Abstract
Molecularly targeted therapies are rapidly growing with respect to their clinical development and impact on cancer treatment due to their highly selective anti-tumor action. However, many aggressive cancers such as triple-negative breast cancer (TNBC) currently lack well-defined therapeutic targets against which such agents can be developed. The identification of tumor-associated antigens and the generation of antibody drug-conjugates represent an emerging area of intense interest and growth in the field of cancer therapeutics. Glycoprotein non-metastatic b (GPNMB) has recently been identified as a gene that is over-expressed in numerous cancers, including TNBC, and often correlates with the metastatic phenotype. In breast cancer, GPNMB expression in the tumor epithelium is associated with a reduction in disease-free and overall survival. Based on these findings, glembatumumab vedotin (CDX-011), an antibody-drug conjugate that selectively targets GPNMB, is currently being investigated in clinical trials for patients with metastatic breast cancer and unresectable melanoma. This review discusses the physiological and potential pathological roles of GPNMB in normal and cancer tissues, respectively, and details the clinical advances and challenges in targeting GPNMB-expressing malignancies.
Keywords: GPNMB, osteoactivin, breast cancer, antibody-drug conjugates, CDX-011
Introduction
Breast cancer is a highly prevalent and devastating disease. Despite clear advances in screening, more accurate prognosis, and disease management over the past few decades, each year more than 1,600,000 cases are diagnosed and 420,000 deaths are attributed to breast cancer worldwide.1 Breast cancer remains the most commonly diagnosed cancer, and one of the most significant causes of cancer-related deaths in women.2
One of the primary challenges associated with the treatment of breast cancer is tumor heterogeneity, which is manifested by the diversity of histopathologies and molecular features associated with this disease. In the early 2000s, genomic studies employed gene expression signatures to classify breast cancer into five distinct subgroups, which include the luminal A, luminal B, HER2+, triple negative/basal-like, and normal breast-like subtypes.3–5 More recently, the evolution of gene expression profiling techniques has allowed further subclassification of breast cancer through the identification of the claudin-low subtype.6,7 The molecular complexity and heterogeneity of these subtypes is continually being refined as additional genomic, epigenomic, and transcriptomic data becomes available.8–12
Gene expression profiling has also proven useful in identifying patients with a high risk of disease progression and distant recurrence.13 In general, patients with basal-like and HER2+ signatures displayed the shortest relapse-free survival rates, while a luminal A classification was associated with the lowest risk of developing distant metastases.4 Breast cancer most commonly metastasizes to bone, followed by lung, liver, and brain, and the site of distant metastasis can predict the likelihood of overall survival. Typically, the presence of visceral metastasis or metastasis in multiple sites is associated with a shorter survival.14 Accordingly, the majority of luminal A breast cancers metastasize exclusively to bone, while HER2 enriched cancers preferentially give rise to liver and lung metastases, and basal-like breast cancers are associated with increased liver and brain metastasis.15,16
The current availability of therapeutic targets varies according to the molecular subtype, which accounts in part for the differences between these groups with respect to survival3–5,17,18 and response to therapy.7,19–22 Luminal breast cancers are associated with the best prognosis and are characterized by the presence of estrogen and progesterone receptors, which makes them amenable to hormonal therapies such as tamoxifen or aromatase inhibitors. Similarly, drugs that target the HER2 receptor, such as trastuzumab, pertuzumab, and lapatinib, are clinically approved for the treatment of HER2+ breast cancers. Triple-negative breast cancers (TNBC) are diagnosed by the lack of ER, PR and HER2 expression and are largely characterized by a basal-like histopathology (basal-like breast cancer [BLBC]). TNBCs account for an estimated 10%–25% of invasive breast cancers, are associated with a high grade and poor prognosis, and, due to a lack of distinct molecular markers, there are currently no targeted interventions for this aggressive subset of the disease. Although poly ADP ribose polymerase (PARP) inhibitors have shown encouraging results for the BRCA-subset of TNBC patients,23,24 their utility in this subtype is not assured.25 At present, chemotherapy remains the primary treatment option for TNBC.
One of the main problems associated with the use of chemotherapy for the treatment of cancer is the off-target action of the drugs on normal cells, which can lead to painful side effects and complications. In an effort to minimize the cytotoxicity of these therapies, approaches that selectively target tumor-associated antigens are emerging as promising therapeutic strategies for TNBC and other cancers. One such approach is the development of antibody-drug conjugates (ADCs), which synergistically combine the specificity of antibodies with the cytotoxic efficacy of chemotherapy. ADCs consist of antibodies bound to highly potent cytotoxins by a chemical linker.26 These antibodies can be designed to target tumor-specific proteins and thereby serve as vehicles that deliver the drug to the cell of interest, often via internalization of the compound.26 Accordingly, the expression pattern of the selected antigen, both in normal and cancer tissues, is an important consideration in predicting response to ADC therapy.
GPNMB has been recently identified as a potential therapeutic target for patients with BLBC and TNBC.27–29 GPNMB expression in the breast tumor epithelium was shown to strongly correlate with disease-free and overall survival. Additionally, GPNMB is highly expressed in BLBC and TNBC and its levels are associated with a poor prognosis and increased risk for recurrence in this subset. These findings, combined with evidence of high GPNMB expression in numerous cancers,27–34 have sparked an interest in investigating GPNMB as a target for antibody-based therapies in TNBC and other cancers.35–37 This review will discuss the suitability of GPNMB as a target for cancer therapy by summarizing our current understanding of GPNMB expression in normal tissues, its role in cancer progression, and the current use of ADCs for the treatment of GPNMB expressing cancers. We will consider GPNMB in the broader context of several cancers; however, when possible, we will emphasize emerging literature regarding GPNMB and breast cancer.
Homology and structure of GPNMB
GPNMB, initially termed glycoprotein non-metastatic gene B (NMB), was first cloned and described in 1995 as a protein highly expressed in a melanoma cell line with low metastatic potential.38 However, since this initial publication, elevated GPNMB expression is observed in numerous cancers and is often associated with the metastatic phenotype.27–34 GPNMB is also known as hematopoietic growth factor inducible, neurokinin-1 type (HGFIN),39 and is located on the small arm of chromosome 7 (7p15). The rat orthologue, termed osteoactivin, is expressed in the long bones of rats bearing a mutation associated with osteopetrosis and shares 65% protein identity with human GPNMB.40 The mouse orthologue, which has 71% protein identity with human GPNMB, was coined dendritic cell heparin integrin ligand (DC-HIL) following its identification in a particular subset of dendritic cells.41
GPNMB belongs to the vertebrate Pmel17/NMB family,42 which encompasses GPNMB, Pmel17 (melanocyte protein 17), and their orthologues. Pmel17 is the main structural component of melanosomes, where it plays a key role in the pigment biogenesis of melanocytes.43 To a lesser extent, GPNMB also shares homology with lysosome-associated membrane protein (LAMP-1) family members,41 which are glycoproteins with potential roles in cell adhesion and metastasis.44
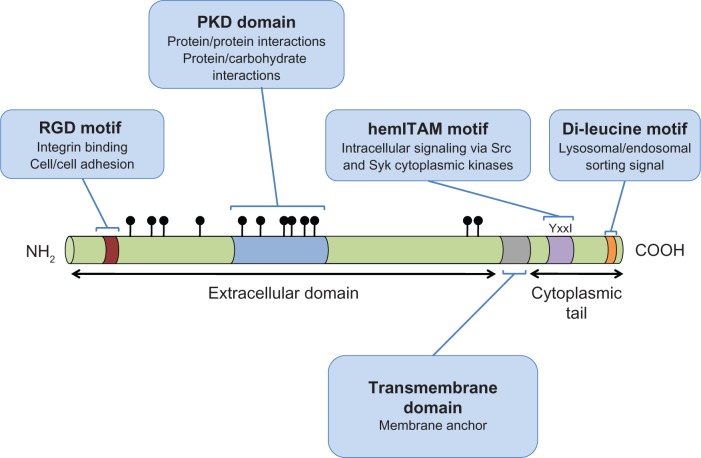
GPNMB is a type I transmembrane protein that contains an N-terminal signal peptide, an integrin-binding (RGD) motif and a polycystic kidney disease (PKD) domain in its extracellular domain (ECD), a single pass transmembrane domain, and a 53 amino acid (AA) cytoplasmic tail45,46 (Figure 1). The cytoplasmic tail harbors a half immunoreceptor tyrosine-based activation motif (hemITAM) and a dileucine motif, which functions as a sorting signal in QNR-71, the quail orthologue of GPNMB.47 In addition to these domains and motifs, there are two known splice variants of GPNMB, comprising a short 560aa and a long 572aa isoform.30 The long isoform contains a 12aa insertion within a poorly conserved region downstream of the PKD-domain.30 To date, there has been no evidence that the short and long isoforms have disparate functions. However, one study reported that the short GPNMB isoform was more frequently expressed in glioma specimens and was significantly correlated with poor survival times whereas the correlation between the long GPNMB isoform and survival times failed to achieve statistical significance.30
Figure 1.
A schematic representation of GPNMB indicating the domains and motifs contributing to GPNMB function.
Notes: The symbols (filled circles) located above the extracellular domain of GPNMB represent glycosylation sites. The RGD sequence comprises an integrin binding domain, where R = Arginine, G = Glycine, D = Aspartic acid. The YxxI sequence constitutes a hemITAM motif, where Y = tyrosine, x = any amino acid, I = isoleucine. The di-leucine motif is a lysosomal/endosomal targeting motif of the D/ExxxLL type, where D = Aspartic acid, E = Glutamic acid, x = any amino acid, L = leucine.
Abbreviations: GPNMB, glycoprotein non-metastatic b; hemiTAM, immunoreceptor tyrosine-based activation motif; PKD, polycystic kidney disease; RGD, integrin-binding.
RGD domain
This motif, comprised of only 3 amino acids, arginine (R), glycine (G), and aspartic acid (D), is found near the N-terminus of the GPNMB ECD and is well characterized in numerous proteins as an integrin-binding motif.48 Integrins are heterodimeric transmembrane proteins expressed on a wide variety of cells, which regulate cell spreading, adhesion, migration, proliferation, and apoptosis.49
PKD domain
The PKD domain belongs to the immunoglobulin-(Ig) like fold superfamily (E-set), which also includes cadherins, protein families containing bacterial Ig-like domains, and several fibronectin type III domain-containing protein families. While the function of the PKD domain is still unclear, based on its structure, it has been proposed to mediate protein-protein or protein-carbohydrate interactions,50 and has been shown to mediate cell-cell adhesion.51
hemiTAM
ITAM (immunoreceptor tyrosine-based activation motif) motifs are commonly found in the cytoplasmic domains of receptors expressed by cells of the hematopoietic systems.52 ITAM motifs are found in antigen receptors, cytokine receptors and toll-like receptors.53 ITAM signaling usually occurs in response to ligand binding, via phosphorylation of the ITAM resident tyrosine residues, primarily by Src-family kinases (ie, Src, Hck, Fgr, Lyn).54
GPNMB is one of several proteins whose cytoplasmic tail contains a highly conserved, single YxxI sequence, which has been referred to as a hemi-ITAM or hemITAM motif.52 Proteins with hemITAMs still exhibit robust ITAM signaling capacity.55 The current view suggests that ligand binding stimulates dimerization of hemITAM-bearing receptors; however, it remains to be seen whether GPNMB is capable of forming such homodimers.
Dileucine sorting motif
GPNMB contains a dileucine motif in its cytoplasmic tail, near the carboxy-terminus, with the sequence EKDPLL. Dileucine-based motifs of this type (D/ExxxLL) are often implicated in rapid receptor internalization from the plasma membrane and lysosomal/endosomal targeting.56 Indeed, when either of these leucine residues is mutated to glycine in quail GPNMB, it is retained at the plasma membrane of HeLa or pigmented quail cells, and not routed to endosomes and lysosomes, as is the case for wild type GPNMB.47 Interestingly, sequences of this type are associated with basolateral targeting in polarized epithelial cells.56
Glycosylation
GPNMB is a heavily glycosylated protein, possessing 12 putative N-glycosylation sites within its extracellular domain, 6 of which are found in the PKD domain.38,45 Glycosidase treatments have confirmed that GPNMB can be N- and O-glycosylated in a variety of cell types.30,57,58 Following immunoblot analyses, human GPNMB is detected as two broad bands that correspond to precursor (P1 ~90 kDa) and mature (M ~115 kDa) GPNMB isoforms.58 In addition, the unglycosylated form of GPNMB (~65 kDa) has been detected in cells, such as osteoclasts (Sheng et al).59 The relative abundance of these bands varies based on the cell type in which GPNMB is expressed.28,58 Studies using N-glycosidases suggest that GPNMB is first N-glycosylated in the ER to yield the P1 isoform, and these N-glycans are further modified during processing in the Golgi apparatus to produce the M-form.58 While both isoforms are susceptible to tyrosine phosphorylation, only the mature form can be proteolytically processed through shedding (discussed below).58,60 A number of studies have linked the glycosylation status of GPNMB to its putative biological functions and will be addressed in the relevant sections described below.
Proteolytic cleavage and ECD shedding
GPNMB is also subject to proteolytic processing, which was first uncovered by the detection of two heavily glycosylated, high molecular weight forms of murine GPNMB (97 kDa, 116 kDa; discussed above) and a stable c-terminal fragment of ~20 kDa.61 It was postulated that GPNMB was susceptible to shedding by members of the matrix metalloproteinase (MMP) family, such as A disintegrin and metalloproteinase (ADAMs), because treatment with a broad-spectrum inhibitor of MMPs (GM6001) reduced the degree to which GPNMB was shed.61 Treatment with a calmodulin inhibitor (W7) or a protein kinase C activator (phorbol myristate acetate [PMA]) enhanced GPNMB shedding, further implicating the ADAMs, as these compounds have both been reported to enhance ADAM-10 and ADAM-17 activity, respectively.58 In agreement with an important role for ADAMs in GPNMB processing, constitutive GPNMB shedding was observed in breast cancer cells and definitively characterized ADAM10 as a sheddase responsible for this cleavage event in breast cancer cells.27 The potential functional implications for this shedding event will be discussed further in subsequent sections.
GPNMB expression and physiological functions in normal tissues
GPNMB mRNA has been detected in the long bones, calvaria, bone marrow, adipose, thymus, skin, placenta, heart, kidney, pancreas, lung, liver, and skeletal muscle;39–41 however, the precise expression patterns varied between these studies. It is also clear that GPNMB can be expressed in multiple cell types within a given tissue, which is evident by its expression in bone osteoblasts40 and osteoclasts,59 for example. Together, these studies clearly demonstrate that GPNMB is expressed in a wide range of tissues and suggest its involvement in a variety of physiological processes.
Brain
Recently, widespread expression of GPNMB has been described within the central nervous system, and shown to be largely specific to the microglia/macrophages of the neural parenchyma.62 Furthermore, GPNMB expression has been described within motor neurons of normal brain tissue. Interestingly, GPNMB upregulation has been observed in the brains of rats following stroke.63 It is also upregulated in the motor neurons and astrocytes of a mouse model of amyotrophic lateral sclerosis (ALS) as well as in the spinal cords of patients with this disease.64 Its over-expression was cytoprotective in ALS-affected tissues; GPNMB secretion from astrocytes induced survival signals in neighboring motor neurons through the PI3K and MEK/Erk pathways and was thus able to ameliorate ALS-induced neuronal degeneration. In this context, GPNMB glycosylation was shown to be important for increasing motor neuron stability, as ubiquitin-mediated degradation of glycosylated GPNMB ultimately triggered motor neuron death.64 However, the presence of GPNMB was cytotoxic in normal neural tissues, suggesting a possible role for GPNMB in maintaining neuronal homeostasis.64
Skin
GPNMB expression in the basal layer of the skin, particularly in melanocytes, has been well documented.58,65–68 During development, GPNMB exhibits a punctate pattern of expression consistent with melanoblast cell populations, which represent precursors of melanocytes.65 In adult melanocytes, it preferentially localizes to late-stage (III and IV) melanosomes, which are characterized by an accumulation of the melanin pigment, suggesting a putative role for GPNMB in melanosome maturation.58,67 Its weak cell-surface expression can be upregulated following UVA irradiation, or stimulation by αMSH, IFNγ and TNFα.67 A separate study showed that GPNMB can be upregulated by UVB and suggested that UVB-mediated formation of early melanosomes is mediated by GPNMB.68 Functionally, GPNMB can mediate melanocyte adhesion to keratinocytes through its RGD domain and is thought to be involved in the transport of late melanosomes to keratinocytes.58,67 GPNMB can be localized at the cell surface and internalized into endosomal and lysosomal compartments in a variety of cell types. In melanocytes and melanoma cells, it is thought that the extensive glycosylation of the GPNMB PKD domain contributes to the differential sorting and localization patterns that are observed between GPNMB and its close homologue, Pmel.69 Indeed, while the PKD domain of Pmel plays an active role in the cellular distribution of Pmel, the degree of glycosylation in GPNMB blocks this sorting function, leading to differential localization of GPNMB.69
Bone
The first link between GPNMB expression and bone physiology was made when it was identified as highly expressed by mature, matrix producing rat osteoblasts in osteopetrotic bones relative to normal bone.40 Subsequent studies have shown that antisense oligonucleotide- or neutralizing antibody-mediated inhibition of GPNMB in developing osteoblasts impairs their differentiation and decreases their ability to produce bone matrix.70,71 Recently, it has been shown that GPNMB addition to a critical-size bone defect model was able to support bone regeneration/formation.72
In addition, GPNMB is abundantly expressed in differentiated osteoclasts73 and was found to play an important role in mediating cell fusion to produce multi-nucleated osteoclasts.59 GPNMB has been shown to physically associate with β1 or β3 containing integrin complexes in osteoclasts and to be an important mediator of osteoclast differentiation/fusion. Interestingly, it was the unglycosylated form of GPNMB that was found in complexes containing β1 or β3 integrins.59 Indeed, neutralizing antibodies against GPNMB reduced osteoclast size and number and decreased their ability to resorb bone.59 Additionally, transgenic mice expressing GPNMB under the control of a tartrate-resistant acid phosphatase (TRAP) promoter displayed evidence of significant bone loss and elevated bone resorption markers compared to non-transgenic controls.74 Osteoclasts isolated from these transgenic mice were twice as large, possessed elevated TRAP activity, exhibited enhanced expression of osteoclast markers, and could resorb bone matrix to a greater degree than osteoclasts isolated from wild-type controls.74 Thus, GPNMB is expressed and contributes to the differentiation and function of both osteoblasts and osteoclasts within the bone microenvironment.
Immune system
The molecular functions of GPNMB are just beginning to be elucidated and perhaps have been best characterized in the immune system. Expression of GPNMB has been detected in leukocytes and antigen presenting cells, including macrophages62,75,76 and dendritic cells41,77 and has been involved in promoting various cell-cell interactions. GPNMB expression on dendritic cells has been shown to mediate their adhesion to endothelial cells through its RGD domain.41 Additionally, the extracellular domain of GPNMB can suppress T-cell activation and proliferation by binding to syndecan-4 on the surface of activated T-cells, and this interaction requires an intact PKD domain.78,79 GPNMB binding to syndecan-4 leads to the recruitment of syntenin and the CD148 protein tyrosine phosphatase, the activation of which occurs following complex formation and is required for syndecan-4 mediated suppression of T-cell activation.80 This ability to modulate adaptive immunity has been documented in a variety of contexts including graft versus host disease (GVHD), where GPNMB suppresses the activity of alloreactive T cells.81
In contrast to these immunosuppressive roles, activation of GPNMB in dendritic cells, either by ligand binding or antibody cross-linking, can induce an innate immune response against fungal antigens. Under these conditions, the hemITAM tyrosine residue of GPNMB became phosphorylated, which induced widespread changes in gene and protein expression, including increased cytokine secretion (TNFα, IL-1β).60 This activation of GPNMB stimulated dendritic cell maturation and augmented their ability to potentiate the activation of naive T-cells.60 While these findings are strongly suggestive of functional hemITAM-based signaling in GPNMB, more research is needed to definitively characterize the role of this motif when GPNMB is expressed in immune or non-immune cells.
In models of cardiomyopathy, liver fibrosis, and kidney disease, increased GPNMB expression was observed in resident and infiltrating macrophages and is thought to serve as a compensatory response to promote tissue repair through autophagy and phagocytosis of cell debris.75,82–84 During tissue repair, GPNMB localizes to LC3-positive lysosomes, which form during autophagy, and mediates degradation of cellular debris by promoting the fusion of autophagosomes to lysosomes.82,84
It is clear from these observations that GPNMB expression is widespread and it is able to regulate a wide range of physiological and pathological processes. Its established roles during normal tissue processes, such as adhesion during transendothelial migration of dendritic cells and autophagy during tissue repair, are also important mechanisms observed during cancer progression and metastasis. Intriguingly, GPNMB expression can be upregulated in pathological conditions, such as chronic liver disease, which can lead to carcinogenesis.31 As discussed below, it is possible that GPNMB expression in infiltrating immune cells may play important roles in supporting the tumor microenvironment. Considering that the mechanisms of action for GPNMB in tumor progression have yet to be fully elucidated, these observations of GPNMB function in normal tissues represent compelling potential roles for GPNMB in cancer and warrant further investigation.
GPNMB and cancer
Tumor suppressive properties
While it has become increasingly clear that the initial designation of GPNMB as “glycoprotein non-metastatic gene B” is inaccurate in the context of melanoma (see below), there are cancers in which GPNMB appears to exert a tumor-suppressive response.
In the vast majority of colorectal carcinomas, GPNMB is epigenetically silenced by promoter methylation and could thus be involved in attenuating aggressiveness and delaying tumor progression.85 Additionally, a recent study examining GPNMB over-expression in prostate carcinoma cell lines reported a reduction in invasion and proliferation in vitro and tumor growth in vivo.86 Upregulation of anti-metastatic genes, including Ndrg1 and maspin, was observed following forced GPNMB expression in this model, and was proposed as a potential mechanism to explain the anti-tumorigenic effects associated with GPNMB expression.86 These findings emphasize the complexity of GPNMB’s role in tumor biology and the need to obtain a more comprehensive understanding of its mechanisms of action.
Tumor promoting properties
Emerging data has generated a more complex picture with respect to GPNMB in cancer progression, and it is now evident that GPNMB can function to promote tumor progression in certain types of cancer and can act as a tumor suppressor in others.46 The literature investigating the relationship between GPNMB and cancer continues to grow, with an increasing number of reports describing positive correlations between GPNMB expression, poor outcomes and pro-invasive/pro-metastatic phenotype in a variety of cancers.
GPNMB expression and function in breast cancer
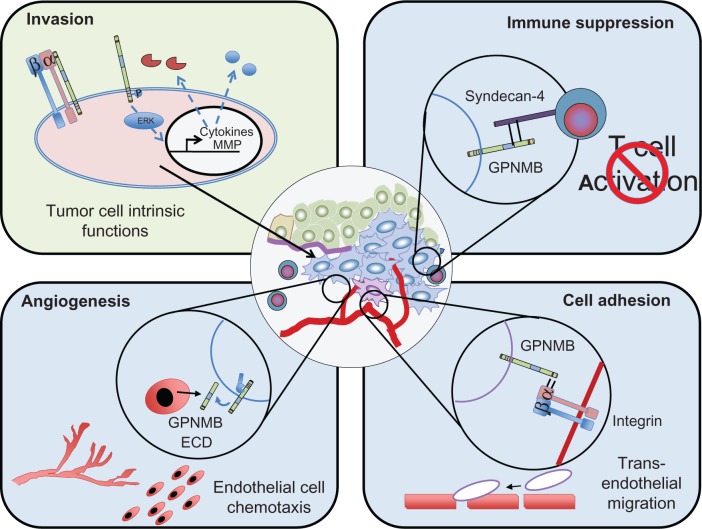
In a screen for metastatic modulators of breast cancer, GPNMB was identified as a gene that is frequently and highly expressed in aggressively metastatic breast cancer cell populations.27,29 Over-expression of GPNMB in weakly metastatic breast cancer cells was shown to drive the acquisition of an invasive phenotype in vitro, characterized by elevated MMP-3 levels, and enhance the bone metastatic potential of these cells.29 A recent study looking at GPNMB over-expression in a murine mammary carcinoma model found that GPNMB could also promote primary mammary tumor growth.27 GPNMB-expressing tumors were characterized by a high endothelial cell density compared to tumors that lacked GPNMB, and in vitro studies revealed that the soluble GPNMB ECD is biologically active as it was capable of inducing endothelial migration.27 These data suggest that GPNMB could regulate the ability of breast cancer cells to recruit vasculature to permit tumor growth and metastasis. Combined, these observations reveal both tumor intrinsic effects of GPNMB that can enhance the invasiveness of tumor cells as well as numerous mechanisms through which GPNMB can facilitate interactions with, and influence the behavior of, cells within the tumor microenvironment to promote the growth and spread of cancer cells (Figure 2).
Figure 2.
Potential mechanisms through which GPNMB promotes malignant cellular phenotypes within cancer cells.
Notes: GPNMB may act cell autonomously (green panel) to induce intracellular signaling, which can influence the expression of multiple targets, including matrix metalloproteinases and cytokines, and enhance the invasiveness of tumor cells. GPNMB may also be important in regulating interactions between tumor cells and cells within the tumor microenvironment (blue panels). It can act as a cell/cell adhesion molecule by engaging integrins expressed on cells in the tumor microenvironment, such as endothelial cells. GPNMB-mediated interactions with syndecan-4 expressed on T cells can block the proliferation and activation of these cells, leading to an immunosuppressive environment favoring tumor growth. Finally, GPNMB may function in a paracrine fashion due to shedding of its extracellular domain, or through its release from cells in the form of microvesicles, leading to endothelial cell recruitment. All of these potential functions of GPNMB can promote tumor growth, invasion, and metastasis in a variety of cancer cells.
Abbreviations: ECD, extracellular domain; GPNMB, glycoprotein non-metastatic b.
In an independent study of GPNMB expression in breast cancer, where the authors employed in situ mRNA hybridization to detect GPNMB in human breast tumors, its expression was reported to be lower in tumors compared to normal tissues.87 GPNMB was also found to be expressed at high levels in immortalized cell lines derived from normal breast epithelium and at low levels in breast cancer cell lines in this study. These studies are in opposition to other published findings and may reflect the fact that the authors did not take the breast cancer subtype into account during their analysis.29,88,89
GPNMB expression and function in other solid malignancies
Brain cancer
The first association of GPNMB with cancer progression was in 2003, when it was reported to promote the invasion of glioma cells.33 These pro-invasive effects were attributed to the ability of GPNMB to enhance the expression of MMP-3 and MMP-9.33 Subsequent studies have confirmed that GPNMB expression is elevated in both benign subependymal giant cell astrocytomas90 as well as malignant glioblastomas.30 Importantly, glioblastoma patients with high levels of GPNMB transcript and protein levels were at significantly higher risk of death.30
Melanoma
The notion that GPNMB is linked to melanomas with low-metastatic potential38 has been dispelled by subsequent studies that report high GPNMB expression in malignant cutaneous melanoma.32,91 In a murine melanoma model, it has been suggested that GPNMB promotes tumor growth via an immunosuppressive mechanism involving a block in T-cell activation.92 Interestingly, this study also reported that GPNMB could be released from melanoma cells in the form of exosomes, and that this dissemination of GPNMB might facilitate systemic immunosuppression of anti-tumor responses.92 It was in the context of cutaneous melanoma that anti-GPNMB therapies were first considered,32,93,94 which is discussed in greater detail below. Interestingly, a recent survey of uveal melanomas revealed that a high percentage of these aggressive tumors also express GPNMB.34
GPNMB function in tumor stroma
GPNMB expression in the stromal compartment of different cancers could also potentially be linked to tumor progression. GPNMB was over-expressed in a subset of CD10-positive cancer associated fibroblasts derived from colon tissue,95 which is in line with previous reports that GPNMB can activate fibroblasts by inducing upregulation of pro-invasive matrix metalloproteases, such as MMP-3 and MMP-9, via Erk-dependent signaling.61,96 In macrophages, treatment with tumor-cell conditioned media induced an 83-fold increase in GPNMB expression.97 Interestingly, these tumor-conditioned macrophages adopted a phenotype similar to the M2-type macrophages,97 which are known for their role in promoting tumor progression.98 In the breast, GPNMB expression is abundant in the stromal compartment of tumor tissue,28 which could be attributed to its expression in a variety of stromal subtypes described above. Taken together, these studies suggest a role for GPNMB in sustaining the tumor microenvironment; however, it remains to be seen if stromal GPNMB can directly influence tumor progression. In this regard, it is interesting to note that GPNMB expression in the tumor epithelium of breast cancers was associated with poorer prognosis, whereas breast cancers that lacked GPNMB or displayed predominantly stromal GPNMB expression displayed better outcomes.28 However, this may be a reflection of the fact that tumor-cell-intrinsic GPNMB expression is required for breast cancer progression and does not necessarily negate an important role for stromal-derived GPNMB in this disease.
GPNMB as a therapeutic target
Given the increasing association between GPNMB expression and a variety of cancers, and the acquisition of aggressive cellular phenotypes in GPNMB-expressing cancer cells, there has been growing interest in the development of GPNMB-targeted therapies.35–37 The pattern of GPNMB expression in normal and cancerous tissues makes it an intriguing target for cancer therapy. Generally speaking, GPNMB localization tends to be restricted to intracellular compartments in normal cells, such as macrophages, melanocytes and pigmented retinal epithelial cells.67,76,99 In contrast, GPNMB expression in tumor cells is enriched on the cell surface.28,32,94 This pattern of sub-cellular localization makes tumor-specific GPNMB more readily available for antibody targeting, thus providing a therapeutic window and making GPNMB a uniquely attractive target for antibody based therapies.
Targeting GPNMB in brain cancers
A single chain antibody coupled to an immunotoxin (F6V-PE38), which is directed against the extracellular domain of GPNMB, has recently been generated for the treatment of glioblastoma multiforme.100 A GPNMB-specific single chain variable fragment (scFv) antibody was first isolated from a phage display library and subsequent mutagenesis/selection of this clone produced a high-affinity GPNMB-specific scFv antibody (F6V). This scFv was then conjugated to a truncated form of Pseudomonas endotoxin A to generate F6V-PE38, which causes protein synthesis inhibition and apoptosis following internalization by GPNMB-expressing target cells. Two xenograft models of malignant glioma (glioblastoma multiforme and medulloblastoma) were subjected to treatment with the anti-GPNMB immunotoxin (F6V-PE38), which resulted in a significant impairment in tumor growth compared to PBS-treated controls.100 Although these findings are preliminary, they address the potential for development of small-size targeted therapeutics against GPNMB, which will penetrate the tumor mass with higher efficiency compared to full-length conjugated antibodies.101
Targeting GPNMB in melanoma and breast cancer
A more developed GPNMB-targeted therapeutic agent is CDX-011, an antibody-drug conjugate also known as CR011-vcMMAE (CR011) or glembatumumab vedotin.94 In the case of CDX-011, the cytotoxin auristatin E, a tubulin destabilizer, is conjugated to an antibody directed against the extracellular domain of GPNMB.94 Upon GPNMB binding and internalization, the drug is released and induces cell cycle arrest and apoptosis of the target cell.
Pre-clinical models
The first evidence of successful therapeutic targeting of GPNMB using this ADC demonstrated that CDX-011 was selectively able to inhibit the growth of GPNMB-expressing metastatic melanoma cells, both in culture and xenograft assays.94 A subsequent study examining the pharmacological properties of this antibody-drug conjugate showed that, at concentrations as low as 2.5 mg/kg, CDX-011 was capable of inducing complete regression in 100% of GPNMB-expressing xenografted SK-Mel-2 and SK-Mel-5 melanoma cells.93 In breast cancer, a single dose of 20 mg/kg CDX-011 was sufficient to induce sustained MDA-MB-468 tumor regression in vivo.28 Numerous studies have reported that cell killing efficacy of CDX-011 is directly proportional to the level of GPNMB expressed on the cell surface.28,32,93,94
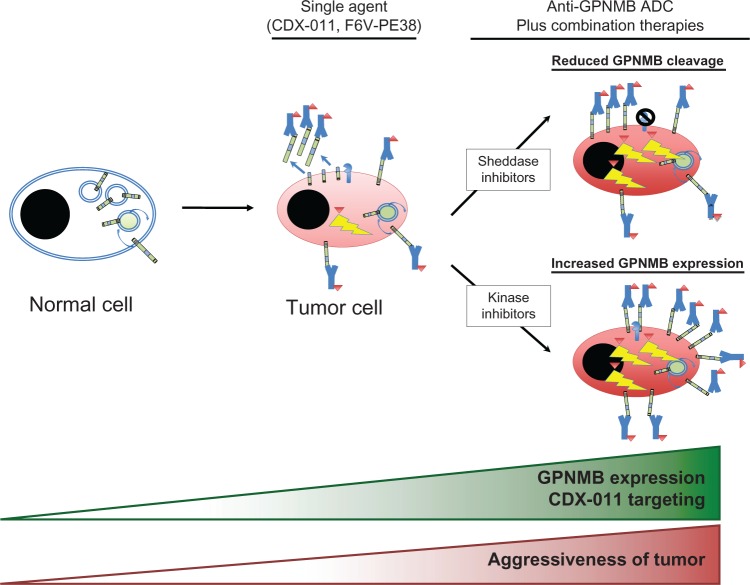
Interestingly, treatment of cancer cells with imatinib or inhibitors of the Erk pathway enhances cell surface expression of GPNMB in cancer cells, which in turn increases sensitivity to CDX-011.32 Additionally, a separate study examining monocyte-derived dendritic cells (moDC) reported that BCR-ABL and Src family kinase inhibitors such as imatinib, dasatinib, and nilotinib increased GPNMB expression and thereby potentiated immune-suppression by moDCs.102 Inhibitors of metalloproteinases, such as GM6001, have also been shown to enhance cell surface GPNMB expression by preventing shedding of its extracellular domain.32,61 In addition to increasing target availability, such inhibitors can minimize the potential for sequestration of CDX-011 by the shed form of GPNMB and thereby increase the targeted killing of GPNMB-expressing tumor cells. However, the effect of these inhibitors on tumor cell sensitivity to CDX-011 has not yet been examined. These findings suggest that combinations with additional targeted therapies (that are capable of enhancing cell surface GPNMB expression) could further enhance the efficacy of CDX-011. Given the pro-invasive and pro-metastatic functions of GPNMB, such a strategy would require careful evaluation in pre-clinical models to ensure that these combination therapies did not increase metastasis of cancer cells that escape CDX-011 mediated killing. (Figure 3).
Figure 3.
Therapeutic strategies employing anti-GPNMB antibody-drug conjugates (ADCs).
Notes: in normal cells, GPNMB is preferentially localized within endosomal/lysosomal compartments, which is not accessible to anti-GPNMB ADCs. In many cancers, including breast, melanoma, and brain cancers, the levels of GPNMB expression increases and a greater proportion is localized on the cell surface. These GPNMB-expressing cancer cells are more susceptible to killing by anti-GPNMB ADCs (CDX-011, F6v-PE38). Evidence suggests that coupling kinase inhibitors (serine/threonine and tyrosine kinase inhibitors), which increase GPNMB expression, may enhance the efficacy of tumor cell killing by anti-GPNMB ADCs. Likewise, inhibiting GPNMB shedding could also lead to greater GPNMB surface expression and more targets for anti-GPNMB ADCs. Thus, GPNMB represents an attractive target due to low surface expression in normal cells and its increased expression in cancer cells, which leads to better tumor cell killing with anti-GPNMB ADCs. Combination therapies have the potential to achieve benefit from enhanced efficacy of the anti-GPNMB ADCs and effects of the coupled inhibitors (kinase inhibitors), but there is the potential risk that those tumor cells not killed by combination treatment may adopt increasing malignant phenotypes due to elevated GPNMB expression.
Abbreviations: ADC, antibody-drug conjugate; CDX-011, glembatumumab vedotin; GPNMB, glycoprotein non-metastatic b.
Clinical trials
CDX-011 was initially tested in two multi-centre phase I/II clinical trials; one for patients with unresectable melanoma103 and the other for patients with locally advanced or metastatic breast cancer.104,105 Tumor shrinkage was reported in 56% of melanoma patients and 62% of breast cancer patients who were treated with a maximum tolerated dose (MTD) of 1.88 mg/kg once every 3 weeks.103,104 GPNMB expression appeared to be a predictive biomarker in the melanoma study. A small subset of melanoma patients with the highest levels of tumoral GPNMB expression (n = 7) had longer median progression-free survival (PFS) times (4.9 months) compared to the median PFS for all patients in the cohort (n = 34; including those with high tumoral GPNMB), which ranged from 1–3.9 months depending on the dose frequency.103 This observation was recapitulated in a subset of breast cancer patients treated with CDX-011. In this study, the median PFS for GPNMB-positive patients (n = 9) was 17.3 weeks compared to 9.1 weeks for all patients (n = 34) treated with the MTD.104 Interestingly, patients with strong GPNMB expression in stromal cells responded to CDX-011 just as well, if not better, than patients with strong GPNMB expression in the tumor epithelium.104 It is conceivable that GPNMB-expressing cells that initially take up CDX-011 can release the drug moiety when the targeted cells die, which can freely diffuse into neighboring cells and kill them regardless of whether they expressed GPNMB. This “bystander” effect has been described with SGN-35, which is an antibody drug conjugate that targets CD30.106
Based on these observations, a subsequent EMERGE (NCT01156753) phase IIb clinical trial was recently carried out to investigate the efficacy and safety of CDX-011 for patients with heavily pre-treated metastatic breast cancer that were positive for GPNMB.104 The final results from this trial were presented at the 2012 San Antonio Breast Cancer Symposium and showed promise for CDX-011 treatment of patients with GPNMB-expressing and triple negative breast cancer.107 The trial enrolled 122 patients and was carried out in a 2:1 randomized fashion where 81 patients received CDX-011 and 41 received investigator’s choice of therapy (IC). Patients treated with IC were allowed to crossover to CDX-011 therapy if they continued to be eligible after confirmation of pharmacodynamics. Eligible patients were required to have GPNMB expression in ≥5% of tumor epithelial and/or stromal tissue, as confirmed by immunohistochemistry on archived tumor samples. Patients were required to have been previously treated with all of the following therapeutic regimens, when indicated, prior to enrollment: taxane, anthracycline, capecitabine, traztuzumab, and lapatinib. Interestingly, 99% of patients tested displayed some level of tumoral GPNMB expression, which was significantly higher than earlier reports of GPNMB expression from breast cancer tissue microarrays.28 To assess the potential for utilizing GPNMB as a predictive marker for CDX-011 therapy, patients were classified as having high or low GPNMB expression based on a threshold cutoff of ≥25% GPNMB positivity, post-hoc.103 The trial reported that 41% of TNBC patients had high GPNMB expression, which was consistent with previous studies, and further confirmed GPNMB as a promising target in this aggressive disease subtype. Partial response was observed in 19% of patients with triple negative disease, compared to 0% with IC, which is an encouraging result for a subgroup of breast cancer patients with currently limited treatment options. The response rate was even higher (33% versus 0%) in the TNBC subset of patients displaying high GPNMB expression, substantiating findings from the melanoma phase I/II trial. Additionally, patients with high GPNMB expression and TNBC had a doubling in progression free survival (3.0 months [n = 12 patients receiving CDX-011] versus 1.5 months [n = 6 patients receiving IC]; P = 0.008) and overall survival (10.0 months [n = 12 patients receiving CDX-011] versus 5.5 months [n = 6 patients receiving IC]; P = 0.003). While the results are encouraging, it must be noted that the sample sizes in these groups are very small. Also, no statistically significant differences were observed across all subtypes between CDX-011 and IC treated patients with high GPNMB expression. However, contrary to reports from previous trials,104 stromal GPNMB expression did not appear to be a predictive marker of response to therapy in the EMERGE study.
In these studies, development of a skin rash was one of the most common side effects experienced by melanoma (57%) and breast cancer patients treated with CDX-011 (48%, 47%).103,104,107 This finding was of great interest, given that GPNMB is expressed in the skin.65,67 Interestingly, melanoma patients who experienced rash within their first cycle of treatment also had significantly longer PFS than CDX-011-treated patients who didn’t develop rash (4.8 versus 1.2 months; P < 0.001), suggesting that rash may be an early indicator of a patient’s ability to tolerate and respond to the drug.103 Additional side effects in the EMERGE study that were worsened in patients treated with CDX-011 compared to IC include other dermatological conditions such as alopecia (hair loss) and pruritus (itch) as well as peripheral neuropathy and vomiting.107 However, patients undergoing CDX-011 therapy witnessed a reduction in hematologic side effects such as neutropenia, leucopenia, and thrombocytopenia. Although GPNMB is largely expressed in intracellular compartments in normal tissues, CDX-011 can adversely affect certain tissue types, which is evident by its ability to induce skin rash.
One tissue that could be susceptible to side effects of CDX-011 treatment includes the bone. The potential use of CDX-011 to target breast cancer bone metastases should be met with caution. Osteoblasts and osteoclasts both express cell-surface localized GPNMB and their targeting by CDX-011 could have detrimental effects on bone turnover. Decreased osteoblast numbers would reduce bone formation, which could lead to an increased risk of fracture for the patients. Conversely, targeted killing of osteoclasts could delay bone healing and lead to osteopetrosis. Indeed, antibodies directed against GPNMB were shown to impair osteoclast formation and function.59 Bone remodeling is a finely-tuned process and tipping the scale in either direction could exacerbate the side effects of CDX-011. These considerations should be kept in mind when choosing patient cohorts for CDX-011 treatment. Overall, in light of the scarcity of treatment options for TNBC patients, these data substantiate further studies investigating the efficacy of CDX-011 in the treatment of metastatic breast cancer.
Conclusions and future perspectives
The development of antibody-based therapeutic agents targeting GPNMB (single chain variable fragment antibodies, antibody drug conjugates) is a promising avenue for several GPNMB-expressing cancers. The latest phase II clinical trial data reinforces the early results from phase I/II trials, which supports the use of CDX-011 in women with triple negative breast cancer. Early efforts to identify potential therapeutic combinations that will increase the efficacy of anti-GPNMB agents will need to be investigated with caution. Enhancing cell surface expression of GPNMB may sensitize tumor cells to more effective killing by agents such as CDX-011; however, the acquisition of malignant phenotypes in cancer cells with elevated levels of GPNMB expression, which are not eliminated, is cause for concern.
A better understanding of the molecular mechanisms through which GPNMB induces aggressive cellular phenotypes, such as enhanced migration and invasion, will be needed in order to fully optimize therapeutic molecules targeting GPNMB. Another aspect that requires further investigation is the contribution of stromal cells within the tumor microenvironment that express GPNMB and how this impacts tumor progression and response to anti-GPNMB therapies.
Acknowledgments
The authors would like to thank Dr Josie Ursini-Siegel and members of the Siegel laboratory for their thoughtful and insightful comments on the manuscript. GM acknowledges studentship support from the Canadian Institutes for Health Research (CIHR). Research conducted in the author’s laboratory, cited in this review, was supported by grants from the CIHR (MOP-119401).
Footnotes
Disclosure
The authors report no conflict of interests in this work.
References:
- 1.Forouzanfar MH, Foreman KJ, Delossantos AM, et al. Breast and cervical cancer in 187 countries between 1980 and 2010: a systematic analysis. Lancet. 2011;378(9801):1461–1484. doi: 10.1016/S0140-6736(11)61351-2. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 4.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100(14):8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herschkowitz JI, Simin K, Weigman VJ, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8(5):R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prat A, Parker JS, Karginova O, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12(5):R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nik-Zainal S, Alexandrov LB, Wedge DC, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149(5):979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nik-Zainal S, Van Loo P, Wedge DC, et al. The life history of 21 breast cancers. Cell. 2012;149(5):994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah SP, Roth A, Goya R, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486(7403):395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stephens PJ, Tarpey PS, Davies H, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486(7403):400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 14.Largillier R, Ferrero JM, Doyen J, et al. Prognostic factors in 1,038 women with metastatic breast cancer. Ann Oncol. 2008;19(12):2012–2019. doi: 10.1093/annonc/mdn424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sihto H, Lundin J, Lundin M, et al. Breast cancer biological subtypes and protein expression predict for the preferential distant metastasis sites: a nationwide cohort study. Breast Cancer Res. 2011;13(5):R87. doi: 10.1186/bcr2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smid M, Wang Y, Zhang Y, et al. Subtypes of breast cancer show preferential site of relapse. Cancer Res. 2008;68(9):3108–3114. doi: 10.1158/0008-5472.CAN-07-5644. [DOI] [PubMed] [Google Scholar]
- 17.Cheang MC, Chia SK, Voduc D, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101(10):736–750. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Z, Fan C, Oh DS, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carey LA, Dees EC, Sawyer L, et al. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13(8):2329–2334. doi: 10.1158/1078-0432.CCR-06-1109. [DOI] [PubMed] [Google Scholar]
- 20.Hugh J, Hanson J, Cheang MC, et al. Breast cancer subtypes and response to docetaxel in node-positive breast cancer: use of an immunohistochemical definition in the BCIRG 001 trial. J Clin Oncol. 2009;27(8):1168–1176. doi: 10.1200/JCO.2008.18.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen TO, Parker JS, Leung S, et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Cancer Res. 2010;16(21):5222–5232. doi: 10.1158/1078-0432.CCR-10-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rouzier R, Perou CM, Symmans WF, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005;11(16):5678–5685. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- 23.Hiller DJ, Chu QD. Current Status of Poly(ADP-ribose) Polymerase Inhibitors as Novel Therapeutic Agents for Triple-Negative Breast Cancer. Int J Breast Cancer. 2012;2012:829315. doi: 10.1155/2012/829315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tutt A, Robson M, Garber JE, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376(9737):235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 25.Balmana J, Domchek SM, Tutt A, Garber JE. Stumbling blocks on the path to personalized medicine in breast cancer: the case of PARP inhibitors for BRCA1/2-associated cancers. Cancer Discov. 2011;1(1):29–34. doi: 10.1158/2159-8274.CD-11-0048. [DOI] [PubMed] [Google Scholar]
- 26.Adair JR, Howard PW, Hartley JA, Williams DG, Chester KA. Antibody-drug conjugates – a perfect synergy. Expert Opin Biol Ther. 2012;12(9):1191–1206. doi: 10.1517/14712598.2012.693473. [DOI] [PubMed] [Google Scholar]
- 27.Rose AA, Annis MG, Dong Z, et al. ADAM10 releases a soluble form of the GPNMB/Osteoactivin extracellular domain with angiogenic properties. PLoS One. 2010;5(8):e12093. doi: 10.1371/journal.pone.0012093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rose AA, Grosset AA, Dong Z, et al. Glycoprotein nonmetastatic B is an independent prognostic indicator of recurrence and a novel therapeutic target in breast cancer. Clin Cancer Res. 2010;16(7):2147–2156. doi: 10.1158/1078-0432.CCR-09-1611. [DOI] [PubMed] [Google Scholar]
- 29.Rose AA, Pepin F, Russo C, Abou Khalil JE, Hallett M, Siegel PM. Osteoactivin promotes breast cancer metastasis to bone. Mol Cancer Res. 2007;5(10):1001–1014. doi: 10.1158/1541-7786.MCR-07-0119. [DOI] [PubMed] [Google Scholar]
- 30.Kuan CT, Wakiya K, Dowell JM, et al. Glycoprotein nonmetastatic melanoma protein B, a potential molecular therapeutic target in patients with glioblastoma multiforme. Clin Cancer Res. 2006;12(7 Pt 1):1970–1982. doi: 10.1158/1078-0432.CCR-05-2797. [DOI] [PubMed] [Google Scholar]
- 31.Onaga M, Ido A, Hasuike S, et al. Osteoactivin expressed during cirrhosis development in rats fed a choline-deficient, L-amino acid-defined diet, accelerates motility of hepatoma cells. J Hepatol. 2003;39(5):779–785. doi: 10.1016/s0168-8278(03)00361-1. [DOI] [PubMed] [Google Scholar]
- 32.Qian X, Mills E, Torgov M, LaRochelle WJ, Jeffers M. Pharmacologically enhanced expression of GPNMB increases the sensitivity of melanoma cells to the CR011-vcMMAE antibody-drug conjugate. Mol Oncol. 2008;2(1):81–93. doi: 10.1016/j.molonc.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rich JN, Shi Q, Hjelmeland M, et al. Bone-related genes expressed in advanced malignancies induce invasion and metastasis in a genetically defined human cancer model. J Biol Chem. 2003;278(18):15951–15957. doi: 10.1074/jbc.M211498200. [DOI] [PubMed] [Google Scholar]
- 34.Williams MD, Esmaeli B, Soheili A, et al. GPNMB expression in uveal melanoma: a potential for targeted therapy. Melanoma Res. 2010 Jun;20(3):184–190. doi: 10.1097/CMR.0b013e3283364a08. [DOI] [PubMed] [Google Scholar]
- 35.Keir CH, Vahdat LT. The use of an antibody drug conjugate, glembatumumab vedotin (CDX-011), for the treatment of breast cancer. Expert Opin Biol Ther. 2012;12(2):259–263. doi: 10.1517/14712598.2012.642357. [DOI] [PubMed] [Google Scholar]
- 36.Naumovski L, Junutula JR. Glembatumumab vedotin, a conjugate of an anti-glycoprotein non-metastatic melanoma protein B mAb and monomethyl auristatin E for the treatment of melanoma and breast cancer. Curr Opin Mol Ther. 2010;12(2):248–257. [PubMed] [Google Scholar]
- 37.Zhou LT, Liu FY, Li Y, Peng YM, Liu YH, Li J. Gpnmb/osteoactivin, an attractive target in cancer immunotherapy. Neoplasma. 2012;59(1):1–5. doi: 10.4149/neo_2012_001. [DOI] [PubMed] [Google Scholar]
- 38.Weterman MA, Ajubi N, van Dinter IM, et al. nmb, a novel gene, is expressed in low-metastatic human melanoma cell lines and xenografts. Int J Cancer. 1995;60(1):73–81. doi: 10.1002/ijc.2910600111. [DOI] [PubMed] [Google Scholar]
- 39.Bandari PS, Qian J, Yehia G, et al. Hematopoietic growth factor inducible neurokinin-1 type: a transmembrane protein that is similar to neurokinin 1 interacts with substance P. Regul Pept. 2003;111(1–3):169–178. doi: 10.1016/s0167-0115(02)00288-4. [DOI] [PubMed] [Google Scholar]
- 40.Safadi FF, Xu J, Smock SL, Rico MC, Owen TA, Popoff SN. Cloning and characterization of osteoactivin, a novel cDNA expressed in osteoblasts. J Cell Biochem. 2001;84(1):12–26. doi: 10.1002/jcb.1259. [DOI] [PubMed] [Google Scholar]
- 41.Shikano S, Bonkobara M, Zukas PK, Ariizumi K. Molecular cloning of a dendritic cell-associated transmembrane protein, DC-HIL, that promotes RGD-dependent adhesion of endothelial cells through recognition of heparan sulfate proteoglycans. J Biol Chem. 2001;276(11):8125–8134. doi: 10.1074/jbc.M008539200. [DOI] [PubMed] [Google Scholar]
- 42.Turque N, Denhez F, Martin P, et al. Characterization of a new melanocyte-specific gene (QNR-71) expressed in v-myc-transformed quail neuroretina. EMBO J. 1996;15(13):3338–3350. [PMC free article] [PubMed] [Google Scholar]
- 43.Yamaguchi Y, Hearing VJ. Physiological factors that regulate skin pigmentation. Biofactors. 2009;35(2):193–199. doi: 10.1002/biof.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saitoh O, Wang WC, Lotan R, Fukuda M. Differential glycosylation and cell surface expression of lysosomal membrane glycoproteins in sublines of a human colon cancer exhibiting distinct metastatic potentials. J Biol Chem. 1992;267(8):5700–5711. [PubMed] [Google Scholar]
- 45.Selim AA. Osteoactivin bioinformatic analysis: prediction of novel functions, structural features, and modes of action. Med Sci Monit. 2009;15(2):MT19–MT33. [PubMed] [Google Scholar]
- 46.Singh M, Del Carpio-Cano F, Belcher JY, et al. Functional roles of osteoactivin in normal and disease processes. Crit Rev Eukaryot Gene Expr. 2010;20(4):341–357. doi: 10.1615/critreveukargeneexpr.v20.i4.50. [DOI] [PubMed] [Google Scholar]
- 47.Le Borgne R, Planque N, Martin P, Dewitte F, Saule S, Hoflack B. The AP-3-dependent targeting of the melanosomal glycoprotein QNR-71 requires a di-leucine-based sorting signal. J Cell Sci. 2001;114(Pt 15):2831–2841. doi: 10.1242/jcs.114.15.2831. [DOI] [PubMed] [Google Scholar]
- 48.Barczyk M, Carracedo S, Gullberg D. Integrins. Cell Tissue Res. 2010;339(1):269–280. doi: 10.1007/s00441-009-0834-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takada Y, Ye X, Simon S. The integrins. Genome Biol. 2007;8(5):215. doi: 10.1186/gb-2007-8-5-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weston BS, Malhas AN, Price RG. Structure-function relationships of the extracellular domain of the autosomal dominant polycystic kidney disease-associated protein, polycystin-1. FEBS Lett. 2003;538(1–3):8–13. doi: 10.1016/s0014-5793(03)00130-3. [DOI] [PubMed] [Google Scholar]
- 51.Ibraghimov-Beskrovnaya O, Bukanov NO, Donohue LC, Dackowski WR, Klinger KW, Landes GM. Strong homophilic interactions of the Ig-like domains of polycystin-1, the protein product of an autosomal dominant polycystic kidney disease gene, PKD1. Hum Mol Genet. 2000;9(11):1641–1649. doi: 10.1093/hmg/9.11.1641. [DOI] [PubMed] [Google Scholar]
- 52.Kerrigan AM, Brown GD. Syk-coupled C-type lectin receptors that mediate cellular activation via single tyrosine based activation motifs. Immunol Rev. 2010;234(1):335–352. doi: 10.1111/j.0105-2896.2009.00882.x. [DOI] [PubMed] [Google Scholar]
- 53.Ivashkiv LB. Cross-regulation of signaling by ITAM-associated receptors. Nat Immunol. 2009;10(4):340–347. doi: 10.1038/ni.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mocsai A, Ruland J, Tybulewicz VL. The SYK tyrosine kinase: a crucial player in diverse biological functions. Nat Rev Immunol. 2010;10(6):387–402. doi: 10.1038/nri2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bradshaw JM. The Src, Syk, and Tec family kinases: distinct types of molecular switches. Cell Signal. 2010;22(8):1175–1184. doi: 10.1016/j.cellsig.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 56.Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 57.Abdelmagid SM, Barbe MF, Rico MC, et al. Osteoactivin, an anabolic factor that regulates osteoblast differentiation and function. Exp Cell Res. 2008;314(13):2334–2351. doi: 10.1016/j.yexcr.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 58.Hoashi T, Sato S, Yamaguchi Y, Passeron T, Tamaki K, Hearing VJ. Glycoprotein nonmetastatic melanoma protein b, a melanocytic cell marker, is a melanosome-specific and proteolytically released protein. FASEB J. 2010;24(5):1616–1629. doi: 10.1096/fj.09-151019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sheng MH, Wergedal JE, Mohan S, Lau KH. Osteoactivin is a novel osteoclastic protein and plays a key role in osteoclast differentiation and activity. FEBS Lett. 2008;582(10):1451–1458. doi: 10.1016/j.febslet.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 60.Chung JS, Yudate T, Tomihari M, Akiyoshi H, Cruz PD, Jr, Ariizumi K. Binding of DC-HIL to dermatophytic fungi induces tyrosine phosphorylation and potentiates antigen presenting cell function. J Immunol. 2009;183(8):5190–5198. doi: 10.4049/jimmunol.0901319. [DOI] [PubMed] [Google Scholar]
- 61.Furochi H, Tamura S, Mameoka M, et al. Osteoactivin fragments produced by ectodomain shedding induce MMP-3 expression via ERK pathway in mouse NIH-3T3 fibroblasts. FEBS Lett. 2007;581(30):5743–5750. doi: 10.1016/j.febslet.2007.11.036. [DOI] [PubMed] [Google Scholar]
- 62.Huang JJ, Ma WJ, Yokoyama S. Expression and immunolocalization of Gpnmb, a glioma-associated glycoprotein, in normal and inflamed central nervous systems of adult rats. Brain Behav. 2012;2(2):85–96. doi: 10.1002/brb3.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buga AM, Scholz CJ, Kumar S, et al. Identification of new therapeutic targets by genome-wide analysis of gene expression in the ipsilateral cortex of aged rats after stroke. PLoS One. 2012;7(12):e50985. doi: 10.1371/journal.pone.0050985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tanaka H, Shimazawa M, Kimura M, et al. The potential of GPNMB as novel neuroprotective factor in amyotrophic lateral sclerosis. Sci Rep. 2012;2:573. doi: 10.1038/srep00573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Loftus SK, Antonellis A, Matera I, et al. Gpnmb is a melanoblast-expressed, MITF-dependent gene. Pigment Cell Melanoma Res. 2009;22(1):99–110. doi: 10.1111/j.1755-148X.2008.00518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Owen TA, Smock SL, Prakash S, et al. Identification and characterization of the genes encoding human and mouse osteoactivin. Crit Rev Eukaryot Gene Expr. 2003;13(2–4):205–220. doi: 10.1615/critreveukaryotgeneexpr.v13.i24.130. [DOI] [PubMed] [Google Scholar]
- 67.Tomihari M, Hwang SH, Chung JS, Cruz PD, Jr, Ariizumi K. Gpnmb is a melanosome-associated glycoprotein that contributes to melanocyte/keratinocyte adhesion in a RGD-dependent fashion. Exp Dermatol. 2009;18(7):586–595. doi: 10.1111/j.1600-0625.2008.00830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang P, Liu W, Zhu C, et al. Silencing of GPNMB by siRNA inhibits the formation of melanosomes in melanocytes in a MITF-independent fashion. PLoS One. 2012;7(8):e42955. doi: 10.1371/journal.pone.0042955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Theos AC, Watt B, Harper DC, et al. The PKD domain distinguishes the trafficking and amyloidogenic properties of the pigment cell protein PMEL and its homologue GPNMB. Pigment Cell Melanoma Res. 2013 Mar 4; doi: 10.1111/pcmr.12084. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abdelmagid SM, Barbe MF, Arango-Hisijara I, Owen TA, Popoff SN, Safadi FF. Osteoactivin acts as downstream mediator of BMP-2 effects on osteoblast function. J Cell Physiol. 2007;210(1):26–37. doi: 10.1002/jcp.20841. [DOI] [PubMed] [Google Scholar]
- 71.Selim AA, Abdelmagid SM, Kanaan RA, et al. Anti-osteoactivin antibody inhibits osteoblast differentiation and function in vitro. Crit Rev Eukaryot Gene Expr. 2003;13(2–4):265–275. doi: 10.1615/critreveukaryotgeneexpr.v13.i24.180. [DOI] [PubMed] [Google Scholar]
- 72.Bateman JP, Safadi FF, Susin C, Wikesjo UM. Exploratory study on the effect of osteoactivin on bone formation in the rat critical-size calvarial defect model. J Periodontal Res. 2012;47(2):243–247. doi: 10.1111/j.1600-0765.2011.01428.x. [DOI] [PubMed] [Google Scholar]
- 73.Ripoll VM, Meadows NA, Raggatt LJ, et al. Microphthalmia transcription factor regulates the expression of the novel osteoclast factor GPNMB. Gene. 2008;413(1–2):32–41. doi: 10.1016/j.gene.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 74.Sheng MH, Wergedal JE, Mohan S, Amoui M, Baylink DJ, Lau KH. Targeted overexpression of osteoactivin in cells of osteoclastic lineage promotes osteoclastic resorption and bone loss in mice. PLoS One. 2012;7(4):e35280. doi: 10.1371/journal.pone.0035280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ramachandran P, Pellicoro A, Vernon MA, et al. Differential Ly-6C expression identifies the recruited macrophage phenotype, which orchestrates the regression of murine liver fibrosis. Proc Natl Acad Sci USA. 2012;109(46):E3186–E3195. doi: 10.1073/pnas.1119964109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ripoll VM, Irvine KM, Ravasi T, Sweet MJ, Hume DA. Gpnmb is induced in macrophages by IFN-gamma and lipopolysaccharide and acts as a feedback regulator of proinflammatory responses. J Immunol. 2007;178(10):6557–6566. doi: 10.4049/jimmunol.178.10.6557. [DOI] [PubMed] [Google Scholar]
- 77.Ahn JH, Lee Y, Jeon C, et al. Identification of the genes differentially expressed in human dendritic cell subsets by cDNA subtraction and microarray analysis. Blood. 2002;100(5):1742–1754. [PubMed] [Google Scholar]
- 78.Chung JS, Dougherty I, Cruz PD, Jr, Ariizumi K. Syndecan-4 mediates the coinhibitory function of DC-HIL on T cell activation. J Immunol. 2007;179(9):5778–5784. doi: 10.4049/jimmunol.179.9.5778. [DOI] [PubMed] [Google Scholar]
- 79.Chung JS, Sato K, Dougherty II, Cruz PD, Jr, Ariizumi K. DC-HIL is a negative regulator of T lymphocyte activation. Blood. 2007;109(10):4320–4327. doi: 10.1182/blood-2006-11-053769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chung JS, Cruz PD, Jr, Ariizumi K. Inhibition of T-cell activation by syndecan-4 is mediated by CD148 through protein tyrosine phosphatase activity. Eur J Immunol. 2011;41(6):1794–1799. doi: 10.1002/eji.201041233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chung JS, Tomihari M, Tamura K, Kojima T, Cruz PD, Jr, Ariizumi K. The DC-HIL ligand syndecan-4 is a negative regulator of T-cell allo-reactivity responsible for graft-versus-host disease. Immunology. 2013;138(2):173–182. doi: 10.1111/imm.12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li B, Castano AP, Hudson TE, et al. The melanoma-associated transmembrane glycoprotein Gpnmb controls trafficking of cellular debris for degradation and is essential for tissue repair. FASEB J. 2010;24(12):4767–4781. doi: 10.1096/fj.10-154757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pahl MV, Vaziri ND, Yuan J, Adler SG. Upregulation of monocyte/macrophage HGFIN (Gpnmb/Osteoactivin) expression in end-stage renal disease. Clin J Am Soc Nephrol. 2010;5(1):56–61. doi: 10.2215/CJN.03390509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Patel-Chamberlin M, Wang Y, Satirapoj B, et al. Hematopoietic growth factor inducible neurokinin-1 (Gpnmb/Osteoactivin) is a biomarker of progressive renal injury across species. Kidney Int. 2011;79(10):1138–1148. doi: 10.1038/ki.2011.28. [DOI] [PubMed] [Google Scholar]
- 85.Mokarram P, Kumar K, Brim H, et al. Distinct high-profile methylated genes in colorectal cancer. PLoS One. 2009;4(9):e7012. doi: 10.1371/journal.pone.0007012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tsui KH, Chang YL, Feng TH, Chang PL, Juang HH. Glycoprotein transmembrane nmb: an androgen-downregulated gene attenuates cell invasion and tumorigenesis in prostate carcinoma cells. Prostate. 2012;72(13):1431–1442. doi: 10.1002/pros.22494. [DOI] [PubMed] [Google Scholar]
- 87.Metz RL, Patel PS, Hameed M, Bryan M, Rameshwar P. Role of human HGFIN/nmb in breast cancer. Breast Cancer Res. 2007;9(5):R58. doi: 10.1186/bcr1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rose AA, Siegel PM. Osteoactivin/HGFIN: is it a tumor suppressor or mediator of metastasis in breast cancer? Breast Cancer Res. 2007;9(6):403. doi: 10.1186/bcr1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tyburczy ME, Kotulska K, Pokarowski P, et al. Novel proteins regulated by mTOR in subependymal giant cell astrocytomas of patients with tuberous sclerosis complex and new therapeutic implications. Am J Pathol. 2010;176(4):1878–1890. doi: 10.2353/ajpath.2010.090950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhao Y, Qiao ZG, Shan SJ, Sun QM, Zhang JZ. Expression of glycoprotein non-metastatic melanoma protein B in cutaneous malignant and benign lesions: a tissue microarray study. Chin Med J (Engl) 2012;125(18):3279–3282. [PubMed] [Google Scholar]
- 92.Tomihari M, Chung JS, Akiyoshi H, Cruz PD, Jr, Ariizumi K. DC-HIL/glycoprotein Nmb promotes growth of melanoma in mice by inhibiting the activation of tumor-reactive T cells. Cancer Res. 2010;70(14):5778–5787. doi: 10.1158/0008-5472.CAN-09-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pollack VA, Alvarez E, Tse KF, et al. Treatment parameters modulating regression of human melanoma xenografts by an antibody-drug conjugate (CR011-vcMMAE) targeting GPNMB. Cancer Chemother Pharmacol. 2007;60(3):423–435. doi: 10.1007/s00280-007-0490-z. [DOI] [PubMed] [Google Scholar]
- 94.Tse KF, Jeffers M, Pollack VA, et al. CR011, a fully human monoclonal antibody-auristatin E conjugate, for the treatment of melanoma. Clin Cancer Res. 2006;12(4):1373–1382. doi: 10.1158/1078-0432.CCR-05-2018. [DOI] [PubMed] [Google Scholar]
- 95.Ghilardi C, Chiorino G, Dossi R, Nagy Z, Giavazzi R, Bani M. Identification of novel vascular markers through gene expression profiling of tumor-derived endothelium. BMC Genomics. 2008;9:201. doi: 10.1186/1471-2164-9-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ogawa T, Nikawa T, Furochi H, et al. Osteoactivin upregulates expression of MMP-3 and MMP-9 in fibroblasts infiltrated into denervated skeletal muscle in mice. Am J Physiol Cell Physiol. 2005;289(3):C697–C707. doi: 10.1152/ajpcell.00565.2004. [DOI] [PubMed] [Google Scholar]
- 97.Solinas G, Schiarea S, Liguori M, et al. Tumor-conditioned macrophages secrete migration-stimulating factor: a new marker for M2-polarization, influencing tumor cell motility. J Immunol. 2010;185(1):642–652. doi: 10.4049/jimmunol.1000413. [DOI] [PubMed] [Google Scholar]
- 98.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9(4):239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bächner D, Schröder D, Gross G. mRNA expression of the murine glycoprotein (transmembrane) nmb (Gpnmb) gene is linked to the developing retinal pigment epithelium and iris. Brain Res Gene Expr Patterns. 2002;1(3–4):159–165. doi: 10.1016/s1567-133x(02)00012-1. [DOI] [PubMed] [Google Scholar]
- 100.Kuan CT, Wakiya K, Keir ST, et al. Affinity-matured anti-glycoprotein NMB recombinant immunotoxins targeting malignant gliomas and melanomas. Int J Cancer. 2011;129(1):111–121. doi: 10.1002/ijc.25645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yokota T, Milenic DE, Whitlow M, Schlom J. Rapid tumor penetration of a single-chain Fv and comparison with other immunoglobulin forms. Cancer Res. 1992;52(12):3402–3408. [PubMed] [Google Scholar]
- 102.Schwarzbich MA, Gutknecht M, Salih J, et al. The immune inhibitory receptor osteoactivin is upregulated in monocyte-derived dendritic cells by BCR-ABL tyrosine kinase inhibitors. Cancer Immunol Immunother. 2012;61(2):193–202. doi: 10.1007/s00262-011-1096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hamid O, Snzol M, Pavlick AC, et al. Frequent dosing and GPNMB expression with CDX-011 (CR011-vcMMAE), an antibody-drug conjugate (ADC), in patients with advanced melanoma. J Clin Oncol (Meeting Abstracts) 2010;28(15s):8525. [Google Scholar]
- 104.Saleh MN, Bendell JC, Rose A, et al. Correlation of GPNMB expression with outcome in breast cancer (BC) patients treated with the antibody-drug conjugate (ADC), CDX-011 (CR011-vcMMAE. J Clin Oncol. 2010;28(15s):1095. [Google Scholar]
- 105.Burris H, Saleh MN, Bendell J, et al. A Phase (Ph) I/II Study of CR011-VcMMAE, an Antibody-Drug Conjugate, in Patients (Pts) with Locally Advanced or Metastatic Breast Cancer (MBC) Cancer Res. 2009;69(24s3):6096. [Google Scholar]
- 106.Okeley NM, Miyamoto JB, Zhang X, et al. Intracellular activation of SGN-35, a potent anti-CD30 antibody-drug conjugate. Clin Cancer Res. 2010;16(3):888–897. doi: 10.1158/1078-0432.CCR-09-2069. [DOI] [PubMed] [Google Scholar]
- 107.Yardley DA, Weaver R, Melisko M, et al. A randomized phase 2 study of the antibody-drug conjugate CDX-011 in advanced GPNMB-overexpressing breast cancer: The EMERGE study. Cancer Res. 2012;72(24s3):P6-10-01. [Google Scholar]