Abstract
Campylobacter jejuni is a major cause of food-borne gastroenteritis worldwide. While mortality is low, morbidity imparted by post-infectious sequelae such as Guillain-Barré syndrome, Reiter syndrome/reactive arthritis and irritable bowel syndrome is significant. In addition, the economic cost is high due to lost productivity. Food animals, particularly poultry, are the main reservoirs of C. jejuni. The over-use of antibiotics in the human population and in animal husbandry has led to an increase in antibiotic-resistant infections, particularly with fluoroquinolones. This is problematic because C. jejuni gastroenteritis is clinically indistinguishable from that caused by other bacterial pathogens, and such illnesses are usually treated empirically with fluoroquinolones. Since C. jejuni is naturally transformable, acquisition of additional genes imparting antibiotic resistance is likely. Therefore, an understanding of the antibiotic resistance mechanisms in C. jejuni is needed to provide proper therapy both to the veterinary and human populations.
Keywords: Campylobacter jejuni, antibiotic resistance, efflux pump, CmeABC, major outer membrane protein, MOMP, porin
Introduction
Campylobacter jejuni is a small, Gram-negative, curved rod and is the most common cause of bacteria-mediated diarrheal disease globally.1 For the first time in 2005, campylobacteriosis exceeded salmonellosis as the most commonly reported zoonosis in the European Union, and the number of cases continues to increase.2,3 Campylobacteriosis is also the most common notifiable disease in New Zealand and Australia.4,5 There is little human-to-human transmission, probably due to its microaerophilic nature. Instead, it is primarily a zoonosis because it is a commensal of food animals, particularly poultry, which serves as the main reservoir for human infection.6 Meat becomes contaminated during the slaughtering process, and C. jejuni survives in the crevices of animal carcasses where oxygen tension is low.7 Although implementation of Hazard Analysis and Critical Control Points (HACCP) in the food industry in the mid 1990s markedly reduced the rate of Campylobacter infections,8C. jejuni remains second only to Salmonella as the cause of food-borne disease in the United States.9 However, other modes of transmission, such as drinking contaminated water, are also important means of disease spread.10
The indiscriminate use of antibiotics in the human population as well as the use of antibiotics in animal husbandry, for treatment, growth promotion and off-label uses, has led to an increase in antibiotic-resistant Campylobacter infections, particularly with regard to fluoroquinolones (FQ).9,11-16 There is evidence to support the hypothesis that resistance patterns in poultry may predict human resistance patterns; this has been most clearly shown with FQ.9,11-20 Although not all cases of Campylobacter infection require treatment,21 many cases of acute diarrhea are empirically treated with FQ, which likely further contributes to the emergence of FQ resistance.
The use of veterinary antibiotics varies greatly throughout the world. Of greatest concern are situations in which antibiotics can be used for growth-promotion purposes (as opposed to therapeutic) because the low levels of antibiotics used in this setting and over long periods of time set the stage for the emergence of resistant bacteria. In some areas including Indonesia, Thailand, India and parts of Africa, veterinary antibiotics can be obtained without prescription or other controls.20,22 In contrast, the general use of antibiotics for growth promotion is banned in the European Union and Japan,23 and FQ cannot be used in food producing animals in Australia.
Although Campylobacter has an extensive formidable restriction modification system that would tend to decrease the uptake of foreign genetic material, it is also naturally transformable, and the acquisition of resistance genes from other organisms has been described.24-35 For all these reasons, the study of the resistance mechanisms present in C. jejuni is important to both human and veterinary health.
The genetic elements that underlie these mechanisms may be chromosomal or plasmid-borne, and represent a combination of endogenous and acquired genes. In general, mechanisms of antibiotic resistance include (Table 1):
Table 1. Antibiotic resistance mechanisms of Campylobacter.
| Antibiotic class | Resistance mechanisms |
|---|---|
| Aminoglycoside |
Modification of the antibiotic by aminoglycoside-modifying enzymes (AphA, AadE, Sat) Contribution of efflux is not clear |
| Beta-Lactam |
Enzymatic inactivation of the antibiotic by β-lactamase (penicillinase, OXA-61) Decreased membrane permeability of most anionic and MW > 360 kDa antibiotics due to MOMP Efflux through CmeABC and possibly others |
| Fluoroquinolone |
Modification of the DNA gyrase target (Thr-86-Ile; also Asp-90-Asn, Ala-70-Thr) Efflux through CmeABC |
| Macrolide |
Mutations in 23S rRNA Contribution of mutations in ribosomal proteins L4/L22 is likely minor Efflux through CmeABC and possibly others Decreased membrane permeability due to MOMP |
| Tetracycline | Modification of the target ribosomal A site by TetO binding Efflux through CmeABC and possibly others Contribution of decreased membrane permeability due to MOMP is not clear |
(1) Modification of the antibiotic’s target and/or its expression (i.e., DNA gyrase mutations)
(2) Inability of the antibiotic to reach its target (i.e., expression of the major outer membrane protein or MOMP)
(3) Efflux of the antibiotic (i.e., multidrug efflux pumps such as CmeABC)
(4) Modification or inactivation of the antibiotic (i.e., β-lactamase production)
In Campylobacter, a recurring theme is synergy between antibiotic efflux and a second mechanism. The best-described multi-drug efflux pump in Campylobacter is CmeABC, consisting of three components: an outer membrane protein (encoded by cmeC), an inner membrane drug transporter (encoded by cmeB), and a periplasmic protein (encoded by cmeA) that bridges CmeB and CmeC.36,37 This efflux pump also contributes to resistance to bile acids.38 Other putative efflux pumps including CmeDEF and CmeG, may also contribute to antibiotic resistance.39,40 Sequencing reveals that C. jejuni has a total of 14 possible efflux pumps, but most have not been characterized functionally.41 In addition to intrinsic resistance mediated by efflux,36,37,39,40,42-44 antibiotic exclusion [via the major outer membrane porin (MOMP),45 lipooligosaccharide and possibly capsule]46 also contribute to intrinsic resistance. Campylobacter exhibits intrinsic resistance to novobiocin, bacitracin and vancomycin, polymyxin/colistin, presumably due to the absence of appropriate targets and/or low affinity binding to targets.47-50 In the case of intrinsic resistance to trimethoprim,26,47,51-53 variant forms of dihydrofolate reductases (encoded by dfr1 most often but also by dfr9) that are not inhibited by trimethoprim are found in > 90% of C. jejuni that have been examined.26
Approximately 90% of Campylobacter infections in humans are caused by C. jejuni (C. coli accounts for ~9%),54 and the majority of the literature on human infection focuses on C. jejuni. Therefore, this review will focus on the antibiotic resistance mechanisms found in C. jejuni for commonly-used antibiotics.
Fluoroquinolone Resistance
Fluoroquinolones manifest concentration-dependent, bactericidal activity against a wide variety of both Gram-negative and Gram-positive organisms, are available in both oral and intravenous forms, are conveniently dosed once or twice daily usually, and are well-tolerated; all these attributes make this a heavily-used class of antibiotic in humans. Nalidixic acid is the parent, non-fluorinated compound of this antibiotic class. The fluoroquinolones include the most commonly used antibiotics (i.e., ciprofloxacin) to treat acute bacterial diarrhea, although macrolides are the drug of choice if campylobacteriosis is strongly suspected.21 However, campylobacteriosis is clinically indistinguishable from other causes of bacterial diarrheal illness, and so without epidemiology suggestive of Campylobacter infection, many cases are treated empirically with FQ. As such, FQ resistance is of great clinical concern.
Worldwide, FQ resistance was unusual in the late 1980s to early 1990s.12,13,55 However, the combination of indiscriminate use of FQ in humans and increased FQ use in the poultry industry in particular, has contributed to an increase in the prevalence of FQ-resistance in both animals and humans.11,12,14 The surveillance of FQ susceptibility in Campylobacter in animals is important not only for purposes of food production, but also because the emergence of resistant strains in animals portends an increase in resistant human infections.11-14,56 Recognition of this connection has supported the limitation or outright banning of FQ for veterinary purposes in many countries including the United States,57 Denmark, and Australia among other countries. Accordingly, FQ-resistance in > 150 Campylobacter isolates from broilers in Australia was reported to be between 0–2.4%,58,59 which likely contributes to the similarly low-level of FQ-resistance (2%) in human isolates.59 Additionally, data from the Danish Integrated Antimicrobial Resistance Monitoring and Research Programme (DANMAP) show that C. jejuni isolates from domestic broilers was 11%, compared with 57% from imported broilers,55 and that while one-third of domestically-acquired Campylobacter infections were FQ-resistant in 2011, 84% of infections acquired abroad were FQ-resistant.55 Similarly, 6.5% of human Campylobacter infections acquired in Norway were FQ-resistant, compared with 67% FQ-resistance in infections acquired abroad.60
However, the veterinary use of FQ varies widely throughout the world depending on the production setting (i.e., commercial vs. family-owned) as well as on a country-wide basis.11,52,53,61 For example, as in many countries, antibiotics are used both prophylactically and therapeutically in industrialized and commercial free-range poultry farms in South Africa, but not in small-scale family farming. The rates of FQ resistance were highest in commercial free-range broilers, at > 95%, but were lower in industrial broiler (16%) and lowest in poultry from family farms (8%).62 Thailand also reports very high rates of FQ resistance in C. jejuni from broilers, upwards of 80%.13,52,63 In Japan, the rate of nalidixic acid resistance in C. jejuni from broiler flocks was 55%, and for a veterinary FQ (enrofloxacin) the rate was 30%.64 The experience in Europe has been quite variable, ranging from very low FQ-resistance (1.2%) in broilers in Norway,60 intermediate in Belgium56 and Poland,65 (44% and 56%, respectively), to alarmingly high in Spain, where it was reported in 2000 that 99% of Campylobacter isolates from broilers were FQ-resistant.66
In the United States in 2004, the Federal Drug Administration reversed its prior approval for the therapeutic use of the veterinary FQ enrofloxacin because of concerns that the rising level of FQ-resistant Campylobacter in poultry was being reflected in increasing FQ resistance in human isolates.12,57 However, it is not yet clear if the elevated rates of FQ resistance will decline after FQ restriction. Studies in several countries have shown that FQ-resistant Campylobacter persists in poultry populations after the withdrawal of FQ,67-70 suggesting that there is little/no fitness cost to FQ-resistance in Campylobacter.70,71 The latest available data from the United States shows that resistance to ciprofloxacin in human isolates of C. jejuni peaked in 2007 at nearly 26%, and has since declined somewhat to ~22% in 2010;54 whether this is a true decline attributable even in part to the enrofloxacin ban is debatable.
In Campylobacter, there are two well-described mechanisms that underlie resistance to FQ: (1) inactivation of the target of FQ; (2) efflux of FQ. These two mechanisms work together synergistically.72-74 In general, the two intracellular enzymatic targets of FQ are DNA gyrase (encoded by gyrA and gyrB) and the structurally related topoisomerase IV (encoded by parC and parE).75 Fluoroquinolones form a stable complex with these enzymes and traps them onto DNA, leading to decreased DNA replication, transcription, and ultimately to cell death.76,77 However, several studies have demonstrated that C. jejuni and C. coli lack the parC and parE genes41,78-80; therefore, they cannot be a source of FQ resistance. Instead, FQ-resistance in C. jejuni and C. coli occurs via specific point mutations in the quinolone resistance-determining region (QRDR) of the gyrA gene, with the Thr-86-Ile mutation being both the most common. This single mutation in gyrA leads to high-level resistance to nalidixic acid and FQ (ciprofloxacin minimum inhibitory concentration (MIC) > 16 g/ml), unlike FQ resistance in E. coli or Salmonella, which requires the accumulation of several point mutations in the QRDR before high-level resistance is achieved.81 Interestingly, the less common Thr-86-Ala mutation confers resistance only to nalidixic acid but not FQ.82 Perhaps since only a single point mutation is required for high level resistance, FQ-resistant mutants appear rapidly both in animals and humans16,67,72,73,83-88 The less common Asp-90-Asn and Ala-70-Thr mutations in gyrA confer intermediate-level resistance to FQ (ciprofloxacin MIC 6–16 g/ml).89,90 While mutations in gyrB have been reported, they do not confer FQ resistance.78,80,91
It is somewhat surprising that the Thr-86-Ile mutation in gyrA seems to increase the fitness of Campylobacter in a chicken model,71 although this observation supported by the previously mentioned studies demonstrating that these resistant strains persist even after FQ are withdrawn from poultry flocks for several years.67-69 Conflicting reports exist about whether this mutation translates into more severe infections in humans.92-97
Another mechanism of FQ-resistance that seems to work in concert with gyrA mutations is efflux via the chromosomally-encoded CmeABC multidrug efflux pump, which reduces the intracellular concentration of FQ and several other antibiotics.36,37 This efflux pump acts synergistically with DNA gyrase mutations to effect high-level FQ-resistance;72-74 for example, strains carrying DNA gyrase mutations that alone lead to intermediate-level FQ resistance manifest high-level resistance when CmeABC is also expressed.73,74 Also, CmeABC assists in the emergence of gyrA mutants that otherwise could not survive selection by even low dose FQ.74 Unlike efflux pump mechanisms in other Gram-negative bacteria which require overexpression to lead to clinically relevant resistance, the basal, constitutive expression of CmeABC is sufficient to mediate FQ resistance (although experimental overexpression does increase the level of FQ-resistance).74
An additional putative efflux pump, CmeG, has also recently been described as conferring both resistance to structurally unrelated antibiotics as well as oxidants.40 Insertional mutagenesis of cmeG led to a 4-fold reduction in ciprofloxacin resistance vs. the wild-type parent, and an 8- to 32-fold increase in resistance to ciprofloxacin and other FQ when cmeG was overexpressed.40
Interestingly, FQ-resistance has emerged on poultry farms even in the absence of FQ administration.64,98 Since the major mechanism of nalidixic acid and FQ-resistance in Campylobacter is via point mutations in gyrA, it is difficult to attribute this phenomenon to co-inheritance of multi-resistance mobile elements. It has been suggested that other antibiotics could select for FQ-resistance in Campylobacter,98 but whether this occurs via expression of the CmeABC pump or other mechanism remains to be clarified.
Macrolide Resistance
Macrolide antibiotics and the closely-related ketolides are large molecules (MW > 700) that inhibit bacterial protein synthesis. The macrolide antibiotic erythromycin is the treatment of choice for campylobacteriosis.21 Other members of this class of antibiotics include clarithromycin, azithromycin, telithromycin (technically a ketolide), tylosin and tilmicosin; the latter two are approved for veterinary use only (erythromycin also has a veterinary indication).99 Macrolides inhibit protein synthesis by binding reversibly to the P site on the 50S subunit of bacterial ribosomes. The main mechanisms of resistance to macrolides in Campylobacter are (1) target modification, (2) efflux and (3) altered membrane permeability. The first two mechanisms act synergistically to confer high-level macrolide resistance.100,101 A fourth mechanism of macrolide resistance, enzymatic modification of macrolides, has not been described in Campylobacter.102
As in other bacteria, point mutations in the peptidyl encoding region in domain V of the 23S rRNA gene at positions 2074 and 2075 (corresponding to positions 2058 and 2059 in E. coli numbering) confer high-level macrolide resistance,101,103-109 with the 2075 substitution being more common.104,110 C. jejuni and C. coli carry three copies of 23s rRNA gene,41,111 all of which are usually mutated in macrolide-resistant strains. However, some strains with lower MICs to macrolides have been found to have only two mutated gene copies, suggesting a gene dosage effect.110,112,113 Strains harboring single mutations in 23S rRNA have not been reported. Mutations (usually insertions) in the ribosomal proteins L4 and L22 leading to macrolide resistance but are not the major means of tetracycline resistance.100,106
The barrier to the generation of macrolide resistance in Campylobacter appears to be much higher than that of FQ-resistance. In two studies, several weeks of tylosin administration to poultry at a growth-promotion dose was necessary to select for macrolide resistance.101,114 In another departure from FQ-resistant Campylobacter, macrolide resistance imparts a fitness cost, at least when analyzed in competition experiments.114-118 These two factors combined with a low spontaneous mutation rate leading to macrolide resistance (~10−10 per cell per generation)101 and clinical efficacy, make macrolides the drug of choice to treat campylobacteriosis. Through 2008, there was concern in the US about increasing macrolide resistance in both C. jejuni and C. coli (2.3% and 10.1%, respectively), but there was a decline over the following two years to 1.2% and 4.3% for these strains.54 As of 2010 in the European Union, the highest rates of macrolide resistance for C. jejuni is in Malta (10%), and Italy for C. coli (33%).119 Unfortunately, rates are much higher in parts of Asia and Africa; for example, in Nigeria, nearly 80% of strains are macrolide-resistant.120 Similar to observations made with FQ-resistance in South Africa, 88% of Campylobacter isolates from poultry raised commercially were erythromycin resistant vs. 0% for those isolates from small-scale family farms.62
The multidrug efflux pump CmeABC also contributes to macrolide resistance36,37,101,107,113,121 and functions synergistically with 23S rRNA mutations to effect high-level macrolide resistance.43,100,122 In mutants that are macrolide-resistant but lack 23S rRNA mutations, gene disruption of cmeB or antisense-mediated gene silencing of cmeA leads to inactivation of the CmeABC transporter and mediates reversion to a macrolide-susceptible phenotype.100,123 The putative efflux pump CmeG may also contribute to macrolide resistance, as insertional mutagenesis of cmeG causes an 8-fold reduction in erythromycin resistance vs. the wild-type parent.40 In addition, there is one study that suggests the existence of a second efflux system that contributes to low-level macrolide resistance, but it has not been further characterized.107
A third mechanism of macrolide resistance involves altered membrane permeability mediated by expression of the major outer membrane porin (MOMP), chromosomally encoded by porA.44,45 In Gram-negative bacteria, porins are outer membrane proteins that form transmembrane pores and allow the passive diffusion of hydrophilic molecules, including many antibiotics. Properties of the pore including its size and charge characteristics underlie the selectivity for what can pass through it. In C. jejuni and C. coli, MOMP forms a cation-selective pore that is smaller than pores typically found in E. coli,124 and therefore should limit the entry of most antibiotics with a molecular weight greater than 360 such as the macrolides (MW > 700).45 However, since macrolides are known to be very effective against Campylobacter, these drugs must be able to cross the outer and cytoplasmic membranes. Whereas porins provide an aqueous environment for the transport of hydrophilic molecules, the relatively hydrophobic macrolides are thought to gain access to the cytoplasm of Gram-negative bacteria via a “hydrophobic pathway”125,126. This pathway seems to be promoted in E. coli and Salmonella strains bearing mutations in lipopolysaccharide (LPS) synthesis genes that yield truncated LPS (lacking hydrophilic O-antigen sugars). The outer membranes of these mutant strains are therefore relatively more hydrophobic than the parent strains, and exhibit increased susceptibility to hydrophobic antibiotics including macrolides.126 Given that Campylobacter naturally expresses lipooligosaccharide (LOS), which lacks the hydrophilic sugars expressed by full-length LPS in other Gram-negative bacteria,127,128 it is reasonable to speculate that this comparatively increased outer membrane hydrophobicity promotes the uptake of macrolides. This is supported by the observation that LOS truncation increases the susceptibility of C. jejuni to erythromycin by 8-fold, an effect that was doubled in C. jejuni mutants also carrying the A2074G mutation.46
β-Lactam Resistance
The β-lactam antibiotics are a diverse class of compounds including penicillins, cephalosporins, carbapenems and monobactams, all of which contain the β-lactam ring required for antimicrobial activity. Individual members of this family are distinguished by various side chains that confer particular properties such as pharmacokinetics, resistance to stomach acid, hydrolysis by β lactamases, etc. By binding to and thereby inactivating the bacterial peptidoglycan transpeptidases (also known as penicillin-binding proteins) required to catalyze the final cross linking step, the resulting bacterial cell walls lack structural integrity and are subject to osmotic swelling and lysis. Exactly how this leads to bacterial cell death is not completely clear, but the unopposed action of autolysins, necessary for normal turnover and remodeling of peptidoglycan, may play a role.129
Three mechanisms mediate β-lactam resistance in Campylobacter: (1) enzymatic inactivation by chromosomally-encoded β-lactamases, (2) reduced uptake due to alterations in outer membrane porins and (3) efflux.
Expression of a penicillinase-type of β-lactamase in Campylobacter confers resistance to amoxicillin, ampicillin and ticarcillin, which can be overcome with the β-lactamase inhibitors tazobactam, clavulanic acid and sulbactam.105 This enzyme does not affect susceptibility to the carbapenems or cephalosporins. More recently, a class D β-lactamase OXA-61, was identified in Campylobacter.130 This enzyme shows similarity to other OXA-type genes in Fusobacterium, Acinetobacter and Pseudomonas, and mediates resistance to penicillin, oxacillin, ampicillin, amoxicillin-clavulanate, piperacillin and carbenicillin.130,131 While OXA-61 is highly prevalent in the veterinary and human populations studied,131 pooled national data on the prevalence of β-lactam resistance in general is not available since the NARMS does not include the β-lactam class for Campylobacter 54. However, it appears that the prevalence of β-lactamase varies widely in both poultry and human populations, but is usually greater than 20%.105,131-134 Finally, two genes encoding a metallo-β-lactamase type of enzyme has been reported, although it is not yet clear if expression actually leads to β-lactamase resistance.131,132,135
As with macrolides, the cation-selective MOMP in C. jejuni and C. coli tend to exclude most β-lactams with a molecular weight greater than 360 or which are anionic.45 The partial positive charge and small size of imipenem (MW 299), ampicillin (MW 333) and cefpirome (MW 347) are consistent with passage through MOMP, and susceptibility to these antibiotics in the absence of a second mechanism such as β-lactamase production. Amoxicillin’s molecular weight of 365 would seem to preclude efficient passage through MOMP, although its partial positive charge might facilitate entry through MOMP; alternatively, a non-MOMP-dependent mechanisms may mediate its entry.45
The CmeABC efflux pump may also contribute to β-lactam resistance. Insertional mutagenesis of cmeB in C. jejuni strain 81-176136 and another strain led to a 32-fold increase in ampicillin susceptibility.36 In another study using NCTC strain 11168, the cmeB mutant was 4 times more susceptible to ampicillin compared with the parent strain, and overexpression of cmeB lead to a 4-fold increase in ampicillin resistance.44 Inactivation of the putative efflux pump CmeDEF by insertional mutagenesis of cmeF only led to a 2-fold increase in ampicillin resistance in strain 11168 and a 2-fold increase in cefotaxime resistance in the well-described, invasive, human outbreak strain 81-176.39 Also, inactivation of the putative efflux pump CmeG did not affect cefotaxime resistance in C. jejuni strain 11168.40 Therefore, at this point it seems that CmeABC is the most potent efflux pump with regard to β-lactams.
Tetracycline Resistance
The tetracyclines were discovered in the 1940s and have activity against Gram-negative and Gram-positive organisms. Due to their heavy use in the past for both human and veterinary indications, widespread resistance has somewhat limited their use today. Commonly used members of this class are tetracycline, doxycycline and minocycline. The tetracyclines are lipophilic protein synthesis inhibitors that likely use a combination of the hydrophobic pathway described for macrolides as well as outer membrane porins to gain access to the bacterial ribosome; exactly how each pathway contributes to tetracycline entry in Campylobacter is not completely clear. Known mechanisms of tetracycline resistance in Campylobacter are (1) alteration of tetracycline’s ribosomal target and (2) efflux.
In other Gram-negatives, tetracyclines form a complex with magnesium, which imparts a positive charge that facilitates passage of the complex through pores formed by OmpC and OmpE.137 Although MOMP of Campylobacter shares an antigenically-related region to OmpC in E. coli,138,139 it is not certain that the high molecular weight of tetracyclines (> 400) allows passage through the relatively small pores (MW exclusion ~360) imparted by MOMP.45 Nevertheless, once inside the bacteria cytoplasm, tetracyclines reversibly bind to the 30S subunit of ribosomes and inhibit protein synthesis by preventing the attachment of charged aminoacyl-tRNA to the ribosomal A site.140,141 The major mechanism of tetracycline resistance in Campylobacter as well as other Gram-negatives is protection of an unoccupied A site by the binding of bacterial protein TetO to that site.142,143 TetO can be encoded on the chromosome,144 or more commonly, on the plasmids pTet in C. jejuni145 and pCC31 in C. coli.146,147 According to 2010 NARMS data, 43% of C. jejuni and 49% of C. coli isolates are tetracycline-resistant,54 making this class of antibiotic of little use in veterinary or human Campylobacter-mediated disease.54,144,148
Although high-level resistance to tetracyclines can be mediated by TetO alone, the contribution of efflux to tetracycline resistance is demonstrated by the increase in tetracycline MIC when efflux pumps are genetically inactivated. For example, disruption of the putative efflux pump cmeG rendered the mutant strain 4-fold more susceptible to tetracycline compared with the wild-type strain.40 Also, inactivation of the CmeABC efflux pump by disruption of cmeB led to an 8-fold decrease in the tetracycline MIC in a TetO-minus poultry isolate,36 and similar findings were described with the NCTC isolate 11168 when cmeB was disrupted.37 In a different study of other C. jejuni strains including 81-176, cmeB disruption rendered strains 16- to 64-fold more susceptible to tetracycline compared with the parent strains.112 These studies also suggested that when both CmeABC and TetO are functional, the impact on tetracycline resistance is synergistic.36,37,112
Aminoglycoside Resistance
Aminoglycosides are protein synthesis inhibitors of many Gram-positive and Gram-negative organisms. They contain amino-modified sugars, are positively charged, water-soluble and have molecular weights ranging from 445 to 600.149,150 Commonly used members of this group include gentamicin, kanamycin, amikacin, neomycin, tobramycin and streptomycin. The initial binding of aminoglycosides to negatively charged bacterial membranes is electrostatic in nature and relatively slow compared with the second phase of rapid but reversible binding to the 30S segment of the ribosome.151 Transfer of aminoglycosides across the bacterial cytoplasmic membranes requires oxygen, an intact electron transport system and ATP.150,152,153 According to 2010 NARMS data, > 99% of C. jejuni and 88% of C. coli isolates are susceptible to aminoglycosides.54 These data suggest that despite Campylobacter’s microaerophilic nature, sufficient oxygen is present for the uptake of aminoglycosides.
There are two major means by which aminoglycosides exert antimicrobial activity: (1) interference with the translocation of the nascent peptide chain from the ribosomal A site to the P site leading to premature termination, and (2) interference with proof-reading, leading to incorporation of incorrect amino acids and dysfunctional protein.154 The main mechanism of aminoglycoside resistance in C. jejuni is via aminoglycoside modifying enzymes, which are usually plasmid-borne.
Aminoglycoside resistance was first detected in C. coli and was mediated by a 3′-aminoglycoside phosphotransferase (encoded by aphA-3) that had been previously described as conferring kanamycin resistance in Streptococcus and Staphylococcus.29 This aphA-3 gene remains the most common source of aminoglycoside resistance in Campylobacter. In some strains, aphA-3 is located downstream of an insertion sequence (IS607*) bearing similarity to IS607 found in H. pylori.27 In other strains, aphA-3 is found with genes encoding streptomycin resistance (encoded by aadE, a 6′-adenylyl transferase) and streptothricin resistance (encoded by sat, an acetyl transferase).27 The existence of a similar resistance cluster in Enterococcus suggests that Campylobacter acquired these genes via horizontal transfer.27 Other Campylobacter strains harbor mosaic plasmids that contain various aminoglycoside resistance genes and insertion or transposon sequences from Gram-negative (i.e., H. pylori, E. coli and Salmonella) and Gram-positive sources (i.e., Enterococcus), along with tetO.24,25,27,28,30-32,34,35 Acquisition of such plasmids by susceptible C. jejuni confers a multi-drug-resistant phenotype and that can present a clinical challenge in both the veterinary and human populations.
Other genes which confer kanamycin resistance include aphA-1 and aphA-7, which were detected on plasmids in C. jejuni.155-157 Unlike aphA-3 and aphA-1, which are thought have been horizontally acquired, aphA-7 has a similar G-C content to C. jejuni chromosomal DNA, suggesting it is intrinsic in Campylobacter.158 Finally, there is a single report of a mutation in ribosomal protein S12 (encoded by rpsL) in C. coli that confers streptomycin resistance, but a similar mutation has not yet been described in C. jejuni.159
The contribution of efflux to aminoglycoside resistance is less clear. In one study, the putative efflux pump inhibitors phenyl-arginine-β-naphthylamide and 1-(1-naphthylmethyl)-piperazine did not affect the MIC of kanamycin in 5 C. jejuni strains.160 Another study which directly measured the effect of the putative efflux pump CmeG by insertional mutagenesis and comparison of MICs of various antibiotics found 16-fold reduction in gentamicin resistance in the mutated CmeG strain vs. the wild-type parent.40 However, no effect on streptomycin resistance was noted, and overexpression did not lead to increased aminoglycoside resistance.40 Therefore, the contribution of efflux to aminoglycoside resistance in Campylobacter is not completely clear but is likely to be less important than the plasmid-borne drug-modifying enzymes described previously.
Conclusions
Antibiotic resistance in C. jejuni is an increasing problem, as it is in many other microorganisms. Due to Campylobacter’s natural competence and hypervariable genomic sequences,41 there is considerable genomic plasticity that supports the emergence of resistant mutants. Because Campylobacter is a commensal of many animal species that are exposed to veterinary antimicrobials, ample opportunity exists for Campylobacter to continue to evolve additional resistance mechanisms. Furthermore, the over-use of antibiotics in the human population is an additional important source of selective pressure. Both scenarios contribute to the current problem of FQ resistance in Campylobacter. In this regard, the lack of a fitness cost (and perhaps even a fitness advantage71) of FQ-resistant C. jejuni is an issue that must be remembered when future resistance mutations arise in C. jejuni against other antimicrobials. Of greatest clinical concern would be the emergence of widespread macrolide resistance, since this class is the current treatment of choice for campylobacteriosis. A better understanding of the mechanism of macrolide entry (possibly via the hydrophobic pathway) may be useful in eventually mitigating the impact of acquired macrolide resistance. Also, the contribution of efflux pumps to antibiotic resistance warrants further study, since this mechanism acts synergistically with other mechanisms of antibiotic resistance to confer high-level resistance in many instances. Furthermore, genome sequencing predicts 14 potential efflux pumps,41 but only CmeABC and CmeG have been studied functionally,40,44,161 making this a fertile area for future research.
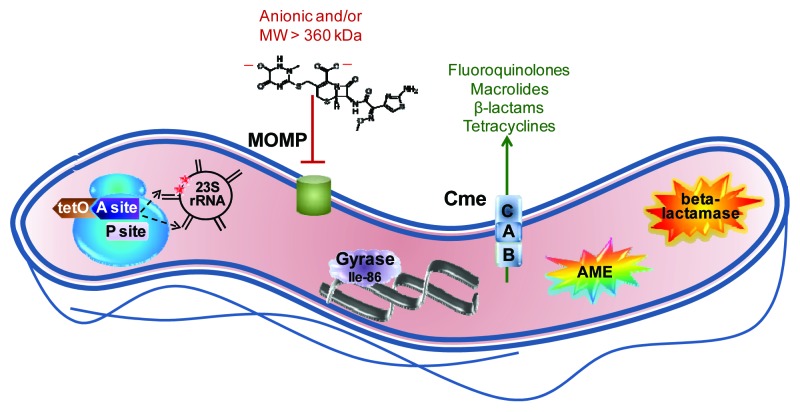
The antibiotic resistance mechanisms discussed herein are summarized in Figure 1.
Figure 1. Summary of major antibiotic resistance mechanisms in Campylobacter. The ribosome, shown in blue at the left, is the site of two major resistance mechanisms. Binding of the TetO protein (shown in brown) to the A site (shown in dark purple) prevents tetracycline from occupying that site but stills allows access of the aminoacyl tRNA so that protein synthesis continues. Point mutations in 23S rRNA in the domain V region (shown in black) at position 2,075 principally and less often at position 2,074 (indicated by red stars) decrease the binding affinity for macrolides and lead to resistance. The major outer membrane protein (MOMP, shown in green), limits the entry of most antibiotics that are negatively charged or with a molecular weight larger than 360 kDa; the structure of the 552 kDa, dianionic antibiotic ceftriaxone is shown as an example. The Thr-86-Ile substitution in DNA gyrase (shown in light purple), is the main means of fluoroquinolone resistance, and this single mutation also confers high level resistance to this antibiotic class. The multi-drug efflux pump CmeABC (shown as stacked blue squares) contributes to resistance against fluoroquinolones, macrolides, β-lactams and tetracyclines, and works synergistically with other resistance mechanisms, often leading to high-level resistance. Aminoglycoside-modifying enzymes (AME; shown as the multi-colored star burst), principally of the aminoglycoside phosphotransferase family, are the main means of aminoglycoside resistance. Finally, β-lactamases (shown as the orange star burst) of the penicillinase type as well as the Ambler class D OXA-61 contribute to β-lactam resistance.
Acknowledgments
I thank Joan Whitlock and Reem Itani for critical reading of this manuscript.
Glossary
Abbreviations:
- FQ
fluoroquinolone
- LOS
lipooligosaccharide
- LPS
lipopolysaccharide
- MIC
minimum inhibitory concentration
- MOMP
major outer membrane protein
- NARMS
National Antimicrobial Resistance Monitoring System
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/virulence/article/23753
References
- 1.Zilbauer M, Dorrell N, Wren BW, Bajaj-Elliott M. Campylobacter jejuni-mediated disease pathogenesis: an update. Trans R Soc Trop Med Hyg. 2008;102:123–9. doi: 10.1016/j.trstmh.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 2.Eurosurveillance editorial team The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2010. Euro Surveill. 2012;17:20113. [PubMed] [Google Scholar]
- 3.EFSA The Community Summary Report on antimicrobial resistance in zoonotic and indicator bacteria from animals and food in the European Union in 2008. Eur Food Safety Auth J. 2010;8:1658–919. [Google Scholar]
- 4.OzFoodNet Working Group Monitoring the incidence and causes of diseases potentially transmitted by food in Australia: annual report of the OzFoodNet Network, 2009. Commun Dis Intell. 2010;34:396–426. [PubMed] [Google Scholar]
- 5.Notifiable and Other Diseases in New Zealand. Annula Report 2011. In: The Institute of Environmental Science and Research Ltd. Porirua, New Zealand; 2012. [Google Scholar]
- 6.Allos BM, Blaser MJ. Campylobacter jejuni and related species. In: Mandell, ed. Principles and Practice of Infectious Diseases. Philadelphia: Churchill Livingstone; 2010:2793-802. [Google Scholar]
- 7.Blaser MJ, Taylor DN, Feldman RA. Epidemiology of Campylobacter jejuni infections. Epidemiol Rev. 1983;5:157–76. doi: 10.1093/oxfordjournals.epirev.a036256. [DOI] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention (CDC) Incidence of foodborne illnesses: preliminary data from the Foodborne Diseases Active Surveillance Network (FoodNet)--United States, 1998. MMWR Morb Mortal Wkly Rep. 1999;48:189–94. [PubMed] [Google Scholar]
- 9.FoodNet - Foodborne Diseases Active Surveillance Network. Centers for Disease Control and Prevention, 2012. (Accessed March 14, 2012, at http://www.cdc.gov/foodnet/)
- 10.Thomas C, Gibson H, Hill DJ, Mabey M. Campylobacter epidemiology: an aquatic perspective. J Appl Microbiol. 1998;85(Suppl 1):168S–77S. doi: 10.1111/j.1365-2672.1998.tb05296.x. [DOI] [PubMed] [Google Scholar]
- 11.Endtz HP, Ruijs GJ, van Klingeren B, Jansen WH, van der Reyden T, Mouton RP. Quinolone resistance in campylobacter isolated from man and poultry following the introduction of fluoroquinolones in veterinary medicine. J Antimicrob Chemother. 1991;27:199–208. doi: 10.1093/jac/27.2.199. [DOI] [PubMed] [Google Scholar]
- 12.Gupta A, Nelson JM, Barrett TJ, Tauxe RV, Rossiter SP, Friedman CR, et al. NARMS Working Group Antimicrobial resistance among Campylobacter strains, United States, 1997-2001. Emerg Infect Dis. 2004;10:1102–9. doi: 10.3201/eid1006.030635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luangtongkum T, Jeon B, Han J, Plummer P, Logue CM, Zhang Q. Antibiotic resistance in Campylobacter: emergence, transmission and persistence. Future Microbiol. 2009;4:189–200. doi: 10.2217/17460913.4.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith KE, Besser JM, Hedberg CW, Leano FT, Bender JB, Wicklund JH, et al. Investigation Team Quinolone-resistant Campylobacter jejuni infections in Minnesota, 1992-1998. N Engl J Med. 1999;340:1525–32. doi: 10.1056/NEJM199905203402001. [DOI] [PubMed] [Google Scholar]
- 15.Threlfall EJ, Ward LR, Frost JA, Willshaw GA. Spread of resistance from food animals to man--the UK experience. Acta Vet Scand Suppl. 2000;93:63–8, discussion 68-74. [PubMed] [Google Scholar]
- 16.van Boven M, Veldman KT, de Jong MC, Mevius DJ. Rapid selection of quinolone resistance in Campylobacter jejuni but not in Escherichia coli in individually housed broilers. J Antimicrob Chemother. 2003;52:719–23. doi: 10.1093/jac/dkg402. [DOI] [PubMed] [Google Scholar]
- 17.Hooper DC. Emerging mechanisms of fluoroquinolone resistance. Emerg Infect Dis. 2001;7:337–41. doi: 10.3201/eid0702.010239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore JE, Barton MD, Blair IS, Corcoran D, Dooley JS, Fanning S, et al. The epidemiology of antibiotic resistance in Campylobacter. Microbes Infect. 2006;8:1955–66. doi: 10.1016/j.micinf.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 19.Kittl S, Kuhnert P, Hächler H, Korczak BM. Comparison of genotypes and antibiotic resistance of Campylobacter jejuni isolated from humans and slaughtered chickens in Switzerland. J Appl Microbiol. 2011;110:513–20. doi: 10.1111/j.1365-2672.2010.04906.x. [DOI] [PubMed] [Google Scholar]
- 20.Monitoring antimicrobial usage in food animals for the protection of human health: Report of a WHO consultation. World Health Organization Department of Communicable Disease, Surveillance and Response 2001.
- 21.Guerrant RL, Van Gilder T, Steiner TS, Thielman NM, Slutsker L, Tauxe RV, et al. Infectious Diseases Society of America Practice guidelines for the management of infectious diarrhea. Clin Infect Dis. 2001;32:331–51. doi: 10.1086/318514. [DOI] [PubMed] [Google Scholar]
- 22.Sarmah AK, Meyer MT, Boxall AB. A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere. 2006;65:725–59. doi: 10.1016/j.chemosphere.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 23.Council regulation (EC) No. 2821/98 of 17 December 1998 amending, as regards withdrawal of the authorization of certain antibiotics, Directive 70/524/EEC concerning additives in feedingstuffs. Official Journal of the European Communities. 1998;351:4–8. [Google Scholar]
- 24.Ansary A, Radu S. Conjugal transfer of antibiotic resistances and plasmids from Campylobacter jejuni clinical isolates. FEMS Microbiol Lett. 1992;70:125–8. doi: 10.1111/j.1574-6968.1992.tb05196.x. [DOI] [PubMed] [Google Scholar]
- 25.Bradbury WC, Munroe DL. Occurrence of plasmids and antibiotic resistance among Campylobacter jejuni and Campylobacter coli isolated from healthy and diarrheic animals. J Clin Microbiol. 1985;22:339–46. doi: 10.1128/jcm.22.3.339-346.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibreel A, Sköld O. High-level resistance to trimethoprim in clinical isolates of Campylobacter jejuni by acquisition of foreign genes (dfr1 and dfr9) expressing drug-insensitive dihydrofolate reductases. Antimicrob Agents Chemother. 1998;42:3059–64. doi: 10.1128/aac.42.12.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibreel A, Sköld O, Taylor DE. Characterization of plasmid-mediated aphA-3 kanamycin resistance in Campylobacter jejuni. Microb Drug Resist. 2004;10:98–105. doi: 10.1089/1076629041310127. [DOI] [PubMed] [Google Scholar]
- 28.Gibreel A, Tracz DM, Nonaka L, Ngo TM, Connell SR, Taylor DE. Incidence of antibiotic resistance in Campylobacter jejuni isolated in Alberta, Canada, from 1999 to 2002, with special reference to tet(O)-mediated tetracycline resistance. Antimicrob Agents Chemother. 2004;48:3442–50. doi: 10.1128/AAC.48.9.3442-3450.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lambert T, Gerbaud G, Trieu-Cuot P, Courvalin P. Structural relationship between the genes encoding 3′-aminoglycoside phosphotransferases in Campylobacter and in gram-positive cocci. Ann Inst Pasteur Microbiol. 1985;136B:135–50. doi: 10.1016/S0769-2609(85)80040-5. [DOI] [PubMed] [Google Scholar]
- 30.Lee MD, Sanchez S, Zimmer M, Idris U, Berrang ME, McDermott PF. Class 1 integron-associated tobramycin-gentamicin resistance in Campylobacter jejuni isolated from the broiler chicken house environment. Antimicrob Agents Chemother. 2002;46:3660–4. doi: 10.1128/AAC.46.11.3660-3664.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nirdnoy W, Mason CJ, Guerry P. Mosaic structure of a multiple-drug-resistant, conjugative plasmid from Campylobacter jejuni. Antimicrob Agents Chemother. 2005;49:2454–9. doi: 10.1128/AAC.49.6.2454-2459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Halloran F, Lucey B, Cryan B, Buckley T, Fanning S. Molecular characterization of class 1 integrons from Irish thermophilic Campylobacter spp. J Antimicrob Chemother. 2004;53:952–7. doi: 10.1093/jac/dkh193. [DOI] [PubMed] [Google Scholar]
- 33.Pinto-Alphandary H, Mabilat C, Courvalin P. Emergence of aminoglycoside resistance genes aadA and aadE in the genus Campylobacter. Antimicrob Agents Chemother. 1990;34:1294–6. doi: 10.1128/AAC.34.6.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sagara H, Mochizuki A, Okamura N, Nakaya R. Antimicrobial resistance of Campylobacter jejuni and Campylobacter coli with special reference to plasmid profiles of Japanese clinical isolates. Antimicrob Agents Chemother. 1987;31:713–9. doi: 10.1128/AAC.31.5.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Velázquez JB, Jiménez A, Chomón B, Villa TG. Incidence and transmission of antibiotic resistance in Campylobacter jejuni and Campylobacter coli. J Antimicrob Chemother. 1995;35:173–8. doi: 10.1093/jac/35.1.173. [DOI] [PubMed] [Google Scholar]
- 36.Lin J, Michel LO, Zhang Q. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob Agents Chemother. 2002;46:2124–31. doi: 10.1128/AAC.46.7.2124-2131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pumbwe L, Piddock LJ. Identification and molecular characterisation of CmeB, a Campylobacter jejuni multidrug efflux pump. FEMS Microbiol Lett. 2002;206:185–9. doi: 10.1111/j.1574-6968.2002.tb11007.x. [DOI] [PubMed] [Google Scholar]
- 38.Lin J, Sahin O, Michel LO, Zhang Q. Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect Immun. 2003;71:4250–9. doi: 10.1128/IAI.71.8.4250-4259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akiba M, Lin J, Barton YW, Zhang Q. Interaction of CmeABC and CmeDEF in conferring antimicrobial resistance and maintaining cell viability in Campylobacter jejuni. J Antimicrob Chemother. 2006;57:52–60. doi: 10.1093/jac/dki419. [DOI] [PubMed] [Google Scholar]
- 40.Jeon B, Wang Y, Hao H, Barton YW, Zhang Q. Contribution of CmeG to antibiotic and oxidative stress resistance in Campylobacter jejuni. J Antimicrob Chemother. 2011;66:79–85. doi: 10.1093/jac/dkq418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–8. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 42.Corcoran D, Quinn T, Cotter L, Fanning S. An investigation of the molecular mechanisms contributing to high-level erythromycin resistance in Campylobacter. Int J Antimicrob Agents. 2006;27:40–5. doi: 10.1016/j.ijantimicag.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 43.Kurincic M, Botteldoorn N, Herman L, Smole Mozina S. Mechanisms of erythromycin resistance of Campylobacter spp. isolated from food, animals and humans. Int J Food Microbiol. 2007;120:186–90. doi: 10.1016/j.ijfoodmicro.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 44.Pumbwe L, Randall LP, Woodward MJ, Piddock LJ. Expression of the efflux pump genes cmeB, cmeF and the porin gene porA in multiple-antibiotic-resistant Campylobacter jejuni. J Antimicrob Chemother. 2004;54:341–7. doi: 10.1093/jac/dkh331. [DOI] [PubMed] [Google Scholar]
- 45.Page WJ, Huyer G, Huyer M, Worobec EA. Characterization of the porins of Campylobacter jejuni and Campylobacter coli and implications for antibiotic susceptibility. Antimicrob Agents Chemother. 1989;33:297–303. doi: 10.1128/AAC.33.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeon B, Muraoka W, Scupham A, Zhang Q. Roles of lipooligosaccharide and capsular polysaccharide in antimicrobial resistance and natural transformation of Campylobacter jejuni. J Antimicrob Chemother. 2009;63:462–8. doi: 10.1093/jac/dkn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karmali MA, De Grandis S, Fleming PC. Antimicrobial susceptibility of Campylobacter jejuni with special reference to resistance patterns of Canadian isolates. Antimicrob Agents Chemother. 1981;19:593–7. doi: 10.1128/AAC.19.4.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keo T, Collins J, Kunwar P, Blaser MJ, Iovine NM. Campylobacter capsule and lipooligosaccharide confer resistance to serum and cationic antimicrobials. Virulence. 2011;2:30–40. doi: 10.4161/viru.2.1.14752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor DE, Ng LK, Lior H. Susceptibility of Campylobacter species to nalidixic acid, enoxacin, and other DNA gyrase inhibitors. Antimicrob Agents Chemother. 1985;28:708–10. doi: 10.1128/AAC.28.5.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Mourik A, Steeghs L, van Laar J, Meiring HD, Hamstra HJ, van Putten JP, et al. Altered linkage of hydroxyacyl chains in lipid A of Campylobacter jejuni reduces TLR4 activation and antimicrobial resistance. J Biol Chem. 2010;285:15828–36. doi: 10.1074/jbc.M110.102061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akhtar SQ. Antimicrobial sensitivity and plasmid-mediated tetracycline resistance in Campylobacter jejuni isolated in Bangladesh. Chemotherapy. 1988;34:326–31. doi: 10.1159/000238587. [DOI] [PubMed] [Google Scholar]
- 52.Boonmar S, Sangsuk L, Suthivarakom K, Padungtod P, Morita Y. Serotypes and antimicrobial resistance of Campylobacter jejuni isolated from humans and animals in Thailand. Southeast Asian J Trop Med Public Health. 2005;36:130–4. [PubMed] [Google Scholar]
- 53.Hoge CW, Gambel JM, Srijan A, Pitarangsi C, Echeverria P. Trends in antibiotic resistance among diarrheal pathogens isolated in Thailand over 15 years. Clin Infect Dis. 1998;26:341–5. doi: 10.1086/516303. [DOI] [PubMed] [Google Scholar]
- 54.National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS). Human Isolates Final Report, 2010. U.S. Department of Health and Human Services, CDC, 2012. (Accessed at http://www.cdc.gov/narms/reports.htm)
- 55.DANMAP. 2011. Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans in Denmark. In: The Danish Integrated Antimicrobial Resistance Monitoring and Research Programme; 2012. [Google Scholar]
- 56.Van Looveren M, Daube G, De Zutter L, Dumont JM, Lammens C, Wijdooghe M, et al. Antimicrobial susceptibilities of Campylobacter strains isolated from food animals in Belgium. J Antimicrob Chemother. 2001;48:235–40. doi: 10.1093/jac/48.2.235. [DOI] [PubMed] [Google Scholar]
- 57.Davidson D. In the matter of enrofloxacin for poultry: Withdrawal of approval of Bayer Corporation's new animal drug application 1 (NADA) 140-828 (Baytril). In: FDA Docket No 00N-1571 2004; 2004. [Google Scholar]
- 58.Miflin JK, Templeton JM, Blackall PJ. Antibiotic resistance in Campylobacter jejuni and Campylobacter coli isolated from poultry in the South-East Queensland region. J Antimicrob Chemother. 2007;59:775–8. doi: 10.1093/jac/dkm024. [DOI] [PubMed] [Google Scholar]
- 59.Unicomb LE, Ferguson J, Stafford RJ, Ashbolt R, Kirk MD, Becker NG, et al. Australian Campylobacter Subtyping Study Group Low-level fluoroquinolone resistance among Campylobacter jejuni isolates in Australia. Clin Infect Dis. 2006;42:1368–74. doi: 10.1086/503426. [DOI] [PubMed] [Google Scholar]
- 60.Norström M, Hofshagen M, Stavnes T, Schau J, Lassen J, Kruse H. Antimicrobial resistance in Campylobacter jejuni from humans and broilers in Norway. Epidemiol Infect. 2006;134:127–30. doi: 10.1017/S0950268805004814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jain D, Sinha S, Prasad KN, Pandey CM. Campylobacter species and drug resistance in a north Indian rural community. Trans R Soc Trop Med Hyg. 2005;99:207–14. doi: 10.1016/j.trstmh.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 62.Bester LA, Essack SY. Observational study of the prevalence and antibiotic resistance of Campylobacter spp. from different poultry production systems in KwaZulu-Natal, South Africa. J Food Prot. 2012;75:154–9. doi: 10.4315/0362-028X.JFP-11-237. [DOI] [PubMed] [Google Scholar]
- 63.Chokboonmongkol C, Patchanee P, Gölz G, Zessin KH, Alter T. Prevalence, quantitative load, and antimicrobial resistance of Campylobacter spp. from broiler ceca and broiler skin samples in Thailand. Poult Sci. 2013;92:462–7. doi: 10.3382/ps.2012-02599. [DOI] [PubMed] [Google Scholar]
- 64.Haruna M, Sasaki Y, Murakami M, Ikeda A, Kusukawa M, Tsujiyama Y, et al. Prevalence and antimicrobial susceptibility of Campylobacter in broiler flocks in Japan. Zoonoses Public Health. 2012;59:241–5. doi: 10.1111/j.1863-2378.2011.01441.x. [DOI] [PubMed] [Google Scholar]
- 65.Wardak S, Szych J, Zasada AA, Gierczynski R. Antibiotic resistance of Campylobacter jejuni and Campylobacter coli clinical isolates from Poland. Antimicrob Agents Chemother. 2007;51:1123–5. doi: 10.1128/AAC.01187-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sáenz Y, Zarazaga M, Lantero M, Gastanares MJ, Baquero F, Torres C. Antibiotic resistance in Campylobacter strains isolated from animals, foods, and humans in Spain in 1997-1998. Antimicrob Agents Chemother. 2000;44:267–71. doi: 10.1128/AAC.44.2.267-271.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Griggs DJ, Johnson MM, Frost JA, Humphrey T, Jørgensen F, Piddock LJ. Incidence and mechanism of ciprofloxacin resistance in Campylobacter spp. isolated from commercial poultry flocks in the United Kingdom before, during, and after fluoroquinolone treatment. Antimicrob Agents Chemother. 2005;49:699–707. doi: 10.1128/AAC.49.2.699-707.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Price LB, Johnson E, Vailes R, Silbergeld E. Fluoroquinolone-resistant Campylobacter isolates from conventional and antibiotic-free chicken products. Environ Health Perspect. 2005;113:557–60. doi: 10.1289/ehp.7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Price LB, Lackey LG, Vailes R, Silbergeld E. The persistence of fluoroquinolone-resistant Campylobacter in poultry production. Environ Health Perspect. 2007;115:1035–9. doi: 10.1289/ehp.10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Q, Lin J, Pereira S. Fluoroquinolone-resistant Campylobacter in animal reservoirs: dynamics of development, resistance mechanisms and ecological fitness. Anim Health Res Rev. 2003;4:63–71. doi: 10.1079/AHR200356. [DOI] [PubMed] [Google Scholar]
- 71.Luo N, Pereira S, Sahin O, Lin J, Huang S, Michel L, et al. Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc Natl Acad Sci U S A. 2005;102:541–6. doi: 10.1073/pnas.0408966102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ge B, McDermott PF, White DG, Meng J. Role of efflux pumps and topoisomerase mutations in fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob Agents Chemother. 2005;49:3347–54. doi: 10.1128/AAC.49.8.3347-3354.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Luo N, Sahin O, Lin J, Michel LO, Zhang Q. In vivo selection of Campylobacter isolates with high levels of fluoroquinolone resistance associated with gyrA mutations and the function of the CmeABC efflux pump. Antimicrob Agents Chemother. 2003;47:390–4. doi: 10.1128/AAC.47.1.390-394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yan M, Sahin O, Lin J, Zhang Q. Role of the CmeABC efflux pump in the emergence of fluoroquinolone-resistant Campylobacter under selection pressure. J Antimicrob Chemother. 2006;58:1154–9. doi: 10.1093/jac/dkl412. [DOI] [PubMed] [Google Scholar]
- 75.Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev. 1997;61:377–92. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shea ME, Hiasa H. Interactions between DNA helicases and frozen topoisomerase IV-quinolone-DNA ternary complexes. J Biol Chem. 1999;274:22747–54. doi: 10.1074/jbc.274.32.22747. [DOI] [PubMed] [Google Scholar]
- 77.Willmott CJ, Critchlow SE, Eperon IC, Maxwell A. The complex of DNA gyrase and quinolone drugs with DNA forms a barrier to transcription by RNA polymerase. J Mol Biol. 1994;242:351–63. doi: 10.1006/jmbi.1994.1586. [DOI] [PubMed] [Google Scholar]
- 78.Bachoual R, Ouabdesselam S, Mory F, Lascols C, Soussy CJ, Tankovic J. Single or double mutational alterations of gyrA associated with fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli. Microb Drug Resist. 2001;7:257–61. doi: 10.1089/10766290152652800. [DOI] [PubMed] [Google Scholar]
- 79.Cooper R, Segal H, Lastovica AJ, Elisha BG. Genetic basis of quinolone resistance and epidemiology of resistant and susceptible isolates of porcine Campylobacter coli strains. J Appl Microbiol. 2002;93:241–9. doi: 10.1046/j.1365-2672.2002.01650.x. [DOI] [PubMed] [Google Scholar]
- 80.Payot S, Cloeckaert A, Chaslus-Dancla E. Selection and characterization of fluoroquinolone-resistant mutants of Campylobacter jejuni using enrofloxacin. Microb Drug Resist. 2002;8:335–43. doi: 10.1089/10766290260469606. [DOI] [PubMed] [Google Scholar]
- 81.Gootz TD, Martin BA. Characterization of high-level quinolone resistance in Campylobacter jejuni. Antimicrob Agents Chemother. 1991;35:840–5. doi: 10.1128/AAC.35.5.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jesse TW, Englen MD, Pittenger-Alley LG, Fedorka-Cray PJ. Two distinct mutations in gyrA lead to ciprofloxacin and nalidixic acid resistance in Campylobacter coli and Campylobacter jejuni isolated from chickens and beef cattle. J Appl Microbiol. 2006;100:682–8. doi: 10.1111/j.1365-2672.2005.02796.x. [DOI] [PubMed] [Google Scholar]
- 83.Farnell MB, Donoghue AM, Cole K, Reyes-Herrera I, Blore PJ, Donoghue DJ. Campylobacter susceptibility to ciprofloxacin and corresponding fluoroquinolone concentrations within the gastrointestinal tracts of chickens. J Appl Microbiol. 2005;99:1043–50. doi: 10.1111/j.1365-2672.2005.02712.x. [DOI] [PubMed] [Google Scholar]
- 84.McDermott PF, Bodeis SM, English LL, White DG, Walker RD, Zhao S, et al. Ciprofloxacin resistance in Campylobacter jejuni evolves rapidly in chickens treated with fluoroquinolones. J Infect Dis. 2002;185:837–40. doi: 10.1086/339195. [DOI] [PubMed] [Google Scholar]
- 85.Takahashi T, Ishihara K, Kojima A, Asai T, Harada K, Tamura Y. Emergence of fluoroquinolone resistance in Campylobacter jejuni in chickens exposed to enrofloxacin treatment at the inherent dosage licensed in Japan. J Vet Med B Infect Dis Vet Public Health. 2005;52:460–4. doi: 10.1111/j.1439-0450.2005.00890.x. [DOI] [PubMed] [Google Scholar]
- 86.Ellis-Pegler RB, Hyman LK, Ingram RJ, McCarthy M. A placebo controlled evaluation of lomefloxacin in the treatment of bacterial diarrhoea in the community. J Antimicrob Chemother. 1995;36:259–63. doi: 10.1093/jac/36.1.259. [DOI] [PubMed] [Google Scholar]
- 87.Segreti J, Gootz TD, Goodman LJ, Parkhurst GW, Quinn JP, Martin BA, et al. High-level quinolone resistance in clinical isolates of Campylobacter jejuni. J Infect Dis. 1992;165:667–70. doi: 10.1093/infdis/165.4.667. [DOI] [PubMed] [Google Scholar]
- 88.Wretlind B, Strömberg A, Ostlund L, Sjögren E, Kaijser B. Rapid emergence of quinolone resistance in Campylobacter jejuni in patients treated with norfloxacin. Scand J Infect Dis. 1992;24:685–6. doi: 10.3109/00365549209054659. [DOI] [PubMed] [Google Scholar]
- 89.Ruiz J, Goñi P, Marco F, Gallardo F, Mirelis B, Jimenez De Anta T, et al. Increased resistance to quinolones in Campylobacter jejuni: a genetic analysis of gyrA gene mutations in quinolone-resistant clinical isolates. Microbiol Immunol. 1998;42:223–6. doi: 10.1111/j.1348-0421.1998.tb02274.x. [DOI] [PubMed] [Google Scholar]
- 90.Wang Y, Huang WM, Taylor DE. Cloning and nucleotide sequence of the Campylobacter jejuni gyrA gene and characterization of quinolone resistance mutations. Antimicrob Agents Chemother. 1993;37:457–63. doi: 10.1128/AAC.37.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Piddock LJ, Ricci V, Pumbwe L, Everett MJ, Griggs DJ. Fluoroquinolone resistance in Campylobacter species from man and animals: detection of mutations in topoisomerase genes. J Antimicrob Chemother. 2003;51:19–26. doi: 10.1093/jac/dkg033. [DOI] [PubMed] [Google Scholar]
- 92.Engberg J, Neimann J, Nielsen EM, Aerestrup FM, Fussing V. Quinolone-resistant Campylobacter infections: risk factors and clinical consequences. Emerg Infect Dis. 2004;10:1056–63. doi: 10.3201/eid1006.030669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Feodoroff FB, Lauhio AR, Sarna SJ, Hänninen ML, Rautelin HI. Severe diarrhoea caused by highly ciprofloxacin-susceptible Campylobacter isolates. Clin Microbiol Infect. 2009;15:188–92. doi: 10.1111/j.1469-0691.2008.02657.x. [DOI] [PubMed] [Google Scholar]
- 94.Helms M, Simonsen J, Olsen KE, Mølbak K. Adverse health events associated with antimicrobial drug resistance in Campylobacter species: a registry-based cohort study. J Infect Dis. 2005;191:1050–5. doi: 10.1086/428453. [DOI] [PubMed] [Google Scholar]
- 95.Nelson JM, Smith KE, Vugia DJ, Rabatsky-Ehr T, Segler SD, Kassenborg HD, et al. Prolonged diarrhea due to ciprofloxacin-resistant campylobacter infection. J Infect Dis. 2004;190:1150–7. doi: 10.1086/423282. [DOI] [PubMed] [Google Scholar]
- 96.Sonnevend A, Kovács J, Pál T, Akawi N, Nagelkerke N, Schneider G. Lack of correlation between the 257C-to-T mutation in the gyrA gene and clinical severity of Campylobacter jejuni infection in a region of high incidence of ciprofloxacin resistance. Scand J Infect Dis. 2011;43:905–11. doi: 10.3109/00365548.2011.603743. [DOI] [PubMed] [Google Scholar]
- 97.Wassenaar TM, Kist M, de Jong A. Re-analysis of the risks attributed to ciprofloxacin-resistant Campylobacter jejuni infections. Int J Antimicrob Agents. 2007;30:195–201. doi: 10.1016/j.ijantimicag.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 98.Asai T, Harada K, Ishihara K, Kojima A, Sameshima T, Tamura Y, et al. Association of antimicrobial resistance in Campylobacter isolated from food-producing animals with antimicrobial use on farms. Jpn J Infect Dis. 2007;60:290–4. [PubMed] [Google Scholar]
- 99.Animal Drugs @ FDA. Center for Veterinary Medicine, 2012. (Accessed at http://www.accessdata.fda.gov/scripts/animaldrugsatfda/index.cfm)
- 100.Cagliero C, Mouline C, Cloeckaert A, Payot S. Synergy between efflux pump CmeABC and modifications in ribosomal proteins L4 and L22 in conferring macrolide resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob Agents Chemother. 2006;50:3893–6. doi: 10.1128/AAC.00616-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lin J, Yan M, Sahin O, Pereira S, Chang YJ, Zhang Q. Effect of macrolide usage on emergence of erythromycin-resistant Campylobacter isolates in chickens. Antimicrob Agents Chemother. 2007;51:1678–86. doi: 10.1128/AAC.01411-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Payot S, Bolla JM, Corcoran D, Fanning S, Mégraud F, Zhang Q. Mechanisms of fluoroquinolone and macrolide resistance in Campylobacter spp. Microbes Infect. 2006;8:1967–71. doi: 10.1016/j.micinf.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 103.Egger R, Korczak BM, Niederer L, Overesch G, Kuhnert P. Genotypes and antibiotic resistance of Campylobacter coli in fattening pigs. Vet Microbiol. 2012;155:272–8. doi: 10.1016/j.vetmic.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 104.Gibreel A, Taylor DE. Macrolide resistance in Campylobacter jejuni and Campylobacter coli. J Antimicrob Chemother. 2006;58:243–55. doi: 10.1093/jac/dkl210. [DOI] [PubMed] [Google Scholar]
- 105.Lachance N, Gaudreau C, Lamothe F, Larivière LA. Role of the beta-lactamase of Campylobacter jejuni in resistance to beta-lactam agents. Antimicrob Agents Chemother. 1991;35:813–8. doi: 10.1128/AAC.35.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lehtopolku M, Kotilainen P, Haanperä-Heikkinen M, Nakari UM, Hänninen ML, Huovinen P, et al. Ribosomal mutations as the main cause of macrolide resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob Agents Chemother. 2011;55:5939–41. doi: 10.1128/AAC.00314-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mamelli L, Prouzet-Mauléon V, Pagès JM, Mégraud F, Bolla JM. Molecular basis of macrolide resistance in Campylobacter: role of efflux pumps and target mutations. J Antimicrob Chemother. 2005;56:491–7. doi: 10.1093/jac/dki253. [DOI] [PubMed] [Google Scholar]
- 108.Pérez-Boto D, López-Portolés JA, Simón C, Valdezate S, Echeita MA. Study of the molecular mechanisms involved in high-level macrolide resistance of Spanish Campylobacter jejuni and Campylobacter coli strains. J Antimicrob Chemother. 2010;65:2083–8. doi: 10.1093/jac/dkq268. [DOI] [PubMed] [Google Scholar]
- 109.Ren GW, Wang Y, Shen Z, Chen X, Shen J, Wu C. Rapid detection of point mutations in domain V of the 23S rRNA gene in erythromycin-resistant Campylobacter isolates by pyrosequencing. Foodborne Pathog Dis. 2011;8:375–9. doi: 10.1089/fpd.2010.0676. [DOI] [PubMed] [Google Scholar]
- 110.Vacher S, Menard A, Bernard E, Santos A, Megraud F. Detection of mutations associated with macrolide resistance in thermophilic Campylobacter spp. by real-time PCR. Microb Drug Resist. 2005;11:40–7. doi: 10.1089/mdr.2005.11.40. [DOI] [PubMed] [Google Scholar]
- 111.Fouts DE, Mongodin EF, Mandrell RE, Miller WG, Rasko DA, Ravel J, et al. Major structural differences and novel potential virulence mechanisms from the genomes of multiple campylobacter species. PLoS Biol. 2005;3:e15. doi: 10.1371/journal.pbio.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gibreel A, Wetsch NM, Taylor DE. Contribution of the CmeABC efflux pump to macrolide and tetracycline resistance in Campylobacter jejuni. Antimicrob Agents Chemother. 2007;51:3212–6. doi: 10.1128/AAC.01592-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Payot S, Avrain L, Magras C, Praud K, Cloeckaert A, Chaslus-Dancla E. Relative contribution of target gene mutation and efflux to fluoroquinolone and erythromycin resistance, in French poultry and pig isolates of Campylobacter coli. Int J Antimicrob Agents. 2004;23:468–72. doi: 10.1016/j.ijantimicag.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 114.Logue CM, Danzeisen GT, Sherwood JS, Thorsness JL, Mercier BM, Axtman JE. Repeated therapeutic dosing selects macrolide-resistant Campylobacter spp. in a turkey facility. J Appl Microbiol. 2010;109:1379–88. doi: 10.1111/j.1365-2672.2010.04765.x. [DOI] [PubMed] [Google Scholar]
- 115.Almofti YA, Dai M, Sun Y, Haihong H, Yuan Z. Impact of erythromycin resistance on the virulence properties and fitness of Campylobacter jejuni. Microb Pathog. 2011;50:336–42. doi: 10.1016/j.micpath.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 116.Han F, Pu S, Wang F, Meng J, Ge B. Fitness cost of macrolide resistance in Campylobacter jejuni. Int J Antimicrob Agents. 2009;34:462–6. doi: 10.1016/j.ijantimicag.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 117.Hao H, Dai M, Wang Y, Peng D, Liu Z, Yuan Z. 23S rRNA mutation A2074C conferring high-level macrolide resistance and fitness cost in Campylobacter jejuni. Microb Drug Resist. 2009;15:239–44. doi: 10.1089/mdr.2009.0008. [DOI] [PubMed] [Google Scholar]
- 118.Zeitouni S, Collin O, Andraud M, Ermel G, Kempf I. Fitness of macrolide resistant Campylobacter coli and Campylobacter jejuni. Microb Drug Resist. 2012;18:101–8. doi: 10.1089/mdr.2011.0188. [DOI] [PubMed] [Google Scholar]
- 119.EFSA The European Union Summary Report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2010. Eur Food Safety Auth J. 2012;10:2598–830. [Google Scholar]
- 120.Smith SI, Sansa TI, Coker AO. Antibiotic susceptibility patterns and beta-lactamase production of animal and human isolates of Campylobacter in Lagos, Nigeria. Z Naturforsch C. 1999;54:583–6. doi: 10.1515/znc-1999-7-820. [DOI] [PubMed] [Google Scholar]
- 121.Gibreel A, Kos VN, Keelan M, Trieber CA, Levesque S, Michaud S, et al. Macrolide resistance in Campylobacter jejuni and Campylobacter coli: molecular mechanism and stability of the resistance phenotype. Antimicrob Agents Chemother. 2005;49:2753–9. doi: 10.1128/AAC.49.7.2753-2759.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Caldwell DB, Wang Y, Lin J. Development, stability, and molecular mechanisms of macrolide resistance in Campylobacter jejuni. Antimicrob Agents Chemother. 2008;52:3947–54. doi: 10.1128/AAC.00450-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jeon B, Zhang Q. Sensitization of Campylobacter jejuni to fluoroquinolone and macrolide antibiotics by antisense inhibition of the CmeABC multidrug efflux transporter. J Antimicrob Chemother. 2009;63:946–8. doi: 10.1093/jac/dkp067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Huyer MP, Hancock T, Page RW. Outer membrane porin protein of Campylobacter jejuni. FEMS Microbiol Lett. 1986;37:247–50. doi: 10.1111/j.1574-6968.1986.tb01803.x. [DOI] [Google Scholar]
- 125.Hancock RE, Bell A. Antibiotic uptake into gram-negative bacteria. Eur J Clin Microbiol Infect Dis. 1988;7:713–20. doi: 10.1007/BF01975036. [DOI] [PubMed] [Google Scholar]
- 126.Vaara M. Outer membrane permeability barrier to azithromycin, clarithromycin, and roxithromycin in gram-negative enteric bacteria. Antimicrob Agents Chemother. 1993;37:354–6. doi: 10.1128/AAC.37.2.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Logan SM, Trust TJ. Structural and antigenic heterogeneity of lipopolysaccharides of Campylobacter jejuni and Campylobacter coli. Infect Immun. 1984;45:210–6. doi: 10.1128/iai.45.1.210-216.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Naess V, Hofstad T. Isolation and chemical composition of lipopolysaccharide from Campylobacter jejuni. Acta Pathol Microbiol Immunol Scand B. 1982;90:135–9. doi: 10.1111/j.1699-0463.1982.tb00095.x. [DOI] [PubMed] [Google Scholar]
- 129.Giesbrecht P, Kersten T, Maidhof H, Wecke J. Staphylococcal cell wall: morphogenesis and fatal variations in the presence of penicillin. Microbiol Mol Biol Rev. 1998;62:1371–414. doi: 10.1128/mmbr.62.4.1371-1414.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Alfredson DA, Korolik V. Isolation and expression of a novel molecular class D beta-lactamase, OXA-61, from Campylobacter jejuni. Antimicrob Agents Chemother. 2005;49:2515–8. doi: 10.1128/AAC.49.6.2515-2518.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Griggs DJ, Peake L, Johnson MM, Ghori S, Mott A, Piddock LJ. Beta-lactamase-mediated beta-lactam resistance in Campylobacter species: prevalence of Cj0299 (bla OXA-61) and evidence for a novel beta-Lactamase in C. jejuni. Antimicrob Agents Chemother. 2009;53:3357–64. doi: 10.1128/AAC.01655-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lachance N, Gaudreau C, Lamothe F, Turgeon F. Susceptibilities of beta-lactamase-positive and -negative strains of Campylobacter coli to beta-lactam agents. Antimicrob Agents Chemother. 1993;37:1174–6. doi: 10.1128/AAC.37.5.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tajada P, Gomez-Graces JL, Alós JI, Balas D, Cogollos R. Antimicrobial susceptibilities of Campylobacter jejuni and Campylobacter coli to 12 beta-lactam agents and combinations with beta-lactamase inhibitors. Antimicrob Agents Chemother. 1996;40:1924–5. doi: 10.1128/aac.40.8.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wright EP, Knowles MA. Beta-lactamase production by Campylobacter jejuni. J Clin Pathol. 1980;33:904–5. doi: 10.1136/jcp.33.9.904-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Alfredson DA, Korolik V. Identification of putative zinc hydrolase genes of the metallo-beta-lactamase superfamily from Campylobacter jejuni. FEMS Immunol Med Microbiol. 2007;49:159–64. doi: 10.1111/j.1574-695X.2006.00197.x. [DOI] [PubMed] [Google Scholar]
- 136.Potter ME, Blaser MJ, Sikes RK, Kaufmann AF, Wells JG. Human Campylobacter infection associated with certified raw milk. Am J Epidemiol. 1983;117:475–83. doi: 10.1093/oxfordjournals.aje.a113565. [DOI] [PubMed] [Google Scholar]
- 137.Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65:232–60. doi: 10.1128/MMBR.65.2.232-260.2001. [second page, table of contents.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bolla JM, Loret E, Zalewski M, Pagés JM. Conformational analysis of the Campylobacter jejuni porin. J Bacteriol. 1995;177:4266–71. doi: 10.1128/jb.177.15.4266-4271.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kervella M, Fauchère JL, Fourel D, Pagès JM. Immunological cross-reactivity between outer membrane pore proteins of Campylobacter jejuni and Escherichia coli. FEMS Microbiol Lett. 1992;78:281–5. doi: 10.1111/j.1574-6968.1992.tb05582.x. [DOI] [PubMed] [Google Scholar]
- 140.Epe B, Woolley P, Hornig H. Competition between tetracycline and tRNA at both P and A sites of the ribosome of Escherichia coli. FEBS Lett. 1987;213:443–7. doi: 10.1016/0014-5793(87)81539-9. [DOI] [PubMed] [Google Scholar]
- 141.Goldman RA, Hasan T, Hall CC, Strycharz WA, Cooperman BS. Photoincorporation of tetracycline into Escherichia coli ribosomes. Identification of the major proteins photolabeled by native tetracycline and tetracycline photoproducts and implications for the inhibitory action of tetracycline on protein synthesis. Biochemistry. 1983;22:359–68. doi: 10.1021/bi00271a020. [DOI] [PubMed] [Google Scholar]
- 142.Manavathu EK, Hiratsuka K, Taylor DE. Nucleotide sequence analysis and expression of a tetracycline-resistance gene from Campylobacter jejuni. Gene. 1988;62:17–26. doi: 10.1016/0378-1119(88)90576-8. [DOI] [PubMed] [Google Scholar]
- 143.Salyers AA, Speer BS, Shoemaker NB. New perspectives in tetracycline resistance. Mol Microbiol. 1990;4:151–6. doi: 10.1111/j.1365-2958.1990.tb02025.x. [DOI] [PubMed] [Google Scholar]
- 144.Dasti JI, Gross U, Pohl S, Lugert R, Weig M, Schmidt-Ott R. Role of the plasmid-encoded tet(O) gene in tetracycline-resistant clinical isolates of Campylobacter jejuni and Campylobacter coli. J Med Microbiol. 2007;56:833–7. doi: 10.1099/jmm.0.47103-0. [DOI] [PubMed] [Google Scholar]
- 145.Taylor DE. Plasmid-mediated tetracycline resistance in Campylobacter jejuni: expression in Escherichia coli and identification of homology with streptococcal class M determinant. J Bacteriol. 1986;165:1037–9. doi: 10.1128/jb.165.3.1037-1039.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Batchelor RA, Pearson BM, Friis LM, Guerry P, Wells JM. Nucleotide sequences and comparison of two large conjugative plasmids from different Campylobacter species. Microbiology. 2004;150:3507–17. doi: 10.1099/mic.0.27112-0. [DOI] [PubMed] [Google Scholar]
- 147.Taylor DE, Courvalin P. Mechanisms of antibiotic resistance in Campylobacter species. Antimicrob Agents Chemother. 1988;32:1107–12. doi: 10.1128/AAC.32.8.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sahin O, Plummer PJ, Jordan DM, Sulaj K, Pereira S, Robbe-Austerman S, et al. Emergence of a tetracycline-resistant Campylobacter jejuni clone associated with outbreaks of ovine abortion in the United States. J Clin Microbiol. 2008;46:1663–71. doi: 10.1128/JCM.00031-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bérdy J. CRC handbook of antibiotic compounds. Boca Raton, Fla.: CRC Press; 1980. [Google Scholar]
- 150.Jana S, Deb JK. Molecular understanding of aminoglycoside action and resistance. Appl Microbiol Biotechnol. 2006;70:140–50. doi: 10.1007/s00253-005-0279-0. [DOI] [PubMed] [Google Scholar]
- 151.Taber HW, Mueller JP, Miller PF, Arrow AS. Bacterial uptake of aminoglycoside antibiotics. Microbiol Rev. 1987;51:439–57. doi: 10.1128/mr.51.4.439-457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bryan LE, Kwan S. Roles of ribosomal binding, membrane potential, and electron transport in bacterial uptake of streptomycin and gentamicin. Antimicrob Agents Chemother. 1983;23:835–45. doi: 10.1128/AAC.23.6.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bryan LE, Van Den Elzen HM. Gentamicin accumulation by sensitive strains of Escherichia coli and Pseudomonas aeruginosa. J Antibiot (Tokyo) 1975;28:696–703. doi: 10.7164/antibiotics.28.696. [DOI] [PubMed] [Google Scholar]
- 154.Hermann T. Aminoglycoside antibiotics: old drugs and new therapeutic approaches. Cell Mol Life Sci. 2007;64:1841–52. doi: 10.1007/s00018-007-7034-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ouellette M, Gerbaud G, Lambert T, Courvalin P. Acquisition by a Campylobacter-like strain of aphA-1, a kanamycin resistance determinant from members of the family Enterobacteriaceae. Antimicrob Agents Chemother. 1987;31:1021–6. doi: 10.1128/AAC.31.7.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tenover FC, Fennell CL, Lee L, LeBlanc DJ. Characterization of two plasmids from Campylobacter jejuni isolates that carry the aphA-7 kanamycin resistance determinant. Antimicrob Agents Chemother. 1992;36:712–6. doi: 10.1128/AAC.36.4.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tenover FC, Filpula D, Phillips KL, Plorde JJ. Cloning and sequencing of a gene encoding an aminoglycoside 6′-N-acetyltransferase from an R factor of Citrobacter diversus. J Bacteriol. 1988;170:471–3. doi: 10.1128/jb.170.1.471-473.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tenover FC, Gilbert T, O’Hara P. Nucleotide sequence of a novel kanamycin resistance gene, aphA-7, from Campylobacter jejuni and comparison to other kanamycin phosphotransferase genes. Plasmid. 1989;22:52–8. doi: 10.1016/0147-619X(89)90035-8. [DOI] [PubMed] [Google Scholar]
- 159.Olkkola S, Juntunen P, Heiska H, Hyytiäinen H, Hänninen ML. Mutations in the rpsL gene are involved in streptomycin resistance in Campylobacter coli. Microb Drug Resist. 2010;16:105–10. doi: 10.1089/mdr.2009.0128. [DOI] [PubMed] [Google Scholar]
- 160.Hannula M, Hänninen ML. Effect of putative efflux pump inhibitors and inducers on the antimicrobial susceptibility of Campylobacter jejuni and Campylobacter coli. J Med Microbiol. 2008;57:851–5. doi: 10.1099/jmm.0.47823-0. [DOI] [PubMed] [Google Scholar]
- 161.Pumbwe L, Piddock LJV. Identification and molecular characterisation of CmeB, a Campylobacter jejuni multidrug efflux pump. FEMS Microbiol Lett. 2002;206:185–9. doi: 10.1111/j.1574-6968.2002.tb11007.x. [DOI] [PubMed] [Google Scholar]