Abstract
It has been suggested that a specific pattern of histone posttranslational modifications and their crosstalk may constitute a code that determines transcriptional outcomes. However, recent studies indicate that histone modifications have context-dependent effects, making their interplay more like a language within the chromatin signaling pathway than a code.
For almost two decades, a primary focus in the field of transcriptional regulation was to identify DNA elements that control the expression of genes. These efforts were in part motivated by the expectation that it would one day be possible to look at a gene and its regulatory sequences and predict when and where a gene was going to be transcribed. We then learned that along with the sequence-specific binding of activators and repressors, there is an additional world of factors that modify, interact and remodel chromatin to regulate gene expression. The identification of a multitude of histone modifications—some correlated with activation, some with repression—led to the proposal that the modifications constitute a code that could be recognized by transcription factors to determine the transcriptional state of a gene. However, additional research has since added layers of complexity, revealing a nuanced and intriguing language, not a strict code, as the basis for transcriptional regulation through chromatin signaling pathway. Here, we review the complex crosstalk among histone modifications, including recent studies that illustrate how the context and timing of these modifications are critical for a particular transcriptional readout.
Histone Crosstalk and Gene Activity
In eukaryotic cells, gene expression can be regulated at the level of chromatin structure. Numerous residues within the histone tails and several residues within the histone globular domains can be modified in a variety of ways, including acetylation, phosphorylation, ubiquitination, and methylation. A well-characterized posttranslational modification regulating chromatin structure is the acetylation of histone N-terminal tails, which is thought to facilitate transcriptional activation either by charge neutralization of the tails’ interaction with DNA or by forming a binding site for bromodomain-containing transcription factors, some of which can remodel nucleosomes. Another well-studied histone modification is the methylation of lysine 4 of histone H3 (H3K4), a modification generally associated with transcriptionally-active genes, and a binding site for a variety of factors that include histone modifying and remodeling activities (Shilatifard, 2006).
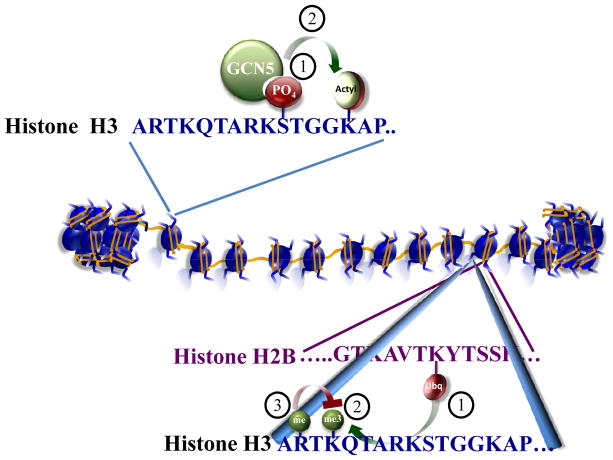
More complex scenarios arise when histone modifications act combinatorially in a context-dependent manner to facilitate or repress chromatin-mediated transcription. In some cases the modification of one residue can alter the ability of a second residue to be implemented by its modifying enzyme(s) (Figure 1). The first example of histone crosstalk falls into this category: the phosphorylation of serine 10 on histone H3 stimulates the ability of Gcn5 to acetylate histone H3 at lysine 14 (H3K14) (Cheung et al., 2000; Lo et al., 2000) (Figure 1A). Another well-characterized example is the requirement of histone H2B monoubiquitination for proper H3K4 methylation by the H3K4 methylase complex COMPASS (Figure 1B) (Shilatifard, 2006). This process, initially discovered in yeast (Shilatifard, 2006) is now known to be highly conserved among eukaryotes (Kim et al., 2009). Additionally, a recent comprehensive mutation analysis of all histone residues reveals that a single point mutation in histone H3K14, a site of acetylation, results in the specific loss of H3K4 trimethylation, but not mono- or dimethlyation (Nakanishi et al., 2008). This screen also demonstrated that H3K4 trimethylation is regulated by both monoubiquitination-dependent and monoubiquitination-independent processes (Nakanishi et al., 2008).
Figure 1. Examples of Histone crosstalk.
(A) The first characterized example of histone crosstalk is the stimulation of acetyltransferase activity of GCN5 towards the histone H3 tail by prior phosphorylation (P) of serine 10. Acetylation, Ac. (B) Crosstalk among histone modifications can span more than one histone. Monoubiquitination of histone H2B can lead to the trimethylation of lysine 4 in the histone 3 tail (H3K4) by Set1/COMPASS. However, H3K4 methylation by COMPASS and COMPASS-like complexes can be blocked if the nearby arginine of H3 is already methylated.
Given that histone-modifying enzymes are often found in multi-subunit complexes, modification of nearby residues can create binding sites for the components of the complex helping to anchor an enzyme to a nucleosome. For example, the PHD finger of Yng1, a subunit of the NuA3 histone acetyltransferase complex, recognizes methylated H3K4 and helps recruit this histone acetyltransferase complex for acetylation of H3K14 (Martin et al., 2006; Taverna et al., 2006). The Yng1-related ING2 can also bind methylated H3K4, however, it is present in a histone deacetylase complex (Shi et al., 2006). Therefore, H3K4 methylation can serve as a landing platform for a variety of histone-modifying enzymes with opposing activities.
Modifications of nearby residues can also prevent the recognition of a substrate by an enzyme, as recently reported to occur when methylation of histone H3 arginine 2 (H3R2) interferes with H3K4 methylation by Set1/COMPASS in yeast and COMPASS-like complexes in mammalian cells (Guccione et al., 2007; Hyllus et al., 2007; Kirmizis et al., 2007) (Figure 1B). Histone modifications can also prevent the recruitment of factors other than enzymes. For example, heterochromatin protein 1 (HP1), which binds methylated H3K9, cannot do so when the adjacent serine 10 (H3S10) is phosphorylated during mitosis or during gene activation (Fischle et al., 2005; Mateescu et al., 2008).
Multiple types of histone crosstalk, involving numerous histone-modifying complexes, can occur at any one gene. A major challenge is to understand the events that regulate changes in gene expression through these modifications/crosstalk. One strategy has been to profile histone modifications genomewide, with the expectation that a given pattern will indicate a transcriptional outcome due to the recruitment of specific proteins by these modifications. However, some recent examples of trans-histone crosstalk illustrate that transcriptional readout depends on context and timing by which these modifications are introduced. Simply put, just looking at the pattern of chromatin modifications at a locus is not sufficient to determine its gene expression status. These studies provide new insight into the language of histone crosstalk.
From histone phosphorylation to transcription elongation
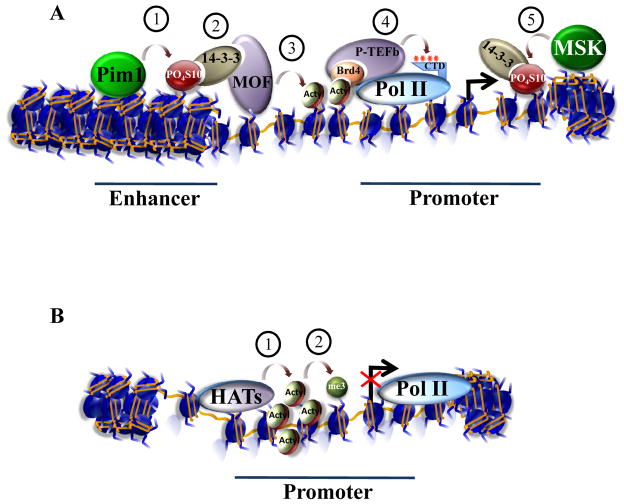
A novel form of crosstalk was recently discovered by Zippo and colleagues studying the transcriptional control of FOSL1, a gene activated in response to serum (Zippo et al., 2009) (Figure 2A). They present evidence for a transcription activation pathway in which the phosphorylation of H3 tails leads to the acetylation of H4 tails. In turn, acetylation of H4 tails is required for the recruitment of the RNA Pol II positive transcription elongation factor, P-TEFb (Figure 2A). Previously, the authors found that activation of the FOSL1 gene requires PIM1, a proto-oncogene whose kinase activity is activated through MAP kinase signaling. Numerous cellular substrates of PIM1 have been identified, including H3S10. Other H3S10 kinases, such as MSK1 and MSK2 (MSK1/2) are also implicated in the phosphorylation of histones at serum responsive genes, including FOSL1. Zippo and colleagues find that the spatiotemporal pattern of H3S10 phosphorylation differs for PIM1 and MSK1/2. MSK1/2 mediates the phosphorylation of H3S10 at the promoter of FOSL1 at early time points of gene expression, whereas PIM1 is required for H3S10 phosphorylation at a FOSL1 enhancer after the MSK1/2-mediated phosphorylation of H3S10 (Figure 2A).
Figure 2. Context dependent outcomes of histone crosstalk.
(A) Zippo and colleagues (Zippo et al., 2009) uncover a new form of histone crosstalk by studying the transcriptional control of FOS1L, a gene activated in response to serum. Activation requires the binding of PIM1 to the FOS1L enhancer. PIM1 is a kinase responsible for phosphorylation (P) of serine 10 on the histone H3 tail (H3S10). Phosphorylated H3S10 creates a binding site for 14-3-3, a phosphoserine binding protein. Acetylation (Ac) of lysine 16 on the H4 (H4K16) tail occurs through interaction of 14-3-3 with the histone acetyltransferase MOF. Acetylated H4K16 can in turn form a binding site for the bromodomain-containing protein Brd4, a component of P-TEFb, a kinase that phosphorylates the C-terminal domain of RNA Pol II to facilitate transcription elongation. However, at an earlier stage of serum stimulation, an MSK1/2 kinase is recruited to the promoter where it phosphorylates H3S10. 14-3-3 is then recruited to the promoter, but unlike the situation at the enhancer, MOF is not recruited to the promoter. Thus, the timing, location, and perhaps identity of the H3 kinase, and not the H3S10 modification alone, determines downstream events. (B) Another example of how the order of implementation of histone modifications can affect transcription comes from work from Wang et al. (2009). They report that despite correlations between histone acetylation and H3K4 methylation, artificially increasing acetylation through treatment of cells with deacetylase inhibitors (HDACs) does not lead to productive transcription, despite the presence of H3K4 methylation and Pol II recruitment. Therefore, patterns of histone modifications cannot simply be “read”, but have distinct effects depending on the cellular context and upstream signaling events.
Screening for other histone modifications specifically associated with the FOSL1 enhancer shows that the acetylation of H4K16 coincides with H3S10 phosphorylation. RNA interference-mediated knockdown of PIM1 results in loss of H4K16 acetylation, suggesting a trans-tail crosstalk from H3S10 phosphorylation to H4K16 acetylation. Zippo and colleagues asked whether 14-3-3 γ, ε and ζ proteins, previously shown to bind phosphorylated H3S10, are recruited to the promoter and the enhancer of FOSL1 in response to serum. They find that 14-3-3ε and 14-3-3ζ are recruited to both the promoter and enhancer of FOSL1 after serum stimulation. However, 14-3-3 is required only for recruiting the H4K16 histone acetyltransferase MOF to the enhancer, and not to the promoter of FOSL1. Recruitment of MOF to the enhancer results in H4K16 acetylation, which can be bound by the bromodomain-containing protein, Brd4. Brd4 is a component of P-TEFb, a kinase that phosphorylates Pol II to facilitate transcription elongation (Figure 2A). Thus, Zippo and colleagues propose that crosstalk between modifications on two different histone tails regulate productive transcription elongation through the sequential recruitment of proteins that bind these modifications.
One question raised by this study is why H3S10 phosphorylation produces different results at the enhancer than at the promoter of FOSL1 even though 14-3-3 is recruited to both sites. At the enhancer, 14-3-3 recruits the histone acetyltransferase MOF. At the promoter, it does not. What is the difference between 14-3-3 at the promoter and at the enhancer? Interestingly, 14-3-3ε and 14-3-3ζ are thought to be regulated via lysine acetylation (Choudhary et al., 2009) and an acetyltransferase, Tip60, is preferentially recruited to the promoter of FOSL1. One possibility is that Tip60 acetylates 14-3-3 and prevents its interaction with MOF.
Another intriguing aspect of the study by Zippo and colleagues is the link between H3S10 phosphorylation and H4K16 acetylation. These two modifications were previously linked in studies of dosage compensation in the fruit fly Drosophila. In Drosophila dosage compensation, MOF is recruited to the coding region of X-linked genes in males where it mediates H4K16 acetylation in a process thought to facilitate transcription elongation. Co-localizing with MOF on the male X chromosome is the JIL-1 kinase, an MSK1/2-related kinase that mediates the phosphorylation of H3S10 on this chromosome. In the case of Drosophila dosage compensation, recruitment of the JIL-1 kinase to the male X chromosome occurs later than H4K16 acetylation (Wang et al., 2001), a reversal of the order of the addition of these marks at FOSL1 in response to serum. Concordantly, the MOF complex that mediates acetylation in coding regions is likely to be distinct from the MOF complex that mediates promoter/enhancer acetylation of genes (Li et al., 2009). Thus, by all appearances, these two examples of the coexistence of both H3S10 phosphorylation and H4K16 acetylation are unrelated in their order of implementation and in their biological meaning. This suggests that descriptions of histone modification patterns, without understanding the mechanisms leading to the implementation of these marks, should be interpreted with caution. Importantly, the study by Zippo and colleagues begins to determine the role of histone modifications in the activation of FOSL1, with a spatial and temporal dissection of how a cascade of histone modifications can lead to a particular transcriptional outcome.
Trimethylation Converses with Acetylation
Another example of trans-tail crosstalk was proposed by Wang et al. (2009). In this case the communication takes place between the H3 and H4 tails, and like the example provided by Zippo et al. (2009) involves H4K16 acetylation and FOSL1 transcription. By analyzing genomewide profiles of several histone acetyltransferases, deacetylases, and modifications these investigators find a link between H3K4 methylation and H4K16 acetylation at some inducible genes, including FOSL1. The authors show that a subset of transcriptionally-quiescent genes, marked by the presence of H3K4 methylation, display a marked increase in histone acetylation at H3K9 and H4K16 after adding deacetylase inhibitors. In contrast, quiescent genes not marked with H3K4 methylation rarely show this increase in acetylation in response to deacetylase inhibitors.
In order to determine whether H3K4 methylation is functionally linked to H4K16 acetylation, Wang and colleagues use RNA interference-mediated knockdown of WDR5, a common component of the Set1 and MLL (mixed-lineage leukemia) COMPASS-like H3K4 methyltransferase complexes. Upon knockdown of WDR5, they observe reduced levels of histone acetylation at the subset of transcriptionally-quiescent genes marked by H3K4 methylation. Based on this information, Wang and colleagues suggest that H3K4 methylation primes certain genes for an increase in H3K9 and H4K16 acetylation. Interestingly, what was not considered by Wang and colleagues is the fact that WDR5 is also a component of complexes that contain the H4K16 acetyltransferase MOF. For example WDR5 is part of the NSL/MSL1v complex (Cai et al., 2010; Li et al., 2009) as well as the ATAC complex, which contains the H3K9 and K14 acetyltransferase GCN5 (Suganuma et al., 2008; Wang et al., 2008). As such, the effect of WDR5 knockdown could be a consequence of WDR5’s role as a subunit of the H3K4 methylases, or WDR5’s role as a subunit of the H3 and H4 acetyltransferase complexes, or a combination of the two. Given that WDR5 is part of the H3 and H4 acetyltransferase complexes, the existence of crosstalk between H3K4 and H4K16 needs to be further characterized.
One surprising finding of the study by Wang and colleagues is that transcription is rarely induced at the genes tested, although Pol II is recruited following treatment with a deacetylase inhibitor (Figure 2B). Thus, Pol II recruitment does not lead to the anticipated increase in transcription, despite the fact that H3K9 and H4K16 acetylation are increased. These modifications coincide with transcriptional activation at FOSL1 upon serum treatment (Zippo et al., 2009). Together with the studies of H3S10 phosphorylation and H4K16 acetylation at the FOSL1 enhancer, it is clear that knowing the mechanism and timing of these modifications is necessary for determining the transcriptional outcome.
The Emerging Grammar of Histone Crosstalk
The existence of a histone modification code was proposed ten years ago as a way to approach the study of the quickly growing number of histone modifications involved in the regulation of gene expression and other DNA-templated processes, such as replication, repair, and recombination. New “words” of histone modifications are being discovered and they continue to appear in interesting combinations. However, discovering the exact roles these modifications play in gene expression has been complicated by finding a counter example for almost every example of crosstalk, such as the case of H3K4 and H3K36 methylation recruiting both histone acetyltransferases and deacetylases.
A common theme of recent research on histone crosstalk is that the order and mechanism of the addition and removal of histone modifications are important for the transcriptional readout of a gene. The recent examples of histone crosstalk that we have addressed here illustrate this point. In one study, the implementation of H3S10 phosphorylation at two different locations, by two different enzymes and at two different times after serum stimulation, had disparate effects on subsequent histone acetylation at the respective locations (Zippo et al., 2009) (Figure 2A). Zippo and colleagues were able to propose a mechanism of gene activation by identifying the histone-modifying enzymes, the histone modifications, and a set of proteins that recognized these modifications on the FOSL1 gene after serum stimulation. In another study, Wang and colleagues found that artificially recreating histone modifications that correlate with gene expression could result in the recruitment of RNA Pol II, but this was not sufficient for transcription (Figure 2B). Thus, simply mapping histone modification patterns without understanding the recruitment, regulation, and interactions of the complexes implementing these marks is not sufficient to understand the mechanisms regulating gene expression. Genomewide profiling techniques have now become widely adopted, providing the ability to map histone modifications, the enzymes implementing these marks, and the factors that recruit them under different experimental conditions.
The study of the regulation of gene expression has grown from identifying transcription factors and their binding sites to include a wide variety of other binding events associated with the modifications of histones that package the DNA. Future progress will require us to learn much more about how the words comprising the dictionary of histone crosstalk are used in a particular order to provide the grammar of this complex language.
Acknowledgments
We thank Dr. E. Park for the critical reading of the manuscript. We also thank L. Shilatifard for editorial assistance. Studies in the Shilatifard laboratory are supported in part by grants from the NIH R01CA89455, R01GM069905 and R01CA150265. We apologize to all of our colleagues whose work we were not able to cite in this minireview due to space limitations.
References
- Cai Y, Jin J, Swanson SK, Cole MD, Choi SH, Florens L, Washburn MP, Conaway JW, Conaway RC. Subunit Composition and Substrate Specificity of a MOF-containing Histone Acetyltransferase Distinct from the Male-specific Lethal (MSL) Complex. J Biol Chem. 2010;285:4268–4272. doi: 10.1074/jbc.C109.087981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, Luscher B, Amati B. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449:933–937. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- Hyllus D, Stein C, Schnabel K, Schiltz E, Imhof A, Dou Y, Hsieh J, Bauer UM. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 2007;21:3369–3380. doi: 10.1101/gad.447007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, Bahler J, Green RD, Kouzarides T. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449:928–932. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wu L, Corsa CA, Kunkel S, Dou Y. Two mammalian MOF complexes regulate transcription activation by distinct mechanisms. Mol Cell. 2009;36:290–301. doi: 10.1016/j.molcel.2009.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo WS, Trievel RC, Rojas JR, Duggan L, Hsu JY, Allis CD, Marmorstein R, Berger SL. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol Cell. 2000;5:917–926. doi: 10.1016/s1097-2765(00)80257-9. [DOI] [PubMed] [Google Scholar]
- Martin DG, Baetz K, Shi X, Walter KL, MacDonald VE, Wlodarski MJ, Gozani O, Hieter P, Howe L. The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol Cell Biol. 2006;26:7871–7879. doi: 10.1128/MCB.00573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateescu B, Bourachot B, Rachez C, Ogryzko V, Muchardt C. Regulation of an inducible promoter by an HP1beta-HP1gamma switch. EMBO Rep. 2008;9:267–272. doi: 10.1038/embor.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi S, Sanderson BW, Delventhal KM, Bradford WD, Staehling-Hampton K, Shilatifard A. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat Struct Mol Biol. 2008;15:881–888. doi: 10.1038/nsmb.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: implicationsin the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Suganuma T, Gutierrez JL, Li B, Florens L, Swanson SK, Washburn MP, Abmayr SM, Workman JL. ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding. Nat Struct Mol Biol. 2008;15:364–372. doi: 10.1038/nsmb.1397. [DOI] [PubMed] [Google Scholar]
- Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, Baker L, Boyle J, Blair LP, Chait BT, et al. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang W, Jin Y, Johansen J, Johansen KM. The JIL-1 tandem kinase mediates histone H3 phosphorylation and is required for maintenance of chromatin structure in Drosophila. Cell. 2001;105:433–443. doi: 10.1016/s0092-8674(01)00325-7. [DOI] [PubMed] [Google Scholar]
- Wang YL, Faiola F, Xu M, Pan S, Martinez E. Human ATAC Is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J Biol Chem. 2008;283:33808–33815. doi: 10.1074/jbc.M806936200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 2009;138:1122–1136. doi: 10.1016/j.cell.2009.07.031. [DOI] [PubMed] [Google Scholar]