Abstract
Rho GDP dissociation inhibitors (RhoGDIs) can inhibit cell motility, invasion, and metastasis in cancer by inactivating the RhoGTPases. A member of RhoGDI family has been consistently shown to interact with estrogen receptor (ER), and change its transcriptional activity. ER is a receptor known to be inversely correlated with cell motility and invasion in breast cancer. The consequence of RhoGDIα activity on migration and invasion of ER+ and ER− breast cancers is not clear. The aim of our study was to investigate the possible opposing effect of RhoGDIα on the migration and invasion of ER+ MCF7 and ER− MDA-MB-231 breast cancer cells. RhoGDIα was downregulated using short interfering RNA (siRNA) and upregulated using GFP-tagged ORF clone of RhoGDIα, and their ability for migration and invasion was assayed using transwell chambers. It was found that the silencing of RhoGDIα in MCF7 and MDA-MB-231 cells significantly increased migration and invasion of these cells into the lower surface of porous membrane of the chambers. Overexpression of RhoGDIα in MCF7 cells suppressed their migration and invasion, but no significant effect was found on MDA-MB-231 cells. Our results indicate that the downregulation of RhoGDIα similarly affects the in vitro migration and invasion of ER+ MCF7 and ER− MDA-MB-231 cells. However, our assays are differently affected by the upregulation of RhoGDIα in these two cell lines and this may be due to the differences in ER expression, primary invasive ability and/or other molecules between these two cell line models which warrant further investigation.
Keywords: MCF7, MDA-MB-231, migration, invasion, transwell
Introduction
The spread of cancer cells from the primary site to a distant organ, or metastasis, accounts for the majority of cancer-related deaths. The molecular mechanisms of metastasis are largely unknown. Recent evidence has indicated that the ability of cancer cells to migrate and invade into surrounding tissues are prerequisite for cancer spread and metastasis, and studies have thus been directed to identify molecules that contribute to cancer cell motility, migration, and invasion.1 Agents that specifically target the motility of tumor cells are not only potentially more effective at treating metastasis but also eliminate the side effects of the current generally acting therapies.
A group of proteins identified in several cancer studies as prognostic-related biomarkers include the Rho GDP dissociation inhibitor (RhoGDI) family of proteins. RhoGDIs were initially identified as negative regulators of RhoGTPases. RhoGDIs, by inhibiting the release of GDP and the loading of GTP from RhoGTPases, sequester RhoGTPases in the cytosol. RhoGTPases exist either in an inactive, GDP-bound form in the cytosol or an active GTP-bound form associated with membranes. RhoGTPases control all aspects of cellular motility, migration, and invasion, including cellular polarity, cytoskeletal organization, in addition to cellular proliferation and apoptosis.2-5
Despite the fact that it was initially thought that RhoGDIs activities are restricted to the inhibition of RhoGTPases, recent data indicate that the function of RhoGDIs is more complex. RhoGDI family of proteins comprises three members: RhoGDIα, RhoGDIβ, and RhoGDIγ.2-5 RhoGDIα is the most abundant and well-known member of the family. In addition to its interaction with several RhoGTPases, it binds to estrogen receptor (ER),3,6-9 a molecule with remarkable impact on breast cancer invasion. Marzouk et al. showed that overexpression of RhoGDIα enhanced ER-mediated transcriptional activation. Gene silencing of the RhoGDIα using small interfering RNA (siRNA) showed that ER protein was increased when RhoGDIα was downregulated. This increase was observed only in the absence of estradiol in culture, but not in the presence of estradiol.7 Su et al. also reported positive relationship between the Rho GDIα and ER activity. In their study, Rho GDIα positively regulated ER transcription by stimulating the transcriptional activity of CBP/p300. Their study concluded that this in turn recruited GRIP1, AF-1, and AF-2 to the ER promoter to increase ER transcription.9 In contrast, Barone et al. found that overexpression of RhoGDIα decreased ER-mediated transcriptional activation.6
In vivo studies suggested that RhoGDIα has prognostic value. Jiang et al. found that RhoGDIα expression was significantly lower in tumor than in normal tissues. They showed that lower RhoGDIα expression was associated with higher grade and nodal involvement, metastases and death in breast cancer.3 In comparing gene expression of primary tumors from tamoxifen-treated patients with no recurrence or progression during the therapy, Barone et al. found RhoGDIα as a highly significant under-expressed protein in the latter group. They subsequently demonstrated that silencing of the RhoGDIα expression in ER+ MCF7 cells conferred them highly metastatic in an animal model.6 However, the consequence of RhoGDIα silencing in ER− cells was not assessed. Others found that higher RhoGDIα levels were significantly contributed to more favorable prognosis in cyclophosphamide, methotrexate, and 5-fluorouracil treated patients. A lower percentage of cases were reported with high RhoGDIα expression on ER− (40% ER− vs. 59% ER+), but the difference did not reach statistical significance (P = 0.07), that might be due to the low number of the patients.10
Two models of ER+ and ER− breast cancers are MCF7 and MDA-MB-231 cell lines, respectively.11 RhoGDIα has been detected in cytosolic fractions of both cell lines using western blotting with no significant different levels of expression.7 In contrast to MDA-MB-231, which is a highly migratory and invasive cell line, MCF7 has a low migratory and invasive activity as assessed using transwell chambers.11,12
Considering the impact of ER on the invasiveness of breast cancer, and direct binding of ER and RhoGDIα,6-8 a partly different role for RhoGDIα in invasion and migration of ER+ and ER− breast cancers is not unlikely. The importance of this theory is underscored by the opinion that effects of RhoGDIs on tumors are evidently multifaceted, and even a single Rho family member can have opposite effects in different types of a tumor.2 Accordingly, the aims of this study were to investigate the consequence of RhoGDIα silencing and overexpression on migration and invasion of MCF7 (ER+) and MDA-MB-231 (ER−) breast cancer cell lines with or without 17β-estradiol (E2).
Results
Downregulation and upregulation of RhoGDIα
Real-time PCR and western blotting using specific antibody revealed no significant difference of RhoGDIα in MCF7 cells compared with MDA-MB-231 cells in both mRNA level and protein expression (Figs. 1A and 2). To assess the possible roles of RhoGDIα in the aggressiveness of ER+ MCF7 and ER− MAD-MB-231 cells, the cells were transiently downregulated using lipofectamine and RhoGDIα siRNA, or upregulated using lipofectamine and RhoGDIα plasmid with or without E2. There was no significant difference in the RhoGDIα mRNA expression with or without E2 (Fig. 1A).
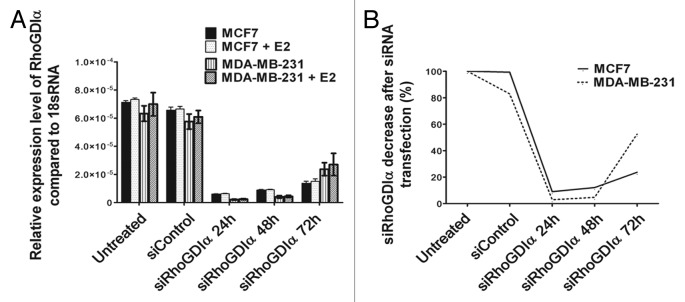
Figure 1. RhoGDIα mRNA expression in MCF7 and MDA-MB-231 cells. (A) Relative RhoGDIα mRNA levels between untreated MCF7 and MAD-MB-231 cells with or without E2 were not significantly different. RhoGDIα expression was decreased at 24, 48, and 72 h after transfection with siRNA. Values represent averages of three independent experiments with error bars indicating the standard deviation. (B) The total mRNA levels in untreated cells were set as 100%. mRNA expression levels showed 90% decrease at 24 and 48 h after siRNA treatment in both cell lines.
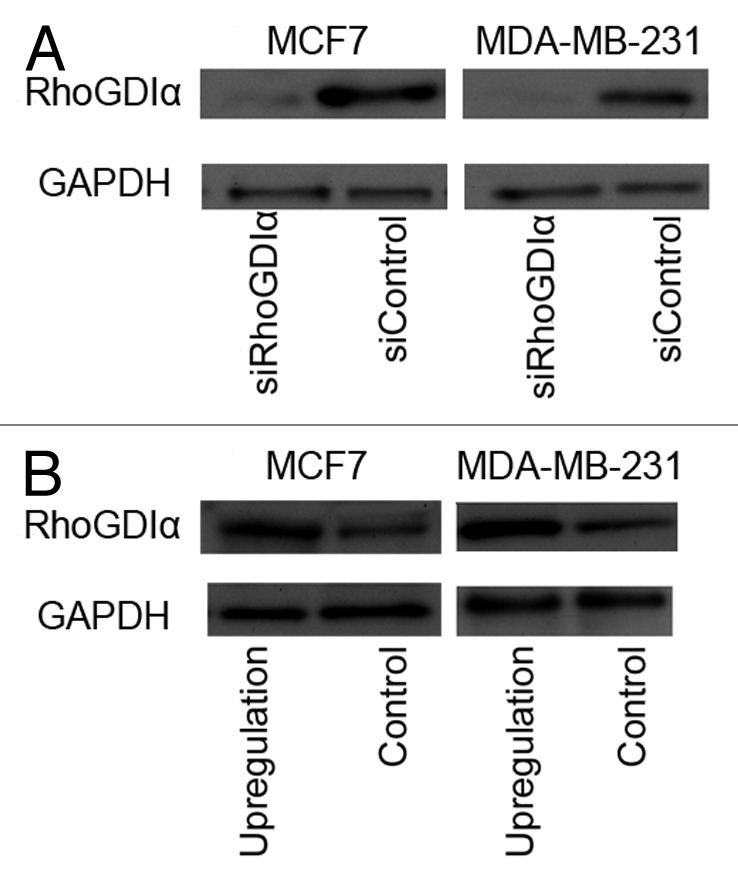
Figure 2. Representative western blot of RhoGDIα in MCF7 and MDA-MB-231cells 48 h after transfection with RhoGDIα-specific siRNA or siRNA control (A) and upregulation of RhoGDIα or control (B). GAPDH was used as loading control.
Real time PCR revealed that more than 90% RhoGDIα expression decreased in both cell lines after 24 and 48 h (Fig. 1B). Immunobloting using specific antibody 48 h post-transfection showed a hardly visible band for RhoGDIα. GAPDH was used as loading control. Representatives of MCF7 and MDA-MB-231 cells are shown in Figure 2.
Transfection efficiency in both breast cancer cell lines, MCF7 and MDA-MB-231 for RhoGDIα upregulation, were confirmed in three independent experiments by fluorescent microscopy and flow cytometryanalysis after 24–72 h post-transfection. The maximum of fluorescent events (M2) was observed 48 h post-infection (72% after 48 h vs. about 50% after 24 h or 72 h). The percentage of cells expressing GFP protein was calculated by dividing the number of fluorescent events by the total events (M1 + M2). Western blotting with specific antibody was performed 48 h after transfection, which indicated upregulation of the RhoGDIα in both cell lines (Fig. 2B). The western blot was quantified using ImageJ software package. Overexpression RhoGDIα level in both cell lines MCF7 and MDA-MB-231, showed more than three times higher than the controls. The western blotting was repeated three times with the same results.
Migration and invasion assay
Transwell chambers without matrigel (migration assay) or with a layer of 25 μg/cm2 matrigel (invasion assay) were employed to explore the possible role of RhoGDIα in migration and invasion of MCF7 and MDA-MB-231 cell line models. We counted the cells that migrated or invaded onto the lower surface of the porous membrane using an inverted microscope Olympus CKX41.
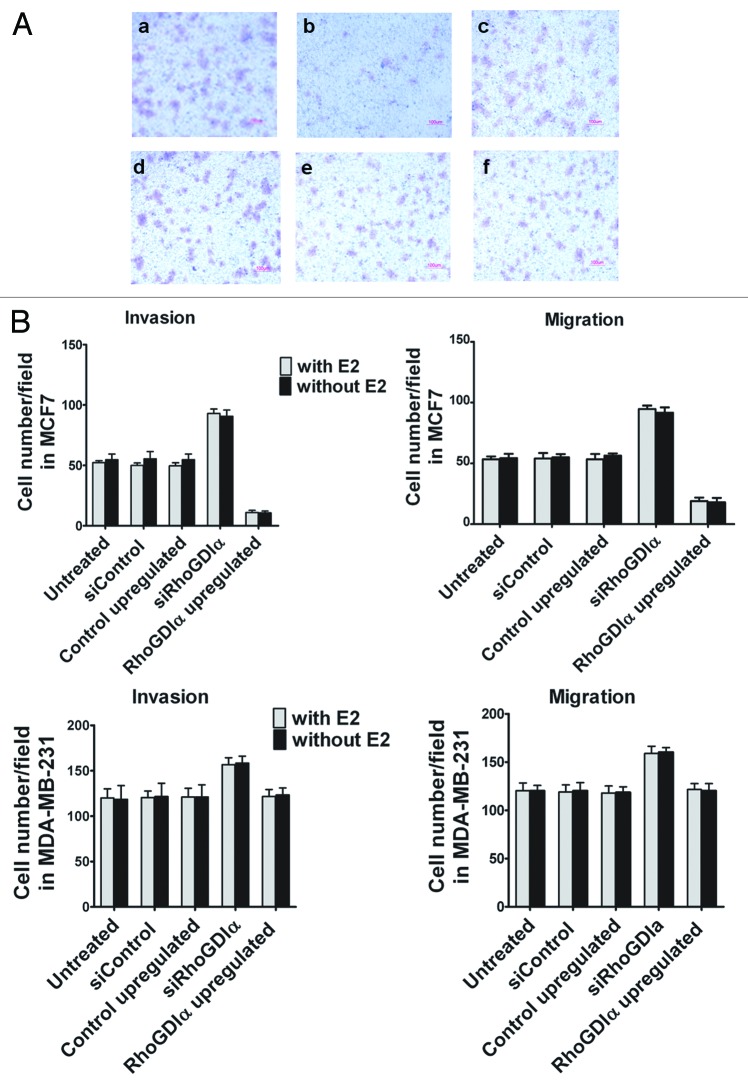
We observed that the numbers of migrated and invaded cells in MCF7 and MDA-MB-231 cells treated with RhoGDIα siRNA were significantly higher than those treated with control siRNA (P < 0.05) (Fig. 3). There was no significant difference in the number of cells between E2 treated and untreated experiments.
Figure 3. (A) Representative photographs of MCF7 and MDA-MB-231 cells invasion assays. Downregulation (a) and upregulation (b) of RhoGDIα, increased and decreased, respectively, the numbers of invaded cells in MCF7 compared with control (c). The numbers of invaded cells in MDA-MB-231 cells treated with RhoGDIα siRNA (d) was increased in comparison to the upregulation (e) and control (f). (B) The mean numbers/per field of traversed cells onto the lower surface of porous membrane of transwell chambers after downregulation and upregulation of RhoGDIα in MCF7 and MDA-MB-231 cells with or without E2. *indicates a significant difference compared with the corresponding control.
As shown in Figure 3B, in contrast to downregulation of RhoGDIα, its overexpression decreased the number of migrated and invaded cells in MCF7 (P < 0.001). However, overexpression of RhoGDIα was not significantly associated with the ability of MDA-MB-231 cells for migration and invasion through the membrane of the transwell chambers (P > 0.05). The presence or absence of E2 did not change the number of migrated or invaded cells.
Proteomic analysis
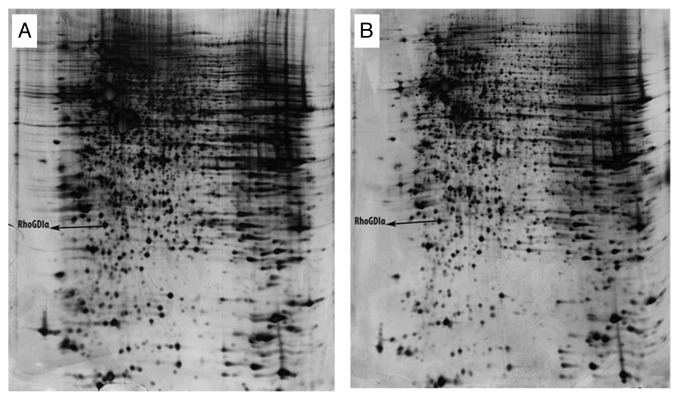
Two differentially expressed protein spots from 2D gels derived from RhoGDIα overexpressed MDA-MB-23 cells that one protein was identified by mass spectrometry as RhoGDIα while other one was not identified. RhoGDIα position on gels is shown in Figure 4.
Figure 4. Representative of 2D gels from MDA-MB-231 cells overexpressed RhoGDIα (A) and controls (B). The differentially expressed proteins have been labeled.
Discussion
It is well known that RhoGDIα binds directly to ER. Knowledge regarding the importance of this interaction in migration and invasion of ER+ and ER− breast cancer is relatively poor. Previously, Jiang et al. showed that RhoGDIα expression was significantly lower in tumor tissues than in normal ones, and that under-expression of RhoGDIα was associated with higher grade and nodal involvement, metastases and death in breast cancer. However, they did not report the ER status in the patients studied.3 In another study, lower RhoGDIα expression was associated with poorer prognosis in cyclophosphamide, methotrexate, and 5-fluorouracil treated breast cancer patients, and an insignificant higher percentage for ER positivity was observed in cases with upregulation of RhoGDIα (59% ER+ vs. 40% ER−) in that subgroup.10
In the present study, the migration and invasion of the ER+ MCF7 and ER− MDA-MB-231 cell lines were assayed after silencing and also upregulating the RhoGDIα gene in the presence or absence of E2.We found that silencing the RhoGDIα gene in ER+ MCF7 and ER− MDA-MB-231 cell line models significantly increased their migration and invasion through transwell membranes. Our results may be interpreted by the well-known role of RhoGDIα as an inhibitor for RhoGTPases such as Rac, Rho, and Cdc42, the strong stimulators of motility, migration, and invasion of cancer cells.4 We cannot disregard the fact that other differences between MCF7 and MDA-MB-231 cell lines cover the possible different effects on invasion of RhoGDIα on ER+ and ER− breast cancer cells. It has been shown that MCF7 cells in which RhoGDIα expression had been silenced had increased levels of activated Rho GTPases including Cdc42, RAc-1, and Rho A, B, and C.6 Such data are not available for ER− cancer cell lines.
Here, the upregulation of RhoGDIα significantly decreased the number of migrated and invaded cells in MCF7 cells compared with control cells, but did not affect those in MDA-MB-231 cells. One can argue that the highly invasive phenotype of MDA-MB-231 cells cannot be affected by the upregulation of just one molecule. Alternatively, the differential expression of ER or other interacting molecules with RhoGDIα between these cell lines might have caused the observed differences of MCF7 and MDA-MB-231 cells in response to RhoGDIα overexpression.
Interestingly, in our study, only 2 spots were reproducibly differentially expressed between 2D gels from MDA-MB-231 cells overexpressed RhoGDIα and controls. One of the proteins was identified as RhoGDIα while the other one was not identified probably because of low concentration. Preliminary data indicate several differentially expressed proteins between other circumstances that deserve further attention for their characterization. Consistent with our findings on 2D gels, it has been previously observed that the number of genes showing more than 2-fold changes was much higher in MCF7 (n = 435) than in MDA-MB-231 (n = 18) in response to the stimulation by protein kinase C (PKC)-activator phorbol 12-myristate 13-acetate (PMA).13 This can partly be explained by differential activity of PKC in ER+ MCF7 and ER− MAD-MB-231 cells, and a high PKC activity in absence of ER.14 PKC has been shown critical role in the RhoGDIα phosphorylation, which leads to dissociation from Rho and Rho activation.15 Whether the insignificant effect of RhoGDIα on MDA-MB- cells in our study might be due to constitutively high PKC activity in these cells need more investigations.
Some other findings in our experiments were consistent or inconsistent with previous reports. Similar to a previous report,11 we found that MDA-MB-231 cells had a significantly higher ability for migration and invasion compared with MCF7 cells. Furthermore, we did not observe any significant difference in MCF7 and MDA-MB-231 migration and invasion with or without E2. In contrast to the general agreement on insignificant effect of E2 on ER− MDA-MB-231 cells, traversing MCF7 cells through transwells was increased or decreased in the presence of E2.16,17 These conflicting results might be explained by different exposure times to E2,16 the passage number of the cells,18 and genotypic differences between MCF7 cells in different laboratories.19
Theoretically, in transwell chambers, the number of the migrated cells might be higher than the invaded cells which traverse from a thicker barrier coated with matrigel. We did not find any significant differences between migration and invasion of either MCF7 cells or MDA-MB-231 cells. Yokotsuka et al. found similar results in the case of MCF7 and MDA-MB-231 cells while SKBR3 and AU565 breast cancer cell lines showed a significantly higher number of penetrated cells in the absence of matrigel (migration) than in the presence of matrigel (invasion).11 The reason for such differences across cell lines has not been well understood.
In conclusion, we found that downregulation of RhoGDIα in MCF7 and MDA-MB-231 cells increased the invasion and migration of these cells, while its upregulation only influenced the MCF7 cells in an inhibitory manner. Although ER has a direct interaction with RhoGDIα, it seems that the differential expression of ER between these two cell lines does not critically affect the activity of RhoGDIα in these two cell line models.
Materials and Methods
Cell culture and treatments
Breast cancer cell lines MDA-MB-231 and MCF7 were purchased from National Cell Bank of Institute Pasteur of Iran. They were cultured in phenol red-containing RPMI-1640 supplemented with 10% fetal bovine serum (FBS), 100 unit/ml penicillin and 100 µg/ml streptomycin under standard conditions (37 °C incubator with 95% humidified air and 5% CO2). In preparation for cell migration and invasion assays, the cells were then subcultured into phenol red-free media (it has been shown that the pH indicator phenol red can act as a weak estrogen20) containing 5% charcoal–dextran treated calf serum (CDCS) and grown for 2 d to allow adaptation.
Transient gene transfection (gene silencing and overexpression)
The maximum transfected cell rates were given by 40–50% cell confluence for gene silencing experiments and 80–90% cell confluence for overexpression experiments. Therefore, 2–2.5 × 105 cells for gene silencing experiments and 4–4.5 × 105 cells for overexpression experiments in 2 mL phenol red-free media containing 3% CDCS without antibiotic were seeded into each well of a 6-well plate. After 24 h, the cells were transfected with either duplexes of RhoGDIα-specific siRNAoligos (00058983 and 00337331, Sigma), scrambled control siRNA (Sigma), green fluorescent protein (GFP)-tagged clone of homo sapience RhoGDP dissociation inhibitor α plasmid (Origene), or control plasmid pCMV-AC-GFP (Origene) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Briefly, for gene silencing, 7 µL of Lipofectamine 2000 and 5 µL of 20 µM siRNA stock solutions were separately diluted in 250 µL RPMI-1640 (Invitrogen). After 5 min incubation at room temperature, they were combined and mixed gently for 20 min at room temperature to allow complex formation. The 500 µL mixture was added drop wise onto a well while rocking the plate back and forth. The plates were then transferred to a 37 °C humidified CO2 incubator. Following 6 h incubation, media were replaced with phenol red-free RPMI 1640 containing 3% CDCS (without antibiotic) for an additional 24–72 h with or without 10 nM E2.
For overexpression experiments, the procedure was the same, except that 3 µg of either test or control plasmid was used instead of siRNAoligos for each well. For confirmation of gene silencing, western blotting and quantitative real-time PCR and for overexpression experiments, fluorescence microscope, flow cytometry, and western blotting were performed.
Quantitative real-time PCR
Quantification of gene expression at mRNA level was performed via quantitative real-time PCR using a Bio-Rad (Chromo4 Real-time PCR Detector, Bio-Rad) system. Total RNA was extracted from breast cancer cell lines using an RNA isolation kit (Roche), and the cDNA was made from the total RNA using revert Aid H Minus First Strand cDNA Synthesis Kit (Fermentaze) according to the manufacturers’ protocols. The primers and probes for RhoGDIα, and 18S rRNA were designed with Primer 3 software (SourceForge, Geeknet Inc., http://sourceforge.net/about).
Primer sets for: RhoGDIα 5′-GCC GTT TCC GCA GAC CCC AA-3′ (forward primer), 5′-TCT CCA GGT CGC CCG TCA GG-3′ (reverse primer), and 5′-FAM- TGA CTG GCC TGA CCC TGG TGT GCA GCT CGG-TAMRA-3′ (probe), and 18S rRNA 5′- CCA CCA GGA GTG GAG CCT GC-3′ (forward primer), 5′- AGA ACG GCC ATG CAC CAC CA-3′ (reverse primer), and 5′-HEX-ACC TCA CCC GGC CCG GAC ACG GAC A-TAMRA-3′ (probe). Each sample was normalized on the basis of 18S rRNA expression. The levels of mRNA were assayed in 24–72 h after transfection.
Immunoblot analysis
For the detection of protein expression levels, MCF7 or MDA-MB-231 cells were lysed in 2× Laemmli buffer containing cocktail protease inhibitors (Roche), and 20 μg of the lystates from each cell line was subjected to electrophoretic separation using 12% SDS–PAGE. The gels were then transferred to PVDF membranes (Bio-Rad) using a semi-dry system (Bio-Rad). The western blots were probed using antibodies directed against RhoGDIα (1/100) (ab93735, Abcam) and anti-actin (Abcam) as loading control. After washing, blots were incubated with secondary antibody horseradish peroxidase conjugated antibody (Abcam). ECL kit (GE Healthcare) was employed for the detection of the bands.
Migration assay
Migration assays were performed using 24-well transwell chambers (BD BioCoat Control Inserts from BD Biosciences) with 8.0-µm pore size polycarbonate membrane according to the manufacturer’s recommendations. Briefly, MCF7 and MAD-MB-231 cells 48 h after treatment with siRNA or plasmid were washed twice with PBS and resuspended in starvation medium (RPMI-1640 plus 1% CDCS). In the lower chamber of each well, 600 µL of the NIH-3T3 conditioned media as the chemoattractant, and in the upper chamber, 100 µL starvation media containing 6 × 104 cells were added. After 20 h incubation at 37 °C, the non-migratory cells on the upper side of the membrane were removed with a cotton swab. The migrated cells on the underside of the membrane were then fixed and stained with a solution containing 2.0% ethanol and 0.2% Crystal Violet. The cells were then counted in 10 randomly separate fields per membrane using an inverted microscope (Olympus CKX41) at 200× magnification. Three independent experiments in triplicate were performed. The numbers of migrated/invaded cells were presented as the mean cells per field ± SD from three independent experiments.
Invasion assay
The invasion assay was identical to the above migration assay except that 1 mg/mL Matrigel (BD Bioscience) was used to coat the inserts. The gels were allowed to polymerize for 2 h at 37 °C with a minimum thickness of 20 μm.
Two-dimensional electrophoresis (2DE)
The lystaes of MDA-MB-231cells overexpressed RhoGDIα and control cells were subjected to two-dimensional electrophoresis (2DE) as previously described.21 Briefly, 250 μg of lystaes was applied per IGP strips (18-cm: pH 3–10 nonlinear) for isoelectric focusing (IEF). After IEF, the focused strips were equilibrated, and run on the top of a second dimension SDS-PAGE (12%). The resulting 2D gels were visualized by both analytical and preparative (MS compatible) silver staining protocols.22 The resulting 2D gels were scanned and analyzed using the Prodigy Same Spots version 1.0 software (Nonlinear Dynamic) according to manufacturer`s instructions. Protein spots that showed >2-fold and P < 0.05 in the average normalized volume between treated and control groups (3 gels in each group), were considered as differentially expressed proteins and were picked from the gels. Spots were then sent for matrix-assisted laser desorption/ionization time-of-flight/time-of-flight (MALDI-TOF/TOF) MS analysis at the Department of Biochemistry, School of Medicine, National University of Singapore.
Statistical analysis
All values are expressed as mean ± SD. Statistical analysis was performed using the Student t test. Two-sided P values less than 0.05 were considered as statistically significant.
Acknowledgments
This study was supported by the Novin Committee of Shiraz University of Medical Sciences, the Institute for Cancer Research (ICR-100-503), and Faculty of Medicine and Health Sciences, University Putra Malaysia. The study was based on a PhD thesis (S. Hooshmand) submitted to Faculty of Medicine and Health Sciences, University Putra Malaysia. The authors would like to thank Mrs Mahsa Yazdanpanah for her help in cloning experiments.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/celladhesion/article/24204
References
- 1.Palmer TD, Ashby WJ, Lewis JD, Zijlstra A. Targeting tumor cell motility to prevent metastasis. Adv Drug Deliv Rev. 2011;63:568–81. doi: 10.1016/j.addr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia-Mata R, Boulter E, Burridge K. The ‘invisible hand’: regulation of RHO GTPases by RHOGDIs. Nat Rev Mol Cell Biol. 2011;12:493–504. doi: 10.1038/nrm3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang WG, Watkins G, Lane J, Cunnick GH, Douglas-Jones A, Mokbel K, et al. Prognostic value of rho GTPases and rho guanine nucleotide dissociation inhibitors in human breast cancers. Clin Cancer Res. 2003;9:6432–40. [PubMed] [Google Scholar]
- 4.Lin M, van Golen KL. Rho-regulatory proteins in breast cancer cell motility and invasion. Breast Cancer Res Treat. 2004;84:49–60. doi: 10.1023/B:BREA.0000018424.43445.f3. [DOI] [PubMed] [Google Scholar]
- 5.Cushman I, Casey PJ. RHO methylation matters: a role for isoprenylcysteine carboxylmethyltransferase in cell migration and adhesion. Cell Adh Migr. 2011;5:11–5. doi: 10.4161/cam.5.1.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barone I, Brusco L, Gu G, Selever J, Beyer A, Covington KR, et al. Loss of Rho GDIα and resistance to tamoxifen via effects on estrogen receptor α. J Natl Cancer Inst. 2011;103:538–52. doi: 10.1093/jnci/djr058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El Marzouk S, Schultz-Norton JR, Likhite VS, McLeod IX, Yates JR, Nardulli AM. Rho GDP dissociation inhibitor alpha interacts with estrogen receptor alpha and influences estrogen responsiveness. J Mol Endocrinol. 2007;39:249–59. doi: 10.1677/JME-07-0055. [DOI] [PubMed] [Google Scholar]
- 8.Su LF, Knoblauch R, Garabedian MJ. Rho GTPases as modulators of the estrogen receptor transcriptional response. J Biol Chem. 2001;276:3231–7. doi: 10.1074/jbc.M005547200. [DOI] [PubMed] [Google Scholar]
- 9.Su LF, Wang Z, Garabedian MJ. Regulation of GRIP1 and CBP Coactivator activity by Rho GDI modulates estrogen receptor transcriptional enhancement. J Biol Chem. 2002;277:37037–44. doi: 10.1074/jbc.M111607200. [DOI] [PubMed] [Google Scholar]
- 10.Ronneburg H, Span PN, Kantelhardt E, Dittmer A, Schunke D, Holzhausen HJ, et al. Rho GDP dissociation inhibitor alpha expression correlates with the outcome of CMF treatment in invasive ductal breast cancer. Int J Oncol. 2010;36:379–86. [PubMed] [Google Scholar]
- 11.Yokotsuka M, Iwaya K, Saito T, Pandiella A, Tsuboi R, Kohno N, et al. Overexpression of HER2 signaling to WAVE2-Arp2/3 complex activates MMP-independent migration in breast cancer. Breast Cancer Res Treat. 2011;126:311–8. doi: 10.1007/s10549-010-0896-x. [DOI] [PubMed] [Google Scholar]
- 12.Sherwood DR. Cell invasion through basement membranes: an anchor of understanding. Trends Cell Biol. 2006;16:250–6. doi: 10.1016/j.tcb.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Lacroix M, Haibe-Kains B, Hennuy B, Laes JF, Lallemand F, Gonze I, et al. Gene regulation by phorbol 12-myristate 13-acetate in MCF-7 and MDA-MB-231, two breast cancer cell lines exhibiting highly different phenotypes. Oncol Rep. 2004;12:701–7. doi: 10.3892/or.12.4.701. [DOI] [PubMed] [Google Scholar]
- 14.Fabbro D, Küng W, Roos W, Regazzi R, Eppenberger U. Epidermal growth factor binding and protein kinase C activities in human breast cancer cell lines: possible quantitative relationship. Cancer Res. 1986;46:2720–5. [PubMed] [Google Scholar]
- 15.Mehta D, Rahman A, Malik AB. Protein kinase C-alpha signals rho-guanine nucleotide dissociation inhibitor phosphorylation and rho activation and regulates the endothelial cell barrier function. J Biol Chem. 2001;276:22614–20. doi: 10.1074/jbc.M101927200. [DOI] [PubMed] [Google Scholar]
- 16.Saji S, Kawakami M, Hayashi S, Yoshida N, Hirose M, Horiguchi S, et al. Significance of HDAC6 regulation via estrogen signaling for cell motility and prognosis in estrogen receptor-positive breast cancer. Oncogene. 2005;24:4531–9. doi: 10.1038/sj.onc.1208646. [DOI] [PubMed] [Google Scholar]
- 17.Rochefort H, Platet N, Hayashido Y, Derocq D, Lucas A, Cunat S, et al. Estrogen receptor mediated inhibition of cancer cell invasion and motility: an overview. J Steroid Biochem Mol Biol. 1998;65:163–8. doi: 10.1016/S0960-0760(98)00010-7. [DOI] [PubMed] [Google Scholar]
- 18.Sakai K, Kurokawa T, Furui Y, Kuronuma Y, Sekiguchi M, Ando J, et al. Invasion of carcinoma cells into reconstituted type I collagen gels: visual real-time analysis by time-lapse microscopy. Biosci Trends. 2011;5:10–6. doi: 10.5582/bst.2011.v5.1.10. [DOI] [PubMed] [Google Scholar]
- 19.Bahia H, Ashman JN, Cawkwell L, Lind M, Monson JR, Drew PJ, et al. Karyotypic variation between independently cultured strains of the cell line MCF-7 identified by multicolour fluorescence in situ hybridization. Int J Oncol. 2002;20:489–94. doi: 10.3892/ijo.20.3.489. [DOI] [PubMed] [Google Scholar]
- 20.Glover JF, Irwin JT, Darbre PD. Interaction of phenol red with estrogenic and antiestrogenic action on growth of human breast cancer cells ZR-75-1 and T-47-D. Cancer Res. 1988;48:3693–7. [PubMed] [Google Scholar]
- 21.Mojtahedi Z, Safaei A, Yousefi Z, Ghaderi A. Immunoproteomics of HER2-positive and HER2-negative breast cancer patients with positive lymph nodes. OMICS. 2011;15:409–18. doi: 10.1089/omi.2010.0131. [DOI] [PubMed] [Google Scholar]
- 22.Sarvari J, Mojtahedi Z, Kuramitsu Y, Malek-Hosseini SA, Shamsi Shahrabadi M, Ghaderi A, et al. Differential expression of haptoglobin isoforms in chronic active hepatitis, cirrhosis and HCC related to HBV infection. Oncol Lett. 2011;2:871–7. doi: 10.3892/ol.2011.321. [DOI] [PMC free article] [PubMed] [Google Scholar]