallographic analysis, to provide insight into how the sulfotransferases recognize their saccharide substrates.
Chemoenzymatic synthesis of HS oligosaccharides
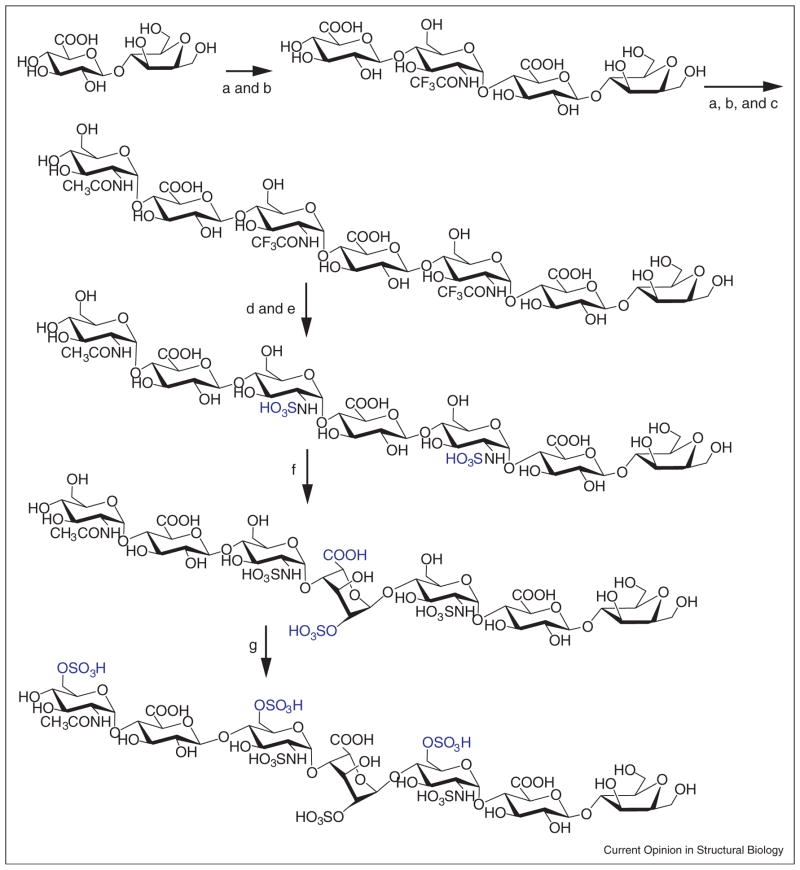
A number of approaches including chemical, enzymatic and cell-based methods are currently being developed for production of HS [5–7,8••,9,10•]. These HS products are essential to probe structure–function relationships in given biological systems, as well as for HS-based drug discovery. To date, the inability to access specific structurally defined HS has severely limited progress in HS-related research. Heparin, a prevalent clinical anticoagulant drug, is a specialized form of HS that contains a higher sulfation level and higher IdoA content. Heparin is currently isolated from porcine intestine or bovine lung through a poorly regulated supply chain. The worldwide distribution of contaminated heparin in 2007 raised concerns over the safety and reliability of animal-sourced heparins [11]. A cost-effective, quality-assured method for preparing synthetic heparin is, therefore, highly desirable. Furthermore, a facile method for preparing heparin enables engineering of HS/heparin with improved anticoagulant efficacy and reduced side effects, as well as exploiting heparin or heparin-like molecules for the development of anticancer, anti-inflammatory and antiviral drugs [12,13•,14,15•]. Although heparin fragments can be synthesized by a purely chemical method, the synthesis is extremely challenging, especially for products larger than a hexasaccharide [5]. Recently, a new chemoenzymatic approach offers a simplified synthetic route for preparing heparin and HS fragments with diversified sulfation patterns [8••]. Using recombinant glycosyltransferases and HS sulfotransferases, the saccharide chain can be extended one sugar at a time, followed by controlled sulfation, to produce the desired homogenous HS. This approach was critical in obtaining a homogeneous heptasaccharide for cocrystallization with 3OST isoform 1, providing insight into HS binding for the production of anticoagulant HS (Figure 1) [16•]. Accordingly, improved knowledge of HS interactions with the biosynthetic enzymes will further enhance the chemoenzymatic method for preparing HS for research and therapeutic purposes.
Figure 1.
Chemoenzymatic synthetic scheme for the synthesis of HS heptasaccharide (a substrate for 3OST-1 [16•]). The starting material is a disaccharide (GlcA-AnMan, where AnMan represents 2,5-anhydromannitol). The synthesis includes nine steps, involving the backbone elongation as well as sulfation and epimerization. Five elongation steps convert a disaccharide to a heptasaccharide (steps a–c), which is achieved by N-acetylglucosaminyl transferase from E. coli K5 strain (for N-trifluoroacetyl glucosamine (GlcNTFA, step a) and GlcNAc transfer, step c) and heparosan synthase 2 of Pasteurella multocida (for GlcA transfer, step b). The conversion of a GlcNTFA to a GlcNS residue is completed in two steps, including detrifluoroacetylation under alkaline conditions (step d) and N-sulfation by NST (step e). The conversion of a GlcA to an IdoA2S reside is carried out by both C5-epimerase and 2OST (step f). The 6-O-sulfation (step g) is completed by the mixture of 6OST-1 and 6OST-3.
Overall fold of HS sulfotransferases
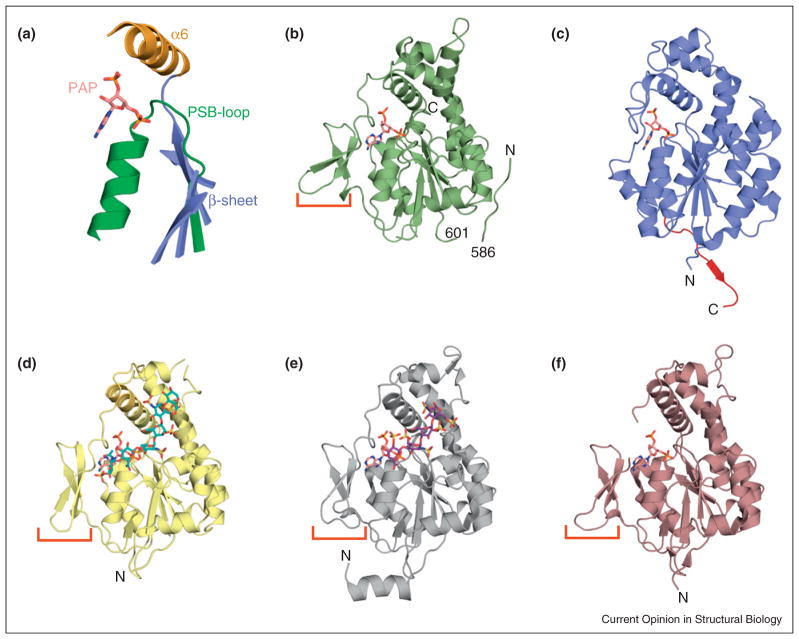
To date, crystal structures exist for the sulfotransferase domains of three of the four HS sulfotransferases: NDST1 (NST-1), 2OST, and three 3OSTs (isoforms 1, 3, and 5) (Figure 2) [16•,17–21]. Currently, no structural information has been published for 6OST. The 3OSTs share 30% sequence identity with NST-1, while these enzymes share less than 15% identity to 2OST. Despite low sequence conservation, they have structurally similar folds and catalytic features. At the heart of these enzymes lies an α/β motif with a core five-stranded parallel β-sheet. Central to this motif is a strand-loop helix structure (PSB-loop) involved in binding the 5′-phosphosulfate of the PAPS cofactor, and a conserved α-helix (α6) that runs across the top of the β-sheet (Figure 2a) [22]. HS sulfotransferases contain a large hydrophilic cleft oriented perpendicular to the plane of the β-sheet. This cleft properly positions large, linear polysaccharide substrates for the proposed catalytic SN2 in-line displacement attack required for sulfo transfer [16•,19]. The catalytic base for 2OST appears to be a conserved histidine (His142), located at the end of the fourth β-strand of the core β-sheet (Figure 3a). NST-1 and 3OSTs lack this residue, instead utilizing a glutamate found on a loop (not present in 2OST) after the last strand of the β-sheet (Figure 3a). Also absent in 2OST is a three-stranded antiparallel β-sheet on the exterior of the central core (Figure 2c). Unlike the 3OSTs and NST-1, 2OST forms a symmetrical trimer that buries nearly 25% of the surface area for each molecular unit (Figure 3c) [21]. This arrangement positions all the N-termini on the same face of the trimer, suggesting its orientation with respect to the Golgi membrane (Figure 3d). A significant contribution to the trimer formation comes from the C-terminal tail that extends away from the core and forms an antiparallel β-sheet arrangement with the edge of the central β-sheet in the substrate binding cleft of a neighboring molecule (Figure 3b, red). Supporting the crystallographic analysis, truncation mutants of this tail have disrupted trimer formation and reduced sulfotransferase activity [21].
Figure 2.
Crystal structures of HS sulfotransferases. (a) Core secondary structural elements of the HS sulfotransferases with bound cofactor product PAP (pink). These enzymes display an α/β motif wherein a central five-stranded parallel β-sheet (blue) is flanked by α-helices. The highly conserved strand-loop-helix (green) which contains the PSB-loop, is critical for binding of the universal sulfate donor PAPS. The α6 helix (orange) is a major structural component of the substrate binding cleft. (b) Binary complex of the sulfotransferase domain of murine NDST1 with bound PAP cofactor (PDB ID: 1NST). (c) Binary complex of chicken 2OST with bound PAP cofactor (PDB ID: 3F5F). The C-terminal tail is colored red. (d) Ternary complex of murine 3OST-1 with PAP and bound chemoenzymatically synthesized heptasaccharide substrate (cyan) (PDB ID: 3UAN). (e) Ternary complex of human 3OST-3 with PAP and a tetrasaccharide substrate (purple) prepared from hydrolase treated heparin (PDB ID: 1T8U). (f) Binary complex of human 3OST-5 with bound PAP cofactor (PDB ID: 3BD9). The small three-stranded antiparallel β-sheet present in NDST1 and the 3OSTs is marked by a red bracket. The PAP cofactor is drawn in stick (pink). These figures were generated in PyMOL [46].
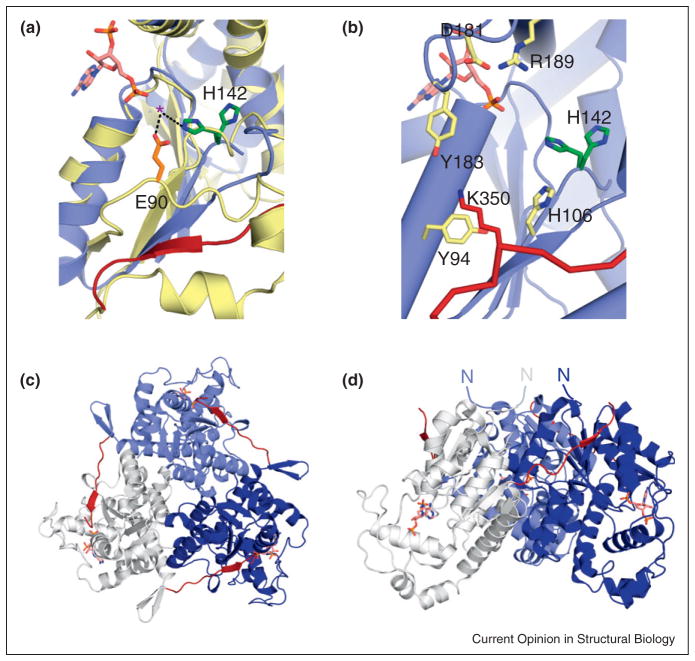
Figure 3.
Key structural characteristics of 2OST. (a) Active site architecture of 2OST, as compared to 3OST-1. Chicken 2OST (blue, PDB ID: 3F5F) was superimposed with murine 3OST-1 (gold, PDB ID: 3UAN), using the PSB-loop as a reference. The residues acting as a catalytic base for each molecule (His142 in 2OST, green; Glu90 in 3OST-1, orange) are shown in stick. Both conformations of His142 present in the crystal structure are shown. The location of the C-terminal tail from a neighboring 2OST monomer in the trimer is displayed in red. A purple asterisk has been placed at the position the acceptor hydroxyl of the heptasaccharide bound to 3OST-1. (b) 2OST residues that were determined by structurally guided mutagenesis to play roles in substrate binding and/or specificity. The proposed catalytic base, His142 (green) displays the estimated location of sulfo transfer. Alanine substitution of Asp181, Tyr183, and/or Lys350 (from a neighboring monomer, red) reduces sulfotransferase activity as a result of impaired substrate binding. Mutations of Tyr94, His106, and Arg189 alter specificity. (c) Trimer of 2OST (white, slate blue, navy blue) with the C-terminal tails shown in red. (d) Proximal grouping of the N-termini from each of the three monomer units suggests the orientation of the trimeric 2OST with respect to the Golgi membrane.
N-deacetylase/N-sulfotransferase
Biological relevance
There are four isoforms of NDST in humans [23–25]. NDSTs catalyze the first HS sulfation event, generating the substrate for subsequent epimerization and sulfation steps (Figure 1 and Supplementary Figure 1). Different expression levels of the isoforms may create varying degrees of N-sulfation in different tissues [26]. NDST-1 is the predominant isoform in most tissues while NDST-2 is the major isoform in mast cells that produce heparin [25]. It has been demonstrated that NDST-1 sulfates HS from the non-reducing end, toward the reducing end along a chain of -GlcA-GlcNAc-, and terminating sulfation five saccharides short of a sulfated GlcNS. This creates patches of sulfated and unsulfated regions [27,28]. NDSTs have two separate domains: the N-deacetylase domain and the sulfotransferase domain (NST). Neither structures of the full length nor N-deacetylase domain of NDST are currently available.
Oligosaccharide binding
The structure of NST (Figure 2b) lacks bound substrate. However, certain mutations along the proposed binding cleft can greatly affect activity, suggesting a role in substrate binding [17,29]. NST-1 contains a six amino acid insertion before the catalytic base not present in the 3OSTs. This region likely contributes to substrate binding as point mutations in this area (F640A and E641A) significantly affect activity [29]. Other residues that line the substrate binding pocket and which appear to contribute to substrate binding are Trp713, His716, and His720 from conserved helix α6 [29].
2-O-sulfotransferase
Biological relevance
Only a single isoform of 2OST exists in humans and its sequence is highly conserved across species [30]. 2OST preferentially sulfates IdoA, but is also capable of sulfating GlcA [31]. IdoA2S is critical for FGF binding, dimerization, and mediating downstream signal transduction pathways [32]. To date, there is no known physiological function for GlcA2S; however, reports indicate that it has been found in HS from adult human brain and in nuclei of rat hepatocytes [33,34].
Oligosaccharide binding
Similar to NST-1 (Figure 2b), the structure of 2OST (Figure 2c) lacks bound substrate; however, some insight into substrate recognition has been derived from site directed mutagenesis. Alanine mutations of Tyr94, His106, Asp181, Tyr183, Arg189, and/or Lys350 all have a significant effect on activity when using two substrates—one containing only IdoA and the other containing only GlcA (Figure 3b) [21]. Single mutations of D181A, Y183A, and K350A result in decreased activity against both Ido-containing and GlcA-containing substrates suggesting a general role in polysaccharide binding. However, the single mutants Y94A and H106A show substantial loss of activity for the GlcA substrate relative to the IdoA substrate, suggesting a role for Tyr94 and His106 in GlcA sulfation. Alternatively, the mutant R189A virtually eliminates sulfation of IdoA while maintaining wild-type activity for the GlcUA substrate which supports a role in IdoA binding. The different ring conformations associated with the two isomers GlcA and IdoA likely places key substituents on the saccharide in different positions, ultimately affecting protein interaction. Future crystal structures of these mutants with different substrates will be essential for understanding their substrate preferences.
3-O-Sulfotransferases
Biological relevance
In humans, there are seven isoforms of 3OSTs: 3OST-1, 2, 3a, 3b, 4, 5, and 6 [35–37]. These isomers have two established substrate preferences. 3OST-1 preferentially sulfates a GlcNS±6S with a GlcA residue on its nonreducing end. 3OST-1-like activity produces anticoagulant HS [38]. The 3-O-sulfation increases the binding affinity of HS to antithrombin III by almost 20,000 fold [39]. 3OST-3 prefers to sulfate a GlcNS±6S with an IdoA2S on the nonreducing end. The product from a 3OST-3-like modification functions as a coreceptor in herpes simplex virus type 1 entry by binding to the glycoprotein D on the viral surface [36,40]. 3OST-2, 3OST-4, and 3OST-6 appear to have substrate preferences similar to 3OST-3, while 3OST-5 is more promiscuous and displays activity for both defined substrates (Supplementary Figure 1) [36–38,41–43].
Oligosaccharide binding
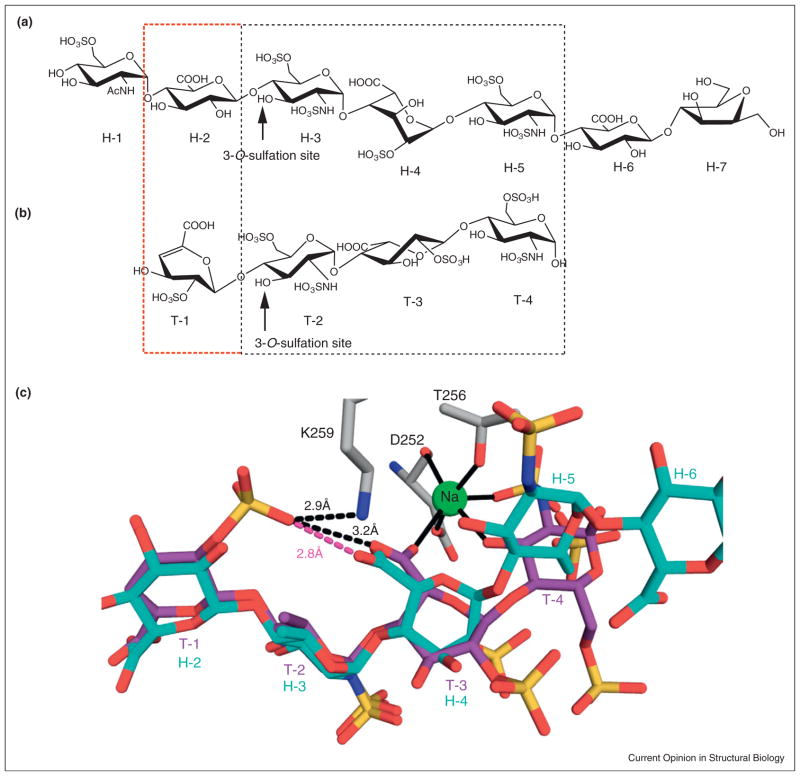
Currently there are two ternary structures of 3OSTs: 3OST-1 with a bound heptasaccharide (Figures 2d, 4a and c) and 3OST-3 with a bound tetrasaccharide (Figures 2e, 4b and c) [16•,19]. These complexes provide a wealth of information on substrate specificity differences between the 3OSTs. Though the two oligosaccharide substrates share a common trisaccharide motif (GlcNS6S-IdoA2S-GlcNS6S) on their reducing end, the IdoA2S residue in the trisaccharide sequence displays significant structural differences when bound to 3OST-1 versus 3OST-3 (Figure 4). IdoA residues in HS can be found in either a 2S0 skew boat or a 1C4 chair conformation [44,45]. In the 3OST-3 structure, the IdoA2S residue of the trisaccharide is found in the 2S0 conformation, while adopting a 1C4 conformation when bound to 3OST-1 (Figure 4c). The conformational difference of the IdoA2S, combined with a kink and rotation in the glycosidic bond to the nonreducing end acceptor GlcNS6S, alters the positioning of the reducing end. In 3OST-3, the binding of the reducing end to the protein is mediated via a metal ion (likely sodium) that forms interactions with this IdoA2S and the terminal GlcNS6S (Figure 4c) [19]. In 3OST-1, the position of the reducing end is mediated by direct interactions with the protein [19].
Figure 4.
Comparison of substrate binding by the 3OSTs. (a) The chemoenzymatically synthesized heptasaccharide substrate crystallized with 3OST-1 (PDB ID: 3UAN). (b) The heparin hydrolase produced tetrasaccharide crystallized with 3OST-3 (PDB ID: 1T8U). Reduction of the uronic acid residue (T-1) is a result of the hydrolase reaction. The saccharide unit determining the substrate specificity for either 3OST-1 (GlcA) or 3OST-3 (IdoA2S) is bracketed in red. The reducing end trisaccharide motif of identical sequence between the two is boxed in black. (c) Conformational differences for oligosaccharide substrates bound to 3OST-1 (cyan) versus 3OST-3 (purple). Superposition of these two substrates provides a structural basis for the preference of a 2-O-sulfated saccharide unit on the nonreducing side of the acceptor GlcNS6S (residue H-4 for 3OST-1 or T-3 for 3OST-3). Residues from 3OST-3 that affect substrate specificity are drawn in gray stick. While 3OST-1 interacts directly with its substrate, 3OST-3 utilizes a metal ion (likely sodium, green) to mediate contacts with the reducing end of its substrate.
It was thought that a conformational difference might exist between the GlcA (H-2 for 3OST-1 substrate) and the ΔUA2S (T-1 for 3OST-3 substrate) that dictates substrate specificity for each isoform (Figure 4a and b, red bracket). Surprisingly, both residues bind to the enzymes in similar orientations (Figure 4c). For 3OST-1, the GlcA is found in the 4C1 chair conformation while 3OST-3 binds the ΔUA2S in a 2H1 half-chair that superimposes well with the GlcA. Despite the similar positioning, the superposition of the GlcA and ΔUA2S suggests how the sugar identity at this position may influence binding preference. The GlcA in the 3OST-1 substrate probably cannot be 2-O-sulfated, as the extra sulfo group would likely be located ~2.8 Å from the carboxylate on the reducing end IdoA2S (H-4), which places two negatively charged groups in close proximity. For the 3OST-3 substrate, the 2-O-sulfo group of the ΔUA2S is located farther away (3.2 Å) from the carboxylate on the IdoA2S (T-3) and within hydrogen bonding distance to Lys259 (Figure 4c). Lys259 may ameliorate the charge repulsion in this area, a role that is supported by severe loss of function that results from alanine substitution. This residue is highly conserved in mammalian 3OST-3s but is not conserved in 3OST-1 [19].
In addition to the residues near the active site, residues located at a remote ‘gate’ influence the activity and specificity of 3OSTs (Supplementary Figure 2) [20]. Manipulation of these residues alters the substrate specificity of 3OSTs. In the 3OST-1 structure, without substrate present (Supplementary Figure 2A) [18], this gate is narrow (6.7 Å) in comparison to 3OST-3 and 5 (14.2 Å for both, Supplementary Figure 2b and c) [19,20]. It has been demonstrated that a double mutation of these gate residues in 3OST-5 (S120E/A306H) to those of 3OST-1, greatly reduces the 3OST-3-like activity while enhancing the 3OST-1 like activity [20]. A double mutation of these gate residues in 3OST-1(H271G/E88G) to 3OST-3 residues increased the relative amount of 3OST-3 activity versus 3OST-1 like activity, decreasing substrate selectivity [20]. The structure of 3OST-1 with heptasaccharide bound suggests that the nonreducing end of the HS can extend into the gate (Supplementary Figure 2d and e) [16•]. It should be noted that upon binding of the substrate, gate residue His271 in 3OST-1 adopts a different rotamer to accommodate the substrate, effectively ‘opening’ the gate. Glu88 becomes disordered in the presence of the substrate, and the apparent motility of these residues leads to speculation as to possible relevance for substrate specificity. Alanine substitution of R268 (located just past the gate) contributes to 3OST-1 activity, emphasizing the importance of this region to 3OST-1 for substrate binding (Supplementary Figure 2d and e). Interestingly, mutations of 3OST-3 gate residues to those of 3OST-1, significantly reduces sulfotransferase activity without affecting substrate specificity. It is clear from these studies that the interactions with the substrate across the entire length of the cleft contribute to binding as well as specificity of the 3OST isoform [16•,20].
Conclusion
The evolving crystal structures of HS sulfotransferases are yielding detailed information on substrate binding, and subsequently providing the blueprint for engineering the sulfotransferases through mutagenesis. These mutants allow for more precisely controlled synthesis of functionally distinct HS. Chemoenzymatic synthesis can supply specific oligosaccharide substrates for more informative crystal structures, revealing the key interactions between proteins and carbohydrates at atomic resolution. Our experience demonstrates that the structural studies and synthesis are intimately linked. The crystal structure of 3OST-1 represents the first example of the power of this synergistic approach. Future efforts will undoubtedly expand the capability of the chemoenzymatic synthesis for production of HS therapeutics as well as significantly contribute to research in HS-related and heparin-related glycomics.
Supplementary Material
Acknowledgments
We thank L.G. Pedersen and R. Gosavi for critical reading of the manuscript. This research was supported by the Division of Intramural Research of the National Institute of Environmental Health Sciences, National Institutes of Health (1 ZIA ES102645-03, to L. Pedersen) and NIH grants AI050050, HL094463 and HL096972 (to J. Liu).
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.sbi.2012. 07.004.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- 2.Kreuger J, Spillmann D, Li JP, Lindahl U. Interactions between heparan sulfate and proteins: the concept of specificity. J Cell Biol. 2006;174:323–327. doi: 10.1083/jcb.200604035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindahl U, Li JP. Interactions between heparan sulfate and proteins-design and functional implications. Int Rev Cell Mol Biol. 2009;276:105–159. doi: 10.1016/S1937-6448(09)76003-4. [DOI] [PubMed] [Google Scholar]
- 4.Kusche-Gullberg M, Kjellen L. Sulfotransferases in glycosaminoglycan biosynthesis. Curr Opin Struct Biol. 2003;13:605–611. doi: 10.1016/j.sbi.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Petitou M, van Boeckel CA. A synthetic antithrombin III binding pentasaccharide is now a drug! What comes next? Angew Chem Int Ed Engl. 2004;43:3118–3133. doi: 10.1002/anie.200300640. [DOI] [PubMed] [Google Scholar]
- 6.Martin JG, Gupta M, Xu Y, Akella S, Liu J, Dordick JS, Linhardt RJ. Toward an artificial Golgi: redesigning the biological activities of heparan sulfate on a digital microfluidic chip. J Am Chem Soc. 2009;131:11041–11048. doi: 10.1021/ja903038d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baik JY, Gasimli L, Yang B, Datta P, Zhang F, Glass CA, Esko JD, Linhardt RJ, Sharfstein ST. Metabolic engineering of Chinese hamster ovary cells: towards a bioengineered heparin. Metab Eng. 2012;14:81–90. doi: 10.1016/j.ymben.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8••.Xu YM, Masuko S, Takieddin M, Xu HM, Liu RP, Jing J, Mousa SA, Linhardt RJ, Liu J. Chemoenzymatic synthesis of homogeneous ultralow molecular weight heparins. Science. 2011;334:498–501. doi: 10.1126/science.1207478. Introduces a new flexible methodology for the production of homogeneous heparins using enzymes to build and modify the polysaccharide chains with specific biological targets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuberan B, Lech MZ, Beeler DL, Wu ZLL, Rosenberg RD. Enzymatic synthesis of antithrombin III-binding heparan sulfate pentasaccharide. Nat Biotechnol. 2003;21:1343–1346. doi: 10.1038/nbt885. [DOI] [PubMed] [Google Scholar]
- 10•.Olson ST, Swanson R, Petitou M. Specificity and selectivity profile of EP217609: a new neutralizable dual-action anticoagulant that targets thrombin and factor Xa. Blood. 2012;119:2187–2195. doi: 10.1182/blood-2011-09-381764. Reports on a novel anticoagulant composed of an antithrombin binding pentasaccharide with both a thrombin inhibitor and a biotin moiety covalently attached for neutralization with avidin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu H, Zhang Z, Linhardt RJ. Lessons learned from the contamination of heparin. Nat Prod Rep. 2009;26:313–321. doi: 10.1039/b819896a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shriver Z, Raguram S, Sasisekharan R. Glycomics: a pathway to a class of new and improved therapeutics. Nat Rev Drug Discov. 2004;3:863–873. doi: 10.1038/nrd1521. [DOI] [PubMed] [Google Scholar]
- 13•.Baleux F, Loureiro-Morais L, Hersant Y, Clayette P, Arenzana-Seisdedos F, Bonnaffe D, Lortat-Jacob H. A synthetic CD4-heparan sulfate glycoconjugate inhibits CCR5 and CXCR4 HIV-1 attachment and entry. Nat Chem Biol. 2009;5:743–748. doi: 10.1038/nchembio.207. Presents a peptide-linked HS with antiviral activity against HIV-1. [DOI] [PubMed] [Google Scholar]
- 14.Rao NV, Prestwich GD, Hoidal JR, Kennedy TP. Low anticoagulant heparins in the treatment of metastasis. In: Murph M, editor. Research on Melanoma — A Glimpse into Current Directions and Future Trends. InTech; 2011. [Google Scholar]
- 15•.Rao NV, Argyle B, Xu X, Reynolds PR, Walenga JM, Prechel M, Prestwich GD, MacArthur RB, Walters BB, Hoidal JR, et al. Low anticoagulant heparin targets multiple sites of inflammation, suppresses heparin-induced thrombocytopenia, and inhibits interaction of RAGE with its ligands. Am J Physiol Cell Physiol. 2010;299:C97–C110. doi: 10.1152/ajpcell.00009.2010. Investigates the mechanism for 2-O, 3-O desulfated low anticoagulant heparin as an anti-inflammatory agent. [DOI] [PubMed] [Google Scholar]
- 16•.Moon AF, Xu Y, Woody SM, Krahn JM, Linhardt RJ, Liu J, Pedersen LC. Dissecting the substrate recognition of 3-O-sulfotransferase for the biosynthesis of anticoagulant heparin. Proc Natl Acad Sci U S A. 2012;109:5265–5270. doi: 10.1073/pnas.1117923109. Crystal structure of 3OST-1 with the bound chemoenzymatically synthesized heptasaccharide reveals the substrate specificity of 3OST-1 for a GlcA-containing polysaccharide precursor to anticoagulant heparin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kakuta Y, Sueyoshi T, Negishi M, Pedersen LC. Crystal structure of the sulfotransferase domain of human heparan sulfate N-deacetylase/N-sulfotransferase 1. J Biol Chem. 1999;274:10673–10676. doi: 10.1074/jbc.274.16.10673. [DOI] [PubMed] [Google Scholar]
- 18.Edavettal SC, Lee KA, Negishi M, Linhardt RJ, Liu J, Pedersen LC. Crystal structure and mutational analysis of heparan sulfate 3-O-sulfotransferase isoform 1. J Biol Chem. 2004;279:25789–25797. doi: 10.1074/jbc.M401089200. [DOI] [PubMed] [Google Scholar]
- 19.Moon AF, Edavettal SC, Krahn JM, Munoz EM, Negishi M, Linhardt RJ, Liu J, Pedersen LC. Structural analysis of the sulfotransferase (3-O-sulfotransferase isoform 3) involved in the biosynthesis of an entry receptor for herpes simplex virus 1. J Biol Chem. 2004;279:45185–45193. doi: 10.1074/jbc.M405013200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu D, Moon AF, Song D, Pedersen LC, Liu J. Engineering sulfotransferases to modify heparan sulfate. Nat Chem Biol. 2008;4:200–202. doi: 10.1038/nchembio.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bethea HN, Xu D, Liu J, Pedersen LC. Redirecting the substrate specificity of heparan sulfate 2-O-sulfotransferase by structurally guided mutagenesis. Proc Natl Acad Sci U S A. 2008;105:18724–18729. doi: 10.1073/pnas.0806975105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kakuta Y, Pedersen LG, Pedersen LC, Negishi M. Conserved structural motifs in the sulfotransferase family. Trends Biochem Sci. 1998;23:129–130. doi: 10.1016/s0968-0004(98)01182-7. [DOI] [PubMed] [Google Scholar]
- 23.Aikawa J, Esko JD. Molecular cloning and expression of a third member of the heparan sulfate/heparin GlcNAc N-deacetylase/N-sulfotransferase family. J Biol Chem. 1999;274:2690–2695. doi: 10.1074/jbc.274.5.2690. [DOI] [PubMed] [Google Scholar]
- 24.Aikawa J, Grobe K, Tsujimoto M, Esko JD. Multiple isozymes of heparan sulfate/heparin GlcNAc N-deacetylase/GlcN N-sulfotransferase. Structure and activity of the fourth member, NDST4. J Biol Chem. 2001;276:5876–5882. doi: 10.1074/jbc.M009606200. [DOI] [PubMed] [Google Scholar]
- 25.Kusche-Gullberg M, Eriksson I, Pikas DS, Kjellen L. Identification and expression in mouse of two heparan sulfate glucosaminyl N-deacetylase/N-sulfotransferase genes. J Biol Chem. 1998;273:11902–11907. doi: 10.1074/jbc.273.19.11902. [DOI] [PubMed] [Google Scholar]
- 26.Grobe K, Ledin J, Ringvall M, Holmborn K, Forsberg E, Esko JD, Kjellen L. Heparan sulfate and development: differential roles of the N-acetylglucosamine N-deacetylase/N-sulfotransferase isozymes. Biochim Biophys Acta. 2002;1573:209–215. doi: 10.1016/s0304-4165(02)00386-0. [DOI] [PubMed] [Google Scholar]
- 27.Sheng J, Liu R, Xu Y, Liu J. The dominating role of N-deacetylase/N-sulfotransferase 1 in forming domain structures in heparan sulfate. J Biol Chem. 2011;286:19768–19776. doi: 10.1074/jbc.M111.224311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy KJ, Merry CL, Lyon M, Thompson JE, Roberts IS, Gallagher JT. A new model for the domain structure of heparan sulfate based on the novel specificity of K5 lyase. J Biol Chem. 2004;279:27239–27245. doi: 10.1074/jbc.M401774200. [DOI] [PubMed] [Google Scholar]
- 29.Kakuta Y, Li L, Pedersen LC, Pedersen LG, Negishi M. Heparan sulphate N-sulphotransferase activity: reaction mechanism and substrate recognition. Biochem Soc Trans. 2003;31:331–334. doi: 10.1042/bst0310331. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi M, Habuchi H, Yoneda M, Habuchi O, Kimata K. Molecular cloning and expression of Chinese hamster ovary cell heparan-sulfate 2-sulfotransferase. J Biol Chem. 1997;272:13980–13985. doi: 10.1074/jbc.272.21.13980. [DOI] [PubMed] [Google Scholar]
- 31.Rong J, Habuchi H, Kimata K, Lindahl U, Kusche-Gullberg M. Substrate specificity of the heparan sulfate hexuronic acid 2-O-sulfotransferase. Biochemistry. 2001;40:5548–5555. doi: 10.1021/bi002926p. [DOI] [PubMed] [Google Scholar]
- 32.Kreuger J, Salmivirta M, Sturiale L, Gimenez-Gallego G, Lindahl U. Sequence analysis of heparan sulfate epitopes with graded affinities for fibroblast growth factors 1 and 2. J Biol Chem. 2001;276:30744–30752. doi: 10.1074/jbc.M102628200. [DOI] [PubMed] [Google Scholar]
- 33.Lindahl B, Eriksson L, Lindahl U. Structure of heparan sulphate from human brain, with special regard to Alzheimer’s disease. Biochem J. 1995;306(Pt 1):177–184. doi: 10.1042/bj3060177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fedarko NS, Conrad HE. A unique heparan sulfate in the nuclei of hepatocytes: structural changes with the growth state of the cells. J Cell Biol. 1986;102:587–599. doi: 10.1083/jcb.102.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shworak NW, Liu J, Petros LM, Zhang L, Kobayashi M, Copeland NG, Jenkins NA, Rosenberg RD. Multiple isoforms of heparan sulfate D-glucosaminyl 3-O-sulfotransferase. Isolation, characterization, and expression of human cdnas and identification of distinct genomic loci. J Biol Chem. 1999;274:5170–5184. doi: 10.1074/jbc.274.8.5170. [DOI] [PubMed] [Google Scholar]
- 36.Xia G, Chen J, Tiwari V, Ju W, Li JP, Malmstrom A, Shukla D, Liu J. Heparan sulfate 3-O-sulfotransferase isoform 5 generates both an antithrombin-binding site and an entry receptor for herpes simplex virus, type 1. J Biol Chem. 2002;277:37912–37919. doi: 10.1074/jbc.M204209200. [DOI] [PubMed] [Google Scholar]
- 37.Xu D, Tiwari V, Xia G, Clement C, Shukla D, Liu J. Characterization of heparan sulphate 3-O-sulphotransferase isoform 6 and its role in assisting the entry of herpes simplex virus type 1. Biochem J. 2005;385:451–459. doi: 10.1042/BJ20040908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Shworak NW, Sinay P, Schwartz JJ, Zhang L, Fritze LM, Rosenberg RD. Expression of heparan sulfate D-glucosaminyl 3-O-sulfotransferase isoforms reveals novel substrate specificities. J Biol Chem. 1999;274:5185–5192. doi: 10.1074/jbc.274.8.5185. [DOI] [PubMed] [Google Scholar]
- 39.Atha DH, Lormeau JC, Petitou M, Rosenberg RD, Choay J. Contribution of monosaccharide residues in heparin binding to antithrombin-III. Biochemistry. 1985;24:6723–6729. doi: 10.1021/bi00344a063. [DOI] [PubMed] [Google Scholar]
- 40.Shukla D, Liu J, Blaiklock P, Shworak NW, Bai X, Esko JD, Cohen GH, Eisenberg RJ, Rosenberg RD, Spear PG. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 1999;99:13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 41.Tiwari V, O’Donnell CD, Oh MJ, Valyi-Nagy T, Shukla D. A role for 3-O-sulfotransferase isoform-4 in assisting HSV-1 entry and spread. Biochem Biophys Res Commun. 2005;338:930–937. doi: 10.1016/j.bbrc.2005.10.056. [DOI] [PubMed] [Google Scholar]
- 42.Mochizuki H, Yoshida K, Shibata Y, Kimata K. Tetrasulfated disaccharide unit in heparan sulfate: enzymatic formation and tissue distribution. J Biol Chem. 2008;283:31237–31245. doi: 10.1074/jbc.M801586200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duncan MB, Chen J, Krise JP, Liu J. The biosynthesis of anticoagulant heparan sulfate by the heparan sulfate 3-O-sulfotransferase isoform 5. Biochim Biophys Acta. 2004;1671:34–43. doi: 10.1016/j.bbagen.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 44.Mulloy B, Forster MJ. Conformation and dynamics of heparin and heparan sulfate. Glycobiology. 2000;10:1147–1156. doi: 10.1093/glycob/10.11.1147. [DOI] [PubMed] [Google Scholar]
- 45.Ferro DR, Provasoli A, Ragazzi M, Casu B, Torri G, Bossennec V, Perly B, Sinay P, Petitou M, Choay J. Conformer populations of L-iduronic acid residues in glycosaminoglycan sequences. Carbohydr Res. 1990;195:157–167. doi: 10.1016/0008-6215(90)84164-p. [DOI] [PubMed] [Google Scholar]
- 46.DeLano WL. The PyMOL Molecular Graphics System, Version 1.4. Schrodinger, LLC; 2012. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.