Abstract
Nonsense-mediated mRNA decay and Staufen1-mediated mRNA decay are mechanistically related pathways that serve distinct purposes. In the present article, we give an overview of each pathway. We describe how a factor that is common to both pathways results in their competition. We also explain how competition between the two pathways contributes to the differentiation of C2C12 myoblasts to multinucleated myotubes.
Keywords: eukaryotic initiation factor 4E (eIF4E), exon junction complex (EJC), homoeostatic control of genes, myogenesis, nonsense-mediated mRNA decay (NMD), Staufen1
Introduction
mRNA decay has long been appreciated to constitute a step that is critical for the proper regulation of gene expression (for recent reviews, see [1,2]). In the present paper, we review what is known about the functional significance of two related mRNA decay pathways: NMD (nonsense-mediated mRNA decay) and SMD [STAU1 (Staufen1)-mediated mRNA decay]. We present evidence for the mechanistic convergence of the two pathways in a way that results in their competition. We also illustrate how competition feeds into the complex network of post-transcriptional regulatory steps that leads to skeletal-muscle myoblast maintenance in an undifferentiated state or differentiation to multinucleated myotubes.
NMD
NMD provides a means by which eukaryotic cells control the quality of gene expression: NMD generally eliminates mRNAs that prematurely terminate translation because they harbour either a frameshift or a nonsense mutation (see, e.g., [3–14]). NMD apparently evolved to protect cells from routine inaccuracies in gene expression that result in the production of truncated proteins, which have the potential to acquire dominant-negative or gain-of-function activities that could be deleterious to cells. For example, approximately one-third of alternatively spliced transcripts are NMD targets [15], which findings indicate are largely generated by mistakes made during pre-mRNA splicing [16]. Therefore NMD provides an important mechanism whereby cells ensure the quality of mRNA function and, as a consequence, the quality of gene expression. NMD also eliminates genomic noise, such as non-functional transcripts that have assimilated transposons or retroviral sequences [17]. Additionally, NMD serves regulatory functions by targeting physiological transcripts that harbour, e.g. an upstream open translational reading frame [ORF (open reading frame)] or an intron within their 3′-UTR (3′-untranslated region) [17–19]. NMD further functions in the homoeostatic control of genes that encode serine-arginine-rich proteins and heterogeneous nuclear RNP (ribonucleoprotein) splicing factors [20–28]. Thus NMD degrades both abnormal and normal transcripts, the latter to establish appropriate levels of gene expression.
The importance of NMD in mammals has been inferred from a number of studies. For example, mouse embryos that lack the NMD factor UPF1 are resorbed shortly after implantation, and mouse blastocysts isolated 3.5 days post-coitum that lack UPF1 undergo apoptosis after a brief growth period [29]. As another example, haemopoietic stem and progenitor cells that lack the NMD factor UPF2 fail to proliferate in part because of the up-regulation of a battery of transcripts that include RNAs deriving from processed pseudogenes, non-productive DNA rearrangements and alternative splicing [18]. However, UPF1, which is a member of the RNA helicase superfamily 1 that manifests RNA-dependent ATP hydrolytic and 5′–3′ ATP-dependent unwinding activities in vitro [30–32], and UPF2, the activity of which is less well characterized, function in pathways other than NMD [6]. For example, both UPF1 and UPF2 regulate the binding of telomeric repeat-containing RNA to telomeres and are enriched at telomeres compared with Alu sequences [33]. Furthermore, amino acids of UPF1 that associate with UPF2 during NMD can alternatively bind to STAU1 during SMD so that inhibiting the efficiency of NMD augments the efficiency of SMD ([34], see below). Therefore, although NMD is unquestionably important for cellular growth and maintenance, it has not yet been possible to tease apart critical contributions of NMD from critical contributions of other UPF factor-dependent pathways.
As a rule, NMD in mammalian cells degrades newly synthesized mRNAs during a `pioneer' round of translation. This round of translation utilizes mRNA that is associated with the mostly nuclear yet shuttling CBP (cap-binding protein) heterodimer CBP80–CBP20: studies demonstrating that translational repression is critical for NMD suggest that the pioneer round involves the loading of not only the ribosome that recognizes the nonsense codon but also at least one 43S pre-initiation complex that can be targeted for repression during NMD (Figure 1) [35–42]. Provided the mRNA derives from pre-mRNA splicing, the pioneer translation–initiation complex would also contain a post-splicing EJC (exon junction complex) situated upstream of each exon–exon junction [5,43,44]. The pioneer round precedes subsequent rounds of `steady-state' translation. Steady-state rounds utilize mRNA that is the remodelled product of CBP80–CBP20-bound mRNA so as to contain eIF (eukaryotic initiation factor) 4E at the cap, and they do not detectably support NMD in mammalian cells. The immunity of eIF4E-bound mRNA to NMD can be explained not only because of the lack of CBP80–CBP20 but also because of the absence of detectable EJCs [35–37,40,45,46], which are removed by the pioneer round of translation ([47,48]; H. Sato and L.E. Maquat, unpublished work).
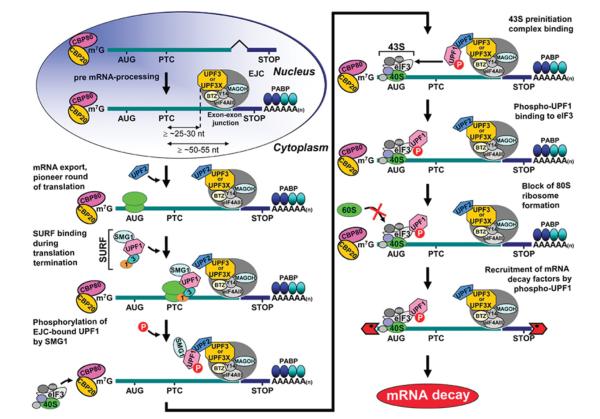
Figure 1. Model for NMD.
In mammals, newly synthesized CPB80–CBP20-bound mRNA is targeted for NMD once mRNA has been generated by pre-mRNA processing and exported from the nucleus to the cytoplasm. During pre-mRNA processing, splicing results in the deposition of an EJC of proteins upstream of mRNA exon–exon junctions. EJC components include eIF4AIII, Y14, MAGOH (mago-nashi homologue), Barentz (BTZ) and many other proteins. The UPF3 (also called UPF3a) or UPF3X (also called UPF3b) NMD factor is mostly nuclear but shuttles to the cytoplasm and is thought to join EJCs in the nucleus so as to be exported with mRNA to the cytoplasm. In the cytoplasm, UPF3 or UPF3X recruits UPF2. The translation of CBP80–CBP20-bound mRNA constitutes the pioneer round. Translation termination during the pioneer round at a premature termination codon (PTC) that is situated ≥50–55 nt upstream of an exon-exon junction (i.e. ≥25–30 nt upstream of an EJC) involves the SURF complex, which consists of the PI3K-related protein kinase that phosphorylates UPF1, SMG1, together with UPF1, eRF1 and eRF3. As a consequence, NMD generally occurs. During the process, UPF1 together with SMG1 is thought to bind EJC-associated UPF2 in a way that is promoted by CBP80. UPF1 binding to the EJC results in UPF1 phosphorylation. Phospho-UPF1 triggers NMD by promoting translational repression of the NMD target. Translational repression involves the binding of phospho-UPF1 to eIF3 within the 43S pre-initiation complex that is poised at the AUG translation initiation codon so as to prevent 60S ribosomal subunit joining. Phospho-UPF1 also promotes NMD by recruiting mRNA degradative activities. Not shown are SMG5, SMG6 and SMG7, which activate UPF1 dephosphorylation and thus recycling. SMG6 appears to additionally function as an endonuclease. Very recently, roles for SMG8 and SMG9 as SMG1-interacting proteins have been defined [49]. Notably, mammalian-cell NMD can also target mRNAs that have not undergone splicing downstream of a PTC, in a mechanism that has been called failsafe NMD or EJC-dependent NMD, provided that they have undergone a splicing event upstream of the PTC [5,8]. Nucleolytic activities are indicated by the red irregular hexagons. PABP, poly(A)-binding protein, where darker shapes specify the largely nuclear PABPN1 and lighter shapes denote the largely cytoplasmic PABPC1; AUG, translation initiation codon; STOP, normal termination codon; 1, eRF1; 3, eRF3.
According to the current model (Figure 1), NMD depends on joining of the PI3K (phosphatidylinositol 3-kinase)-related protein kinase SMG1 and UPF1 with the two translation termination factors eRF1 and eRF3 to form the SURF complex at the site of translation termination [39,49]. If an EJC exists sufficiently downstream of the termination event, then SURF-derived SMG1 and UPF1 are thought to bind to that EJC, possibly via SURF, forming a bridge between the termination codon and EJC, so as to trigger UPF1 phosphorylation. UPF1 phosphorylation then results in translational repression by augmenting UPF1 binding to the eIF3 constituent of a 43S pre-initiation complex that is poised at the mRNA translation initiation codon in a way that inhibits 60S ribosomal subunit joining [38]. UPF1 phosphorylation also activates the recruitment of ribonucleolytic activities [38] that degrade NMD targets from either end, beginning with decapping or deadenylation [50–54]. Recently, NMD has been shown to additionally involve some degree of SMG6-mediated endonucleolytic cleavage [55–57]. Furthermore, NMD can target mRNAs that occur in the absence of splicing occurring downstream of a PTC provided that splicing occurred upstream of the PTC, presumably reflecting the need for a post-splicing EJC situated 5′ to the PTC [8,45].
SMD
SMD, unlike NMD, targets both CBP80–CBP20-bound and eIF4E-bound mRNAs that harbour an SBS (STAU1-binding site) sufficiently downstream of their normal termination codon (Figure 2) and it occurs independently of splicing [36,58]. During SMD, when translation terminates sufficiently upstream of an SBS, UPF1 binding to the SBS via the dsRNA (double-stranded RNA)-binding protein STAU1 is thought to trigger mRNA decay [58,59]. Thus a STAU1-associated SBS functions during SMD analogously to how an EJC-associated exon–exon junction functions during NMD: translation–termination sufficiently upstream of, respectively, SBS-bound STAU1 or the EJC triggers mRNA decay. Since UPF1 binds directly to STAU1, SMD, unlike NMD, does not require EJC constituents (including UPF2, UPF3 or UPF3X) or CBP80–CBP20 [36,58]. Steps in the SMD pathway after UPF1 binds to an SBS via STAU1 may be similar to those in the NMD pathway after UPF1 binds to an EJC via UPF2.
Figure 2. Model for SMD.
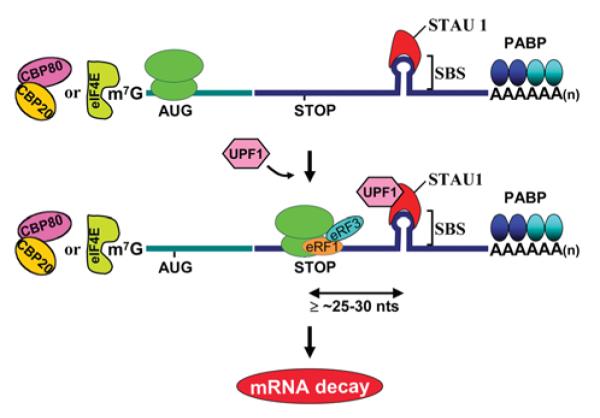
In mammals, SMD targets both newly synthesized CBP80–CBP20-bound mRNAs and the corresponding steady-state eIF4E-bound mRNAs provided they contain an SBS ≥25–30 nt downstream of the normal termination codon. According to current thinking, when translation terminates sufficiently upstream of SBS-bound STAU1, UPF1 binds STAU1. UPF1 binding then triggers mRNA decay, presumably analogously to how UPF1 binding to an EJC during NMD triggers mRNA decay. AUG, translation initiation codon; STOP, normal translation termination codon; PABP, PABPN1 and/or PABPC1 depending on the cap-binding complex.
In contrast with NMD, which serves largely as a quality-control mechanism, SMD provides a means to conditionally down-regulate the expression of genes encoding mRNAs that contain an SBS in their 3′-UTRs. Conditionally regulated pathways that modulate mRNA half-life would be expected to target eIF4E-bound mRNA, which constitutes the bulk of cellular mRNA. While SMD targets were initially identified by microarray analyses of transcripts that bind STAU1 [58], once it was realized that STAU1 binding to an mRNA can elicit the decay of that mRNA, it became apparent that a more sensitive approach to identify SMD targets would use microarrays to assay for mRNAs that are up-regulated upon STAU1 depletion. In three independently performed experiments, conservatively 1.1% of the 11569 HeLa-cell transcripts that were analysed were found to be up-regulated upon STAU1 down-regulation, and a number of these have been proven to be SMD targets based on studies that demonstrate STAU1 binding to their 3′-UTRs in a way that shortens mRNA half-life [59]. To date, the SBS of human ARF1 (ADP-ribosylation factor 1) mRNA is the best-characterized SBS, consisting of a 19-bp stem-loop structure in which base-pairing appears to be more critical for STAU1 binding than the precise constitution of the base-paired region [59]. Nevertheless, the exact nature of this and other SBSs has yet to be defined.
As for NMD, it is difficult to tease apart the importance of SMD to mammalian-cell metabolism since, as noted, the efficiency of SMD is influenced by the efficiency of NMD. Furthermore, STAU1 functions not only in SMD but also in other capacities. As examples, STAU1 is involved in dendritic RNP localization that when inhibited correlates with impaired dendritic outgrowth and spine formation [60]. Additionally, STAU1 that has been engineered to bind a 5′-UTR can facilitate mRNA translation [61]. Thus, although STAU1 has been demonstrated to have an essential role in embryonic stem cell differentiation [62], it is unclear how SMD contributes to this essential role.
Evidence for competition between NMD and SMD
Remarkably, the function of UPF1 in both SMD and NMD results in competition between the two pathways (Figure 3) [34]. STAU1- and UPF2-binding sites within UPF1 not only overlap but are also mutually exclusive so that the IP (immunoprecipitation) of STAU1 precludes the detectable co-IP of UPF2 and vice versa. Furthermore, down-regulating the cellular abundance of STAU1, which inhibits SMD, increases the efficiency of NMD, whereas down-regulating the cellular abundance of UPF2, which inhibits NMD, increases the efficiency of SMD. Competition under physiological conditions is exemplified during the differentiation of mouse C2C12 myoblasts to myotubes, which is accompanied by a smaller decrease in the level of STAU1 relative to UPF1 than the level of UPF2 relative to the level of UPF1: during differentiation, the efficiency of SMD increases while the efficiency of NMD decreases, consistent with the finding that more STAU1 but less UPF2 binds UPF1 in differentiated myotubes compared with undifferentiated myoblasts. The functional significance of the remarkable balance between SMD and of NMD becomes apparent with the finding that PAX3 mRNA, whose decay promotes myogenesis [63], is an SMD target, and myogenin mRNA, which encodes a protein that is required for myogenesis [64], is an NMD target.
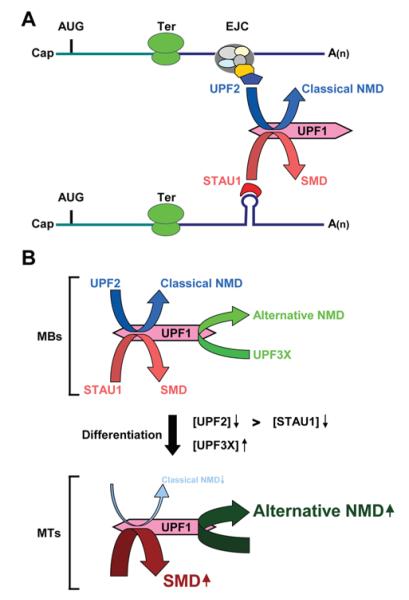
Figure 3. Model for competition between SMD and NMD: competition contributes to the differentiation of C2C12 myoblasts to myotubes.
(A) UPF2, which is an EJC constituent and functions in the classical NMD pathway, and STAU1, which is an RNA-binding protein that functions in the SMD pathway, compete for binding to UPF1 and thus the recruitment of UPF1 to mRNA. UPF1 functions in both pathways to elicit mRNA decay when translation terminates sufficiently upstream of an EJC, in the case of NMD, or STAU1 that is associated with an SBS, in the case of SMD. Ter can be either a premature or a normal termination codon.
(B) As a consequence of C2C12-cell differentiation from myoblasts (MBs) to myotubes (MTs), the efficiency of SMD increases, the efficiency of classical, i.e. UPF2-dependent, NMD decreases, and the efficiency of an alternative NMD pathway that relies on UPF3X, but not appreciably on UPF2, increases. During myogenesis, a larger decrease in the abundance of UPF2, which drops to almost undetectable levels relative to STAU1, permits STAU1 to out-compete UPF2 for binding to UPF1, and a ~4-fold increase in the level of UPF3X supports an increase in the efficiency of the alternative NMD pathway.
To further complicate the picture, myogenesis is also accompanied by an increase in the cellular level of UPF3X [34]. This results in an increase in the efficiency of an alternative NMD pathway that, unlike `classical' NMD is largely insensitive to UPF2 down-regulation [25,65]. As a consequence, myogenesis brings about a decreased level of SC1.7 and SC1.6 mRNAs, which are two well-characterized targets of alternative NMD that encode splicing variants of the SC35 splicing factor [34].
Summary
Future studies of SMD and classical and alternative NMD pathways during myogenesis and other cellular processes are certain to elucidate additional targets of post-transcriptional regulation that contribute, along with mechanisms of transcriptional control, to the complex and interdependent network of regulatory events that are required for the maintenance of or progression to distinct cell types.
Acknowledgments
Funding Work in the Maquat laboratory is supported by National Institutes of Health R01 grants [grant numbers GM 074593 and GM059614] to L.E.M. C.G. is additionally supported by an Elon Huntington Hooker Graduate Student Fellowship.
Abbreviations used
- CBP
cap-binding protein
- eIF
eukaryotic initiation factor
- EJC
exon junction complex
- IP
immunoprecipitation
- NMD
nonsense-mediated mRNA decay
- PI3K
phosphoinositide 3-kinase
- SMD
STAU1 (Staufen1)-mediated mRNA decay
- SBS
STAU1-binding site
- RNP
ribonucleoprotein
- 3′-UTR
3′-untranslated region
References
- 1.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 2.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Behm-Ansmant I, Kashima I, Rehwinkel J, Sauliere J, Wittkopp N, Izaurralde E. mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 2007;581:2845–2853. doi: 10.1016/j.febslet.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 4.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 5.Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 6.Isken O, Maquat LE. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat. Rev. Genet. 2008;9:699–712. doi: 10.1038/nrg2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGlincy NJ, Smith CW. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem. Sci. 2008;33:385–393. doi: 10.1016/j.tibs.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 8.M ühlemann O, Eberle AB, Stalder L, Zamudio Orozco R. Recognition and elimination of nonsense mRNA. Biochim. Biophys. Acta. 2008;1779:538–549. doi: 10.1016/j.bbagrm.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 9.Neu-Yilik G, Kulozik AE. NMD: multitasking between mRNA surveillance and modulation of gene expression. Adv. Genet. 2008;62:185–243. doi: 10.1016/S0065-2660(08)00604-4. [DOI] [PubMed] [Google Scholar]
- 10.Sánchez-Sánchez F, Mittnacht S. Nonsense-mediated decay: paving the road for genome diversification. BioEssays. 2008;30:926–928. doi: 10.1002/bies.20825. [DOI] [PubMed] [Google Scholar]
- 11.Shyu AB, Wilkinson MF, van Hoof A. Messenger RNA regulation: to translate or to degrade. EMBO J. 2008;27:471–481. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silva AL, Romão L. The mammalian nonsense-mediated mRNA decay pathway: to decay or not to decay! Which players make the decision? FEBS Lett. 2009;583:499–505. doi: 10.1016/j.febslet.2008.12.058. [DOI] [PubMed] [Google Scholar]
- 13.Stalder L, Mühlemann O. The meaning of nonsense. Trends Cell Biol. 2008;18:315–321. doi: 10.1016/j.tcb.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Rebbapragada I, Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr. Opin. Cell Biol. 2009;21:394–402. doi: 10.1016/j.ceb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 15.Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. U.S.A. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan Q, Saltzman AL, Kim YK, Misquitta C, Shai O, Maquat LE, Frey BJ, Blencowe BJ. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes Dev. 2006;20:153–158. doi: 10.1101/gad.1382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 18.Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Monch K, Thoren LA, Nielsen FC, Jacobsen SE, Nerlov C, Porse BT. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 2008;22:1381–1396. doi: 10.1101/gad.468808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wittmann J, Hol EM, Jäck HM. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol. Cell. Biol. 2006;26:1272–1287. doi: 10.1128/MCB.26.4.1272-1287.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jumaa H, Nielsen PJ. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 1997;16:5077–5085. doi: 10.1093/emboj/16.16.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- 22.Lejeune F, Cavaloc Y, Stévenin J. Alternative splicing of intron 3 of the serine/arginine-rich protein 9G8 gene: identification of flanking exonic splicing enhancers and involvement of 9G8 as a trans-acting factor. J. Biol. Chem. 2001;276:7850–7858. doi: 10.1074/jbc.M009510200. [DOI] [PubMed] [Google Scholar]
- 23.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, Shiue L, Clark TA, Blume JE, Ares M., Jr Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol. Cell. Biol. 2008;28:4320–4330. doi: 10.1128/MCB.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spellman R, Llorian M, Smith CW. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol. Cell. 2007;27:420–434. doi: 10.1016/j.molcel.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sureau A, Gattoni R, Dooghe Y, Stévenin J, Soret J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 2001;20:1785–1796. doi: 10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wollerton MC, Gooding C, Wagner EJ, Garcia-Blanco MA, Smith CW. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell. 2004;13:91–100. doi: 10.1016/s1097-2765(03)00502-1. [DOI] [PubMed] [Google Scholar]
- 29.Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum. Mol. Genet. 2001;10:99–105. doi: 10.1093/hmg/10.2.99. [DOI] [PubMed] [Google Scholar]
- 30.Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA. 2000;6:1226–1235. doi: 10.1017/s1355838200000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat. Struct. Mol. Biol. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- 32.Cheng Z, Muhlrad D, Lim MK, Parker R, Song H. Structural and functional insights into the human Upf1 helicase core. EMBO J. 2007;26:253–264. doi: 10.1038/sj.emboj.7601464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 34.Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE. SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev. 2009;23:54–66. doi: 10.1101/gad.1717309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiu SY, Lejeune F, Ranganathan AC, Maquat LE. The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes Dev. 2004;18:745–754. doi: 10.1101/gad.1170204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hosoda N, Kim YK, Lejeune F, Maquat LE. CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol. 2005;12:893–901. doi: 10.1038/nsmb995. [DOI] [PubMed] [Google Scholar]
- 37.Ishigaki Y, Li X, Serin G, Maquat LE. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 38.Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133:314–327. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lejeune F, Ishigaki Y, Li X, Maquat LE. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J. 2002;21:3536–3545. doi: 10.1093/emboj/cdf345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lejeune F, Ranganathan AC, Maquat LE. eIF4G is required for the pioneer round of translation in mammalian cells. Nat. Struct. Mol. Biol. 2004;11:992–1000. doi: 10.1038/nsmb824. [DOI] [PubMed] [Google Scholar]
- 42.Singh G, Lykke-Andersen J. New insights into the formation of active nonsense-mediated decay complexes. Trends Biochem. Sci. 2003;28:464–466. doi: 10.1016/S0968-0004(03)00176-2. [DOI] [PubMed] [Google Scholar]
- 43.Le Hir H, Andersen GR. Structural insights into the exon junction complex. Curr. Opin. Struct. Biol. 2008;18:112–119. doi: 10.1016/j.sbi.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 44.Tange TO, Nott A, Moore MJ. The ever-increasing complexities of the exon junction complex. Curr. Opin. Cell Biol. 2004;16:279–284. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 45.Matsuda D, Hosoda N, Kim YK, Maquat LE. Failsafe nonsense-mediated mRNA decay does not detectably target eIF4E-bound mRNA. Nat. Struct. Mol. Biol. 2007;14:974–979. doi: 10.1038/nsmb1297. [DOI] [PubMed] [Google Scholar]
- 46.Woeller CF, Gaspari M, Isken O, Maquat LE. NMD resulting from encephalomyocarditis virus IRES-directed translation initiation seems to be restricted to CBP80/20-bound mRNA. EMBO Rep. 2008;9:446–451. doi: 10.1038/embor.2008.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dostie J, Dreyfuss G. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr. Biol. 2002;12:1060–1067. doi: 10.1016/s0960-9822(02)00902-8. [DOI] [PubMed] [Google Scholar]
- 48.Gehring NH, Lamprinaki S, Kulozik AE, Hentze MW. Disassembly of exon junction complexes by PYM. Cell. 2009;137:536–548. doi: 10.1016/j.cell.2009.02.042. [DOI] [PubMed] [Google Scholar]
- 49.Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, Katsuhata Y, Muramatsu R, Morita T, Iwamatsu A, Hachiya T, et al. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 2009;23:1091–1105. doi: 10.1101/gad.1767209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lehner B, Sanderson CM. A protein interaction framework for human mRNA degradation. Genome Res. 2004;14:1315–1323. doi: 10.1101/gr.2122004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lejeune F, Li X, Maquat LE. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell. 2003;12:675–687. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- 52.Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 2002;22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Unterholzner L, Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol. Cell. 2004;16:587–596. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 54.Yamashita A, Kashima I, Ohno S. The role of SMG-1 in nonsense-mediated mRNA decay. Biochim. Biophys. Acta. 2005;1754:305–315. doi: 10.1016/j.bbapap.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 55.Eberle AB, Lykke-Andersen S, Mühlemann O, Jensen TH. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat. Struct. Mol. Biol. 2009;16:49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- 56.Glavan F, Behm-Ansmant I, Izaurralde E, Conti E. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J. 2006;25:5117–5125. doi: 10.1038/sj.emboj.7601377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA. 2008;14:2609–2617. doi: 10.1261/rna.1386208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 59.Kim YK, Furic L, Parisien M, Major F, DesGroseillers L, Maquat LE. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J. 2007;26:2670–2681. doi: 10.1038/sj.emboj.7601712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vessey JP, Macchi P, Stein JM, Mikl M, Hawker KN, Vogelsang P, Wieczorek K, Vendra G, Riefler J, Tubing F, et al. A loss of function allele for murine Staufen1 leads to impairment of dendritic Staufen1-RNP delivery and dendritic spine morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 2008;105:16374–16379. doi: 10.1073/pnas.0804583105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dugre-Brisson S, Elvira G, Boulay K, Chatel-Chaix L, Mouland AJ, DesGroseillers L. Interaction of Staufen1 with the 5′ end of mRNA facilitates translation of these RNAs. Nucleic Acids Res. 2005;33:4797–4812. doi: 10.1093/nar/gki794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gautrey H, McConnell J, Lako M, Hall J, Hesketh J. Staufen1 is expressed in preimplantation mouse embryos and is required for embryonic stem cell differentiation. Biochim. Biophys. Acta. 2008;1783:1935–1942. doi: 10.1016/j.bbamcr.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 63.Epstein JA, Lam P, Jepeal L, Maas RL, Shapiro DN. Pax3 inhibits myogenic differentiation of cultured myoblast cells. J. Biol. Chem. 1995;270:11719–11722. doi: 10.1074/jbc.270.20.11719. [DOI] [PubMed] [Google Scholar]
- 64.Wright WE, Sassoon DA, Lin VK. Myogenin, a factor regulating myogenesis, has a domain homologous to MyoD. Cell. 1989;56:607–617. doi: 10.1016/0092-8674(89)90583-7. [DOI] [PubMed] [Google Scholar]
- 65.Gehring NH, Kunz JB, Neu-Yilik G, Breit S, Viegas MH, Hentze MW, Kulozik AE. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol. Cell. 2005;20:65–75. doi: 10.1016/j.molcel.2005.08.012. [DOI] [PubMed] [Google Scholar]