Summary
Thrombospondins are secreted, extracellular matrix (ECM) proteins that are upregulated in the heart and other tissues in response to ischemic injury and myocardial stress. Roles for thrombospondins after they are secreted have been examined in a variety of disease models, including myocardial pathology. However, a recent study published in the journal Cell by Lynch et al1 shifts this paradigm by focusing on roles for intracellular thrombospondins; these authors showed that thrombospondin-4 (Thbs4) can function from within cells to protect the heart by enhancing adaptive aspects of the endoplasmic reticulum (ER) stress response that are mediated by activating transcription factor 6, ATF6. Although this study was carried out in the cardiac context, the results add to our understanding of protein folding and quality control in all tissues. Moreover, the findings underscore the potential widespread therapeutic benefit of enhancing adaptive responses that are regulated by ATF6.
Keywords: thrombospondin-4, ATF6, GRP78, ER stress, unfolded protein, cardioprotection
The study by Lynch et al1, which was carried out in the laboratory of Dr Jeffery Molkentin, employed a series of molecular approaches, including in vivo gain- and loss-of-function models in mice to demonstrate new roles for Thbs4 in the management of ER protein quality control in the heart. The ER is the site of synthesis and folding of most secreted and membrane proteins, which constitute at least 35% of all proteins2. Secreted and membrane proteins are synthesized by ER-bound ribosomes, co-translationally translocated across the ER membrane, and folded in the lumen of the ER, after which they are transported to the Golgi, where they are sorted to their final destinations3, 4. The ER lumen is populated with chaperones, protein disulfide isomerases, protein oxidoreductases, and other proteins that are responsible for the proper folding of secreted and membrane proteins. This elaborate machinery requires an optimal ER environment for efficient protein folding. Conditions such as myocardial ischemia, hypertrophy, and heart failure alter this environment in ways that reduce protein folding, leading to the accumulation of misfolded, potentially toxic5 proteins that can cause ER stress6.
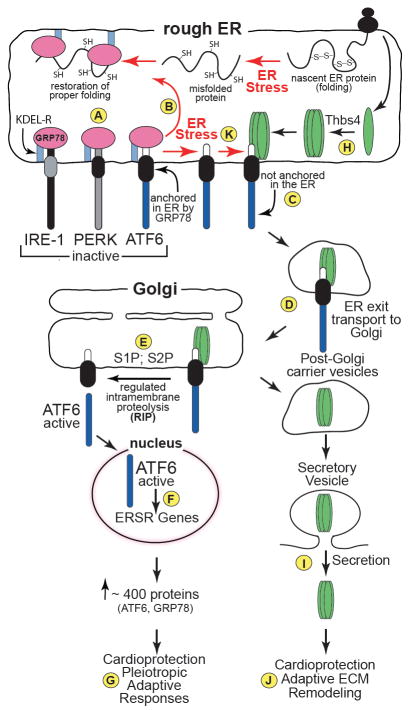
The ER stress response averts the potential proteotoxicity associated with the accumulation of misfolded proteins in the ER during ER stress7-9. Three ER transmembrane proteins, PERK (protein kinase RNA-like ER kinase), IRE-1 (inositol-requiring protein-1), and ATF6 (activating transcription factor 6) serve as sensors of misfolded proteins in the ER as well as effectors of the response to ER stress (Fig. A). Although the mechanisms by which these sensors detect misfolded proteins are not completely understood, one of the first mechanisms to be described involves the ubiquitous ER-luminal chaperone, glucose regulated protein 78 kD (GRP78), which plays numerous roles in ER protein folding and quality control10, 11. When protein folding in the ER is optimal, GRP78, which has a C-terminal ER retention motif, or KDEL, binds to the ER-luminal domains of the three proximal sensors and keeps them inactive12, 13 (Fig. A). Upon ER stress, GRP78 relocates from the proximal sensors to misfolded proteins in efforts to fold them (Fig. B). This relocation of GRP78 has different effects on the three sensors. In the case of ATF6, which is a major topic of the study by Lynch et al, the relocation of GRP78 releases ATF6 from the ER, permitting its transport to the Golgi (Fig. C), which is facilitated by Golgi localization signals13. In the Golgi, Site-1 and Site-2 proteases (S1P and S2P) cleave the trans-ER membrane region of ATF6 (Fig. D). This liberates the N-terminal, cytosolic domain of ATF6, which translocates to the nucleus, where it binds to and induces nearly 400 genes in the myocardium14 (Fig. E); many of these genes are known to contribute to restoring ER protein folding capacitymuy and enhancing cardioprotection15 via pleiotropic adaptive responses14 (Fig. F). Thus, the conditional interaction of ATF6 with the ER-resident GRP78 is a determinant of ATF6 location and, thus, ATF6 activity. Lynch et al found that ATF6 location and activity can also be determined by its interaction with Thbs4.
Figure. Roles of GRP78 and Thrombospondin-4 in ATF6 Activation.
The ER-transmembrane proteins, IRE-1, PERK, and ATF6, are sensors of protein folding in the ER. When ER protein folding is optimal, the ER luminal domains of each sensor are associated with the ER chaperone, GRP78 (A). GRP78 is an ER resident protein by virtue of its C-terminal KDEL sequence, which facilitates its retrieval from the Golgi to the ER via its binding to the KDEL receptor. Because it binds to GRP78, ATF6 is anchored in the ER, which keeps ATF6 from being activated. However, when ER protein folding is impaired, for example, during ER stress (red arrows), GRP78 relocates to misfolded proteins as they accumulate (B); since, under these conditions, GRP78 is no longer bound to ATF6 in the ER, ATF6 is free to relocate to the Golgi (C). In the Golgi, ATF6 is cleaved by S1P and S2P (D) to form an active transcription factor that induces ER stress response genes (E), including ATF6 and GRP78, which primarily mediate pleiotropic adaptive, or protective responses (F). Thrombospondin-4 (Thbs4) is synthesized as monomers in the ER, which assemble into pentamers (G) that transit from the ER lumen to the Golgi (H), then to secretory vesicles, from which Thbs4 is eventually secreted (I). Extracellular Thbs4 binds to structural ECM proteins and modulates the ECM in response to tissue damage, providing cardioprotective and adaptive ECM remodeling (J). The study by Lynch et al showed that Thbs4 can bind to ATF6 (K) and facilitate its activation, most likely by facilitating its translocation to the Golgi (L). Since Thbs4 is retained in the ER during some ER stress treatments that are known to stimulate ATF6 translocation to the Golgi (see text), it may be that there are some conditions under which ATF6 relocation from the ER to the Golgi, and thus, ATF6 activation can take place in a Thbs4-independent manner (C and D).
The thrombospondin (Thbs) family is composed of five members (Thbs1-5) whose expression and secretion from numerous cell types, including cardiac myocytes, is increased during pathology and tissue damage16. Thrombospondins form two subfamilies. Thrombospondins 1 and 2 assemble as homo- and heterotrimers, while the remaining Thsb family members form homo- or heteropentamers. Most studies have focused on the functions of Thbs after its secretion, upon which it interacts with various structural ECM proteins and contributes to matrix remodeling during the course of pathology and response to tissue injury. Under most conditions, Thbs is expressed at low levels in cardiac myocytes, but is expressed at moderate levels in other cell types in the heart 17. Thbs1 and Thbs2 knockout mouse studies have shown that these thrombospondins protect against myocardial infarction (MI) injury and overload-induced hypertrophy18. Thbs4 has captured recent attention because it is expressed primarily in cardiac and skeletal muscle, and is further upregulated upon myocardial infarction, hypertrophy, and heart failure19, 20.
The synthesis and assembly of Thbs4 has not been studied in detail; however, by analogy to studies on other Thbs family members17, 21, 22, it can be inferred that Thbs4 monomers (~900 AA, ~100 kD each), which have no transmembrane motifs, are synthesized on ER-bound ribosomes, followed by folding and assembly into hetero- or homopentameric oligomers in the ER lumen (Fig. G; shown as homopentamers). Since it does not have a C-terminal KDEL motif to facilitate ER retention, Thbs4 presumably travels rapidly from the ER to the Golgi (Fig. H) on its way to secretion via the constitutive secretory pathway (Fig. I), a process by which most secreted proteins are released from cells at rates dictated primarily by their expression levels3, 23. Although Thbs secretion from cardiac myocytes has not been studied in detail, Thbs has been shown to be released from endothelial cells within 60 min of its synthesis21, consistent with rapid, constitutive secretion. To fold properly thrombospondins must be glycosylated. In endothelial cells, the impairment of protein glycosylation in the ER by tunicamycin leads to Thbs misfolding, which results in its retention in the ER and inhibition of secretion21. After its secretion, Thbs contributes to cardioprotection through processes such as adaptive extracellular matrix remodeling (Fig. J).
Targeted disruption of Thbs4 in the mouse heart has been shown to increase the maladaptive effects of pressure overload; these effects were attributed to extracellular Thbs418, 24, 25. However, the study by Lynch et al represents a paradigm shift, because it identifies a new function for intracellular Thbs4, linking it to ATF6-mediated cardioprotection. To determine the role of Thbs4 in the heart, Lynch et al overexpressed Thbs4 in mouse hearts and showed that it enhanced survival and preserved cardiac function after MI. The effects of pressure overload were unclear, since there was no apparent loss of cardiac function in either control or Thbs4 transgenic mice subjected to trans-aortic constriction. The authors performed a gene expression analysis to gain further insight into the mechanism of Thbs4-mediated cardioprotection; although the data were not shown, the authors concluded that many genes previously shown to be increased in the hearts of transgenic mice expressing activated ATF6 in cardiac myocytes14 were also upregulated in the hearts of Thbs4 transgenic mice. The authors showed that Thbs4 increased the levels of cleaved ATF6, as well as numerous ATF6-inducible proteins. This finding led to the authors’ hypothesis that Thbs4 might exert at least some of its protective effects by facilitating ATF6 activation. In support of this hypothesis, the authors showed that Thbs4 interacted with ATF6. To examine whether Thbs4-mediated activation of ATF6 required translocation to the Golgi, they generated a form of Thbs4 with a C-terminal ER retention KDEL motif (Thbs- KDEL), which should convert Thbs4 into an ER resident protein. Unlike native Thbs4, Thbs4-KDEL did not increase the levels of activated ATF6, presumably because it resided in the ER and was thus unable to facilitate ATF6 transport to the Golgi. The study also showed that the ATF6 activation observed in the hearts of wild-type mice subjected to overload-induced hypertrophy and MI was lost in Thbs4 knockout mouse hearts. Moreover, compared to wild-type mice, the hearts of the Thbs4 knockout mice exhibited reduced cardiac function and increased injury in the hypertrophy and MI models, respectively. These results are consistent with the hypothesis that Thbs4 mediates protection from cardiac pathology, at least partly by activating ATF6 and the subsequent expression of genes downstream of ATF6.
The results of this study have broad impact because they suggest that, in addition to its roles in ECM remodeling after secretion, Thbs4 has cardioprotective functions before its secretion. Moreover, since Lynch et al showed that Thbs1, which is in a different subfamily, and structurally somewhat different than Thbs4, also facilitated ATF6 activation, it is apparent that other members of the Thbs family activate ATF6. In terms of determining the relative importance of intra- and extracellular Thbs, Lynch et al showed that, in contrast to overexpressed Thbs4, the addition of recombinant Thbs4 to culture media did not increase activated ATF6, consistent with the idea that only intracellular Thbs4 can bind to and activate ATF6. However, the relative contributions of intra- and extracellular Thbs4 in the cardioprotection that was observed in vivo remain unknown.
As a liberator of ATF6 from the ER, Thbs4 is a counterpart of GRP78, which anchors ATF6 in the ER. The mechanism responsible for these opposing actions is likely to be complex and will require further study to delineate. However, coupled with previous studies, the study by Lynch et al supports several different mechanisms. For example, since overexpressing Thbs4 activates ATF6, and since ATF6 activation requires its liberation from the ER, then overexpressed Thbs4 must liberate ATF6 from the ER by overcoming the ability of GRP78 to anchor ATF6 in the ER. This could happen if Thbs4 is able to physically displace GRP78 from ATF6. The relatively close proximity of the GRP78 binding sites on ATF6, which were described previously13, to the Tbhs4 binding sites on ATF6, described by Lynch et al, supports this hypothesis, leading to the concept that the winner of the competition serves as a determinant of ATF6 location. Moreover, the site on ATF6 to which Lynch et al showed Thbs4 binds was previously shown to serve as a Golgi localization sequence, which is masked when GRP78 binds to ATF613. This is consistent with the hypothesis that GRP78 dislocation from ATF6 unmasks a Golgi localization sequence to which Thbs4 can bind (Fig. K), which then facilitates ATF6 translocation to the Golgi (Fig. L). Thus, the relative amounts of GRP78 and Thbs4 could determine whether ATF6 remains in the ER or relocates to the Golgi. Furthermore, it is possible that Thbs4 overexpression may additionally contribute to ATF6 translocation to the Golgi by attracting GRP78 away from ATF6 to assist in the folding of nascent Thbs4. In support of this possibility are previous studies showing that GRP78 binds to Thbs while it is being synthesized in the ER26, 27.
A provocative result from this study was the finding that overexpressed Thbs4 increased the secretion of both atrial natriuretic factor (ANF) and mesencephalic astrocyte-derived neurotrophic factor (MANF)1 from neonatal cardiac myocytes. The authors suggested that this may be due to the ability of Thbs4 to enhance the secretory capacity of the ER. While this may be a contributing factor, the ability of Thbs4 to coordinately increase the secretion of both of these proteins is unexpected, since ANF and MANF are secreted under different conditions and via different mechanisms. In the case of ANF, since its secretion from ventricular myocytes is constitutive, and therefore dictated primarily by its expression levels, increased levels of secreted ANF are a reflection of increased cellular levels of ANF28. Thus, perhaps Thbs4 overexpression increased cellular ANF, which resulted in increased secretion. In contrast to ANF, MANF secretion from ventricular myocytes is not constitutive because, like ATF6, MANF binds to GRP78, which conditionally retains MANF in the ER. Disruption of MANF binding to GRP78 has been shown to release MANF from the ER and, since it has no transmembrane domains, MANF is secreted29. Similar to ATF6, it is the conditional association of MANF with GRP78 that dictates its location. Accordingly, it may be that Thbs4 increases MANF secretion by liberating it from the ER-anchoring effects of GRP78, much like it does with ATF6.
The study by Lynch et al did not address whether Thbs4 is required for the activation of ATF6 during ER stress. However, previous studies have shown that when ER stress is activated, either by inhibiting protein glycosylation with tunicamycin, or by decreasing ER calcium, which are conditions under which ATF6 translocates from the ER to the Golgi where it is cleaved and activated, Thbs exit from the ER and secretion are blocked21, 30 and Thbs is degraded. Accordingly, at least under these conditions of ER stress, the binding of Thbs4 to ATF6 may not be required for ATF6 translocation and activation (Fig. L). Thbs4-mediated activation of ATF6 in the absence of ER stress is consistent with the hypothesis that ATF6 may have functions beyond its known roles as a first responder to ER protein misfolding31, 32. In support of this hypothesis is a study by Wu et al33 which showed that ATF6 was activated during physiological skeletal muscle exercise, and that this activation was required for metabolic adaptation to exercise training. This supports roles for ATF6-mediated adaptive responses beyond those that take place upon acute ER protein misfolding. Perhaps, like it does in skeletal muscle, ATF6 contributes to adaptive responses to physiological conditions in cardiac muscle that are not necessarily associated with overt activation of ER stress. Moreover, the findings reported by this study raise awareness to the potentially beneficial effects of Thbs4 and ATF6 in treating a vast array of pathologies associated with protein misfolding in the ER.
Acknowledgments
Sources of Funding: National Institutes of Health, HL-075573, HL-085577, and HL104535 to CCG, and by grants and fellowships from the Rees-Stealy Research Foundation, the San Diego Chapter of the Achievement Rewards for College Scientists (ARCS) Foundation, the American Heart Association (Predoctoral Fellowship 10PRE3410005), and the Inamori Foundation, to SD.
Footnotes
Disclosures: None
MANF was called ARMET (arginine-rich, mutated in early stage tumors) by Lynch et al.
References
- 1.Lynch JM, Maillet M, Vanhoutte D, Schloemer A, Sargent MA, Blair NS, Lynch KA, Okada T, Aronow BJ, Osinska H, Prywes R, Lorenz JN, Mori K, Lawler J, Robbins J, Molkentin JD. A thrombospondin-dependent pathway for a protective ER stress response. Cell. 2012;149(6):1257–1268. doi: 10.1016/j.cell.2012.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palade GE, Siekevitz P. Liver microsomes; an integrated morphological and biochemical study. J Biophys Biochem Cytol. 1956;2(2):171–200. doi: 10.1083/jcb.2.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halban PA, Irminger JC. Sorting and processing of secretory proteins. Biochem J. 1994;299(Pt 1):1–18. doi: 10.1042/bj2990001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doroudgar S, Glembotski CC. The cardiokine story unfolds: ischemic stress-induced protein secretion in the heart. Trends Mol Med. 2011;17(4):207–214. doi: 10.1016/j.molmed.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roth DM, Balch WE. Modeling general proteostasis: proteome balance in health and disease. Curr Opin Cell Biol. 2011;23(2):126–134. doi: 10.1016/j.ceb.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glembotski CC. Endoplasmic reticulum stress in the heart. Circ Res. 2007;101(10):975–984. doi: 10.1161/CIRCRESAHA.107.161273. [DOI] [PubMed] [Google Scholar]
- 7.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334(6059):1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 8.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13(2):89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 9.Rutkowski DT, Hegde RS. Regulation of basal cellular physiology by the homeostatic unfolded protein response. J Cell Biol. 2010;189(5):783–794. doi: 10.1083/jcb.201003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ni M, Zhang Y, Lee AS. Beyond the endoplasmic reticulum: atypical GRP78 in cell viability, signalling and therapeutic targeting. Biochem J. 2011;434(2):181–188. doi: 10.1042/BJ20101569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gething MJ. Role and regulation of the ER chaperone BiP. Semin Cell Dev Biol. 1999;10(5):465–472. doi: 10.1006/scdb.1999.0318. [DOI] [PubMed] [Google Scholar]
- 12.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2(6):326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 13.Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3(1):99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 14.Belmont PJ, Chen WJ, San Pedro MN, Thuerauf DJ, Gellings Lowe N, Gude N, Hilton B, Wolkowicz R, Sussman MA, Glembotski CC. Roles for endoplasmic reticulum-associated degradation and the novel endoplasmic reticulum stress response gene Derlin-3 in the ischemic heart. Circ Res. 2010;106(2):307–316. doi: 10.1161/CIRCRESAHA.109.203901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martindale JJ, Fernandez R, Thuerauf D, Whittaker R, Gude N, Sussman MA, Glembotski CC. Endoplasmic reticulum stress gene induction and protection from ischemia/reperfusion injury in the hearts of transgenic mice with a tamoxifen-regulated form of ATF6. Circ Res. 2006;98(9):1186–1193. doi: 10.1161/01.RES.0000220643.65941.8d. [DOI] [PubMed] [Google Scholar]
- 16.Adams JC, Lawler J. The thrombospondins. Cold Spring Harb Perspect Biol. 2011;3(10) doi: 10.1101/cshperspect.a009712. a009712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schellings MW, van Almen GC, Sage EH, Heymans S. Thrombospondins in the heart: potential functions in cardiac remodeling. J Cell Commun Signal. 2009;3(3-4):201–213. doi: 10.1007/s12079-009-0070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frangogiannis NG. Matricellular proteins in cardiac adaptation and disease. Physiol Rev. 2012;92(2):635–688. doi: 10.1152/physrev.00008.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rysa J, Leskinen H, Ilves M, Ruskoaho H. Distinct upregulation of extracellular matrix genes in transition from hypertrophy to hypertensive heart failure. Hypertension. 2005;45(5):927–933. doi: 10.1161/01.HYP.0000161873.27088.4c. [DOI] [PubMed] [Google Scholar]
- 20.Mustonen E, Aro J, Puhakka J, Ilves M, Soini Y, Leskinen H, Ruskoaho H, Rysa J. Thrombospondin-4 expression is rapidly upregulated by cardiac overload. Biochem Biophys Res Commun. 2008;373(2):186–191. doi: 10.1016/j.bbrc.2008.05.164. [DOI] [PubMed] [Google Scholar]
- 21.Vischer P, Beeck H, Voss B. Synthesis, intracellular processing and secretion of thrombospondin in human endothelial cells. Eur J Biochem. 1985;153(3):435–443. doi: 10.1111/j.1432-1033.1985.tb09321.x. [DOI] [PubMed] [Google Scholar]
- 22.Frangogiannis NG. Matricellular proteins in cardiac adaptation and disease. Physiol Rev. 2011;92(2):635–688. doi: 10.1152/physrev.00008.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly RB. Pathways of protein secretion in eukaryotes. Science. 1985;230(4721):25–32. doi: 10.1126/science.2994224. [DOI] [PubMed] [Google Scholar]
- 24.Frolova EG, Sopko N, Blech L, Popovic ZB, Li J, Vasanji A, Drumm C, Krukovets I, Jain MK, Penn MS, Plow EF, Stenina OI. Thrombospondin-4 regulates fibrosis and remodeling of the myocardium in response to pressure overload. FASEB J. 2012;26(6):2363–2373. doi: 10.1096/fj.11-190728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cingolani OH, Kirk JA, Seo K, Koitabashi N, Lee DI, Ramirez-Correa G, Bedja D, Barth AS, Moens AL, Kass DA. Thrombospondin-4 is required for stretch-mediated contractility augmentation in cardiac muscle. Circ Res. 2011;109(12):1410–1414. doi: 10.1161/CIRCRESAHA.111.256743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prabakaran D, Kim PS, Dixit VM, Arvan P. Oligomeric assembly of thrombospondin in the endoplasmic reticulum of thyroid epithelial cells. Eur J Cell Biol. 1996;70(2):134–141. [PubMed] [Google Scholar]
- 27.Kuznetsov G, Chen LB, Nigam SK. Multiple molecular chaperones complex with misfolded large oligomeric glycoproteins in the endoplasmic reticulum. J Biol Chem. 1997;272(5):3057–3063. doi: 10.1074/jbc.272.5.3057. [DOI] [PubMed] [Google Scholar]
- 28.De Young MB, Keller JC, Graham RM, Wildey GM. Brefeldin A defines distinct pathways for atrial natriuretic factor secretion in neonatal rat atrial and ventricular myocytes. Circ Res. 1994;74(1):33–40. doi: 10.1161/01.res.74.1.33. [DOI] [PubMed] [Google Scholar]
- 29.Glembotski CC, Thuerauf DJ, Huang C, Vekich JA, Gottlieb RA, Doroudgar S. Mesencephalic Astrocyte-derived Neurotrophic Factor Protects the Heart from Ischemic Damage and Is Selectively Secreted upon Sarco/endoplasmic Reticulum Calcium Depletion. J Biol Chem. 2012;287(31):25893–25904. doi: 10.1074/jbc.M112.356345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veliceasa D, Ivanovic M, Hoepfner FT, Thumbikat P, Volpert OV, Smith ND. Transient potential receptor channel 4 controls thrombospondin-1 secretion and angiogenesis in renal cell carcinoma. FEBS J. 2007;274(24):6365–6377. doi: 10.1111/j.1742-4658.2007.06159.x. [DOI] [PubMed] [Google Scholar]
- 31.Wu J, Kaufman RJ. From acute ER stress to physiological roles of the Unfolded Protein Response. Cell Death Differ. 2006;13(3):374–384. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- 32.Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GD, Kaufman RJ. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13(3):351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 33.Wu J, Ruas JL, Estall JL, Rasbach KA, Choi JH, Ye L, Bostrom P, Tyra HM, Crawford RW, Campbell KP, Rutkowski DT, Kaufman RJ, Spiegelman BM. The unfolded protein response mediates adaptation to exercise in skeletal muscle through a PGC-1alpha/ATF6alpha complex. Cell Metab. 2011;13(2):160–169. doi: 10.1016/j.cmet.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]