Introduction
Commensal bacteria have been shown to be active participants in the development of the structure and function of host tissues (Gordon and Pesti, 1971; Hooper et al., 2001; Macpherson and Harris, 2004; Umesaki and Setoyama, 2000; Xu and Gordon, 2003). For example, commensal bacteria are required for the complete development of Peyer’s patches, the lamina propria, and the intraepithelial spaces, all three of the main immune elements found in the intestine (Duncan and Edberg, 1995; Falk et al., 1998). The intestinal tissue morphology is also altered by commensal colonization such that the villi of the small intestine are longer and the crypts are shorter. In addition, studies in germ-free mice have revealed that the commensal bacteria induce angiogenesis, thereby contributing to the development of the complex vascular beds found just underneath the mucosal surface (Stappenbeck et al., 2002). Furthermore, it has been found that intestinal commensal bacteria contribute to intestinal epithelial cell homeostasis through TLR recognition pathways providing protection from epithelial cell injury (Rakoff-Nahoum et al., 2004). Finally, a state of “controlled” inflammation that normally exists in the intestine has been attributed to both the quality and quantity of intestinal commensal microorganisms (Cebra, 1999; Chadwick and Anderson, 1992).
However, little is known concerning the contribution of the oral commensal microbiome to periodontal tissue structure and function. This is apparently due to the fact that in contrast to intestinal tissue, few structural or functional differences have been reported when GF and SPF mice have been compared (Curtis et al., 2011). This is surprising since one of the main mechanisms by which periodontal tissue regulates the numbers of oral commensal bacteria that reside on the tooth and its root surface depends on the constant transit of neutrophils through gingival tissue and into the gingival crevice (Attström and Schroeder, 1979; Carrassi et al., 1989; Hart et al., 1994; Page et al., 1987; Waldrop et al., 1987). This process involves a highly orchestrated expression of select innate host defense mediators that safely guide extravasated neutrophils through connective tissue, and finally through the junctional epithelium, a loosely organized specialized epithelial tissue which connects the tooth surface to periodontal tissue and provides the first line of defense to bacterial invasion (Gemmell et al., 1994; Moughal et al., 1992; Nylander et al., 1993; Tonetti, 1997; Tonetti et al., 1994). The requirement for neutrophil transit through the periodontium for the maintenance of healthy periodontal tissue is well documented in both humans (Attström and Schroeder, 1979; Carrassi et al., 1989; Hart et al., 1994; Page et al., 1987; Waldrop et al., 1987) and mice (Niederman et al., 2001).
The regulation of the number of neutrophils that migrate through periodontal tissue is important in both health and disease. Defective recruitment of neutrophils to the periodontal tissue, (Nussbaum and Shapira, 2011), or excessive recruitment of neutrophils, as seen in the absence of the regulatory endothelial protein Del-1(Eskan et al., 2012) are conditions that both lead to periodontitis. Hence periodontal health is fundamentally dependent on a critical level of neutrophil recruitment to the gingival tissues: inappropriate under or over recruitment to this critical level can lead to disease. The regulation of neutrophil recruitment is therefore a key determinant of periodontal health.
Here we demonstrate that mouse chemokine receptor CXCR2 is the major mediator of neutrophil recruitment to periodontal tissue. We then show that two different CXCR2 ligands, CXCL1 and CXCL2, are expressed in germ-free tissues. However, CXCL2 but not CXCL1, is significantly increased in SPF mice, is controlled by the MyD88 activation pathway and can account for the increased neutrophil recruitment in the SPF periodontal tissue. These data demonstrate the select usage of CXCR2 ligands in periodontal tissue and indicate that the different usage of CXCR2 ligands is one mechanism by which the periodontal tissue regulates neutrophil migration in response to commensal bacterial colonization.
Results
GF and SPF gingival tissues display relatively minor differences in cytokine protein expression levels
In an initial pilot study, we have previously reported a small but statistically significant increase in gingival tissue IL-1β protein levels in SPF mice as compared to GF mice (Dixon et al., 2004). Therefore, we initially sought to confirm this observation and determine if other host cytokines in gingival tissue were significantly modulated by commensal colonization. ELISAs were performed on the homogenized tissues obtained from GF and SPF mice for IL-1β, TNF, INF-γ and CXCL2. Although a trend for increases in IL-1β and CXCL2 and decreases in TNFα and INF-γ were observed when SPF mice were compared to GF mice, the data was not statistically significant when assays performed from different tissue collections were compared. We suspect that the inability to accurately collect a standardized gingival tissue sample combined with the highly localized expression of select cytokines and chemokines precluded our ability to obtain statistically significant differences in the levels of these cytokines. These data demonstrate that the effects of commensal colonization on the expression levels of these cytokines are not sufficiently significant to be detected in ELISA assays with whole gingival tissues.
Commensal colonization significantly increases the number of neutrophils found in the junctional epithelium
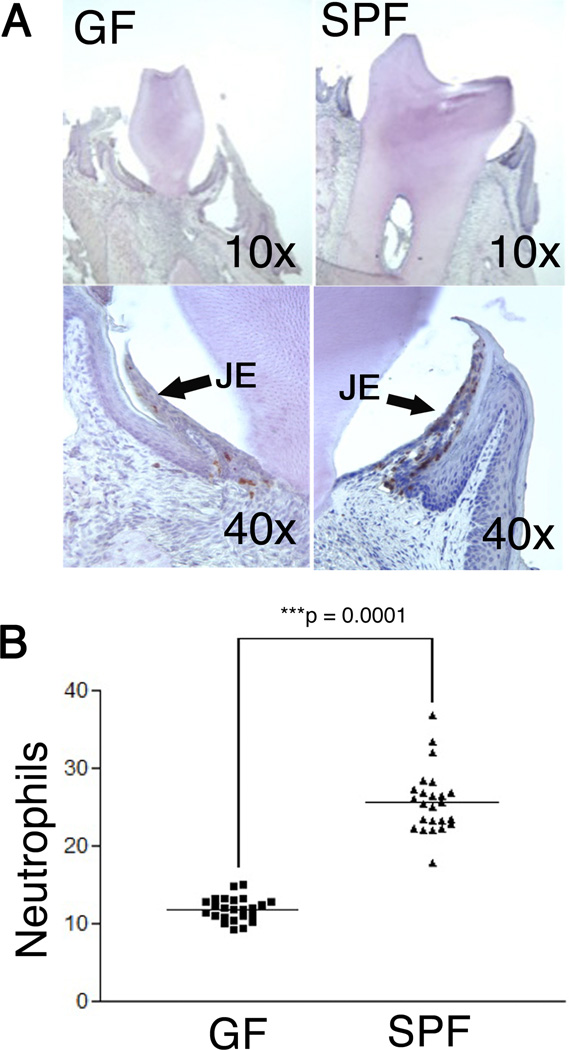
Recently it has been reported that SPF mice contain a higher number of neutrophils in the junctional epithelium when compared to GF mice (Tsukamoto et al., 2012). The method employed in the previous study reported the number of neutrophils in a defined junctional epithelium area. This structural difference was not examined in the strain of mice used for the current study and therefore, the totoal number of neutrophils found in the junctional epithelium was determined. Neutrophils were stained with a neutrophil specific antibody and they were found to be concentrated in the junctional epithelia of both mouse groups with only occasional staining in the vasculature and no staining in the oral and sulcular epithelia or the underlying connective tissue (Fig 1A). Four different mice were examined in each group of GF and SPF mice such that 100 sections / group were counted for neutrophils. Analysis of the results revealed that there was no statistical difference in neutrophil numbers within a group (p > 0.01, Mann-Whitney Test). However, in contrast, GF mice contained significantly (p < 0.0001, Mann-Whitney Test) fewer neutrophils than age- and strain- matched SPF controls (Fig. 1B). These data demonstrate that oral commensal bacteria significantly contribute to increased neutrophil migration to the junctional epithelium.
Fig 1. GF and SPF mice contain major neutrophil number differences in gingival tissue.
A. Neutrophil staining of GF and SPF mouse gingival tissue (×10 and ×40 mag). Most neutrophils were found in the junctional epithelium (JE), however, a few neutrophils were also detected in both the gingival and pulp vasculature (data not shown). B. Neutrophils, stained and quantified shown here as average neutrophils per section, were mainly located in the junctional epithelial (junctional epithelium) tissue. Quantification includes 25 sections per mouse. *** denotes statistically significant difference (Mann-Whitney test).
CXCR2 is the major mouse periodontal neutrophil homing receptor
CXCR2 is a major neutrophil chemokine receptor in mice, which facilitates neutrophil migration into host tissues (Cacalano et al., 1994; Shuster et al., 1995) and contributes to neutrophil homeostasis by maintaining normal neutrophil numbers in the circulation (Mei et al., 2012). CXCR2 KO mice have also been shown to develop spontaneous periodontitis (Hajishengallis et al., 2011; Yu et al., 2007) which is suspected to be due the lack of neutrophil migration to periodontal tissue. Since, multiple mechanisms of neutrophil chemotaxis may occur at sites of bacterial colonization (Phillipson and Kubes, 2011), the contribution of CXCR2 to neutrophil migration to the junctional epithelium was examined in CXCR2 KO mice (Fig. 2). The junctional epithelium was nearly completely void of neutrophils, of the over 50 tissue sections examined; only a rare and apparently transient neutrophil was ever identified in the junctional epithelium. There were however, neutrophils identified in the vessels and occasionally in the sub-epithelial vasculature. These data demonstrate that while neutrophils are present in the vessels, without proper signaling to the CXCR2 receptor the neutrophils are unable to migrate to the junctional epithelium, in contrast to the normal neutrophil recruitment in strain-matched wild-type mice (Fig. 1)
Fig 2. Lack of neutrophils in the gingival tissue of CXCR2KO mice.
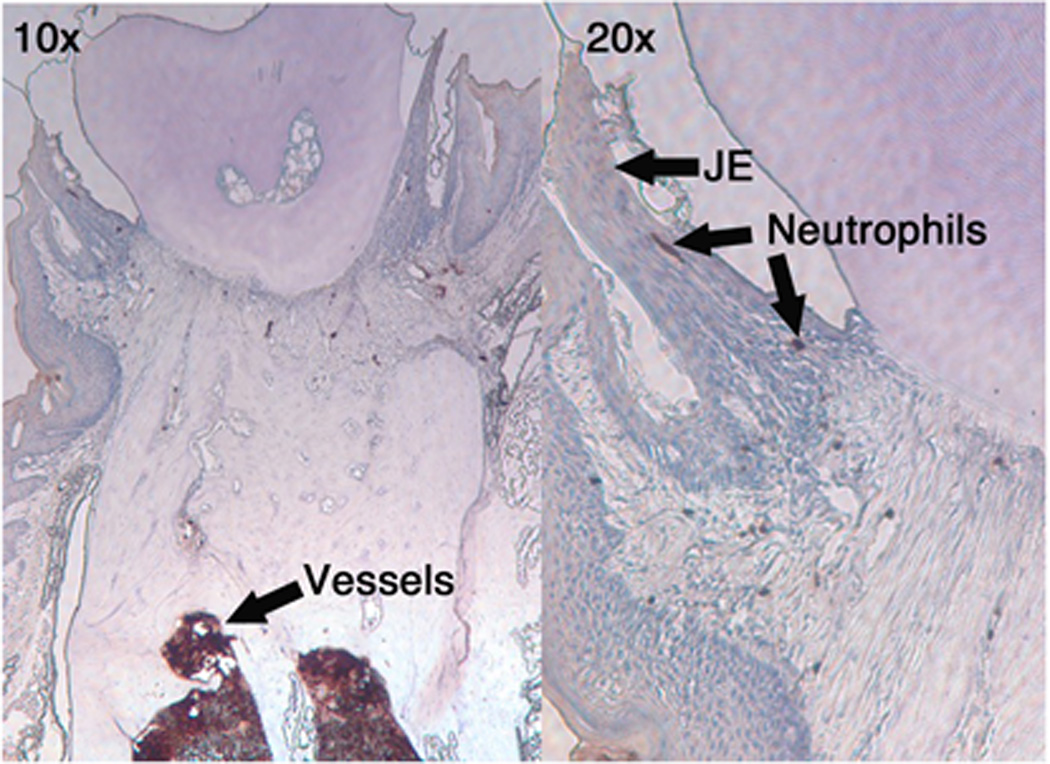
Gingival tissue obtained from CXCR2 KO mice were stained for neutrophils as described in the text. (10×, 20×). Most neutrophils were detected in vessels while occasional neutrophils were observed in the vasculature and only rarely in junctional epithelium.
CXCL2 but not CXCL1, expression in the junctional epithelium is significantly dependent on commensal colonization
Next, the expression of two major ligands for CXCR2, CXCL1 and CXCL2, was examined since CXCR2 was essential for neutrophil migration to the junctional epithelium. CXCL1 and CXCL2 antibody concentrations for immunohistochemical analysis were optimized for maximum sensitivity (Fig. 3). The slides were then blindly scored and the expression level for each chemokine was then presented as the relative staining intensity on a scale of 0 (weakest) to 3 (strongest). A multi-level regression analysis for CXCL1 expression revealed that there was no significant difference in the relative expression levels between GF and its strain matched SPF strain (GF versus SPF, p=0.9; N=120). More specifically, nearly identical percentages of relative intensity scores were found for both GF and SPF mice. In contrast to CXCL1, CXCL2 expression levels were significantly different (GF versus SPF, p=0.0001; N=120) in the junctional epithelium. These data demonstrate that CXCL2 expression, but not CXCL1, is modulated by oral commensal bacterial colonization.
FIG 3. SPF mice express significantly higher levels of CXCL2 in the junctional epithelium.
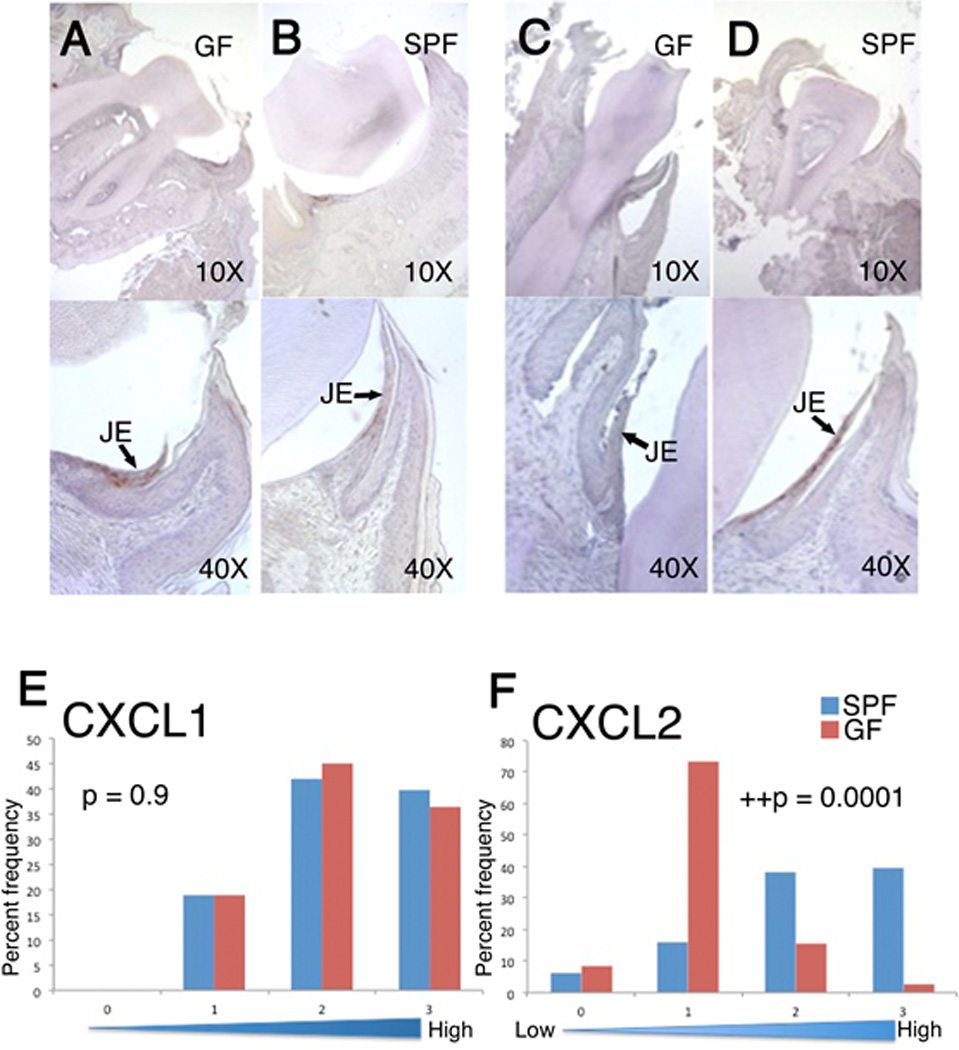
CXCL1 and CXCL2 immunohistochemical (IHC) staining of oral tissue. CXCL1 (A and B) and CXCL2 (C and D) expression was determined in GF (A and C) and SPF mice (B and D). Local structures are indicated as well as magnification with neutrophil staining detected in the junctional epithelium. The junctional epithelium appears detached from the tooth surface due to the fixing procedure. Lower panels represent a semi-quantitative analysis of CXCL1 (E) and CXCL2 (F) expression as described in the text: Y axes indicate the percentage of sections examined; X axes indicate staining intensity scores. ++, denotes statistically significant difference (Multi-level logistic regression test, N=120 tissue sections).
Neutrophils and CXCL2 but not CXCL1 expression in the junctional epithelium is significantly dependent on the MyD88 signaling pathway
MyD88 is a co-adaptor protein facilitating host responses after bacterial recognition through TLRs and three members of the IL-1 receptor family (Kawai and Akira, 2005). Therefore, to determine if commensal bacteria may utilize a TLR-dependent activation pathway to augment neutrophil migration in SPF mice as shown in Fig 1, the number of neutrophils in the junctional epithelium of SPF MyD88KO and strain matched wild type mice was determined as described above. A similar staining pattern of neutrophil accumulation in the junctional epithelium was observed in both MyD88 KO and wild type SPF mice (see Fig. 1 for example). Quantitative analysis performed as described above for GF and SPF comparisons revealed a significant decrease in the number of neutrophils in the junctional epithelium in the MyD88 KO mice when compared to the wild type strain (p<0.0001, Mann Whitney Test, Fig. 4). This result demonstrates that a MyD88 dependent signaling pathway is required for optimal neutrophil migration in SPF mice. Furthermore, a multilevel regression analysis (Fig. 4) for CXCL1 expression revealed that there was no significant difference in the relative expression levels between MyD88KO and its strain matched wild type strain (wild type versus MyD88KO, p=0.9, N=120). In contrast, CXCL2 expression levels were significantly different (MyD88KO versus WT p<0.0001, N=120: multilevel regression analysis). These data demonstrate that CXCL2 expression, but not CXCL1, is modulated by the MyD88 activation pathway.
Fig. 4. Myd88 mice display defects in neutrophil migration and CXCL2 expression.
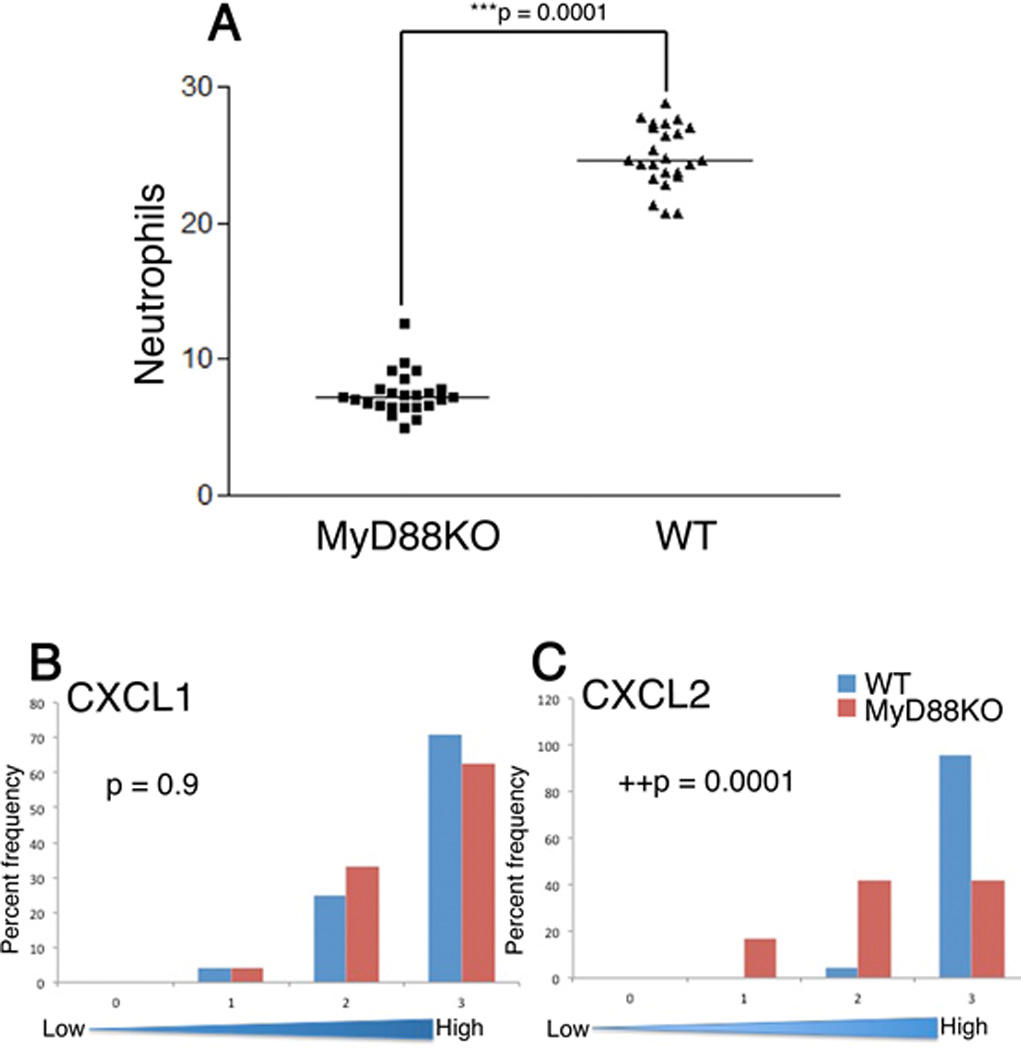
Neutrophil staining and CXCL1 and CXCL2 expression in MyD88KO and strain matched WT mice. The number of neutrophils in the junctional epithelium (A) were determined as described in the text. ***, denotes statistically significant difference (Mann-Whitney test). Semi-quantitative analysis of CXCL1 (B) and CXCL2 (C) expression was performed as described in the text., ++, denotes statistically significant difference (Multi-level logistic regression test, N=120 tissue sections)
Discussion
This manuscript found that in contrast to intestinal tissue, gingival tissue did not display readily observable differences in tissue structure when GF and SPF mice were compared. The analysis of select cytokine expression levels also did not reveal major alterations due to oral colonization of commensals in SPF mice. These data are consistent with an early report that described neutrophils in the periodontium of germ free rats (Rovin et al., 1966) and more recent analyses that have shown that GF and SPF mice express comparable levels of SLPI (Hayashi et al., 2010) and CEACAM-1 (Heymann et al., 2001), two highly specialized host molecules expressed by junctional epithelial cells involved in neutrophil crosstalk and specialized adhesion junctions, respectively. In addition, immunohistological staining revealed that both GF and SPF mice expressed CXCL1 and CXCL2, two ligands which contribute to neutrophil migration. These data provide evidence that bacterial colonization of the oral cavity is not required for neutrophil migration through the junctional epithelium, a key function of innate defense of periodontal tissue. Furthermore, these data demonstrate that, in contrast to intestinal tissue, bacteria-free periodontal tissue can develop the apparently appropriate structural and functional attributes normally associated with a tissue in close contact with commensal bacteria. The mechanisms which tailor the structure and function of this tissue in the absence of a bacterial signal are not known.
The contribution of oral commensal bacteria to the structure and function of periodontal tissue appears to be more subtle than that observed in intestinal tissue. It required quantitative analysis of neutrophil numbers present in the highly specialized junctional epithelial tissue in the work presented here and from a recent publication (Tsukamoto et al., 2012) to demonstrate that SPF mice contain a significant increase in neutrophils numbers when compared to their GF counterparts. Furthermore, commensal colonization altered the expression of CXCL2, a CXCR2 ligand associated with neutrophil chemotaxis. This difference was detected using a semi-quantitative immunohistochemical analysis where the expression levels of CXCL2, but not CXCL1 were significantly increased in SPF mice when compared to GF mice. Similarly, ELISA analysis of gingival tissue trended towards higher CXCL2 expression in SPF mice but was not significantly different from GF mice (Fig. 1). However, the ELISA data relied on tissue collected from around the teeth which additionally included connective and vascular tissues (determined by FACS analysis, data not shown) where cytokine expression levels could vary within the different tissue environments. This could explain why significant differences in the highly localized junctional epithelial tissue were not detected. A recent report employing laser capture microdissection of junctional epithelial tissue has shown that the message for both CXCL1 and CXCL2 is greater in SPF mice when compared to GF animals (Hayashi et al., 2010) which would appear to contradict our findings on the increased expression of CXCL2 but not CXCL1. However, the protein levels were not compared in that study and it is possible that similar to our previous work (Dixon et al., 2004) mRNA levels may not always correlate to protein expression. It is particularly appropriate to consider mismatched transcriptional and translational expression patterns with respect to chemokine receptor ligands and other mediators of inflammation, where instability sequences in the mRNA sequence regulate protein expression levels (Tebo et al., 2000). Therefore, both the subtle effects of commensal colonization and the technical challenges associated with characterizing small amounts of highly specialized tissues in the periodontium, such as the junctional epithelium, have contributed to the lack of an adequate description of how oral commensal bacteria contribute to periodontal tissue structure and function.
The use of MyD88 KO mice identified this adaptor protein as required for both the increase in neutrophil numbers and CXCL2 expression observed in SPF mice. MyD88 is a required adaptor protein for all TLR’s except TLR3 and for three members of the IL-1 receptor family (Kawai and Akira, 2005). These data, from the MyD88KO mice, provide strong evidence that oral commensal bacteria either directly through TLR activation or indirectly through IL-1β expression facilitate the increase in both neutrophil migration and CXCL2 expression associated with SPF mice. The MyD88 activation pathway did not modulate the level of CXCL1 expression consistent with its expression being independent of commensal colonization.
The data presented in this manuscript provide the first evidence for the selective use of ligand CXCL2 in healthy periodontal tissue. CXCR2 has been shown to be a critical component maintaining periodontal homeostasis (Yu et al., 2007). In this manuscript, we demonstrate that CXCR2 KO mice fail to properly recruit neutrophils to the junctional epithelium revealing this receptor as the major mechanism of neutrophil transmigration into periodontal tissue. Previous studies in different inflammatory models of disease in mice have shown that the selective use of chemokine ligands facilitate highly specific temporal and tissue selective neutrophil homing responses (Borregaard, 2010; Lee et al., 1995; McDonald and Kubes, 2010; Ritzman et al., 2010; Rovai et al., 1998; Sadik et al., 2011; Wuyts et al., 1996). Consistent with the findings that CXCL2, but not CXCL1, was regulated by commensal bacteria and the MyD88 activation pathway, GF mice displayed stronger expression of CXCL1 compared to CXCL2, whereas SPF mice expressed both of these CXCR2 ligands equally. Therefore, the select usage of CXCR2 ligands may allow the host to homeostatically regulate neutrophil migration to the periodontal tissue in response to– and for immune surveillance of– commensal bacterial colonization.
Materials and Methods
Animal resource
All animal procedures described in this study were approved by the institutional animal care and use committees, in compliance with established federal and state policies. Germ-free (GF) C3H/Orl mice (Charles River Laboratories International) or CXCR2−/− mice on BALB/c genetic background (The Jackson Laboratory) were maintained in isolators at the Royal Veterinary College, University of London. The sterility of GF animals was examined by aerobic and anaerobic culture of oral swabs and fecal pellets on nonselective media and by PCR using universal 16S primers. Specific-pathogen-free (SPF) mice (below) were maintained in individually ventilated cages at the animal care facilities of Queen Mary University of London. Original GF mice were divided; half were raised and propagated in conventional cages, thus creating the SPF. MyD88KO mice and wild-type littermate controls (C57BL/6) mice were raised under specific-pathogen-free conditions in individually ventilated cages at the animal care facility of the University of Louisville.
Histology
All mice were sacrificed between 12 and 14-weeks of age. 5 mice per group were dissected and mandibles and maxillas were prepared for immunohistochemistry. Tissues were fixed in Bouin's solution for 24 h, rinsed with 70% ethanol and demineralized (for post-27 dpc tissues) in acetic acid/formalin/sodium chloride solution. Tissues were processed according to standard histological procedures and embedded in paraffin. Each tooth was sectioned serially in a buccolingual (frontal) orientation (6mm) using a microtome and mounted as numbered serial sections on charged glass slides. This resulted in approximately 100 sections/tooth. High-resolution digital images were captured using Metavue software (Molecular Devices, Sunnyvale, Calif., USA) with a SPOT CCD camera (Diagnostic Instruments, Sterling Heights, Mich., USA).
Immunohistochemistry (IHC)
IHC was performed on mouse mandible and maxilla tissues. Every fourth section of serially sectioned tooth was stained resulting in approximately 25 stained sections/tooth for each primary antibody. Tissues were deparaffinized in xylene and rehydrated using decreased graded dilutions of ethanol. Tissue specimens were blocked by incubation in 1.5% H202 in methanol solution for 30 minutes. Primary antibodies (neutrophil elastase (Santa Cruz Biotechnology, sc-71674), KC (abcam, ab17882) and MIP2 (abcam ab9950) were used with biotinylated secondary antibodies against rabbit primary antibodies, as appropriate, and slides were developed using a 3-amino-9-ethylcarbazole substrate kit The Abcam IHC-P staining kits were used for immunohistochemical staining. Positive controls included staining in WT tissues, where immunolocalization of target proteins were well characterized. Negative controls were performed without a primary antibody.
ELISA
Oral tissues were extracted by previous published method (Dixon et al., 2004). Extracted tissues were dissected from mandible and maxilla, stored in RNAlater (Qaigen) as per manufacturer instructions. The tissues were then sonicated with microtip in PBS on ice for 15s on, 15s off for total of 3min on. Serial dilutions of the supernatents were made on each ELISA and BioRad protein assay (500-0001). The ELISAs employed were as follows: MIP2, R&D Systems Cat Num MM200; TNF-α, eBioscience 88-7324-22; IL-1β, ThermoScientific 1858003; and IFN-γ, eBioscience 88-8314-22. All ELISA kits were used as per manufacturer directions.
Statistical Analyses
Initial experiments revealed that there were no significant differences between mandibular and maxillary teeth within each group (p<0.001, random mixed model test). Therefore, quantitative analysis of neutrophil numbers in the junctional epithelium of GF and SPF mice was performed by obtaining sections surrounding either the maxillary first molar or mandibular first molar in each mouse. Individual neutrophils were counted in each section and totaled for each mouse. The differences between WT and KO groups were tested in PRISM by the parametric Student t test and nonparametric Mann-Whitney test and the Multi-level logistic regression test. The Multi-level logistic regression test was employed since there was variability in staining intensity of CXCL1 and CXCL2 in the different sections obtained from the same mouse, and in sections obtained from different mice in the same group as well as the differences noted between the different mouse groups.
IHCKC and MIP2
4 mice per group were examined by serial sectioning, 100 sections/group. Each slide was blindly rated 0–3 for intensity. Semi-quantitative analysis for staining intensity was performed using multilevel logistic regression.
Acknowledgments
This work was supported by NIH DE018274 to Richard Darveau, NIH DE015254 to George Hajishengallis, MRC MR/J011118/1 to Mike Curtis, and the Changhai Hospital 1255 project # 125543000 to Xiao Long Luo.
References
- Attström R, Schroeder HE. Effect of experimental neutropenia on initial gingivitis in dogs. Scand J Dent Res. 1979;87:7–23. doi: 10.1111/j.1600-0722.1979.tb01935.x. [DOI] [PubMed] [Google Scholar]
- Borregaard N. Neutrophils, from marrow to microbes. Immunity. 2010;33:657–670. doi: 10.1016/j.immuni.2010.11.011. [DOI] [PubMed] [Google Scholar]
- Cacalano G, Lee J, Kikly K, Ryan AM, Pitts-Meek S, Hultgren B, Wood WI, Moore MW. Neutrophil and B cell expansion in mice that lack the murine IL-8 receptor homolog. Science. 1994;265:682–684. doi: 10.1126/science.8036519. [DOI] [PubMed] [Google Scholar]
- Carrassi A, Abati S, Santarelli G, Vogel G. Periodontitis in a patient with chronic neutropenia. J Periodontol. 1989;60:352–357. doi: 10.1902/jop.1989.60.6.352. [DOI] [PubMed] [Google Scholar]
- Cebra JJ. Influences of microbiota on intestinal immune system development. Am J Clin Nutr. 1999;69:1046S–1051S. doi: 10.1093/ajcn/69.5.1046s. [DOI] [PubMed] [Google Scholar]
- Chadwick VS, Anderson RP. Microorganisms and their products in inflammatory bowel disease. In: MacDermott RP, Stenson WF, editors. Inflammatory Bowel Disease. Amsterdam: Elsevier Science; 1992. pp. 241–258. [Google Scholar]
- Curtis MA, Zenobia C, Darveau RP. The relationship of the oral microbiotia to periodontal health and disease. Cell Host Microbe. 2011;10:302–306. doi: 10.1016/j.chom.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DR, Reife RA, Cebra JJ, Darveau RP. Commensal bacteria influence innate status within gingival tissues: a pilot study. J Periodontol. 2004;75:1486–1492. doi: 10.1902/jop.2004.75.11.1486. [DOI] [PubMed] [Google Scholar]
- Duncan HE, Edberg SC. Host-microbe interaction in the gastrointestinal tract. Crit Rev Microbiol. 1995;21:85–100. doi: 10.3109/10408419509113535. [DOI] [PubMed] [Google Scholar]
- Eskan MA, Jotwani R, Abe T, Chmelar J, Lim JH, Liang S, Ciero PA, Krauss JL, Li F, Rauner M, et al. The leukocyte integrin antagonist Del-1 inhibits IL-17-mediated inflammatory bone loss. Nat Immunol. 2012;13:465–473. doi: 10.1038/ni.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk PG, Hooper LV, Midtvedt T, Gordon JI. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol Mol Biol Rev. 1998;62:1157–1170. doi: 10.1128/mmbr.62.4.1157-1170.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmell E, Walsh LJ, Savage NW, Seymore GJ. Adhesion molecule expression in chronic inflammatory periodontal disease tissue. J Periodontal Res. 1994;29:46–53. doi: 10.1111/j.1600-0765.1994.tb01090.x. [DOI] [PubMed] [Google Scholar]
- Gordon HA, Pesti L. The gnotobiotic animal as a tool in the study of host microbial relationships. Bacteriol Rev. 1971;35:390–429. doi: 10.1128/br.35.4.390-429.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, McIntosh ML, Alsam A, Kirkwood KL, Lambris JD, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10:497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart TC, Shapira L, Van Dyke TE. Neutrophil defects as risk factors for periodontal diseases. J Periodontol. 1994;65:521–529. doi: 10.1902/jop.1994.65.5s.521. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Matsunaga T, Yamamoto G, Nishii K, Usui M, Yamamoto M, Tachikawa T. Comprehensive analysis of gene expression in the junctional epithelium by laser microdissection and microarray analysis. J Periodontal Res. 2010;45:618–625. doi: 10.1111/j.1600-0765.2010.01276.x. [DOI] [PubMed] [Google Scholar]
- Heymann R, Wroblewski J, Terling C, Midtvedt T, Obrink B. The characteristic cellular organization and CEACAM1 expression in the junctional epithelium of rats and mice are genetically programmed and not influenced by the bacterial microflora. J Periodontol. 2001;72:454–460. doi: 10.1902/jop.2001.72.4.454. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. Toll-like receptor downstream signaling. Arthritis research & therapy. 2005;7:12–19. doi: 10.1186/ar1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Cacalano G, Camerato T, Toy K, Moore MW, Wood WI. Chemokine binding and activities mediated by the mouse IL-8 receptor. J Immunol. 1995;155:2158–2164. [PubMed] [Google Scholar]
- Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- McDonald B, Kubes P. Chemokines: sirens of neutrophil recruitment-but is it just one song? Immunity. 2010;33:148–149. doi: 10.1016/j.immuni.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Mei J, Liu Y, Dai N, Hoffmann C, Hudock KM, Zhang P, Guttentag SH, Kolls JK, Oliver PM, Bushman FD, et al. Cxcr2 and Cxcl5 regulate the IL-17/G-CSF axis and neutrophil homeostasis in mice. J Clin Invest. 2012;122:974–986. doi: 10.1172/JCI60588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moughal NA, Adonogianaki E, Thornhill MK, Kinane DF. Endothelial cell leukocyte adhesion molecule-1(ELAM-1) on endothelial cells in experimental gingivitis in humans. J Periodont. 1992;64:355–357. doi: 10.1111/j.1600-0765.1992.tb01746.x. [DOI] [PubMed] [Google Scholar]
- Niederman R, Westernoff T, Lee C, Mark LL, Kawashima N, Ullman-Culler M, Dewhirst FE, Paster BJ, Wagner DD, Mayadas T, et al. Infection-mediated early-onset periodontal disease in P/E-selectin-deficient mice. J Clin Periodontol. 2001;28:569–575. doi: 10.1034/j.1600-051x.2001.028006569.x. [DOI] [PubMed] [Google Scholar]
- Nussbaum G, Shapira L. How has neutrophil research improved our understanding of periodontal pathogenesis? J Clin Periodontol. 2011;38(Suppl 11):49–59. doi: 10.1111/j.1600-051X.2010.01678.x. [DOI] [PubMed] [Google Scholar]
- Nylander K, Danielsen B, Fejerskov O, Dabelsteen E. Expression of the endothelial leukocyte adhesion molecule-1 (ELAM-1) on endothelial cells in experimental gingivitis in humans. J Periodontol. 1993;64:355–357. doi: 10.1902/jop.1993.64.5.355. [DOI] [PubMed] [Google Scholar]
- Page RC, Beatty P, Waldrop TC. Molecular basis for the functional abnormality in neutrophils from patients with generalized prepubertal periodontitis. J Periodontal Res. 1987;22:182–183. doi: 10.1111/j.1600-0765.1987.tb01562.x. [DOI] [PubMed] [Google Scholar]
- Phillipson M, Kubes P. The neutrophil in vascular inflammation. Nature medicine. 2011;17:1381–1390. doi: 10.1038/nm.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Ritzman AM, Hughes-Hanks JM, Blaho VA, Wax LE, Mitchell WJ, Brown CR. The chemokine receptor CXCR2 ligand KC (CXCL1) mediates neutrophil recruitment and is critical for development of experimental Lyme arthritis and carditis. Infect Immun. 2010;78:4593–4600. doi: 10.1128/IAI.00798-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovai LE, Herschman HR, Smith JB. The murine neutrophil-chemoattractant chemokines LIX, KC, and MIP-2 have distinct induction kinetics, tissue distributions, and tissue-specific sensitivities to glucocorticoid regulation in endotoxemia. Journal of leukocyte biology. 1998;64:494–502. doi: 10.1002/jlb.64.4.494. [DOI] [PubMed] [Google Scholar]
- Rovin S, Costich ER, Gordon HA. The influence of bacteria and irritation in the initiation of periodontal disease in germfree and conventional rats. J Periodontal Res. 1966;1:193–204. doi: 10.1111/j.1600-0765.1966.tb01860.x. [DOI] [PubMed] [Google Scholar]
- Sadik CD, Kim ND, Luster AD. Neutrophils cascading their way to inflammation. Trends in immunology. 2011;32:452–460. doi: 10.1016/j.it.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster DE, Kehrli ME, Jr, Ackermann MR. Neutrophilia in mice that lack the murine IL-8 receptor homolog. Science. 1995;269:1590–1591. doi: 10.1126/science.7667641. [DOI] [PubMed] [Google Scholar]
- Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci U S A. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebo JM, Datta S, Kishore R, Kolosov M, Major JA, Ohmori Y, Hamilton TA. Interleukin-1-mediated stabilization of mouse KC mRNA depends on sequences in both 5'- and 3'-untranslated regions. J Biol Chem. 2000;275:12987–12993. doi: 10.1074/jbc.275.17.12987. [DOI] [PubMed] [Google Scholar]
- Tonetti MS. Molecular factors associated with compartmentalization of gingival immune responses and transepithelial neutrophil migration. J Periodontal Res. 1997;32:104–109. doi: 10.1111/j.1600-0765.1997.tb01389.x. [DOI] [PubMed] [Google Scholar]
- Tonetti MS, Imboden MA, Gerber L, Lang NP, Laisue J, Mueller C. Localized expression of mRNA for phagocyte-specific hemotactic cytokines in human periodontal infections. Infect and Immun. 1994;62:4005–4014. doi: 10.1128/iai.62.9.4005-4014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y, Usui M, Yamamoto G, Takagi Y, Tachikawa T, Yamamoto M, Nakamura M. Role of the junctional epithelium in periodontal innate defense and homeostasis. J Periodontal Res. 2012 doi: 10.1111/j.1600-0765.2012.01490.x. [DOI] [PubMed] [Google Scholar]
- Umesaki Y, Setoyama H. Structure of the intestinal flora responsible for development of the gut immune system in a rodent model. Microbes Infect. 2000;2:1343–1351. doi: 10.1016/s1286-4579(00)01288-0. [DOI] [PubMed] [Google Scholar]
- Waldrop TC, Anderson DC, Hallmon WW, Schmalstieg FC, Jacobs RL. Periodontal manifestations of the heritable Mac-1, LFA-1, deficiency syndrome. Clinical, histopathologic and molecular characteristics. J Periodontol. 1987;58:400–416. doi: 10.1902/jop.1987.58.6.400. [DOI] [PubMed] [Google Scholar]
- Wuyts A, Haelens A, Proost P, Lenaerts JP, Conings R, Opdenakker G, Van Damme J. Identification of mouse granulocyte chemotactic protein-2 from fibroblasts and epithelial cells. Functional comparison with natural KC and macrophage inflammatory protein-2. J Immunol. 1996;157:1736–1743. [PubMed] [Google Scholar]
- Xu J, Gordon JI. Inaugural Article: Honor thy symbionts. Proc Natl Acad Sci U S A. 2003;100:10452–10459. doi: 10.1073/pnas.1734063100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JJ, Ruddy MJ, Wong GC, Sfintescu C, Baker PJ, Smith JB, Evans RT, Gaffen SL. An essential role for IL-17 in preventing pathogen-initiated bone destruction: recruitment of neutrophils to inflamed bone requires IL-17 receptor-dependent signals. Blood. 2007;109:3794–3802. doi: 10.1182/blood-2005-09-010116. [DOI] [PMC free article] [PubMed] [Google Scholar]