Abstract
Biliary epithelial cells line the intrahepatic biliary network, a complex three-dimensional network of conduits. The loss of differentiated biliary epithelial cells is the primary cause of many congenital liver diseases. We identified a zebrafish snapc4 (small nuclear RNA-activating complex polypeptide 4) mutant in which biliary epithelial cells initially differentiate but subsequently disappear. In these snapc4 mutant larvae, the biliary epithelial cells undergo apoptosis, leading to the degeneration of the intrahepatic biliary network. Consequently, in snapc4 mutant larvae, biliary transport of ingested fluorescent lipids to the gallbladder is blocked. Snapc4 is the largest subunit of the protein complex that regulates small nuclear RNA (snRNA) transcription. The snapc4s445 mutation causes a truncation of the C-terminus thereby deleting the domain responsible for a specific interaction with Snapc2, a vertebrate specific subunit of the SNAP complex. This mutation leads to a hypomorphic phenotype, as only a subset of snRNA transcripts are quantitatively altered in snapc4s445 mutant larvae. snapc2 knockdown also disrupts the intrahepatic biliary network in a similar fashion as in snapc4s445 mutant larvae. These data indicate that the physical interaction between Snapc2 and Snapc4 is important for the expression of a subset of snRNAs and biliary epithelial cell survival in zebrafish.
Keywords: Liver, snRNA, SNAP190, SNAPC2, biliary epithelial cells, vanishing bile duct
Introduction
The liver is a major digestive organ responsible for many important physiological functions, including bile synthesis and secretion. Bile is synthesized in hepatocytes and transported to the intestine through the biliary network. The biliary network is a complex of three-dimensional conduits lined by specialized cells called biliary epithelial cells. Many congenital liver diseases are associated with biliary network malformation (Alvaro, 1999; Lazaridis et al., 2004; Lemaigre and Zaret, 2004; Strazzabosco et al., 2005; Raynaud et al., 2011). Despite their etiological heterogeneity and difference in degree of inflammatory response, one end result of many biliary disorders is the loss of biliary cells (Strazzabosco et al., 2000; Lazaridis et al., 2004). Thus, understanding the molecular mechanisms underlying the loss of biliary epithelial cells could lead to novel insights into the pathogenesis of various biliary disorders, and may provide the basis for novel therapeutic strategies.
The small nuclear RNA (snRNA)-activating protein complex (SNAPc) is a transcription factor essential for transcription of the snRNA genes, including U1, U2, U4, U5, and U6 spliceosomal snRNAs (Jawdekar and Henry 2008, Ma and Hernandez, 2001; Yoon et al., 1995). SNAPc is composed of at least five proteins; Snapc1 (also know as SNAP43 or PTFγ) (Yoon et al., 1996), Snapc2 (SNAP45 or PTFδ) (Sadowski et al., 1996), Snapc3 (SNAP50 or PTFβ) (Henry et al., 1996), Snapc4 (SNAP190 or PTFα) (Wong et al., 1998), and Snapc5 (SNAP19) (Henry et al., 1998). Snapc4 is the largest subunit, and serves as a backbone to assemble SNAPc. All subunits except for Snapc2 assemble on the N-terminus of Snapc4. Snapc2 instead binds to a domain called Snapc2 binding domain within the C-terminus of Snapc4 (Ma and Hernandez, 2001). Interestingly, a partial SNAPc containing Snapc1, Snapc3 and the N-terminal third of Snapc4 are fully sufficient for target DNA sequence binding and snRNA gene transcription in vitro (Mittal et al., 1999). Thus, neither the C-terminal two-thirds of Snapc4 nor Snapc2 are required for in vitro basal snRNA transcription. The same three subunits required for the partial SNAPc are widely conserved in Drosophila, trypanosomes and plants (Li et al., 2004; Das and Bellofatto, 2003; Schimanski et al., 2005). In contrast, the Snapc2 and Snapc5 subunits are not conserved in invertebrates or plants, and these subunits are, at least in vitro, dispensable for vertebrate snRNA transcription (Mittal et al., 1999). Although Snapc2 binding changes SNAPc target recognition, the in vivo function of this vertebrate specific subunit is not known.
Zebrafish express genes encoding all 5 subunits of SNAPc and contain vertebrate specific organs such as the liver, making it a suitable genetic model to study functions of the vertebrate specific subunits of SNAPc. In zebrafish, the formation and function of the biliary network has been characterized (Lorent et al., 2004; Matthews et al., 2004; Sakaguchi et al., 2008). The small size of the larval liver enables visualization of the three-dimensional branching structure of the intrahepatic biliary network (Sakaguchi et al., 2008). Imaging studies of the intrahepatic biliary network have been facilitated by the recent development of a notch activity reporter transgenic line in which GFP is specifically expressed in the biliary network (Parsons et al., 2009; Lorent et al., 2010).
In this study, we identified a mutation in the zebrafish snapc4 gene which truncates the C-terminal third of the protein and deletes a Snapc2 binding domain. The mutant form of Snapc4 is similar to that of invertebrate Snapc4 (Hernandez Jr et al., 2006; Kim et al., 2010) or the mini-Snapc4 used for in vitro studies (Ma and Hernandez, 2001). As expected from these in vitro studies, the truncated mutant form of Snapc4 still can regulate the expression of most of its target snRNAs, and thus can be considered to be a hypomorphic allele. We further show that this snapc4 hypomorph unexpectedly causes the loss of differentiated biliary epithelial cells in zebrafish.
Results
Zebrafish s445 mutants exhibit intrahepatic biliary network defects
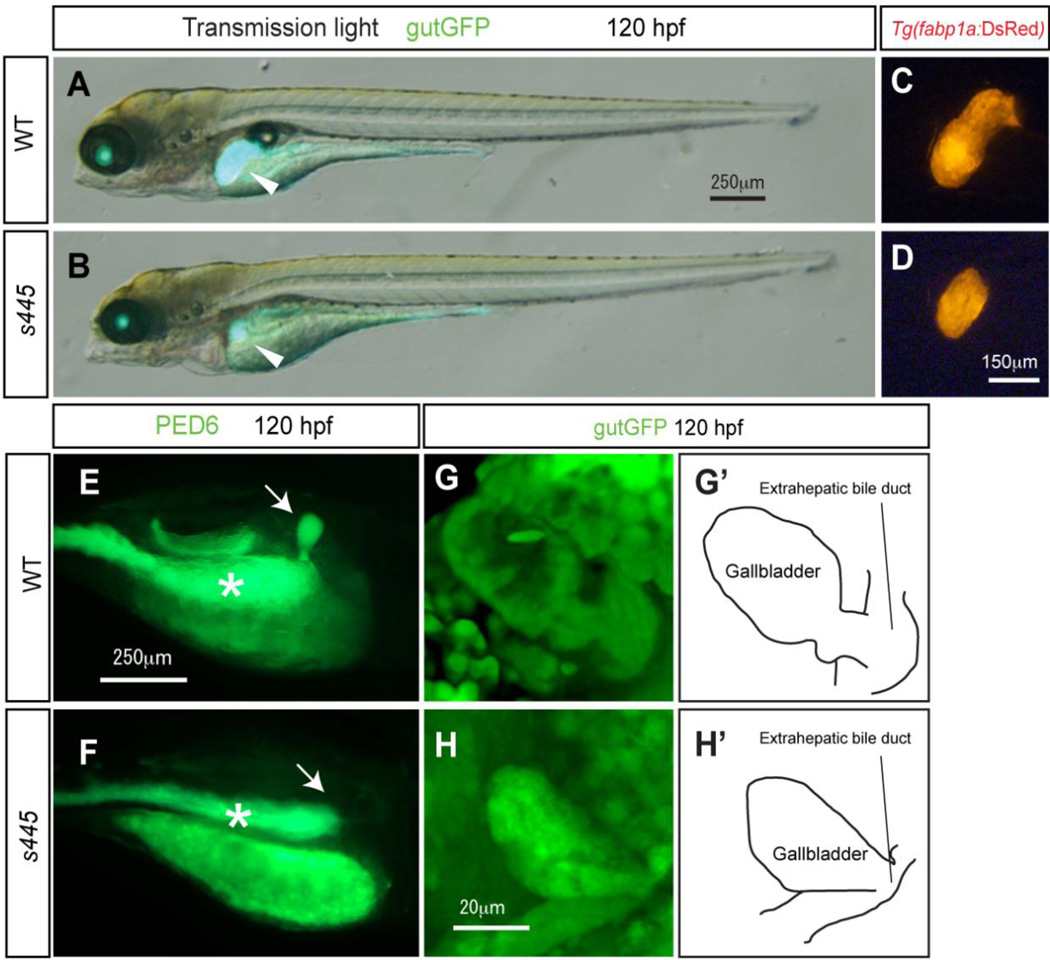
We identified a recessive lethal mutation, s445, in a forward genetic screen for genes that regulate liver organogenesis in zebrafish, Danio rerio (Ober et al., 2006). At 120 hours post-fertilization (hpf), homozygous s445 mutant larvae are morphologically similar to wild-type siblings (Fig. 1A and B). However their liver, which is visualized by Tg(gutGFP)s854 expression (Field et al., 2003), is noticeably smaller (Fig. 1B; Supplementary fig. 1). We also compared the size of the regions marked by hepatocyte specific Tg(fabp1a:DsRed)gz15 expression, which is clearly smaller in s445 mutant larvae as compared to wild-type at 120 hpf (Fig. 1C and D), supporting the observation that the liver is the smaller in s445 mutant larvae.
Figure 1.
Morphological and physiological phenotype of s445 mutant larvae at 120 hpf. (A and B) Bright-field images of wild-type (A) and s445 mutant (B) larvae at 120 hpf are overlaid with Tg(gutGFP)s854 expression (green). The physiological appearance of s445 mutant larvae is normal, however their liver size is smaller at 120 hpf. Lateral views, anterior to the left. White arrowheads point to the liver. (C and D) Fluorescent image of wild-type (C) and s445 mutant (D) Tg(fabp1a:DsRed)gz15 larvae at 120 hpf. Liver size visualized by Tg(fabp1a:DsRed)gz15 expression is smaller in s445 mutant larvae. Lateral views, anterior to the left. (E and F) Fluorescent images of wild-type (E) and s445 mutant (F) larvae soaked in PED6. PED6 is processed by phospholipase and fluorescence is detected in the intestine in both wild-type and s445 mutant larvae. However, PED6 fluorescence is missing from the s445 mutant gallbladder. Lateral views, anterior to the right. Asterisks indicate the intestine. (G and H) Projected confocal image of the gallbladders of wild-type (G) and s445 mutant (H) Tg(gutGFP)s854 larvae. The gallbladder and the extrahepatic bile duct visualized by Tg(gutGFP)s854 expression are schematically presented in (G’ and H’). Ventral views, anterior to the top.
We then made use of the PED6 fluorescence assay to test in vivo the function of the digestive organs in s445 mutants. In this assay, larvae are fed the quenched fluorescent phospholipid PED6, which is cleaved in the intestine, activating the fluorescent signal (Farber et. al., 2001). The fluorescent metabolites are then transported through the biliary system to the gallbladder (Fig. 1E). PED6 fluorescence can be observed in the intestines of s445 mutant larvae (Fig. 1F, asterisk). However, in s445 mutant larvae, no PED6 signal is observed in the gallbladder, despite the presence of a morphologically identifiable gallbladder and extrahepatic bile duct (Fig. 1H). Based on these data, we hypothesized that the defect in PED6 metabolite transportation is due to an impairment of the intrahepatic biliary network.
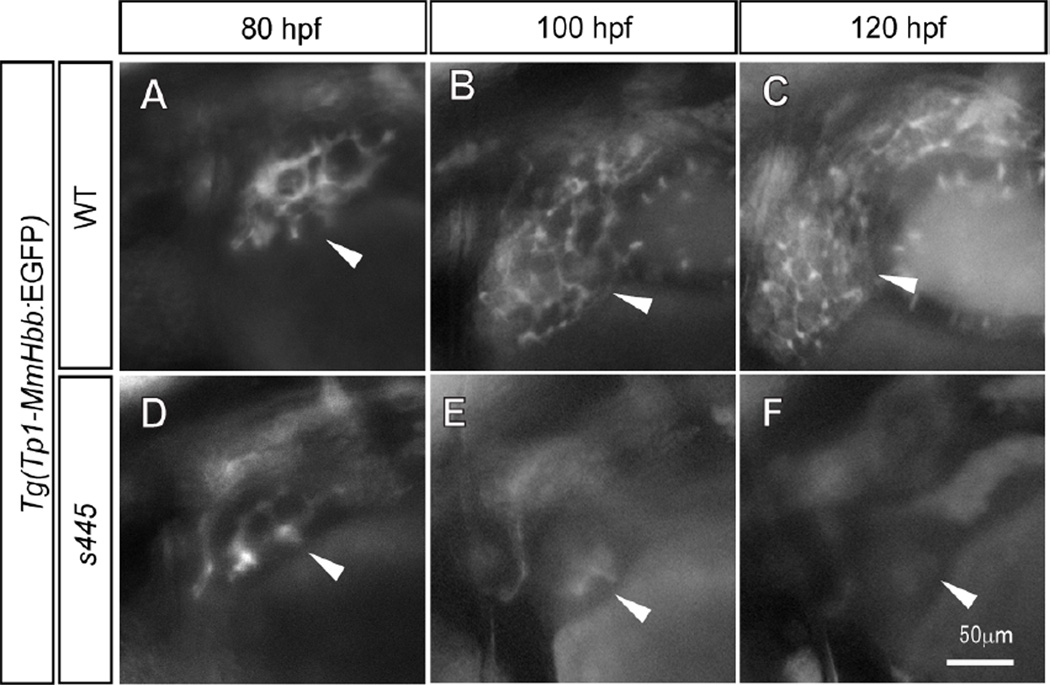
To test this hypothesis, we undertook time course observations of intrahepatic biliary network formation in s445 mutants. Tg(Tp1-MmHbb:EGFP)um14 is expressed by intrahepatic biliary epithelial cells in the zebrafish liver (Parsons et al., 2009, Lorent et al., 2010). At 80 hpf, we did not observe any difference in Tg(Tp1-MmHbb:EGFP)um14 expression in the s445 mutant liver (Fig. 2A and D), suggesting that the intrahepatic biliary network initially formed. At 100 hpf, the Tg(Tp1-MmHbb:EGFP)um14 positive intrahepatic biliary network is highly branched in wild-type larvae. However in contrast, by 100 hpf in s445 mutant larvae, Tg(Tp1-MmHbb:EGFP)um14 expression largely disappears from the liver, with only weak fluorescence detected in the central part of the liver (Fig. 2E). At 120 hpf, Tg(Tp1-MmHbb:EGFP)um14 fluorescence cannot be detected in the s445 mutant liver (Fig. 2F). These data suggest that the intrahepatic biliary network forms initially but disappears by 120 hpf in s445 mutant larvae.
Figure 2.
Time course observation of intrahepatic biliary network development. (A–C) Tg(Tp1-MmHbb:EGFP)um14 expression in the wild-type intrahepatic biliary network at 80 (A), 100 (B) and 120 (C) hpf; same larva is shown at all stages. (D–F) Tg(Tp1-MmHbb:EGFP)um14expression in the s445 mutant intrahepatic biliary network at 80 (A), 100 (B) and 120 (C) hpf; same larva is shown at all stages. At 80 hpf, Tg(Tp1-MmHbb:EGFP)um14 expression in the s445 mutant intrahepatic biliary network is indistinguishable from that of wild-type, however it disappears after 100 hpf. Lateral views, anterior to the left. White arrowheads point to the liver in each image.
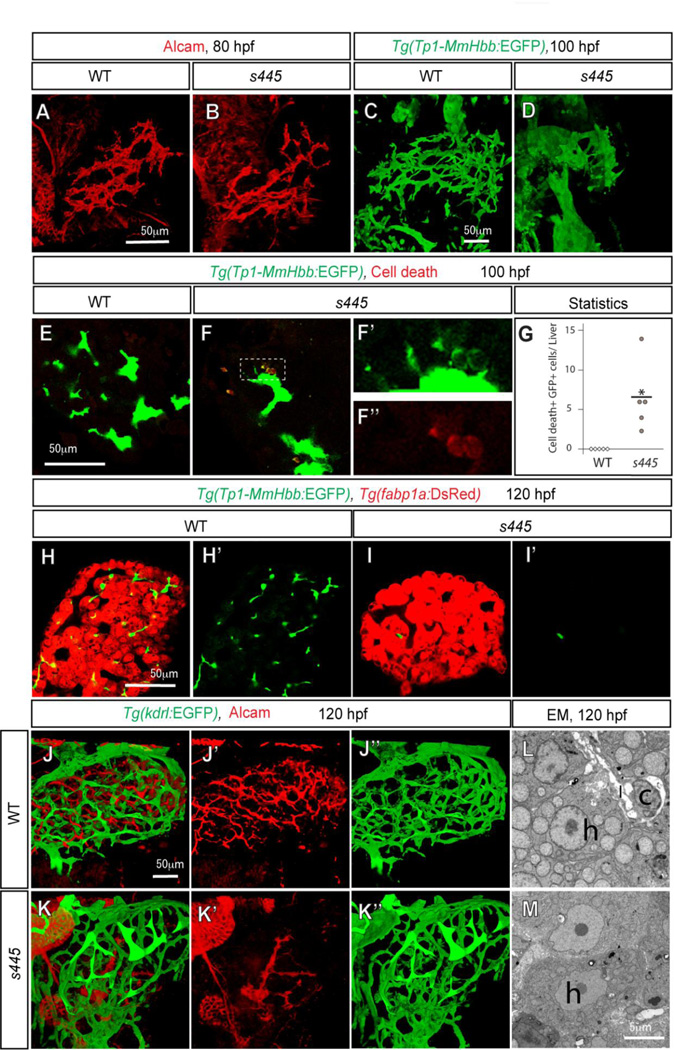
To better understand the defects caused by the s445 mutation, we visualized the three-dimensional structure of the intrahepatic biliary network. Immunofluorescence staining for Alcam, the intrahepatic biliary network marker (Sakaguchi et al., 2008), was used to visualize the intrahepatic biliary network (Fig. 3A). Consistent with the time course observations, we did not observe any difference in Alcam or Tg(Tp1-MmHbb:EGFP)um14 expression in s445 mutant larvae at 80 hpf (Fig. 3A and B; Supplementary Fig. 2), supporting the idea that biliary epithelial cells initially differentiate in s445 mutant larvae. However, in s445 mutant larvae at 100 hpf, Tg(Tp1-MmHbb:EGFP)um14 expression in the liver was significantly different from wild-type siblings (Fig. 3D). In the wild-type livers, the Tg(Tp1-MmHbb:EGFP)um14 expressing intrahepatic biliary network added new branches (Fig. 3C). In contrast, the intrahepatic biliary network appears to be degenerating in the s445 mutant livers (Fig. 3D).
Figure 3.
The intrahepatic biliary network disappears in s445 mutant larvae. Projected (A–D and J–K) and z-plane (E–F and H–I) confocal images of the liver at 80 (A and B), 100 (C–F) and 120 (H–K) hpf. Ventral views, anterior to the top. (A and B) Wild-type (A) and s445 mutant (B) larvae visualized for Alcam expression. The Alcam-positive intrahepatic biliary network in s445 mutant larvae is indistinguishable from that of wild-type at 80 hpf. (C and D) Wild-type (C) and s445 mutant (D) Tg(Tp1-MmHbb:EGFP)um14 larvae visualized for GFP expression. Tg(Tp1-MmHbb:EGFP)um14 expression in the liver is restricted to the intrahepatic biliary network at 100 hpf. The intrahepatic biliary network in s445 mutant appears to be degenerating at this stage. (E and F) Wild-type (E) and s445 mutant (F) Tg(Tp1-MmHbb:EGFP)um14 larvae stained for red fluorescent TUNEL. GFP expression and TUNEL signals in the outlined area are magnified and shown separately in F’ and F”, respectively. (G) Number of TUNEL positive Tg(Tp1-MmHbb:EGFP)um14 expressing cells in the wild-type (open square) and s445 mutant (closed circle) liver. *, p < 0.05. (H and I) Wild-type (H) and s445 (I) mutant Tg(Tp1-MmHbb:EGFP)um14; Tg(fabp1a:DsRed)gz15 larvae visualized for GFP and DsRed expression. Tg(Tp1-MmHbb:EGFP)um14 expressing biliary epithelial cells have largely disappeared from s445 mutant livers by 120 hpf, while Tg(fabp1a:DsRed)gz15expressing hepatocytes remain. GFP expression is shown separately in H’ and I’. (J and K) Wild-type (J) and s445 mutant (K) Tg(kdrl:EGFP)s843 larvae visualized for GFP (green) and Alcam (red) expression. The Tg(kdrl:EGFP)s843-expressing intrahepatic vascular network is relatively unaffected while the Alcam-expressing intrahepatic biliary network is severely affected in s445 mutant larvae. Alcam and GFP expressions are shown separately in (J’ and K’) and (J” and K”), respectively. (L and M) Electron micrographs of the wild-type (L) and s445 mutant (M) liver at 120 hpf. h, hepatocyte; c, cholangiocyte; l, the lumen of the cholangiocyte.
In s445 mutant larvae, we detected fluorescent TUNEL-positive/Tg(Tp1-MmHbb:EGFP)um14-positive cells in the liver (Average 6.6 cells per liver, SD= 4.9, n=5, p < 0.05) at 100 hpf (Fig. 3F and G), while no Tg(Tp1-MmHbb:EGFP)um14 expressing cells in the wild-type liver (n=5) showed TUNEL signal at the same stage (Fig. 3E and G). These data suggest that biliary epithelial cells are initially present but undergo cell death in s445 mutant larvae. In contrast, there is no significance difference in the number of TUNEL-positive/Tg(Tp1-MmHbb:EGFP)um14-negative cells in the liver between wild-type (average 3.8 cells per liver; SD = 2.16; n = 5) and s445 mutant (average 2.6 cells per liver; SD = 1.81; n = 5; p > 0.05) larvae, suggesting that cell death is significantly increased only in biliary epithelial cells in the s445 mutant liver. Consistent with this observation, at 120 hpf, there were very few Tg(Tp1-MmHbb:EGFP)um14 expressing biliary epithelial cells in s445 mutant livers (Fig. 3I), while the Tg(fabp1a:DsRed)gz15 expressing hepatocytes were sustained (Fig. 3H and I). The Tg(kdrl:EGFP)s843 (Jin et al., 2005) expressing intrahepatic vascular network is a highly branched conduit (Fig. 3J), which is intertwined with the intrahepatic biliary network in zebrafish (Fig. 3J; Sakaguchi et al., 2008). In s445 mutant larvae, the intrahepatic vascular network was less affected, maintaining its branched structures (Fig. 3K”), while the Alcam-expressing intrahepatic biliary network had largely degenerated (Fig. 3K’). Differentiated biliary epithelial cells and their lumens are evident in electron micrographs of the wild-type liver at 120 hpf (Fig. 3L). However at the same stage, no biliary epithelial cells can be morphologically confirmed in electron micrographs of the s445 mutant (Fig. 3M). Overall, these data suggest that the s445 mutation preferentially affects the intrahepatic biliary network.
s445 mutation truncates the C-terminal third of zebrafish Snapc4
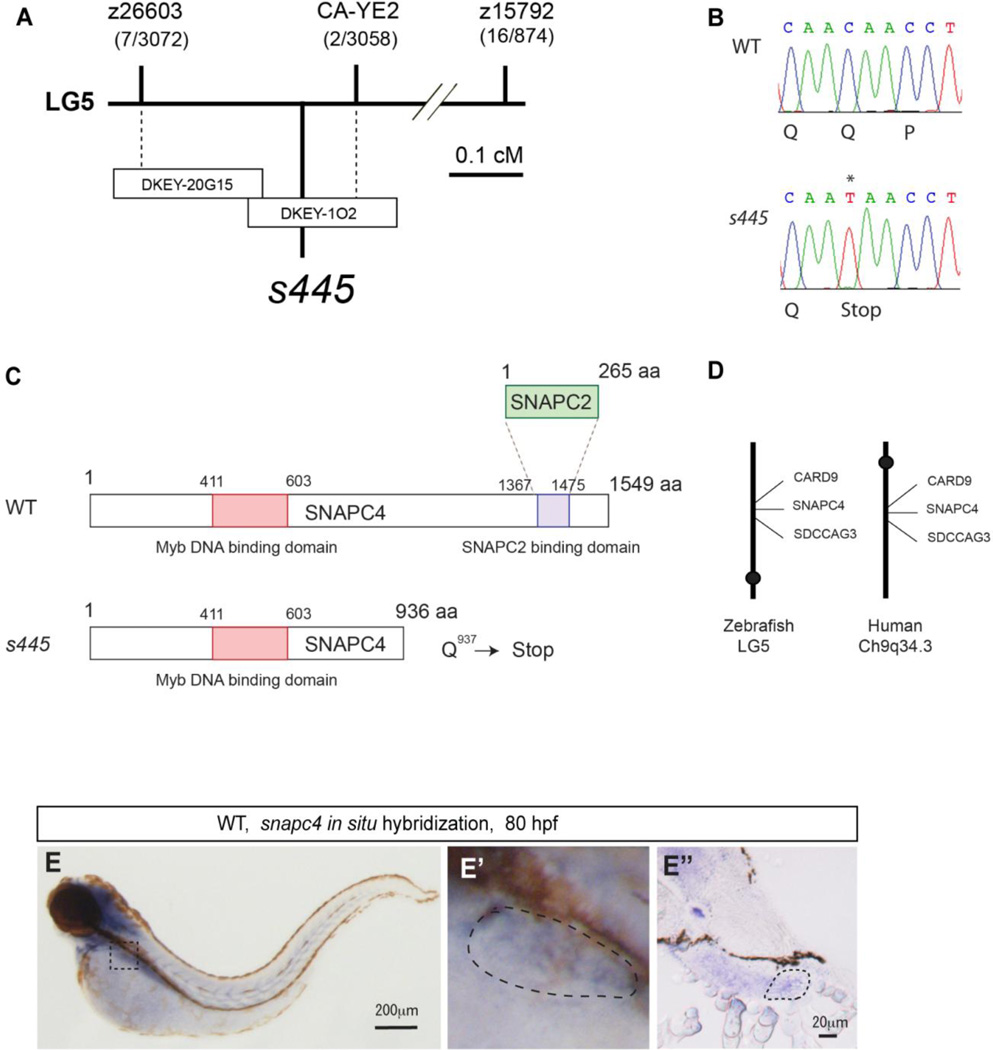
To identify the s445 gene, we employed a positional cloning strategy. Bulk segregant analyses using markers from the zebrafish genetic map placed the s445 locus on zebrafish chromosome 5 (Fig. 4A). In subsequent steps, we defined a critical region surrounding the s445 locus, which was flanked by markers 0.2 cM (z26603) and 0.07 cM (CA-YE2) from the s445 locus (Fig. 4A). Two partially overlapping BACs, DKEY-20G15 and DKEY-1O2, spanned the critical region (Fig. 4A). Injecting a DKEY-102, but not DKEY-20G15 BAC into s445 homozygous mutants partially rescued its phenotype (data not shown), indicating that the DKEY-102 BAC contains the s445 locus.
Figure 4.
Positional cloning of the s445 mutant locus. (A) Schematic of the 445 locus. s445 is located on chromosome 5 in between two CA-repeat markers, z26603 (0.2 cM; 7 recombinants out of 3072 meioses) and CA-YE2 (0.07 cM; 2 recombinants out of 3056 meioses). The critical region of the s445 locus was spanned by two partially overlapping BAC clones, DKEY-20G15 and DKEY-1O2. (B) A single nucleotide mutation (C to T) that results in a premature stop of the snapc4 gene was identified in s445 mutants (0 recombinants out of > 3000 meioses). (C) Schematic of zebrafish Snapc4. Zebrafish Snapc4 contains a conserved Myb DNA binding domain and a Snapc2 binding domain. The s445 mutation generates a premature stop codon in the zebrafish snapc4 cDNA. The truncated mutant Snapc4 protein is approximately two-thirds the length of wild-type Snapc4 and lacks the Snapc2 binding domain. (D) s445 is located on zebrafish chromosome 5 in a region syntenic to a region of human chromosome 9q34.3. (E) snapc4 expression in wild-type larvae at 80 hpf. snapc4 expression is detected in the liver at this stage. The outlined area is magnified and shown in E’. Lateral views, anterior to the left. Cross section of the stained larva in the region of the liver is shown in E”. The liver is outlined in E’ and E”.
There are four predicted genes between CA-YE2 and the end of DKEY-102. Sequence analysis of the four candidate cDNAs from s445 mutant and wild-type larvae identified only a single point mutation in the zebrafish snapc4 (snRNA-activating protein complex peptide 4) gene (Fig. 4B). This C to T transition generated a premature stop codon in the zebrafish snapc4 cDNA (Fig. 4C), indicating that this lesion is responsible for the s445 phenotype.
Snapc4 is the largest subunit of the transcription factor SNAPc (snRNA activating protein complex; Wong et al., 1998), which is responsible for snRNA gene transcription. Zebrafish Snapc4 contains conserved Myb DNA binding and Snapc2 binding domains (Fig. 4C). The s445 mutation deletes the Snapc2 binding domain while the Myb DNA binding domain remains intact (Fig. 4C). To further confirm that the snapc4 gene was responsible for the s445 mutant phenotype, we injected antisense morpholino oligonucleotides (MOs) to knock down snapc4 gene function. A splice blocking MO(A) designed to truncate the C-terminal third of the Snapc4 protein phenocopied the s445 mutant phenotype (n=35, Supplementary fig. 3C and D). These data further support the hypothesis that truncation of the C-terminal third of the Snapc4 protein is the cause of the s445 mutant phenotype. Injecting another splice blocking MO(B), which is designed to block splicing in front of the Myb DNA binding domain, generated a more severe morphological phenotype than the s445 mutants (n > 50, Supplementary fig. 3E and F), suggesting that s445 is a hypomorphic mutation in which the s445 mutant phenotype is milder than a complete loss-of-function phenotype. Injecting a snapc4 MO(C) which targets the translation initiation codon induced developmental arrest within 18 to 24 hpf (n=38, data not shown). Since splice blocking MOs target only zygotic snpac4 mRNA but the translation initiation blocking MO targets both maternal and zygotic snapc4 mRNA, the complete loss of snapc4 function severely affects embryonic development.
Since the region surrounding zebrafish snapc4 is syntenic to human chromosome 9q34.3 where human SNAPC4 is located (Fig. 4D), zebrafish snapc4 is likely the orthologue of human SNAPC4.
Consistent with the s445 mutant phenotypes, the snapc4 gene is expressed in the liver at 80 hpf (Fig. 4E). Strong snapc4 expression is also detected in the brain, eyes and branchial arches at 80 hpf (Fig. 4E).
snRNA expression in s445 mutant larvae
SNAPc is required for the transcription of snRNA genes in vitro (Jawdekar and Henry, 2008). We thus examined snRNA gene expression in s445 mutant larvae by real-time quantitative PCR and Northern blotting. Unexpectedly, there were no significant differences in U1, U2 and U6 snRNA expression levels in s445 mutant larvae at 100 hpf (Fig. 5 A and B). Meanwhile, in s445 mutant larvae, U4 and U5 snRNA expression levels were downregulated approximately 50 and 75 percent, respectively (Fig. 5A and B). These data support the hypothesis that the s445 mutation is hypomorphic, affecting only a subset of its downstream target snRNA genes.
Figure 5.
snRNA expression in s445 mutant larvae. (A) Real-time PCR to compare snRNA gene expression between wild-type and s445 mutant larvae at 100 hpf. Data are represented as fold change relative to expression levels in wild-type larvae. The expression levels of U1, U2, and U6 snRNAs are not significantly changed, while those of U4 (average 0.59 fold; s.d. = 0.22; p = 0.037) and U5 (average 0.36 fold; s.d. = 0.29; p = 0.024) snRNAs are down-regulated in s445 mutant larvae. *, p < 0.05; N.S., not significant (p > 0.05). (B) Northern blotting of snRNAs. Total RNA from wild-type and s445 mutant larvae at 100 hpf were blotted with snRNA probes to the indicated genes. A probe for the 28S ribosomal RNA was used as a loading control.
Snapc2 is required for intrahepatic biliary network formation
Since the Snapc2 binding domain is deleted from Snapc4 in s445 mutants, we hypothesized that the absence of the Snapc2/Snapc4 interaction is responsible for downregulation of U4 and U5 snRNA transcription as well as the degenerate intrahepatic biliary network phenotype in s445 mutant larvae. To test this hypothesis, we knocked down the zebrafish snapc2 gene. We designed two non-overlapping splice blocking snapc2 MOs (MO(A) and MO(B)). Injecting both snapc2 MOs generated the same phenotype (Fig. 6B), suggesting that these MOs are both working in a target-specific manner. snapc2 MOs-injected larvae had smaller livers and slight pericardial edema at 100 hpf (Fig. 6B), while their body length was normal. The snapc2 gene is expressed in the liver at 80 hpf (Fig. 6C), and its expression pattern is similar to that of the snapc4 gene (Compare Fig, 6C and Fig. 4E). Similar to s445 mutant larvae, in snapc2 MO(A)-injected larvae at 100 hpf, Tg(Tp1-MmHbb:EGFP)um14 expression in the liver was missing (Fig. 6E), while hepatocyte marker Tg(fabp1a:DsRed)gz15 expression was sustained (Fig. 6G). These data suggest that, similar to s445 mutant larvae, the intrahepatic biliary network is preferentially affected in snapc2 MO(A) injected larvae. In snapc2 MO(A) injected larvae, a 52 bp deletion in the snacp2 cDNA was detected (Fig. 6H), further confirming that snapc2 MO(A) works in a target specific manner. We also found that in snapc2 MO injected larvae, U4 and U5 snRNA expression levels were downregulated (Fig. 6I) in a similar manner to s445 mutant larvae. In snapc2 MO injected larvae, U2 snRNA expression was also slightly down-regulated (Fig. 6I). These data suggest that Snapc2 is required for intrahepatic biliary network maintenance, as well as snRNA transcription.
Figure 6.
Zebrafish Snapc2 is essential for intrahepatic biliary network formation. (A and B) Bright-field images of wild-type (A) and snapc2 morpholino injected (B) larvae at 100 hpf are overlaid with Tg(gutGFP)s854 fluorescence (green). snapc2 morpholino injected larvae show smaller livers and mild pericardial edema. (C) snapc2 expression in wild-type larvae at 80 hpf. snapc2 expression is detected in the liver at this stage. The outlined area is magnified and shown in E’. The liver is outlined in E’. (D and E) Tg(Tp1bglob:EGFP)um14 expression in the intrahepatic biliary network of wild-type (D) and snapc2 morpholino injected (E) larvae at 100 hpf. Tg(Tp1-MmHbb:EGFP)um14 expression is absent from the snapc2 morpholino injected liver. (F and G) Tg(fabp1a:DsRed)gz15 expression in the hepatocytes of wild-type (F) and snapc2 morpholino injected (G) larvae at 100 hpf. Lateral views, anterior to the left. White arrowheads point to the liver in each image. (H) Reverse transcriptase-PCR analysis shows that 52 bp are deleted from snapc2 mRNA in snapc2 MO(A) injected larvae. (I) Realtime PCR to compare snRNA gene expression between wild-type and snapc2 MO(A) injected larvae at 60 hpf. Data are represented as fold change relative to expression levels in wild-type larvae. The expression levels of U1 and U6 snRNAs are not significantly changed, while those of U2 (average 0.75 fold; s.d. = 0.153), U4 (average 0.657 fold; s.d. = 0.02) and U5 (average 0.368 fold; s.d. = 0.218) snRNAs are down-regulated in s445 mutant larvae. *, p < 0.05; N.S., not significant (p > 0.05). (J) Bright-field image of snapc2 binding domain mRNA injected larvae at 100 hpf. Schematic of the Snapc2 binding domain of Snapc4 is shown in (J’). (K) Tg(Tp1bglob:EGFP)um14 expression in the intrahepatic biliary network of snapc2 binding domain mRNA injected larvae at 100 hpf.
Since the snapc2 binding domain of Snapc4 is known to interact physically with Snapc2, we hypothesized that over-expressing only the Snapc2 binding domain might inhibit Snapc2 binding to endogenous Snapc4. To test this hypothesis, we cloned the Snapc2 binding domain of Snapc4, and attached translation initiation and stop codons in the beginning and the end of it, respectively (Figure 6J’ and materials and methods). When we injected synthesized mRNA encoding the Snapc2 binding domain, injected larvae showed similar phenotype (26/33) to that of larvae injected with Snapc2 MOs at 100 hpf (compare Figure 6B and 6J). Tg(Tp1bglob:EGFP)um14 expression in the liver was missing in the Snapc2 binding domain mRNA injected larvae at 100 hp (compare Figure 6D and 6K). These data further support the hypothesis that physical interaction between Snapc4 and Snapc2 is important for the maintenance of the intrahepatic biliary network.
Discussion
In this study, we identified a zebrafish snapc4 mutant in which differentiated biliary epithelial cells initially develop but later degenerate. To our knowledge, this is the first in vivo evidence showing that Snapc4 is required for a specific biological process, the maintenance of the intrahepatic biliary network.
The C-terminal region of Snapc4 has an inhibitory function
The snRNA-activating protein complex, SNAPc, is a multi-subunit transcription factor that is required for transcription of the snRNA genes (Jawdekar and Henry, 2008). Snapc4, the largest subunit of the SNAPc, forms the backbone of the complex (Wong et al., 1998). In vitro studies have demonstrated that the C-terminal region of Snapc4 possesses an inhibitory effect on its target DNA binding and transcriptional abilities (Mittal et al., 1999). Binding of Snapc2 to the C-terminal portion of Snapc4 masks the inhibitory effect, and enhances the DNA binding of Snapc4 to the target DNA in vitro (Mittal et al., 1999). In fact, removing the C-terminal two-thirds of Snapc4 improves its DNA binding and transcriptional abilities. Thus, in vitro, omission of Snapc2, which releases an inhibitory effect on the C-terminal portion of Snapc4, impairs Snapc4 function to a greater degree than deletion of the C-terminal two-thirds of Snapc4 (Mittal et al., 1999; Ma and Hernandez, 2001). These in vitro data are fully consistent with our zebrafish in vivo data. The depletion of Snapc2 in larvae injected with the snapc2 MO results in a more severe morphological phenotype than that of s445 mutant larvae, in which the C-terminal third of Snapc4 is deleted (Compare Fig. 1B and Fig. 6B).
In vitro, the N-terminal third of Snapc4 is sufficient to regulate basal levels of U1 and U6 snRNA transcription (Mittal et al., 1999). Consistent with these data, in s445 mutant larvae, we detected no significant change in U1, U2 and U6 snRNA expression levels (Fig. 5). Blockage of U2 snRNA function is known to induce early developmental arrest in zebrafish (König et al., 2007), suggesting that snRNA genes are indispensable for zebrafish development. Consistently, we detected developmental arrest after translational blockage in snapc4 MO(C) injected larvae, in which most of Snapc4 protein should be depleted. This result suggests that full length Snapc4 is required for early developmental progress. Thus, we conclude that the s445 mutation is hypomorphic, affecting only a subset of target snRNA transcription. To our knowledge, this is the first in vivo genetic evidence that, consistent with in vitro data, the C-terminal third of Snapc4 is not required for U1and U6 snRNA transcription.
Zebrafish and human Snapc4
Since the synteny is conserved (Fig. 4D), zebrafish snapc4, which is affected by the s445 mutation, is most likely an orthologue of human SNAPC4. Zebrafish Snapc4 contains conserved Myb DNA binding and Snapc2 binding domains. However, an Oct-1 binding domain, which is conserved across mammalian Snapc4 genes, is not found in zebrafish Snapc4. This lack of a recognizable Oct-1 binding domain does not exclude the possibility that a different sequence in zebrafish Snapc4 is responsible for Oct-1 binding. To our surprise, zebrafish snapc4 shows tissue specific expression at 80 hpf (Fig. 4E), at a time when U1 snRNA is expressed ubiquitously (data not shown). We assume that highly proliferating cells require additional snRNA transcriptional regulators to maintain constant snRNA levels. However, we cannot exclude the possibility that the SNAPc possesses additional roles in organogenesis beyond snRNA transcriptional regulation.
Human SNAPC4 is located on chromosome 9q34.3. Interestingly, the deletion of 9q34.3 results in complicated clinical syndromes (Iwakoshi et al., 2004; Harada et al., 2006; Neas et al., 2005; Pointon et al., 2010). The fact that a hypomorphic snapc4 mutation can cause tissue or cell type specific phenotypes in zebrafish could help identify the responsible genes for these human diseases.
Vanishing bile duct syndrome is an irreversible human liver disorder caused by biliary duct destruction (Pass et al., 2008). In the vanishing bile duct syndrome, differentiated biliary epithelial cells undergo apoptotic cell death and disappear (Strazzabosco et al., 2000). It is noteworthy that the phenotype we observe in s445 mutant larvae - that biliary epithelial cells initially differentiate but subsequently disappear due to apoptosis - is similar to that observed in vanishing bile duct syndrome.
Degenerating biliary epithelial cells in s445 mutant larvae
The livers of s445 mutant larvae are indistinguishable from those of wild-type siblings at 80 hpf (Fig. 2A and D; Fig. 3A and B). However, at 100 hpf, the intrahepatic biliary network shows drastic differences in s445 mutant larvae (Fig. 3C and D). In s445 mutant larvae, many biliary branches are contracted, displaying prominent cell death in Tg(Tp1-MmHbb:EGFP)um14 positive biliary epithelial cells (Fig. 3F). Thus at 100 hpf, biliary epithelial cells are more vulnerable to the s445 mutation than other cell types in the liver (Fig. 3I and K). However, it should be noted that, at 120 hpf, hepatocytes also decrease in number (Fig. 1D) and appear to have morphological differences (Fig. 3M) in s445 mutant larvae. s445 mutant larvae generally die at 8 dpf.
Since snapc2 knock down leads to a similar biliary phenotype, we posit that the absence of the Snapc2-Snapc4 interaction is responsible for the loss of biliary epithelial cells in s445 mutant larvae. Indeed, blocking the Snapc2-Snapc4 interaction by overexpression of the Snapc2 binding domain also impairs intrahepatic biliary network formation (Fig. 6J and K). We have therefore demonstrated at least two novel functions for the Snapc2-Snapc4 interaction: proper transcription of the U4 and U5 snRNA genes and survival of biliary epithelial cells. We predict that downregulation of the snRNA genes affects biliary epithelial cell survival. However, since Snapc2 plays two roles in the cell, one as a subunit of the SNAPc and another as a factor for mitotic regulation (Shanmugam and Hernandez, 2008), we cannot exclude the possibility that Snapc2 regulates biliary epithelial cell survival independent of SNAPc and snRNA gene transcription.
Materials and Methods
Zebrafish husbandry
Zebrafish (Danio rerio) larvae were obtained from natural crosses of wild-type or heterozygous mutant fish, raised at 28.5°C and staged according to age (hours post-fertilization (hpf) at 28.5 °C) as previously described. The following transgenic and mutant lines were used in this study: Tg(fabp1a:DsRed; ela3l:EGFP)gz15 (Korzh et al., 2008); Tg(Tp1-MmHbb:EGFP)um14 (Parsons et al., 2009); Tg(kdrl:EGFP)s843 (Jin et al., 2005); Tg(gutGFP)s854 (also known as Tg(XlEef1a1:GFP)s854 ) (Field et al., 2003).
Immunohistochemistry, in situ cell detection and in situ hybridization
Immunohistochemistry was performed as previously described (Sakaguchi et al., 2008). Confocal images were obtained in a z-series by a TCS SP5 confocal microscope (Leica Microsystems). All confocal images were processed using Volocity software (PerkinElmer). In situ hybridization was performed as previously described (Sakaguchi et al., 2008).
Bulk segregant and linkage analysis
The heterozygous s445 fish carrying the Tg(gutGFP)s854 transgene were crossed to SJD strain to create a map cross. s445 mutant larvae were collected from the offspring of the map cross fish based on their Tg(gutGFP)s854 expression. Genomic DNA was isolated from individual mutant larvae by Proteinase K digestion. Using bulk segregant analysis, we initially mapped the s445 locus to linkage group 5 (between z21461 and z4299). We then collected more than 2000 mutants and performed linkage and recombinant analysis. We tested specific and potential polymorphic markers on linkage group 5 by PCR using genomic DNA of individual mutant larvae. We identified a polymorphic marker, CA-YE2 (5’- tcaaaggggagctaattattctgac-3’ and 5’-ggtgcactctttgcaaatcata-3’), as the most tightly linked marker to the s445 locus. The zebrafish snapc4 cDNA was amplified by PCR and cloned using PCRIITOPO (Invitrogen).
Genotyping of s445 mutants
In order to genotype s445 mutants, we used dCAPS primers (Neff et al., 1998) to introduce an s445 mutant allele specific restriction site in PCR products. PCR was performed with s445 dCAPS primers (5’-tcaacaacaatctctaccaccttctcat-3’ and 5’-cactaaatgttgcacttccc-3’) and the PCR product was digested with MseI.
Transmission electron micrograph (TEM)
Wild-type and s445 mutant larvae at 120 hpf were fixed in 4% glutaraldehyde in 0.2M sodium cacodylate (pH7.4) overnight at 4 °C. Samples were post-fixed in 1% osmium tetroxide for 1 h and incubated in 1% uranyl acetate in Maleate buffer for 1 h. Samples were dehydrated through an ethanol series, and embedded. Ultrathin sections of 85 nm were cut with diamond knife, and stained with uranyl acetate and lead citrate. Images were obtained with Philips CM12 electron microscope operated at 60 kV.
Real-time quantitative reverse transcriptase-PCR
Total RNA was isolated by the RNeasy kit (Qiagen) and cDNA was synthesized by miScript (Qiagen) according to the manufacturer’s instructions. SYBR green qPCR mix (SA Bioscience) was used to monitor real-time PCR on a 7500 real-time PCR system (Applied Biosystems). We used following primers: U1 (5’-TTACCTGGCAGGGGAGACACCA-3’ and 5’-CAGTCGAGATTCCCACATTTGG-3’); U2 (5’-TATCTGATACGTGCCCTACCCG-3’ and 5’-GGGTGCACCGTTCCCGGAAGTA-3’); U4 (5’-AGCTTTGCGCAGTGGCAGTATCG-3’ and 5’-TATCAGTCTCTGTCGAGACCGTC-3’); U6 (5’-CGCTACGGTGGCACATATAC-3’ and 5’- GAGCGCTTCACGGATTTG-3’); Large subunit ribosomal RNA (EF417169: 5’-TTGGGAATGCAGCCCAAAGCGG −3’ and 5’- ACCTCTCAACGGTTTCACGCCC −3’). The ΔΔCt method was used for relative quantification.
Northern blotting
Northern blotting was performed using a NorthernMax kit (Applied Biosystems) according to the manufacturer’s instruction.
Morpholino injection
Morpholinos were prepared and injected as previously described (Sakaguchi et al., 2006). We used following morpholinos in this study: 2 ng/embryo of snapc4 MO(A) 5’-AATCAACGCTATCAGGCTTACTTTG-3’; 2 ng/embryo snapc4 MO(B) 5’-GCATGCTGTCTTAATACGTACATCT-3’; 8 ng/embryo of snapc4 MO(C) 5’-CGCTTGTAAGTCGTCAGATGCCATT- 3’; 1 ng/embryo of snapc2 MO(A) 5’-TTCTCATACACACCTGGGAAAAGGC-3’; 1ng/embryo of snapc2 MO(B) 5’-TGTGAGCTTACCTGTTTGAGAGAAC-3’.
Morpholino specificity test
RT-PCR was used to test snapc2 MO(A) and snapc4 MO(B) specificity. Wild-type, snapc2 MO(A) and snapc4 MO(B) injected larvae were collected at 100 hpf and total RNA was isolated using the RNeasy kit (Qiagen). RT reactions were performed using SuperScriptII RT-PCR kit (Gibco BRL) according to the manufacturer’s recommendations. PCR was performed using following primer pairs: snapc2 (5’-AAGTTCCCATTGAAACCTGGG-3’ and 5’-TTAGGGGTGTGAGCAGGGTAAAGA-3’) and snapc4 (5’-CAGGCAACTCTATCTTCTCA −3’ and 5’-TTGATCGTCTTTCAGGGTTC-3’) to amplify snapc2 and snpac4 cDNA fragments, respectively.
Cloning and overexpression of the Snapc2 binding domain of Snapc4
cDNA corresponding to the snapc2 binding domain of snapc4 was amplified using following primers: snapc2BDF, GAATTCACCATGGAGTGGATGGATGGGAAAGGTG; snapc2BDR, CTCTCGAGCTAACTCAATAACTGACGTTCCT. The amplified PCR product was subcloned into the EcoRI-XhoI sites of pCS2+. Corresponding mRNA was synthesized as previously described (Sakaguchi et al., 2006). 100 pg per embryo of snapc2 binding domain mRNA was injected at the 1-cell stage.
Supplementary Material
Research Highlights.
We identified a novel zebrafish mutation, snapc4s445.
The intrahepatic biliary network degenerates in snapc4s445 mutant larvae.
The snapc4s445 mutation deletes the Snapc2 interaction domain.
The Snapc4-Snapc2 interaction is important for the maintenance of the intrahepatic biliary network.
Acknowledgements
We thank Laura Nagy, Michele Pritchard, Allyson McCledon and Arthur McCullough for critical reading of the manuscript. We also thank Mei Yin and the Cleveland Clinic Imaging Core for TEM. This work was supported in part by an APS postdoctoral fellowship, an NIH K99/R00 award (DK078138), and a startup package from the Cleveland Clinic Foundation to T.F.S. This work was also supported by grants from the NIH (NIDDK) and Packard foundation to D.Y.R.S. The snapc4s445 mutation was identified and positionally cloned at UCSF by T.F.S. when he was a postdoctoral fellow in D.Y.R.S.’s lab; phenotypic analysis and further characterization of the snapc4s445 mutant was carried out both at UCSF and at Cleveland Clinic where T.F.S. is now an independent investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvaro D. Biliary epithelium: a new chapter in cell biology. Ital J Gastroenterol Hepatol. 1999;31:78–83. [PubMed] [Google Scholar]
- Das A, Bellofatto V. RNA polymerase II-dependent transcription in trypanosomes is associated with a SNAP complex-like transcription factor. Proc Natl Acad Sci U S A. 2003;100:80–85. doi: 10.1073/pnas.262609399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber SA, Pack M, Ho SY, Johnson ID, Wagner DS, Dosch R, Mullins MC, Hendrickson HS, Hendrickson EK, Halpern ME. Genetic analysis of digestive physiology using fluorescent phospholipid reporters. Science. 2001;292:1385–1388. doi: 10.1126/science.1060418. [DOI] [PubMed] [Google Scholar]
- Field HA, Ober EA, Roeser T, Stainier DYR. Formation of the digestive system in zebrafish. I. Liver morphogenesis. Developmental Biology. 2003;253:279–290. doi: 10.1016/s0012-1606(02)00017-9. [DOI] [PubMed] [Google Scholar]
- Harada N, Visser R, Dawson A, Fukamachi M, Iwakoshi M, Okamoto N, Kishino T, Niikawa N, Matsumoto N. A 1-Mb critical region in six patients with 9q34.3 terminal deletion syndrome. J Hum Genet. 2004;49:440–444. doi: 10.1007/s10038-004-0166-z. [DOI] [PubMed] [Google Scholar]
- Henry RW, Ma B, Sadowski CL, Kobayashi R, Hernandez N. Cloning and characterization of SNAP50, a subunit of the snRNA-activating protein complex SNAPc. EMBO J. 1996;15:7129–7136. [PMC free article] [PubMed] [Google Scholar]
- Henry RW, Mittal V, Ma B, Kobayashi R, Hernandez N. SNAP19 mediates the assembly of a functional core promoter complex (SNAPc) shared by RNA polymerases II and III. Genes Dev. 1998;12:2664–2672. doi: 10.1101/gad.12.17.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez G, Jr, Valafar F, Stumph WE. Insect small nuclear RNA gene promoters evolve rapidly yet retain conserved features involved in determining promoter activity and RNA polymerase specificity. Nucleic Acids Res. 2007;35:21–34. doi: 10.1093/nar/gkl982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakoshi M, Okamoto N, Harada N, Nakamura T, Yamamori S, Fujita H, Niikawa N, Matsumoto N. 9q34.3 deletion syndrome in three unrelated children. Am J Med Genet A. 2004;126A:278–283. doi: 10.1002/ajmg.a.20602. [DOI] [PubMed] [Google Scholar]
- Jawdekar GW, Henry RW. Transcriptional regulation of human small nuclear RNA genes. Biochim Biophys Acta. 2008;1779:295–305. doi: 10.1016/j.bbagrm.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SW, Beis D, Mitchell T, Chen JN, Stainier DY. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development. 2005;132:5199–5209. doi: 10.1242/dev.02087. [DOI] [PubMed] [Google Scholar]
- Kim MK, Kang YS, Lai HT, Barakat NH, Magante D, Stumph WE. Identification of SNAPc subunit domains that interact with specific nucleotide positions in the U1 and U6 gene promoters. Mol Cell Biol. 2010;30:2411–2423. doi: 10.1128/MCB.01508-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig H, Matter N, Bader R, Thiele W, Muller F. Splicing segregation: the minor spliceosome acts outside the nucleus and controls cell proliferation. Cell. 2007;131:718–729. doi: 10.1016/j.cell.2007.09.043. [DOI] [PubMed] [Google Scholar]
- Korzh S, Pan X, Garcia-Lecea M, Winata CL, Wohland T, Korzh V, Gong Z. Requirement of vasculogenesis and blood circulation in late stages of liver growth in zebrafish. BMC Dev Biol. 2008;8:84. doi: 10.1186/1471-213X-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis KN, Strazzabosco M, Larusso NF. The cholangiopathies: disorders of biliary epithelia. Gastroenterology. 2004;127:1565–1577. doi: 10.1053/j.gastro.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Lemaigre F, Zaret KS. Liver development update: new embryo models, cell lineage control, and morphogenesis. Curr Opin Genet Dev. 2004;14:582–590. doi: 10.1016/j.gde.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Li C, Harding GA, Parise J, McNamara-Schroeder KJ, Stumph WE. Architectural arrangement of cloned proximal sequence element-binding protein subunits on Drosophila U1 and U6 snRNA gene promoters. Mol Cell Biol. 2004;24:1897–1906. doi: 10.1128/MCB.24.5.1897-1906.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorent K, Moore JC, Siekmann AF, Lawson N, Pack M. Reiterative Use of the Notch Signal During Zebrafish Intrahepatic Biliary Development. Developmental Dynamics. 2010;239:855–864. doi: 10.1002/dvdy.22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Hernandez N. A map of protein-protein contacts within the small nuclear RNA-activating protein complex SNAPc. J Biol Chem. 2001;276:5027–5035. doi: 10.1074/jbc.M009301200. [DOI] [PubMed] [Google Scholar]
- Matthews RP, Lorent K, Russo P, Pack M. The zebrafish onecut gene hnf-6 functions in an evolutionarily conserved genetic pathway that regulates vertebrate biliary development. Dev Biol. 2004;274:245–259. doi: 10.1016/j.ydbio.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Mittal V, Ma B, Hernandez N. SNAP(c): a core promoter factor with a built-in DNA-binding damper that is deactivated by the Oct-1 POU domain. Genes Dev. 1999;13:1807–1821. doi: 10.1101/gad.13.14.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neas KR, Smith JM, Chia N, Huseyin S, St Heaps L, Peters G, Sholler G, Tzioumi D, Sillence DO, Mowat D. Three patients with terminal deletions within the subtelomeric region of chromosome 9q. Am J Med Genet A. 2005;132:425–430. doi: 10.1002/ajmg.a.30496. [DOI] [PubMed] [Google Scholar]
- Neff MM, Neff JD, Chory J, Pepper AE. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J. 1998;14:387–392. doi: 10.1046/j.1365-313x.1998.00124.x. [DOI] [PubMed] [Google Scholar]
- Ober EA, Verkade H, Field HA, Stainier DY. Mesodermal Wnt2b signalling positively regulates liver specification. Nature. 2006;442:688–691. doi: 10.1038/nature04888. [DOI] [PubMed] [Google Scholar]
- Parsons MJ, Pisharath H, Yusuff S, Moore JC, Siekmann AF, Lawson N, Leach SD. Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mechanisms of Development. 2009;126:898–912. doi: 10.1016/j.mod.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pass AK, McLin VA, Rushton JR, Kearney DL, Hastings CA, Margolin JF. Vanishing bile duct syndrome and Hodgkin disease: a case series and review of the literature. J Pediatr Hematol Oncol. 2008;30:976–980. doi: 10.1097/MPH.0b013e31818b37c4. [DOI] [PubMed] [Google Scholar]
- Pointon JJ, Harvey D, Karaderi T, Appleton LH, Farrar C, Stone MA, Sturrock RD, Brown MA, Wordsworth BP. Elucidating the chromosome 9 association with AS; CARD9 is a candidate gene. Genes Immun. 2010;11:490–496. doi: 10.1038/gene.2010.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud P, Tate J, Callens C, Cordi S, Vandersmissen P, Carpentier R, Sempoux C, Devuyst O, Pierreux CE, Courtoy P, et al. A classification of ductal plate malformations based on distinct pathogenic mechanisms of biliary dysmorphogenesis. Hepatology. 2011;53:1959–1966. doi: 10.1002/hep.24292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski CL, Henry RW, Kobayashi R, Hernandez N. The SNAP45 subunit of the small nuclear RNA (snRNA) activating protein complex is required for RNA polymerase II and III snRNA gene transcription and interacts with the TATA box binding protein. Proc Natl Acad Sci U S A. 1996;93:4289–4293. doi: 10.1073/pnas.93.9.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi T, Kikuchi Y, Kuroiwa A, Takeda H, Stainier DY. The yolk syncytial layer regulates myocardial migration by influencing extracellular matrix assembly in zebrafish. Development. 2006;133:4063–4072. doi: 10.1242/dev.02581. [DOI] [PubMed] [Google Scholar]
- Sakaguchi TF, Sadler KC, Crosnier C, Stainier DY. Endothelial signals modulate hepatocyte apicobasal polarization in zebrafish. Curr Biol. 2008;18:1565–1571. doi: 10.1016/j.cub.2008.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimanski B, Nguyen TN, Gunzl A. Characterization of a multisubunit transcription factor complex essential for spliced-leader RNA gene transcription in Trypanosoma brucei. Mol Cell Biol. 2005;25:7303–7313. doi: 10.1128/MCB.25.16.7303-7313.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam M, Hernandez N. Mitotic functions for SNAP45, a subunit of the small nuclear RNA-activating protein complex SNAPc. J Biol Chem. 2008;283:14845–14856. doi: 10.1074/jbc.M800833200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strazzabosco M, Spirli C, Okolicsanyi L. Pathophysiology of the intrahepatic biliary epithelium. J Gastroenterol Hepatol. 2000;15:244–253. doi: 10.1046/j.1440-1746.2000.02091.x. [DOI] [PubMed] [Google Scholar]
- Strazzabosco M, Fabris L, Spirli C. Pathophysiology of cholangiopathies. J Clin Gastroenterol. 2005;39:S90–S102. doi: 10.1097/01.mcg.0000155549.29643.ad. [DOI] [PubMed] [Google Scholar]
- Wong MW, Henry RW, Ma B, Kobayashi R, Klages N, Matthias P, Strubin M, Hernandez N. The large subunit of basal transcription factor SNAPc is a Myb domain protein that interacts with Oct-1. Mol Cell Biol. 1998;18:368–377. doi: 10.1128/mcb.18.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JB, Murphy S, Bai L, Wang Z, Roeder RG. Proximal sequence element-binding transcription factor (PTF) is a multisubunit complex required for transcription of both RNA polymerase II- and RNA polymerase III-dependent small nuclear RNA genes. Mol Cell Biol. 1995;15:2019–2027. doi: 10.1128/mcb.15.4.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JB, Roeder RG. Cloning of two proximal sequence element-binding transcription factor subunits (gamma and delta) that are required for transcription of small nuclear RNA genes by RNA polymerases II and III and interact with the TATA-binding protein. Mol Cell Biol. 1996;16:1–9. doi: 10.1128/mcb.16.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.