Abstract
Understanding rhythmic behavior at the developmental and genetic levels has important implications for neurobiology, medicine, evolution, and robotics. We studied rhythmic behavior—larval crawling—in the genetically and developmentally tractable organism, Drosophila melanogaster. We used narrow-diameter channels to constrain behavior to simple, rhythmic crawling. We quantified crawling at the organism, segment, and muscle levels. We showed that Drosophila larval crawling is made up of a series of periodic strides. Each stride consists of two phases. First, while most abdominal segments remain planted on the substrate, the head, tail, and gut translocate; this “visceral pistoning” moves the center of mass. The movement of the center of mass is likely powered by muscle contractions in the head and tail. Second, the head and tail anchor while a body wall wave moves each abdominal segment in the direction of the crawl. These two phases can be observed occurring independently in embryonic stages before becoming coordinated at hatching. During forward crawls, abdominal body wall movements are powered by simultaneous contraction of dorsal and ventral muscle groups, which occur concurrently with contraction of lateral muscles of the adjacent posterior segment. During reverse crawls, abdominal body wall movements are powered by phase-shifted contractions of dorsal and ventral muscles; and ventral muscle contractions occur concurrently with contraction of lateral muscles in the adjacent anterior segment. This work lays a foundation for use of Drosophila larva as a model system for studying the genetics and development of rhythmic behavior.
Introduction
Rhythmic movements such as respiration, digestion, circulation, and locomotion are fundamental to animal life. Many rhythmic movements—the digestive gastric mill and pyloric rhythms of the crustacean stomatogastric nervous system, or swimming in leech and lamprey—are well understood on the cellular and network levels (Marder and Bucher, 2007; Mullins et al., 2011). However, much less is known about the genetics and development of circuitry underlying rhythmic movements in these systems. To overcome this obstacle we are developing a genetically tractable model to study a rhythmic locomotor behavior: crawling in Drosophila larvae.
In Drosophila larvae, many behaviors have been studied such as phototaxis, learning, and navigation (Iyengar et al., 1999; Freeman et al., 2010; Luo et al., 2010). These and other behaviors are based on regulating locomotion in response to stimuli or experience. Drosophila larval locomotion includes a repertoire of many different types of movements, including turns, burrowing, linear crawling, and other movements (Green et al., 1983; Wang et al., 1997; Hwang et al., 2007). The relative complexity of larval locomotor behavior has precluded detailed analysis of the neuromuscular mechanism underlying any one type of movement, such as linear crawling. However, understanding the motor pattern that drives behavior is a requisite step in understanding its cellular basis (Marder and Calabrese, 1996).
In this paper we present a detailed kinematic study of Drosophila larval linear crawling, with the goal of linking muscle contraction patterns to behavior. We simplify the larval behavioral repertoire by using narrow channels that constrain larval behavior to linear forward or reverse crawling. We developed methods to quantify these two types of crawling at the organism, segment, and muscle levels. At each of these levels we identified parameters that describe normal rhythmic crawling behavior. These data will serve as a basis for assessing the normality of crawling movements in other genetic backgrounds, or other preparations, such as fictive preparations. In addition, these data can help predict possible characteristics of the neural mechanisms underlying behavior. Together, the results of this work provide a set of tools and a conceptual foundation that can be used for the study of Drosophila larval crawling as a new and potentially powerful model for rhythmic behavior.
Materials and Methods
Channel fabrication
Channels were fabricated using standard soft lithographic methods (Fig. 1A; McCormick et al., 2011). Full resolution electronic files of photomasks are available upon request. Silicon wafer masters were created by exposing 100,150 and 200 μm layers of SU-8 2000 (MicroChem) through a transparency mask with channel widths varying from 100 to 300 μm, at 50 μm increments, and then dissolving away unexposed material. Masters were treated with chlorotrimethylsilane (Aldrich). Before use, individual channels were cast using 3% agarose (Life Technologies) in distilled water, and agarose-coated coverslips (VWR) were placed on top of individual channels.
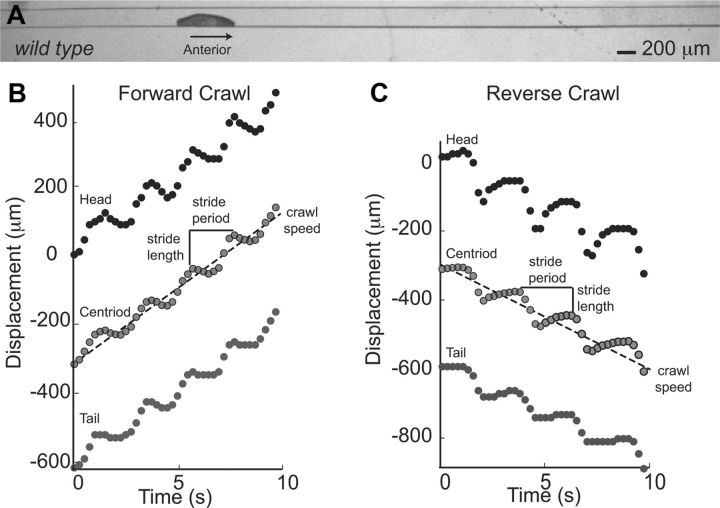
Figure 1.
Linear crawling in channels is a periodic behavior that can be divided into two phases. A, We constructed agarose channels to constrains Drosophila larval behavior to linear crawling. A representative frame from a movie shows a newly hatched, wild-type Drosophila larva crawling through a 200 μm wide, 200 μm deep channel. Anterior is to the right. B, C, We plotted displacement of the head (black dots), centroid (gray dots with black outline), and tail (dark gray dots) over time for forward (B) and reverse (C) crawls. Initial head position was considered 0 μm. Positive values were used for forward movement, and negative values for reverse movement. Crawls were composed of repeated cycles of motion, termed strides. Note that each stride can be divided into two phases: head, centroid, and tail each move slowly; then head, centroid and tail move rapidly in the direction of the crawl. We measured the stride length, as the net displacement of the centroid during a stride, and the stride period as the time to complete a stride. Speed and direction of each crawl was determined using the slope of a regression line fit to displacement of centroid over time.
Genetic strains and behavior
Drosophila strains used include: yw, MHC-GFP (YD0783) (Quiñones-Coello et al., 2007). wg-gal4 (Bloomington Stock Center, Bloomington IN) was crossed to UAS-mCD8-GFP (Lee and Luo, 1999) to generate wg>GFP genotype. Newly hatched larvae <1000 μm in average length were transferred from 24 h collections (non-yeasted, apple juice agar plates).
Whole-organism, bright-field behavior
Wild-type larvae, yw, of either sex were used for all experiments.
Open, flat surfaces.
Larvae were placed on apple juice agar caps (15 mm diameter) and imaged for 3–5 min.
Channels.
Larvae were gently placed ventral side down in 200 × 200 μm channels and imaged for 3–5 min (Fig. 1A).
Segmental and muscle assays
wg>GFP (segmental assay) and MHC-GFP (muscle assay) larvae were mounted on their side in narrow diameter channels, usually 100 × 150 μm, and imaged for 10 min. For the wg>GFP embryo assay, embryos were staged (17 e/f) based on air-filled trachea (Pereanu et al., 2007). Embryos were manually dechorionated, and dorsal side was placed on double-sided tape (Scotch Brand, 3M) in oxygenated Halocarbon oil 27 (Sigma-Aldrich) on a glass slide (VWR). Embryos were recorded for variable lengths of time, all movements discussed in the text occurred within 15 min of hatching.
Video capture, segmentation and registration
Whole-organism assay
Behavioral recordings were acquired on an Olympus stereo microscope using variable magnification (8–20×). The microscope was equipped with a Scion 1394 Camera, using Scion VisiCapture software. Images were acquired at 4 Hz. A custom ImageJ (NIH) plug-in used Gaussian blurring and k-means clustering to segment the image; The centroid estimates position of the larval centroid, r(Centroid); the right- and left-most points estimates head and tail position respectively, r(Head) and r(Tail) (Table 1).
Table 1.
Parameters measured for organism, segments, and muscles
| Variable description | Variable name |
|---|---|
| Whole-organism behavioral | |
| Head position | r(Head) |
| Centroid position | r(Centroid) |
| Tail position | r(Tail) |
| Segmental movement | |
| Mouth hook position | r(Mouth Hooks) |
| Tail position | r(A8) |
| Gut position | r(Gut) |
| Segment position | r(T2, T3, A1-A7) |
| Individual muscle contractions | |
| Length of muscle DO1 on segments A2–A7 | l(DO1) |
| Length of muscle LT1 on segments A2–A7 | l(LT1) |
| Length of muscle VL1 on segments A2–A7 | l(VL1) |
Segmental and muscle assays
Behavioral recording were acquired on a McBain spinning disc confocal microscope equipped with a Hamamatsu EM-CCD camera, using a 10× 0.3 NA (numerical aperture) or 20× 0.70 NA lens (Zeiss) and Volocity software (PerkinElmer). Images were acquired at 10 Hz. Image segmentation and registration were done as follows. For wg>GFP, we drew a line on the first frame of each movie that went from the head to the tail of each larva or embryo. We used ImageJ (NIH) MultipleKymograph plug-in to generate kymographs. This was followed by a combination of automatic thresholding and manual annotation to identify the position of each structure: r(Mouth Hooks), r(Tail), r(Gut), r(T2, T3, A1-A7) (Table 1). For MHC-GFP we manually annotated position of muscle insertion sites, i.e., r(DO1_A1-A7). We wrote custom scripts in Ruby and MATLAB (MathWorks) to determine length of each muscle, i.e., l(DO1_A2-A7), l(LT1_A2-A7), l(VL1_A1-A7) in each frame of the movie (Table 1).
Data analysis
Whole-organism behavior
Open surfaces.
Distance of each crawl, Δr(Open_Crawl) was determined using our ImageJ plug-in. Crawl duration, Δt(Open_Crawl) was manually measured, and speed = Δr(Open_Crawl)/Δt(Open_Crawl). Number of reverse crawls (three or more reverse strides) and strides per crawl were scored manually.
Channel.
We wrote custom scripts in python and excel to plot centroid position over time, r(Centroid)/t, as well as r(Head)/t and r(Tail)/t (Fig. 1B,C). For this bright-field dataset we used only crawls that were made up of five or more strides. Regression analysis to determine speed and identification of local maxima to determine strides was done using custom written python scripts. These scripts also calculated stride period and stride length.
Segmental assay
Montages of larval crawling were made from movies using ImageJ, and representative frames were traced in Adobe Illustrator (Fig. 2). Custom MATLAB (MathWorks) scripts plotted the position of wg<GFP labeled structures over time: r(Mouth Hooks), r(Tail), r(Gut), and r(T2, T3, A1-A7)/t (Figs. 3, 4). MATLAB (MathWorks) was used to convert these data into polar plots (Figs. 3, 4). Briefly, time of initiation of each stride (i(Stride Cycle[n])) was determined as movement of tail segment in the direction of the crawl. Stride period is (Δt(Stride Cycle) = i(Stride Cycle [n+1]) − i(Stride Cycle [n])). For each stride, initiation of the stride was set to time = 0 (i(Stride Cycle[n])) = 0). Times after initiation (t) were adjusted accordingly, and times were then divided by stride period and multiplied by 100 to convert to percentage of stride cycle elapsed (e.g., (t − i(Stride Cycle[n]))/(Δt(Stride cycle) * 100)). Velocity was calculated by taking the difference in position and dividing that by time elapsed (e.g., Δr(Mouth Hooks)/Δt(Mouth Hooks)). Only positive velocity was plotted on phase plots. For all other calculations, initiation (e.g., i(A3)), duration (e.g., Δt(A3)), and displacement of each segment in the same direction as the crawl (e.g., Δr(A3)), was calculated for segments A3 and A6 (Fig. 5B). Stride period (Δt(Stride cycle)) was calculated based on time between initiations, averaging both A3 and A6 for at least three strides (Fig. 4). Intersegmental phase lag = (abs[i(A6) − i(A3)]/Δt(Stride cycle))/(100/3) (Fig. 4). For embryos timing of embryonic wave phases and embryonic visceral piston phases was manually scored (Fig. 5).
Figure 2.
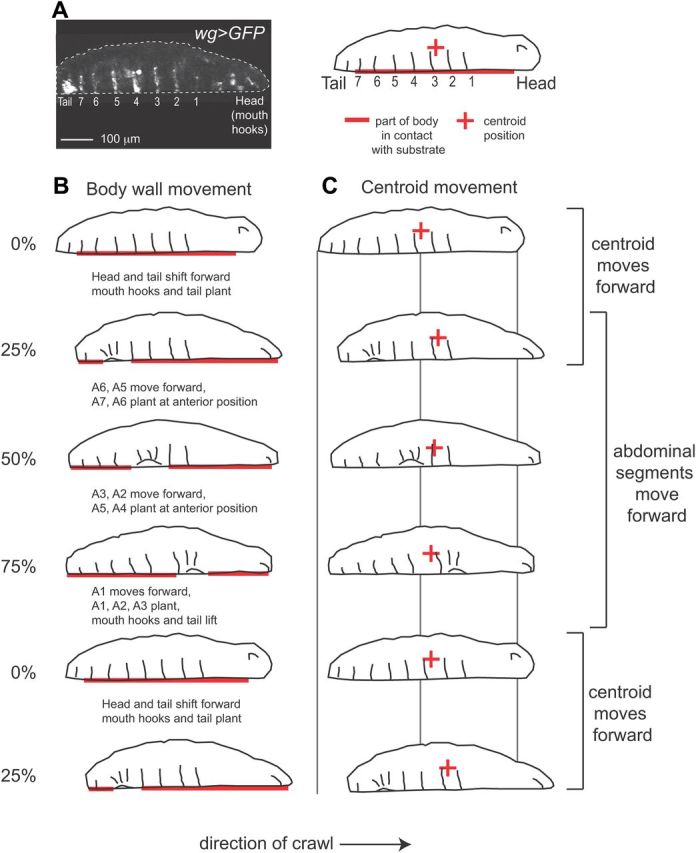
Larval abdominal segments move asynchronously with larval centroid (center of mass). A, At left is a frame from a movie of a first instar Drosophila larva crawling through a channel. Anterior is to the right and is ventral down. The position of the tail, and abdominal segments A1–A7 (1–7) were genetically labeled using wg>GFP. The tail, as well as a single row of cells, are fluorescent in each segment, and the mouth hooks in the head are autofluorescent. At left is a tracing of a wg>GFP larva. The bold, red line indicates the portion of the larvae in contact with the substrate, and the red + symbol indicates the centroid (approximation of center of mass). B, C, We traced representative frames from a movie of a larva crawling forward. For wg>GFP data we considered a forward stride cycle to begin and end with movement of the tail in the direction of the crawl. Frames represent the larval body at 0%, 25%, 50%, 75% and 100% (or 0%) stride cycle completion. Text (in B) describes the movements that happened between frames. B, At the start of the stride cycle, 0%, the abdominal segments are in contact with the substrate. The head and tail shift forward, while abdominal segments remain in contact with the substrate. Next, 25–100%, each abdominal segment is picked up off the substrate, moved forward, and placed back on the substrate (see bracket at right). This happens sequentially beginning with segments in the posterior. C, The centroid moves out of phase with movement of abdominal segments. The centroid (+) moves in two phases: First, the centroid moves rapidly in the direction of the crawl, between 0 and 25% (see brackets at right); this occurs as the head and tail shift forward, and abdominal segments are in contact with the substrate. Second, the centroid moves slowly in the direction opposite to the crawl, between 25 and 100%; this occurs as the abdominal segments move forward.
Figure 3.
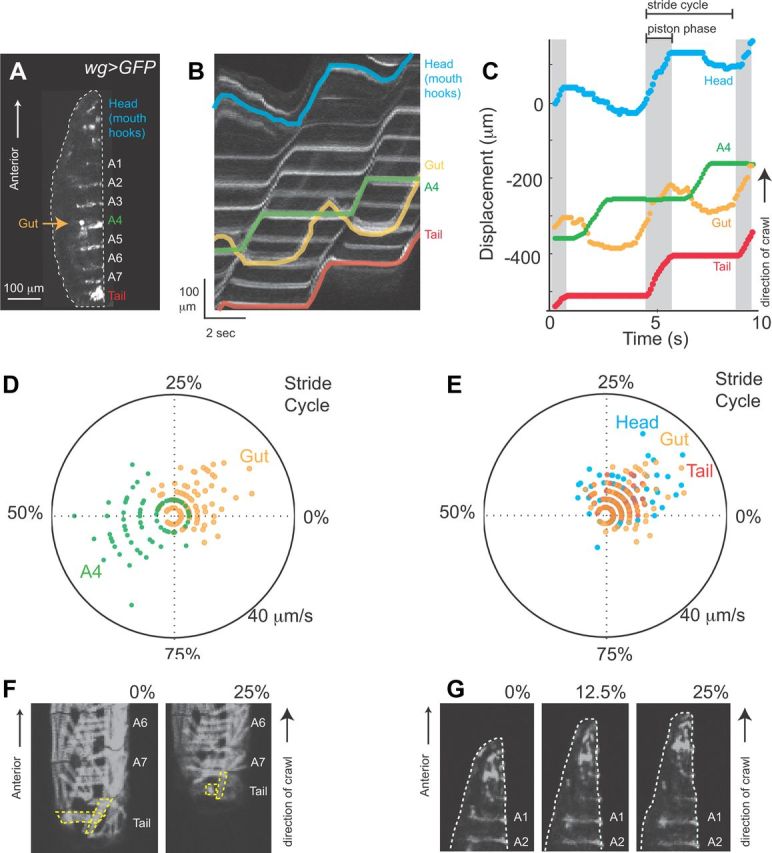
Characterizing the visceral pistoning phase of larval crawling. A, wg>GFP can be used to visualize the relative movement of gut and external larval body parts. This genotype labels tail, abdominal segments, and mouth hooks in the head; in addition autofluorescence in wg>GFP (arrow), labels a portion of the gut that resides near A4 when larvae are at rest. This image is a side view of a newly hatched, wg>GFP larva with anterior to the top and ventral to the left. B, Relative movement of gut and external larval body parts during a forward crawl is shown on a kymograph. To generate these kymographs a line was drawn from head to tail in the first frame of a movie. ImageJ multiple kymograph plug-in was then used to compute the intensity along the line in each frame of the movie, and to plot each line along an x-axis. The resulting display shows position in space on the y-axis and time on the x-axis. We colored the position of each of the following structures in the kymograph: head (blue), gut (orange), abdominal segment A4 (green), and tail (red). Anterior to the top. C, We plotted displacement of head (blue), gut (orange), abdominal segment A4 (green), and tail (red) over time. Note movement of the head, gut and tail in the direction of the crawl occur nearly simultaneously. We term this part of the stride cycle, the visceral piston phase. We estimated the visceral piston phase as the time during which the tail moves in the same direction as the crawl (gray, piston phase). Note movement of segment A4 is phase shifted with respect to movement of the gut. Brackets indicate the stride cycle, which begins and ends with the movement of the tail in the direction of the crawl. D, E, We created composite representations of the forward stride cycle using polar plots. The start/end of a forward stride cycle was considered as the time at which the tail moves in the direction of the crawl, 0% of the stride cycle elapsed, and corresponds to the start of the visceral piston phase Next we determined stride period, and calculated the part of the stride cycle during which each structure moved. The angle represents the percent of the stride cycle elapsed. Position along the radial axis represents the speed; note for clarity we only plotted speed of the structure moving the same direction as the crawl. We plotted data from crawls performed at three different stride periods in three different larvae on each polar plot. D, The movement of the gut (orange) and abdominal segment A4 (green) phase shifted during the stride cycle, although at rest they occupy a similar anteroposterior position along the larval body (see A). E, The movement of the head (blue), gut (orange), and tail (red) show extensive temporal overlap. F, Muscles in the tail and abdominal segments A7, A6 were visualized in the MHC-GFP genetic background. During the visceral piston phase of the forward stride cycle (0–25%) muscles in the tail (yellow outlines) shorten. G, A close up of the larval anterior visualized with wg>GFP during the visceral piston phase of the forward stride cycle (0–25%) shows head and abdominal segments A1, A2. During the visceral piston the head elongates. Note the changing shape of the head, and that segments A1 and A2 remain stationary because they are planted on the substrate.
Figure 4.
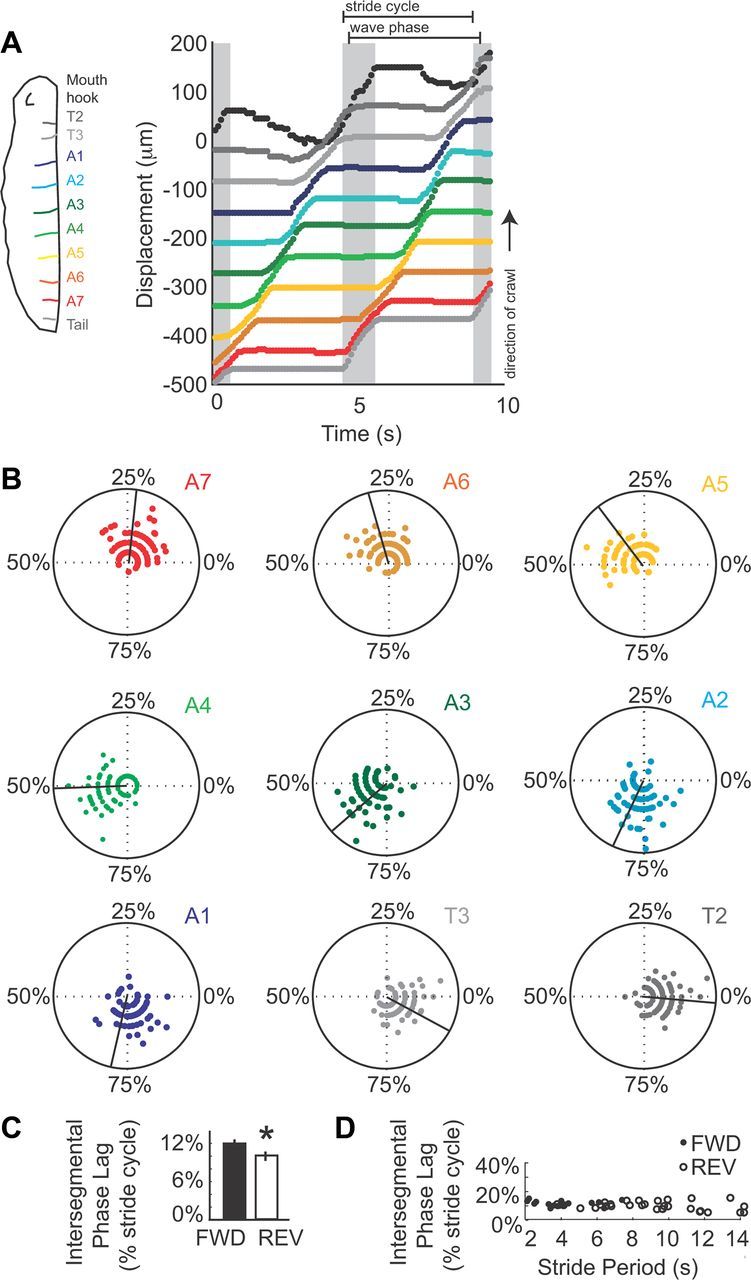
Characterizing the wave phase of larval crawling. A, Left, A drawing of a larva shows the color scheme used in this figure: mouth hooks in the head (black), segment T2 (dark gray), T3 (light gray), A1 (dark blue), A2 (light blue), A3 (dark green), A4 (light green), A5 (orange), A6 (brown), A7 (red), tail (gray). Right, A displacement plot show the position of head, tail, abdominal segments and thoracic segments over time. Gray shading indicates the visceral piston phase of the stride cycle, estimated as the movement of the tail in the direction of the crawl. Brackets indicate the stride cycle, beginning and ending with the movement of the tail in the direction of the crawl. The wave phase of the stride cycle, beginning with movement of A7 in the direction of the crawl, and ending with movement of T2 in the direction of the crawl, is also indicated by brackets. B, Composite representations of positive velocity for abdominal and thoracic segments are shown on polar plots. Angle represents percentage stride cycle elapsed, where 0% is initial movement of the tail in direction of the crawl. The black line indicates the mean. Radial position represents magnitude of velocity. These plots combine data from three different larvae crawling at three different stride periods. Note the temporal overlap in movement of adjacent segments, and that posterior segments such as A7 move earlier in the stride cycle than more anterior segments. C, D, Intersegmental phase lag was calculated by determining intersegmental travel time ([absolute latency between A3 and A6 movement divided by three, the number of segments traveled], divided by stride period), for both forward (black) and reverse (outline) crawls. C, We plotted average intersegmental phase lag for forward (black) and reverse crawls (outline) on a bar graph. Error bars represent SD. Note the slight, but significant difference between sample means (*p < 0.05, t test). D, Intersegmental phase lag was plotted against stride period. Note intersegmental phase lag did not vary over a range of stride periods indicating it was phase constant for both forward (black circles) and reverse (open circles) stride cycles.
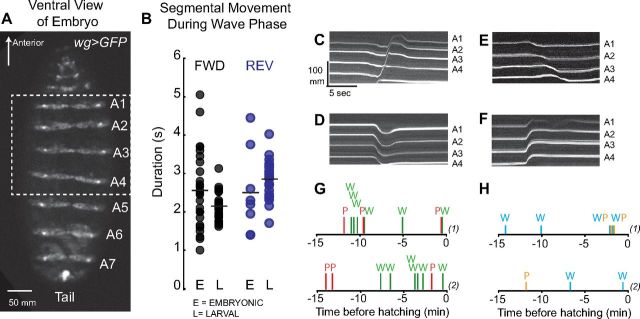
Figure 5.
Embryonic wave phase and embryonic visceral piston phases can occur independently in embryos. A, We used wg>GFP to monitor segmental movements in embryonic stages. A representative image of a wg>GFP embryo at stage 17e/f, after the trachea have been inflated is shown. Body wall segments A1–A7 are labeled; the dashed box shows the region of the embryo viewed in C–F. View is ventral with anterior to the top. B, We measured the time it took for a single segment to move in the direction of the wave phase during embryonic and larval wave phases. Duration of movement during forward embryonic wave phase (black dots in E column), during forward larval wave phase (black dots in L column), during reverse embryonic wave phase (blue dots in E column), and during reverse larval wave phase (blue dots in L column) are plotted. Means are indicated with black lines. Note for both forward and reverse wave phases the ranges overlap, with the embryonic ranges more wide-spread. C–F, Kymographs show embryonic wave phases (C, E) and embryonic visceral piston phases (D, F). During an embryonic forward wave phase (C) each segment moves toward the head. During an embryonic reverse wave phase (E) each segment moves toward the tail. There are two embryonic visceral piston phases: in one type (F) all segments moved in unison toward the head, which is likely to correspond to a larval reverse visceral piston phase because the center of mass cannot translocate across the substrate when the embryo is encased in the egg. In the other type (D), all segments moved in unison toward the tail, which is likely to be the embryonic equivalent of a larval forward visceral piston phase. We noted that each of these phases could occur independently, without any other segmental movement within 5 s. G, H, We scored the sequence of tail to head embryonic waves (green bars, W), and head to tail embryonic piston phases (red bars, P), which likely correspond segmental movements seen during a forward larval crawl. We also scored the sequence of head to tail embryonic waves (blue bars, W), and tail to head embryonic piston phases (orange bars, P), which likely correspond to segmental movements seen during a reverse larval crawl. Examples from two embryos [denoted as (1) and (2)] are shown. Note that each movement can occur independently.
Muscle assay
Muscle length was plotted versus time (e.g., l(DO1_A2-A7)/t) using custom written MATLAB (MathWorks) scripts (Fig. 6). Three features of these plots were extracted using custom written MATLAB (MathWorks) scripts: Contraction time (t(C)) was estimated by taking the time at which a muscle reach half minimum muscle length, relaxation time (t(R)) was estimated by taking the time at which a muscle reached half maximal muscle length; peak contraction time (t(P)) was estimated as the time at which muscles reached minimum length. Stride period, Δt(Stride) was estimated as the time between contractions of each muscle (Δt(C)) for at least two muscle groups on three adjacent segments for three strides. Latency was calculated by taking difference between peak contractions time (t(P)) of two different muscles (e.g., t(P[VL1]), t(P[DO1]), t(P[LT1])). V-D = (t(P[VL1]) − t(P[DO1])); V-T = (t(P[VL1]) − t(P[LT1])); D-T = (t(P[DO1]) − t(P[LT1])); V-T(+1) = (t(P[VL1 An]) − t(P[LT1 An+1])); V-T(−1) = (t(P[VL1 An]) − t(P[LT1 An-1])). Average latency per crawl and SD was calculated using measurements from segments A4–A6 for three strides.
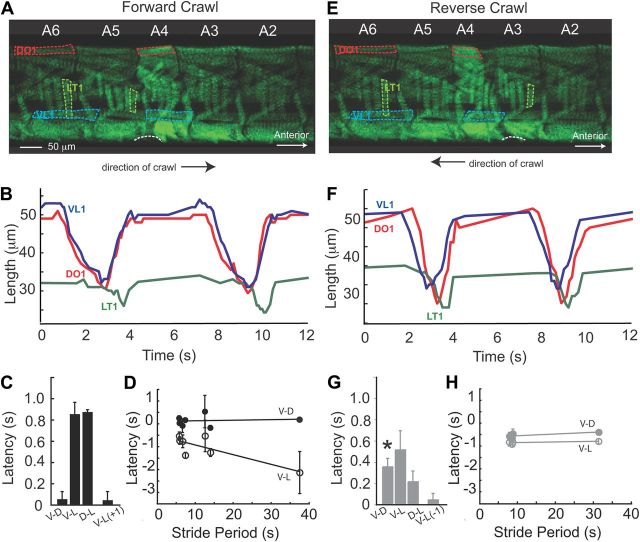
Figure 6.
Muscle contraction pattern in abdominal segments during forward and reverse Drosophila larval crawling. A, E, Representative frames from movies of a larva crawling forward (A) or backward (E) through a narrow diameter channel. Segments A2–A6 are labeled (white numbers above each segment). In segment A6 relaxed muscles are outlined (D muscle, DO1, red; V muscle, VL1, blue; L muscle, LT1, green). Direction of crawl is indicated by arrow beneath each image. Side views are shown with anterior to the right. Note for both forward (A) and reverse (E) crawls, D muscles (red dashes in segment A4), and V muscles (blue dashes in segment A4) were contracting. For forward crawls (A), D/V muscle contraction occurred as L muscles in A5 were contracting (green dashes). In contrast for reverse crawls (E), D/V muscle contraction occurred as L muscles in A3 were contracting (green dashes). A region of the abdominal body wall lifted off the substrate is indicated (white dashed line). B, F, Plots of the length of a V muscle (VL1, blue), a D muscle (DO1, red), and a L muscle (LT1, green) over time are shown for muscles in segment A5. During forward crawls (B) D and V muscles contract in phase, and before L muscles of the same segment. During reverse crawls (F) D and V muscles contract with a slight phase shift, and before L muscles of the same segment. C, G, Average latency between muscle contractions was calculated using data from segments A4–A6 (n = 7 forward crawls, E, black; n = 6 reverse crawls, G, gray). V-D is latency between VL1 and DO1 peak contractions; V-L is latency between VL1 and LT1 peak contractions; D-L is latency between DO1 and LT1 peak contractions. V-L(+1) is latency between peak contractions of VL1 and LT1 of the adjacent posterior segment. V-L(−1) is latency between peak contractions of VL1 and LT1 of adjacent anterior segment. Note, the V-D delay in forward (C) versus reverse (G) crawls is slightly but significantly different (t test, *p < 0.05). Error bars show SD. D, H, Average latency for each crawl forward (black) or reverse (gray) crawl is plotted against stride period. Note V-D latency (closed circles) for both forward and reverse crawls does not change with stride period, nor does V-L (open circles) latency for reverse crawls. In contrast V-L latency (open circles) for forward crawls does vary with stride period (FWD V-L regression line y = −0.04 x − 0.51, r2 = 0.68).
Statistics
Unless otherwise mentioned Student's t tests, to test to compare sample means, linear regression analysis, and calculation of Pearson's correlation coefficients were performed using MATLAB (MathWorks) software.
Results
Channels constrain locomotor behavior to linear crawling
In this report we study two Drosophila larval locomotor behaviors: linear forward and linear reverse crawling. To reliably observe these behaviors we engineered narrow channels that force larvae to crawl in a straight line (Fig. 1A). Compared with crawling on flat, open surfaces wild-type larvae in channels could crawl forward at the same average speeds, with bouts of linear crawling extending for longer durations, and larvae often crawling in reverse (data not shown). Channels can be used for both low-resolution recordings to observe organismal behavior (Fig. 1A), and high-resolution recording to observe segmental and muscle movements during behavior (Figs. 3, 6). In addition, in channels larvae can be viewed from any orientation—top, bottom or side. In conclusion, building channels was a key advance for our work, because it allows us to study extended bouts of linear crawling at multiple levels of detail.
Crawling is composed of periodic strides
Our ultimate goal is to use Drosophila larval crawling as a model for studying rhythmic behavior. Thus, we examined the periodicity of the crawling behavior of larvae in channels. Previous studies of maggot (dipterian insect larvae) locomotion, including third-instar Drosophila melanogaster larvae, showed that forward crawling is composed of repetitive cycles of motion called strides (Berrigan and Pepin, 1995). We confirmed and extended these findings by showing that both forward and reverse crawls in first instar Drosophila melanogaster larvae were composed of strides as well (Fig. 1B,C). Specifically, we acquired bright-field movies of larvae crawling through channels, and measured the position of the larval head, tail and centroid in each frame of a movie (Table 1). We generated displacement plots to visualize the movement of head, centroid and tail over time, and see periodic cycles of motion in each trace (Fig. 1B,C). To reliably recognize a cycle of motion in these data, we identified local maxima of centroid displacement, and used the interval between local maxima to calculate stride period (Fig. 1B,C). We found that the average stride period in our bright-field recordings ranged from 0.6 to 2.1 s for forward crawls, and from 1.2 to 3.3 s for reverse crawls. In summary, both forward and reverse linear crawling in channels are rhythmic movements.
Crawling speed correlates with stride period rather than stride length
Next we wanted to understand the variability in behavior exhibited by wild-type larvae crawling through channels by measuring organism speed. We measured speed by taking the slope of the regression line describing centroid displacement of a crawl of five strides or more (Fig. 1B,C). All regression models fit the data with an R2 value of at least 0.95 (data not shown). This method was based on the observation that, once initiated, larvae maintained a given speed until the crawl was terminated (data not shown). We find that larvae crawl at a range of speeds (forward 39.5–126.5 μm/s [n = 14]; reverse 21.2–88.0 μm/s [n = 17]), and, on average, forward crawls were significantly faster than reverse crawls (forward 88.7 ± 28.8 μm/s; reverse 62.3 ± 21.3 μm/s, p < 0.05). We noted that any given larva could crawl at multiple speeds in both forward and reverse directions (data not shown); furthermore an individual larvae always crawled faster traveling forward than traveling in reverse (data not shown). Thus, an individual larva is capable of producing both forward and reverse crawls at a range of speeds.
We wanted to understand the relationship between speed and stride cycle. In organisms such as earthworms, animals increase speed primarily by increasing the length of each stride (Quillin, 1999). Other soft-bodied animals, such as caterpillars, increase speed primarily by increasing stride frequency (or decreasing stride period) (Casey, 1991). Thus, we asked, as Drosophila larvae crawl faster do they take larger strides or take faster strides? First, we analyzed our bright-field population data. We performed regression analysis between average stride period and crawling speed, as well as average stride length and crawling speed. For both forward and reverse crawls, crawling speed correlated much better with average stride period than average stride length (Table 2). Next, to control for non-neural factors, for example, staging, hormone level, genotype, or environment, we compared forward crawls produced by individual larvae that crawl at multiple speeds. For a given larva we did pairwise comparisons between every possible combination of crawls to determine the difference in average stride period, average stride length and crawling speed. We did regression analysis to examine the relationship between these three values, and find that 92% of the variation in forward crawling speed could be due to stride period (Table 2). Thus, Drosophila larvae largely regulate crawling speed by regulating stride period as do many limbed organisms.
Table 2.
Regression analysis shows crawling speed is best correlated with stride period
| X-axis | Y-axis | Slope | Intercept | R2 |
|---|---|---|---|---|
| Forward crawl | ||||
| Stride period | Crawling speed | −71 | +188 | 0.87 |
| Stride length | Crawling speed | 1.0 | −29 | 0.59 |
| Reverse crawl | ||||
| Stride period | Crawling speed | −34 | +128 | 0.88 |
| Stride length | Crawling speed | 0.78 | −22 | 0.47 |
| Forward crawl (comparing difference in crawls from individuals) | ||||
| Change in stride period | Change in crawling speed | 57 | +1.0 | 0.92 |
During a stride the centroid and abdominal body wall move asynchronously
Because crawling speed can be understood in terms of strides, we focused the remaining analysis in this paper at the level of a stride cycle. In our bright-field data, we noted that in each stride there were two phases of movement. The centroid moved slowly (almost always) in a direction opposite to the crawl direction; and the centroid moved rapidly in the direction of the crawl. These two phases of movement were also easily detectable in the displacement plots of head and tail movements, and were seen for both forward and reverse crawls (Fig. 1B,C). To understand in more detail how each segment of the Drosophila larval body moved during these two phases of movement we used wg-gal4 to express UAS-mCD8-GFP (wg>GFP) in a single row of cells in each segment of the abdominal and thoracic body wall, as well as the tail (Fig. 2A). In addition, mouth hooks in the head could be tracked by following autofluorescence (Fig. 2A, Table 1). We placed wg>GFP larvae in channels and recorded crawling behavior. For this fluorescent dataset (wg>GFP), we used the tail movement rather than the centroid movement to determine the bounds of the stride cycle; we considered stride cycle to begin with movement of the tail in the direction of the crawl. We traced frames that represent the larval body at 0%, 25%, 50%, 75% and 100%/0% stride cycle completion (Fig. 2). For each frame we estimated which part of the larval body was in contact with the substrate (Fig. 2B, horizontal red line), and calculated the position of the larval centroid (Fig. 2C, red plus sign). Surprisingly, we found that that the centroid and abdominal body wall moved asynchronously. As larval centroid moved forward rapidly the abdominal segments remained stationary with respect to the substrate (Fig. 2, 0–25% of stride cycle). In contrast, when each abdominal segment was lifted off of the substrate, moved forward, and replaced back on the substrate in a more anterior position, the centroid remained stationary or moved slightly backward (Fig. 2, 25–0% of stride cycle). Thus, we conclude that a single stride is composed of two phases of movement: a phase of abdominal body wall advance and a phase of centroid advance.
Drosophila larvae use a visceral locomotor piston mechanism to move body mass
A crawling cycle consisting of two phases of movement, centroid advance and abdominal body wall advance has been recently documented in Manduca sexta caterpillar forward crawling (Simon et al., 2010a); this mechanism of movement is termed “visceral locomotor pistoning” (Simon et al., 2010a). So we tested whether Drosophila larvae also use a visceral piston mechanism for crawling. Specifically, we tested whether the centroid advance phase of the Drosophila larval crawling stride cycle is a “visceral piston phase.” To be considered a visceral piston phase, we used Manduca as a reference to lay out two criteria. First, the tail and gut should move together. Second, as the gut and tail move, abdominal segments should be stationary (Simon et al., 2010a). We examined the movement of the Drosophila larval gut relative to movement of the tail, and abdominal segment A4 (representing the center of the abdomen), using wg>GFP larvae. In this genotype, autofluorescence allowed us to visualize a portion of the gut found near abdominal segment A4 when the larvae are at rest (Fig. 3A). From movie data we constructed kymographs (see figure legend for details) (Fig. 3B), and from kymographs we determined the position of each structure (Table 1). We found that the Drosophila larval stride cycle contains a visceral piston phase. The gut and tail moved together, while abdominal segments remained stationary. First, this can be seen by examining the movements of gut, tail, and segment A4, on a displacement plot (gray shading, Fig. 3C). Second, we determined the speed of each structure over time, and calculated Pearson's correlation coefficient for gut speed versus tail speed, as well as gut speed versus segment A4 speed. We found that on average the tail and gut speed has a strong positive correlation (forward r = 0.67 ± 0.16 [n = 8], reverse r = 0.56 ± 0.18 [n = 3]), but gut and segment A4 speed were negatively correlated (forward r = −0.39 ± 0.23 [n = 8], reverse r = −0.41 ± 0.15 [n = 3]). Last, we found that gut movement and segment A4 movement were almost perfectly out of phase. This was seen on polar plots (see figure legend for details), which show high gut speeds occurred between 0 and 25% of the stride cycle, whereas high abdominal segment A4 speeds occurred between 50 and 75% of the stride cycle (Fig. 3D). In contrast, movement of gut and tail showed a large amount of temporal overlap (Fig. 3E). We conclude that the first part of the Drosophila larval stride cycle—rapid movement of the head, gut, tail and centroid in the direction of the crawl, is a visceral piston phase.
Role of head and tail during the visceral piston phase
Because it is a requisite step in understanding the cellular basis of behavior, we wanted to understand the muscle contractions powering the visceral piston phase. During the visceral piston phase we saw concurrent movement of tail and gut (Fig. 3B,C,E). Thus, we hypothesized that active movements of the tail could provide a source of power for larval translocation. We imaged muscles in the tail using MHC-GFP reporter larvae in which all somatic muscles are labeled (Fig. 3F). During visceral piston phase we saw shortening of tail musculature as the tail moved forwards (Fig. 3F, n = 3 crawls). Thus, contraction of muscles in the tail could power forward movement during the visceral piston phase.
We also noted that during the visceral piston phase the head moved forward (Fig. 3B,C,E). This observation is consistent with the idea that the head moved passively forward; alternatively, this observation is consistent with the idea that active contraction of head muscles provide a source of power for larval translocation. The head musculature in our MHC-GFP reporter line was not labeled distinctly enough to enable direct examination of muscle contraction (data not shown). Thus, to distinguish between these possibilities, we used an indirect measure, the movement of the head and mouth hooks which are controlled by head musculature (Bate, 1993; Hanslik et al., 2010). We saw the mouth hooks move forward, exhibiting a distinct up and down motion before planting into the substrate (n = 8 crawls), which is consistent with a prior report showing mouth hooks engage with the substrate (Wang et al., 1997). In addition, the head region elongated in shape as segments A1, and A2 remained in place (Figs. 3F, n = 8 crawls). These observations support the idea that the movement of the head was not merely passive. Our data are consistent with the hypothesis that the forward visceral piston phase is powered by muscle contractions in both the head and tail.
Characterization of the wave phase of the stride cycle
We found that the stride cycle is composed of two phases: the visceral piston phase, described above, and the wave phase in which the abdominal body wall advances. A number of studies on Drosophila larval forward crawling have documented peristaltic waves that travel from tail to head (Hughes and Thomas, 2007; Song et al., 2007; Dixit et al., 2008; Lahiri et al., 2011), and we characterize this wave phase in more detail here. We found that during forward crawling, there are sequential movements of abdominal segments. Moreover these movements were initiated in posterior segments and traveled toward the anterior (Fig. 4A,B). These observations are in agreement with previous studies (Hughes and Thomas, 2007; Song et al., 2007; Dixit et al., 2008; Lahiri et al., 2011). In addition, we found that for reverse crawling, the wave phase was characterized by sequential movements of abdominal segments that travel from head to tail (data not shown). In both forward and reverse crawls the movements that occurred during the wave phase can be termed “direct,” because they traveled along the body axis in the same direction as the crawl (Elder, 1980). Thus, consistent with previous work on Drosophila larval behavior, we found that direct, sequential movements of external body segments occur during the wave phase of larval crawling.
Because we are interested in determining the neural basis of Drosophila crawling we wanted to understand the intersegmental phase lag, which describes the phase relationship of segmental movements as a percentage of the stride cycle. Much experimental and theoretical work has gone into understanding the neural mechanisms contributing to intersegmental phase lag in preparations such as crayfish swimmerete beating (Friesen and Cang, 2001). Using our wg>GFP dataset, we measured the absolute latency between the onset of movement in segment A3 and onset of movement in segment A6, and divided by the 3, the number of segments traversed. This value gave us an approximation of the intersegmental travel time (ISTT) for segmental movements for each crawl. We found that ISTT varied linearly with stride duration (data not shown). Next, we normalized ISTT to stride period to determine the intersegmental phase lag. For forward crawls (n = 20) we found an average intersegmental phase lag of 12 ± 0.2% of the stride cycle/segment (Fig. 4C). For reverse crawls (n = 33) we found an average intersegmental phase lag of 10 ± 0.3% of the stride cycle/segment (Fig. 4C). Notably, by examining displacement plots of our wg>GFP data, we found that for both forward and reverse crawls up to three segments could be moving in the direction of the crawl at any time point (Fig. 4A,B; data not shown). In conclusion, we have determined parameters of the wave phase that will be useful in further study of the neural basis of crawl behavior.
In many rhythmic behaviors such as leech and lamprey swimming, intersegmental phase lags are constant over different cycle periods; it is thought that this helps maintain the body form (Skinner and Mulloney, 1998). It has been postulated that for terrestrial forms of locomotion such as leech crawling, that it is less important to maintain a specific body form to move efficiently (Cacciatore et al., 2000). Thus, we wanted to determine whether the intersegmental phase lag was constant over a range of stride periods in Drosophila larvae. We plotted intersegmental phase lag for both forward and reverse crawling against stride period, and found it did not vary over a range of stride periods (Fig. 4D). An additional observation that supports the idea that Drosophila larvae maintain intersegmental phase relationships over differing stride periods comes from examine composite polar plots (Fig. 4B). To generate these plots, speed of each segment was normalized to stride period, and three crawls performed at different stride periods were overlaid. The composite data well recapitulates data from a single crawl; for example, on composite plots up to three segments can be moving at any point in the crawl cycle. Together these data suggest that for both forward and reverse crawling Drosophila larvae maintain their body form even as they crawl at different speeds.
Visceral piston phase and wave phases can be uncoupled in embryo
During forward larval crawling both the visceral piston and wave phase began as the tail moved forward (Figs. 3, 4). This observation caused us to ask whether these two phases were causally linked. We decided to look for evidence of decoupling between wave phase and visceral piston phase in embryos. If we could see such uncoupling, this would suggest that the visceral piston phases may not be causally linked, but rather distinct movements.
We looked at late stage embryos just before hatching, because previous studies have shown fully mature waves of segmental contraction occur by this stage (Suster and Bate, 2002; Pereanu et al., 2007; Crisp et al., 2008), but to our knowledge embryonic visceral piston movements not have been reported. One of the advantages of our wg>GFP assay used to monitor segmental movements is that this assay allows for a quantitative comparison between segmental movements in newly hatched larval and segmental movements in the embryo (Fig. 5A). To ensure that we were examining healthy embryos, we recorded embryos until they hatched, and sampled segmental movements that occurred within 15 min of initiation of hatching.
First, we asked whether we could find an embryonic wave phase, equivalent to larval wave phase. Many studies have documented wave-like movements in the embryo (Suster and Bate, 2002; Pereanu et al., 2007; Crisp et al., 2008). Consistent with previous work, in wg>GFP embryos we two found patterns of movement that were similar to forward and reverse wave phases observed in larvae. In one pattern, we found each abdominal and thoracic body wall segment moved toward the head; these movements were initiated in posterior segments, and moved sequentially with up to three segments moving at one time (Fig. 5C). This pattern was similar to the pattern of segmental movement seen in forward larval wave phases, and we term it an embryonic forward wave phase. The other pattern was similar to the pattern seen in reverse larval wave phases (Fig. 5E), and we term it an embryonic reverse wave phase. To further compare the embryonic wave phases and larval wave phases, we measured how long it took for a single segment to travel toward the head during each type of wave phase; we saw the ranges overlap with a wider range seen in the embryo (Fig. 5B). Thus, we found patterns of movement in the embryo similar to larval wave phases.
Next, we asked whether equivalents of larval visceral piston phases were present in embryos. We observed patterns of segmental movement in the embryo that bear similarity to larval visceral piston phase. First, both larval and embryonic visceral piston phases showed a movement of the viscera relative to the abdominal body wall (data not shown). Notably, we did not expect abdominal segments to be stationary during an embryonic visceral piston phase because embryos are encased in a series of membranes and cannot make contact with the substrate. Second, as in larvae, we observed two types of embryonic visceral piston phases, those that simultaneously moved segments toward the tail and those that simultaneously moved segments toward the head (Fig. 5D,F). Third, embryonic visceral piston phases had significantly shorter duration than the duration of embryonic wave phases (Fig. 5C–F). Thus, we find likely embryonic equivalents to both forward and reverse larval visceral piston phases.
Finally, we wanted to determine whether the embryonic visceral piston phase could be uncoupled from embryonic wave phase. We scored the sequence of embryonic wave phases and embryonic visceral piston phases that occurred in the 15 min before embryo hatching (Fig. 5G,H). We found clear examples where embryonic wave phases and embryonic visceral piston phases did not occur within five seconds of other segmental movements (15/30 “forward” embryonic wave phases, 7/20 forward embryo visceral piston phases, 6/11 “reverse” embryonic wave phases, 1/6 reverse embryo visceral piston phases) (Fig. 5C–F). For example, two wave phases without an intervening visceral piston phase, or a piston phase with out an associated wave phase (>5 s apart). In conclusion we found evidence to suggest that visceral piston phases could be considered distinct from the wave phase.
Intrasegmental phase relationship of dorsal and ventral muscles
A relatively well accepted idea of how insect larvae, such as caterpillars, use muscles to crawl was proposed first in 1937 by Barth (Barth, 1937; Wigglesworth, 1950; Champman, 1998). In insects, muscles fall into the three groups: dorsal muscles (D), ventral muscles (V) and lateral muscles (L) (Snodgrass, 1935); these three muscle groups are found in Drosophila larvae (Bate, 1993). Barth's model suggests that within a segment, D muscles contract before L muscles and L muscles contract before V muscles. Thus a defining feature of Barth's model is that D muscles and V muscles within a segment contract asynchronously. This motor pattern was thought to help lift each segment off the substrate and then replace it. Recently, however, a study in Manduca sexta caterpillars has challenged Barth's model (Simon et al., 2010b). As Manduca sexta caterpillars crawl forward, D and V muscles contract simultaneously (Simon et al., 2010b). Thus, we wanted to understand the relationship of D muscle contractions versus V muscle contractions in Drosophila larvae. In Drosophila larvae, do D and V muscles contract simultaneously, as seen during Manduca crawling, or with a phase shift as suggested by Barth?
MHC-GFP larvae that express GFP in all somatic muscles were imaged as they crawled forward or in reverse through channels (Fig. 6). We selected three muscles for detailed comparison: from the D group—DO1 (Dorsal Oblique 1 [muscle 9], dorsal-most), from the L group—LT1 (Lateral Transverse 1 [muscle 21] posterior-most), and from the V group, VL1 (Ventral Longitudinal 1 [muscle 6]; Fig. 6A,E). First, we analyzed coordination of D and V muscle contractions during forward crawling. We examined movies of MHC-GFP larvae crawling, frame-by-frame. We noted that within a given segment D and V muscles contracted simultaneously (Fig. 6A). Next, we plotted VL1 and DO1 muscle length over time (Fig. 6B), and we found that D and V muscles contracted in synchrony (n = 7 crawls, 4 segments each). For each segment examined there was no significant difference in timing of muscle contraction or muscle relaxation (data not shown). In addition, we measured the average latency between peak V and D contractions. To compute V-D latency we took the difference between peak contraction time of VL1 and peak contraction time of DO1 for three stride cycles on three consecutive segments. We found a V-D latency of 0.05 ± 0.07 s. (Fig. 6C), and this latency did not vary with stride period (Fig. 6C). In summary, we find that a feature of the muscle contraction pattern for forward crawling in Drosophila larvae is that D and V muscles within the same segment contract simultaneously.
Next we asked whether during reverse crawling in Drosophila larvae D and V muscles also contracted simultaneously. We found that VL1 muscles contracted slightly, but significantly, before the DO1 muscles of the same segment (Fig. 6E,F). We found an average latency V-D latency of 0.35 ± 0.08 s (Fig. 6G), and this latency was constant over a range of stride periods. In summary, we find the phase-shifted relationship between V and D muscle contractions during reverse crawling in Drosophila larvae, which is slightly different from that seen for forward crawling.
Intrasegmental phase relationship of ventral and lateral muscle groups
Having defined the phase relationships of D and V muscle contractions during forward and reverse larval crawling in Drosophila larvae, we next we looked at the lateral (L) muscle contractions. We found that V/D muscles contract before L muscles of the same segment (Fig. 6B). We measured V-L latency by determining the time between peak VL1 contraction and peak LT1 contraction, and found and average of 0.85 ± 0.11 s (Fig. 6C). Similarly, we measured D-L latency by determining the time between peak DO1 and peak LT1 muscle contractions, and found an average of 0.87 ± 0.02 s (Fig. 6C). These data are consistent with the finding that D and V muscle groups contract simultaneously. We find that V-L latency varied with stride period: with longer stride period we found longer V-L latencies (Fig. 6D). In summary, we find that in Drosophila larvae during forward crawls the muscles contraction pattern has three distinguishing features: D and V muscles contract simultaneously; D/V muscle contract before L muscles of the same segment; V-L latency changes with changing stride period.
We next assayed reverse crawls. Here too we found the L muscles contract after V and D muscles within the same segment. We found a V-L latency of 0.51 ± 0.18 s and a D-L latency of 0.21 ± 0.1 s (Fig. 6F,G). The V-L latency was significantly longer than the D-L latency (t test, p < 0.05), consistent with the idea that V and D muscle contractions are phase shifted relative to each other. In contrast to forward crawls (Fig. 6H), during reverse crawls the V-L latency did not vary with changing stride period. In summary, we observe the pattern of muscle contractions within a given segment during reverse crawls in Drosophila larvae to have two distinct features. First, the sequence of muscle contraction was V muscles, followed by D muscles, and finally contraction of L muscles. Second, the latency between contractions of different muscle groups did not depend on stride period.
Intersegmental phase relationship between ventral and lateral muscle groups
Crawling in soft-bodied organisms such as leech and earthworm is driven by waves of muscle contractions in functionally antagonistic groups of muscles— circumferential and longitudinal muscles. Contraction waves pass from head to tail sequentially; as the circumferential wave passes through a segment causing elongation, the longitudinal wave is passing through a more anterior segment causing shortening (Gray and Lissmann, 1938; Baader and Kristan, 1995). Thus, for Drosophila larval crawling we wanted to define the intersegmental relationship between functionally antagonistic muscle groups. We chose to examine contraction of the V muscle, VL1, which is oriented along the larval anteroposterior axis, and contraction of the L muscle, LT1, which is oriented along the dorsoventral axis (Fig. 6A). We found that during forward crawling, V muscles contract at nearly the same time as L muscles in the adjacent posterior segment (n = 7 crawls, 4 segments). For example VL1 in segment A4 contracted as LT1 in segment A5 contracted (Fig. 6). To quantify this relationship we calculated the V-L(+1) latency by determining the time difference between peak contraction of VL1 in segment n, and peak contraction of LT1 in segment n+1, and found an average latency 0.04 ±0.06 s. (Fig. 6C). This latency did not vary with stride period (data not shown). In summary, a defining feature of forward crawling in Drosophila larvae is that V muscles and L muscles in adjacent posterior segments contract in phase, nearly simultaneously.
We analyzed the relationship between contractions of V and L muscle groups during reverse crawls. We saw V muscles contract at nearly the same time as L muscles in the adjacent anterior segment (n = 6 crawls, 4 segments). For example, VL1 in segment A4 contracted while LT1 in segment A3 contracted (Fig. 6E). In addition, we calculated the V-L(−1) latency by determining the time difference between peak contraction of VL1 in segment n and LT1 in segment n-1. We found the average V-L(−1) latency to 0.04 ± 0.08 s (Fig. 6G), and this did not vary with stride periods (data not shown). In conclusion, a defining feature of reverse crawling in Drosophila larvae is that the contractions of V muscles and L muscles in adjacent anterior segments are coordinated to temporally coincide.
Discussion
Comparison between datasets
One advantage of using channels to elicit extended bouts of rhythmic crawling was that we could easily record and compare the same larval behavior in three differentially labeled populations of Drosophila larvae. In all datasets we saw individual larvae perform multiple crawls at a range of speeds in both directions, reverse crawls were always performed with a longer stride period (data not shown), and we saw direct, sequential segmental movements during the wave phase of the stride cycle (data not shown). In addition, the curves describing motion of the tail and head movements in our bright-field and wg>GFP dataset were similar (Figs. 1, 3, 4); the temporal overlap between segmental movements in wg>GFP and MHC-GFP datasets were similar (data not shown). We note, however, that in wg>GFP and MHC-GFP recordings the average stride period was significantly longer than in bright-field recordings. These differences could come from many sources: differences in the details of behavioral recordings (i.e., channel dimensions, microscope stage and illumination conditions), differences in genetic background, or biased sample selection of wg>GFP and MHC-GFP for slower crawls. In summary, we believe that the fundamental crawl motion is similar in all datasets.
Muscle contractions during forward crawling
One of our stated goals was to link muscle contraction patterns with Drosophila larval linear crawling behavior. We discuss this below.
Visceral piston phase
During the visceral piston phase movement the larval center of mass (centroid) moves in the direction of the crawl. During the visceral piston phase, tail muscles shorten (Fig. 3F). In Drosophila larvae, the tail consists of modified abdominal segments (Hartenstein, 1993). When the tail contracts, it could have two actions: moving the tail forward, and pushing the gut forward via “gut suspension” muscles (Bate, 1993; Sink, 2006). We speculate that tail contraction provides both functions during the forward crawl.
In addition, during the visceral piston phase, the head elongates and the mouth hooks move (Fig. 3). The external head of a Drosophila larva is an extension of the thorax (T1) (Hartenstein, 1993). There are no muscles connecting the external head and gut in Drosophila making it unlikely that external head muscles play a direct role in pulling the viscera forward (Bate, 1993; Sink, 2006). It is more likely that contraction of external head muscles move the mouth hooks to a more anterior location where they act as a point of anchorage.
Wave phase
During the wave phase of crawl a wave of muscle contractions serves to pick up each abdominal and thoracic body wall segment sequentially, move each segment in the direction of the crawl, and then replace each segment on the substrate. During a forward crawl we identified two defining features of the abdominal motor pattern: D and V muscles within a given segment contract simultaneously, and these contractions occur as L muscles in the adjacent posterior segment contract (Fig. 6). How does this pattern of muscle contraction lift each segment of the abdominal and thoracic body wall off of the substrate, and move it to a more anterior position (Fig. 2)? D and V muscles attach at the anterior and posterior borders of a segment (Crossley, 1978; Bate, 1993; Fig. 6). When D/V muscles contract in a given segment, for example A4, the segment shortens along the anteroposterior axis (Snodgrass, 1935; Fig. 6). Shortening of A4 could do two things. First, if the adjacent anterior segment was anchored to the substrate, shortening of A4 would move segment A5 toward the head. This is what occurs during a forward crawl (Fig. 6). Second, if the adjacent posterior segment was anchored to the substrate, shortening of A4 would move segment A3 toward the tail. This is what occurs during a reverse crawl (Fig. 6). Thus, the function of D/V muscle contraction depends on the context in which it occurs.
Contraction of D/V muscles in a given segment occurs in the context of movement of other segments. First, during forward crawls D/V muscles contract as L muscles in the adjacent posterior segment contract (Fig. 6A). L muscles attach at dorsoventral positions along the abdominal and thoracic body wall, and their contraction causes body wall to lift off of the substrate (Snodgrass, 1935, Crossley, 1978; Bate, 1993; Fig. 6A). Thus, during a forward crawl D/V muscles contract, as the adjacent posterior segment is lifted from the substrate. Second, segmental movements temporally overlap. For example, at the time that segment A4 begins to move, the adjacent anterior segment is stationary, whereas the adjacent posterior segment is moving (Fig. 4). This further contributes to a situation during forward crawling in which D/V contractions occur as the adjacent posterior segment is not in contact with the substrate. Thus, during forward crawling D/V muscles shorten a segment and move the adjacent posterior segment toward the head.
Comparison of Drosophila larval crawling with other well studied forms of crawling in soft-bodied animals
Crawling in soft-bodied animals has been studied most extensively in annelids, such as earthworms and leeches. There are four features that differ between annelid and Drosophila larval crawling. First, during an annelid crawl cycle, the head and tail move out of phase (Stern-Tomlinson et al., 1986; Quillin, 1999). For Drosophila, the head and tail move together during a limited portion of the stride cycle (Figs. 1, 3). Second, during annelid crawling retrograde waves of muscle contraction pass along the body axis in the direction opposite to locomotion (Gray and Lissmann, 1938; Gray et al., 1938); whereas Drosophila larvae use direct waves of muscle contraction that pass in the same direction as the direction of locomotion (Fig. 4). Third, in leech, waves of contractions of antagonistic muscle groups travel down the body axis at differing rates, and this phenomenon only becomes more extreme as the step duration increases (Cacciatore et al., 2000). In Drosophila, the relationship between antagonistic muscles groups remains constant over the range of stride durations we examined (Fig. 6; data not shown). Fourth, during earthworm crawling the animal mass is moved in the direction of the crawl gradually over the stride cycle. This is because in the earthworm each segment is separated by internal septa, and has a fixed volume (Trueman, 1975). Therefore as muscles of a segment contract, and move the segment, they also move the internal mass of the segment. In contrast, Drosophila the center of mass (centroid) moves forward rapidly at the start of the stride cycle (Fig. 2). This occurs because the internal organs move asynchronously with surrounding abdominal movements (Fig. 3). Thus, crawling in Drosophila larva represents an alternative type of soft-bodied crawling.
Footnotes
This work was supported by American Heart Association Fellowship # 0920025G to E.S.H. We thank Ryan Kersch, Jourdain Roberts-Richart, Karen Sotillie, Parker Coleman, Serge Faumont, and Ann Wehman.
The authors declare no competing financial interests.
References
- Baader AP, Kristan WB., Jr Parallel pathways coordinate crawling in the medicinal leech, Hirudo medicinalis. J Comp Physiol A. 1995;176:715–726. doi: 10.1007/BF00192620. [DOI] [PubMed] [Google Scholar]
- Barth R. Barth: Muskulatur und bewegungsart der raupen. Zool Jb Physiol. 1937;62:507–566. [Google Scholar]
- Bate M. The mesoderm and its derivatives. In: Bate M, Martinez Arias A, editors. The development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor; 1993. pp. 1013–1090. [Google Scholar]
- Berrigan D, Pepin D. How maggots move: allometry and kinematics of crawling in larval Diptera. J Insect Physiol. 1995;41:329–337. [Google Scholar]
- Cacciatore TW, Rozenshteyn R, Kristan WB., Jr Kinematics and modeling of leech crawling: evidence for an oscillatory behavior produced by propagating waves of excitation. J Neurosci. 2000;20:1643–1655. doi: 10.1523/JNEUROSCI.20-04-01643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey TM. Energetics of caterpillar locomotion: biomechanical constraints of a hydraulic skeleton. Science. 1991;252:112–114. doi: 10.1126/science.252.5002.112. [DOI] [PubMed] [Google Scholar]
- Champman R. The insects. New York: Cambridge UP; 1998. [Google Scholar]
- Crisp S, Evers JF, Fiala A, Bate M. The development of motor coordination in Drosophila embryos. Development. 2008;135:3707–3717. doi: 10.1242/dev.026773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley A. The morphology and development of the Drosophila muscular system. In: Ashburner M, Wright T, editors. The genetics and biology of Drosophila. New York: Academic; 1978. pp. 499–560. [Google Scholar]
- Dixit R, Vijayraghavan K, Bate M. Hox genes and the regulation of movement in Drosophila. Dev Neurobiol. 2008;68:309–316. doi: 10.1002/dneu.20589. [DOI] [PubMed] [Google Scholar]
- Elder H. Peristaltic mechanisms. In: Elder HY, Trueman ER, editors. Aspects of animal movement. New York: Cambridge UP; 1980. pp. 71–92. [Google Scholar]
- Freeman A, Bowers M, Mortimer AV, Timmerman C, Roux S, Ramaswami M, Sanyal S. A new genetic model of activity-induced Ras signaling dependent pre-synaptic plasticity in Drosophila. Brain Res. 2010;1326:15–29. doi: 10.1016/j.brainres.2010.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen WO, Cang J. Sensory and central mechanisms control intersegmental coordination. Curr Opin Neurobiol. 2001;11:678–683. doi: 10.1016/s0959-4388(01)00268-9. [DOI] [PubMed] [Google Scholar]
- Gray J, Lissmann H. Studies in animal locomotion. VII. Locomotory reflexes in the earthworm. J Exp Biol. 1938;15:506–517. [Google Scholar]
- Gray J, Lissmann H, Pumphrey R. The mechanism of locomotion in the leech (Hirudo medicinalis Ray) J Exp Biol. 1938;15:408–430. [Google Scholar]
- Green C, Burnet B, Connolly K. Organization and patterns of inter-and intraspecific variation in the behaviour of Drosophila larvae. Anim Behav. 1983;31:282–291. [Google Scholar]
- Hanslik U, Schoofs A, Niederegger S, Heinzel HG, Spiess R. The thoracic muscular system and its innervation in third instar Calliphora vicina larvae. I. Muscles of the pro- and mesothorax and the pharyngeal complex. J Morphol. 2010;271:960–968. doi: 10.1002/jmor.10848. [DOI] [PubMed] [Google Scholar]
- Hartenstein V. Atlas of Drosophila development. Cold Spring Harbor, NY: Cold Spring Harbor; 1993. [Google Scholar]
- Hughes CL, Thomas JB. A sensory feedback circuit coordinates muscle activity in Drosophila. Mol Cell Neurosci. 2007;35:383–396. doi: 10.1016/j.mcn.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang RY, Zhong L, Xu Y, Johnson T, Zhang F, Deisseroth K, Tracey WD. Nociceptive neurons protect Drosophila larvae from parasitoid wasps. Curr Biol. 2007;17:2105–2116. doi: 10.1016/j.cub.2007.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar B, Roote J, Campos AR. The tamas gene, identified as a mutation that disrupts larval behavior in Drosophila melanogaster, codes for the mitochondrial DNA polymerase catalytic subunit (DNApol-gamma125) Genetics. 1999;153:1809–1824. doi: 10.1093/genetics/153.4.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri S, Shen K, Klein M, Tang A, Kane E, Gershow M, Garrity P, Samuel ADT. Two alternating motor programs drive navigation in Drosophila larva. PLoS One. 2011;6:e23180. doi: 10.1371/journal.pone.0023180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Luo L, Gershow M, Rosenzweig M, Kang K, Fang-Yen C, Garrity PA, Samuel ADT. Navigational decision making in Drosophila thermotaxis. J Neurosci. 2010;30:4261–4272. doi: 10.1523/JNEUROSCI.4090-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marder E, Bucher D. Understanding circuit dynamics using the stomatogastric nervous system of lobsters and crabs. Annu Rev Physiol. 2007;69:291–316. doi: 10.1146/annurev.physiol.69.031905.161516. [DOI] [PubMed] [Google Scholar]
- Marder E, Calabrese RL. Principles of rhythmic motor pattern generation. Physiol Rev. 1996;76:687. doi: 10.1152/physrev.1996.76.3.687. [DOI] [PubMed] [Google Scholar]
- McCormick KE, Gaertner BE, Sottile M, Phillips PC, Lockery SR. Microfluidic devices for analysis of spatial orientation behaviors in semi-restrained Caenorhabditis elegans. PLoS One. 2011;6:e25710. doi: 10.1371/journal.pone.0025710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins OJ, Hackett JT, Buchanan JT, Friesen WO. Neuronal control of swimming behavior: comparison of vertebrate and invertebrate model systems. Prog Neurobiol. 2011;93:244–269. doi: 10.1016/j.pneurobio.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereanu W, Spindler S, Im E, Buu N, Hartenstein V. The emergence of patterned movement during late embryogenesis of Drosophila. Dev Neurobiol. 2007;67:1669–1685. doi: 10.1002/dneu.20538. [DOI] [PubMed] [Google Scholar]
- Quillin KJ. Kinematic scaling of locomotion by hydrostatic animals: ontogeny of peristaltic crawling by the earthworm Lumbricus terrestris. J Exp Biol. 1999;202:661–674. doi: 10.1242/jeb.202.6.661. [DOI] [PubMed] [Google Scholar]
- Quiñones-Coello AT, Petrella LN, Ayers K, Melillo A, Mazzalupo S, Hudson AM, Wang S, Castiblanco C, Buszczak M, Hoskins RA, Cooley L. Exploring strategies for protein trapping in Drosophila. Genetics. 2007;175:1089–1104. doi: 10.1534/genetics.106.065995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MA, Woods WA, Jr, Serebrenik YV, Simon SM, van Griethuijsen LI, Socha JJ, Lee WK, Trimmer BA. Visceral-locomotory pistoning in crawling caterpillars. Curr Biol. 2010a;20:1458–1463. doi: 10.1016/j.cub.2010.06.059. [DOI] [PubMed] [Google Scholar]
- Simon MA, Fusillo SJS, Colman KK, Trimmer BAB. Motor patterns associated with crawling in a soft-bodied arthropod. CORD Conference Proceedings. 2010b;213:2303–2309. doi: 10.1242/jeb.039206. [DOI] [PubMed] [Google Scholar]
- Sink H. New York: Landes; 2006. Muscle development in Drosophila (Molecular Biology Intelligence Unit) [Google Scholar]
- Skinner FK, Mulloney B. Intersegmental coordination in invertebrates and vertebrates. Curr Opin Neurobiol. 1998;8:725–732. doi: 10.1016/s0959-4388(98)80114-1. [DOI] [PubMed] [Google Scholar]
- Snodgrass RE. Principles of insect morphology. Ithaca, NY: Cornell UP; 1935. [Google Scholar]
- Song W, Onishi M, Jan LY, Jan YN. Peripheral multidendritic sensory neurons are necessary for rhythmic locomotion behavior in Drosophila larvae. Proc Natl Acad Sci U S A. 2007;104:5199–5204. doi: 10.1073/pnas.0700895104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Tomlinson W, Nusbaum MP, Perez LE, Kristan WB., Jr A kinematic study of crawling behavior in the leech, Hirudo medicinalis. J Comp Physiol A. 1986;158:593–603. doi: 10.1007/BF00603803. [DOI] [PubMed] [Google Scholar]
- Suster ML, Bate M. Embryonic assembly of a central pattern generator without sensory input. Nature. 2002;416:174–178. doi: 10.1038/416174a. [DOI] [PubMed] [Google Scholar]
- Trueman ER. The locomotion of soft-bodied animals. London: Edward Arnold; 1975. [Google Scholar]
- Wang JW, Sylwester AW, Reed D, Wu DA, Soll DR, Wu CF. Morphometric description of the wandering behavior in Drosophila larvae: aberrant locomotion in Na+ and K+ channel mutants revealed by computer-assisted motion analysis. J Neurogenet. 1997;11:231–254. doi: 10.3109/01677069709115098. [DOI] [PubMed] [Google Scholar]
- Wigglesworth V. The principles of insect physiology. London: Chapman and Hall; 1950. [Google Scholar]