Abstract
Efficient cell migration is central to the normal development of tissues and organs and is involved in a wide range of human diseases, including cancer metastasis, immune responses, and cardiovascular disorders. Mesenchymal migration is modulated by focal-adhesion proteins, which organize into large integrin-rich protein complexes at the basal surface of adherent cells. Whether the extent of clustering of focal-adhesion proteins is actually required for effective migration is unclear. We recently demonstrated that the depletion of major focal-adhesion proteins, as well as modulation of matrix compliance, actin assembly, mitochondrial activity, and DNA recombination, all converged into highly predictable, inter-related, biphasic changes in focal adhesion size and cell migration. Herein, we further discuss the role of focal adhesions in controlling cell spreading and test their potential role in cell migration.
Keywords: focal adhesions, cell migration, mechanosensing, high-throughput phenotyping, systems biology
Introduction
A myriad of proteins play a role in cell migration, including cytoskeletal, motor, mechanosensing, and scaffolding proteins as well as regulatory kinases and phosphatases. In particular, a defined subset of cytoplasmic and membrane-bound proteins that cluster into focal adhesions at the basal surface of adherent cells regulate cell migration, sensation of mechanical stimuli, signal transduction through the cell membrane, and cell adhesion.1-3 Morphology and dynamics of focal adhesions, such as size, shape, molecular density and activity, turnover rate, and spatial distribution, strongly depend on the cell type and matrix properties such as dimensionality, topology, and compliance.3-7 Here a systems-biological approach uncovers a universal biphasic relationship between focal adhesion size and cell migration speed.8 Based on this data, we found that focal adhesion size uniquely predicts cell adhesion and morphology.9-11
Recapitulation of Biphasic Relationship between Focal Adhesion Size and Cell Migration Speed
Fast-moving fish keratocytes, human leukocytes, and Dictyostelium discoideum cells display small focal adhesions at their basal surface, while slow-moving fibroblasts and endothelial cells display large focal adhesions.12-14 Therefore, a superficial comparison among migratory cells suggests that cells that feature small focal adhesions migrate more rapidly than cells that feature large focal adhesions. This disparate data suggests that the extent of clustering of focal-adhesion proteins into basal adhesion plaques would inversely correlate with cell migration. However, a rigorous assessment of the role of focal-adhesion clustering in the migration of isotypic cells has been lacking.
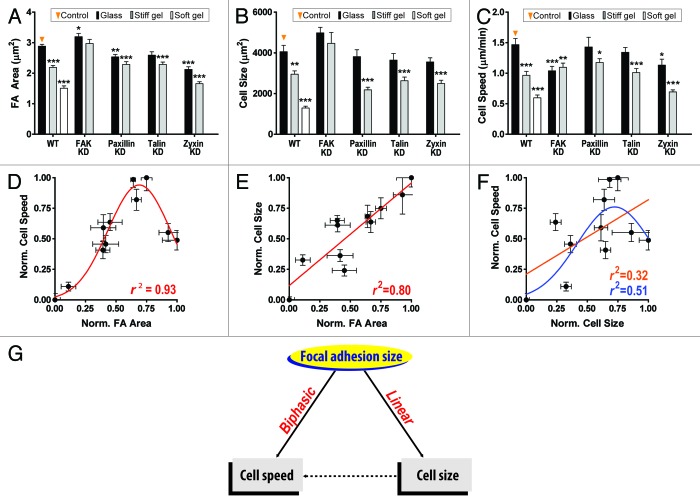
To assess the potential interplay between focal adhesion formation and cell migration, we measured the speed and persistence of migration of control mouse embryonic fibroblasts (MEFs) and MEFs depleted of major focal adhesion proteins (focal adhesion kinase, paxillin, talin, and zyxin), spontaneously migrating on flat substrates of controlled mechanical compliance, and determined these cells’ ability to form focal adhesions. These proteins and mechanical stimuli were chosen because they were known to affect the organization of focal adhesions and/or modulate cell migration15-24 (Fig. 1A–C). High-throughput quantitative live-cell microscopy revealed that the mean size of focal adhesions and mean cell migration speed were biphasically related (Fig. 1D), i.e., as focal adhesion size increased, cell moved more rapidly; past a maximum threshold speed, cell migration decreased for increasing focal adhesion size. Importantly, neither the shape of focal adhesions, nor their number or the relative cell surface occupied by focal adhesions, nor the molecular composition of focal adhesions seems to predict cell migration.8
Figure 1. Focal adhesion size is a unique predictor of cell migration speed. (A–C) Effect of changes in substrate compliance—rigid glass (black), stiff (gray), and soft (white) polyacrylamide gels coated with collagen I, and depletion of focal adhesion proteins (FAK, paxillin, talin, and zyxin) on focal adhesion size (A), cell size (B), and cell migration speed (C). At least 30 cells per condition were analyzed to assess focal-adhesion and cell morphology and >50 cells per condition were tracked to assess cell motility. Error bars represent SEM. Multiple comparison to the control (i.e., WT cells on stiff substrates) was performed by 1-way analysis of variance (ANOVA) using the Dunnett post test. Significant statistical difference are shown as follows, ***P < 0.001, **P < 0.005, *P < 0.01. (D–F) Assessment of regression among focal adhesion size, cell size, and cell speed. Mean size of focal adhesion is biphasically and linearly correlated with cell speed (D) and cell size (E), respectively, while cell size is weakly correlated with cell speed either biphasically (r2 = 0.51) or linearly (r2 = 0.32). Gaussian (nonlinear) and linear models were tested to the data set ranged between 0 and 1 after normalization as (x-xmin)/(xmax-xmin). Error bars represent SEM. Note that cell size is not statistically related to cell speed. (G) Schematic of prediction of cell speed by focal adhesion size. Cell speed is predicted by the mean size of focal adhesion not through regulation of cell size. Panels (A, C, and D) were reprinted from ref. 8.
To test the predictive power of this biphasic relation between focal adhesion size and cell migration speed, we manipulated the expression and activity of proteins that were (spatially and functionally) progressively further away from focal adhesion complexes. For instance, disassembly of actin filaments to block actomyosin-mediating force relay25 and depletion of the F-actin-crosslinking protein α-actinin, which is functionally associated with force transduction between adhesion site and cytoskeleton,26,27 induce changes in cell speed that are robustly predicted by corresponding changes in focal adhesion size. Deactivation of mitochondria and DNA recombination, which had not been previously reported to play a role in cell migration or in the formation of focal adhesions,28-30 modulated focal-adhesion formation, and cell migration in ways quantitatively predicted by the pre-established biphasic relation. Finally, the biphasic relationship established with MEFs was further validated with HT-1080 cells, a highly tumorigenic human fibrosarcoma cell line. Together these results establish that focal-adhesion size uniquely and robustly predicts cell migration across cell types and extracellular conditions.8
The Interplay between Cell Migration and Spreading
The adhesion between an adherent cell and its underlying substrate regulates cell migration speed biphasically.31 Cell-matrix adhesion strength may depend on the contact area between the cell and its adhesive substrate (i.e., cell spreading size),32 cell mechanics and contractility,33 the level of expression and activation of adhesion molecules (integrins),34 and presumably, their extent of clustering into focal adhesions, and the affinity of individual integrin molecules with their matrix molecules. Current experimental approaches such as estimation of cell spreading area or fraction of remaining adherent cells after centrifugation35 or shearing in microfluidic devices,36 and measurement of single-bond rupture force by atomic force microscopy37,38 have severe limitations, since they do not decipher the various contributors to global cell adhesion, that are intertwined with each other, and may indirectly or directly influence cell-matrix adhesion.
Since cell speed depends biphasically on focal adhesion size8 and biphasically on cell adhesion,31 focal adhesion size may correlate linearly with cell-matrix adhesion. The migratory speed, focal adhesion morphology, and spreading (cell size) of MEFs subjected to genetic manipulations and different mechanical stimuli were systematically compared (Fig. 1A–F). As predicted, the extent of cell spreading increases linearly with focal adhesion size (Fig. 1E); however, cell migration and cell spreading are poorly correlated, as assessed by linear and nonlinear fits (Fig. 1F and G). Hence, more work is needed to establish the relation between cell spreading and cell-adhesion strength.
Conclusions
Through a validated correlative analysis between descriptors of focal adhesion morphology (size, shape, and density) and descriptors of cell migration, we have addressed a long-standing question in cell biology: whether morphology of focal adhesions is functionally related to cell migration. The power of such analysis is increased substantially by using a combination of genetic and mechanical perturbations as well as blind tests. Results from this analysis show that: (1) the mean size of focal adhesions—not their shape or their number per cell—predicts cell migration across cell types and (2) the mean size of focal adhesions predicts cell spreading, while cell spreading does not predict cell migration.
These results may have important implications in biomedical research: defects in organ and tissue development or disease resulting from the onset of or defects in cell migration may occur through misregulated changes in focal adhesion size. This provides for a conceptually new pharmacological target of disease: not a specific molecular target, but a morphological descriptor of an organelle—focal adhesion size.
Acknowledgments
Work in our lab is supported by grants of the National Institutes of Health (GM084204, CA143868, and CA85839).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/celladhesion/article/24804
References
- 1.Sastry SK, Burridge K. Focal adhesions: a nexus for intracellular signaling and cytoskeletal dynamics. Exp Cell Res. 2000;261:25–36. doi: 10.1006/excr.2000.5043. [DOI] [PubMed] [Google Scholar]
- 2.Riveline D, Zamir E, Balaban NQ, Schwarz US, Ishizaki T, Narumiya S, et al. Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J Cell Biol. 2001;153:1175–86. doi: 10.1083/jcb.153.6.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim DH, Khatau SB, Feng YF, Walcott S, Sun SX, Longmore GD, et al. Actin cap associated focal adhesions and their distinct role in cellular mechanosensing. Sci Rep. 2012;2:555. doi: 10.1038/srep00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berginski ME, Vitriol EA, Hahn KM, Gomez SM. High-resolution quantification of focal adhesion spatiotemporal dynamics in living cells. PLoS One. 2011;6:e22025. doi: 10.1371/journal.pone.0022025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fraley SI, Feng YF, Giri A, Longmore GD, Wirtz D. Dimensional and temporal controls of three-dimensional cell migration by zyxin and binding partners. Nat Commun. 2012;3:719. doi: 10.1038/ncomms1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walcott S, Kim DH, Wirtz D, Sun SX. Nucleation and decay initiation are the stiffness-sensitive phases of focal adhesion maturation. Biophys J. 2011;101:2919–28. doi: 10.1016/j.bpj.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim DH, Wirtz D. Recapitulating cancer cell invasion in vitro. Proc Natl Acad Sci U S A. 2011;108:6693–4. doi: 10.1073/pnas.1103983108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim DH, Wirtz D. Focal adhesion size uniquely predicts cell migration. FASEB J. 2013;27:1351–61. doi: 10.1096/fj.12-220160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watanabe J, Eriguchi T, Ishihara K. Cell adhesion and morphology in porous scaffold based on enantiomeric poly(lactic acid) graft-type phospholipid polymers. Biomacromolecules. 2002;3:1375–83. doi: 10.1021/bm025652p. [DOI] [PubMed] [Google Scholar]
- 10.Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, et al. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil Cytoskeleton. 2005;60:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- 11.Hakkinen KM, Harunaga JS, Doyle AD, Yamada KM. Direct comparisons of the morphology, migration, cell adhesions, and actin cytoskeleton of fibroblasts in four different three-dimensional extracellular matrices. Tissue Eng Part A. 2011;17:713–24. doi: 10.1089/ten.tea.2010.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J, Jacobson K. The composition and dynamics of cell-substratum adhesions in locomoting fish keratocytes. J Cell Sci. 1997;110:2833–44. doi: 10.1242/jcs.110.22.2833. [DOI] [PubMed] [Google Scholar]
- 13.Hogg N, Laschinger M, Giles K, McDowall A. T-cell integrins: more than just sticking points. J Cell Sci. 2003;116:4695–705. doi: 10.1242/jcs.00876. [DOI] [PubMed] [Google Scholar]
- 14.Nagasaki A, Kanada M, Uyeda TQ. Cell adhesion molecules regulate contractile ring-independent cytokinesis in Dictyostelium discoideum. Cell Res. 2009;19:236–46. doi: 10.1038/cr.2008.318. [DOI] [PubMed] [Google Scholar]
- 15.Pelham RJ, Jr., Wang Yl. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci U S A. 1997;94:13661–5. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–43. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 17.Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 18.Prager-Khoutorsky M, Lichtenstein A, Krishnan R, Rajendran K, Mayo A, Kam Z, et al. Fibroblast polarization is a matrix-rigidity-dependent process controlled by focal adhesion mechanosensing. Nat Cell Biol. 2011;13:1457–65. doi: 10.1038/ncb2370. [DOI] [PubMed] [Google Scholar]
- 19.Sieg DJ, Hauck CR, Schlaepfer DD. Required role of focal adhesion kinase (FAK) for integrin-stimulated cell migration. J Cell Sci. 1999;112:2677–91. doi: 10.1242/jcs.112.16.2677. [DOI] [PubMed] [Google Scholar]
- 20.Dumbauld DW, Michael KE, Hanks SK, García AJ. Focal adhesion kinase-dependent regulation of adhesive forces involves vinculin recruitment to focal adhesions. Biol Cell. 2010;102:203–13. doi: 10.1042/BC20090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turner CE, Glenney JR, Jr., Burridge K. Paxillin: a new vinculin-binding protein present in focal adhesions. J Cell Biol. 1990;111:1059–68. doi: 10.1083/jcb.111.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilmore AP, Burridge K. Regulation of vinculin binding to talin and actin by phosphatidyl-inositol-4-5-bisphosphate. Nature. 1996;381:531–5. doi: 10.1038/381531a0. [DOI] [PubMed] [Google Scholar]
- 23.Calderwood DA, Ginsberg MH. Talin forges the links between integrins and actin. Nat Cell Biol. 2003;5:694–7. doi: 10.1038/ncb0803-694. [DOI] [PubMed] [Google Scholar]
- 24.Hirota T, Morisaki T, Nishiyama Y, Marumoto T, Tada K, Hara T, et al. Zyxin, a regulator of actin filament assembly, targets the mitotic apparatus by interacting with h-warts/LATS1 tumor suppressor. J Cell Biol. 2000;149:1073–86. doi: 10.1083/jcb.149.5.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wakatsuki T, Schwab B, Thompson NC, Elson EL. Effects of cytochalasin D and latrunculin B on mechanical properties of cells. J Cell Sci. 2001;114:1025–36. doi: 10.1242/jcs.114.5.1025. [DOI] [PubMed] [Google Scholar]
- 26.Choi CK, Vicente-Manzanares M, Zareno J, Whitmore LA, Mogilner A, Horwitz AR. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat Cell Biol. 2008;10:1039–50. doi: 10.1038/ncb1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esue O, Tseng Y, Wirtz D. Alpha-actinin and filamin cooperatively enhance the stiffness of actin filament networks. PLoS One. 2009;4:e4411. doi: 10.1371/journal.pone.0004411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cloos J, Reid CBA, van der Sterre MLT, Tobi H, Leemans CR, Snow GB, et al. A comparison of bleomycin-induced damage in lymphocytes and primary oral fibroblasts and keratinocytes in 30 subjects. Mutagenesis. 1999;14:87–93. doi: 10.1093/mutage/14.1.87. [DOI] [PubMed] [Google Scholar]
- 29.Byrnes RW, Petering DH. DNA double-strand breakage by bleomycin in Ehrlich ascites tumor cells as measured by nondenaturing filter elution. Radiat Res. 1994;137:162–70. doi: 10.2307/3578807. [DOI] [PubMed] [Google Scholar]
- 30.Li NY, Ragheb K, Lawler G, Sturgis J, Rajwa B, Melendez JA, et al. Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. J Biol Chem. 2003;278:8516–25. doi: 10.1074/jbc.M210432200. [DOI] [PubMed] [Google Scholar]
- 31.DiMilla PA, Barbee K, Lauffenburger DA. Mathematical model for the effects of adhesion and mechanics on cell migration speed. Biophys J. 1991;60:15–37. doi: 10.1016/S0006-3495(91)82027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshikawa HY, Kawano T, Matsuda T, Kidoaki S, Tanaka M. Morphology and Adhesion Strength of Myoblast Cells on Photocurable Gelatin under Native and Non-native Micromechanical Environments. J Phys Chem B. 2013;117:4081–8. doi: 10.1021/jp4008224. [DOI] [PubMed] [Google Scholar]
- 33.Hale CM, Sun SX, Wirtz D. Resolving the role of actoymyosin contractility in cell microrheology. PLoS One. 2009;4:e7054. doi: 10.1371/journal.pone.0007054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gallant ND, Michael KE, García AJ. Cell adhesion strengthening: contributions of adhesive area, integrin binding, and focal adhesion assembly. Mol Biol Cell. 2005;16:4329–40. doi: 10.1091/mbc.E05-02-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maheshwari G, Brown G, Lauffenburger DA, Wells A, Griffith LG. Cell adhesion and motility depend on nanoscale RGD clustering. J Cell Sci. 2000;113:1677–86. doi: 10.1242/jcs.113.10.1677. [DOI] [PubMed] [Google Scholar]
- 36.Lu H, Koo LY, Wang WM, Lauffenburger DA, Griffith LG, Jensen KF. Microfluidic shear devices for quantitative analysis of cell adhesion. Anal Chem. 2004;76:5257–64. doi: 10.1021/ac049837t. [DOI] [PubMed] [Google Scholar]
- 37.Bajpai S, Feng Y, Krishnamurthy R, Longmore GD, Wirtz D. Loss of alpha-catenin decreases the strength of single E-cadherin bonds between human cancer cells. J Biol Chem. 2009;284:18252–9. doi: 10.1074/jbc.M109.000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bajpai S, Correia J, Feng Y, Figueiredo J, Sun SX, Longmore GD, et al. alpha-Catenin mediates initial E-cadherin-dependent cell-cell recognition and subsequent bond strengthening. Proc Natl Acad Sci U S A. 2008;105:18331–6. doi: 10.1073/pnas.0806783105. [DOI] [PMC free article] [PubMed] [Google Scholar]