Abstract
The production of extracellular poly-β-1,6-N-acetyl-d-glucosamine (PNAG) by Staphylococcus epidermidis is the principal determinant of biofilm formation on indwelling medical devices. Enzymes that degrade PNAG therefore provide an attractive strategy for biofilm removal and for the manufacture of biofilm-resistant coatings. Here we present methods that allow the identification of PNAG-degrading enzymes with the ability to detach biofilms. Our protocol includes the preparation of soluble PNAG from S. epidermidis cultures, the incubation of soluble PNAG with candidate enzymes and assays that detect the release of N-acetyl-d-glucosamine using high-pH anion-exchange chromatography (HPAEC) followed in parallel by pulsed amperometric detection (PAD) and online electrospray ionization mass spectrometry (ESI-MS). We validated our procedures using dispersin B, which is currently the only known PNAG-degrading enzyme.
Keywords: Staphylococcus epidermidis; biofilm; exopolysaccharide; high-pH anion-exchange chromatography; poly-β-1,6-N-acetyl-d-glucosamine
Introduction
Biofilms on indwelling medical devices represent a significant cause of morbidity and mortality in hospitalized patients, and are responsible for nosocomial conditions such as ventilator-associated pneumonia (VAP), catheter-associated urinary tract infection (CAUTI), catheter-related bloodstream infection (CRBSI) and prosthetic implant infection (PII).1 Although microbial communities in biofilms tend to be highly diverse, Staphylococcus epidermidis is considered to play a key role in the colonization of medical devices.2-4 Most nosocomial strains of this commensal inhabitant of human skin and mucous membranes carry the icaADBC operon, and are able to produce an extracellular linear polysaccharide consisting of β-1,6-linked N-acetyl-d-glucosamine (Fig. 1).5,6 Poly-β-1,6-N-acetyl-d-glucosamine (PNAG) was originally described as polysaccharide intercellular adhesin (PIA) and appears to be a major constituent of many biofilms.7 During or after secretion into the extracellular space, approximately 16% of the N-acetylglucosamine residues in PNAG are deacetylated by the icaB gene product, which is non-covalently attached to the cell surface.8 The resulting glucosamine residues make the exopolysaccharide more cationic, which is essential for electrostatic interactions with the cell envelope and confers resistance to the innate immune system by repelling cationic antimicrobial peptides.9 In addition to S. epidermidis, PNAG is also synthesized by biofilm-producing strains of S. aureus.10-12 Sequences homologous to the icaADBC genes are also found in many pathogenic Gram-negative bacteria, and the production of PNAG has been confirmed biochemically and/or immunologically in Acinetobacter baumannii, Actinobacillus pleuropneumoniae, Aggregatibacter actinomycetemcomitans, Bordetella spp, Burkholderia spp, Escherichia coli and Yersinia pestis.13,14
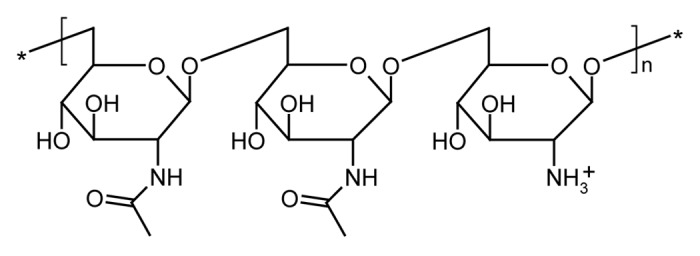
Figure 1. Chemical structure of PNAG. The residues are partly deacetylated, thus conferring a positive charge upon the polymer.
Currently, the only enzyme known to degrade PNAG is dispersin B, which is derived from the periodontal disease-associated bacterium A. actinomycetemcomitans. The dispersin B gene was originally discovered by transposon mutagenesis, generating a bacterial strain that formed rough colonies and was deficient in the release of cells from biofilms.15 Dispersin B was then shown specifically to cleave the β-1,6-glycosidic bond between the N-acetylglucosamine units of PNAG.16 Experiments using natural PNAG isolated from E. coli suggested that dispersin B initially cleaves the glycosidic linkages endolytically, producing larger oligosaccharides,16 but the results from another study using synthetic β-1,6-linked N-acetyl-d-glucosamine oligomers (dimers, tetramers and hexamers) labeled with a p-methoxyphenyl group at the reducing end instead indicated exolytic activity.17 In naturally-occurring biofilms, dispersin B probably acts as a trigger for the detachment of cells and cell aggregates allowing the bacteria to spread and colonize new surfaces. Biotechnological applications of dispersin B could include the production of cleaning agents, coatings or wound gels.18 Dispersin B was recently integrated into polymer matrices, producing biocompatible hydrogels that were largely resistant to colonization by S. epidermidis.19
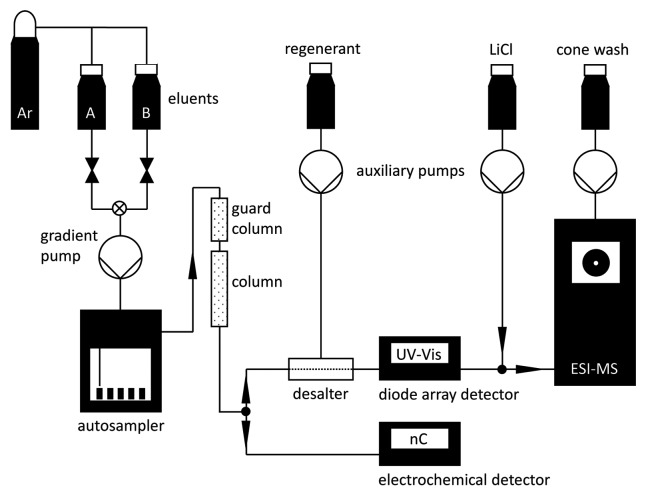
We have initiated a systematic search for further PNAG-degrading enzymes that could be used to prevent the formation of biofilms or to remove them from medical devices. Testing for biofilm degradation is frequently achieved by treating experimental biofilms in multi-well plates or on other surfaces with the candidate enzymes, followed by microscopy and/or staining of the residual biofilm material with dyes such as crystal violet or safranin. Using such assays, lysostaphin and DNase I have also been shown to degrade S. epidermidis biofilms.20,21 However, these enzymes lack PNAG-degrading activity suggesting that a more specific assay is necessary. Therefore we established an analytical system for the detection of N-acetylglucosamine monomers produced by the degradation of PNAG. N-acetylglucosamine monomers and oligomers have previously been detected by thin-layer chromatography and matrix-assisted laser desorption/ionization-time-of-flight (MALDI-TOF) MS, respectively.16 In addition, the Morgan-Elson assay has been used for the colorimetric quantitation of total hexosamine released from biofilm-producing S. epidermidis cells after treatment with dispersin B.22 We have developed a novel approach using high-pH anion-exchange chromatography (HPAEC) followed in parallel by pulsed amperometric detection (PAD) and online electrospray ionization mass spectrometry (ESI-MS) for the specific and sensitive detection of N-acetylglucosamine (Fig. 2). The HPAEC technique exploits the weakly-acidic nature of the hydroxyl groups of carbohydrates, which are separated on an anion-exchange column using NaOH as the mobile phase.23 The eluting carbohydrates are detected with an electrochemical device that measures the current generated by their oxidation on a gold electrode at pH ≥ 12. A pulsed electrical potential is used to clean and regenerate the electrode during each measuring cycle. In parallel, the carbohydrates are analyzed as H2O and Li+ adducts by ESI-MS, which requires the removal of NaOH by desalting and the addition of LiCl solution prior to injection into the mass spectrometer.
Figure 2. Schematic drawing of the experimental setup for carbohydrate analysis by HPAEC-PAD-ESI-MS.
As expected, the HPAEC-PAD-ESI-MS approach allowed us to detect N-acetylglucosamine in the supernatants of S. epidermidis biofilms growing in 24-well plates after treatment with dispersin B. However, the signal intensity was low, and large concentrations of dispersin B were required. Therefore, we investigated the use of soluble PNAG as a substrate instead of the intact pre-formed biofilms to develop a more sensitive and robust assay. The chemical synthesis of β-1,6-linked N-acetyl-d-glucosamine oligomers that resemble PNAG has been reported.17,24,25 Although a well-defined synthetic substrate would be desirable, such a material is not available in sufficient quantities due to the complicated synthesis process. The production of PNAG from cultures of S. epidermidis and other bacteria has also been reported, including purification by anion-exchange chromatography (obtaining PNAG in the flow-through) and/or size exclusion chromatography (separating PNAG into the high-molecular-mass polysaccharide fraction).7,10,26,27 We developed a much less complicated procedure based on the treatment of S. epidermidis biofilms with ultrasound under conditions that dissolve the extracellular polymer without disrupting the cell walls, resulting in a crude soluble PNAG preparation. Using this approach, we were able to measure the release of N-acetylglucosamine after incubation with dispersin B at concentrations as low as 1 µg/mL.
In the protocol described below, we set out experimental procedures that will allow researchers to test enzymes for their ability to degrade S. epidermidis biofilms and to investigate whether this activity is related to the degradation of PNAG. Because dispersin B is not commercially available, we first describe how a recombinant enzyme can be produced and purified in a single step by immobilized metal ion affinity chromatography (IMAC). We then describe a 24-well plate assay that allows the visual confirmation of biofilm-detaching activity, and explain how N-acetylglucosamine can be detected in the supernatants from this assay. Next we present our procedure for the preparation of soluble PNAG, and finally we discuss how this material is used as substrate in a sensitive PNAG-degradation assay.
Materials
Reagents
General reagents
(1) Ethanol ≥ 99.8% p.a. (Carl Roth, 9065.4)
(2) HCl 25% (Carl Roth, X897.1) !CAUTION Corrosive
(3) NaCl (Carl Roth, 9265.2)
(4) NaH2PO4·2H2O (Carl Roth, T879.1)
(5) Na2HPO4·2H2O (Carl Roth, 4984.2)
(6) NaOH (Carl Roth, 6771.1) !CAUTION Corrosive
(7) pH indicator paper (Carl Roth, KH69.1)
(8) Tris(hydroxymethyl)aminomethane (TRIS base) (Carl Roth, AE15.1)
Reagents for recombinant dispersin B production
(1) Expression vector pASK-IBA33plus (IBA, Göttingen)
(2) One Shot TOP10 chemically competent E. coli cells (Invitrogen)
(3) Terrific Broth medium (Fluka, T0918)
(4) Glycerol (Carl Roth, 3783.2)
(5) Ampicillin (Carl Roth, K029.1)
(6) Anhydrotetracycline (IBA, 2-0401-001)
(7) Imidazole (Carl Roth, 3899.3)
(8) TALON Superflow Co2+ resin, supplied as a 50% slurry in 20% ethanol (Clontech Laboratories)
Reagents for the biofilm-detachment assay and preparation of soluble PNAG
(1) S. epidermidis RP62A (ATCC 35984) !CAUTION Biosafety Level 2
(2) Tryptic Soy Broth (TSB) medium (Fluka, T8907)
(3) Agar-agar (Carl Roth, 2266.3)
(4) Crystal violet (Merck, 1.15940.0100)
(5) 2 n HCl (Fluka, 653799-500ML)
Enzymes to be used as controls in the biofilm detachment-assay
(1) Lysostaphin (Sigma-Aldrich, L9043-5MG)
(2) Hen egg-white lysozyme (Carl Roth, 8259.2)
(3) DNase I (Roche Diagnostics, 11284932001)
Reagents for HPAEC-PAD-ESI-MS
(1) Argon 5.3, 1-L cylinder (Linde)
(2) Helium 6.0, 50-L cylinder (Linde) !CAUTION Secure cylinder to prevent falling and breaking
(3) NaOH 50–52% in water (Fluka, 72064). Do not use NaOH pellets to prepare this solution. !CAUTION Corrosive
(4) LiCl (Sigma-Aldrich, 20367-10G)
Standards for HPAEC-PAD-ESI-MS
(1) Glucose (Carl Roth, HN06.3)
(2) N-Acetylglucosamine (Sigma-Aldrich, A8625-5G)
(3) Glucosamine (Sigma-Aldrich, G4875-25G)
Equipment
General equipment
(1) 1-L Erlenmeyer flasks with baffles and non-hermetic stainless steel caps (VGKL, 192010054, 192901224).
(2) 1.5-mL polypropylene microcentrifuge tubes (Eppendorf, 0030125.215)
(3) 50-mL polypropylene conical tubes (Greiner Bio-One, 227261)
(4) Autoclave 3850 EL (Tuttnauer)
(5) Centrifuge tubes (Beckman Coulter, 355607 and 357003)
(6) Class II microbiological safety cabinet SafeFAST Elite 212 S (Faster)
(7) High-performance centrifuge Avanti J-26XP with rotors JA-10 and JA-25.50 (Beckman Coulter)
(8) Microbiological incubator Multitron (Infors)
(9) Microcentrifuge MICRO 220R (Hettich)
(10) pH meter Seven Multi (Mettler-Toledo)
(11) Ultra-low temperature freezer GFL-6384 (GFL)
(12) Ultrasonic bath Sonorex RK 514 H (Bandelin)
(13) Ultrasonic homogenizer with generator GM 2200, ultrasonic convertor UW 2200, booster horn SH 213 G, 6-mm diameter tapered tip probe KE 76, 13-mm diameter extended probe VS 70 T, and Rosett Cell RZ 5 (Bandelin)
(14) Water purification system with 0.2 µm outlet filter Pacific 3 UP and GenPure UV/UF (TKA)
Equipment for recombinant dispersin B production
(1) 0.22-µm disposable vacuum filtration units (Nalgene, 164-0020)
(2) Low pressure liquid chromatography system ÄKTAprime plus (GE Healthcare)
(3) Empty column XK 26/20 (GE Healthcare)
(4) Spectrophotometer SPECORD 210 (Analytik Jena)
Equipment for the biofilm-detachment assay and preparation of soluble PNAG
(1) Roti-Store Cryotubes (Carl Roth, P730.1)
(2) 24-well tissue culture plates (Greiner Bio-One, 662160)
(3) Cell scraper (Greiner Bio-One, 541070)
(4) Cell culture centrifuge ROTINA 420R with 4723 swing-out rotor, 4750 round buckets, 4751 aerosol-tight lids, and 4770 adapters for 50-mL tubes (Hettich)
(5) 0.3-mL glass reaction vials with Teflon septum (Thermo Scientific, TS-13220)
(6) Heating block MBT 250-3 (ETG)
(7) Centrifugal ultrafiltration devices with 3-kDa molecular mass cut-off Nanosep 3K Omega (Pall Life Sciences, OD010233)
Equipment for HPAEC-PAD-ESI-MS
(1) CarboPac PA20 separator (3 × 150 mm) and guard (3 × 30 mm) column, packed with 6-µm microporous, pellicular anion exchange resin (Dionex, 060142 and 060144)
(2) Low-pressure mixing quaternary gradient pump with vacuum degasser ICS-3000 (Dionex, 061706)
(3) Corrosion-resistant Eluent Organizer (Dionex, 062628)
(4) Regulator Accessory with pressure regulator and gauge assembly (Dionex, 062345)
(5) Autosampler with sample tray (Dionex, 069542 and 055057)
(6) Detector/Chromatography Module DC-2 (Dionex, 063946/11)
(7) Electrochemical detector with electrochemical cell, gold working electrode and Ag/AgCl reference electrode (Dionex, 061719, 061757, 061875 and 061879)
(8) 2-mm anion self-regenerating suppressor ASRS 300 (Dionex, 064555)
(9) Auxiliary peristaltic pump ISM596D (Ismatec)
(10) Two auxiliary single-piston pumps AXP (Dionex, 063973)
(11) ESI-MS MSQ Plus (Dionex, 063116)
(12) Rotary pre-vacuum pump E2M30 (BOC Edwards)
(13) Nitrogen generator LCMS 20-1 (Parker Domnick Hunter)
(14) Two T-pieces (Dionex, 048227)
(15) Seven 2-L plastic eluent reservoirs (Dionex, 044129)
(16) 1-L polypropylene measuring cylinder (Carl Roth, 1669.1)
Optional equipment for HPAEC-PAD-ESI-MS
(1) Diode array detector DAD-3000 with 13-µL optical cell (Dionex, 5082.0010 and 6086.0400)
(2) Conductivity detector (Dionex, 061830)
(3) Automation Manager (Dionex, 062214-01)
Reagent Setup
Synthetic gene construct encoding dispersin B
A synthetic gene, codon adapted for E. coli K12 and encoding the entire dispersin B amino acid sequence (GenBank accession number AAP31025) can be obtained by custom gene synthesis (e.g., from Eurofins MWG Operon). The sequences 5′-ATGGTAGGTCTCAAATG-3′ and 5′-AGCGCTGAGACCTACCAT-3′ must be added to the 5′ and 3′ ends of the coding sequence, respectively, for insertion into the expression vector via the two BsaI restriction sites. The recombinant dispersin B sequence will be extended at the C-terminus with the vector-derived sequence SARGSHHHHHH.
IMAC buffer A (100 mM NaCl, 30 mM TRIS-HCl, pH 7.5)
Dissolve 5.84 g NaCl and 36.3 g TRIS base in 900 mL H2O. Adjust pH to 7.5 with HCl. Make up to 1 L with H2O. Degas by bubbling with helium in an ultrasonic bath for 15 min.
IMAC buffer B (200 mM imidazole, 100 mM NaCl, 30 mM TRIS-HCl, pH 7.5)
Dissolve 13.6 g imidazole, 5.84 g NaCl and 36.3 g TRIS base in 900 mL H2O. Adjust pH to 7.5 with HCl. Make up to 1 L with H2O. Degas by bubbling with helium in an ultrasonic bath for 15 min.
PNAG buffer (150 mM NaCl, 10 mM TRIS-HCl, pH 7.5)
Dissolve 8.77 g NaCl and 1.21 g TRIS base in 900 mL H2O. Adjust pH to 7.5 with HCl. Make up to 1 L with H2O.
50 mM sodium phosphate buffer, pH 5.9
Mix 3.95 mL of 1 M Na2HPO4 with 46.05 mL of 1 M NaH2PO4. Dilute the combined 1 M stock solutions to 1 L with H2O. Adjust pH to 5.9 with NaOH.
Standards for HPAEC-PAD-ESI-MS
Prepare 1 mM solutions of glucose, glucosamine and N-acetylglucosamine. Dilute 1:20 in water (final concentration 50 µM). Store at –20°C.
Eluent A (2 mM NaOH) and eluent B (250 mM NaOH) for HPAEC-PAD-ESI-MS
Eluent A is prepared by adding 320 µL 50% NaOH to 2,000 mL water, whereas eluent B is prepared by adding 40 mL 50% NaOH to 1,960 mL water. Use reagent-grade water with a conductivity of 0.055 µS/cm filtered through a 0.2 µm filter. Only use plasticware to handle NaOH.
In detail, the eluents are prepared using the following procedure: Set apart two 2-L plastic eluent reservoirs and a 1-L polypropylene measuring cylinder, which must not be used for other purposes. Fill each of the 2-L plastic reservoirs with 1.5 L water and degas by bubbling with helium in an ultrasonic bath for 15 min. Fill the 1-L polypropylene measuring cylinder with 500 mL of the degassed water and add 320 µL or 40 mL of 50% NaOH, respectively. Remove the reservoir for the corresponding eluent from the Eluent Organizer in the chromatography system after closing the stopcock valve of the argon supply and removing the lid with the tubing. Discard residual eluent and rinse the reservoir with degassed water. Pour the pre-diluted NaOH solution from the measuring cylinder into the reservoir and add the appropriate volume of degassed water to make up to 2,000 mL. Screw the lid on and carefully invert the reservoir to mix. Place the reservoir back into the Eluent Organizer. Open the stopcock valve to resupply the headspace with argon.
Use the following precautions to handle the 50% NaOH stock: To reduce carbonate contamination that may occur when NaOH reacts with CO2 in the air, avoid mixing the 50% NaOH stock solution. Pipette the aliquots used for eluent preparation from approximately 2.5 cm below the surface because any sodium carbonate precipitate will sink. Discard the 50% NaOH solution when approximately 2/3 of the volume has been used.
Equipment Setup
HPAEC-PAD-ESI-MS instrumentation
A chromatography system with a chemically-inert, metal-free, polyether ether ketone (PEEK)-based flow path is used in order to avoid corrosion by the NaOH eluent (Fig. 2). The eluents are delivered from two 2-L plastic reservoirs held in a corrosion-resistant Eluent Organizer. In order to minimize carbonate contamination, the headspace of the eluent reservoirs is filled with argon gas (~0.3 bar) supplied by the Regulator Accessory with a pressure regulator, gauge assembly and outputs for connections to up to four eluent reservoirs. The argon supply tubing is connected via stopcock valves on the lids of the eluent reservoirs. After the separator column, a T-piece is introduced as 1:1 flow splitter, one path leading to the electrochemical detector, the other via the desalter and the diode array detector to the ESI-MS. An anion self-regenerating suppressor is used for desalting, operating in auto-suppression external water mode (PROBLEM 1). The water regenerant is delivered from a 2-L reservoir using a peristaltic pump. The diode array detector is integrated after the desalter in order to avoid damage to the optical cell after prolonged exposure to NaOH. The use of a diode array detector is not necessarily required for carbohydrate analysis but allows the detection of unrelated compounds. Neutral carbohydrates are detected as Li+ adducts by delivering 0.5 mM LiCl to the flow path prior to injection into the MS from a 2-L reservoir using an AXP auxiliary pump and a T-piece. The entrance cone of the MS is washed with water delivered from a 2-L reservoir with an AXP pump. Nitrogen serving as a sheath and nebulizing gas is provided from the nitrogen generator. Chromeleon chromatography data system software (version 6.8, SR6) is used for instrument control, data acquisition and processing. The MS is calibrated and tuned using Xcalibur (version 2.0.7) and MSQ 2.0 SP1 tune program.
HPAEC-PAD-ESI-MS chromatography conditions
Chromatography is performed under isocratic conditions at a flow rate of 0.4 mL/min (resulting in a system pressure of 160–170 bar) using the following protocol: separation, 0–15 min, 14.4 mM NaOH (95% eluent A, 5% eluent B); column cleaning, 15–20 min, 76.4 mM NaOH (70% eluent A, 30% eluent B); column reconditioning, 20–27 min, 14.4 mM NaOH (95% eluent A, 5% eluent B).
The auxiliary pumps are set as follows: regenerant peristaltic pump, 25 rpm (~1 mL/min); LiCl addition pump, 0.05 mL/min; MS entrance cone wash pump, 0.05 mL/min.
The waveform settings of the electrochemical detector are set as follows: 0.10 V (0.00–0.20 sec, 0.20–0.40 sec GainRegion on, Integration on), ramp to –2.00 V (0.40–0.41 sec), –2.00 V (0.41–0.42 sec), ramp to 0.60 V (0.42–0.43 sec), ramp to –0.10 V (0.43–0.44 sec), –0.10 V (0.44–0.50 sec). GainRegion and Integration are off except for the 0.20–0.40 sec time frame.
The suppressor current is set to 8 mA during the separation and column reconditioning step (0–15 min and 25–27 min), and to 100 mA during the column cleaning step (15–25 min); these settings take into account the delay caused mainly by the column void volume.
The mass spectrometer is operated in positive ion mode with the following settings: ESI probe temperature, 349°C; ESI needle voltage, 3 kV; detector voltage, 1,106 V; nitrogen gas pressure, 5 bar.
For the detection of N-acetylglucosamine H2O and Li+ adducts in selected ion-monitoring (SIM) mode, the following settings are used: mass, 246 Da; cone voltage, 100 V; RF-lens, –1.0 V; mass span, 0.5 Da; scan time, 0.33 sec. For the detection of glucose H2O and Li+ adducts in SIM mode (as a control for system performance) the following settings are used: mass, 205 Da; cone voltage, 75 V; RF-lens, –1.0 V; mass span, 0.5 Da; scan time, 0.33 sec.
If not in use, the system is maintained in a standby mode with the following settings: gradient pump, 0.025 mL/min, 95% eluent A, 5% eluent B; LiCl addition pump, off; MS entrance cone wash pump, off; regenerant peristaltic pump, 5 rmp; suppressor current, 1 mA; ESI probe temperature, off; cone voltage, 0 V; ESI needle voltage, 0 V. Vacuum and nitrogen supply remain connected.
Procedure
Production of recombinant dispersin B
(1) Prepare six 1-L Erlenmeyer flasks each containing 400 mL Terrific Broth medium supplemented with 300 µg/mL ampicillin.
(2) Introduce the synthetic gene construct into competent E. coli cells following the protocol provided by the supplier.
(3) Use the bacterial suspension obtained from the transformation procedure directly to inoculate the culture flasks (50 µL for each flask). Inoculation from a plate culture may result in the selection of clones that produce the recombinant protein inefficiently.
(4) Incubate the cultures at 30°C and 250 rpm overnight until the OD600nm reaches ~2.
(5) Induce the expression of recombinant dispersin B by adding 40 µL of a 2 mg/mL stock solution of anhydrotetracycline in ethanol to each flask (final concentration 200 ng/mL).
(6) Continue incubation at 37°C and 250 rpm for 2 h.
(7) Harvest the cells by centrifugation (10,000 g, 10 min, 4°C, JA-10 rotor) and discard the supernatant.
(8) The cell pellets can be stored at –80°C (PAUSE POINT). Before continuing with protein purification, pack, wash and equilibrate the IMAC column using the low pressure chromatography system.
(9) Pack a 2.6 × 4 cm TALON Superflow Co2+ column (~20 mL column bed volume) by pouring 40 mL of the slurry into the empty XK 26/20 column. Wash with IMAC buffer A at a flow rate of 5 mL/min until the resin has completely settled. For column assembly and packing, follow the manufacturer’s instructions.
(10) Wash the column with 100 mL of IMAC buffer B at a flow rate of 5 mL/min. The imidazole will cause the column to take on a purplish hue.
(11) Equilibrate the column with IMAC buffer A until the normal pink color is restored.
(12) Column equilibration can be performed overnight at a low flow rate (0.5 mL/min). The equilibrated column can be removed from the chromatography system and stored at 4°C for several days (PAUSE POINT).
(13) To continue the protein purification, resuspend the E. coli cell pellets in 200 mL of IMAC buffer A.
(14) Transfer the suspension to the Rosett Cell packed in an ice-water bath and disrupt the E. coli cells by ultrasonication for 10 min (30% pulse time, 70% amplitude) using the ultrasonic homogenizer equipped with the 6-mm tapered tip probe.
(15) Remove cell debris by centrifugation (75,000 g, 30 min, 10°C, JA-25.50 rotor) and filter the supernatant through a 0.22-µm filter.
(16) Load the cleared lysate onto the equilibrated column applying a flow rate of 5 mL/min (all subsequent steps are performed at the same flow rate).
(17) Wash the column with IMAC buffer A until absorption at 280 nm remains constant.
(18) Wash the column with 50 mL of IMAC buffer containing 10 mM imidazole (95% IMAC buffer A, 5% IMAC buffer B).
(19) Elute dispersin B with 100 mL IMAC buffer B. Observe absorption at 280 nm and collect fractions of ~5 mL.
(20) Measure the absorption of the individual fractions at 280 nm. Calculate the protein concentration, assuming that a 1 mg/mL solution results in an absorption of 1.25 (http://web.expasy.org/protparam/).
(21) Combine the main fractions, prepare small aliquots in polypropylene microcentrifuge tubes, shock-freeze in a dry ice-methanol bath or liquid nitrogen, and store at –80°C. Avoid repeated freeze-thaw cycles (PROBLEM 2).
Biofilm-detachment assay
(22) Prepare stocks of cryopreserved S. epidermidis RP62A cells from a fresh overnight culture using Roti-Store Cryotubes according to the manufacturer’s instructions.
(23) Streak a glass pellet with cryopreserved S. epidermidis RP62A cells from a Roti-Store Cryotube over a TSB plate containing 1.5% agar-agar, and incubate at 37°C for 72 h.
(24) Scratch with a 1-mL serological plastic pipette through the bacteria grown on the plate and inoculate 25 mL TSB medium in a 50-mL polypropylene conical tube.
(25) Incubate the tube for 5 h at 37°C without shaking and without aeration.
(26) Transfer 1 mL of bacterial suspension from the tube to each well of a 24-well tissue culture plate, and incubate for 24 h at 37°C without shaking.
(27) Before continuing with washing the biofilm formed on the bottom of the wells (described in the next step) prepare the samples to be tested in the assay. Prepare a 1:10 dilution series from 1,000 µg/mL to 0.01 µg/mL of dispersin B in 1 mL 50 mM sodium phosphate (pH 5.9) in polypropylene microcentrifuge tubes. The following enzymes, prediluted in the indicated buffers, may be also tested as controls: lysostaphin (50 mM TRIS-HCl, pH 7.5), lysozyme (50 mM TRIS-HCl, pH 7.5) DNase I (150 mM NaCl, 100 mM TRIS-HCl, 25 mM MgCl2, 5 mM CaCl2, pH 7.5).
(28) Aspirate the medium from the wells of the 24-well plate with a pipette and carefully wash the biofilm with 1 mL water. Avoid scratching the biofilm by inclining the plate and placing the pipet tip on the wall of the wells.
(29) Add 300 µL of the prediluted enzyme samples to each well. Incubate at 28°C with gentle shaking (50 rpm) for 2 h.
(30) Carefully transfer the supernatants to polypropylene microcentrifuge tubes.
(31) Wash the remaining biofilm in the wells of the 24-well plate with 1 mL water and stain with 400 µL 0.1% crystal violet solution in water for 5 min. Remove the dye solution and wash with 1 mL water. Allow to air-dry, and photograph the plate for documentation.
(32) Pass the supernatants from the individual wells through centrifugal ultrafiltration devices with a 3-kDa molecular mass cut-off. Collect the flow-through for HPAE-PAD-ESI-MS analysis.
(33) The samples can be stored at –20°C (PAUSE POINT).
Preparation of soluble PNAG
(34) Prepare 15 1-L Erlenmeyer flasks each containing 400 mL of TSB medium.
(35) Inoculate each flask with one glass pellet containing cryopreserved cells from a Roti-Store Cryotube.
(36) Cover the flasks with non-hermetic caps and incubate without shaking at 37°C for 20 h.
(37) Discard the culture liquid and harvest the biofilm formed on the wall and bottom of the flasks with a cell scraper in a small volume of residual culture medium.
(38) Transfer the suspension into 50-mL polypropylene conical tubes and collect the biofilm material by centrifugation in a swing-out rotor (4,000 g, 10 min, 4°C).
(39) Discard the supernatant and wash the pellets three times with water by centrifugation (4,000 g, 10 min, 4°C).
(40) Suspend the combined pellets in 20 mL PNAG buffer in a 50-mL polypropylene conical tube.
(41) Place the tube in an ice-water bath and solubilize the PNAG from the biofilm material by ultrasonication for 30 sec (50% pulse time, 50% amplitude) using the ultrasonic homogenizer equipped with the 13-mm extended probe VS 70 T. Repeat sonication five times, cooling for 60 sec between treatments.
(42) Remove the cells by centrifugation in a swing-out rotor (4,000 g, 10 min, 4°C).
(43) Clear the supernatant containing the solubilized PNAG using a second centrifugation step (14,000 g, 15 min, 4°C, JA-25.50 rotor).
(44) Prepare aliquots of the supernatant in polypropylene microcentrifuge tubes and store at –20°C (PAUSE POINT).
Analysis of the PNAG monosaccharide composition
(45) Take samples of 1 µL, 3 µL, 10 µL and 30 µL from the soluble PNAG preparation.
(46) Transfer the samples to 0.3-mL glass reaction vials with Teflon septa, make up to 100 µL with water, and combine with 100 µL of 1 n HCl (diluted from 2 n HCl).
(47) Heat the vials to 110°C for 2 h in a heating block. !CAUTION Vials are under pressure during heating. Open after cooling on an ice bath. Wear protective goggles.
(48) Neutralize the content of the vials by adding small aliquots of 2 M NaOH; check pH by spotting 5-µL aliquots on indicator paper.
(49) Pass the contents of the individual vials through centrifugal ultrafiltration devices with a 3-kDa molecular mass cut-off. Collect the flow-trough for HPAE-PAD-ESI-MS analysis.
(50) The samples can be stored at –20°C (PAUSE POINT).
Digestion of soluble PNAG with dispersin B
(51) Using polypropylene microcentrifuge tubes, combine 100-µL aliquots of the soluble PNAG preparation with 100 µL dispersin B solution prediluted in sodium phosphate buffer (pH 5.9) and incubate at 37°C for 2 h.
(52) Pass the contents of the individual tubes through centrifugal ultrafiltration devices with a 3-kDa molecular mass cut-off. Collect the flow-trough for HPAE-PAD-ESI-MS analysis.
(53) The samples can be stored at –20°C (PAUSE POINT).
HPAE-PAD-ESI-MS analysis
(54) Program a sequence for sample analysis. Start with a blank, a water control and the standards. In order to check for carry-over from the standards, run a water control before analyzing the test samples. The sample injection volume is 10 µL.
(55) Transfer 200 µl of each sample to 0.3-mL polypropylene sample vials and load the autosampler.
(56) Start the analysis. The analysis is typically run overnight (PAUSE POINT).
Problem Handling
Problem 1: NaOH breakthrough caused by suppressor failure
Injection of NaOH into the MS must be avoided under any circumstances. For optimal performance, the suppressor should be operated at a backpressure of 2.0–2.7 bar. This requires empirical selection of the lengths and diameters of the tubing for the two parallel detector lines. The suppressor backpressure can be determined by operating the gradient pump at 0.4 mL/min and testing the actual flow rate (~0.2 mL/min) over the MS line by measuring the volume of eluent released from the disconnected MS inlet tubing. Plug the outlet of the flow splitter T-piece that leads to the electrochemical detector and turn on the gradient pump at the appropriate MS line flow rate. Read the system pressure before and after disconnecting the eluent outlet of the suppressor. Suppressor performance also depends on the correct settings for the water regenerant flow rate and suppressor current. Refer to the manual provided with the suppressor if chromatography conditions other than those described here are required.
Check the pH value of the eluent released from the MS inlet tubing with indicator paper. It should be neutral. Suppressor failure during system operation will increase the absorption at 205 nm registered by the diode array detector during the column cleaning step. In addition, the carbohydrates will be detected by the MS as Na+ instead of Li+ adducts. If NaOH has accidentally been injected, disassembly and thorough cleaning of the MS will be necessary.
Optionally, a conductivity detector and an Automation Manager (not shown in Fig. 2) can be inserted after the suppressor in order to detect NaOH breakthrough. The Automation Manager can be programmed to switch the eluent flow to the waste outlet via the high-pressure valve if the conductivity increases to > 1 mS/cm.
Problem 2: Instability of dispersin B
Dispersin B appears to be intrinsically unstable, and it has not been possible thus far to produce the recombinant protein without a His6 tag. Accordingly, our initial attempts to produce untagged dispersin B failed, apparently due to precipitation on an anion exchange column. The purified His6-tagged dispersin B can also precipitate after freeze-thaw. Nevertheless, the samples remained active in the biofilm detachment assay after one freeze-thaw cycle. The stability of dispersin B could potentially be improved by adding detergent during purification as described.28
Anticipated Results
Production of recombinant dispersin B
Expression of a synthetic, codon-optimized dispersin B gene in E. coli is achieved using the vector pASK-IBA33plus, which features an anhydrotetracycline-inducible promoter system, a multiple cloning site flanked by two BsaI restriction sites and a C-terminal His6 tag sequence. Cloning via the BsaI restriction sites eliminates the need to append non-native amino acid residues to the N-terminus of the recombinant enzyme. The promoter system results in minimal residual recombinant protein synthesis in the absence of the inducer. A 2.4-L E. coli culture yields ~150 mg of dispersin B after purification by IMAC (Fig. 3).
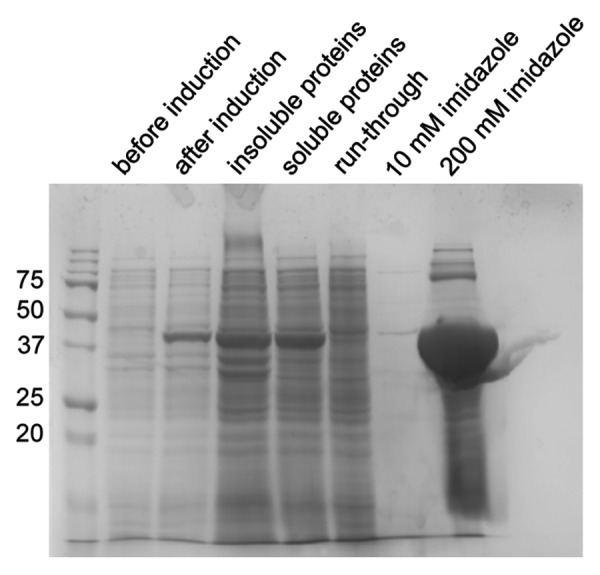
Figure 3. Production of recombinant dispersin B in E. coli. The His6-tagged enzyme was purified from the soluble fraction of the E. coli cells by chromatography on a Co2+ column and eluted using an imidazole step-gradient. Samples were analyzed by sodium dodecyl sulfate PAGE on a 12% gel under reducing conditions. The gel was stained with Coomassie brilliant blue.
Biofilm detachment assay
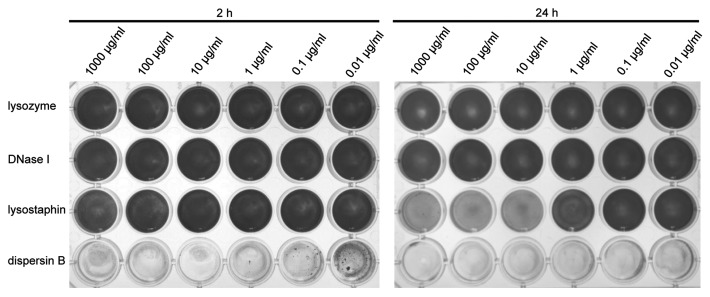
S. epidermidis biofilms can be generated reproducibly in 24-well plates using the procedure described here. It is not necessary to start with a defined number of suspended bacterial cells as previously suggested.22 We also found no advantage to increasing the NaCl or glucose concentration in the culture medium, which was previously reported to enhance biofilm formation.29 Incubation with dispersin B at concentrations of ≥ 0.1 µg/mL leads to almost complete removal of the biofilm (Fig. 4). Lysostaphin also has a noticeable impact, particularly if the incubation time is extended to 24 h, but biofilm removal is incomplete and leaves a residual opaquely-stained layer. No detectable biofilm degradation is achieved with DNase I or hen egg-white lysozyme. HPAE-PAD-ESI-MS analysis allows the detection of N-acetylglucosamine in the supernatants from biofilms treated with dispersin B but not those treated with the other enzymes (Fig. 5).
Figure 4. Biofilm-detachment assay. Biofilms obtained by growing S. epidermidis in 24-well tissue culture plates were incubated for 2 h or 24 h with the candidate enzymes at different concentrations. The wells were rinsed with water and the remaining biofilms stained with crystal violet.
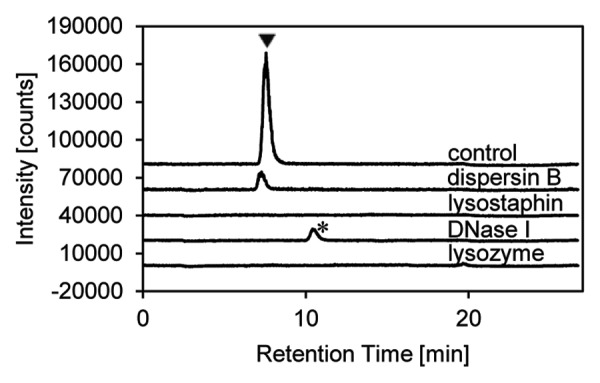
Figure 5. Analysis of supernatants from the biofilm-detachment assay by HPAEC-PAD-ESI-MS. Online detection by ESI-MS revealed the presence of N-acetylglucosamine (arrowhead) after treatment with dispersin B, but not after treatment with lysostaphin, DNase I or lysozyme (1,000 µg/mL of each enzyme). An unidentified peak (asterisk) appeared after DNase I treatment. A 50 µM N-acetylglucosamine solution was used as control.
The minimal anti-biofilm activity of lysostaphin and the absence of N-acetylglucosamine in the supernatant reflects the mode of action of this enzyme, which is known to cleave the cross-linking pentaglycine bridges in the staphylococcal cell wall,20,30 but not to degrade extracellular polysaccharide components, thus lysing the bacterial cells within the polysaccharide matrix instead of detaching the biofilm. The inability of DNase I to degrade biofilms is consistent with previous findings that only biofilms less than 6 h old are detached by this enzyme, but not biofilms grown for 12 or 24 h.21,22 The acetylation of N-acetylmuramic acid at the O-6 position makes peptidoglycans produced by pathogenic staphylococci resistant to the muramidase activity of lysozyme.31
Preparation of soluble PNAG
Soluble PNAG can be prepared by ultrasound treatment starting with biofilm material collected from S. epidermidis cultures in Erlenmeyer flasks. For analytical characterization, small samples of such preparations are heated with HCl to hydrolyze PNAG into its monosaccharide components and to deacetylate N-acetylglucosamine to glucosamine.32 Accordingly, glucosamine is identified as the major component of the hydrolysate by HPAE-PAD-ESI-MS (Fig. 6). Glucosamine, because of its cationic properties, is removed by the desalter and is therefore recorded only by the electrochemical detector.
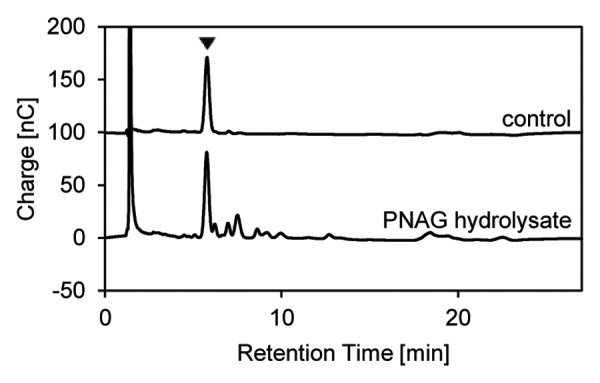
Figure 6. Characterization of soluble PNAG prepared from S. epidermidis biofilm material. A 3-µL sample of the PNAG preparation was hydrolyzed with HCl and analyzed by HPAEC-PAD-ESI-MS. Glucosamine (arrowhead) from the deacetylation of N-acetylglucosamine monomers was recorded by PAD. A 50 µM authentic N-acetylglucosamine solution was subjected to the same procedure as a control.
Digestion of soluble PNAG with dispersin B
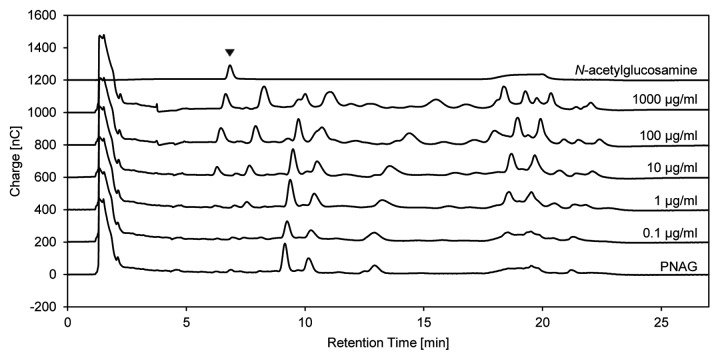
Incubation of soluble PNAG with dispersin B and subsequent HPAE-PAD-ESI-MS analysis causes a marked increase in the N-acetylglucosamine signal intensity compared with the supernatants from intact biofilms on 24-well plates treated with dispersin B (Fig. 7). Although 100 µg/mL of dispersin B is required to release detectable N-acetylglucosamine in the plate assay, 1 µg/mL is sufficient with soluble PNAG as the substrate. As well as detecting N-acetylglucosamine by online ESI-MS, changes in the total carbohydrate profile can be recorded by PAD. With this detection method, a peak correlating with N-acetylglucosamine can be observed when soluble PNAG is digested with dispersin B at concentrations of ≥ 10 µg/mL (Fig. 8). Several unassigned peaks at higher retention times may indicate larger PNAG degradation products or strongly negatively-charged components that may also be present in the exopolysaccharide.
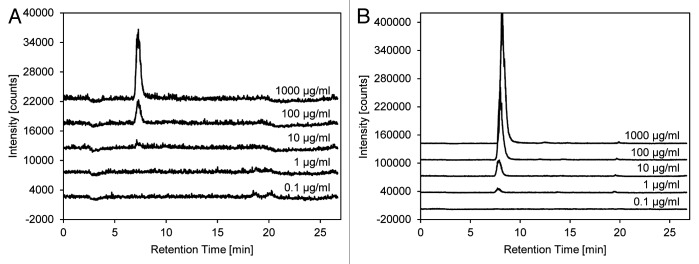
Figure 7. Determination of N-acetylglucosamine release after the treatment of intact biofilms and soluble PNAG with dispersin B. Supernatants from the 24-well plate biofilm-detachment assay (A) and the reaction products obtained by digesting soluble PNAG (B) with dispersin B at the indicated concentrations were analyzed by HPAEC-PAD-ESI-MS. Chromatograms were recorded by online ESI-MS. The intensity scale in panel A is magnified 10-fold.
Figure 8. Total carbohydrate profile of PNAG treated with dispersin B. Soluble PNAG was incubated with the indicated concentrations of dispersin B and analyzed by HPAEC-PAD-ESI-MS. Chromatograms were recorded by PAD. Untreated PNAG and 50 µM N-acetylglucosamine were used as controls. The peak corresponding to N-acetylglucosamine is labeled with an arrowhead.
Implementation in screening programs
Our procedure is based on the measurement of N-acetylglucosamine released from soluble PNAG, providing a specific functional assay for the detection of dispersin B at concentrations as low as 1 µg/mL. Remarkably, pre-formed S. epidermidis biofilms on 24-well plates can be detached with dispersin B concentrations at least 10-fold lower. As a possible explanation, only partial endolytic cleavage of PNAG may be sufficient for biofilm detachment, whereas the appearance of detectable amounts of N-acetylglucosamine monomers may require more extensive digestion. This suggests that screening programs for new PNAG-degrading enzymes should be performed in two steps, the first using a multi-well plate biofilm-detachment assay followed by the testing of potential hits for their ability to release N-acetylglucosamine from soluble PNAG.
Acknowledgments
We wish to thank Wilma Ziebuhr for helpful discussions and for providing the S. epidermidis strain we used. The work was supported by the German federal state of Hessen as part of the LOEWE-Schwerpunkt Insektenbiotechnologie.
Glossary
Abbreviations:
- ESI-MS
electrospray ionization mass spectrometry
- HPAEC
high-pH anion-exchange chromatography
- IMAC
immobilized metal ion affinity chromatography
- p.a.
pro analysi (reagent grade)
- PAD
pulsed amperometric detection
- PNAG
poly-β-1,6-N-acetyl-d-glucosamine
- SIM
selected ion-monitoring
- TSB
tryptic soy broth
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/virulence/article/23560
References
- 1.Rosenthal VD, Bijie H, Maki DG, Mehta Y, Apisarnthanarak A, Medeiros EA, et al. INICC members International Nosocomial Infection Control Consortium (INICC) report, data summary of 36 countries, for 2004-2009. Am J Infect Control. 2012;40:396–407. doi: 10.1016/j.ajic.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 2.Uçkay I, Pittet D, Vaudaux P, Sax H, Lew D, Waldvogel F. Foreign body infections due to Staphylococcus epidermidis. Ann Med. 2009;41:109–19. doi: 10.1080/07853890802337045. [DOI] [PubMed] [Google Scholar]
- 3.Schoenfelder SM, Lange C, Eckart M, Hennig S, Kozytska S, Ziebuhr W. Success through diversity - how Staphylococcus epidermidis establishes as a nosocomial pathogen. Int J Med Microbiol. 2010;300:380–6. doi: 10.1016/j.ijmm.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 4.Vandecandelaere I, Matthijs N, Van Nieuwerburgh F, Deforce D, Vosters P, De Bus L, et al. Assessment of microbial diversity in biofilms recovered from endotracheal tubes using culture dependent and independent approaches. PLoS One. 2012;7:e38401. doi: 10.1371/journal.pone.0038401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Götz F. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol. 1996;20:1083–91. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- 6.Kozitskaya S, Olson ME, Fey PD, Witte W, Ohlsen K, Ziebuhr W. Clonal analysis of Staphylococcus epidermidis isolates carrying or lacking biofilm-mediating genes by multilocus sequence typing. J Clin Microbiol. 2005;43:4751–7. doi: 10.1128/JCM.43.9.4751-4757.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mack D, Fischer W, Krokotsch A, Leopold K, Hartmann R, Egge H, et al. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J Bacteriol. 1996;178:175–83. doi: 10.1128/jb.178.1.175-183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vuong C, Kocianova S, Voyich JM, Yao Y, Fischer ER, DeLeo FR, et al. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J Biol Chem. 2004;279:54881–6. doi: 10.1074/jbc.M411374200. [DOI] [PubMed] [Google Scholar]
- 9.Wiesner J, Vilcinskas A. Antimicrobial peptides: the ancient arm of the human immune system. Virulence. 2010;1:440–64. doi: 10.4161/viru.1.5.12983. [DOI] [PubMed] [Google Scholar]
- 10.Joyce JG, Abeygunawardana C, Xu Q, Cook JC, Hepler R, Przysiecki CT, et al. Isolation, structural characterization, and immunological evaluation of a high-molecular-weight exopolysaccharide from Staphylococcus aureus. Carbohydr Res. 2003;338:903–22. doi: 10.1016/S0008-6215(03)00045-4. [DOI] [PubMed] [Google Scholar]
- 11.Archer NK, Mazaitis MJ, Costerton JW, Leid JG, Powers ME, Shirtliff ME. Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence. 2011;2:445–59. doi: 10.4161/viru.2.5.17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atshan SS, Nor Shamsudin M, Sekawi Z, Lung LT, Hamat RA, Karunanidhi A, et al. Prevalence of adhesion and regulation of biofilm-related genes in different clones of Staphylococcus aureus. J Biomed Biotechnol. 2012;2012:976972. doi: 10.1155/2012/976972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi AH, Slamti L, Avci FY, Pier GB, Maira-Litrán T. The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-beta-1-6-N-acetylglucosamine, which is critical for biofilm formation. J Bacteriol. 2009;191:5953–63. doi: 10.1128/JB.00647-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yakandawala N, Gawande PV, LoVetri K, Cardona ST, Romeo T, Nitz M, et al. Characterization of the poly-β-1,6-N-acetylglucosamine polysaccharide component of Burkholderia biofilms. Appl Environ Microbiol. 2011;77:8303–9. doi: 10.1128/AEM.05814-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaplan JB, Ragunath C, Ramasubbu N, Fine DH. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous beta-hexosaminidase activity. J Bacteriol. 2003;185:4693–8. doi: 10.1128/JB.185.16.4693-4698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itoh Y, Wang X, Hinnebusch BJ, Preston JF, 3rd, Romeo T. Depolymerization of beta-1,6-N-acetyl-D-glucosamine disrupts the integrity of diverse bacterial biofilms. J Bacteriol. 2005;187:382–7. doi: 10.1128/JB.187.1.382-387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerrigan JE, Ragunath C, Kandra L, Gyémánt G, Lipták A, Jánossy L, et al. Modeling and biochemical analysis of the activity of antibiofilm agent Dispersin B. Acta Biol Hung. 2008;59:439–51. doi: 10.1556/ABiol.59.2008.4.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan JB. Therapeutic potential of biofilm-dispersing enzymes. Int J Artif Organs. 2009;32:545–54. doi: 10.1177/039139880903200903. [DOI] [PubMed] [Google Scholar]
- 19.Pavlukhina SV, Kaplan JB, Xu L, Chang W, Yu X, Madhyastha S, et al. Noneluting enzymatic antibiofilm coatings. ACS Appl Mater Interfaces. 2012;4:4708–16. doi: 10.1021/am3010847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu JA, Kusuma C, Mond JJ, Kokai-Kun JF. Lysostaphin disrupts Staphylococcus aureus and Staphylococcus epidermidis biofilms on artificial surfaces. Antimicrob Agents Chemother. 2003;47:3407–14. doi: 10.1128/AAC.47.11.3407-3414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin Z, Ou Y, Yang L, Zhu Y, Tolker-Nielsen T, Molin S, et al. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology. 2007;153:2083–92. doi: 10.1099/mic.0.2007/006031-0. [DOI] [PubMed] [Google Scholar]
- 22.Izano EA, Amarante MA, Kher WB, Kaplan JB. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl Environ Microbiol. 2008;74:470–6. doi: 10.1128/AEM.02073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGuire JM, Stewart YM, Smith KD. The effect of pH on the high pH anion-exchange chromatography elution of monosaccharides. Chromatographia. 1999;49:699–702. doi: 10.1007/BF02466915. [DOI] [Google Scholar]
- 24.Yudina ON, Gening ML, Tsvetkov YE, Grachev AA, Pier GB, Nifantiev NE. Synthesis of five nona-β-(1→6)-d-glucosamines with various patterns of N-acetylation corresponding to the fragments of exopolysaccharide of Staphylococcus aureus. Carbohydr Res. 2011;346:905–13. doi: 10.1016/j.carres.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fekete A, Borbás A, Gyémánt G, Kandra L, Fazekas E, Ramasubbu N, et al. Synthesis of β-(1→6)-linked N-acetyl-D-glucosamine oligosaccharide substrates and their hydrolysis by Dispersin B. Carbohydr Res. 2011;346:1445–53. doi: 10.1016/j.carres.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maira-Litrán T, Kropec A, Abeygunawardana C, Joyce J, Mark G, 3rd, Goldmann DA, et al. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect Immun. 2002;70:4433–40. doi: 10.1128/IAI.70.8.4433-4440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sadovskaya I, Vinogradov E, Flahaut S, Kogan G, Jabbouri S. Extracellular carbohydrate-containing polymers of a model biofilm-producing strain, Staphylococcus epidermidis RP62A. Infect Immun. 2005;73:3007–17. doi: 10.1128/IAI.73.5.3007-3017.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yakandawala N, Gawande PV, LoVetri K, Romeo T, Kaplan JB, Madhyastha S. Enhanced expression of engineered ACA-less beta-1, 6-N-acetylglucosaminidase (dispersin B) in Escherichia coli. J Ind Microbiol Biotechnol. 2009;36:1297–305. doi: 10.1007/s10295-009-0613-0. [DOI] [PubMed] [Google Scholar]
- 29.Rachid S, Ohlsen K, Witte W, Hacker J, Ziebuhr W. Effect of subinhibitory antibiotic concentrations on polysaccharide intercellular adhesin expression in biofilm-forming Staphylococcus epidermidis. Antimicrob Agents Chemother. 2000;44:3357–63. doi: 10.1128/AAC.44.12.3357-3363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar JK. Lysostaphin: an antistaphylococcal agent. Appl Microbiol Biotechnol. 2008;80:555–61. doi: 10.1007/s00253-008-1579-y. [DOI] [PubMed] [Google Scholar]
- 31.Bera A, Biswas R, Herbert S, Götz F. The presence of peptidoglycan O-acetyltransferase in various staphylococcal species correlates with lysozyme resistance and pathogenicity. Infect Immun. 2006;74:4598–604. doi: 10.1128/IAI.00301-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith RL, Gilkerson E. Quantitation of glycosaminoglycan hexosamine using 3-methyl-2-benzothiazolone hydrazone hydrochloride. Anal Biochem. 1979;98:478–80. doi: 10.1016/0003-2697(79)90170-2. [DOI] [PubMed] [Google Scholar]