Summary
V(D)J recombination is initiated by a specialized transposase consisting of RAG-1 and RAG-2. The ability of antigen receptor gene segments to undergo V(D)J recombination is correlated with spatially- and temporally-restricted chromatin modifications. We have found that RAG-2 binds specifically to histone H3 and that this binding is absolutely dependent on di- or trimethylation at lysine 4 (H3K4me2 or H3K4me3). The interaction requires a non-canonical plant homeodomain (PHD) that had previously been described within the non-core region of RAG-2. Binding of the RAG-2 PHD finger to chromatin across the IgH D–JH-C locus showed a strong correlation with the distribution of trimethylated histone H3 K4. Mutation of a conserved tryptophan residue in the RAG-2 PHD finger abolished binding to H3K4me3 and greatly impaired recombination of extrachromosomal and endogenous immunoglobulin gene segments. Together, these findings are consistent with the interpretation that recognition of hypermethylated histone H3 K4 promotes efficient V(D)J recombination in vivo.
Introduction
The antigen receptor genes of lymphocytes are encoded in separate DNA segments that are brought together during lymphoid development by V(D)J recombination to generate a diverse immunologic repertoire (Fugmann et al., 2000a; Gellert, 2002). RAG-1 and RAG-2, the sole lymphoid-specific components of the recombinase machinery, initiate V(D)J recombination by cleaving participating gene segments at specific recombination signal sequences (RSSs), producing two signal ends, terminating in flush, double-stranded breaks, and two coding ends, terminating in hairpin structures (Gellert, 2002). Recombination is then completed by components of the general cellular machinery for non-homologous DNA end joining (Fugmann et al., 2000a; Gellert, 2002).
V(D)J recombination is directed toward specific antigen receptor loci in an ordered fashion during lymphoid development. In the B lineage, for example, the immunoglobulin heavy chain (IgH) locus is rearranged before the light chain loci, and within the IgH locus D-to-JH joining preceeds VH-to-DJH joining (Alt et al., 1984). This specificity has been proposed to be enforced by epigenetic mechanisms that regulate access of antigen receptor loci to RAG-1 and RAG-2 (Cobb et al., 2006). The ability of antigen receptor gene segments to undergo V(D)J recombination is correlated with spatially- and temporally-restricted chromatin modifications, including histone acetylation (Chowdhury and Sen, 2001; Chowdhury and Sen, 2003; Goldmit et al., 2005; Johnson et al., 2003; McMurry and Krangel, 2000) and methylation (Goldmit et al., 2005; Johnson et al., 2004; Morshead et al., 2003; Perkins et al., 2004).
Recombinationally active regions are typically marked by hyperacetylated histones H3 and H4, as well as lysine 4-methylated histone H3 (Chowdhury and Sen, 2001; Chowdhury and Sen, 2003; Goldmit et al., 2005; Johnson et al., 2003; Johnson et al., 2004; McMurry and Krangel, 2000; Morshead et al., 2003; Perkins et al., 2004). In pre-B cells the preferential activation of an Ig κ allele for V(D)J recombination is accompanied by increased methylation of H3 K4 (Goldmit et al., 2005), and peaks of histone H3, dimethylated at lysine 4 (H3K4me2) have been shown to flank the D–JH cluster in pro-B cells poised to undergo D-to-JH rearrangement (Morshead et al., 2003). In contrast, dimethylation of histone H3 on lysine 9 (H3K9me2) is associated with silent chromatin and is correlated with inhibition of V(D)J recombination (Johnson et al., 2004; Osipovich et al., 2004). Targeting of the G9a histone H3 K9 methyltransferase to the TCR Dβ1 promoter resulted in inhibition of Dβ-to-Jβ recombination; this effect was associated with an elevated local density of Me H3 K9 and other epigenetic modifications, including reduced H3 K9 acetylation and increased DNA methylation (Osipovich et al., 2004). Moreover, H3 K9 is hypermethylated over VH segments in hematopoietic progenitors and in non-B lineage hematopoietic cells; removal of this modification in the B lineage requires Pax5, consistent with an inhibitory role in V(D)J recombination (Johnson et al., 2004).
The full length RAG-1 and RAG-2 proteins are 1040 and 527 amino acid residues long. Residues 384 through 1008 of RAG-1 constitute the core fragment, which contains the catalytic site for DNA nicking and transesterification (Fugmann et al., 2000b; Kim et al., 1999; Landree et al., 1999), mediates binding to recombination signal sequences (Akamatsu and Oettinger, 1998; Difilippantonio et al., 1996; Swanson and Desiderio, 1998) and contacts the coding flank near the scissile bond (Eastman et al., 1999; Swanson and Desiderio, 1999). Although RAG-2 contains no known catalytic residues and has no intrinsic DNA binding activity, it is essential for V(D)J recombination. The core RAG-2 fragment, consisting of residues 1 through 387, stabilizes and extends interactions of RAG-1 with the RSS; its presence in RAG-DNA complexes is essential for helical distortion near the scissile bond, a possible prerequisite for transesterification (Akamatsu and Oettinger, 1998; Difilippantonio et al., 1996; Hiom and Gellert, 1997; Swanson and Desiderio, 1998). Accordingly, mutations that impair recombinase-mediated cleavage and joining have been identified in core RAG-2 (Qiu et al., 2001).
Residues 387 through 527 of RAG-2, while phylogenetically conserved, are dispensable for recombination of extrachromosomal substrates in vivo and for DNA cleavage by the RAG proteins in vitro. Nonetheless, removal of the non-core region in its entirety has been reported to reduce the efficiency of recombination within extrachromosomal substrates (Cuomo and Oettinger, 1994; Kirch et al., 1998; McMahan et al., 1997; Sadofsky et al., 1994; Sadofsky et al., 1993; Steen et al., 1999), to increase the production of hybrid joints (Sekiguchi et al., 2001), to impede VH-to-DJH joining at endogenous loci (Akamatsu et al., 2003; Kirch et al., 1998; Liang et al., 2002) and to promote aberrant recombination (Talukder et al., 2004). The mechanisms underlying these effects are not well understood and may be complex, as removal of the entire non-core region is expected to have multiple functional consequences.
Residues within the non-core region of RAG-2, including a phosphorylation site for cyclin A/Cdk2 at Thr490 and the interval spanning amino acids 499 – 508, couple V(D)J recombination to the G1 cell cycle phase by supporting Skp2-SCF-dependent polyubiquitylation and destruction of RAG-2 at the G1-S transition (Jiang et al., 2005; Lee and Desiderio, 1999; Li et al., 1996). Periodic destruction of RAG-2 may protect against aberrant joining of RAG-induced DNA breaks generated outside of G1.
More recently, sequence alignment (Callebaut and Mornon, 1998) and solution structural analysis (Elkin et al., 2005) have revealed a non-canonical plant homeodomain (PHD) finger within residues 419 through 481 of the non-core region of RAG-2. The canonical PHD finger is widespread among eukaryotes, particularly in proteins associated with epigenetic regulation of gene expression (Bienz, 2006; Mellor, 2006), and specifically binds H3K4me2 or histone H3, trimethylated at lysine 4 (H3K4me3) (Li et al., 2006; Pena et al., 2006; Shi et al., 2006; Wysocka et al., 2006). Indeed, a carboxy-terminal fragment of RAG-2, spanning the non-canonical PHD finger, was reported to interact with histones (West et al., 2005). Moreover, mutations within this domain are associated with immunodeficiency disorders (Gomez et al., 2000; Noordzij et al., 2002; Schwarz et al., 1996; Villa et al., 2001).
In this communication we show that the PHD finger of RAG-2 specifically binds H3K4me2 or H3K4me3 and that this interaction mediates association of RAG-2 with rearranging gene segments. We go on to demonstrate that in the context of full-length RAG-2, the PHD finger is critical for efficient rearrangement of extrachromosomal V(D)J recombination substrates and of endogenous immunoglobulin gene segments.
Results
Binding of RAG-2 to histone H3 dimethylated or trimethylated at lysine 4
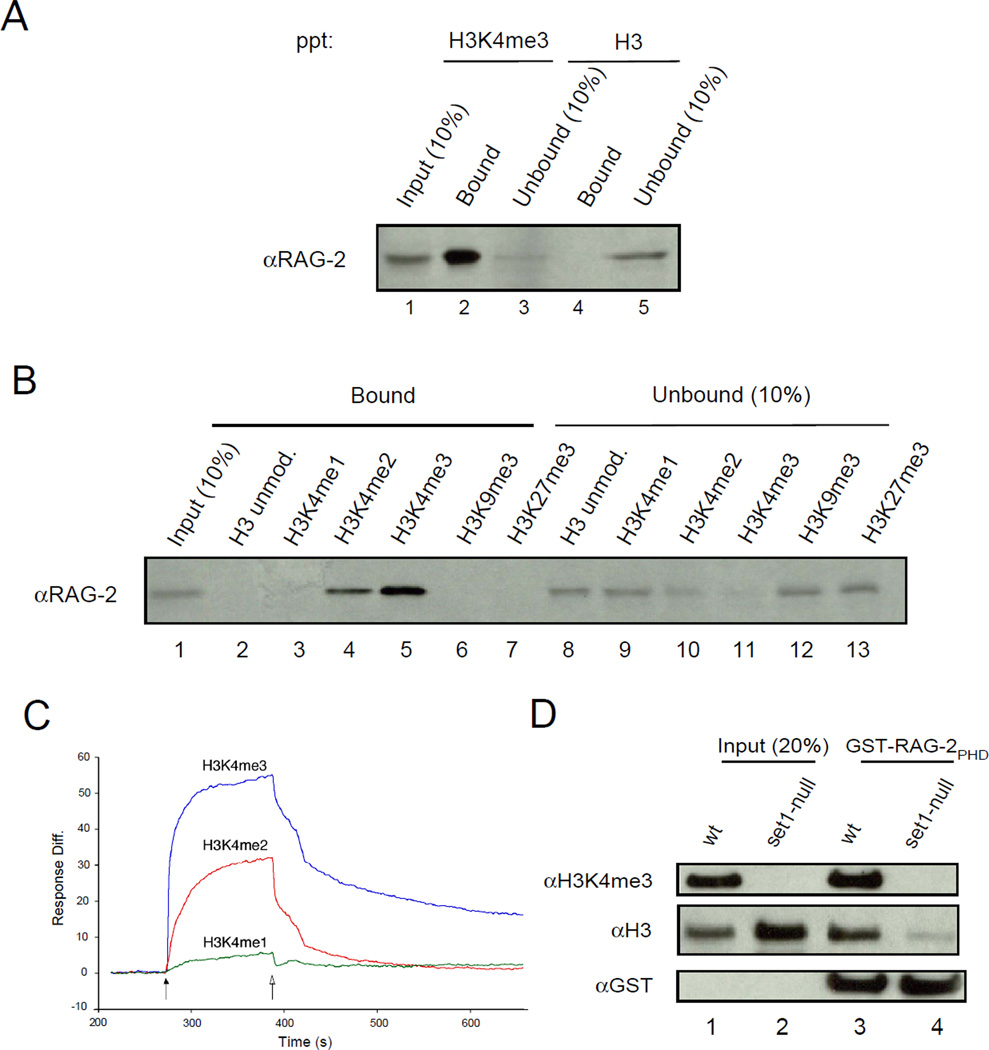
To determine whether intact RAG-2 could bind specifically to H3K4me3, we first assayed binding to synthetic peptides derived from histone H3. A lysate of 293T cells expressing wild-type, murine RAG-2 was incubated with beads bearing an unmodified peptide corresponding to residues 1 – 21 of histone H3 or with beads bearing a similar peptide that was trimethylated at lysine 4. RAG-2 was precipitated by beads attached to the H3K4me3 peptide, but not by beads bound to the unmodified peptide (Fig 1A, compare lanes 2 and 4). To probe the specificity of this interaction, a series of histone H3-derived peptides was assayed for binding. RAG-2 was precipitated by beads bearing the H3K4me3 peptide and less efficiently by the corresponding H3K4me2 peptide (Fig. 1B, lanes 4 and 5). No association was detected between RAG-2 and the unmodified histone H3 peptide (Fig. 1B, lane 2), histone H3 monomethylated at lysine 4 (H3K4me1) (Fig. 1B, lane 3) or the corresponding peptide trimethylated at lysine 9 (Fig. 1B, lane 6). RAG-2 also failed to bind to a peptide spanning residues 21 – 44 of histone H3 which was trimethylated at lysine 27 (Fig 1B, lane 7).
Figure 1.
Specific binding of RAG-2 to histone H3 containing di- or trimethylated lysine 4. (A) Whole-cell lysates of 293T cells expressing full-length, wild-type RAG-2 were incubated with streptavidin bead-bound, biotinylated peptides corresponding to residues 1 – 21 of histone H3. Peptides were unmodified (H3) or trimethylated at lysine 4 (H3K4me3). Bead-bound protein was fractionated by SDS-PAGE and RAG-2 was detected by immunoblotting. Portions (10%) of the lysate (Input) or unbound protein were assayed in parallel. (B) Lysates of 293T cells expressing wild-type RAG-2 were incubated with unmodified or modified histone H3 peptides, affixed to beads. Modifications are indicated above. All peptides correspond to residues 1 – 21 of histone H3 except for the H3K27me3 peptide, which corresponds to residues 21 – 44. Bound protein was fractionated as in (A) and RAG-2 was detected by immunobloting. Portions (10%) of the lysate (Input) or unbound protein were assayed in parallel. (C) Surface plasmon resonance binding curves for GST-RAG-2PHD association with histone H3 peptides as a function of lysine 4 methylation. Biotin-tagged H3 peptides (residues 1 – 21) were immobilized at 0.06 ng each on flow cells of a sensor chip. GST-RAG-2PHD (5 µM) was injected (solid arrow) for 2 min, followed by injection of buffer alone (open arrow). Binding curves were deduced after subtracting the response of the reference surface. (D) Methylation at lysine 4 is required for binding of the RAG-2 PHD finger to intact histone H3 in vitro. GST-RAG-2PHD was immobilized on glutathione-coated beads and incubated with histones isolated from wild-type or set1-null S. cerevisiae. Histone H3 trimethylated at lysine 4, bulk H3 and GST were detected in input and bound fractions by immunoblotting.
These results were consistent with specific binding of RAG-2 to H3K4me2 and H3K4me3, with stronger binding to the trimethylated derivative. This was confirmed using surface plasmon resonance (SPR). Biotinylated peptides corresponding to H3K4me1, H3K4me2 and H3K4me3 were affixed to streptavidin-coated sensor chips at similar densities and assayed for binding to a glutathione-S transferase (GST) fusion protein (GST-RAG-2PHD) containing residues 388 – 494 of RAG-2, spanning the non-canonical PHD finger and the linker that connects the PHD finger to the RAG-2 core (Callebaut and Mornon, 1998). SPR response curves were consistent with affinities of the RAG-2 non-core region for H3 peptides in the order H3K4me3 > H3K4me2 >> H3K4me1 (Fig 1C).
We proceeded to test the dependence of binding of GST-RAG-2PHD to intact histone H3. In budding yeast, deletion of SET1 abolishes methylation of histone H3 on lysine 4 (Briggs et al., 2001). Histones were isolated by acid extraction from wild-type and set1-null yeast strains and assayed for binding to GST-RAG-2PHD. Histone H3 from wild-type yeast was efficiently precipitated by GST-RAG-2PHD; binding to H3 from cells lacking the Set1 H3K4 methyltransferase was greatly reduced (Fig 1D, compare lanes 2 and 4). Taken together, these results indicate that methylation of histone H3 at lysine 4 supports binding to RAG-2 in vitro.
Binding to di- and trimethyl lysine 4 of histone H3 mediated by the non-canonical PHD finger
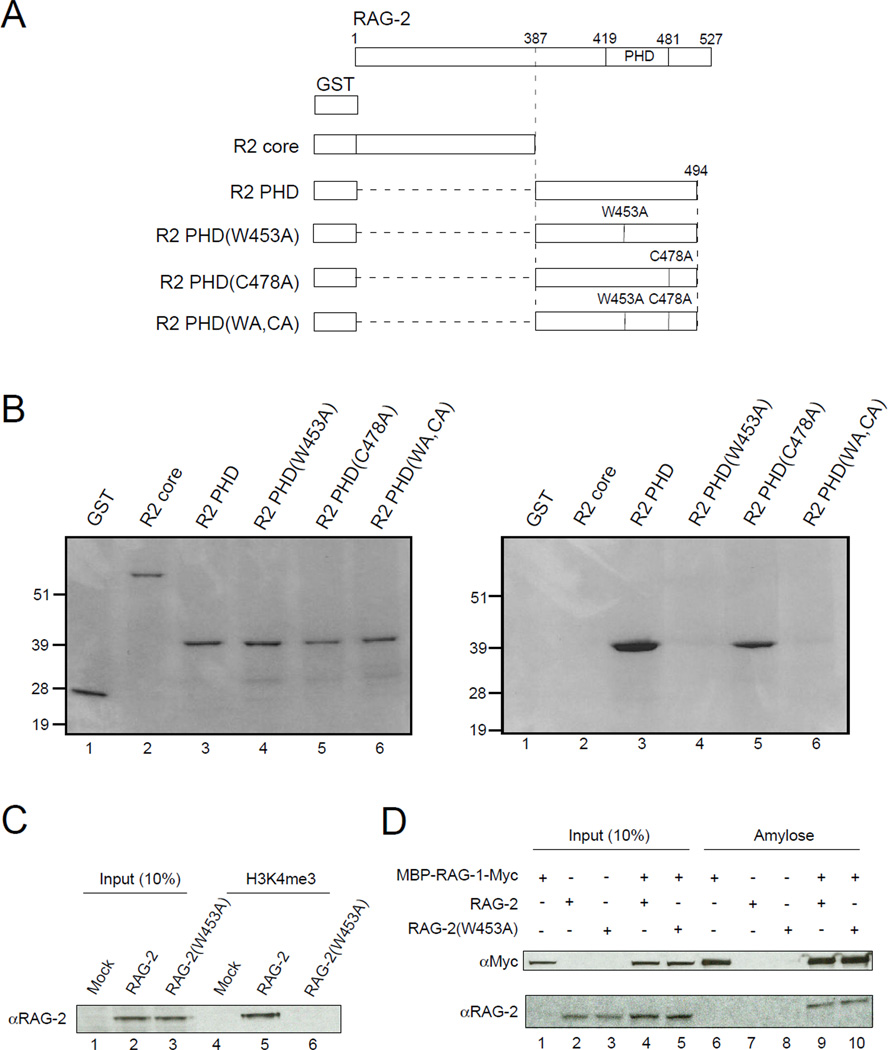
To confirm that RAG-2 binding to H3K4me2 and H3K4me3 is mediated by the non-canonical PHD finger, we tested a series of RAG-2 truncation mutants (Fig. 2A) in the peptide precipitation assay. GST fusion proteins were expressed in bacteria, purified and used in binding assays at similar concentrations (Fig. 2B, left). As shown above, the GST-RAG-2PHD fusion bound the H3K4me3 peptide, while a GST fusion to the RAG-2 core (residues 1 – 387) did not (Fig. 2B, right, lanes 2 and 3). Sequence comparison reveals that residues corresponding to W453 and C478 of RAG-2 are highly conserved among PHD fingers (Ruthenburg et al., 2007). In crystal structures of PHD domains from ING2 (Pena et al., 2006) and NURF (Li et al., 2006), a residue corresponding to RAG-2 W453 recognizes the trimethylammonium group of H3K4me3, while a residue corresponding to RAG-2 C478 coordinates Zn2+. Moreover, mutations at the homologous positions are associated with immunodeficiency syndromes in humans (Gomez et al., 2000; Noordzij et al., 2002; Schwarz et al., 1996; Villa et al., 2001). We introduced single and double alanine substitutions at W453 and C478 of the GST-RAG-2PHD fusion. The W453A mutation, but not the C478A mutation, abolished binding to the H3K4me3 peptide (Fig. 2B, right, lanes 4 and 5); the W453A, C478A double mutant, like the W453A single mutant, failed to bind. These data are consistent with a direct interaction between the non-canonical PHD domain of RAG-2 and histone H3 trimethylated at lysine 4.
Figure 2.
Binding of RAG-2 to tri-Me H3K4 is mediated by the PHD finger. (A) Representation of proteins used in binding assays. All constructs carry an N-terminal GST tag. Full-length RAG-2 is shown at top; vertical lines indicate the C-terminal end of the core domain (387), the boundaries of the PHD domain (419 – 481) and the positions of amino acid substitutions (W453A, C478A). (B) The RAG-2 PHD finger mediates direct binding to tri-Me H3K4. Proteins described in (A) were expressed in E.coli, purified on glutathione-agarose and assayed for binding to biotinylated H3K4me3 peptide immobilized on streptavidin-linked beads. Bound protein (left) or 10% input protein (right) was fractionated by SDS-PAGE and detected by Coomassie blue. (C) Lysates of mock transfected 293T cells or cells expressing full-length wild-type RAG-2 or RAG-2(W453A) were assayed for binding to the biotinylated H3K4me3 peptide as above (lanes 4 – 6). Bound protein was fractionated by SDS-PAGE and detected by immunoblotting for RAG-2. Input protein (10%) was analyzed in parallel (lanes 1 – 3). (D) RAG-2(W453A) retains the ability to associate with RAG-1. Lysates of 293T cells coexpressing a c-myc-tagged MBP-RAG-1 fusion and RAG-2 or RAG-2(W453A) were adsorbed to amylose beads. Bound protein (lanes 6 – 10) was fractionated by SDS PAGE and detected by immunoblotting for c-myc (top) or RAG-2 (bottom). Input protein (10%) was assayed in parallel (lanes 1 – 5).
The effects of these mutations were also tested in the context of full-length RAG-2. Cells were transfected with wild-type RAG-2 or the RAG-2(W453A) mutant and protein was precipitated from cell lysates by H3K4me3 peptide affixed to beads. The W453A mutation abolished binding to H3K4me3 peptide in this assay (Fig. 2C, compare lanes 5 and 6). Importantly, the RAG-2(W453A) mutant retained the ability to associate with RAG-1 in transfected cells (Fig 2D), suggesting that this mutation does not disrupt the overall structure of the protein.
Association between endogenous RAG-2 and hypermethylated histone H3 K4 from primary pro-B cells
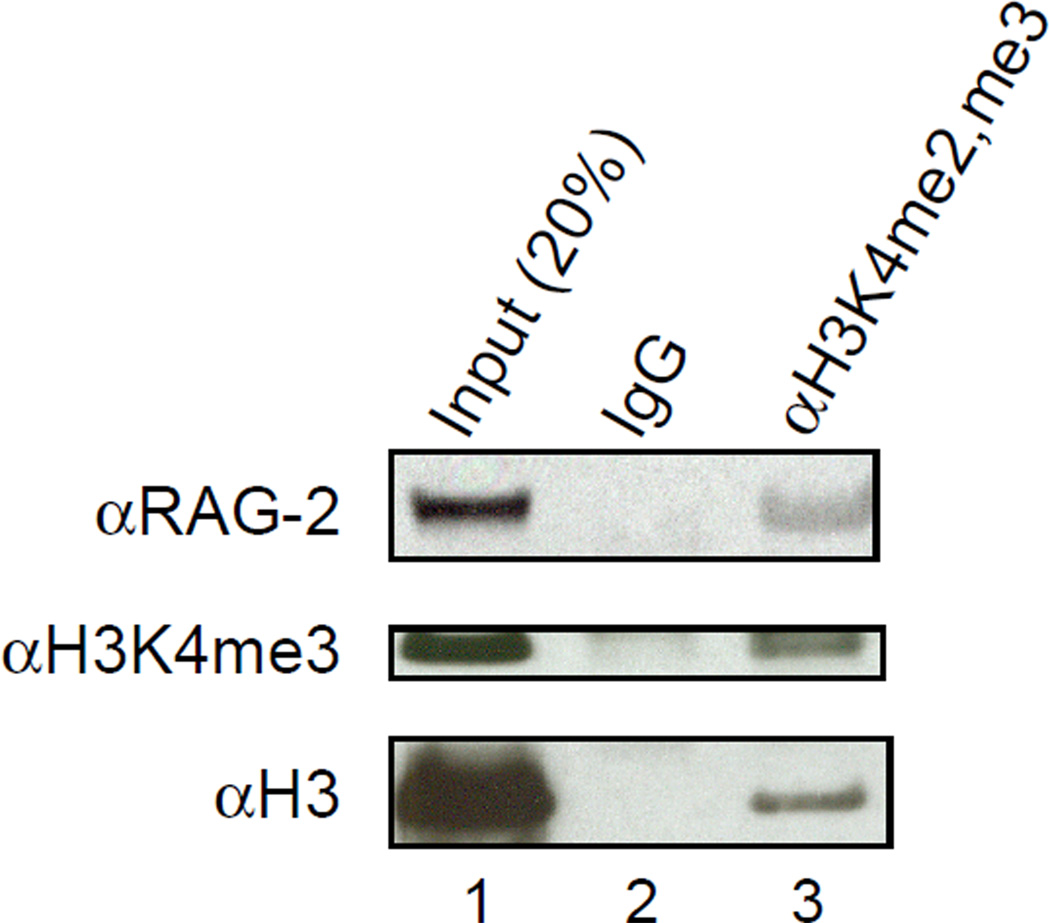
We next sought to detect an association between endogenous RAG-2 and histone H3 containing di- or trimethylated lysine 4. In mice bearing a homozygous, targeted mutation of the IgM transmembrane region (μMT−/−), RAG-expressing B cell progenitors accumulate at the pro-B-to-pre-B transition (Kitamura et al., 1991). Bone marrow cells were collected from 6-week-old, μMT−/− mice and incubated in the presence of IL-7 for 10 d to obtain a homogeneous culture of CD43+B220+ pro-B cells. Protein was immunoprecipitated from cell lysates with an antibody specific for the di- and tri-Me K4 forms of histone H3. Precipitation of histone H3 bearing trimethylated K4 was monitored by immunoblotting (Fig. 3, middle, lane 3). Endogenous RAG-2 co-immunoprecipitated with histone H3 bearing di- and tri-methyl K4, but was not detected in a control immunoprecipitate (Fig. 3, top, lanes 2 and 3). We infer that a portion of RAG-2 is associated in vivo with histone H3 containing di- or trimethylated lysine 4.
Figure 3.
Pro-B cell lysates contain RAG-2 bound to histone H3 di- or trimethylated at lysine 4. Bone marrow cells were cultured in IL-7 for 10 d to generate B220+CD43+ pro-B cells. Protein was immunoprecipitated from pro-B cell lysates with an antibody specific for H3K4me2 and H3K4me3 or with an isotype control. Immunoprecipitates (lanes 2 and 3) were fractionated by SDS-PAGE and assayed by immunoblotting for RAG-2 (top), H3K4me3 (middle) and histone H3 (bottom). Input protein (20%) was assayed as a reference (lane 1).
Binding of the RAG-2 PHD finger to chromatin in the IgH D–JH-C region is correlated with the density of trimethylated histone H3 K4
Hypermethylated histone H3 K4marks active chromatin domains and is associated with antigen receptor gene segments that are poised to undergo V(D)J recombination (Goldmit et al., 2005; Morshead et al., 2003). The ability of the RAG-2 PHD finger to bind H3K4me2 and H3K4me3 suggested that such modifications might direct association of the PHD finger with chromatin in the context of an antigen receptor locus. To determine whether the RAG-2 PHD domain recognized modified histones associated with recombinationally active antigen receptor loci, we used a modified chromatin immunoprecipitation (ChIP) procedure. Sheared chromatin was precipitated with purified GST-RAG-2PHD or GST-RAG-2PHD(W453A), followed by quantitative PCR analysis with primers directed to specific regions of the germline IgH locus (Fig. 4A). We chose a set of amplicons that showed distinct association patterns with H3K4me2 and H3K4me3, reasoning that such a set may also test the selectivity of the RAG-2 PHD domain for each modification. Using chromatin prepared from the RAG-2-deficient pro-B cell line 63-12 (Shinkai et al., 1992), we found that DFL16.1 and Cμ amplicons were enriched in anti-H3K4me2 immunoprecipitates compared to amplicons centered over JH2 and JH4 (Fig. 4B, left). Conversely, JH2 and JH4 amplicons were enriched in anti-H3K4me3 immunoprecipitates (Fig. 4B, middle). Cγ3 and γ-actin promoter primers served as negative and positive controls. In these experiments we observed a striking correspondence between the pattern of amplicon enrichment in chromatin precipitated with GST-RAG-2PHD and anti-H3K4me3 immunoprecipitation (Fig. 4B, right). We observed no specific binding to the GST-RAG-2PHD(W453A) mutant. These results are consistent with a higher affinity of the RAG-2 PHD finger for trimethylated versus dimethylated histone H3 lysine 4 in the context of cellular chromatin, and suggest that this modification may play a role in directing the recombinase to poised antigen receptor loci. Taken together our observations suggest that trimethylation of H3 K4 plays a dominant role in defining the specificity with which the RAG-2 PHD finger binds cellular chromatin.
Figure 4.
Binding of the RAG-2 PHD finger to chromatin in the IgH D–JH-C region is correlated with the density of trimethylated histone H3 K4. (A) Schematic representation of the IgH D–JH-C region through the Cγ3 exons. Gray boxes, D gene segments. Black vertical lines, JH gene segments; open boxes, Cμ, Cδ and Cγ3 exons. Black oval, Eμ intronic enhancer. Black dots below indicate the approximate locations of primer pairs for the segments indicated. (B) Precipitation of chromatin from the RAG-2-deficient 63-12 cell line with an anti-H3K4me2 antibody (left), an anti-H3K4me3 (middle) antibody, wild-type GST-RAG-2PHD (right, open bars) or the mutant GST-RAG-2PHD(W453A) (right, filled bars). DNA recovered from chromatin precipitates was analyzed by quantitative real-time PCR (qRT-PCR). The fold enrichment of each amplified DNA fragment, indicated below as in (A), was determined relative to input DNA and then normalized to that of α-actin. The promoter region of γ-actin was amplified as a positive control. Error bars indicate standard deviations (n = 3).
Disruption of H3K4me3 binding by the RAG-2 PHD finger is associated with impaired recombination of endogenous Ig gene segments and replicative extrachromosomal substrates
Having found that the W453A mutation abolished binding of the RAG-2 PHD finger to H3K4me3 and to chromatin at the D–JH-C locus, we wished to test the effect of this mutation on V(D)J recombination. Because endogenous antigen receptor gene rearrangements are subject to epigenetic constraints, we tested the ability of the RAG-2(W453A) mutant to support endogenous IgH gene rearrangement. We expressed wild-type RAG-2 or RAG-2(W453A) in a RAG-2-deficient pro-B cell line using a dual-expression lentiviral vector that produces GFP from a cassette residing downstream of an internal ribosomal entry site (Fig. 5A). To assess recombination of the endogenous IgH locus in transduced pro-B cells, genomic DNA was harvested 8 d after infection and joining of DFL16.1 and DSP2 gene segments to JH1 through 4 was detected using the polymerase chain reaction (PCR) and Southern hybridization as described. Rearrangements of both D segment families were readily detected in cells transduced with wild-type RAG-2 (Fig. 5B, lanes 3). In contrast, DFL16.1-to-JH rearrangements were undetectable and DSP2-to-JH rearrangements were profoundly reduced in cells transduced with RAG-2(W453A) (Fig. 5B, lanes 4). To eliminate the possibility that W453A renders RAG-2 catalytically inactive, we tested the ability of RAG-2(W453A) to support RSS-dependent DNA cleavage in vitro (Bergeron et al., 2006). In the presence of core RAG-1, equivalent amounts of wild-type RAG-2 and RAG-2(W453A) supported similar levels of nicking and transesterification on 12-RSS and 23-RSS substrates (Fig. 5C). The correlation between loss of H3K4me2 and H3K4me3 binding and impaired rearrangement of endogenous Ig gene segments observed for the RAG-2(W453A) mutant is consistent with the interpretation that recognition of hypermethylated histone H3 K4 is required for the support of efficient V(D)J recombination by full-length RAG-2.
Figure 5.
A mutation abolishing binding of H3K4me3 by the RAG-2 PHD finger impairs recombination of endogenous immunoglobulin gene segments in pro-B cells. (A) RAG-2(W453A) retains catalytic activity in vitro. Core RAG-1 (cR1) and complexes of core RAG-1 with core RAG-2 (cR2), full-length RAG-2 (fR2) or full-length RAG-2(W453A) (fR2[WA]) were assayed in vitro for cleavage of 12-RSS (lanes 1 – 5) or 23-RSS (lanes 6 – 10) substrates; (−), no protein added. Positions of nicked and hairpin products are indicated. (B) EGFP expression in infected cells. RAG-2−/− R2FL63-12 pro-B cells were infected with control lentivirus or with a lentivirus expressing wild-type RAG-2 or RAG-2(W453A). Infection was confirmed by flow cytometric detection of EGFP, which was encoded by all three lentiviruses, at 2 d after infection. Numbers indicate percentages of cells in gated areas. RAG-2 mRNA was assayed by qRT-PCR amplification of reverse transcripts from total RNA isolated 8 d after infection. The amounts of RAG-2 mRNA in cells infected with control, RAG-2 or RAG-2(W453A) lentiviruses, relative to γ-actin mRNA, were 0.00, 1.00 and 3.47, respectively. (C) Detection of completed D-to-JH rearrangements. D-to-JH rearrangements were assayed 8 d after infection by PCR using genomic DNA from control- or RAG-2-infected pro-B cells as template as indicated above. Amplification was performed in the absence of template in parallel (- template). Forward primers were specific for DFL16.1 (top panel) or DSP2 (middle panel); the reverse primer initiates synthesis 3’ of JH4. Products were separated by gel electrophoresis and detected by hybridization to a radiolabeled, locus-specific probe that recognizes all JH segments. Genomic DNA from C57BL/6 mouse spleen was used as a positive control for D-to-JH rearrangements. The β-globin locus (bottom panel) was amplified as a control for gel loading.
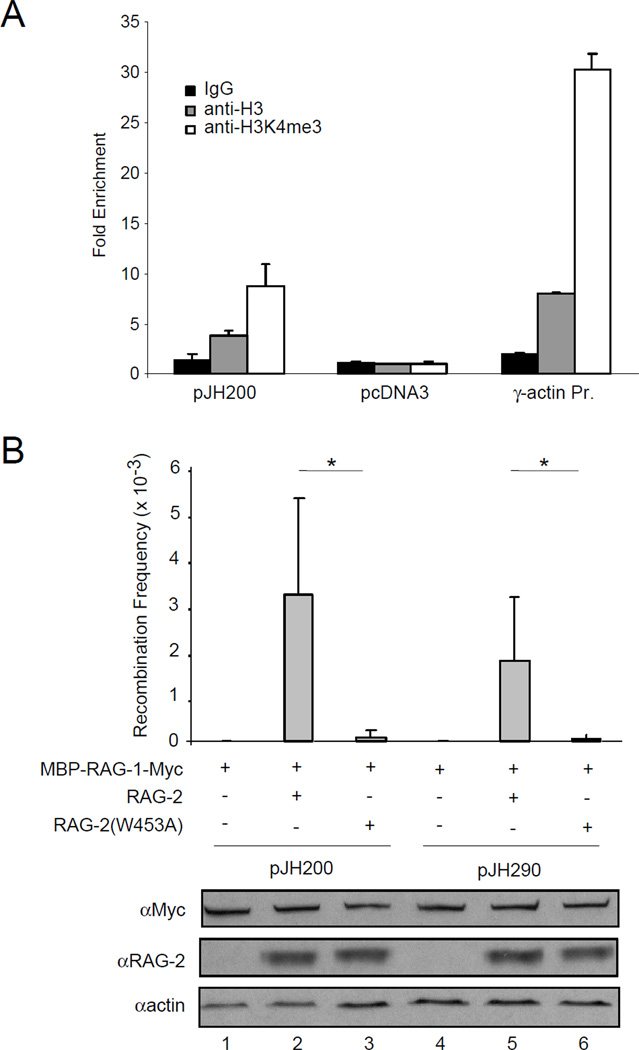
We also examined the effect of the W453A mutation on rearrangement of the extrachromosomal substrates pJH200 and pJH290, which report formation of signal joints and coding joints, respectively (Hesse et al., 1987; Lieber et al., 1988), in NIH3T3 cells. In cells of murine origin these plasmids replicate and are organized into chromatin (Baumann et al., 2003). Moreover, using ChIP we found that in NIH3T3 cells pJH200 is associated with histone H3 and H3K4me3, while a non-replicative plasmid, pcDNA3, showed no such association (Fig. 6A). The RAG-2(W453A) mutation resulted in a 30-fold decrease in the frequency of signal (pJH200, p < 0.03) and coding (pJH290, p < 0.04) joint formation (Fig. 6B, upper panel). This is in agreement with the effect of a W453R mutation on rearrangement of extrachromosomal substrates (Elkin et al., 2005). In contrast, complete removal of the RAG-2 non-core region reduced rearrangement of pJH200 by only 3.8-fold and of pJH290 by 2.1-fold (data not shown), consistent with published results (Cuomo and Oettinger, 1994; Sadofsky et al., 1994). Possible reasons for the difference in activity between core RAG-2 and RAG-2(W453A) are discussed in the next section. Moreover, the accumulation of RAG-2(W453A) (Fig. 6B, bottom panel) and its association with RAG-1 were similar to wild-type RAG-2. Together with the ability of RAG-2(W453A) to catalyze DNA cleavage in vitro, these observations suggest that the effect of the W453A mutation on recombination is more directly related to its debilitating effect on binding to hypermethylated histone H3 lysine 4.
Figure 6.
A mutation abolishing binding of H3K4me3 by the RAG-2 PHD finger impairs recombination of extrachromosomal substrates. (A) Association of pJH200 with histone H3 and H3K4me3 in NIH3T3 cells. Chromatin was precipitated from transfected cells with control IgG (filled bar), an anti-H3 antibody (shaded bar) or an anti-H3K4me3 antibody (open bar) and detected by qRT-PCR. Plasmid pJH200 was detected by amplification across the 12-spacer RSS; the cotransfected, non-replicative plasmid pcDNA3 was assayed by amplification of neo. The endogenous promoter region of γ-actin was amplified as a positive control. The fold enrichment of each amplified DNA fragment was determined relative to input DNA and then normalized to that of the f1 origin region of pcDNA3. Error bars indicate standard deviations (n = 2). (B) Plasmids encoding a c-myc-tagged MBP-RAG-1 fusion and RAG-2 or RAG-2(W453A) were transiently cotransfected with the recombination substrate pJH200 or pJH290 into NIH3T3 cells. In control experiments plasmid pcDNA3 was transfected instead of the RAG-2 encoding plasmid. Plasmids were introduced in the combinations indicated. At 48 hr after transfection plasmids were recovered and transfected into E. coli; transfectants were scored for resistance to ampicillin (Ampr) or ampicillin and chloramphenicol (Ampr + Camr). Recombination frequency (mean ± S.D., n = 2) was calculated as the ratio of double to single resistant colonies (Ampr + Camr/Ampr). (*), P < 0.04, two-tailed t-test. Cell lysates were analyzed by immunoblotting for RAG-1 (c-myc), RAG-2 or actin as indicated.
Discussion
Trimethylation of histone H3 at lysine 4 is an evolutionarily conserved posttranslational modification associated with transcription start sites in yeast and metazoans (Sims and Reinberg, 2006). H3K4me3 is specifically bound by a methyl-lysine-binding domain, the PHD finger, which has been found in subunits of a number of multiprotein complexes that execute the posttranslational modification of histones. These include the ACF1 component of the ATP-dependent chromatin assembly factor (ACF) chromatin remodeling complex (Eberharter et al., 2004), the histone acetyltransferases CBP and p300 (Kalkhoven et al., 2002), ING1 and 2, which are associated with Sin3/HDAC1/2 histone deacetylase complexes (Shi et al., 2006), and ING 3, which is a part of the NuA4 histone acetyltransferase complex (Doyon et al., 2006). In these instances the PHD finger may promote region-specific chromatin modifications through recognition of H3K4me3 (Ruthenburg et al., 2007).
We have shown here that a non-canonical PHD finger within RAG-2 is also capable of specific binding to H3K4me3, and have provided evidence that this interaction supports the specific association of RAG-2 with subregions within the IgH locus in chromatin from cells poised to undergo D-to-JH rearrangement. Our results indicate that the patterns of H3K4me2 and H3K4me3 over the IgH locus in pro-B cells are distinct, and suggest that the density of H3K4me3 determines binding of the RAG-2 PHD finger to chromatin at the IgH D–JH-C locus. Moreover, a point mutation that abolishes specific binding to hypermethylated H3 K4 also profoundly impairs recombination of endogenous gene segments in pro-B cells. These results shed light on the contributions of the RAG-2 non-core region to V(D)J recombination, and raise several questions concerning the function of the RAG-2 PHD finger. We will consider these issues below in the context of locus specificity.
At the level of unchromatinized DNA, the V(D)J recombinase is targeted to antigen receptor gene segments by means of specific interactions with flanking recombination signal sequences (RSS), and this recognition does not require the non-core regions of RAG-1 or RAG-2. Not all RSSs support recombination with the same efficiency, and sequence variation among RSSs can affect gene segment usage (Feeney, 2000). Nonetheless, these differences do not account for the tightly regulated locus specificity that V(D)J recombination exhibits with respect to lymphoid lineage and developmental stage. Rather, ordered rearrangement of antigen receptor gene segments is associated with the imposition or relief of epigenetic marks including changes in histone methylation. As in the case of transcriptional activation, different methylation states may have distinct functional consequences. In particular, the relative distributions of H3K4me2 and H3K4me3 at antigen receptor loci as a function of recombination activity have not yet been examined systematically.
Several amino acid substitutions identified as causes of severe combined immune deficiency (SCID) or Omenn syndrome in humans (Gomez et al., 2000; Noordzij et al., 2002; Schwarz et al., 1996; Villa et al., 1998; Villa et al., 2001), including W453R, N474S, C478F and H481P, reside within the RAG-2 PHD finger. One of these mutations, W453R, unambiguously impairs signal and coding joint formation in an extrachromosomal assay for recombination (Elkin et al., 2005). We have shown here that an alanine substitition at W453 eliminates binding of the RAG-2 PHD finger to hypermethylated H3 K4 and correspondingly to chromatin at the IgH D–JH-C locus in pro-B cells. The W453A mutation, like the W453R mutation, impairs recombination of extrachromosomal substrates. Importantly, we observed a similar impairment of D-to-JH joining at endogenous loci in pro-B cells, despite the fact that RAG-2(W453A) retains catalytic function in vitro, and that accumulation of this mutant and its association with RAG-1 are similar to wild-type RAG-2. The susceptibility of extrachromosomal substrates to the debilitating effect of the W453A mutation may be explained by the fact that these substrates replicate in permissive cells, where they are associated with chromatin (Baumann et al., 2003) and more specifically with histone H3 bearing trimethylated lysine 4. Thus we favor the interpretation that impaired recombination in the context of full-length RAG-2 is due to loss of H3K4me3 binding.
This result seemed paradoxical for several reasons. First, complete removal of the RAG-2 non-core region impairs recombination by only 2-to-3-fold in extrachromosomal assays (Cuomo and Oettinger, 1994; Sadofsky et al., 1994), as we confirmed. Second, although a knock-in mutation that removes the RAG-2 non-core region is associated with impaired rearrangement of VH gene segments, there was little or no effect on D-to-JH joining (Akamatsu et al., 2003; Liang et al., 2002). Third, enforced expression of core RAG proteins in R2FL63-12 pro-B cells rescues the defect in D-to-JH joining (Kirch et al., 1998), in contrast to the impaired ability of the full-length RAG-2(W453A) mutant to support recombination.
An economical way to reconcile the behavior of core RAG-2 and the RAG-2(W453A) mutant is to propose that the non-core region contains an inhibitory domain whose function is relieved by binding of the PHD finger to H3K4me3. In this model, impairment of binding by the PHD finger, such as occurs in the RAG-2(W453A) mutant, would result in constitutive inhibition of RAG-2 activity at chromatinized substrates in vivo. Conversely, wholesale removal of the non-core region, aside from its effects on programmed degradation, would produce a constitutively active form of RAG-2 in which activity is uncoupled from interaction with H3K4me3. In this model, hypermethylated H3 K4 would not simply represent a docking site for the RAG proteins, but would play an active role through allosteric activation of recombinase function.
The ability of the RAG-2 PHD finger to bind hypermethylated histone H3 K4 in general is in agreement with its ability to interact with chromatin at loci other than antigen receptor genes, such as γ-actin. Clearly, modes of regulation other than recognition of hypermethylated histone H3 K4 must determine the locus specificity of recombinase activity. One possibility is that RSS recognition by the RAG heterodimer ensures productive binding only to antigen receptor gene segments. Another possibility, not exclusive of the first, is that specificity is determined by chromatin modifications other than or in addition to hypermethylation on H3 K4. Regardless of such additional specifiers, the recruitment of the RAG complex to the vicinity of poised antigen receptor loci by hypermethylated H3 K4 may itself enhance recombination through allosteric effects on activity against chromatinized substrates, as argued above, but also by increasing the local availability of the recombinase.
Experimental Procedures
Antibodies, Expression Constructs, Cell Culture, GST fusion proteins and Surface Plasmon Resonance assays are described under Supplemental Data.
Protein binding assays
Biotinylated peptides derived from histone H3 were purchased from Upstate Biotechnology. Whole cell lysates were prepared from 293T cells by lysis in a buffer containing 50 mM Tris (pH 7.5), 300 mM NaCl, 1 mM PMSF, 1% NP-40, 1% deoxycholic acid, 0.1% SDS, and a cocktail of protease inhibitors (Roche). After centrifugation at 16,000 g for 15 min, the supernatant was diluted 10-fold in binding buffer (50 mM Tris [pH 7.5], 300 mM NaCl, 1 mM PMSF and protease inhibitors). The diluted supernatant, containing 300 µg protein, was precleared with 50 µl of a 50% streptavidin agarose slurry (Novagen) and incubated with 5 µg peptide prebound to streptavidin agarose for 4 hr at 4°C. Beads were washed three times with binding buffer supplemented with 0.1% NP-40.
Yeast histones were prepared by acid extraction (Edmondson et al., 1996) from wild-type or set1-null strains of S. cerevesiae (Park et al., 2002), provided by Dr. Jef D. Boeke. For binding assays to yeast histone H3, 15 µg of acid extracted yeast histones were incubated for 4 hr at 4°C in binding buffer supplemented with 0.1% NP-40 with 2.5 µg wild-type or mutant GST-tagged RAG-2PHD protein that had been pre-adsorbed to glutathione-agarose. Beads were washed three times with binding buffer supplemented with 0.1% NP-40.
To detect association of wild-type and mutant RAG-2 with MBP-tagged RAG-1-myc-His, 293T cells transfected with the corresponding expression constructs were lysed and complexes were isolated by amylose affinity chromatography as described (Jiang et al., 2004).
Immunoprecipitation
Primary pro-B cells were lysed at 4°C for 1 hr in a buffer containing 50 mM Tris (pH 7.5), 300 mM NaCl, 1 mM PMSF, 1% NP-40, 1% deoxycholic acid, 0.1% SDS, and a cocktail of protease inhibitors (Roche). After centrifugation for 15 min at 16,000 g the supernatant, containing about 500 µg protein, was diluted 10-fold in immunoprecipitation buffer (50 mM Tris [pH 7.5], 300 mM NaCl, 1 mM PMSF and protease inhibitors) and precleared with 50 µl protein A/G agarose (50% slurry). The precleared supernatant was incubated for 4 hr at 4°C with 10 µg antibody. Protein A/G agarose beads (50 µl of a 50% slurry) were added and incubation, with rocking, was continued for another 2 hr at 4°C. Following centrifugation for 1 min at 200 g, beads were collected and washed three times for 10 min each with 1 ml immunoprecipitation buffer containing 0.1% NP-40. Pellets were suspended in SDS loading buffer, heated to 95°C for 5 min and fractionated by SDS-PAGE on a 4 – 12% Bis-Tris gel in NuPAGE MES running buffer (Invitrogen).
Chromatin precipitation assays
Assays were performed using a Chromatin Immunoprecipitation Assay Kit (Upstate, 17–295) according to the manufacturer’s instructions. Between 4 – 5 × 106 63-12 pro-B cells were used for each assay. For precipitation by the RAG-2 PHD domain, 10 µg of purified GST fusion protein and 50 µl glutathione-agarose (75% slurry) were used in each assay. For chromatin immunoprecipitation (ChIP) experiments, 10 µg antibody and 60 µl protein A/G agarose (50% slurry) were used in each assay. For ChIP assays of plasmid DNA, 10 µg pcDNA3 and 4 µg of pJH200 were cotransfected into NIH3T3 cells. At 40 – 48 hr after transfection, ChIP assays were performed as described above using 1 × 106 cells for each assay. Recovered DNA was amplified by quantitative real-time PCR with a SYBR Green PCR Master Mix (Bio-Rad), using a Bio-Rad iCycler iQ. Primer sequences are given in Supplemental Data.
In vitro DNA cleavage assays
RAG fusion proteins were purified by amylose affinity chromatography as described (Bergeron et al., 2006). DNA cleavage reactions were performed in Mg2+ using the 12-RSS substrate DAR39/DAR40 or the 23-RSS substrate DAR61/DAR62 as described (Bergeron et al., 2006) except that core RAG-1 and full-length RAG-2 or RAG-2(W453A) were added at 400 ng each. Reactions were carried out at 37°C for 30 min.
Extrachromosomal assays for V(D)J recombination
Assays were performed as described (Hesse et al., 1987) with slight modification. Briefly, plasmids (10 µg) encoding MBP-RAG-1-myc-His and RAG-2 or RAG-2(W453A) were cotransfected with 4 µg pJH200 or pJH290 into NIH3T3 cells and plasmid DNA was isolated 40 – 48 hr after transfection. E. coli DH5α was transformed with 1 µl (about 40 ng) plasmid DNA and 1.7% of each transformation mix was plated on LB agar containing 50 µg/ml ampicillin; the remainder was plated on LB agar containing 50 µg/ml ampicillin and 12.5 µg/ml chloramphenicol. Plates were scored after 14 – 16 hr at 37°C.
Retroviral transduction
Lentiviral particles coexpressing EGFP and RAG-2 or RAG-2(W453A) were generated as described (Stewart et al., 2003). Briefly, 293T cells were seeded 24 hr before transfection at 2 × 106 cells per 10 cm plate. The empty pWPI vector, pWPI-RAG-2 or pWPI-RAG-2(W453A) were cotransfected with the helper plasmids pΔ8.2R and pVSVG using FuGene 6 (Roche). Medium was replaced 24 hr after transfection and supernatant was collected at 48 and 72 hr after transfection. Virus was concentrated by ultracentrifugation over a cushion of 20% sucrose in a SW32.1Ti rotor for 2 hr at 25,000 rpm and 20°C. RAG-2−/− R2FL63-12 pro-B cells were infected with freshly prepared control, RAG-2-expressing or RAG-2(W453A)-expressing lentivirus by spin inoculation in the presence of 10 µg/ml polybrene. Infection was confirmed by flow cytometric detection of EGFP at 2 d after infection. Genomic DNA and total RNA were isolated from infected cells at 8 d after infection (DNeasy and RNeasy, Qiagen).
Assay for endogenous D-to-JH recombination
A PCR-based assay was used to detect endogenous D-to-JH recombination. Genomic DNA from transduced cells (90 ng) was amplified using forward primers specific for DFL16.1 or DSP2 and a reverse primer that initiates synthesis from a site 3’ of JH4. Genomic DNA from C57BL/6 mouse spleen was used as a positive control template. Amplification was performed using the following protocol: 1 cycle at 95°C for 5 min; 33 cycles of 94°C for 1 min, 65°C for 1 min and 72°C for 3 min; 1 cycle at 72°C for 7 min. Products were detected by Southern hybridization to a degenerate oligonucleotide probe that recognizes all JH segments. The genomic DNA samples were normalized for quality and quantity by PCR followed by Southern hybridization to a probe specific for mouse β-globin. Primers and probes are defined in Supplemental Data.
Supplementary Material
Acknowledgements
We thank Jef Boeke (Johns Hopkins University) for providing set1-null S. cerevesiae, Robert Weinberg (Whitehead Institute) for providing the lentiviral expression system and Yuan Lin (Johns Hopkins University) for assistance with surface plasmon resonance measurements. This work was supported by grant CA16519 from the National Cancer Institute, by a gift to the Institute for Cell Engineering at the Johns Hopkins University School of Medicine and by the Intramural Research Program of the National Institute on Aging, Baltimore, MD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akamatsu Y, Monroe R, Dudley DD, Elkin SK, Gartner F, Talukder SR, Takahama Y, Alt FW, Bassing CH, Oettinger MA. Deletion of the RAG2 C terminus leads to impaired lymphoid development in mice. Proc Natl Acad Sci U S A. 2003;100:1209–1214. doi: 10.1073/pnas.0237043100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akamatsu Y, Oettinger MA. Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequences. Mol Cell Biol. 1998;18:4670–4678. doi: 10.1128/mcb.18.8.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt FW, Yancopoulos GD, Blackwell TK, Wood C, Thomas E, Boss M, Coffman R, Rosenberg N, Tonegawa S, Baltimore D. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 1984;3:1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann M, Mamais A, McBlane F, Xiao H, Boyes J. Regulation of V(D)J recombination by nucleosome positioning at recombination signal sequences. EMBO J. 2003;22:5197–5207. doi: 10.1093/emboj/cdg487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron S, Anderson DK, Swanson PC. RAG and HMGB1 proteins: purification and biochemical analysis of recombination signal complexes. Methods Enzymol. 2006;408:511–528. doi: 10.1016/S0076-6879(06)08032-3. [DOI] [PubMed] [Google Scholar]
- Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci. 2006;31:35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callebaut I, Mornon JP. The V(D)J recombination activating protein RAG2 consists of a six-bladed propeller and a PHD fingerlike domain, as revealed by sequence analysis. Cell Mol Life Sci. 1998;54:880–891. doi: 10.1007/s000180050216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury D, Sen R. Stepwise activation of the immunoglobulin mu heavy chain gene locus. EMBO J. 2001;20:6394–6403. doi: 10.1093/emboj/20.22.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury D, Sen R. Transient IL-7/IL-7R signaling provides a mechanism for feedback inhibition of immunoglobulin heavy chain gene rearrangements. Immunity. 2003;18:229–241. doi: 10.1016/s1074-7613(03)00030-x. [DOI] [PubMed] [Google Scholar]
- Cobb RM, Oestreich KJ, Osipovich OA, Oltz EM. Accessibility control of V(D)J recombination. Adv Immunol. 2006;91:45–109. doi: 10.1016/S0065-2776(06)91002-5. [DOI] [PubMed] [Google Scholar]
- Cuomo CA, Oettinger MA. Analysis of regions of RAG-2 important for V(D)J recombination. Nucleic Acids Res. 1994;22:1810–1814. doi: 10.1093/nar/22.10.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio MJ, McMahan CJ, Eastman QM, Spanopoulou E, Schatz DG. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell. 1996;87:253–262. doi: 10.1016/s0092-8674(00)81343-4. [DOI] [PubMed] [Google Scholar]
- Doyon Y, Cayrou C, Ullah M, Landry AJ, Cote V, Selleck W, Lane WS, Tan S, Yang XJ, Cote J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Eastman QM, Villey IJ, Schatz DG. Detection of RAG protein-V(D)J recombination signal interactions near the site of DNA cleavage by UV cross-linking. Mol Cell Biol. 1999;19:3788–3797. doi: 10.1128/mcb.19.5.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A, Vetter I, Ferreira R, Becker PB. ACF1 improves the effectiveness of nucleosome mobilization by ISWI through PHD-histone contacts. EMBO J. 2004;23:4029–4039. doi: 10.1038/sj.emboj.7600382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson DG, Smith MM, Roth SY. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- Elkin SK, Ivanov D, Ewalt M, Ferguson CG, Hyberts SG, Sun ZY, Prestwich GD, Yuan J, Wagner G, Oettinger MA, Gozani OP. A PHD finger motif in the C terminus of RAG2 modulates recombination activity. J Biol Chem. 2005;280:28701–28710. doi: 10.1074/jbc.M504731200. [DOI] [PubMed] [Google Scholar]
- Feeney AJ. Factors that influence formation of B cell repertoire. Immunol Res. 2000;21:195–202. doi: 10.1385/IR:21:2-3:195. [DOI] [PubMed] [Google Scholar]
- Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol. 2000a;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- Fugmann SD, Villey IJ, Ptaszek LM, Schatz DG. Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol Cell. 2000b;5:97–107. doi: 10.1016/s1097-2765(00)80406-2. [DOI] [PubMed] [Google Scholar]
- Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- Goldmit M, Ji Y, Skok J, Roldan E, Jung S, Cedar H, Bergman Y. Epigenetic ontogeny of the Igk locus during B cell development. Nat Immunol. 2005;6:198–203. doi: 10.1038/ni1154. [DOI] [PubMed] [Google Scholar]
- Gomez CA, Ptaszek LM, Villa A, Bozzi F, Sobacchi C, Brooks EG, Notarangelo LD, Spanopoulou E, Pan ZQ, Vezzoni P, et al. Mutations in conserved regions of the predicted RAG2 kelch repeats block initiation of V(D)J recombination and result in primary immunodeficiencies. Mol Cell Biol. 2000;20:5653–5664. doi: 10.1128/mcb.20.15.5653-5664.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse JE, Lieber MR, Gellert M, Mizuuchi K. Extrachromosomal DNA substrates in pre-B cells undergo inversion or deletion at immunoglobulin V-(D)-J joining signals. Cell. 1987;49:775–783. doi: 10.1016/0092-8674(87)90615-5. [DOI] [PubMed] [Google Scholar]
- Hiom K, Gellert M. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell. 1997;88:65–72. doi: 10.1016/s0092-8674(00)81859-0. [DOI] [PubMed] [Google Scholar]
- Jiang H, Chang FC, Ross AE, Lee J, Nakayama K, Nakayama K, Desiderio S. Ubiquitylation of RAG-2 by Skp2-SCF links destruction of the V(D)J recombinase to the cell cycle. Mol Cell. 2005;18:699–709. doi: 10.1016/j.molcel.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Jiang H, Ross AE, Desiderio S. Cell cycle-dependent accumulation in vivo of transposition-competent complexes between recombination signal ends and full-length RAG proteins. J Biol Chem. 2004;279:8478–8486. doi: 10.1074/jbc.M311219200. [DOI] [PubMed] [Google Scholar]
- Johnson K, Angelin-Duclos C, Park S, Calame KL. Changes in histone acetylation are associated with differences in accessibility of V(H) gene segments to V-DJ recombination during B-cell ontogeny and development. Mol Cell Biol. 2003;23:2438–2450. doi: 10.1128/MCB.23.7.2438-2450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K, Pflugh DL, Yu D, Hesslein DG, Lin KI, Bothwell AL, Thomas-Tikhonenko A, Schatz DG, Calame K. B cell-specific loss of histone 3 lysine 9 methylation in the V(H) locus depends on Pax5. Nat Immunol. 2004;5:853–861. doi: 10.1038/ni1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkhoven E, Teunissen H, Houweling A, Verrijzer CP, Zantema A. The PHD type zinc finger is an integral part of the CBP acetyltransferase domain. Mol Cell Biol. 2002;22:1961–1970. doi: 10.1128/MCB.22.7.1961-1970.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DR, Dai Y, Mundy CL, Yang W, Oettinger MA. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 1999;13:3070–3080. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirch SA, Rathbun GA, Oettinger MA. Dual role of RAG2 in V(D)J recombination: catalysis and regulation of ordered Ig gene assembly. EMBO J. 1998;17:4881–4886. doi: 10.1093/emboj/17.16.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura D, Roes J, Kuhn R, Rajewsky K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature. 1991;350:423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- Landree MA, Wibbenmeyer JA, Roth DB. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13:3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Desiderio S. Cyclin A/CDK2 regulates V(D)J recombination by coordinating RAG-2 accumulation and DNA repair. Immunity. 1999;11:771–781. doi: 10.1016/s1074-7613(00)80151-x. [DOI] [PubMed] [Google Scholar]
- Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI Z, Dordai DI, Lee J, Desiderio S. A conserved degradation signal regulates RAG-2 accumulation during cell division and links V(D)J recombination to the cell cycle. Immunity. 1996;5:575–589. doi: 10.1016/s1074-7613(00)80272-1. [DOI] [PubMed] [Google Scholar]
- Liang HE, Hsu LY, Cado D, Cowell LG, Kelsoe G, Schlissel MS. The "dispensable" portion of RAG2 is necessary for efficient V-to-DJ rearrangement during B and T cell development. Immunity. 2002;17:639–651. doi: 10.1016/s1074-7613(02)00448-x. [DOI] [PubMed] [Google Scholar]
- Lieber MR, Hesse JE, Lewis S, Bosma GC, Rosenberg N, Mizuuchi K, Bosma MJ, Gellert M. The defect in murine severe combined immune deficiency: joining of signal sequences but not coding segments in V(D)J recombination. Cell. 1988;55:7–16. doi: 10.1016/0092-8674(88)90004-9. [DOI] [PubMed] [Google Scholar]
- McMahan CJ, Difilippantonio MJ, Rao N, Spanopoulou E, Schatz DG. A basic motif in the N-terminal region of RAG1 enhances V(D)J recombination activity. Mol Cell Biol. 1997;17:4544–4552. doi: 10.1128/mcb.17.8.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurry MT, Krangel MS. A role for histone acetylation in the developmental regulation of VDJ recombination. Science. 2000;287:495–498. doi: 10.1126/science.287.5452.495. [DOI] [PubMed] [Google Scholar]
- Mellor J. It takes a PHD to read the histone code. Cell. 2006;126:22–24. doi: 10.1016/j.cell.2006.06.028. [DOI] [PubMed] [Google Scholar]
- Morshead KB, Ciccone DN, Taverna SD, Allis CD, Oettinger MA. Antigen receptor loci poised for V(D)J rearrangement are broadly associated with BRG1 and flanked by peaks of histone H3 dimethylated at lysine 4. Proc Natl Acad Sci U S A. 2003;100:11577–11582. doi: 10.1073/pnas.1932643100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordzij JG, de Bruin-Versteeg S, Verkaik NS, Vossen JM, de Groot R, Bernatowska E, Langerak AW, van Gent DC, van Dongen JJ. The immunophenotypic and immunogenotypic B-cell differentiation arrest in bone marrow of RAG-deficient SCID patients corresponds to residual recombination activities of mutated RAG proteins. Blood. 2002;100:2145–2152. [PubMed] [Google Scholar]
- Osipovich O, Milley R, Meade A, Tachibana M, Shinkai Y, Krangel MS, Oltz EM. Targeted inhibition of V(D)J recombination by a histone methyltransferase. Nat Immunol. 2004;5:309–316. doi: 10.1038/ni1042. [DOI] [PubMed] [Google Scholar]
- Park JH, Cosgrove MS, Youngman E, Wolberger C, Boeke JD. A core nucleosome surface crucial for transcriptional silencing. Nat Genet. 2002;32:273–279. doi: 10.1038/ng982. [DOI] [PubMed] [Google Scholar]
- Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins EJ, Kee BL, Ramsden DA. Histone 3 lysine 4 methylation during the pre-B to immature B-cell transition. Nucleic Acids Res. 2004;32:1942–1947. doi: 10.1093/nar/gkh523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JX, Kale SB, Yarnell Schultz H, Roth DB. Separation-of-function mutants reveal critical roles for RAG2 in both the cleavage and joining steps of V(D)J recombination. Mol Cell. 2001;7:77–87. doi: 10.1016/s1097-2765(01)00156-3. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Sadofsky MJ, Hesse JE, Gellert M. Definition of a core region of RAG-2 that is functional in V(D)J recombination. Nucleic Acids Res. 1994;22:1805–1809. doi: 10.1093/nar/22.10.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadofsky MJ, Hesse JE, McBlane JF, Gellert M. Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res. 1993;21:5644–5650. doi: 10.1093/nar/21.24.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz K, Gauss GH, Ludwig L, Pannicke U, Li Z, Lindner D, Friedrich W, Seger RA, Hansen-Hagge TE, Desiderio S, et al. RAG mutations in human B cell-negative SCID. Science. 1996;274:97–99. doi: 10.1126/science.274.5284.97. [DOI] [PubMed] [Google Scholar]
- Sekiguchi JA, Whitlow S, Alt FW. Increased accumulation of hybrid V(D)J joins in cells expressing truncated versus full-length RAGs. Mol Cell. 2001;8:1383–1390. doi: 10.1016/s1097-2765(01)00423-3. [DOI] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Reinberg D. Histone H3 Lys 4 methylation: caught in a bind? Genes Dev. 2006;20:2779–2786. doi: 10.1101/gad.1468206. [DOI] [PubMed] [Google Scholar]
- Steen SB, Han JO, Mundy C, Oettinger MA, Roth DB. Roles of the "dispensable" portions of RAG-1 and RAG-2 in V(D)J recombination. Mol Cell Biol. 1999;19:3010–3017. doi: 10.1128/mcb.19.4.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson PC, Desiderio S. V(D)J recombination signal recognition: distinct, overlapping DNA-protein contacts in complexes containing RAG1 with and without RAG2. Immunity. 1998;9:115–125. doi: 10.1016/s1074-7613(00)80593-2. [DOI] [PubMed] [Google Scholar]
- Swanson PC, Desiderio S. RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol Cell Biol. 1999;19:3674–3683. doi: 10.1128/mcb.19.5.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukder SR, Dudley DD, Alt FW, Takahama Y, Akamatsu Y. Increased frequency of aberrant V(D)J recombination products in core RAG-expressing mice. Nucl. Acids Res. 2004;32:4539–4549. doi: 10.1093/nar/gkh778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa A, Santagata S, Bozzi F, Giliani S, Frattini A, Imberti L, Gatta LB, Ochs HD, Schwarz K, Notarangelo LD, et al. Partial V(D)J recombination activity leads to Omenn syndrome. Cell. 1998;93:885–896. doi: 10.1016/s0092-8674(00)81448-8. [DOI] [PubMed] [Google Scholar]
- Villa A, Sobacchi C, Notarangelo LD, Bozzi F, Abinun M, Abrahamsen TG, Arkwright PD, Baniyash M, Brooks EG, Conley ME, et al. V(D)J recombination defects in lymphocytes due to RAG mutations: severe immunodeficiency with a spectrum of clinical presentations. Blood. 2001;97:81–88. doi: 10.1182/blood.v97.1.81. [DOI] [PubMed] [Google Scholar]
- West KL, Singha NC, De Ioannes P, Lacomis L, Erdjument-Bromage H, Tempst P, Cortes P. A direct interaction between the RAG2 C terminus and the core histones is required for efficient V(D)J recombination. Immunity. 2005;23:203–212. doi: 10.1016/j.immuni.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.