Abstract
Believed to have been critical to the origin of life on Earth 1, the hydrosulfide ion (HS−) and its undissociated form, hydrogen sulfide (H2S), continue to play a prominent role in physiology and cellular signaling 2. As a major metabolite in anaerobic bacterial growth, hydrogen sulfide is a product of both assimilatory and dissimilatory sulfate reduction 2–4. These pathways can reduce various oxidized sulfur compounds including sulfate, sulfite and thiosulfate. The dissimilatory sulfate reduction pathway uses this molecule as the terminal electron acceptor for anaerobic respiration, where it produces excess amounts of H2S4. The reduction of sulfite is a key intermediate step in all sulfate reduction pathways. In Clostridium and Salmonella, an inducible sulfite reductase is directly linked to the regeneration of NAD+, which has been suggested to play a role in energy production and growth, as well as in the detoxification of sulfite 3. Above a certain concentration threshold, both H2S and HS− nhibit cell growth by binding the metal centers of enzymes and cytochrome oxidase5, necessitating a release mechanism for the export of this toxic metabolite from the cell 5–9. Through a combination of genetic, biochemical and functional approaches, we have identified a hydrosulfide ion channel (HSC) in the pathogen Clostridium difficile. The HS− channel is a member of the formate-nitrite-transport (FNT) family, in which ~50 HSC genes form a third subfamily alongside those for formate (FocA) 10,11 and for nitrite (NirC) 12. In addition to HS− ions, HSC is also permeable to formate and nitrite. Such polyspecificity can be explained by the conserved ion selectivity filter observed in the HSC crystal structure. The channel has a low open probability and is tightly regulated, to avoid decoupling of the membrane proton gradient.
As a weak acid with a pKa of 6.8 and a second pKa of 19, over 80 percent of the H2S in a biological system will exist in its ionized form as HS− 2,13. To account for membrane permeation of intracellularly produced hydrogen sulfide 6,9, a channel for HS− or H2S has been suggested 5,7, with the water channel aquaporins (AQPs) as possible candidates 8. While studies in planar lipid bilayers indicate that the aquaporin from Archaeoglobus fulgidus is not permeable to H2S and that the lipid bilayer provides little resistance to H2S permeation 9, in the ventworm Riftia pachyptila it has been shown that HS− ions, but not H2S, are selectively transported through its outer epithelium into its vasculature5.
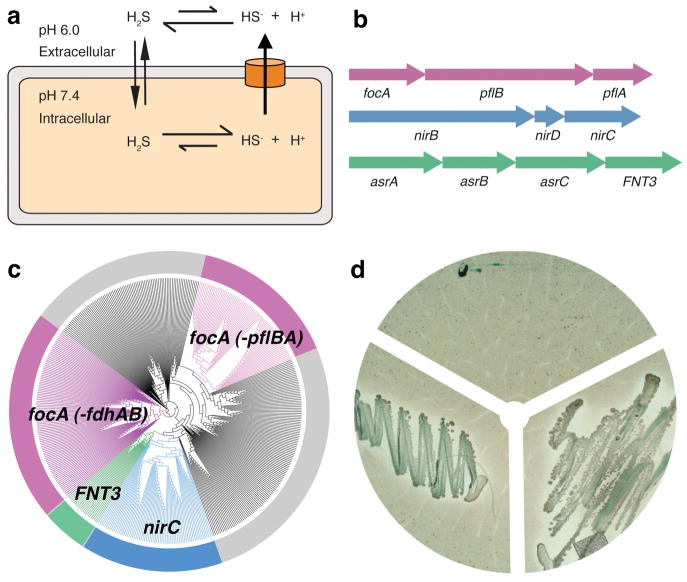
Despite its high permeability across the lipid bilayer, we argue that the weak acidity of H2S necessitates a release mechanism for its ionized form, HS− (Fig. 1a). As a weak acid with a pKa of 6.8, H2S can diffuse readily across the lipid bilayer. Therefore, the distribution of H2S and HS− on both sides of the cell membrane is directly proportional to the pH differential as described by the Henderson-Hasselbalch equation 14. Since cellular pH is kept at neutral levels, and extracellular pH is typically one to two units more acidic, at equilibrium the HS− concentration will be 10–100 fold greater on the inside of the cell in the absence of a release mechanism for the HS− anions. Therefore, given its significant toxicity, it would be beneficial to directly expel this ion across the membrane by the quickest mechanism available—an ion channel.
Fig. 1.
Genetic analyses and functional characterization of the FNT3/HSC gene and the asrABC operon. a, Model of the intracellular anion concentrative effect for the weak acid H2S. b, Genomic organization of FNT and their metabolically related reductase genes. Representatives are shown for the focA, nirC and FNT3/HSC genes with their respectively linked operons. c, Phylogentic tree of 474 bacterial and archaeal members of the FNT family. Branches are colorized based on genetic linkage to metabolic enzymes: pyruvate formate lyase (pflAB) or formate dehydrogenase (fdhAB) linked genes are colored pink, nitrite reductase (nirBD) linked genes are colored blue, and sulfite reductase linked genes (asrABC) are colored green. The FocA protein in archaea is encoded by the fdhC gene. The gray areas represent FNT family members with no assigned function based on genetic linkage. d, Bismuth sulfite agar plate assay. Top: vector control, left: asrA, asrB, asrC, right: asrA, asrB, asrC, FNT3.
Recently, a novel family of anion channels for short-chain acids has been functionally characterized 11,15,16. Members of the FNT family transport various anions during anaerobic bacterial growth 10,12,17. Based on sequence homology, however, only about half of its two thousand identified members clearly belong to either the FocA or NirC subfamily (Figs. 1b&c, Supplementary Figs. 1&2). Intriguingly, for both FocA and NirC, each channel gene is genetically linked to its reductase partner: FocA to the formate reductase pflB 10 and NirC to the nitrate reductase NirBD 12 (Fig. 1b). Our phylogenetic analysis of bacterial genes suggests the existence of additional channel subfamilies in the FNT family (Fig. 1c, Supplementary Fig. 1). One such subfamily is a group of ~50 homologous genes, temporarily termed FNT3, found in various species of Clostridium, major human pathogens grown under strictly anaerobic conditions. As it is linked with a reductase gene, the asrABC operon 18 (Fig. 1b), we hypothesized that FNT3 might encode a channel for HS− or related ions. AsrABC reduces sulfite (SO3−2) into sulfide (S−2), which becomes instantly converted into H2S and HS− under physiological conditions.
asrABC from Salmonella is the only such operon that has been characterized thus far. Its overexpression in E. coli, which lacks this biochemical pathway, was previously shown to reduce SO3−2 to H2S 19. To first verify whether the homologous asrABC operon from C. difficile also encodes an SO3−2 reductase, we tested the growth of E. coli transformed with the C. difficile asrABC genes, plated on bismuth sulfite agar 19,20. When C. difficile asrABC was induced, E. coli were able to overcome the expected growth inhibition by reducing SO3−2 to H2S (Fig. 1d, Supplementary Fig. 3). Darkening along the edges of the colonies, formed by bismuth/iron sulfide precipitation, further indicated that H2S gas was being produced. This showed that the asrABC genes from C. difficile do indeed encode for an SO3−2 reductase, which in turn supports the notion that the FNT3 gene in the asrABC operon may indeed encode a channel for SO3−2, or its reduced product, HS−.
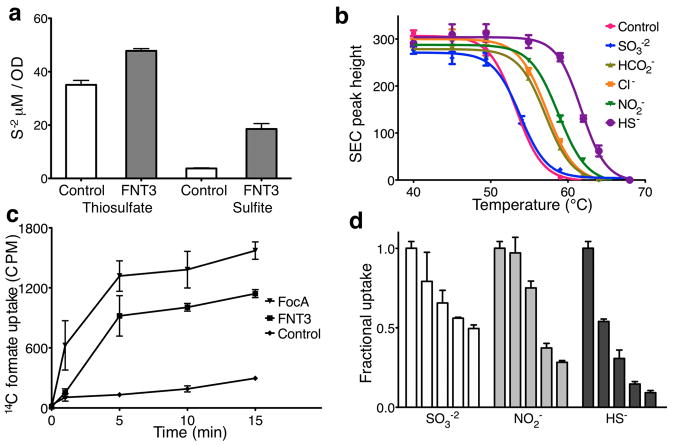
To determine whether the FNT3 protein from C. difficile functions as a channel for HS− or SO3−2, we then carried out whole cell transport, ion – protein binding in solution and transport-inhibition assays in reconstituted proteoliposomes. First, we transformed sulfide-producing S. typhimurium, which lacks an HS− channel, with the C. difficile FNT3 gene. In SO3−2 supplemented minimal medium, the FNT3-expressing S. typhimurium released a high level of HS− ions into the culture medium (Fig. 2a, Supplementary Fig. 4). This increase in extracellular HS− was specific for HS− produced by the endogenous cytoplasmic AsrABC; when the other sulfide-producing reductase in the cell, the periplasmic thiosulfate reductase, was engaged with its substrate thiosulfate, little effect on the extracellular concentration of HS− was observed. This indicates that the measured HS− was generated by SO3−2 reduction by the cytoplasmic AsrABC and then exported. Such an FNT3-linked increase in HS− production in S. typhimurium can be attributed to either increased import of SO3−2 or increased HS−-export from the cell. It follows that FNT3 could be an ion channel for SO3−2 or HS−.
Fig 2.
Binding and transport activity of HSC channel in reconstituted proteoliposomes. a, Measurements of sulfide concentrations in the media of salmonella transformed with vector control or vector encoding FNT3. Minimal media was supplemented with either sulfite or thiosulfate to induce hydrogen sulfide production from either periplasmic thiosulfate reductase or cytoplasmic sulfite reductase. b, Binding of detergent solubilized and purified HSC protein to various anions was determined by using thermostability coupled size exclusion chromatography. Peak heights of recovered samples were plotted against temperature and fitted to a boltzman-sigmoidal model to determine nominal melting temperatures. c, Radiolabeled formate uptake in proteoliposomes reconstituted with purified FNT at pH 8.0 was monitored in a concentrative uptake assay and compared to FocA activity or vesicle controls. d, Inhibition of radiolabeled concentrative uptake of formate by the addition of various anions at increasing concentrations. The plotted bar graphs represent the amount of radiolabeled formate measured at the 10-minute time point for each concentration of anion tested. The concentration of the competing anions were, 0 mM, 0.15 mM, 0.6 mM, 3 mM and 15 mM. Error bars represent s.e.m. (N = 3).
An ion channel protein is often stabilized by its permeating ions through their direct interaction with its selectivity filter 21. Therefore, we tested whether purified FNT3 protein interacts with the monovalent HS− or the divalent SO3−2. Using size-exclusion chromatography of purified protein samples incubated at elevated temperatures, we found that at pH 8.0 the presence of HS− ions was able to increase the nominal melting temperature of the FNT3 protein by 8 °C, whereas SO3−2 had little effect (Fig. 2b, Supplementary Table 1). This result is consistent with the hypothesis that FNT3 functions as an HS− channel but is probably impermeable to SO3−2. Intriguingly, three other monovalent anions, formate, nitrite and chloride, also increased the melting temperature of the FNT3 protein, but to a lesser degree (3 – 4 °C).
We went on to identify the permeable ion(s) of the FNT3 protein in vitro. As channels in the FNT family are expected to have slow conductance rates 11, we measured FNT3 transport activity using a concentrative uptake assay 11,22. Given that HS− ions at concentrations needed to set up a sufficient electrochemical gradient (~150 mM) are severely disruptive to the membrane, we chose to measure the permeability of FNT3 using another interacting ion (Fig. 2b), formate. Proteoliposomes reconstituted at pH 8.0 from purified C. difficile FNT3 protein were found to be permeable to formate, although at a lower rate than FocA from Vibrio cholerae (Fig. 2c). This permeability to formate by FNT3 allowed us to search for other ions that directly compete with formate for transport. Indeed, when HS− or nitrite was present in the buffer outside the proteoliposomes, the uptake of formate was dramatically inhibited (Fig. 2d). In the physiological range of 1—2 mM sulfide, ~70% inhibition was observed, while at the highest concentration tested (15 mM), HS− and nitrite inhibited the uptake of formate by 90% and 70%, respectively.
Such direct competitive inhibition (Fig. 2c), coupled with whole cell transport assays (Fig. 2a) and in vitro binding assays (Fig. 2b), collectively indicates that the FNT3 protein from C. difficile is likely to be a channel for the hydrosulfide ion, HS−. Although the channel is also permeable to formate and nitrite ions, the FNT3 gene is directly linked to the asrABC sulfite reductase operon (Fig. 1b); we have therefore tentatively named this protein a hydrosulfide ion channel (HSC). A definite understanding of the physiological role of these channels will need to be confirmed by in vivo experiments such as genetic deletion studies.
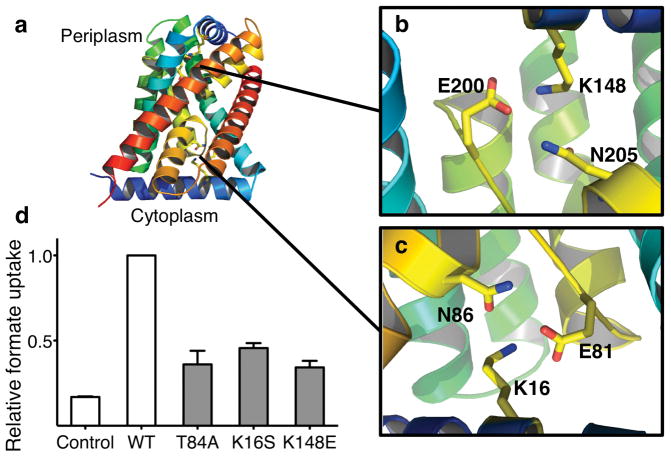
To understand the transport mechanism of the HSC/FNT3 channel from C. difficile (Supplementary Fig. 5), we determined its crystal structure at high-pH (pH = 9.0) to 2.2 Å resolution (Fig. 3a, Supplementary Table 2). The HSC protein adopts the AQP/FocA fold 11,23, and forms a pentamer like FocA, with the five protomers having identical structure (Supplementary Figs. 6–8). The twofold inverted symmetry between the first and second half of the protein goes beyond that observed in FocA and extends to the pore-lining helices TM2b and TM5b (Supplementary Fig. 8b), two salt bridge triads on the periplasmic and the cytoplasmic side (Lys148-Glu200-Asn205 and Lys16-Glu81-Asn86) (Figs. 4a–c) and the two short helices parallel to the membrane (helix P on the periplasmic surface and helix N at the N-terminus on the cytoplasmic surface) (Supplementary Fig. 8b). This helix N, via the Lys16-Glu81-Asn86 salt bridge triad, buttresses TM2b and TM5a, making the cytoplasmic side of the protein more closed and rigid than that in FocA 11.
Fig 3.
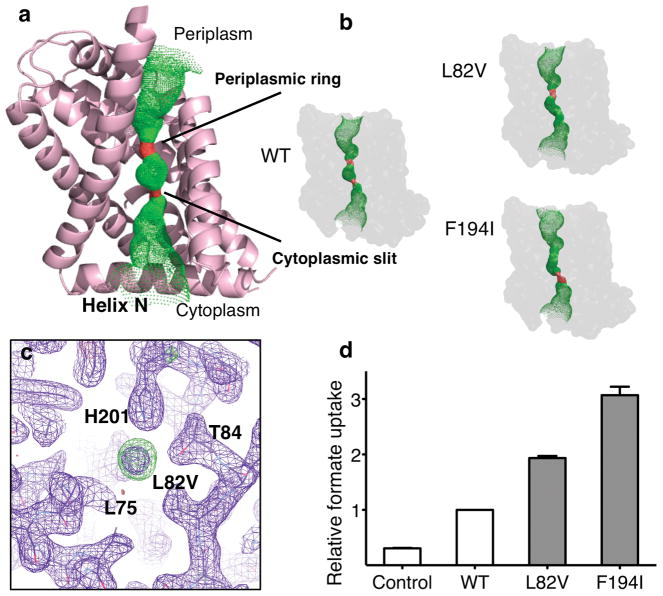
Structural and functional characterization of the ion permeation pathway. a, Structure of a protomer of HSC overlaid with the pore diameter calculations from HOLE, colorized to indicate the radius of water, where green is permeable to water and red is impermeable. Transmembrane helix-2 has been removed for clarity. b, Pore diameter calculations from HOLE of the crystal structures for each of the two permeation pathway mutations, Leu82Val and Phe194Ile and wild-type (WT). c, Close up of the electron density observed at the selectivity filter for the Leu82Val mutant. Purple is the 2Fo – Fc electron density map contoured at 1.1σ and green is the Fo – Fc difference density map contoured at 3σ. d, Relative uptake of proteoliposomes reconstituted with purified HSC protein and pathway mutations. The bar graph represents the 10-minute time point of each concentrative uptake experiment. Error bars represent s.e.m. (N = 3).
Fig 4.
Structural and functional characterization of possible gating mechanisms. a, Structure of the HSC protomer representing possible gating regions. The structure is colorized to show the two-fold inverted topology. b&c, The Glu-Lys-Asn salt bridge triads, related by a pseudo-twofold symmetry, help to stabilize the helix-P at the periplasmic side and helix-N at the cytoplasmic side of the protein. The rotamer that each residue adopts is conserved. d, Relative uptake of proteoliposomes reconstituted with purified HSC protein and gating mutations. The bar graph represents the 10-minute time point of each concentrative uptake experiment. Error bars represent s.e.m. (N = 3).
The pore for ion permeation, located at the center of each protomer, contains the selectivity filter formed by a cytoplasmic slit and a periplasmic ring in the middle of the membrane (Figs. 3a&b). The radius of the cytoplasmic slit measures 0.8 Å which is too small for an HS− ion (radius 1.7 Å 24) to pass through (Supplementary Figs. 9–11). Therefore, the HSC channel in the crystal structure is probably in a closed state. As expected from the sequence conservation in the selectivity filter with that of FocA (Supplementary Fig. 2), the positions or orientations of key residues changed only slightly in HSC (Supplementary Fig. 10). Such conservation explains the permeability of HSC to HS−, as well as to formate and nitrite. When a residue at the cytoplasmic slit or the periplasmic ring is mutated into a smaller residue—Leu82->Val or Phe194->Ile—the pore size is widened, as shown in the crystal structures of the mutant protein (Fig. 3b, Supplementary Table 2), and the formate transport rate of the channel is significantly increased (Fig. 3d). Interestingly, in the crystal structure of the Leu82Val mutant, electron density was observed in the cytoplasmic slit (Fig. 3c). This density, heavier than a water molecule—judged by a B-factor fourfold lower than the surrounding atoms—could be accounted for by a Cl− or an HS− ion. Due to their similar sizes and chemical properties, it has been previously observed that Cl− and HS− ions often bind to the same site in protein crystal structures 25.
The observed polyspecificity of HSC is typical for anion channels 26–28, most likely shared by the FocA and NirC channels as well. Such polyspecific anion recognition is at least partially due to the diffused nature of the electron cloud of anions 29, in contrast to the point-like charge properties of cations. In the meantime, each type of HSC channel in this ancient protein family, coupled with its specific metabolic pathway, is expected to have its own preference for permeating ions, and that preference is probably due to the small structural variations in the selectivity filter that we observed here (Supplementary Fig. 10).
The expulsion of toxic levels of HS− from the cytoplasm through HSC needs to be balanced against the potential risk of the uncoupling of the membrane proton gradient. Freely diffusible H2S and other weak, short-chain acids will dissociate into an anion and a proton in the cytoplasm (Fig. 1a). As the anion is expelled, there will be an intracellular net accumulation of protons. Therefore, the ground state of HSC is likely to be closed and its opening tightly regulated.
Indeed, the HSC structure we determined at high pH is in its closed state (Fig. 3, Supplementary Figs. 9–11). We then asked the question, how easily could this channel be opened? As the homologous FocA channel can be opened by pH or cytoplasmic formate concentration 11,16, we first investigated possible pH-dependent structural changes of HSC by determining its crystal structure at pH 7.5 (3.2 Å resolution) and at pH 4.5 (3.0 Å) (Supplementary Table 2). The protein structures obtained at these neutral and low pHs were the same as the 2.2 Å structure at pH 9.0 (Supplementary Fig. 12), and the channel remained closed. We then studied the two salt bridge triads, both of which contain a glutamate residue that could potentially function as a pH sensor (Fig. 4a–c). When either of the salt bridges was broken, by mutating Lys16->Ser or Lys148->Glu, the HSC crystal structure did not change and the channel still remained closed (Supplementary Fig. 13, Supplementary Table 2). Consistent with these structural results, their transport rate did not increase (Fig. 4d).
To determine whether HSC is gated by anion concentration in a similar manner as the V. Cholerae FocA 11, in which the competition of formate with Thr90 from the Ω loop for hydrogen bonding to His208 in the selectivity filter opens the channel, we mutated the equivalent threonine in HSC, Thr84, to an alanine and determined its 2.4 Å crystal structure (Supplementary Table 2). This mutant structure did not differ from the closed wild-type channel, and no increase in transport was detected (Fig. 4d & Supplementary Fig. 14). As Thr84, Lys16 and Lys148 all line the ion permeation pathway, mutation of these residues slightly reduced the ion permeation rate of the channel. Therefore, we conclude that the opening of HSC is indeed tightly regulated and the channel has a much lower open probability than FocA 11,16.
The low open probability of HSC observed here is consistent with its physiological role in the cell. The opening of HSC probably requires significant movement of helix 2b. As helix 2b is “held” in space by the cytoplasmic helix N (Fig. 4a), the movement of the latter, for example, by direct interaction with a cytoplasmic enzyme in the same metabolic pathway, can trigger the opening of the channel. Of course, we have no evidence for such a hypothesis, and its validation can only be achieved by further experimentation, which we expect would reveal greater variations in both the gating mechanism and anionic preference of FNT channels.
METHODS
Phylogenetic analysis
Phylogenetic data for 474 bacterial and archaeal members of the Formate-Nitrite Transport family were obtained from the HOGENOM database 30, aligned with program Jalview 31 and plotted with program Archaeopteryx 32. Genomic sequences and operon annotations were derived from the NCBI Genbank nucleotide database 33.
Bismuth sulfite Agar plate assay
The FNT3/HSC gene was isolated from Clostridium difficile strain 630. The genes for Clostridium difficile asrA (NCBI ID: 4915353) and asrB (ID: 4915352) were cloned into pCDFDuet-1 (EMD Biosciences) and the genes for C. difficile asrC (ID: 4915351) and HSC (ID: 4915350) were cloned into pACYCDuet-1 (EMD Biosciences). Bismuth sulfite agar plates 20 were freshly prepared following the manufacturer’s protocol and supplemented with 1 mM Isopropyl β-D-1-thiogalactopyranoside and antibiotics chloramphenical and spectinomycin. E. coli BL21 (DE3) cells were transformed with the indicated vectors and streaked, and colonies were allowed to grow at 37 °C for 48 hours. The bismuth sulfite agar 20 contained the ATP-synthase-inhibitor brilliant green 19 and high concentrations of SO3−2. When C. difficile asrABC was induced, E. coli were able to overcome the growth inhibition by producing ATP via glycolysis by reducing SO3−2 to H2S.
Whole cell in vivo transport assay
Salmonella typhimurium LT2 was transformed with the HSC gene cloned into a pBAD vector (Invitrogen). Cells were adapted in MOPS medium supplemented with 10 mM glucose and thiamine at 1 μg/ml. A fraction of each culture (5 ml) was centrifuged and resuspended in fresh media supplemented with ampicillin, 0.2% arabinose and either 6 mM sodium sulfite or 6 mM sodium thiosulfate. Aliquots were incubated at 37°C for 6 hours and then centrifuged to pellet bacteria. Media was gently poured into an equal volume of sulfide antioxidant buffer. Exported sulfide was measured using a combination silver-sulfide electrode (Cole-Palmer). The concentration of sulfide in the medium was determined using standards of known sodium sulfide concentrations in the same antioxidant buffer.
Protein purification
The gene encoding HSC was cloned into a modified pBAD vector (Invitrogen) creating a C-terminal TEV-Flag-10xHis fusion protein and transformed into E. coli BL21 plysS (Sigma-Aldrich) cells 11,34,35. Transformed cells were grown at 37 oC to an OD600 of 1.0 and induced with 0.2 % of arabinose for 4 hours. Cells were collected and lysed by passing twice through an Emulsiflex cell disrupter (Avestin). Lysate was clarified by a low speed spin at 12,000 × g and solubilized with 1% β-dodecyl-maltopyranoside (DDM, Anatrace). Solubilized lysate was incubated with Ni+2 –NTA resin (Qiagen) and the bound protein was eluted with buffer containing 1.1% β-octyl-glucopyranoside (OG, Anatrace), 50 mM Tris 8.0, 200 mM NaCl, 300 mM imidazole and 10% glycerol. The protein sample was incubated with TEV protease overnight to remove the tags, followed by size exclusion chromatography in Superdex 200 in 25 mM TRIS, pH 6.8, 100 mM NaCl and 1.1% OG.
Mass Spectrometry
The mass of purified HSC protein was determined using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry in the laboratory of Dr. T. Neubert following published protocols 36–38.
Protein-ion interaction assay
Ion-binding-induced thermostability of purified protein samples was measured using size exclusion chromatography. Aliquots of 100 mg of Ni+2 –NTA purified protein were incubated for 10 minutes in a thermocycler at increasing temperatures. Samples were injected onto a Shodex KW804 analytical size exclusion chromatography column (Thomson, Clear Brook, VA) on HPLC (Shimadzu, Columbia, MA) equilibrated in 200 mM Na2SO4, 50 mM Tris pH 8.0, 3 mM NaN3 and 0.05 % DDM. The height of monodisperse peak was used to quantify the amount of remaining sample. The nominal melting temperature Tm, defined as the temperature at which 50% of the protein remained soluble, was calculated by fitting the data to a Boltzmann sigmoidal model in Prism5 (GraphPad Software, La Jolla, CA). For screening of various compounds, 20 mM of each sodium salt was added prior to incubation.
Proteoliposome reconstitution and in vitro transport assay
Transport of radiolabeled formate was measured in a concentrative uptake assay 11,22. E. coli total lipids were aliquoted in chloroform and dried under nitrogen. Following resuspension in intraliposomal buffer (150 mM sodium formate and 50 mM Tris pH 8.0), lipids were overlayed with nitrogen, freeze thawed 10 times, vortexed and sonicated to homogeneity. Proteoliposomes were formed by the addition of 1% OG and Ni+2 –NTA purified HSC at a protein to lipid ratio of 1:10,000 (w/w). OG detergent was removed by incubation with 400·mg·ml−1 Bio-Beads SM2 (Bio-Rad) overnight. Proteoliposomes were then subjected to extrusion through 400-nm membrane filter and then frozen at −20 °C until use. Concentrated reuptake of formate was initiated by buffer exchanging 200 ml of thawed proteoliposomes over 2 ml of G50 Sephadex swollen in extraliposomal buffer (150 mM glutamate, 50 mM Tris pH 8.0). 450 μM 14Clabeled sodium formate (American Radiolabeled Chemical) was added to the collected proteoliposomes, and the assays were terminated by centrifuging a 20 ml sample at 1000 × g for 1 minute at each time point through a G-50 Probequant™ Micro-column (GE Healthcare) to remove the external radiolabeled formate.
The amount of transported radiolabeled formate was quantified by adding 1 ml of scintillation fluid to each sample and emitted photons were counted using a Wallac scintillation counter. For competition assays, increasing concentrations of sodium sulfite, sodium nitrite or sodium hydrosulfide at 0, 0.15, 0.6, 3 and 15 mM, were added prior to the addition of radiolabeled formate.
Protein crystallization
High pH, primitive orthorhombic crystals of HSC were grown using the vapor diffusion method, by mixing protein at a concentration of 11 mg/ml in a 1:1 ratio with reservoir solution containing 25–27% (v/v) PEG 400, 100 mM Tris pH 7.0–9.0, 100 mM sodium sulfite, with or without 50 mM sodium hydrosulfide. Medium pH, monoclinic crystals were grown in reservoir substituted with 100 mM sodium nitrite, pH 7.5. Low pH, C-centered orthorhombic crystals were grown in buffer containing 100 mM sodium sulfite, 100 mM sodium nitrite, 8–10% (w/v) PEG 6000, 100 mM zinc acetate and 100 mM sodium acetate pH 4.5.
Structure determination
X-ray diffraction data were collected at beamlines X25 and X29 in the National Synchrotron Light Source of the Brookhaven National Laboratory and 23-ID in the Advanced Photon Source of Argonne National Laboratory. Images were processed and scaled using the HKL2000 suite 39. The structure was solved by molecular replacement using the V. cholerae FocA structure (PDB code 3KLY) 11 as a search model. Models were built using Phenix Autobuild 40 and manually adjusted using COOT 41. Refinement was carried out using Phenix Refine 40. Pore size of the channel was calculated using HOLE 42, and structure figures were generated using the program PYMOL 43
Supplementary Material
Acknowledgments
We are grateful the staff at beamlines X25 and X29 of the National Synchrotron Light Source in the Brookhaven National Laboratory and at the 23ID at the Advanced Photon Source at the Argonne National Laboratory for assistance in X-ray diffraction experiments. We thank A.B, Waight for suggesting the project, J.J. Marden for assistance with cloning of mutants, T. Neubert and S. Blais for mass spectrometry measurements, the 2010 CCP4 Workshop for assistance in processing diffraction data, and A. David, H. Jackson, N.K. Karpowich, J.J. Marden, R.L. Mancusso, Y. Pan and M. Zhou for helpful discussions. This work was financially supported by the NIH (R01-GM093825, R01-DK073973, R01-MH083840 and U54-GM075026). B.K.C. was partially supported by an NIH Supplement Grant to Promote Diversity in Health-Related Research (R01-DK053973-08A1S1) and an NIH predoctoral fellowship (F31-AI086072).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions: B.K.C did the experiments. B.K.C. and D.N.W. wrote the manuscript.
The atomic coordinates and structure factors for high-, medium- and low-pH have been deposited under access codes 3TDO, 3TDR and 3TDP, respectively, and those of the K16S, L82V, T84A, K148E and F194I mutants under the codes 3TE2, 3TDX, 3TE1, 3TE0and 3TDS, respectively.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
The authors dedicate this work to the memory of Lennart Philipson.
References
- 1.Wachtershauser G. Groundworks for an evolutionary biochemistry: the iron-sulphur world. Prog Biophys Mol Biol. 1992;58:85–201. doi: 10.1016/0079-6107(92)90022-x. [DOI] [PubMed] [Google Scholar]
- 2.Kabil O, Banerjee R. Redox biochemistry of hydrogen sulfide. J Biol Chem. 2010;285:21903–21907. doi: 10.1074/jbc.R110.128363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dhillon A, Goswami S, Riley M, Teske A, Sogin M. Domain evolution and functional diversification of sulfite reductases. Astrobiology. 2005;5:18–29. doi: 10.1089/ast.2005.5.18. [DOI] [PubMed] [Google Scholar]
- 4.Rabus R, Hansen T, Widdel F. Dissimilatory sulfate- and sulfur-reducing prokaryotes. In: Dworkin M, et al., editors. The Prokaryotes: Ecophysiology and Biochemistry. Vol. 2. Springer; Heidelberg: 2006. pp. 659–768. [Google Scholar]
- 5.Goffredi SK, Childress JJ, Desaulniers NT, Lallier FJ. Sulfide acquisition by the vent worm Riftia pachyptila appears to be via uptake of HS−, rather than H2S. J Exp Biol. 1997;200:2609–2616. doi: 10.1242/jeb.200.20.2609. [DOI] [PubMed] [Google Scholar]
- 6.Jacques AG. The kinetics of penetration: Xii. Hydrogen sulfide. J Gen Physiol. 1936;19:397–418. doi: 10.1085/jgp.19.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freytag JK, et al. A paradox resolved: sulfide acquisition by roots of seep tubeworms sustains net chemoautotrophy. Proc Natl Acad Sci U S A. 2001;98:13408–13413. doi: 10.1073/pnas.231589498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JK, et al. Structural basis for conductance by the archaeal aquaporin AqpM at 1.68 Å. Proc Natl Acad Sci U S A. 2005;102:18932–18937. doi: 10.1073/pnas.0509469102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mathai JC, et al. No facilitator required for membrane transport of hydrogen sulfide. Proc Natl Acad Sci U S A. 2009;106:16633–16638. doi: 10.1073/pnas.0902952106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suppmann B, Sawers G. Isolation and characterization of hypophosphite--resistant mutants of Escherichia coli: identification of the FocA protein, encoded by the pfl operon, as a putative formate transporter. Mol Microbiol. 1994;11:965–982. doi: 10.1111/j.1365-2958.1994.tb00375.x. [DOI] [PubMed] [Google Scholar]
- 11.Waight AB, Love J, Wang DN. Structure and mechanism of a pentameric formate channel. Nature structural & molecular biology. 2010;17:31–37. doi: 10.1038/nsmb.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jia W, Tovell N, Clegg S, Trimmer M, Cole J. A single channel for nitrate uptake, nitrite export and nitrite uptake by Escherichia coli NarU and a role for NirC in nitrite export and uptake. Biochem J. 2009;417:297–304. doi: 10.1042/BJ20080746. [DOI] [PubMed] [Google Scholar]
- 13.Hughes MN, Centelles MN, Moore KP. Making and working with hydrogen sulfide: The chemistry and generation of hydrogen sulfide in vitro and its measurement in vivo: a review. Free Radic Biol Med. 2009;47:1346–1353. doi: 10.1016/j.freeradbiomed.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 14.Hirshfield IN, Terzulli S, O’Byrne C. Weak organic acids: a panoply of effects on bacteria. Sci Prog. 2003;86:245–269. doi: 10.3184/003685003783238626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, et al. Structure of the formate transporter FocA reveals a pentameric aquaporin-like channel. Nature. 2009;462:467–472. doi: 10.1038/nature08610. [DOI] [PubMed] [Google Scholar]
- 16.Lu W, et al. pH-dependent gating in a FocA formate channel. Science. 2011;332:352–354. doi: 10.1126/science.1199098. [DOI] [PubMed] [Google Scholar]
- 17.Das P, Lahiri A, Chakravortty D. Novel role of the nitrite transporter NirC in Salmonella pathogenesis: SPI2-dependent suppression of inducible nitric oxide synthase in activated macrophages. Microbiology. 2009;155:2476–2489. doi: 10.1099/mic.0.029611-0. [DOI] [PubMed] [Google Scholar]
- 18.Crane BR, Getzoff ED. The relationship between structure and function for the sulfite reductases. Curr Opin Struct Biol. 1996;6:744–756. doi: 10.1016/s0959-440x(96)80003-0. [DOI] [PubMed] [Google Scholar]
- 19.Hallenbeck PC, Clark MA, Barrett EL. Characterization of anaerobic sulfite reduction by Salmonella typhimurium and purification of the anaerobically induced sulfite reductase. J Bacteriol. 1989;171:3008–3015. doi: 10.1128/jb.171.6.3008-3015.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson WJ, EMB A combination of bismuth and sodium sulphite affording an enrichment and selective medium for the typhoid-paratyphoid groups of bacteria. J Pathol Bacteriol. 1926;29:310–311. [Google Scholar]
- 21.Doyle DA, et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 22.Middleton RE, Pheasant DJ, Miller C. Purification, reconstitution, and subunit composition of a voltage-gated chloride channel from Torpedo electroplax. Biochemistry. 1994;33:13189–13198. doi: 10.1021/bi00249a005. [DOI] [PubMed] [Google Scholar]
- 23.Savage DF, O’Connell JD, 3rd, Miercke LJ, Finer-Moore J, Stroud RM. Structural context shapes the aquaporin selectivity filter. Proc Natl Acad Sci U S A. 2010;107:17164–17169. doi: 10.1073/pnas.1009864107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feth S, Gibbs GV, Boisen MB, Jr, Myers RH. Promolecule radii for nitrides, oxides, and sulfides. A comparison with effective ionic and crystal radii. J Phys Chem. 1993;97:11445–11450. [Google Scholar]
- 25.Tai CH, et al. Characterization of the allosteric anion-binding site of O-acetylserine sulfhydrylase. Biochemistry. 2001;40:7446–7452. doi: 10.1021/bi015511s. [DOI] [PubMed] [Google Scholar]
- 26.Hille B. Ionic Channels of Excitable Membranes. Sinauer; Sunderland: 1992. [Google Scholar]
- 27.Yasui M, et al. Rapid gating and anion permeability of an intracellular aquaporin. Nature. 1999;402:184–187. doi: 10.1038/46045. [DOI] [PubMed] [Google Scholar]
- 28.Rychkov GY, Pusch M, Roberts ML, Jentsch TJ, Bretag AH. Permeation and block of the skeletal muscle chloride channel, ClC-1, by foreign anions. J Gen Physiol. 1998;111:653–665. doi: 10.1085/jgp.111.5.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simons J, Jordan KD. Ab initio electronic structure of anions. Chem Rev. 1987;87:535–555. [Google Scholar]
- 30.Penel S, et al. Databases of homologous gene families for comparative genomics. BMC Bioinformatics. 2009;10 (Suppl 6):S3. doi: 10.1186/1471-2105-10-S6-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 32.Han MV, Zmasek CM. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics. 2009;10:356. doi: 10.1186/1471-2105-10-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2011;39:D32–37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Auer M, et al. High-yield expression and functional analysis of Escherichia coli glycerol-3-phosphate transporter. Biochemistry. 2001;40:6628–6635. doi: 10.1021/bi010138+. [DOI] [PubMed] [Google Scholar]
- 35.Wang DN, et al. Practical aspects of overexpressing bacterial secondary membrane transporters for structural studies. Biochim Biophys Acta. 2003;1610:23–36. doi: 10.1016/s0005-2736(02)00709-5. [DOI] [PubMed] [Google Scholar]
- 36.Cadene M, Chait B. A robust, detergent friendly method for mass spectrometry analysis of integral membrane proteins. Anal Chem. 2000;72:5655–5658. doi: 10.1021/ac000811l. [DOI] [PubMed] [Google Scholar]
- 37.Li XD, et al. Monomeric state and ligand binding of recombinant GABA transporter from Escherichia coli. FEBS Lett. 2001;494:165–169. doi: 10.1016/s0014-5793(01)02334-1. [DOI] [PubMed] [Google Scholar]
- 38.Safferling M, et al. The TetL tetracycline efflux protein from Bacillus subtilis is a dimer in the membrane and in detergent solution. Biochemistry. 2003;42:13969–13976. doi: 10.1021/bi035173q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otwinowski Z, Miror W. Processing of X-ray diffraction data collected in oscillation mode. Meth Enzym. 1997;276(Part A):307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 40.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14:354–360. 376. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 43.DeLano WL. The PyMOL User’s Manual. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.