Abstract
Purpose
Farnesoid X receptor (Fxr) is a ligand-activated nuclear receptor critical for liver function. Reports indicate that functions of Fxr in the liver may overlap with those of hepatocyte nuclear factor 4α (Hnf4α), but studies of their precise genome-wide interaction to regulate gene transcription are lacking. Thus, we compared the genome-wide binding of Fxr and Hnf4α in the liver of mice and characterized their cooperative activity on binding to and activating target gene transcription.
Methods and Results
ChIP-Seq of mouse livers revealed that nearly 50% binding sites of Fxr and Hnf4α overlap. Co-immunoprecipitation assays showed a direct Fxr-Hnf4α protein interaction dependent on Fxr activity. Hnf4α bound to shared target sites upstream and in close proximity to Fxr. Moreover, genes co-bound by Fxr and Hnf4α are enriched in complement and coagulation cascades and drug metabolism. Furthermore, transcriptional and binding assays suggest that Hnf4α increases Fxr transcriptional activity; however, binding of Hnf4α can be either Fxr-dependent or -independent at different sites.
Conclusion
Our results showed that Fxr cooperates with Hnf4α in the liver to modulate gene transcription. This study provides the first evidence on a genome-wide scale of both cooperative and independent interactions between Fxr and Hnf4α in regulating gene transcription.
Keywords: Fxr, Hnf4α, ChIP-Seq, co-regulation, nuclear receptor interaction
INTRODUCTION
Farnesoid X receptor (FXR in humans/Fxr in rodents) is a member of the group II nuclear receptor superfamily, activated by bile acids (FXR’s endogenous ligands), highly expressed in the liver and intestine, and a master regulator of the enterohepatic circulation of bile acids (1–6). Hepatocyte nuclear factor 4 alpha (HNF4α in humans/Hnf4α in rodents) is a highly conserved orphannuclear receptor that is also essential for liver development, differentiation, and organism survival (7). Fxr and Hnf4α have been shown to regulate the expression of an overlapping set of genes, including apolipoprotein C-III (ApoC-III), cholesterol 7 alpha-hydroxylase (Cyp7a1), and bile acid-CoA:amino acid N-acyltransferase (Baat protein) (8–12), suggesting an overlap of Fxr and Hnf4α functions in the liver. Despite this overlap, no studies have yet determined how Fxr and Hnf4α interact in the liver on a genome-wide scale to regulate gene transcription. However, studies have shown that HNF4α is capable of enhancing the liver-specific functions of group II nuclear receptors. For example, HNF4α cooperatively enhances the transcriptional activity of constitutive androstane receptor (CAR) and pregnane X receptor (PXR) at the CYP3A4 promoter (13). The effects of HNF4α on FXR activity are largely unknown.
In addition to its role in bile acid homeostasis, Fxr also regulates other metabolic processes such as lipid homeostasis, glucose metabolism, insulin sensitivity, and gastrointestinal cancer development and therefore has become a very promising target for the treatment or prevention of cholestasis, hyperlipidemia, fatty liver, type II diabetes, liver and colon cancers (10, 14–22). Recent genome-wide binding studies have shown that Fxr displays a very high degree of tissue-specific binding, which is likely regulated by other tissue-specific co-factors (23). Motif analysis of genome-wide Fxr binding in the liver revealed a nuclear receptor half site (AGGTCA) associated with the Fxr response element, an inverted repeat separated by one nucleotide (IR-1; AGGTCAnTGACCT) (23, 24), indicating the involvement of orphan nuclear receptors in regulating tissue-specific functions of Fxr.
In hepatocytes, the orphan nuclear receptor HNF4α localizes mainly to the nucleus, binds DNA exclusively as a homodimer, and recognizes response elements consisting of direct repeats, namely, direct repeats separated by one nucleotide (DR-1)(25). Hnf4α regulates a myriad of liver-specific functions, including production of clotting factors, apolipoprotein synthesis, and drug metabolism (25). In addition, Hnf4α directly regulates the transcription of Cyp7a1, the rate-limiting enzyme in bile acid synthesis, suggesting that Hnf4α also plays a regulatory role in bile acid homeostasis (8, 12).
Due to reports of overlapping function of Fxr and Hnf4α in liver and evidence suggesting an uncharacterized orphan nuclear receptor co-regulates the transcriptional function of Fxr, we hypothesized that Hnf4α could be responsible for mediating Fxr function in the liver. To test our theory, this study compared the genome-wide binding of Fxr and Hnf4α in mouse liver and characterized these two factors’ cooperation in binding to target gene regions and in activating gene transcription, using chromatin immunoprecipitation (ChIP), massive parallel sequencing, quantitative polymerase chain reaction (qPCR) analysis, co-immunoprecipitation (Co-IP) assays, and luciferase assays.
MATERIALS AND METHODS
Animals
All mice were maintained at an American Animal Associations Laboratory Animal Care-accredited facility at the University of Kansas Medical Center. Animal protocols and procedures were approved by the Institutional Animal Care and Use Committee. For Hnf4α ChIP-qPCR studies, four-month-old fasted male wild-type (WT) and whole body Fxr–knockout (Fxr KO) (5) mice (n=4 per group) were used. WT mice were orally gavaged with vehicle (1% methylcellulose, 1% Triton-100 in PBS) or GW4064 (75 mg/kg) twice a day for 24 h period. GW4064 is an FXR agonist (26) synthesized by the Chemical Discovery Laboratory at the University of Kansas (Lawrence, KS). Fxr KO mice were only gavaged with vehicle. Livers were collected 4 h after the second dose and prepared for Hnf4α chromatin immunoprecipitation followed by qPCR analysis (ChIP-qPCR). Hepatocyte-specific Hnf4α-null (Hnf4α-HNull) mice were generated as previously described (27) and were fed the same rodent chow as the WT control mice. Livers from 45-day-old male Hnf4α-HNull mice and from their WT control littermates (n=4 per group) were used for Fxr ChIP-qPCR assays. For Co-IP assays, 4-month-old C57BL/6 and Fxr KO mice (n=3 per group) we refed a control diet or a diet supplemented with 1% (w:w) cholic acid (CA) for 5 days. Liver whole-cell lysates were prepared and used for Co-IP analysis.
ChIP Followed by Massive Parallel Sequencing (ChIP-Seq)
ChIP-Seq analysis of Fxr and Hnf4α in mouse liver was done to determine the degree of genome-wide overlapping in binding. Original ChIP-Seq data were obtained from mouse livers generated as previously described (23, 28). Raw Fxr and Hnf4α ChIP-Seq data from single end sequencing on an Illumina Genome Analyzer, obtained from in-house or online databases, were re-analyzed using Model-based Analysis of ChIP-Seq (MACS) (29). Total Fxr binding sites were compared with total Hnf4α binding sites in the liver. The binding frequency of Hnf4α, or number of Hnf4α binding events, relative to the distance of the Fxr binding site within shared target genes was analyzed using BEDTools (30). Histograms of Fxr and Hnf4α binding to the Nr0b2 (small heterodimer partner, Shp) gene were generated using the UCSC Genome Browser (University of California, Santa Cruz) (31).
Peaks identified in ChIP-Seq data that were shared by Fxr and Hnf4α in the liver of mice were analyzed for pathway enrichment using the Functional Annotation Tool in the Database for Annotation, Visualization, and Integrated Discovery (32). P-value less than or equal to 0.05 were considered statistically significant.
ChIP-qPCR
ChIP-qPCR analysis was done on shared Fxr and Hnfα binding regions identified by ChIP-Seq analysis to validate genome-wide analysis and to determine degree of cooperative binding of these two factors. ChIP-quality antibodies for mouse Fxr and Hnf4α were obtained from Santa Cruz Biotechnology (H-130 and C-19). Antibody specificity for Fxr has been shown in previous genome-wide binding analysis (23), and for Hnf4α is demonstrated in Supplemental Fig. 1. For Hnf4α ChIP-qPCR assays, we used livers from WT and Fxr KO mice treated with or without GW4064 and Fxr KO mice treated with vehicle control (n=4). For Fxr ChIP-qPCR assays, we used livers from WT and Hnf4α-HNull mice (n=4) as previously described (23). Purified IP DNA fragments were analyzed by qPCR with primers amplifying shared Fxr and Hnf4α binding sites: Apoc3, Apoe, Baat, Nr0b2 promoter and 3′ regulatory region, and Sqstm1. We also analyzed Fxr and Hnf4α binding to fragments of genes involved in complement and coagulation cascades: C2 (−50 to 0 bp upstream TSS), C3 (−225 to −275 bp upstream TSS), F2 (−425 to −475 bp upstream TSS), C4b (−17125 to −17175 bp upstream TSS), Cfb (−150 to −200 bp upstream TSS), Fga (−200 to −250 bp upstream TSS), and Plg (−125 to −175 bp upstream TSS). Fxr has previously been shown to bind within the second intron of the Fgf15 gene (1880 to 1980 bp downstream TSS) in mouse intestine but not the liver (23). This region has also been shown not to be a binding region of Hnf4α by ChIP-Seq analysis. Therefore this region was originally used as a negative control for Fxr-Hnf4α co-localization experiments. These above target regions were selected for ChIP-qPCR validation and analysis because they belong to pathways highly co-bound by Fxr and Hnf4α, as revealed by ChIP-Seq analysis, and due to their physiologically significant roles in bile acid, lipid, and coagulation pathways. All primers used for ChIP-qPCR are presented in Supplemental Table I. Quantitative PCR reactions were carried out using Maxima™ SYBR Green (Fermentas Molecular Biology Tools). Data were analyzed as fold change over values from vehicle-treated WT mice.
Co-Immunoprecipitation
To investigate whether Fxr and Hnf4α have a protein-protein interaction, we performed Co-IP assays using a kit from Invitrogen on whole-cell liver extracts from WT mice fed with or without 1% CA and from Fxr KO mice fed a control diet. Whole-cell lysates from mouse livers were prepared according to protocol and then immunoprecipitated using an antibody against Fxr (H-130, n=3 each group). Immunoprecipitates from each group were pooled and analyzed by standard Western blot using antibody sc-6556 to detect Hnf4α (Santa Cruz Biotechnology).
Construction of Plasmids for Reporter Gene Luciferase Assay
Luciferase assays were done to determine the transcriptional effects of FXR and Hnf4α on shared target regions. Specifically, the transcriptional activity of FXR/retinoid x receptor alpha (RXRα) and Hnf4α were tested on reporter vectors containing shared Fxr-Hnf4α binding regions within the promoter and the downstream regulator region of Nr0b2 (Shp), the first intron of scavenger receptor class B type 1 (Scarb1; Sr-b1), and the downstream regulatory region of Sqstm1 (p62). Supplemental Table II lists the location and relative Fxr and Hnf4α binding counts in these regions. Reporter vectors of Shp promoter and downstream regulatory region were cloned as previously reported (33). An active Hnf4α binding site located 70 bp upstream of the Baat(Bat) gene transcriptional start site (TSS), previously reported (9), was used as a positive control for HNF4α transcriptional activity. For this study, this 600 bp region upstream of the Bat gene, a region shown to have high Hnf4α activity (9), was amplified from mouse genomic DNA by PCR using pairs of primers containing XhoI and HindIII restriction enzyme sites and cloned upstream of the luciferase gene within the pGL4.23 firefly luciferase vector (Promega). The primers used to generate Bat reporter vector were Forward: 5′-CACAACTCGAGAATGGCTAAGACTATAGAT-3′ and Reverse: 5′-CTGAGGAAGCTTTCTTAGTATTTCCCTCCTC-3′. A 600 bp region around Fxr and Hnf4α binding sites within the first intron of Sr-b1 located 10.7 and 21.5 Kb downstream of the TSS, respectively, has been previously reported to be a Fxr binding site (34). These regions were amplified from mouse genomic DNA by PCR using pairs of primers containing XhoI and BglII restriction enzyme sites. The PCR products, Sr-b1 #1 and Sr-b1 #2, were subcloned upstream of the luciferase gene into the pGL4.23 firefly luciferase vector as previously reported (34). A 2 Kb region of the p62 gene containing Fxr and Hnf4α binding sites, located around 13.1 Kb downstream of the p62 gene TSS, has recently been determined to be an Fxr binding site (35). This region was cloned into a pGL4-TK luciferase vector (Promega) as previously reported (35). All constructs were confirmed by DNA sequencing.
Cell Culture, Transient Transfection, and Luciferase Reporter Gene Assays
Chinese hamster ovary (CHO) cells were cultured at 70–90% cell density in high-glucose Dulbecco’s modified Eagle medium supplemented with 1% penicillin/streptomycin, 1% L-proline (50 μg/mL), and 10% fetal bovine serum (Omega Scientific) and were transiently transfected by reverse transfection methods using TurboFect (Fermentas Molecular Biology Tools) with the various reporter gene constructs as well as pCMV-ICIS human FXR and/or pCMV-SPORT6 mouse Hnf4α (Open Biosystems), PSG5 human RXRα, and phRG-TK-Renilla luciferase vector (Promega, no longer available; see pGL4.74) according to protocol. Human FXR is highly homologous to mouse Fxr and has often been used to test transcriptional activity on mouse gene binding sites (33, 35, 36). After 24 h, cells were treated with 100 nM GW4064 or 0.1% dimethyl sulfoxide(control); cells from the Hnf4α-alone groups were not treated. Firefly luciferase and Renilla luciferase activities were quantified 24 h post-treatment using a Dual-Glo Luciferase Assay System (Promega).
FXR/RXRα expression vectors were co-transfected with increasing amounts (3, 10, and 30 ng) of Hnf4α expression vector with 100 nM GW4064, a FXR synthetic ligand. We used the promoter and downstream regions of Shp, the intron of Sr-b1, and the downstream region of p62 cloned into luciferase expression vectors to assess the effects of Hnf4α on the transcriptional activity of FXR. The transcriptional activity of increasing amounts of Hnf4α expression vector (10, 50, and 100 ng or 10 and 100 ng) on these regions, as well as on a positive control gene, Bat, was also measured by luciferase assay. The firefly luciferase activity value was normalized as a ratio over Renilla luciferase and expressed as firefly luciferase activity/Renilla. The data are presented as the average of six wells ± SE, and the experiments were repeated at least twice.
Statistical Analysis
All data are presented as mean ± SE. Statistical difference between the two groups was analyzed by Student’s t-test. P-values ≤ 0.05 were considered significant.
RESULTS
Genome-wide Fxr and Hnf4α Binding Sites in Mouse Liver
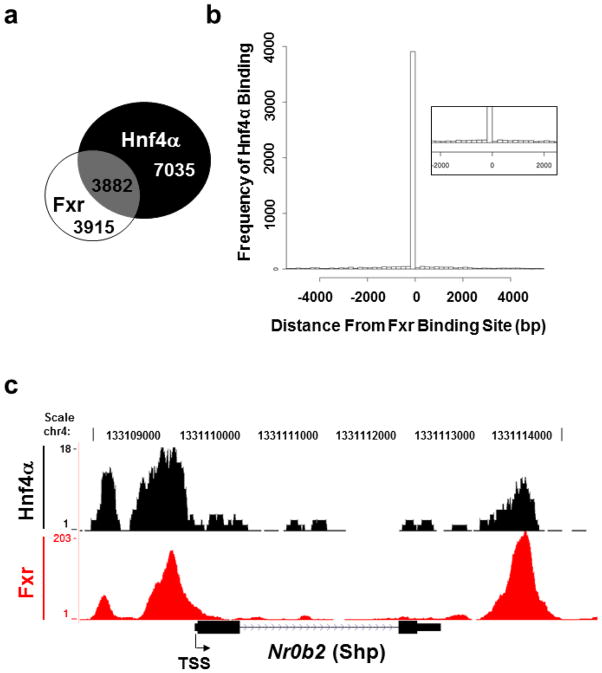
ChIP-Seq binding data of Fxr and Hnf4α in mouse liver from previous reports (23, 28) were re-analyzed using MACS. We found 10917 total binding sites for Hnf4α and 7797 for Fxr, of which 3882 overlap; 50% of total Fxr binding sites co-localize with Hnf4α (Fig 1a). Hnf4α and Fxr do not bind to same site; rather, the frequency (y-axis) of Hnf4α binding to shared target genes was greatest when bound upstream and in close proximity to an Fxr binding site (x-axis; location of collective Fxr binding sites are represented by “0”; Fig. 1b).
Fig. 1.
Genome-wide binding of Fxr and Hnf4α in mouse liver. Previously reported Fxr and Hnf4α ChIP-Seq data was reanalyzed using MACS (23, 28). (a) Venn diagram of total Hnf4α and Fxr binding sites in mouse liver as revealed by chromatin immunoprecipitation followed by massively parallel sequencing (ChIP-Seq) analysis. We found 10917 total Hnf4α binding sites and 7797 total Fxr binding sites in mouse liver, of which 3882 (nearly 50%) overlapped. (b) Histogram of the binding frequency, or number of binding events, of Hnf4α (y-axis) in relation to distance from the Fxr binding site (x-axis) to the shared target genes in mouse liver. Collective Fxr binding sites are represented by “0.” (c) Histogram of Fxr (red) and Hnf4α (black) binding to the Nr0b2 (Shp) gene in mouse liver as determined by ChIP-Seq analysis. This histogram was generated using the UCSC Genome Browser (University of California, Santa Cruz).
Pathway analysis of shared Fxr and Hnf4α target genes revealed pathways involving complement and coagulation cascades had the highest number of genes targeted by Fxr and Hnf4α (Table I). P-values and Bonferonni scores illustrate the degree of significance of Fxr and Hnf4α co-localization within the pathway. P-values ≤ 0.05 were considered statistically significant. Table II lists the shared target genes categorized by the Kyoto Encyclopedia of Genes and Genomes pathway maps as part of complement and coagulation cascades, the locations of Fxr and Hnf4α binding sites in relation to the genes’ TSS and the relative binding events (counts) of each factor.
Table I.
Pathways enriched by both FXR and Hnf4α binding in mouse livers
| Pathway | Genes* | % bound by FXR and HNF4α | P-Value | Bonferroni |
|---|---|---|---|---|
| Complement and coagulation cascades | 17 | 2.24 | 1.38E-07 | 2.25E-05 |
| Drug metabolism | 16 | 2.11 | 8.33E-07 | 1.36E-04 |
| PPAR signaling pathway | 16 | 2.11 | 1.67E-06 | 2.73E-04 |
| Metabolism of xenobiotics by cytochrome P450 | 13 | 1.71 | 2.97E-05 | 4.83E-03 |
| Insulin signaling pathway | 12 | 1.58 | 4.55E-02 | 0.999 |
| Glycine, serine and threonine metabolism | 9 | 1.18 | 6.15E-05 | 9.97E-03 |
| Steroid hormone biosynthesis | 9 | 1.18 | 7.58E-04 | 0.116 |
| Adipocytokine signaling pathway | 9 | 1.18 | 9.80E-03 | 0.799 |
| Pyruvate metabolism | 8 | 1.05 | 2.07E-03 | 0.287 |
| Fatty acid metabolism | 8 | 1.05 | 3.59E-03 | 0.443 |
| Drug metabolism | 8 | 1.05 | 5.19E-03 | 0.572 |
| Retinol metabolism | 8 | 1.05 | 3.20E-02 | 0.995 |
| Cysteine and methionine metabolism | 7 | 0.92 | 3.10E-03 | 0.397 |
| Linoleic acid metabolism | 7 | 0.92 | 1.61E-02 | 0.929 |
| Biosynthesis of unsaturated fatty acids | 6 | 0.79 | 6.32E-03 | 0.644 |
| Starch and sucrose metabolism | 6 | 0.79 | 2.13E-02 | 0.97 |
| ABC transporters | 6 | 0.79 | 4.99E-02 | 1 |
| Selenoamino acid metabolism | 5 | 0.66 | 1.80E-02 | 0.948 |
| Alanine, aspartate and glutamate metabolism | 5 | 0.66 | 4.36E-02 | 0.999 |
| Primary bile acid biosynthesis | 4 | 0.53 | 2.74E-02 | 0.989 |
Note:
indicates the number of genes bound by FXR and Hnf4α in this pathway
Table II.
List of target genes within complement and coagulation cascades bound by both FXR and Hnf4α
| Official Gene Symbol | Binding Site Location from TSS | FXR Total Counts* | HNF4α Total Counts* |
|---|---|---|---|
| Plg | −144, 1992, 6986, 9608 | 99, 155, 127, 95 | 46, 39, 31, 33 |
| Fga | −225, −5561 | 150, 105 | 40, 53 |
| Cpb2 | −9013, 9 | 52, 34 | 38, 19 |
| Serpina1e | −4534, 3769 | 76, 35 | 62, 27 |
| Fgg | −345, −4534, 1904, 2607 | 70, 197, 20, 28 | 9, 47, 26, 25 |
| F2 | −436 | 129 | 67 |
| Mbl1 | −44, 9558 | 192, 156 | 24, 64 |
| Cfh | 21 | 109 | 21 |
| Kng2 | −125, −10481 | 69, 40 | 39, 17 |
| Serpine1 | −507 | 115 | 33 |
| Cfb | −183 | 217 | 78 |
| Serpinf2 | −66 | 41 | 22 |
| C4b | −17142 | 93 | 83 |
| C2 | −4 | 68 | 46 |
| C3 | −236, −2276, −2788, −5187 | 884, 75, 69, 175 | 46, 15, 10, 55 |
| Proc | −1366 | 77 | 53 |
| Kng1 | −124, −1946 | 96. 24 | 35, 21 |
Counts are the number of binding events recorded for FXR and HnfF4α at each location.
Fig. 1c is a histogram of binding of Fxr (red) and Hnf4α (black) to the Nr0b2 gene in mouse liver, generated by the UCSC Genome Browser (31). Both the promoter and downstream FXR binding sites co-localized with those of HNF4α. The binding of Hnf4α to the Nr0b2 promoter has previously been described (37). However, the binding of Hnf4α to the 3′ end of the Nr0b2 gene and the co-localization with Fxr at these regions are novel findings. Sequence analysis of these regions by NUBIScan (38) showed a putative HNF4α binding motif, DR-1, located in the promoter of the Nr0b2 gene (within −320 to −220 bp upstream of TSS) but not in the downstream regulatory region (data not shown), whereas a classical FXR binding motif, IR-1, has been identified in both of these regions (23, 33).
Dependence of Fxr and Hnf4α for Binding to Shared Target Genes
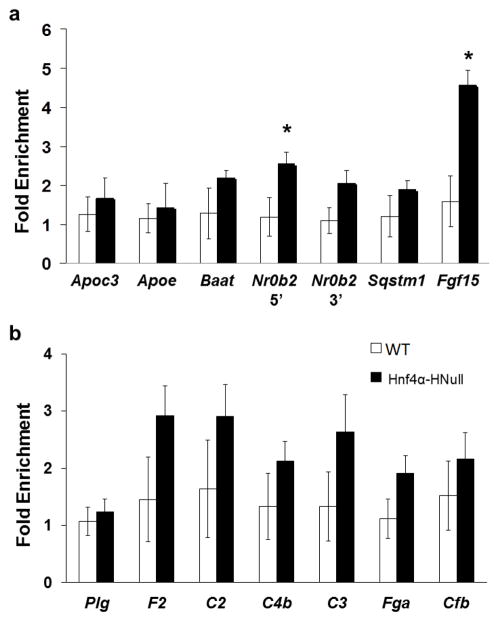
Supplemental Table II summarizes the binding site locations and counts of Fxr and Hnf4α to select shared target genes, including to the 5′ and 3′ end of Nr0b2, revealed by ChIP-Seq analysis. These regions were assessed for Fxr and Hnf4α binding by ChIP-qPCR and luciferase assay. Fxr binding increased nearly 2-fold in Hnf4α-HNull mice at sites located within the Baat gene promoter, the 5′ and 3′ regions of the Nr0b2 gene, the downstream regulatory region of Sqstm1 gene, and a non-shared target site within the Fgf15 gene (Fig. 2a). This increase was only statistically significant for Fxr binding at the 5′ end of Nr0b2 and Fgf15 (*P-value ≤ 0.05). Fgf15 was thought to be an FXR target gene in mouse intestine and not liver (23), and was shown to not be bound by Hnf4α. Therefore, this region was originally used as a negative control region for co-localization. Fxr binding to promoters of Apoc3 and Apoe did not change with Hnf4α deficiency.
Fig. 2.
ChIP-qPCR analysis of Fxr binding to shared target genes. QPCR analysis was performed on DNA fragments immunoprecipitated with Fxr antibody. ChIP-qPCR data are reported as fold increase (y-axis) of Fxr binding in Hnf4α-HNull (black bar) mouse liver compared to WT (white bar) mouse liver. *P-value ≤ 0.05. (a) ChIP-qPCR results of Fxr binding to shared target regions in WT and Hnf4α-HNull mouse liver. Regions within the Apoc3 promoter, the Apoe promoter, the Baat promoter, the promoter and 3′ region of Nr0b2, and downstream of the Sqstm1 TSS are shared target sites of Fxr and HNF4α as revealed by ChIP-Seq analysis of mouse liver (Supplemental Table II). Fgf15 is a target gene of Fxr (but not Hnf4α) in the intestine but not the liver and therefore was originally used as a negative control. (b) ChIP-qPCR results of Fxr binding to shared target regions of genes categorized within complement and coagulation cascades in WT and Hnf4α-HNull mouse liver. These binding sites are located within the promoters (within 500 bp upstream of TSS) of Plg, F2, C2, C3, Fga, Cfb, and −17125 to −17175 bpupstream of C4b gene TSS (Table II).
We also analyzed Fxr binding to genes involved in complement and coagulation cascades (Fig. 2b) in WT versus Hnf4α-HNull mouse liver. Fxr did bind to shared regions in genes of the complement and coagulation cascade within both WT and Hnf4α-HNull mouse liver. Fxr binding events increased in Hnf4α-deficient mice 1.6–2-fold at genes Fga, C3, C4b, C2, and F2 but did not change at binding sites within Cfb and Plg.
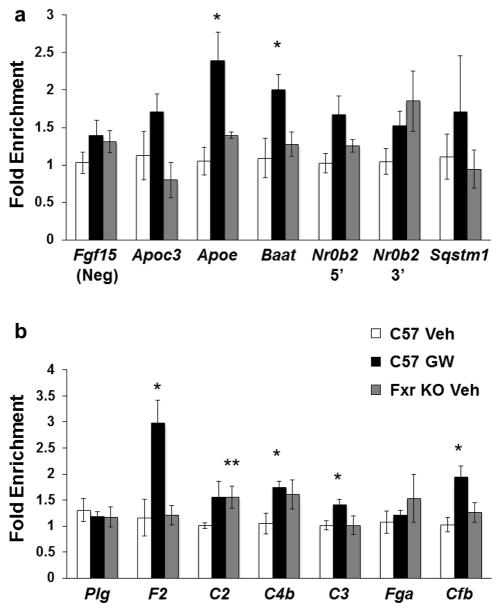
The Hnf4α binding pattern to shared target genes in WT mouse liver treated with or without Fxr ligand GW4064 and in Fxr KO mouse liver, varied at different target sites (Fig. 3). When compared with vehicle-treated WT liver, Hnf4α binding events in WT mice treated with GW4064 increased at shared target sites within Apoc3 (1.5-fold), Apoe (2.3-fold), Baat (1.8-fold), Nr0b2 (1.6- and 1.5-fold), and Sqstm1 (1.5-fold) but not in the negative control region (Fgf15) (Fig. 3a; *P-value < 0.05). Overall, Hnf4α binding to Apoe, Baat, Nr0b2 5′, and Sqstm1 regions did not decrease below baseline in Fxr KO mouse livers. There was a slight non-significant reduction in Apoc3 binding, and binding to Nr0b2 3′ remained elevated in Fxr KO mouse liver. Hnf4α binding increased in mouse liver treated with GW4064 at genes F2 (2.6-fold), C2 (1.6-fold), C4b (1.7-fold), C3 (1.4-fold), and Cfb (1.9-fold) (Fig. 3b; *P-value < 0.05). As seen with the previous regions above, Hnf4α binding to F2, C3, and Cfb did not decrease below baseline in FxrKO mouse liver. However, Hnf4α binding remained elevated at regions within C2 and C4b (**P-value < 0.05). Hnf4α binding was not regulated by Fxr activity at binding sites within Plg or Fga.
Fig. 3.
ChIP-qPCRof Hnf4α binding to shared target genes.(a) Fxr shares the Hnf4α binding regions within the Apoc3 promoter, the Apoe promoter, the Baat promoter, the promoter and 3′ region of Nr0b2, and downstream of the Sqstm1 TSS, revealed by ChIP-qPCR (Supplemental Table II). The Fgf15 binding site was determined not to be an Hnf4α binding site and therefore was used as a negative control. Hnf4α binding to these regions was investigated in WT and Fxr KO vehicle and WT GW4064 treated mouse liver. Data are reported as fold enrichment (y-axis) of Hnf4α binding in WT GW (black bar) or Fxr KO veh (gray bar) treated mouse liver normalized to WT veh (white bar) treated mouse liver. (b) ChIP-qPCR data of Hnf4α binding to shared target regions within genes categorized as part of complement and coagulation cascade. These binding sites are located within the promoters (within 500 bp upstream of TSS) of Plg, F2, C2, C3, Fga, Cfb, and −17125 to −17175 bp upstream of C4b gene TSS (Table II). Data are reported as described in part (a). *P-value≤0.05 of C57 GW treated group compared to C57 veh group. **P-value ≤ 0.05 in Fxr KO vehicle group compared to C57 veh group.
Interaction between Fxr and Hnf4α and Dependence of FXR and Hnf4α for Activating Target Genes
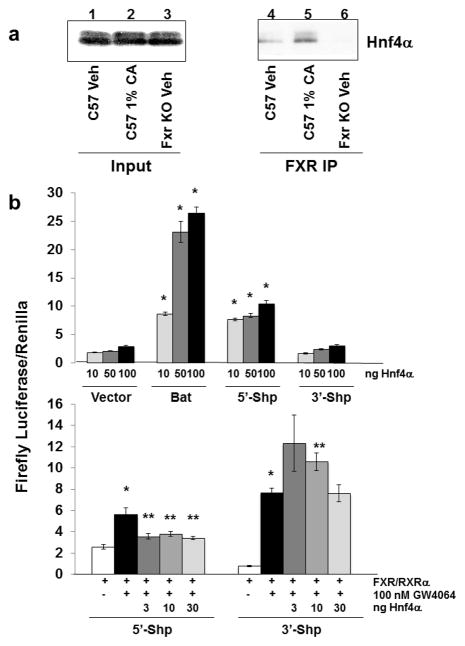
A modest Fxr-Hnf4α protein-protein interaction was detected in WT mice fed a control diet (Fig. 4a). This interaction increased in mice fed a 1% CA diet but was nearly undetectable in Fxr KO mice (Fig. 4a).
Fig. 4.
Co-IP of Fxr and Hnf4α and luciferase assays of Shp regulatory regions. (a) Co-IP of WT mice fed control or 1% CA diet and Fxr KO mice fed control diet. Whole cell liver lysates were prepared and immunoprecipitated using an antibody against Fxr. Liver lysates (input) and IP fractions were pooled and analyzed by Western blot analysis using antibody against Hnf4α. Lanes 1–3 show the levels of Hnf4α within 15 μg of pooled whole-cell lysates from WT control diet (Lane 1), WT 1% CA diet (Lane 2), and Fxr KO control diet (Lane 3) groups. Lane 4–6 show levels of Hnf4α detection within WT control diet (Lane 4), WT 1% CA diet (Lane 5), and Fxr KO control diet (Lane 6) liver lysates immunoprecipitated with an Fxr antibody. (b) Luciferase expression assays showing the effects of increasing amounts of mouse Hnf4α expression vector (10, 50, 100 ng) on transcriptionally activating regulatory regions within the Bat gene promoter, the 5′ and 3′ regulatory regions of the Shp gene, or the luciferase vector control (top panel). Bottom panel shows the effects of increasing mouse Hnf4α expression vector amounts (3, 10, and 30 ng) on FXR-induced transcription of the 5′ and 3′ regulatory regions of the Shp gene after activation of FXR with 100 nM of GW4064. Results are reported as a ratio of firefly luciferase activity over Renilla luciferase activity (y-axis). *P-value ≤ 0.05 of HNF4α transfected groups when compared to vector or FXR/RXRα GW treated groups compared to veh control. **P-value ≤ 0.05 of Hnf4α transfected GW treated groups compared to FXR/RXRα alone GW treated groups.
Hnf4α binding to Shp promoter and downstream regulatory region was detected by ChIP-Seq analysis (Fig. 2a). These binding sites were analyzed for Hnf4α transcriptional activity using luciferase reporter assays. Transcriptional activity of Hnf4α on the Bat gene promoter, which has already been characterized (9), served as a positive control. Results showed that although Hnf4α significantly increased luciferase activities of the Shp and Bat promoter when compared to vector control, with 100 ng Hnf4α having the highest activity (*P-value ≤ 0.05). Hnf4α did not affect the downstream regulatory region of Shp (Fig. 4b, top panel).
FXR has previously been shown to transcriptionally regulate the promoter and downstream regulatory region of the Shp gene (33). Our results confirm that FXR significantly increases luciferase activity at both of these sites (*P-value ≤ 0.05; Fig. 4b, bottom panel). In addition, Hnf4α increased transcriptional activity of FXR on Shp3′ region nearly 2- and 1.4 fold (**P-value ≤ 0.05) at 3 and 10 ng. However, Hnf4α appears to have slightly and significantly decreased transcriptional activity of FXR at the Shp promoter (**P-value ≤ 0.05).
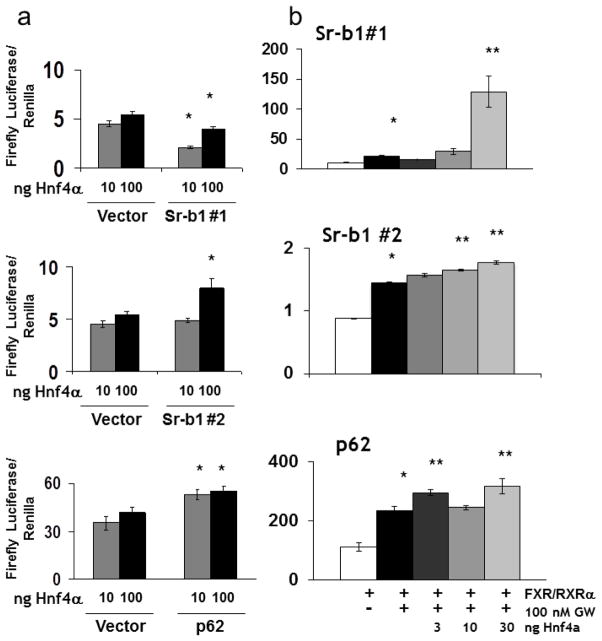
Hnf4α significantly reduced luciferase activity of the Sr-b1#1 (10.7 Kb regulatory region downstream of the TSS, Supplemental Table II), 2-fold for 10 ng and 1.3-fold for 100 ng of Hnf4α expression vector (*P-value ≤ 0.05; Fig 5a top). Conversely, Hnf4α significantly increased luciferase activity of the Sr-b1#2 site (21.5 Kb regulatory region downstream of the TSS, Supplemental Table II) and p62 in a dose-dependent manner (*P-value ≤ 0.05 at 100 ngfor Sr-b1 #2 and at 10 and 100 ng for p62; Fig 5a middle and bottom). Next, the effects of Hnf4α on FXR transcriptional activity of binding sites within Sr-b1and p62 were tested (Fig. 5b). FXR significantly increased transcriptional activity of two binding sites within the Sr-b1gene (#1 and #2) and downstream regulatory region of p62 (*P-value ≤ 0.05), which is consistent with previous reports (34, 35). Hnf4α increased FXR-induced transcriptional activity at each of these sites. Interestingly, even though Hnf4α alone moderately decreased luciferase activity at Sr-b1#1 (Fig. 5a, top), Hnf4α synergistically and significantly enhanced the FXR activity nearly 20-fold in this region at 30 ng of Hnf4α expression vector (**P-value ≤ 0.05 for 3 and 30 ng; Fig. 5b, top). Hnf4α only moderately enhanced FXR’s transcriptional activity at Sr-b1 #2 in a weakly additiveand dose-dependent manner (**P-values ≤ 0.05; Fig. 5b, middle). Finally, Hnf4α significantly enhanced FXR transcriptional activity of p62, although this effect was saturated at 3 ng of Hnf4α expression vector and again was weakly additive (**P-value ≤ 0.05 for 3 and 30 ng; Fig. 5b, bottom). Collectively, these results indicate that Hnf4α can enhance FXR activity in an additive and synergistic manner.
Fig. 5.
Transcriptional effects of Hnf4α alone and with FXR at shared target genes by luciferase assays. (a) Luciferase expression assays showing the effects of increasing amounts Hnf4α expression vector (10 and 100 ng) on transcriptionally activating regulatory regions within 10.6 Kb (#1; top) and 21.5 Kb (#2; middle) downstream of theSr-b1TSS, in the downstream regulatory region of p62 (bottom), and in the luciferase vector controls. (b) Effects of increasing amounts of Hnf4α expression vector (3, 10, and 30 ng) on FXR-induced transcription of regulatory regions within the Sr-b1 gene (#1 (top) and #2 (middle)) and p62 (bottom) after activation of FXR with 100 nM of GW4064. Results of luciferase assays (both [a] and [b]) are reported as a ratio of firefly luciferase activity over Renilla luciferase activity (y-axis). *P-value ≤ 0.05 of Hnf4α transfected groups when compared to vector or FXR/RXRα GW treated groups compared to veh control. **P-value ≤ 0.05 of Hnf4α transfected GW treated groups compared to FXR/RXR alone GW treated groups.
DISCUSSION
Our findings demonstrate that genome-wide Hnf4α and Fxr DNA-binding sites have a very high degree of overlap in mouse liver and that these shared target genes are highly enriched within the genes involved in complement and coagulation cascades. Furthermore, within shared target regions, these two nuclear receptors bind in close proximity and exhibit a protein-protein interaction dependent on Fxr activation. Deficiency of Fxr or Hnf4α affects the binding of either to target genes in a gene-selective manner. We conclude that Fxr and Hnf4α likely regulate transcription of some shared target genes independently of each other, as seen with apolipoprotein genes, but cooperate to regulate transcription of other shared target genes, such as genes involved in bile acid homeostasis.
There are several potential explanations for why Fxr binding increases at select shared target genes in the absence of functional Hnf4α, which is opposite to the original hypothesis. Hnf4α and Fxr may compete for the same binding site to regulate transcription of the target gene, as illustrated in previous studies showing that Fxr displaces Hnf4α binding to the Apo C-III promoter and inversely regulates transcription of this gene (10). However, in the current study, ChIP-Seq analysis on a genome-wide scale shows Hnf4α binding not at the same location as Fxr but rather upstream of the Fxr binding site. We think that increased FXR binding is due to the increase in endogenous ligands of Fxr in the Hnf4α mice, because loss of Hnf4α leads to a marked increase in bile acid concentration (8, 9), which likely further activates Fxr. This is illustrated by the increase in Fxr binding within a region of Fgf15, which is not typically an Fxr target gene in the liver under normal conditions. Therefore, our results suggest complex interactions among nuclear receptors via direct interaction at the chromatin level or via the modification of endogenous ligands.
Furthermore, we found that increased Hnf4α binding to shared target genes depended on Fxr activity at some target regions, including the promoter of the Baat gene and the 3′ regulatory region of Nr0b2. Interestingly, baseline binding of Hnf4α to the 3′ end of the Nr0b2 gene increased in Fxr KO mice compared with WT mice. This trend was also seen at C4b and C2. This observance possibly indicates the ability of Hnf4α to compensate for the loss of FXR at these regions. The dependence of Hnf4α binding on Fxr activity within complement and coagulation genes also showed differential regulation. Indeed, Hnf4α binding increased at binding sites within F2, C2, C3, C4b, and Cfb genes, but not at Plg or Fga genes, after Fxr activation. The increased binding at these five regions was shown to depend on the activation of Fxr at F2, C3, and Cfb. The mechanism responsible for the differential binding of Hnf4α in the absence of Fxr is unknown but could be due to direct modification of Hnf4α binding to chromatin, post-translational modification of Hnf4α, a change in Hnf4α ligand availability, and/or as mentioned, the compensation of Hnf4α for Fxr loss. Further studies will be needed to clarify the underlying mechanism.
Similarly, Hnf4α to transcriptional assays indicated that Hnf4α binding detected by ChIP-Seq analysis is not directly correlated to transcriptional regulation of genes. For example, Hnf4α showed a low level of binding to the 3′ end of the Nr0b2 gene as well as to sites 10.7 Kb downstream of the Sr-b1 TSS. However, neither of these sites was transcriptionally activated by Hnf4α. Although Hnf4α alone did not elicit transcription of these sites, it did moderately enhance FXR’s transcriptional activity in both of these regions, suggesting that Hnf4α regulates FXR activity or other factors, possibly via modifying the chromatin structure. Furthermore, Hnf4α alone induced transcriptional activity of the Nr0b2 promoter but seemed to have a slight, insignificant inhibitory effect on FXR’s transcriptional activity in this region.
Future studies will determine whether HNF4α interacts with FXR to stabilize localized chromatin environments surrounding gene loci. A recent study has shown that Fxr binding to the 5′ and 3′ end of Nr0b2 mediates a head-to-tail chromatin loop around the gene (33). This may be an essential process required for the efficient transcription of the Nr0b2 gene in response to Fxr activation. ChIP-Seq data demonstrates that Hnf4α also co-localizes with Fxr to the 5′ and 3′ regions of Nr0b2, suggesting that Hnf4α may be important for mediating the Fxr-induced head-to-tail chromatin loop around the Nr0b2 gene.
Other studies have demonstrated that Fxr and Hnf4α have opposite effects on gene transcription of shared target genes. Studies show that Hnf4α binding increases the transcription of ApoC-III, a well-characterized Hnf4α target gene (11, 39). Fxr inhibits the transcription of ApoC-III by binding to its promoter region (10). However, none of these studies have examined the transcriptional effect of Hnf4α and Fxr on shared targets genes on a genome-wide scale. Our analysis suggests these two factors can have cooperative, compensatory, or independent effects on the transcription of target genes. In addition, although not completely demonstrated here, these factors can have an antagonistic effect on gene transcription as previously reported (10).
It is interesting to note that the degree of induction in binding in experimental groups was small or lacked statistical significance. We have seen increasing numbers of genes being regulated in this way despite strong binding of transcription factors (TFs) revealed by ChIP-Seq analysis (23), and due to valid negative control comparisons, we do not believe this to be a result of false positive binding. One explanation for this observance could be the inability of antibody-TF interactions from experimental technology to enrich small fractions of desired TF-DNA interactions from whole tissues. Furthermore, previous publications from our group have reported a similar phenomenon with Fxr (23). In this study, it was argued due to potential constitutive TF binding; ligand activation of the TF does not necessarily result in increased localization of TF to target DNA, but rather changes the recruitment of co-repressors to co-activators.
Nevertheless, this study provides the first line of evidence for interactions between Fxr and other nuclear receptors on a genome-wide scale in mouse liver. FXR and HNF4α have been shown to be highly homologous between mouse and human (36, 40), and have similar functions between these species (41). Therefore, information gained from these mouse models will likely reveal similar FXR-HNF4α interactions in human. It was originally thought that HNF4α could regulate FXR activity similar to how forkhead box protein A1 (FOXA1), otherwise known as hepatocyte nuclear factor 3-alpha, directs estrogen receptor alpha genome-wide binding (42, 43). However, this study revealed a more complex Fxr-Hnf4α interaction in mouse liver that was both Fxr-dependent and –independent, illustrated anindirect cross-talk resulting from disruption of bile acid homeostasis in Fxr and Hnf4α deficient mice, and implicated chromatin remodeling as a mechanism of cooperative activity between these two factors. These studies help broaden our understanding of nuclear receptor function and the complicated interactions they have with other transcriptional machinery that is necessary to fine-tune target gene transcription.
CONCLUSION
In summary, our results reveal a high percentage of co-localized Fxr binding to Hnf4α in mouse liver. We conclude that Fxr and Hnf4α cooperate to a moderate extent to regulate gene transcription and share a direct protein interaction. They likely regulate transcription of target genes in both a dependent and independent manner and can cooperate or antagonize the activity of the other. Our findings suggest that both factors can compensate for the other’s deficiency at certain sites and this compensation may be a mechanism important for maintaining cellular integrity and homeostasis. Despite a direct Fxr-Hnf4α interaction, it is unlikely that Hnf4α is a major determining orphan nuclear receptor responsible for directing tissue-specific binding of Fxr. Nonetheless, the Fxr-Hnf4α interaction could play a critical role in certain diseased systems and/or within specific cellular pathways such as complement and coagulation cascades or drug metabolism and should be further investigated.
Supplementary Material
Acknowledgments
This work was supported by CPRIT Training Grants RP101502 to The University of Texas MD Anderson Cancer Center (AMT), DK031343 (GLG), DK090036 (GLG), and a Madison and Lila Self Graduate Fellowship from the University of Kansas (SNH).
ABBREVIATIONS
- ApoC-III
apolipoprotein C-III
- CA
cholic acid
- ChIP-Seq
chromatin immunoprecipitation followed by massively parallel sequencing
- ChIP-qPCR
chromatin immunoprecipitation followed by quantitative polymerase chain reaction
- Co-IP
co-immunoprecipitation
- Cyp7a1
cholesterol 7 alpha-hydroxylase
- DR-1
direct hexanucleotide repeat separated by 1 nucleotide
- FXR/Fxr
farnesoid X receptor
- HNF4α/Hnf4α
hepatocyte nuclear factor 4 alpha
- IR-1
inverted hexanucleotide repeat separated by 1 nucleotide
- KO
knockout
- RXRα
retinoid x receptor alpha
- Shp
small heterodimer partner
- Sr-b1
scavenger receptor class B type 1
- TSS
transcriptional start site
References
- 1.Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, et al. Bile Acids: Natural Ligands for an Orphan Nuclear Receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- 2.Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, et al. Identification of a Nuclear Receptor for Bile Acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- 3.Wang H, Chen J, Hollister K, Sowers LC, Forman BM. Endogenous Bile Acids Are Ligands for the Nuclear Receptor Fxr/Bar. Mol Cell. 1999;3:543–553. doi: 10.1016/s1097-2765(00)80348-2. [DOI] [PubMed] [Google Scholar]
- 4.Seol W, Choi HS, Moore DD. Isolation of Proteins That Interact Specifically with the Retinoid X Receptor: Two Novel Orphan Receptors. Mol Endocrinol. 1995;9:72–85. doi: 10.1210/mend.9.1.7760852. [DOI] [PubMed] [Google Scholar]
- 5.Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, et al. Targeted Disruption of the Nuclear Receptor Fxr/Bar Impairs Bile Acid and Lipid Homeostasis. Cell. 2000;102:731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 6.Kok T, Hulzebos CV, Wolters H, Havinga R, Agellon LB, et al. Enterohepatic Circulation of Bile Salts in Farnesoid X Receptor-Deficient Mice. J Biol Chem. 2003;278:41930–41937. doi: 10.1074/jbc.M306309200. [DOI] [PubMed] [Google Scholar]
- 7.Chen WS, Manova K, Weinstein DC, Duncan SA, Plump AS, et al. Disruption of the Hnf-4 Gene, Expressed in Visceral Endoderm, Leads to Cell Death in Embryonic Ectoderm and Impaired Gastrulation of Mouse Embryos. Genes Dev. 1994;8:2466–2477. doi: 10.1101/gad.8.20.2466. [DOI] [PubMed] [Google Scholar]
- 8.Inoue Y, Yu AM, Yim SH, Ma X, Krausz KW, et al. Regulation of Bile Acid Biosynthesis by Hepatocyte Nuclear Factor 4alpha. J Lipid Res. 2006;47:215–227. doi: 10.1194/jlr.M500430-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inoue Y, Yu AM, Inoue J, Gonzalez FJ. Hepatocyte Nuclear Factor 4alpha Is a Central Regulator of Bile Acid Conjugation. J Biol Chem. 2004;279:2480–2489. doi: 10.1074/jbc.M311015200. [DOI] [PubMed] [Google Scholar]
- 10.Claudel T, Inoue Y, Barbier O, Duran-Sandoval D, Kosykh V, et al. Farnesoid X Receptor Agonists Suppress Hepatic Apolipoprotein Ciii Expression. Gastroenterology. 2003;125:544–555. doi: 10.1016/s0016-5085(03)00896-5. [DOI] [PubMed] [Google Scholar]
- 11.Shih DQ, Dansky HM, Fleisher M, Assmann G, Fajans SS, et al. Genotype/Phenotype Relationships in Hnf-4alpha/Mody1: Haploinsufficiency Is Associated with Reduced Apolipoprotein (Aii), Apolipoprotein (Ciii), Lipoprotein(a), and Triglyceride Levels. Diabetes. 2000;49:832–837. doi: 10.2337/diabetes.49.5.832. [DOI] [PubMed] [Google Scholar]
- 12.Stroup D, Chiang JY. Hnf4 and Coup-Tfii Interact to Modulate Transcription of the Cholesterol 7alpha-Hydroxylase Gene (Cyp7a1) J Lipid Res. 2000;41:1–11. [PubMed] [Google Scholar]
- 13.Tirona RG, Lee W, Leake BF, Lan LB, Cline CB, et al. The Orphan Nuclear Receptor Hnf4alpha Determines Pxr- and Car-Mediated Xenobiotic Induction of Cyp3a4. Nat Med. 2003;9:220–224. doi: 10.1038/nm815. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Lee FY, Barrera G, Lee H, Vales C, et al. Activation of the Nuclear Receptor Fxr Improves Hyperglycemia and Hyperlipidemia in Diabetic Mice. Proc Natl Acad Sci U S A. 2006;103:1006–1011. doi: 10.1073/pnas.0506982103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe M, Houten SM, Wang L, Moschetta A, Mangelsdorf DJ, et al. Bile Acids Lower Triglyceride Levels Via a Pathway Involving Fxr, Shp, and Srebp-1c. J Clin Invest. 2004;113:1408–1418. doi: 10.1172/JCI21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Staels B, Kuipers F. Bile Acid Sequestrants and the Treatment of Type 2 Diabetes Mellitus. Drugs. 2007;67:1383–1392. doi: 10.2165/00003495-200767100-00001. [DOI] [PubMed] [Google Scholar]
- 17.Cariou B, van Harmelen K, Duran-Sandoval D, van Dijk TH, Grefhorst A, et al. The Farnesoid X Receptor Modulates Adiposity and Peripheral Insulin Sensitivity in Mice. J Biol Chem. 2006;281:11039–11049. doi: 10.1074/jbc.M510258200. [DOI] [PubMed] [Google Scholar]
- 18.Ma K, Saha PK, Chan L, Moore DD. Farnesoid X Receptor Is Essential for Normal Glucose Homeostasis. J Clin Invest. 2006;116:1102–1109. doi: 10.1172/JCI25604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maran RR, Thomas A, Roth M, Shen Z, Esterly N, et al. Fxr Deficiency in Mice Leads to Increased Intestinal Epithelial Cell Proliferation and Tumor Development. J Pharmacol Exp Ther. 2008 doi: 10.1124/jpet.108.145409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Modica S, Murzilli S, Salvatore L, Schmidt DR, Moschetta A. Nuclear Bile Acid Receptor Fxr Protects against Intestinal Tumorigenesis. Cancer Res. 2008;68:9589–9594. doi: 10.1158/0008-5472.CAN-08-1791. [DOI] [PubMed] [Google Scholar]
- 21.Kim I, Morimura K, Shah Y, Yang Q, Ward JM, et al. Spontaneous Hepatocarcinogenesis in Farnesoid X Receptor-Null Mice. Carcinogenesis. 2007;28:940–946. doi: 10.1093/carcin/bgl249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang F, Huang X, Yi T, Yen Y, Moore DD, et al. Spontaneous Development of Liver Tumors in the Absence of the Bile Acid Receptor Farnesoid X Receptor. Cancer Res. 2007;67:863–867. doi: 10.1158/0008-5472.CAN-06-1078. [DOI] [PubMed] [Google Scholar]
- 23.Thomas AM, Hart SN, Kong B, Fang J, Zhong XB, et al. Genome-Wide Tissue-Specific Farnesoid X Receptor Binding in Mouse Liver and Intestine. Hepatology. 2010;51:1410–1419. doi: 10.1002/hep.23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chong HK, Infante AM, Seo Y-K, Jeon T-I, Zhang Y, et al. Genome-Wide Interrogation of Hepatic Fxr Reveals an Asymmetric Ir-1 Motif and Synergy with Lrh-1. Nucleic Acids Res. 2010;38:6007–6017. doi: 10.1093/nar/gkq397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez FJ. Regulation of Hepatocyte Nuclear Factor 4 Alpha-Mediated Transcription. Drug Metab Pharmacokinet. 2008;23:2–7. doi: 10.2133/dmpk.23.2. [DOI] [PubMed] [Google Scholar]
- 26.Maloney PR, Parks DJ, Haffner CD, Fivush AM, Chandra G, et al. Identification of a Chemical Tool for the Orphan Nuclear Receptor Fxr. J Med Chem. 2000;43:2971–2974. doi: 10.1021/jm0002127. [DOI] [PubMed] [Google Scholar]
- 27.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte Nuclear Factor 4alpha (Nuclear Receptor 2a1) Is Essential for Maintenance of Hepatic Gene Expression and Lipid Homeostasis. Mol Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, et al. Five-Vertebrate Chip-Seq Reveals the Evolutionary Dynamics of Transcription Factor Binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, et al. Model-Based Analysis of Chip-Seq (Macs) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quinlan AR, Hall IM. Bedtools: A Flexible Suite of Utilities for Comparing Genomic Features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, et al. The Human Genome Browser at Ucsc. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, et al. David: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 33.Li G, Thomas AM, Hart SN, Zhong X, Wu D, et al. Farnesoid X Receptor Activation Mediates Head-to-Tail Chromatin Looping in the Nr0b2 Gene Encoding Small Heterodimer Partner. Mol Endocrinol. 2010;24:1404–1412. doi: 10.1210/me.2010-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li G, Thomas AM, Williams JA, Kong B, Liu J, et al. Farnesoid X Receptor Induces Murine Scavenger Receptor Class B Type I Via Intron Binding. PLoS One. 2012;7:e35895. doi: 10.1371/journal.pone.0035895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams JA, Thomas AM, Li G, Kong B, Zhan L, et al. Tissue Specific Induction of P62/Sqstm1 by Farnesoid X Receptor. PLoS One. 2012;7:e43961. doi: 10.1371/journal.pone.0043961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huber RM, Murphy K, Miao B, Link JR, Cunningham MR, et al. Generation of Multiple Farnesoid-X-Receptor Isoforms through the Use of Alternative Promoters. Gene. 2002;290:35–43. doi: 10.1016/s0378-1119(02)00557-7. [DOI] [PubMed] [Google Scholar]
- 37.Li T, Chiang JY. Rifampicin Induction of Cyp3a4 Requires Pregnane X Receptor Cross Talk with Hepatocyte Nuclear Factor 4alpha and Coactivators, and Suppression of Small Heterodimer Partner Gene Expression. Drug Metab Dispos. 2006;34:756–764. doi: 10.1124/dmd.105.007575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Podvinec M, Kaufmann MR, Handschin C, Meyer UA. Nubiscan, an in Silico Approach for Prediction of Nuclear Receptor Response Elements. Mol Endocrinol. 2002;16:1269–1279. doi: 10.1210/mend.16.6.0851. [DOI] [PubMed] [Google Scholar]
- 39.Sladek FM, Zhong WM, Lai E, Darnell JE., Jr Liver-Enriched Transcription Factor Hnf-4 Is a Novel Member of the Steroid Hormone Receptor Superfamily. Genes Dev. 1990;4:2353–2365. doi: 10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]
- 40.Bolotin E, Schnabl JM, Sladek FM, et al. IYDe. Hnf4a: The Transcription Factor Encyclopedia. Genome Biol. 2012;13:R24. doi: 10.1186/gb-2012-13-3-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Modica S, Gadaleta RM, Moschetta A. Deciphering the Nuclear Bile Acid Receptor Fxr Paradigm. Nucl Recept Signal. 2010;8:e005. doi: 10.1621/nrs.08005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. Foxa1 Is a Key Determinant of Estrogen Receptor Function and Endocrine Response. Nat Genet. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, et al. Foxa1 Translates Epigenetic Signatures into Enhancer-Driven Lineage-Specific Transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1.Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, et al. Bile Acids: Natural Ligands for an Orphan Nuclear Receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- 2.Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, et al. Identification of a Nuclear Receptor for Bile Acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- 3.Wang H, Chen J, Hollister K, Sowers LC, Forman BM. Endogenous Bile Acids Are Ligands for the Nuclear Receptor Fxr/Bar. Mol Cell. 1999;3:543–553. doi: 10.1016/s1097-2765(00)80348-2. [DOI] [PubMed] [Google Scholar]
- 4.Seol W, Choi HS, Moore DD. Isolation of Proteins That Interact Specifically with the Retinoid X Receptor: Two Novel Orphan Receptors. Mol Endocrinol. 1995;9:72–85. doi: 10.1210/mend.9.1.7760852. [DOI] [PubMed] [Google Scholar]
- 5.Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, et al. Targeted Disruption of the Nuclear Receptor Fxr/Bar Impairs Bile Acid and Lipid Homeostasis. Cell. 2000;102:731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 6.Kok T, Hulzebos CV, Wolters H, Havinga R, Agellon LB, et al. Enterohepatic Circulation of Bile Salts in Farnesoid X Receptor-Deficient Mice. J Biol Chem. 2003;278:41930–41937. doi: 10.1074/jbc.M306309200. [DOI] [PubMed] [Google Scholar]
- 7.Chen WS, Manova K, Weinstein DC, Duncan SA, Plump AS, et al. Disruption of the Hnf-4 Gene, Expressed in Visceral Endoderm, Leads to Cell Death in Embryonic Ectoderm and Impaired Gastrulation of Mouse Embryos. Genes Dev. 1994;8:2466–2477. doi: 10.1101/gad.8.20.2466. [DOI] [PubMed] [Google Scholar]
- 8.Inoue Y, Yu AM, Yim SH, Ma X, Krausz KW, et al. Regulation of Bile Acid Biosynthesis by Hepatocyte Nuclear Factor 4alpha. J Lipid Res. 2006;47:215–227. doi: 10.1194/jlr.M500430-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inoue Y, Yu AM, Inoue J, Gonzalez FJ. Hepatocyte Nuclear Factor 4alpha Is a Central Regulator of Bile Acid Conjugation. J Biol Chem. 2004;279:2480–2489. doi: 10.1074/jbc.M311015200. [DOI] [PubMed] [Google Scholar]
- 10.Claudel T, Inoue Y, Barbier O, Duran-Sandoval D, Kosykh V, et al. Farnesoid X Receptor Agonists Suppress Hepatic Apolipoprotein Ciii Expression. Gastroenterology. 2003;125:544–555. doi: 10.1016/s0016-5085(03)00896-5. [DOI] [PubMed] [Google Scholar]
- 11.Shih DQ, Dansky HM, Fleisher M, Assmann G, Fajans SS, et al. Genotype/Phenotype Relationships in Hnf-4alpha/Mody1: Haploinsufficiency Is Associated with Reduced Apolipoprotein (Aii), Apolipoprotein (Ciii), Lipoprotein(a), and Triglyceride Levels. Diabetes. 2000;49:832–837. doi: 10.2337/diabetes.49.5.832. [DOI] [PubMed] [Google Scholar]
- 12.Stroup D, Chiang JY. Hnf4 and Coup-Tfii Interact to Modulate Transcription of the Cholesterol 7alpha-Hydroxylase Gene (Cyp7a1) J Lipid Res. 2000;41:1–11. [PubMed] [Google Scholar]
- 13.Tirona RG, Lee W, Leake BF, Lan LB, Cline CB, et al. The Orphan Nuclear Receptor Hnf4alpha Determines Pxr- and Car-Mediated Xenobiotic Induction of Cyp3a4. Nat Med. 2003;9:220–224. doi: 10.1038/nm815. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Lee FY, Barrera G, Lee H, Vales C, et al. Activation of the Nuclear Receptor Fxr Improves Hyperglycemia and Hyperlipidemia in Diabetic Mice. Proc Natl Acad Sci U S A. 2006;103:1006–1011. doi: 10.1073/pnas.0506982103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe M, Houten SM, Wang L, Moschetta A, Mangelsdorf DJ, et al. Bile Acids Lower Triglyceride Levels Via a Pathway Involving Fxr, Shp, and Srebp-1c. J Clin Invest. 2004;113:1408–1418. doi: 10.1172/JCI21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Staels B, Kuipers F. Bile Acid Sequestrants and the Treatment of Type 2 Diabetes Mellitus. Drugs. 2007;67:1383–1392. doi: 10.2165/00003495-200767100-00001. [DOI] [PubMed] [Google Scholar]
- 17.Cariou B, van Harmelen K, Duran-Sandoval D, van Dijk TH, Grefhorst A, et al. The Farnesoid X Receptor Modulates Adiposity and Peripheral Insulin Sensitivity in Mice. J Biol Chem. 2006;281:11039–11049. doi: 10.1074/jbc.M510258200. [DOI] [PubMed] [Google Scholar]
- 18.Ma K, Saha PK, Chan L, Moore DD. Farnesoid X Receptor Is Essential for Normal Glucose Homeostasis. J Clin Invest. 2006;116:1102–1109. doi: 10.1172/JCI25604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maran RR, Thomas A, Roth M, Shen Z, Esterly N, et al. Fxr Deficiency in Mice Leads to Increased Intestinal Epithelial Cell Proliferation and Tumor Development. J Pharmacol Exp Ther. 2008 doi: 10.1124/jpet.108.145409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Modica S, Murzilli S, Salvatore L, Schmidt DR, Moschetta A. Nuclear Bile Acid Receptor Fxr Protects against Intestinal Tumorigenesis. Cancer Res. 2008;68:9589–9594. doi: 10.1158/0008-5472.CAN-08-1791. [DOI] [PubMed] [Google Scholar]
- 21.Kim I, Morimura K, Shah Y, Yang Q, Ward JM, et al. Spontaneous Hepatocarcinogenesis in Farnesoid X Receptor-Null Mice. Carcinogenesis. 2007;28:940–946. doi: 10.1093/carcin/bgl249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang F, Huang X, Yi T, Yen Y, Moore DD, et al. Spontaneous Development of Liver Tumors in the Absence of the Bile Acid Receptor Farnesoid X Receptor. Cancer Res. 2007;67:863–867. doi: 10.1158/0008-5472.CAN-06-1078. [DOI] [PubMed] [Google Scholar]
- 23.Thomas AM, Hart SN, Kong B, Fang J, Zhong XB, et al. Genome-Wide Tissue-Specific Farnesoid X Receptor Binding in Mouse Liver and Intestine. Hepatology. 2010;51:1410–1419. doi: 10.1002/hep.23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chong HK, Infante AM, Seo Y-K, Jeon T-I, Zhang Y, et al. Genome-Wide Interrogation of Hepatic Fxr Reveals an Asymmetric Ir-1 Motif and Synergy with Lrh-1. Nucleic Acids Res. 2010;38:6007–6017. doi: 10.1093/nar/gkq397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez FJ. Regulation of Hepatocyte Nuclear Factor 4 Alpha-Mediated Transcription. Drug Metab Pharmacokinet. 2008;23:2–7. doi: 10.2133/dmpk.23.2. [DOI] [PubMed] [Google Scholar]
- 26.Maloney PR, Parks DJ, Haffner CD, Fivush AM, Chandra G, et al. Identification of a Chemical Tool for the Orphan Nuclear Receptor Fxr. J Med Chem. 2000;43:2971–2974. doi: 10.1021/jm0002127. [DOI] [PubMed] [Google Scholar]
- 27.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte Nuclear Factor 4alpha (Nuclear Receptor 2a1) Is Essential for Maintenance of Hepatic Gene Expression and Lipid Homeostasis. Mol Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, et al. Five-Vertebrate Chip-Seq Reveals the Evolutionary Dynamics of Transcription Factor Binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, et al. Model-Based Analysis of Chip-Seq (Macs) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quinlan AR, Hall IM. Bedtools: A Flexible Suite of Utilities for Comparing Genomic Features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, et al. The Human Genome Browser at Ucsc. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, et al. David: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 33.Li G, Thomas AM, Hart SN, Zhong X, Wu D, et al. Farnesoid X Receptor Activation Mediates Head-to-Tail Chromatin Looping in the Nr0b2 Gene Encoding Small Heterodimer Partner. Mol Endocrinol. 2010;24:1404–1412. doi: 10.1210/me.2010-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li G, Thomas AM, Williams JA, Kong B, Liu J, et al. Farnesoid X Receptor Induces Murine Scavenger Receptor Class B Type I Via Intron Binding. PLoS One. 2012;7:e35895. doi: 10.1371/journal.pone.0035895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams JA, Thomas AM, Li G, Kong B, Zhan L, et al. Tissue Specific Induction of P62/Sqstm1 by Farnesoid X Receptor. PLoS One. 2012;7:e43961. doi: 10.1371/journal.pone.0043961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huber RM, Murphy K, Miao B, Link JR, Cunningham MR, et al. Generation of Multiple Farnesoid-X-Receptor Isoforms through the Use of Alternative Promoters. Gene. 2002;290:35–43. doi: 10.1016/s0378-1119(02)00557-7. [DOI] [PubMed] [Google Scholar]
- 37.Li T, Chiang JY. Rifampicin Induction of Cyp3a4 Requires Pregnane X Receptor Cross Talk with Hepatocyte Nuclear Factor 4alpha and Coactivators, and Suppression of Small Heterodimer Partner Gene Expression. Drug Metab Dispos. 2006;34:756–764. doi: 10.1124/dmd.105.007575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Podvinec M, Kaufmann MR, Handschin C, Meyer UA. Nubiscan, an in Silico Approach for Prediction of Nuclear Receptor Response Elements. Mol Endocrinol. 2002;16:1269–1279. doi: 10.1210/mend.16.6.0851. [DOI] [PubMed] [Google Scholar]
- 39.Sladek FM, Zhong WM, Lai E, Darnell JE., Jr Liver-Enriched Transcription Factor Hnf-4 Is a Novel Member of the Steroid Hormone Receptor Superfamily. Genes Dev. 1990;4:2353–2365. doi: 10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]
- 40.Bolotin E, Schnabl JM, Sladek FM, et al. IYDe. Hnf4a: The Transcription Factor Encyclopedia. Genome Biol. 2012;13:R24. doi: 10.1186/gb-2012-13-3-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Modica S, Gadaleta RM, Moschetta A. Deciphering the Nuclear Bile Acid Receptor Fxr Paradigm. Nucl Recept Signal. 2010;8:e005. doi: 10.1621/nrs.08005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. Foxa1 Is a Key Determinant of Estrogen Receptor Function and Endocrine Response. Nat Genet. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, et al. Foxa1 Translates Epigenetic Signatures into Enhancer-Driven Lineage-Specific Transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.