Abstract
Nonsense-mediated mRNA decay (NMD) largely functions to ensure the quality of gene expression. However, NMD is also crucial to regulating appropriate expression levels for certain genes and for maintaining genome stability. Furthermore, just as NMD serves cells in multiple ways, so do its constituent proteins. Recent studies have clarified that UPF and SMG proteins, which were originally discovered to function in NMD, also have roles in other pathways, including specialized pathways of mRNA decay, DNA synthesis and cell-cycle progression, and the maintenance of telomeres. These findings suggest a delicate balance of metabolic events — some not obviously related to NMD — that can be influenced by the cellular abundance, location and activity of NMD factors and their binding partners.
Nonsense-mediated mRNA decay (NMD) is best known for its role in providing eukaryotic cells with a means to degrade abnormal mRNAs that prematurely terminate translation1–3. Such transcripts can arise owing to genomic frameshift or nonsense mutations, many of which cause diseases because of a failure to produce functional proteins. NMD targets can also arise as a result of error in cellular processes; such targets include inefficiently spliced pre-mRNAs as well as transcripts from T-cell-receptor genes, immunoglobulin genes and other antigen-receptor genes that undergo error-prone somatic-cell DNA rearrangements and hypermutations during lymphocyte development. The potential hazards of failing to degrade mRNAs that encode truncated proteins are exemplified by the numerous recessively inherited diseases that acquire a dominant-negative phenotype when NMD fails to target transcripts that prematurely terminate translation4.
There is a growing appreciation that NMD and proteins that are key players in the NMD pathway have important functions other than mRNA quality control. These functions include regulation of the expression of certain classes of genes, roles in specialized pathways of mRNA decay, functions in DNA synthesis and cell-cycle progression, and contributions to the maintenance of telomeres. In this Review, we first summarize the features of NMD targets and the trans-acting factors that recognize them, then we discuss how NMD can influence gene expression, focusing on the roles of NMD in splicing-factor production, nutrient homeostasis, protection of cells from oxidative stress, and maintenance of chromosome structure and function. We go on to present evidence that two central NMD-pathway proteins — up-frameshift 1 (UPF1; also known as regulator of nonsense transcripts 1, RENT1) and the phosphatidylinositol 3-kinase-related protein kinase (PIKK) SMG1 — are important players in a number of cellular pathways other than NMD, many of which have no apparent connection to NMD other than broadly controlling gene expression. We discuss UPF1 function in specialized mRNA-decay pathways, including Staufen 1 (STAU1)-mediated mRNA decay (SMD) and replication-dependent histone mRNA decay. We also review evidence that SMG1 has a role in stress-response pathways and DNA repair, and that UPF1 influences DNA synthesis and cell-cycle progression. Finally, we present data implicating SMG- and UPF-factor function in telomere maintenance. Whenever possible, we clarify if the NMD pathway or individual NMD factors function directly or indirectly in the cellular process under consideration.
Cis- and trans-acting NMD determinants
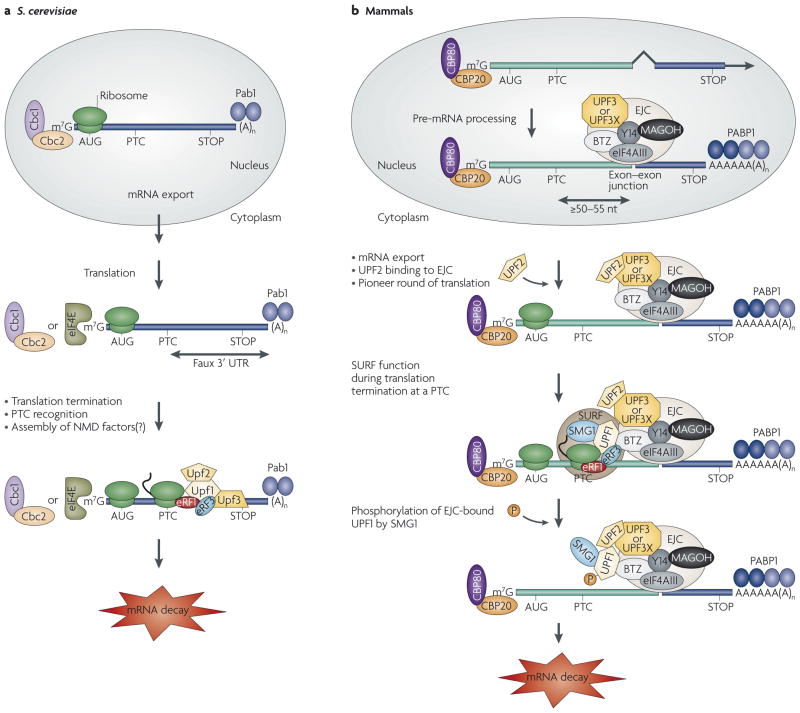
NMD is usually triggered when translation terminates prematurely at one of the three nonsense codons, that is, at premature termination codons (PTCs). The RNA features and protein factors that are generally necessary for a nonsense codon to trigger NMD can vary depending on the organism. For example, in Saccharomyces cerevisiae, an abnormally long distance between a termination event and the 3′ poly(A) tail, as defined by the presence of poly(A)-binding protein 1 (Pab1; PABP1 in mammals), seems to be sufficient to elicit NMD3,5 (FIG. 1a) but might not always be necessary6. In mammals, NMD usually requires at least one intron within pre-mRNA that results in the deposition of a post-splicing exon-junction complex (EJC) of proteins situated more than ~25–30 nucleotides downstream of a termination codon1–3 (FIG. 1b). The dependence on an EJC at least partly explains the apparent restriction of NMD in mammals to a pioneer round of translation. The pioneer round involves newly synthesized mRNA that is bound by the RNA cap-binding protein (CBP) heterodimer CBP80–CBP20 (also known as NCBP1–NCBP2), which has a mostly nuclear localization. Evidence that CBP80–CBP20-bound mRNA supports translation derives from a number of findings, including its co-immunoprecipitation with cytoplasmic polysomes in association with known eukaryotic translation initiation factors (eIFs) and other positive effectors of translation, including eIF4G, eIF3, eIF2A and PABP1, and the insensitivity of NMD to eIF4E-binding protein 1 (4EBP1), which inhibits the translation of eIF4E-bound mRNA but not of CBP80–CBP20-bound mRNA7–12. By the time eIF4E replaces CBP80–CBP20 at the cap, mRNAs are not detectably associated with EJCs; if an mRNA is not targeted for NMD before eIF4E replaces CBP80–CBP20 at the cap, then it becomes immune to NMD7,13–15. This is true even when translation initiates from a viral internal ribosome entry site when the site functionally depends on eIF3, the functional inhibition of which results in a step of translational repression that is required for NMD12,16. In S. cerevisiae, Cbc1–Cbc2-bound mRNA, which is orthologous to mammalian CBP80–CBP20-bound mRNA, can also be translated17, although Cbc1 is not essential for translation18,19. However, in contrast to the situation in mammals, NMD in S. cerevisiae targets not only Cbc1–Cbc2-bound mRNA, but also eIF4E-bound mRNA20.
Figure 1. Models for nonsense-mediated mRNA decay in Saccharomyces cerevisiae and mammals.
a | In Saccharomyces cerevisiae, newly synthesized mRNAs that contain a premature termination codon (PTC) and that are bound to the RNA cap-binding protein heterodimer Cbc1–Cbc2 and to steady-state mRNAs that are bound by the cap-binding protein eukaryotic translation initiation factor 4E (eIF4E) are targeted for nonsense-mediated mRNA decay (NMD) once the mRNA is exported from the nucleus to the cytoplasm. In at least one mechanism, an abnormally long or ‘faux’ 3′ UTR results in inefficient translation termination. As a consequence, termination involves not only the eukaryotic release factor 1 (eRF1) and eRF3 translation-termination factors, which fail to effectively mediate the release of the nascent polypeptide because of an inefficient interaction between eRF3 and poly(A)-binding protein 1 (Pab1), but probably also the up-frameshift 1 (Upf1), Upf2 and Upf3 NMD factors. These factors then recruit and/or activate mRNA degradative activities. Although Upf1 is a phosphoprotein, whether Upf1 undergoes a cycle of phosphorylation and dephosphorylation during NMD in S. cerevisiae is unknown. b | In mammals, newly synthesized PTC-containing mRNA that is bound to the RNA cap-binding protein heterodimer CPB80–CBP20 is targeted for NMD once the mRNA has been generated by pre-mRNA splicing and exported from the nucleus to the cytoplasm. Notably, pre-mRNA splicing results in the deposition of an exon-junction complex (EJC) of proteins upstream of mRNA exon–exon junctions. Core EJC components consist of eIF4AIII, RNA-binding-motif protein Y14, mago nashi homologue (MAGOH) and Barentsz (BTZ; also known as cancer susceptibility candidate 3, CASC3). The UPF3 or UPF3X NMD factor, which shuttles to the nucleus, is thought to be recruited to EJCs in the nucleus and is exported with the mRNA to the cytoplasm. UPF3 or UPF3X then recruits UPF2, which is primarily cytoplasmic. The translation of mRNA that is bound to CBP80–CBP20 is termed the pioneer round. Translation termination at a PTC during the pioneer round involves the SURF complex, which consists of the phosphatidylinositol 3-kinase-related protein kinase (PIKK) SMG1 together with UPF1, eRF1 and eRF3. Generally, if translation terminates more than ~50–55 nucleotides (nt) upstream of an exon–exon junction (that is, more than ~25–30 nt upstream of an EJC), then NMD will occur. UPF1, together with SMG1, is thought to bind EJC-associated UPF2 in a way that is promoted by CBP80 (not shown). UPF1 binding to the EJC triggers UPF1 phosphorylation and NMD by promoting translational repression and recruiting mRNA degradative activities. Not shown are SMG5, SMG6 and SMG7, which seem to recruit protein phosphatase 2A (PP2A) and function in UPF1 dephosphorylation and, thus, recycling. AUG, translation initiation codon; STOP, normal termination codon.
EJC function in NMD seems to be restricted to mammals and plants1–3,21. However, proteins that are similar to the mammalian core EJC constituents — eIF4AIII, RNA-binding-motif protein Y14, mago nashi homologue (MAGOH) and Barentsz (BTZ; also known as cancer susceptibility candidate 3, CASC3)22,23 — are present in invertebrates, including Drosophila melanogaster and Caenorhabditis elegans, even though they have no apparent role in NMD24–28.
Regardless of the species, in terms of trans-acting NMD proteins, the core set of NMD factors consists of UPF1, UPF2 (also known as RENT2) and UPF3 (TABLE 1); UPF3 consists of two isoforms in mammals that are encoded by distinct genes. In humans, UPF3 is encoded by an autosomal gene, whereas UPF3X (also known as UPF3B) is encoded by an X-linked gene29. UPF3 seems to partially substitute for UPF3X when it is absent from cells30–32. For example, UPF3 might substitute for UPF3X in certain patients with syndromic and non-syndromic mental retardation who lack detectable UPF3X32. UPF3X shuttles between the nucleus and the cytoplasm (UPF3 was not tested) UPF2 is cytoplasmic and can be seen concentrated along the cytoplasmic side of the nuclear envelope, and UPF1, which is an RNA-dependent ATPase and RNA helicase, is primarily cytoplasmic but also localizes to the nucleus when its export to the cytoplasm is blocked using leptomycin B29,33–35 (TABLE 1). UPF1 forms a stable complex with CBP80 (REF. 15).
Table 1.
Proteins that function in mammalian NMD
| Protein | Role in NMD | Cellular localization | Functional characteristics | Non-NMD roles | S. cerevisiae | C. elegans | Refs |
|---|---|---|---|---|---|---|---|
| UPF1 | Component of the SURF complex; joins EJC when translation terminates sufficiently upstream of EJC to undergo SMG1-mediated phosphorylation; negatively regulates efficient translation termination; cycles of phosphorylation–dephos- phorylation are necessary for NMD | Primarily cytoplasmic, but also shuttles | ATPase; 5′-to-3′ ATP-dependent helicase | Functions in STAU1-mediated mRNA decay and replication- dependent histone mRNA decay; implicated to function in DNA replication; enriched at telomeres in vivo; involved in maintenance of telomere integrity; negatively regulates TERRA association with chromatin | Upf1 | SMG-2 | 33,39,40, 43,45,50, 91,97,98, 103,105, 113,116, 118,127, 128, |
| UPF2 | Recruited to EJC by interacting with either UPF3X or UPF3; recruits UPF1 to the EJC; promotes UPF1 phosphorylation | Mainly cytoplasmic | – | Enriched at telomeres in vivo; involved in telomere maintenance | Upf2 | SMG-3 | 29,33,36, 40,42,44, 118,129, 130 |
| UPF3X (also known as UPF3B) | Usually the first UPF protein to associate with the EJC by Y14–MAGOH; recruits UPF2 to EJC | Mainly nuclear, but shuttles | – | Enriched at telomeres in vivo; involved in telomere maintenance; mutations in UPF3X gene cause syndromic and non-syndromic mental retardation | – | – | 30–32,36, 39,40,44, 118,130, 131,132 |
| UPF3 (also known as UPF3A) | Like UPF3X, usually the first UPF protein to associate with the EJC by Y14–MAGOH although less efficiently then UPF3X; recruits UPF2 to EJC | Mainly nuclear, but shuttles | – | Enriched at telomeres in vivo; involved in telomere maintenance | Upf3 | SMG-4 | 30–32,36, 39,40,44, 118,130, 131 |
| SMG1 | UPF1 kinase; dominant-negative variants inhibit NMD; downregulation inhibits NMD | Predominantly cytoplasmic, but shuttles | Related to phosphatidylinositol 3-kinase | Involved in maintenance of genome stability, stress response and DNA repair | None | SMG-1 | 39,40,42, 45,49, 108,118, 127 |
| SMG5, SMG6, SMG7 | SMG5 and SMG7 interact with PP2A; all are proposed to be involved in UPF1 dephosphorylation | Predominantly cytoplasmic; SMG7 is also present in the nucleus | Tethering of SMG7 to mRNA recruits mRNA-decay activities independently of translation | SMG5 and SMG6 interact with telomerase that is enriched at telomeres in vivo; negatively regulate TERRA association with chromatin | None | SMG-5, SMG-6, SMG-7 | 43,45,47, 91,116, 118,128 |
| SMGL1, SMGL2 | Required for NMD in C. elegans and human cells | – | – | Essential for proper embryonic development in C. elegans | None | SMGL-1, SMGL-2 | 28 |
| Y14– MAGOH heterodimer | Core EJC component; recruits UPF3X or, less efficiently, UPF3 | Predominantly nuclear, but shuttles | – | Stable heterodimer; accompanies mRNA into polysomes; ultimately removed by translation (as, probably, are other EJC constituents) | None | None | 32,33,36, 121,122, 126,131, 133 |
| eIF4AIII | Core EJC component; thought to bind to the exon–exon junction directly; anchors Y14– MAGOH and CASC3 to the EJC | Nuclear and cytoplasmic, but shuttles | – | Found in splicing complexes and spliceosomes containing pre-mRNA; closely related to eIF4AI and eIF4AII | None | None | 36,123, 126, 133–135 |
| BTZ (also known as CASC3) | Core EJC component; forms complex with eIF4AIII and Y14– MAGOH | Predominantly cytoplasmic, but shuttles | – | Functions in mRNA localization in Drosophila overexpressed in breast cancer; functions in stress- granule assembly | None | None | 36, 124–126 |
BTZ, Barentsz; CASC3, cancer susceptibility candidate 3; eIF, eukaryotic translation initiation factor; EJC, exon-junction complex; MAGOH, mago nashi homologue; NMD, nonsense-mediated mRNA decay; PP2A, protein phosphatase 2A; SMG, phosphatidylinositol 3-kinase-related protein kinase SMG; SMGL, SMG lethal; STAU1, Staufen1; SURF, complex containing SMG1, UPF1 and eukaryotic release factor 1 and 3; TERRA, telomeric repeat-containing RNA; UPF, up-frameshift.
Although early genetic and biochemical data indicated that UPF1, UPF2 and one or both of UPF3X and UPF3 function together, more recent results demonstrate that certain NMD targets in mammals can manifest differential sensitivity to either UPF2 or UPF3X depletion, the latter occurs both with and without the concomitant depletion of UPF3 (REFS 32,36,37). Remarkably, NMD targets also exist that are insensitive to the simultaneous downregulation of UPF2 and UPF3X38. Whether NMD can occur either in a mechanism that is less sensitive to UPF2 and/or UPF3X downregulation or independently of either or both UPF proteins remains to be determined.
The role of the UPF factors in NMD is arguably best understood for mammals (FIG. 1b). It is thought that the premature termination of translation of CBP80–CBP20- bound mRNA involves the so-called SURF complex, which consists of SMG1, UPF1 and the translation termination factors eukaryotic release factor 1 (eRF1) and eRF3 (REF. 39). Currently, the relationship between CBP80-bound UPF1 and UPF1 in SURF is unclear. The presence of UPF1 in SURF seems to promote inefficient translation termination by inhibiting eRF1 and eRF3 function. In contrast, PABP1 — which competes with UPF1 for binding to eRF3 — stimulates efficient translation termination40. This scenario is analogous to how Pab1 stimulates translation termination in S. cerevisiae41.
If translation terminates more than ~25–30 nucleotides upstream of an EJC, then UPF1 and SMG1 are thought to join the EJC and trigger SMG1-mediated UPF1 phosphorylation in a mechanism that seems to be promoted by CBP80 (REFS 15,16,42,43). The recruitment of UPF1 and SMG1 to the EJC is generally mediated by UPF2 and UPF3X, which are stably associated with the EJC. UPF2 and UPF3X stimulate the ATPase and RNA helicase activities of UPF1 in vitro44, and these proteins promote UPF1 phosphorylation in vivo39,42. UPF1 phosphorylation in turn triggers translational repression and recruitment of NMD degradative activities16. Additional NMD factors — SMG5, SMG6 and SMG7 — subsequently promote UPF1 dephosphorylation43,45–48, and SMG7 also serves to recruit mRNA degradative activities47. Notably, SMG1 and SMG7 are present in both nuclear and cytoplasmic compartments, whereas SMG5 and SMG6 are thought to localize to the cytoplasm47,49 (TABLE 1). Supporting the existence of a UPF2-independent NMD pathway as mentioned above, a UPF1 variant that fails to detectably bind UPF2 was recently shown to retain the ability to undergo phosphorylation, which is presumably promoted by SMG1 (REF. 40).
As is the case in mammals, cycles of UPF1 phosphorylation and dephosphorylation are required for NMD in C. elegans43,46,50,51 and D. melanogaster27, although SMG1 seems to promote rather than be required for NMD in D. melanogaster52,53. Whereas bona fide SMG proteins have yet to be found in S. cerevisiae, Upf1 is a phosphoprotein in this species54, as it is in mammals, and the Ebs1 protein enhances NMD and shares structural similarities primarily to SMG7 but also to SMG5 and SMG6 (REF. 55). Potential orthologues of one or more, but not all of, SMG1, SMG5, SMG6 and SMG7 can be found in those non-mammalian organisms that have been examined to date56 (TABLE 1). Notably, two novel NMD factors, named SMG lethal 1 (SMGL-1) and SMGL-2, have recently been identified in C. elegans. These proteins are required for NMD in C. elegans and humans and for embryonic development in C. elegans, although their exact functions — and their cellular locations — are currently unknown28 (TABLE 1).
NMD-mediated gene regulation
Apart from cases that arise owing to gene mutations or to metabolic errors, there are a number of cases in which NMD regulates gene expression so that the protein encoded by the NMD target is produced at a level that is appropriate for cellular function.
Gene expression profiles of S. cerevisiae, D. melanogaster and human cells that completely or partially lack an NMD factor have indicated that a significant fraction of cellular transcripts (1–10%) are upregulated and thus affected by NMD37,42,57–61. However, these calculations are probably overestimates of the number of genuine NMD targets because they are derived from microarray analyses that pool together direct and indirect effects.
Efforts to define genuine NMD targets in S. cerevisiae have also used microarray analyses. One study identified mRNAs that either decreased in abundance after the inhibition and subsequent reactivation of NMD or that copurified with Upf1, suggesting that Upf1 seems to selectively associate with NMD targets in S. cerevisiae61,62, as it also does in C. elegans51 but not in mammals15 (see below). Results revealed that 395 (42%) of mRNAs that were decreased in abundance when NMD was reactivated were among the 991 mRNAs that associated with Upf1, the remainder possibly being indirect NMD targets62. This percentage of candidate direct NMD targets is comparable to the ~45% of transcripts that manifest an increased half-life in microarray studies of S. cerevisiae strains lacking Upf1 (REF. 63).
In some cases, NMD silences genomic noise, such as non-functional transcripts that have acquired transposons or retroviral sequences59. In other cases, NMD targets serve regulatory functions. These targets include: functional genes that have acquired intronic transposons or retroviral sequences that, once spliced into mRNA, introduce nonsense codons; other types of expressed pseudogenes; genes harbouring an upstream ORF, an intron within their 3′ UTR or a programmed frameshifting site; genes encoding mRNAs that are subject to leaky scanning; and genes encoding selenoprotein mRNA37,42,59,60,62–68. We will now discuss several situations in which NMD contributes to proper cellular functions.
Role of NMD in the production of RNA-binding proteins
NMD functions in the homeostatic control of the expression of genes that encode serine–arginine (SR)-rich proteins and heterogeneous nuclear ribonucleoprotein (hnRNP) splicing factors (TABLE 2). A splicing activator can promote exon inclusion and a splicing repressor can promote intron inclusion, in some cases resulting in autogenous splicing-activated NMD — that is, by targeting the pre-mRNA from which the splicing effector derives — which is a regulated unproductive splicing and translation (RUST) mechanism69. In each case, the splicing-promoted inclusion or exclusion of specific sequences in the mRNA results in the generation of a termination codon that triggers NMD. Examples of proteins involved in autogenous alternative splicing-activated NMD include: the SC35 splicing factor, which mediates the activation of intron removal downstream of the normal termination codon of SC35 pre-mRNA70; polypyrimidine tract-binding protein (PTB, also known as hnRNPI), which promotes skipping of exon 11 of PTB pre-mRNA71; the SRp20 splicing factor, which enhances the inclusion of exon 4 of SRp20 pre-mRNA72; and the 9G8 splicing factor, which mediates the removal of intron 3 of 9G8 pre-mRNA73. Remarkably, genes for the PTB paralogues nPTB, which is neuronally restricted, and regulator of differentiation 1 (ROD1), which is expressed in haematopoietic cells, are also subject to alternative splicing-activated NMD, which is promoted by PTB (in the case of nPTB) or by both PTB and nPTB (in the case of ROD1), suggesting there is crossregulation and regulatory redundancy74,75. Support for the physiological relevance of this crossregulation derives from the finding that microRNA miR-124-mediated downregulation of PTB during neuronal differentiation results in the accumulation of productively spliced nPTB mRNA, the product of which functions in the differentiation of progenitor cells to mature neurons75.
Table 2.
Homoestatic control of splicing-factor gene expression by alternative pre-mRNA splicing coupled to NMD
| Splicing factor gene | Splicing factor | Alternative splicing event | Refs |
|---|---|---|---|
| SFRS3 | SRp20 | SRp20-mediated exon inclusion and/or flanking intron retention that results in PTC-containing SRp20 mRNA; this event is antagonized by ASF/SF2 | 67,72,76 |
| SFRS7 | 9G8 | 9G8-mediated exon inclusion and/or flanking intron retention that results in PTC-containing 9G8 mRN7A | 67,73,76 |
| SFRS9 | SRp30c | Intron retention that results in PTC-containing SRp30c mRNA | 76 |
| FUSIP1 | SRp38 | Alternative 5′ splice-site selection followed by inclusion of an alternative exon that results in PTC-containing SRp38 mRNA | 76 |
| SFRS5 | SRp40 | Exon inclusion and/or flanking intron retention that results in PTC-containing SRp40 mRNA | 76 |
| SFRS11 | SRp54 | Exon inclusion and/or flanking intron retention that results in PTC-containing SRp54 mRNA | 67,76 |
| SFRS6 | SRp55 | Exon inclusion and/or flanking intron retention that results in PTC-containing SRp55 mRNA | 67,76 |
| SFRS4 | SRp75 | Exon inclusion and/or flanking intron retention that results in PTC-containing SRp75 mRNA | 76 |
| SFRS2 | SC35 | SC35-mediated alternative splicing that generates exon–exon junctions downstream of the normal termination codon within SC35 mRNA | 70,76 |
| SFRS2B | SRp46 | Alternative splicing that generates an exon–exon junction downstream of normal termination codon within SRp46 mRNA | 76 |
| SFRS1 | ASF/SF2 | Alternative splicing that generates an exon–exon junction downstream of normal termination codon within ASF/SF2 mRNA | 67,76 |
| PTBP1 | PTB | PTB-mediated exon-skipping that results in PTC-containing PTB mRNA | 71 |
| PTBP2 | nPTB | PTB-mediated exon-skipping that results in PTC-containing nPTB mRNA | 67,74,75 |
| PTBP3 | ROD1 | PTB- and nPTB-mediated exon-skipping that results in PTC-containing ROD1 mRNA | 75 |
ASF/SF2, alternate splicing factor, splicing factor 2; FUSIP1, FUS interacting protein (serine/arginine-rich) 1; NMD, nonsense-mediated mRNA decay; nPTB, neuronally-restricted PTB; PTB, polypyrimidine tract-binding protein; PTC, premature termination codon; ROD1, regulator of differentiation 1; SFRS, splicing factor arginine–serine rich.
Other examples of alternative splicing-activated NMD that are subject to feedback regulation — autoregulatory and/or crossregulatory — have been proposed on the basis of the presence of exonic ultraconserved elements that often contain or overlap with a nonsense codon in RNA splicing-activator transcripts. Beside transcripts encoding the SC35, SRp20 and 9G8 splicing factors, these transcripts include those encoding the RNA-binding proteins TRA2A, TRA2B, SRp55, CAPERα and TIAR — all but TIAR function in splicing67,76. Notably, transcripts encoding the core spliceosome component U1 small nuclear ribonucleoprotein (snRNP)-specific 70kDa protein, the common snRNP Sm proteins SmB and B′, and the SF1, PRPF18 and SMNDC1 proteins that are involved in spliceosome formation are also regulated by alternative splicing events that introduce PTCs and that are most probably autoregulated, although not necessarily directly, as was shown for survival motor neuron domain containing 1 (SMNDC1) transcripts38. The homeostatic control of splicing-factor gene expression by NMD seems to be a conserved response. For example, a number of SR-protein genes in C. elegans have been shown to be upregulated following NMD downregulation77. Furthermore, SR-protein genes in plants and D. melanogaster also contain highly conserved sequences that support autoregulation or crossregulation78–80.
Despite evidence that unproductive splicing can provide a physiologically significant means to downregulate splicing-factor gene expression by NMD, most of the estimated one-third or more of alternatively spliced transcripts that are NMD targets in humans seem to be mistakes that were made during pre-mRNA splicing81. In support of this idea, most NMD targets are expressed at low levels in diverse tissues and cell types, unlike splice variants that produce functional protein isoforms81. NMD targets in S. cerevisiae and D. melanogaster might also be largely the consequence of transcriptional or post-transcriptional mistakes, on the basis of their overall low abundance57,58,60,82.
Role of NMD in nutrient homeostasis
NMD targets can serve regulatory functions. One example that typifies regulatory roles of NMD in S. cerevisiae is the cryptic unstable transcript that reads through the SER3 regulatory gene (SRG1) and the serine requiring 3 gene (SER3). Synthesis of the SRG1–SER3 read-through transcript is initiated upstream of the biosynthetic SER3 gene, and thus represses SER3 gene expression by transcriptional interference in a serine-dependent manner. Presumably, this transcript undergoes NMD because it directs translation initiation upstream of the SER3 translation-initiation codon, but terminates translation prematurely83,84. In fact NMD, which occurs in the cytoplasm, might contribute, along with nuclear decay pathways, to the degradation of many regulatory transcripts that under particular cellular conditions are synthesized from sites upstream of genes involved in nutrient homeostasis84 (G. Chanfreau, personal communication). These transcripts are predicted to inhibit gene expression by removing a transcriptional activator and/or occluding the promoter.
The CPA1 gene, which encodes a small subunit of carbamoyl phosphate synthetase (a protein that is required for arginine biosynthesis), is another gene that is targeted for NMD and that exemplifies the role of NMD in nutrient homeostasis in S. cerevisiae. In this case, arginine addition promotes ribosome stalling and translation termination at the termination codon of an upstream ORF of CPA1 mRNA85.
Amino-acid starvation in eukaryotes results in a global inhibition of cellular translation that is mediated by eIF2A phosphorylation86, and eIF2A phosphorylation inhibits mammalian-cell NMD7. Amino-acid starvation in cultured human cells was found to preferentially upregulate transcripts that are required for amino-acid homeostasis59. In fact, microarray analyses demonstrated that of the 80 transcripts analysed that encode proteins that function under the ontologic category of ‘amino-acid transport’, ‘amino-acid biosynthesis’ or ‘amino-acid activation’, a disproportionately high percentage (15%, which is threefold higher than the corresponding fraction for all transcripts assayed) was upregulated when NMD was inhibited by downregulating UPF1 (REF. 59). Studies using S. cerevisiae also found that certain transcripts involved in amino-acid homeostasis were upregulated when UPF1 was downregulated58. Whether the aforementioned human and yeast transcripts functioning in amino-acid metabolism are direct or indirect NMD targets remains to be determined. Given the block in general cellular translation, it also remains to be determined if the preferential stabilization of NMD targets during amino-acid starvation would contribute in significant ways to the preferential production of the encoded protein. Conceivably, the preferential stabilization of those targets involved in amino-acid homeostasis would contribute to a fast restoration of cellular metabolism once the cell returns to a state of amino-acid sufficiency.
Role of NMD in protecting cells from oxidative stress
There are also indications that NMD modulates the transcriptional response to oxidative stress in Schizosaccharomyces pombe, although the exact mechanism is uncertain61. Upf1 stabilizes atf1 mRNA, which encodes a subunit of a bZIP transcription-factor complex that is required for the expression of specific genes involved in the response to oxidative stress and thereby protects cells from excessive accumulation of reactive oxygen species and other damaging consequences61. Another protein, Csx1, which is an RNA-binding protein, has also been implicated in the response to oxidative stress in S. pombe, and has been suggested to function in conjunction with Atf1 to regulate gene expression. A model has been proposed in which Upf1, through its NMD function, regulates Csx1-dependent gene expression to target a negative effector of atf1 mRNA stabilization61. The finding that Upf2 is also required for survival during oxidative stress implicates an NMD-related function, rather than an NMD-independent function, for Upf1 in this context. Microarray analyses of changes in mRNA abundance following Upf1 depletion indicated that Upf1, possibly through its role in NMD, might additionally be involved in stabilizing pcr1 mRNA, which encodes a protein that together with Atf1 forms a heteromeric transcription factor61.
A recent study using cultured human cells demonstrated that NMD is inhibited during hypoxia, which results in eIF2A phosphorylation87. Furthermore, transcripts manifesting longer half-lives when UPF1 is downregulated include hypoxia-induced mRNAs that are known to have significant roles in the integrated stress response. These mRNAs encode the transcription factors ATF4, ATF3 and CHOP; ATF3 and CHOP are upregulated by ATF4 (REF. 87). At least, ATF4 mRNA seems to be a direct target of NMD owing to its 5′ upstream ORFs. As was discussed in the case of amino-acid starvation, it is possible that preferential stabilization of NMD targets during hypoxia would contribute in significant ways to preferential production of the encoded protein either during the stress response or during recovery from the stress response, although this remains unproven.
Roles of NMD in chromosome structure and function
A significant fraction of mRNAs that have been identified as possible direct targets of NMD-mediated regulation encode proteins involved chromosome structure and behaviour, including telomere maintenance and replication, chromatin-mediated silencing, and the transmission of chromosomes during cell division. Consistent with the importance of NMD to telomere function, S. cerevisiae cells lacking Upf1, Upf2 or Upf3 have telomeres that are ~100 bp shorter than normal and show a loss of chromatin silencing at telomeres88,89. In fact, transcripts encoding the Est2 catalytic subunit of telomerase, the Est1, Est3, Stn1 and Ten1 telomerase regulators, and Sas2 and Orc5, which influence chromatin structure at telomeres, are upregulated following UPF1 gene deletion89. The finding that deletion of each UPF protein led to the same telomere abnormalities suggests that particular NMD targets, rather than NMD-independent UPF-protein contributions, are required for proper telomere function. The best studied of the transcripts that were upregulated following UPF1 gene deletion is STN1 mRNA, but currently there is mixed evidence that it is a direct NMD target: although STN1 mRNA abundance increases when NMD is inhibited90 and STN1 mRNA seems to bind Upf1 (REF. 62), STN1 mRNA half-life does not detectably increase in strains lacking Upf1 (REF. 89). The finding that overexpressing the EST1A transcript, and thus the Est1A protein, results in telomeric fusions and telomere shortening91 suggests that NMD, either directly or indirectly, suppresses telomere abnormalities.
In summary of these findings it is possible that NMD widely regulates the expression of multiple gene products that function in a single cellular process59,61,63. However, it is important to differentiate direct from indirect NMD targets as well as the precise role of an NMD factor in the cellular process under study. For example, Upf1 function in telomere maintenance is notable because Upf1, along with other NMD factors, is unexpectedly enriched at telomeres for purposes other than NMD (see below).
Specialized mRNA-decay pathways
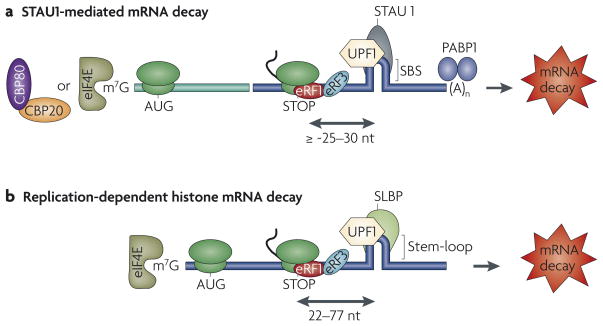
In addition to its role in mRNA decay by NMD, UPF1 also functions in two specialized non-NMD mRNA-decay pathways in mammalian cells that do not require the mRNA to be derived from splicing. In both pathways, UPF1 is recruited downstream of the normal termination codon by a specific mRNA-binding protein and triggers mRNA decay most probably when translation terminates. One pathway targets mRNAs that microarray data indicate might be hundreds of mammalian-cell mRNAs that bind the double-stranded (ds) RNA-binding protein STAU1, which interacts directly with UPF1. The other pathway targets the 65 non-allelic mammalian mRNAs that encode the four core histones and histone H1, all of which contain a binding site for stem-loop binding protein (SLBP) within their 3′ UTRs. SLBP also interacts directly with UPF1. Therefore, transcripts bound within their 3′ UTR by STAU1, like transcripts bound within their 3′ UTR by SLBP, constitute a group of ‘regulons’, which are defined as transcripts subjected to a common regulatory pathway that might be fine-tuned by mRNA-specific binding proteins92. It is conceivable that UPF1 conditionally mediates the decay of other regulons provided that additional RNA-binding proteins recruit UPF1 either directly or indirectly to mRNA 3′ UTRs.
UPF1 and SMD
STAU in Drosophila functions in the transport, localization and translational control of certain mRNAs during oogenesis and during the development of the central nervous system93. Orthologues to Drosophila STAU in mammals form mRNA-containing granules that are transported to the ends of dendrites during neurogenesis94 or are encapsidated together with HIV-1 RNA in virus particles95,96. Thus, it initially came as a surprise to find that human STAU1 interacts directly with human UPF1, which had no apparent connection to known STAU1 functions97. STAU1 binding to the 3′ UTR of particular mRNAs triggers SMD in a way that depends on mRNA translation and UPF1 but not the other UPF factors97. SMD is similar to NMD as it requires translation termination upstream of the site of UPF1 recruitment; however, the site of translation termination in SMD is generally the normal termination codon rather than a PTC (FIG. 2a). Furthermore, because SMD involves UPF1 recruitment by STAU1 rather than by an EJC, SMD can, unlike NMD, target mRNAs that derive from intron-less genes97,98. Additionally, SMD, unlike NMD, targets not only newly synthesized CBP80–CBP20-bound mRNA but also eIF4E-bound mRNA because STAU1, in contrast to EJCs, associates with both newly synthesized and steady-state mRNA15 (Y.K. Kim and L.E.M., unpublished observations). The finding that SMD targets eIF4E-bound mRNA, which produces the bulk of cellular protein, is consistent with the idea that SMD functions as a conditionally regulated pathway. Whether SMG proteins function in SMD has yet to be tested.
Figure 2. UPF1 function in specialized mRNA-decay pathways.
a | Staufen 1 (STAU1)-mediated mRNA decay targets particular newly synthesized mRNAs that contain a STAU1 binding site (SBS) within their 3′ UTR and that are bound to the cap-binding protein heterodimer CBP80–CBP20 and to the corresponding steady-state mRNAs that are bound to the cap-binding protein eukaryotic translation initiation factor 4E (eIF4E). SBS-bound STAU1 recruits the NMD factor up-frameshift 1 (UPF1), which presumably undergoes phosphorylation by a protein kinase (yet to be identified) when translation terminates more than ~20–25 nucleotides (nt) upstream of the SBS. b | Replication-dependent histone mRNAs are degraded at the end of S phase of the cell cycle or following replication stress. These histone mRNAs contain a binding site for the stem-loop binding protein (SLBP) within their 3′ UTR. 3′-UTR-bound SLBP recruits UPF1, which seems to undergo phosphorylation mediated by ataxia-telangiectasia mutated and Rad 3-related (ATR) or DNA-dependent protein kinase (DNA-PK) when translation terminates anywhere in the 22–77 nucleotide region upstream of the SLBP-binding site. The 77-nucleotide limit is because of the need for SLBP to be situated sufficiently close to the termination codon to function in translation termination. Notably, it is primarily eIF4E-bound histone mRNA that is targeted for decay at the end of S phase owing to the concomitant downregulation of histone gene transcription. AUG, translation initiation codon; eRF, eukaryotic release factor; STOP, normal translation termination codon: PABP1, poly(A)-binding protein 1.
Microarray studies demonstrated that 1.1% of transcripts from 11,569 human HeLa cell genes were upregulated at least twofold when the cellular abundance of STAU1 was downregulated using small interfering RNAs (siRNAs)97. Experimentally verified examples of bona fide SMD targets include the mRNAs that encode ADP ribosylation factor 1 (ARF1), serpin peptidase proteinase inhibitor clade E (nexin plasminogen activator inhibitor type 1) member 1 (SERPINE1), interleukin 7 receptor (IL7R), jun oncogene (JUN), and growth associated protein 43 (GAP43)97,98. These SMD targets encode proteins that are not obviously functionally related. Thus, SMD probably regulates gene expression in parallel with other post-transcriptional processes that vary depending on the particular transcript and cellular conditions. Our finding that the efficiency of SMD increases during the differentiation of C2C12 myoblasts to myotubes suggests that SMD might be important for myogenesis98. The role of SMD in other cellular processes has yet to be examined.
Human STAU2 shares 51% identity with STAU1 but is encoded by a different gene99. Considering that hemagglutinin (HA)-tagged STAU2 isoforms immunoprecipitate with mRNAs from the human cell line HEK293T99,100, it is possible that STAU2 functions analogously to STAU1 and SLBP to recruit UPF1 to the 3′ UTRs of particular mRNAs and mediates the decay of these mRNAs when translation terminates normally. Countering this possibility, however, UPF1 and STAU2 failed to co-immunoprecipitate in neuronal-cell extracts100.
UPF1 and replication-dependent histone mRNA decay
Metazoan replication-dependent histone mRNAs are degraded at the end of the S phase of the cell cycle or when DNA replication is inhibited using pharmacologic agents — in each case when histone protein synthesis is no longer needed for newly synthesized chromatin formation101. These mRNAs are distinct from the rest of cellular mRNAs because they lack a 3′ poly(A) tail and instead they end in a conserved stem-loop structure. The co-ordination of histone mRNA decay with DNA replication requires histone mRNA translation and recruitment of the UPF1 NMD factor to histone mRNA by SLBP102 (FIG. 2b). Data indicate that SLBP functions analogously to an EJC or STAU1 after translation terminates at the normal termination codon102,103 (FIG. 2b).
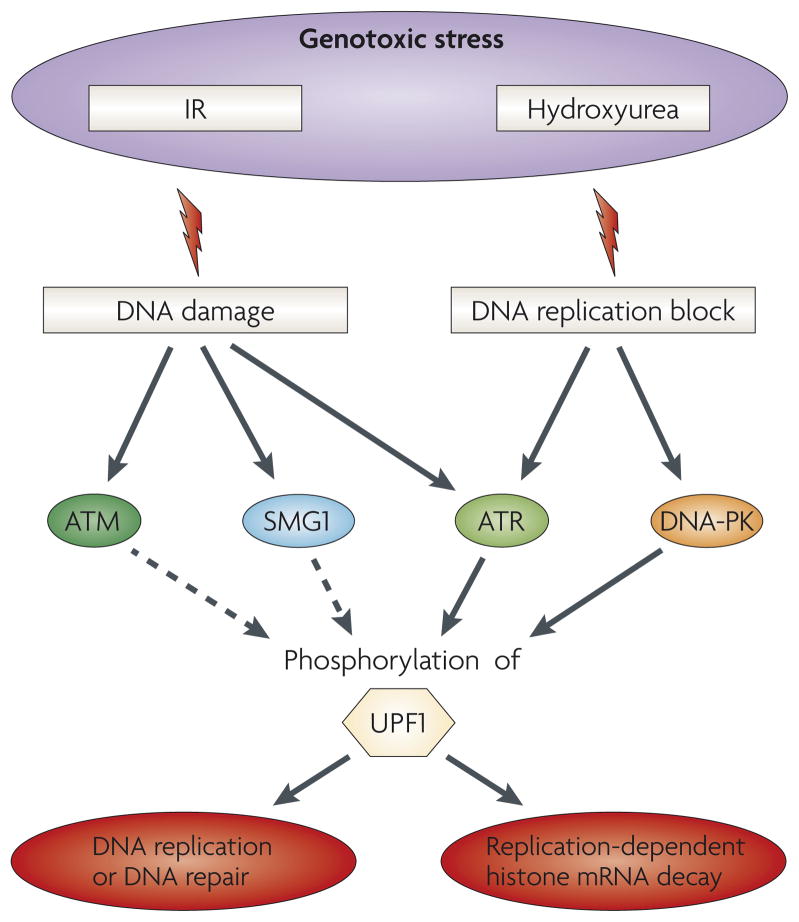
The UPF1-mediated decay of histone mRNA is controlled by the PIKK ATR (ataxia-telangiectasia mutated and Rad 3-related)103, which is a key regulator of the DNA damage-induced checkpoint pathway that is activated during replication stress. For example, exposing human U2OS osteosarcoma cells to DNA-damaging reagents that activate the ATR pathway — including not only hydroxyurea, which inhibits ribonucleotide reductase and arrests cells in the S phase of the cell cycle, but also aphidicolin, which inhibits DNA polymerase, as well as UV light, which causes thymidine-dimer formation — results in histone mRNA decay103 (FIG. 3). Consistent with the possibility that ATR phosphorylates UPF1, the treatment of HeLa cells with hydroxyurea causes UPF1 phosphorylation but not SLBP phosphorylation104. Furthermore, histone mRNA decay following a hydroxyurea-induced block in DNA synthesis is prevented by the expression of a kinase-inactive ATR in U2OS cells or by the pretreatment of U2OS cells with caffeine, which inhibits PIKKs (including ATR)103. By contrast, ATR does not seem to function in NMD105, suggesting that UPF1 phosphorylation by different PIKKs can variously influence UPF1 function (see also below).
Figure 3. Phosphorylated UPF1 functions when DNA is damaged or replication is otherwise blocked.
Exposure of cells to different types of genotoxic stress such as ionizing radiation (IR) or hydroxyurea induces DNA damage and/or a block in DNA replication. These abnormalities activate one or more of the phosphatidylinositol 3-kinase-related protein kinase (PIKK) signal transducers ataxia-telangiectasia mutated (ATM), ataxia-telangiectasia mutated and Rad 3-related (ATR), SMG1 or DNA-dependent protein kinase (DNA-PK), which stimulate phosphorylation of target proteins, such as the NMD factor up-frameshift 1 (UPF1). The resulting phosphorylated UPF1 then functions in DNA replication and repair as well as in histone mRNA decay. The multiple roles of UPF1 in these processes, all presumably by its phosphorylation by PIKKs, ensures that histone production is closely coupled to the cellular need for newly synthesized chromatin.
In addition to ATR, the PIKK DNA-dependent protein kinase (DNA-PK) also signals histone mRNA decay, as indicated by the treatment of HeLa cells with the DNA-PK inhibitor LY294002 and by the fact that DNA-PK phosphorylates UPF1 but not SLBP104. However, exposing U2OS cells to ionizing radiation (IR), which induces the formation of dsDNA breaks and activates the PIKK ataxia-telangiectasia mutated (ATM), did not result in histone mRNA decay103. Because IR enhances not only ATM-mediated but also SMG1-mediated UPF1 phosphorylation49, SMG1 might not have a major role in UPF1-triggered histone mRNA decay. The importance of SMG5, SMG6 or SMG7 in this pathway has yet to be tested. Notably, IR fails to inhibit NMD except when SMG1 is downregulated as indicated by studies of human Calu6 cells, which produce PTC-containing p53 tumour suppressor protein mRNA that was used to assay NMD49. These and other data suggest that IR treatment when the cellular abundance of SMG1 is limiting activates the ATM-mediated phosphorylation of UPF1, which precludes UPF1 function in NMD.
NMD factors and genome stability
NMD factors can function in cellular mechanisms that do not involve NMD or other mRNA-decay pathways. Genome integrity is essential to maintain genome function and to preclude changes that might lead to a neoplastic transformed state. The maintenance of genomic integrity is achieved by a combination of processes, including DNA repair and telomere maintenance. Although DNA repair is the basic response to genotoxic stress, when DNA damage reaches an extent that overwhelms the response for cell survival, apoptic cell death can result106. Effective DNA repair requires not only a manageable amount of stress but also constructive coordination with other cellular processes, including DNA synthesis, gene transcription and cell-cycle checkpoint controls. As outlined below, several lines of evidence implicate NMD factors in the maintenance of genome stability.
SMG1, stress-response pathways and DNA repair
SMG1 not only phosphorylates UPF1, but it also phosphorylates other cellular proteins. As a consequence, SMG1 not only functions in NMD, but it also responds to cell stress induced by DNA-damaging agents such as IR, which as noted above induces dsDNA breaks, and UV-B light, which also induces dsDNA breaks, although to a lesser extent than IR49. In fact, an early name for human SMG1 was ‘ATX’, on the basis of its resemblance primarily to ATM and secondarily to ATR, both of which function in the cellular response to DNA damage107. SMG1 mediates p53 stabilization and phosphorylation at serine 15 in IR-damaged U2OS cells49, as do ATM and ATR. Additionally, SMG1, like ATM and ATR, contributes to IR-induced G2 checkpoint signalling, which delays cell-cycle progress and allows time for DNA repair, suggesting that all three of these PIKKs function in parallel to optimize activation of the DNA-damage response. The finding that inhibiting SMG1 activity in Atm−/− primary thymocytes using 5 μM wortmannin, which affects the activity of ATM but not of other known PIKKs, reduces p53 phosphorylation induced by the chemopreventative antioxidant tempol suggests that SMG1 is also activated by reactive oxygen species in a way that elicits cytoprotection108. SMG1, unlike ATR, also phosphorylates p53 at serine 15 during the early stages of hyperoxia to contribute to G1 checkpoint signalling; by contrast, ATM functions primarily after 24 hours to maintain p53 phosphorylation and G1 checkpoint signalling109.
In support of a role for SMG1 in genome surveillance or repair, or both, the siRNA-mediated downregulation of SMG1 in U2OS cells results in the accumulation of dsDNA breaks and activation of ATM- and/or ATR-mediated checkpoint responses, as indicated by an accumulation of G2–M-phase cells49. These findings lend credence to the idea that SMG1 is necessary for the DNA-damage response even without experimentally induced DNA damage, although the possibility that accumulated NMD targets could be the cause of the response has yet to be ruled out.
Surprisingly, downregulating SMG1 but not UPF1 in U2OS cells using siRNAs increases cell sensitivity to apoptosis induced by tumour necrosis factor alpha (TNFα) or tumour necrosis factor (ligand) superfamily, member 10 (TNFSF10; also known as TRAIL)110. However, no increase was observed in non-malignant human mammary epithelial MCF-10A cells111. Nevertheless, the finding that SMG1 localizes in U2OS cells to the outer mitochondrial membrane raises the possibility that SMG1 prevents mitochondrial damage and, by so doing, suppresses activation of apoptosis in certain cancer cells, making it a potential target for anti-cancer therapeutics111. SMG1 was also suggested as a possible therapeutic target in acute myeloid leukaemia on the basis of its abnormally high level of expression in certain patient samples and the finding that inhibiting PIKKs in myeloid leukaemias promotes apoptosis112.
UPF1, DNA synthesis and cell-cycle progression
It is possible that UPF1 regulates cell-cycle progression by regulating the decay of histone mRNA at the end of S phase (see above). However, UPF1 also seems to have a direct role in DNA synthesis. Downregulating UPF1 in HeLa cells causes cell-growth arrest early in S phase in a DNA-damage response that is primarily mediated by ATR105. By contrast, HeLa cells in which UPF2 was down-regulated progressed normally through the cell cycle105. Chromatin immunoprecipitation performed after HeLa cells were subjected to formaldehyde crosslinking demonstrate that ~2% of DNA associates with UPF1, and this amount increases approximately fourfold after release from a hydroxyurea-induced block in S phase105. Normally, UPF1 binding to chromatin occurs at a low level in mitosis and in early G1 phase, starts to increase in mid-G1 phase and is highest in S phase, consistent with a role in DNA synthesis. UPF1 binding to chromatin also increases above the level observed in asynchronously growing cells in an ATR-dependent mechanism following IR treatment, suggesting a role in DNA repair. Consistent with data indicating that DNA damage induces UPF1 phosphorylation104, chromatin-bound UPF1 is hyperphosphorylated105. Conversely, the bulk of mammalian-cell UPF1 is hypophosphorylated16,39,43,45,113. Whereas ATR, SMG1 and ATM augment UPF1 binding to chromatin after IR treatment, ATR seems to control UPF1 binding to chromatin in untreated cells105.
The idea that chromatin-bound UPF1 is a component of DNA synthesis and repair pathways is supported by the finding that UPF1 co-immunoprecipitates with DNA polymerase δ114 primarily during S phase, when UPF1 is hyperphosphorylated105 (FIG. 3). By contrast, UPF2 does not detectably co-immunoprecipitate with DNA polymerase δ105.
SMG and UPF factors and telomere maintenance
Telomeres consist of protein-associated tandem double-stranded repeats of 5′-TTAGGG-3′ on the 3′-overhanging strand at the ends of eukaryotic chromosomes, and they stabilize the genome by preventing the physical ends of chromosomes from being recognized as damage-induced DNA breakpoints that could be fused to other ends, or that could be improperly repaired or degraded115. Human SMG5 and SMG6 interact with telomerase, which maintains telomere length by adding telomeric repeats to the 3′ ends of chromosomes, and overexpressing SMG5 results in telomere shortening and chromosome-end fusions91,116,117. SMG6 contains a high affinity but low specificity RNA-binding domain and also interacts with telomerase reverse transcriptase polypeptide, suggesting that it contacts both RNA and protein within the telomerase RNP117. Although telomeres were previously thought to be transcriptionally silent, subtelomeric and telomeric regions of HeLa cells have recently been shown to be transcribed into telomeric repeat-containing RNA (TERRA)118. TERRA consists of heterogenous nuclear transcripts that are synthesized from subtelomeric regions into the telomeric repeats. TERRA molecules bind to chromatin at telomeres in a way that de-protects chromosome ends118.
A series of findings implicate NMD factors in telomere maintenance: the binding of TERRA to telomeres is negatively regulated by UPF1, SMG1, SMG6 and to some extent UPF2; depletion of UPF1, SMG1 and SMG6 and to a lesser extent UPF2 and SMG7 results in telomere loss; depletion of UPF1, SMG1 and SMG6 generates chromosome and chromatid breaks at both subtelomeric and intrachromosomal regions; and UPF1, UPF2, UPF3, SMG1, SMG5, SMG6 and SMG7 are enriched at telomeres compared to Alu sequences118. These findings have led to the hypothesis that TERRA facilitates telomeric heterochromatin assembly in a mechanism that is coordinated by UPF and SMG factors.
Conclusions
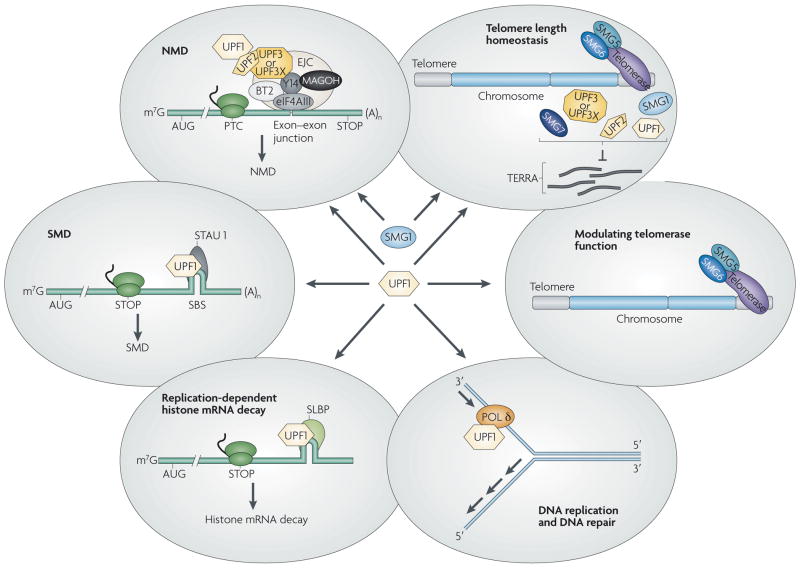
By degrading mRNAs that prematurely terminate translation, NMD controls the quality of gene expression. As we have seen in this Review, by using the same mechanism of target recognition, NMD also functions in a variety of ways to regulate the expression of normal cellular genes. So, like alternative splicing, the NMD pathway and its components add to the regulatory potential of eukaryotic genomes. Beyond their functions as NMD factors, incompletely defined combinations of UPF and SMG proteins demonstrate unexpected roles in specialized mRNA-decay pathways, in stress-response pathways and DNA repair, in DNA synthesis and cell-cycle progression, and in telomere maintenance (FIG. 4). Therefore, these proteins can be viewed as components of overarching RNA- and DNA-surveillance processes and as multitasking players in the intricate network of signal transduction pathways that respond to genetic and acquired errors in nucleic-acid metabolism.
Figure 4. UPF1 and SMG1 are at the interface of processes important for gene and genome regulation in mammalian cells.
(Anticlockwise from the upper left). Up-frameshift 1 (UPF1) and phosphatidylinositol 3-kinase-related protein kinase SMG1 are involved in multiple RNA and DNA surveillance pathways. UPF1 functions during nonsense-mediated mRNA decay (NMD), in which translation termination that occurs sufficiently upstream of a splicing-generated exon-junction complex7 (EJC), usually at a premature termination codon (PTC), results in SMG1-mediated UPF1 phosphorylation and, as a consequence, mRNA decay. UPF1 is also instrumental to the RNA-binding protein Staufen 1 (STAU1)-mediated mRNA decay (SMD) and replication-dependent histone mRNA decay, during which it is recruited to specific 3′ UTRs by STAU1 (in SMD) or stem-loop binding protein (SLBP) (in replication-dependent histone mRNA decay). Although the UPF1 kinase involved in SMD is unknown, ataxia-telangiectasia mutated and Rad 3-related (ATR), ataxia-telangiectasia mutated (ATM) and DNA-dependent protein kinase (DNA-PK) seem to phosphorylate UPF1 during histone mRNA decay. Additionally, UPF1 associates with DNA polymerase (Pol) δ and is essential for human cells to complete DNA replication and repair in a process that involves ATR if not other phosphatidylinositol 3-kinase-related protein kinases (PIKKs). Finally, UPF and SMG proteins seem to function in genome stability to modulate telomerase function and to regulate telomere length: they are enriched at telomeres so as to negatively regulate telomeric repeat-containing RNA (TERRA) association with telomeric chromosomes. AUG, translation initiation codon; BTZ, Barentsz (also known as cancer susceptibility candidate 3, CASC3); eIF4AIII, eukaryotic initiation factor 4AIII; MAGOH, mago nashi homologue; PTC, premature translation termination codon; SBS, STAU1 binding site; STOP, normal termination codon; Y14, RNA-binding motif-protein Y14.
It will be interesting to understand how pathways that use the same UPF and/or SMG factor(s) achieve a balance with one another so that their roles in regulating the expression or organization of genetic material are effective. As an example of the importance of balance, it is reasonable to assume that NMD and SMD could be competitive pathways in which UPF2 and STAU1 compete for binding to UPF1 (C. Gong, Y.K. Kim and L.E.M, unpublished observations). A number of other cellular proteins have likewise been shown to demonstrate multiple functions that must also be balanced.
It is unclear how cells direct NMD factors to one or another function. Protein function can certainly be influenced by binding partners. Relevant to this Review, for example, the binding of UPF1 to an EJC, to SLBP or to STAU1 determines if it functions in NMD, in histone mRNA decay or in SMD, respectively. Furthermore, protein modifications such as phosphorylation might also signal the specific association of distinct protein– protein or protein–nucleic acid (DNA or RNA) interactions, thereby determining the different roles of NMD factors in DNA or RNA metabolism. Notably, the differential phosphorylation of, for example, UPF1 by distinct kinases most probably contributes to the correct balance of its various functions.
Future studies will undoubtedly reveal other unforeseen roles for UPF and SMG factors. As one example, the recent identification of UPF1 as a component of microRNA-containing Argonaute 1 (AGO1) or AGO2 complexes in HeLa cells suggests that UPF1 might somehow function in AGO-mediated gene silencing119. Additionally, while this manuscript was in preparation, UPF1 was reported to be a component of HIV-1 RNP and to increase the level of nuclear HIV-1 RNA in a way that depends on translation of the GAG ORF but not on UPF1 function in NMD120.
In conclusion, the multitasking of SMG and UPF factors exemplifies cellular ‘unity in multiplicity’. The more we learn about individual cellular pathways, the clearer it becomes that a true understanding will only be achieved when each pathway is placed in the context of the complicated web of processes that constitute the whole of cellular metabolism.
Acknowledgments
We thank C. Woeller for reading the manuscript and help with its formatting and G. Chanfreau for conversations. The Maquat lab is supported by NIH R01 grants GM074593 and GM059514 to L.E.M.
Glossary
- Telomeres
Condensed repetitive DNA sequences at chromosomal ends in most eukaryotes that compensate for incomplete semi-conservative DNA replication by protecting against homologous recombination and non-homologous end joining, thereby conferring genomic stability
- Oxidative stress
A disturbance in the normal redox state of a cell that is due to an imbalance between the production of reactive oxygen and either detoxification of the resulting reactive intermediates (for example, peroxides and free radicals) or repair of the consequential damage. Oxidative-stress damage can potentially occur to all components of the cell, including proteins, lipids and DNA
- Phosphatidylinositol 3-kinase-related protein kinase
(PIKK). Examples include ATM, ATR, SMG1, DNA-PK and mTOR. PIKKs constitute a subfamily of serine and threonine kinases that resemble lipid kinase phosphatidylinositol 3-kinases (PI-3 kinases). PIKKs transduce signals in cell-growth and stress-response pathways
- Histone
Protein component of chromatin that functions to regulate gene expression and is synthesized coordinately with DNA replication. Two each of the core histones H2A, H2B, H3 and H4 make up an octameric nucleosome, around which DNA winds. The linker histone H1 binds the nucleosome, locking the DNA into place
- Premature termination codon
(PTC). UAA, UGA or UAG codon (that is, a nonsense codon, which generally does not encode an amino acid) within mRNA. PTCs are situated upstream of the normal termination codon and direct the premature termination of mRNA translation, which usually results in nonsense-mediated mRNA decay
- Exon-junction complex
(EJC). Complex of proteins that is deposited ~20–25 nucleotides upstream of the exon–exon junctions of newly synthesized spliced mRNAs. Despite its potentially ancient origin, the EJC functions in mammalian-cell and plant-cell NMD but not detectably in NMD in other organisms studied
- RNA cap-binding protein (CBP)
Protein that binds the 7-methyl guanosine cap structure at the 5′ end of mRNAs. In mammalian cells, mRNA is bound first by the mostly nuclear but shuttling CBP heterodimer CBP80–CBP20. Subsequently, CBP80–CBP20 is replaced by eukaryotic translation initiation factor 4E (eIF4E), a state that typifies the bulk of cellular mRNA
- NMD degradative activities
Decapping and 5′-to-3′ exonucleolytic activities as well as deadenylating and 3′-to-5′ exosome-mediated activities
- Selenoprotein mRNA
One of a number of mRNAs, exemplified by glutathione peroxidase 1 mRNA, that harbour one or more UGA codons that direct the incorporation of the twenty-first amino acid, selenocysteine. Incorporation, which competes with translation termination, requires at least one cis-acting selenocysteine insertion element that resides in the mRNA 3′ UTR
- Serine–arginine (SR)-rich protein
Member of an evolutionarily conserved family of essential pre-mRNA splicing factors in metazoans. Individual SR proteins have distinct but occasionally overlapping abilities to promote 5′ splice-site usage. They also function in post-transcriptional steps that occur after pre-mRNA splicing
- Heterogeneous nuclear ribonucleoprotein (hnRNP) splicing factor
hnRNP proteins that, in metazoans, can either repress or enhance splicing by antagonizing or promoting splice-site selection. Cooperative interactions between pre-mRNA-bound hnRNP proteins might enhance splicing between specific pairs of splice sites while at the same time inhibiting other combinations of splice-site usage
- Alternative splicing
Used to generate the multiple mRNAs that derive from at least 75% of human genes, often in a tissue-specific manner, as a means to generate more than one protein isoform per gene. An estimated one-third of alternatively spliced human mRNAs are targeted for NMD
- Pre-mRNA splicing
Nuclear process whereby one or more introns, or intervening sequences, are removed from a primary gene transcript, or a pre-mRNA, and the resulting exons are coordinately ligated together to form mRNA
- Upstream ORF
An open translational reading frame that exists upstream of a protein-encoding reading frame. Upstream ORFs are usually short and are often regulatory, lacking the potential to encode a functional protein. Translation termination at an upstream ORF can result in NMD
- Telomerase
A reverse transcriptase that uses its constituent RNA as a template to add specific DNA repeats — TTAGGG in all vertebrates — to the 3′-extended ends of telomeres, which are shortened after each replication cycle
Footnotes
DATABASES
UniProtKB: http://ca.expasy.org/sprot
Atf1 | ATR | CBP20 | CBP80 | Csx1 | eIF4E | PABP1 | SLBP | SMG1 | SMG5 | SMG6 | SMG7 | STAU1 | STAU2 | UPF1 | UPF2 | UPF3 | UPF3X
FURTHER INFORMATION
The Macquat laboratory: http://dbb.urmc.rochester.edu/labs/maquat/maquat_lab_people.htm
References
- 1.Behm-Ansmant I, et al. mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 2007;581:2845–2853. doi: 10.1016/j.febslet.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 2.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 3.Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. A comprehensive review of translation-dependent mRNA surveillance mechanisms, with a focus on NMD. [DOI] [PubMed] [Google Scholar]
- 4.Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nonsense-mediated decay approaches the clinic. Nature Genet. 2004;36:801–808. doi: 10.1038/ng1403. [DOI] [PubMed] [Google Scholar]
- 5.Amrani N, Sachs MS, Jacobson A. Early nonsense: mRNA decay solves a translational problem. Nature Rev Mol Cell Biol. 2006;7:415–425. doi: 10.1038/nrm1942. A comprehensive overview of NMD in S. cerevisiae. [DOI] [PubMed] [Google Scholar]
- 6.Meaux S, van Hoof A, Baker KE. Nonsense-mediated mRNA decay in yeast does not require PAB1 or a poly(A) tail. Mol Cell. 2008;29:134–140. doi: 10.1016/j.molcel.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiu SY, Lejeune F, Ranganathan AC, Maquat LE. The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes Dev. 2004;18:745–754. doi: 10.1101/gad.1170204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lejeune F, Ranganathan AC, Maquat LE. eIF4G is required for the pioneer round of translation in mammalian cells. Nature Struct Mol Biol. 2004;11:992–1000. doi: 10.1038/nsmb824. [DOI] [PubMed] [Google Scholar]
- 9.Sato H, Hosoda N, Maquat LE. Efficiency of the pioneer round of translation affects the cellular site of nonsense-mediated mRNA decay. Mol Cell. 2008;29:255–262. doi: 10.1016/j.molcel.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 10.Matsuda D, Hosoda N, Kim YK, Maquat LE. Failsafe nonsense-mediated mRNA decay does not detectably target eIF4E-bound mRNA. Nature Struct Mol Biol. 2007;14:974–979. doi: 10.1038/nsmb1297. [DOI] [PubMed] [Google Scholar]
- 11.Hosoda N, Lejeune F, Maquat LE. Evidence that poly(A) binding protein C1 binds nuclear pre-mRNA poly(A) tails. Mol Cell Biol. 2006;26:3085–3097. doi: 10.1128/MCB.26.8.3085-3097.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woeller CF, Gaspari M, Isken O, Maquat LE. NMD resulting from encephalomyocarditis virus IRES-directed translation initiation seems to be restricted to CBP80/20-bound mRNA. EMBO Rep. 2008;9:446–451. doi: 10.1038/embor.2008.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishigaki Y, Li X, Serin G, Maquat LE. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 14.Lejeune F, Ishigaki Y, Li X, Maquat LE. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J. 2002;21:3536–3545. doi: 10.1093/emboj/cdf345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hosoda N, Kim YK, Lejeune F, Maquat LE. CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nature Struct Mol Biol. 2005;12:893–901. doi: 10.1038/nsmb995. [DOI] [PubMed] [Google Scholar]
- 16.Isken O, et al. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133:314–327. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fortes P, et al. The yeast nuclear cap binding complex can interact with translation factor eIF4G and mediate translation initiation. Mol Cell. 2000;6:191–196. [PubMed] [Google Scholar]
- 18.Das B, Guo Z, Russo P, Chartrand P, Sherman F. The role of nuclear cap binding protein Cbc1p of yeast in mRNA termination and degradation. Mol Cell Biol. 2000;20:2827–2838. doi: 10.1128/mcb.20.8.2827-2838.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baron-Benhamou J, Fortes P, Inada T, Preiss T, Hentze MW. The interaction of the cap-binding complex (CBC) with eIF4G is dispensable for translation in yeast. RNA. 2003;9:654–662. doi: 10.1261/rna.5100903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao Q, Das B, Sherman F, Maquat LE. Cap-binding protein 1-mediated and eukaryotic translation initiation factor 4E-mediated pioneer rounds of translation in yeast. Proc Natl Acad Sci USA. 2005;102:4258–4263. doi: 10.1073/pnas.0500684102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park NI, Muench DG. Biochemical and cellular characterization of the plant ortholog of PYM, a protein that interacts with the exon junction complex core proteins Mago and Y14. Planta. 2007;225:625–639. doi: 10.1007/s00425-006-0385-y. [DOI] [PubMed] [Google Scholar]
- 22.Ballut L, et al. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nature Struct Mol Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- 23.Tange TO, Shibuya T, Jurica MS, Moore MJ. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA. 2005;11:1869–1883. doi: 10.1261/rna.2155905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gatfield D, Izaurralde E. REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J Cell Biol. 2002;159:579–588. doi: 10.1083/jcb.200207128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Longman D, Johnstone IL, Caceres JF. The REF/Aly proteins are dispensable for mRNA export and development in Caenorhabditis elegans. RNA. 2003;9:881–891. doi: 10.1261/rna.5420503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawano T, Kataoka N, Dreyfuss G, Sakamoto H. Ce-Y14 and MAG-1, components of the exon–exon junction complex, are required for embryogenesis and germline sexual switching in Caenorhabditis elegans. Mech Dev. 2004;121:27–35. doi: 10.1016/j.mod.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 2003;22:3960–3970. doi: 10.1093/emboj/cdg371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Longman D, Plasterk RH, Johnstone IL, Caceres JF. Mechanistic insights and identification of two novel factors in the C. elegans NMD pathway. Genes Dev. 2007;21:1075–1085. doi: 10.1101/gad.417707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4) Mol Cell Biol. 2001;21:209–223. doi: 10.1128/MCB.21.1.209-223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE. Y14 and hUpf3b form an NMD-activating complex. Mol Cell. 2003;11:939–949. doi: 10.1016/s1097-2765(03)00142-4. [DOI] [PubMed] [Google Scholar]
- 31.Kunz JB, Neu-Yilik G, Hentze MW, Kulozik AE, Gehring NH. Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. RNA. 2006;12:1015–1022. doi: 10.1261/rna.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tarpey PS, et al. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nature Genet. 2007;39:1127–1133. doi: 10.1038/ng2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lykke-Andersen J, Shu MD, Steitz JA. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 34.Mendell JT, Medghalchi SM, Lake RG, Noensie EN, Dietz HC. Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol Cell Biol. 2000;20:8944–8957. doi: 10.1128/mcb.20.23.8944-8957.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mendell JT, ap Rhys CM, Dietz HC. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science. 2002;298:419–422. doi: 10.1126/science.1074428. [DOI] [PubMed] [Google Scholar]
- 36.Gehring NH, et al. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol Cell. 2005;20:65–75. doi: 10.1016/j.molcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 37.Chan WK, et al. An alternative branch of the nonsense-mediated decay pathway. EMBO J. 2007;26:1820–1830. doi: 10.1038/sj.emboj.7601628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saltzman A, et al. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol Cell Biol. 2008;28:4320–4330. doi: 10.1128/MCB.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kashima I, et al. Binding of a novel SMG-1–Upf1–eRF1–eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 2008;27:736–747. doi: 10.1038/emboj.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amrani N, et al. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- 42.Wittmann J, Hol EM, Jack HM. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol Cell Biol. 2006;26:1272–1287. doi: 10.1128/MCB.26.4.1272-1287.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohnishi T, et al. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell. 2003;12:1187–1200. doi: 10.1016/s1097-2765(03)00443-x. [DOI] [PubMed] [Google Scholar]
- 44.Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nature Struct Mol Biol. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- 45.Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001;15:2215–2228. doi: 10.1101/gad.913001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grimson A, O’Connor S, Newman CL, Anderson P. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA decay in Caenorhabditis elegans. Mol Cell Biol. 2004;24:7483–7490. doi: 10.1128/MCB.24.17.7483-7490.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Unterholzner L, Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell. 2004;16:587–596. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 48.Glavan F, Behm-Ansmant I, Izaurralde E, Conti E. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J. 2006;25:5117–5125. doi: 10.1038/sj.emboj.7601377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brumbaugh KM, et al. The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol Cell. 2004;14:585–598. doi: 10.1016/j.molcel.2004.05.005. This paper implicates SMG1, the newest member of the family of protein serine-threonine kinases, in stress-induced signalling pathways. [DOI] [PubMed] [Google Scholar]
- 50.Page MF, Carr B, Anders KR, Grimson A, Anderson P. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol Cell Biol. 1999;19:5943–5951. doi: 10.1128/mcb.19.9.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johns L, Grimson A, Kuchma SL, Newman CL, Anderson P. Caenorhabditis elegans SMG-2 selectively marks mRNAs containing premature translation termination codons. Mol Cell Biol. 2007;27:5630–5638. doi: 10.1128/MCB.00410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Z, Smith KR, Batterham P, Robin C. Smg1 nonsense mutations do not abolish nonsense-mediated mRNA decay in Drosophila melanogaster. Genetics. 2005;171:403–406. doi: 10.1534/genetics.105.045674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Metzstein MM, Krasnow MA. Functions of the nonsense-mediated mRNA decay pathway in Drosophila development. PLoS Genet. 2006;2:e180. doi: 10.1371/journal.pgen.0020180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang W, Cajigas IJ, Peltz SW, Wilkinson MF, Gonzalez CI. Role for Upf2p phosphorylation in Saccharomyces cerevisiae nonsense-mediated mRNA decay. Mol Cell Biol. 2006;26:3390–3400. doi: 10.1128/MCB.26.9.3390-3400.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luke B, et al. Saccharomyces cerevisiae Ebs1p is a putative ortholog of human Smg7 and promotes nonsense-mediated mRNA decay. Nucleic Acids Res. 2007;35:7688–7697. doi: 10.1093/nar/gkm912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lynch M, Hong X, Scofield DS. In: Nonsense-Mediated mRNA Decay. Maquat LE, editor. pp. 197–211. Landes Bioscience, Georgetown, 2006). A beautifully written treatise on the association of NMD, the EJC and the location of introns within genes on the basis of phylogenetic analyse. [Google Scholar]
- 57.Lelivelt MJ, Culbertson MR. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol Cell Biol. 1999;19:6710–6719. doi: 10.1128/mcb.19.10.6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He F, et al. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell. 2003;12:1439–1452. doi: 10.1016/s1097-2765(03)00446-5. [DOI] [PubMed] [Google Scholar]
- 59.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nature Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 60.Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA. 2005;11:1530–1544. doi: 10.1261/rna.2160905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodriguez-Gabriel MA, Watt S, Bahler J, Russell P. Upf1, an RNA helicase required for nonsense-mediated mRNA decay, modulates the transcriptional response to oxidative stress in fission yeast. Mol Cell Biol. 2006;26:6347–6356. doi: 10.1128/MCB.00286-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johansson MJ, He F, Spatrick P, Li C, Jacobson A. Association of yeast Upf1p with direct substrates of the NMD pathway. Proc Natl Acad Sci USA. 2007;104:20872–20877. doi: 10.1073/pnas.0709257105. This paper and reference 63 use microarray analyses to identify direct targets of NMD in S. cerevisiae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guan Q, et al. Impact of nonsense-mediated mRNA decay on the global expression profile of budding yeast. PLoS Genet. 2006;2:e203. doi: 10.1371/journal.pgen.0020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moriarty PM, Reddy CC, Maquat LE. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codo-nmediated decay of cytoplasmic mRNA. Mol Cell Biol. 1998;18:2932–2939. doi: 10.1128/mcb.18.5.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wen W, Weiss SL, Sunde RA. UGA codon position affects the efficiency of selenocysteine incorporation into glutathione peroxidase-1. J Biol Chem. 1998;273:28533–28541. doi: 10.1074/jbc.273.43.28533. [DOI] [PubMed] [Google Scholar]
- 66.Mitrovich QM, Anderson P. mRNA surveillance of expressed pseudogenes in C. elegans. Curr Biol. 2005;15:963–967. doi: 10.1016/j.cub.2005.04.055. [DOI] [PubMed] [Google Scholar]
- 67.Ni JZ, et al. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. In parallel with reference 76, this paper identifies RNA-binding transcripts containing ultraconserved sequences that are subject to alternative splicing-activated NMD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weischenfeldt J, et al. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 2008;22:1381–1396. doi: 10.1101/gad.468808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sureau A, Gattoni R, Dooghe Y, Stevenin J, Soret J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 2001;20:1785–1796. doi: 10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wollerton MC, Gooding C, Wagner EJ, Garcia-Blanco MA, Smith CW. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol Cell. 2004;13:91–100. doi: 10.1016/s1097-2765(03)00502-1. [DOI] [PubMed] [Google Scholar]
- 72.Jumaa H, Nielsen PJ. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 1997;16:5077–5085. doi: 10.1093/emboj/16.16.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lejeune F, Cavaloc Y, Stevenin J. Alternative splicing of intron 3 of the serine/arginine-rich protein 9G8 gene. Identification of flanking exonic splicing enhancers and involvement of 9G8 as a trans-acting factor. J Biol Chem. 2001;276:7850–7858. doi: 10.1074/jbc.M009510200. [DOI] [PubMed] [Google Scholar]
- 74.Spellman R, Llorian M, Smith CW. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol Cell. 2007;27:420–434. doi: 10.1016/j.molcel.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- 77.Morrison M, Harris KS, Roth MB. smg mutants affect the expression of alternatively spliced SR protein mRNAs in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1997;94:9782–9785. doi: 10.1073/pnas.94.18.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kalyna M, Lopato S, Barta A. Ectopic expression of atRSZ33 reveals its function in splicing and causes pleiotropic changes in development. Mol Biol Cell. 2003;14:3565–3577. doi: 10.1091/mbc.E03-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kalyna M, Lopato S, Voronin V, Barta A. Evolutionary conservation and regulation of particular alternative splicing events in plant SR proteins. Nucleic Acids Res. 2006;34:4395–4405. doi: 10.1093/nar/gkl570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kumar S, Lopez AJ. Negative feedback regulation among SR splicing factors encoded by Rbp1 and Rbp1-like in Drosophila. EMBO J. 2005;24:2646–2655. doi: 10.1038/sj.emboj.7600723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pan Q, et al. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes Dev. 2006;20:153–158. doi: 10.1101/gad.1382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rehwinkel J, Raes J, Izaurralde E. Nonsense-mediated mRNA decay: Target genes and functional diversification of effectors. Trends Biochem Sci. 2006;31:639–646. doi: 10.1016/j.tibs.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 83.Martens JA, Wu PY, Winston F. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 2005;19:2695–2704. doi: 10.1101/gad.1367605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thompson DM, Parker R. Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:92–101. doi: 10.1128/MCB.01023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gaba A, Jacobson A, Sachs MS. Ribosome occupancy of the yeast CPA1 upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol Cell. 2005;20:449–460. doi: 10.1016/j.molcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 86.Pain VM. Translational control during amino acid starvation. Biochimie. 1994;76:718–728. doi: 10.1016/0300-9084(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 87.Gardner LB. Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response. Mol Cell Biol. 2008;28:3729–3741. doi: 10.1128/MCB.02284-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lew JE, Enomoto S, Berman J. Telomere length regulation and telomeric chromatin require the nonsense-mediated mRNA decay pathway. Mol Cell Biol. 1998;18:6121–6130. doi: 10.1128/mcb.18.10.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dahlseid JN, et al. mRNAs encoding telomerase components and regulators are controlled by UPF genes in Saccharomyces cerevisiae. Eukaryotic Cell. 2003;2:134–142. doi: 10.1128/EC.2.1.134-142.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Enomoto S, Glowczewski L, Lew-Smith J, Berman JG. Telomere cap components influence the rate of senescence in telomerase-deficient yeast cells. Mol Cell Biol. 2004;24:837–845. doi: 10.1128/MCB.24.2.837-845.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Reichenbach P, et al. A human homolog of yeast Est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr Biol. 2003;13:568–574. doi: 10.1016/s0960-9822(03)00173-8. This paper and reference 116 implicate SMG6 as a telomerase-associated factor that controls telomere length. [DOI] [PubMed] [Google Scholar]
- 92.Keene JD. RNA regulons: coordination of post-transcriptional events. Nature Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 93.St Johnston D. The beginning of the end. EMBO J. 2001;20:6169–6179. doi: 10.1093/emboj/20.22.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Roegiers F, Jan YN. Staufen: a common component of mRNA transport in oocytes and neurons? Trends Cell Biol. 2000;10:220–224. doi: 10.1016/s0962-8924(00)01767-0. [DOI] [PubMed] [Google Scholar]
- 95.Mouland AJ, et al. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: evidence for a role in genomic RNA encapsidation. J Virol. 2000;74:5441–5451. doi: 10.1128/jvi.74.12.5441-5451.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chatel-Chaix L, et al. Identification of Staufen in the human immunodeficiency virus type 1 Gag ribonucleoprotein complex and a role in generating infectious viral particles. Mol Cell Biol. 2004;24:2637–2648. doi: 10.1128/MCB.24.7.2637-2648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′ UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. This is the first demonstration that UPF1 can be recruited to particular mRNAs by a mechanism other than NMD to trigger mRNA decay when translation terminates normally. [DOI] [PubMed] [Google Scholar]
- 98.Kim YK, et al. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J. 2007;26:2670–2681. doi: 10.1038/sj.emboj.7601712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Furic L, Maher-Laporte M, DesGroseillers L. A genome-wide approach identifies distinct but overlapping subsets of cellular mRNAs associated with Staufen1- and Staufen2-containing ribonucleoprotein complexes. RNA. 2008;14:324–335. doi: 10.1261/rna.720308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Monshausen M, Gehring NH, Kosik KS. The mammalian RNA-binding protein Staufen2 links nuclear and cytoplasmic RNA processing pathways in neurons. Neuromolecular Med. 2004;6:127–144. doi: 10.1385/NMM:6:2-3:127. [DOI] [PubMed] [Google Scholar]
- 101.Kaygun H, Marzluff WF. In: Nonsense-Mediated mRNA Decay. Maquat LE, editor. Landes Bioscience; Georgetown: 2006. pp. 237–253. [Google Scholar]
- 102.Kaygun H, Marzluff WF. Translation termination is involved in histone mRNA degradation when DNA replication is inhibited. Mol Cell Biol. 2005;25:6879–6888. doi: 10.1128/MCB.25.16.6879-6888.2005. This is an important demonstration that SLBP recruits UPF1 to the 3′ UTR of replication-dependent histone mRNAs to elicit histone mRNA decay at the end of the S phase of the cell cycle. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kaygun H, Marzluff WF. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nature Struct Mol Biol. 2005;12:794–800. doi: 10.1038/nsmb972. [DOI] [PubMed] [Google Scholar]
- 104.Muller B, Blackburn J, Feijoo C, Zhao X, Smythe C. DNA-activated protein kinase functions in a newly observed S phase checkpoint that links histone mRNA abundance with DNA replication. J Cell Biol. 2007;179:1385–1398. doi: 10.1083/jcb.200708106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Azzalin CM, Lingner J. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr Biol. 2006;16:433–439. doi: 10.1016/j.cub.2006.01.018. A significant piece of work demonstrating that UPF1 is required for human cells to progress completely through S phase and that ATR-mediated UPF1 phosphorylation promotes UPF1 binding to chromatin. [DOI] [PubMed] [Google Scholar]
- 106.van Heemst D, den Reijer PM, Westendorp RG. Ageing or cancer: a review on the role of caretakers and gatekeepers. Eur J Cancer. 2007;43:2144–2152. doi: 10.1016/j.ejca.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 107.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nature Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 108.Erker L, et al. Cancer chemoprevention by the antioxidant tempol acts partially via the p53 tumor suppressor. Hum Mol Genet. 2005;14:1699–1708. doi: 10.1093/hmg/ddi181. [DOI] [PubMed] [Google Scholar]
- 109.Gehen SC, Staversky RJ, Bambara RA, Keng PC, O’Reilly MA. hSMG-1 and ATM sequentially and independently regulate the G(1) checkpoint during oxidative stress. Oncogene. 2008 Mar 10; doi: 10.1038/onc.2008.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Oliveira V, et al. A protective role for the human SMG-1 kinase against tumor necrosis factor-alpha-induced apoptosis. J Biol Chem. 2008;283:13174–13184. doi: 10.1074/jbc.M708008200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Abraham RT, Oliveira V. In: Nonsense-Mediated mRNA Decay. Maquat LE, editor. Landes Bioscience; Georgetown: 2006. pp. 215–229. [Google Scholar]
- 112.Neben K, et al. Distinct gene expression patterns associated with FLT3- and NRAS-activating mutations in acute myeloid leukemia with normal karyotype. Oncogene. 2005;24:1580–1588. doi: 10.1038/sj.onc.1208344. [DOI] [PubMed] [Google Scholar]
- 113.Pal M, Ishigaki Y, Nagy E, Maquat LE. Evidence that phosphorylation of human Upfl protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA. 2001;7:5–15. doi: 10.1017/s1355838201000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Carastro LM, et al. Identification of delta helicase as the bovine homolog of HUPF1: demonstration of an interaction with the third subunit of DNA polymerase delta. Nucleic Acids Res. 2002;30:2232–2243. doi: 10.1093/nar/30.10.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cheung AL, Deng W. Telomere dysfunction, genome instability and cancer. Front Biosci. 2008;13:2075–2090. doi: 10.2741/2825. [DOI] [PubMed] [Google Scholar]
- 116.Snow BE, et al. Functional conservation of the telomerase protein Est1p in humans. Curr Biol. 2003;13:698–704. doi: 10.1016/s0960-9822(03)00210-0. [DOI] [PubMed] [Google Scholar]
- 117.Redon S, Reichenbach P, Lingner J. Protein RNA and protein protein interactions mediate association of human EST1A/SMG6 with telomerase. Nucleic Acids Res. 2007;35:7011–7022. doi: 10.1093/nar/gkm724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. A landmark paper showing that mammalian telomeres are transcribed, and telomere integrity is regulated, by NMD factors. [DOI] [PubMed] [Google Scholar]
- 119.Hock J, et al. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ajamian L, et al. Unexpected roles for UPF1 in HIV-1 RNA metabolism and translation. RNA. 2008;14:914–927. doi: 10.1261/rna.829208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kataoka N, Diem MD, Kim VN, Yong J, Dreyfuss G. Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon–exon junction complex. EMBO J. 2001;20:6424–6433. doi: 10.1093/emboj/20.22.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dostie J, Dreyfuss G. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr Biol. 2002;12:1060–1067. doi: 10.1016/s0960-9822(02)00902-8. [DOI] [PubMed] [Google Scholar]
- 123.Chan CC, et al. eIF4A3 is a novel component of the exon junction complex. RNA. 2004;10:200–209. doi: 10.1261/rna.5230104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Degot S, et al. Association of the breast cancer protein MLN51 with the exon junction complex via its speckle localizer and RNA binding module. J Biol Chem. 2004;279:33702–33715. doi: 10.1074/jbc.M402754200. [DOI] [PubMed] [Google Scholar]
- 125.Baguet A, et al. The exon-junction-complex-component metastatic lymph node 51 functions in stress-granule assembly. J Cell Sci. 2007;120:2774–2784. doi: 10.1242/jcs.009225. [DOI] [PubMed] [Google Scholar]
- 126.Palacios IM, Gatfield D, St Johnston D, Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427:753–757. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- 127.Denning G, Jamieson L, Maquat LE, Thompson EA, Fields AP. Cloning of a novel phosphatidylinositol kinase-related kinase: characterization of the human SMG-1 RNA surveillance protein. J Biol Chem. 2001;276:22709–22714. doi: 10.1074/jbc.C100144200. [DOI] [PubMed] [Google Scholar]
- 128.Fukuhara N, et al. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell. 2005;17:537–547. doi: 10.1016/j.molcel.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 129.Kadlec J, Guilligay D, Ravelli RB, Cusack S. Crystal structure of the UPF2-interacting domain of nonsense-mediated mRNA decay factor UPF1. RNA. 2006;12:1817–1824. doi: 10.1261/rna.177606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kadlec J, Izaurralde E, Cusack S. The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nature Struct Mol Biol. 2004;11:330–337. doi: 10.1038/nsmb741. [DOI] [PubMed] [Google Scholar]
- 131.Kim VN, et al. The Y14 protein communicates to the cytoplasm the position of exon–exon junctions. EMBO J. 2001;20:2062–2068. doi: 10.1093/emboj/20.8.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chiu SY, Serin G, Ohara O, Maquat LE. Characterization of human Smg5/7a: a protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA. 2003;9:77–87. doi: 10.1261/rna.2137903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Le Hir H, Gatfield D, Braun IC, Forler D, Izaurralde E. The protein Mago provides a link between splicing and mRNA localization. EMBO Rep. 2001;2:1119–1124. doi: 10.1093/embo-reports/kve245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shibuya T, Tange TO, Sonenberg N, Moore MJ. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nature Struct Mol Biol. 2004;11:346–351. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- 135.Ferraiuolo MA, et al. A nuclear translation-like factor eIF4AIII is recruited to the mRNA during splicing and functions in nonsense-mediated decay. Proc Natl Acad Sci USA. 2004;101:4118–4123. doi: 10.1073/pnas.0400933101. [DOI] [PMC free article] [PubMed] [Google Scholar]