Abstract
Substrate-attached materials (SAMs) are cellular feet that remain on substrates after the treatment of adherent cells with EGTA. SAMs are thought to contain cell adhesion machineries, but their biochemical properties have not been addressed in detail. To gain insight into the molecular mechanisms operating in cell adhesions, we comprehensively identified the protein components of SAMs by liquid chromatography coupled with tandem mass spectrometry, followed by immunoblot analysis. We found that the tetraspanins CD9, CD81, and CD151 were enriched in SAMs along with other transmembrane proteins that are known to associate with tetraspanins. Notably, integrins were detected in SAMs, but the components of focal adhesions were scarcely detected. These observations are reminiscent of the “footprints” that remain on substrates when the retraction fibers at the rear of migrating cells are released, because such footprints have been reported to contain tetraspanins and integrins but not focal adhesion proteins. In support of this hypothesis, the formation of SAMs was attenuated by inhibitors of ROCK, myosin II and dynamin, all of which are known to participate in rear-end retraction in migrating cells. Furthermore, SAMs left on collagen-coated substrates were found by electron microscopy to be fewer and thinner than those on laminin-coated substrates, reflecting the thin and fragile retraction fibers of cells migrating on collagen. Collectively, these results indicate that SAMs closely resemble the footprints and retraction fibers of migrating cells in their protein components, and that they are yielded by similar mechanisms.
Keywords: focal adhesion, integrin, CD9, ganglioside, cell migration, proteomics
Introduction
Cell adhesions on extracellular matrices (ECMs) are involved in the regulation of a variety of biological processes, including cell growth, differentiation, survival, and migration. The interaction of cells with ECMs is mediated mainly by integrins, heterodimers composed of non-covalently associated α- and β-subunits.1 The binding of integrins to ECMs stimulates their clustering and the formation of multiprotein complexes called focal adhesions. Focal adhesions perform both mechanical and signaling functions that control cytoskeletal rearrangements and various cellular responses.2,3 The mechanical functions are mediated by focal adhesion components such as talin, kindlin, α-actinin, and vinculin, which connect the actin cytoskeleton to the ECM. The signaling functions are mediated by components such as FAK, Src, paxillin, and ILK, which transmit intracellular signals via the regulation of protein and lipid kinases and GTPases.1,2 Defects in the regulation of integrin-mediated cell adhesions result in various diseases such as cancer, immune disorders, thrombosis, and skeletal muscle dystrophy.4-6
Cell migration is a coordinated process that involves the formation and disassembly of cell adhesion sites together with dynamic changes in the actin cytoskeleton.7,8 To migrate efficiently, cells must possess an asymmetric morphology with defined leading and trailing edges. Nascent focal adhesions are newly formed at the leading edge, following which they either mature into stabilized adhesions or turnover. In contrast, focal adhesions at the trailing edge are disassembled during rear-end retraction, which allows the cell body to translocate. The regulation of adhesion disassembly at the trailing edge is less well understood than the maturation and turnover of adhesions at the leading edge.
In the 1970s, substrate-attached materials (SAMs) were found to remain tightly bound to substrates when adherent cells were detached with the Ca2+-specific chelator, EGTA.9 Morphological investigations revealed that SAMs are composed of cell-surface regions rich in adhesion sites.10,11 Biochemical analyses showed that SAMs contain relatively large amounts of cell surface components that participate in cell adhesion such as cellular fibronectin,12 proteoglycans,13,14 and gangliosides.15,16 Detailed analysis of the molecular composition of SAMs may therefore provide insight into the molecular mechanisms underlying the cellular events regulated by cell adhesion, but the molecular properties of SAMs have remained largely uninvestigated over the past two decades.
In the present study, we sought to define the molecular composition of SAMs by proteomic analysis using liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) to investigate the mechanisms underlying the regulation of cellular responses by cell–ECM adhesions. We found that SAMs contain large amounts of tetraspanins and their associated proteins, but not focal adhesion proteins, and thus resemble the footprints and retraction fibers of migrating cells that are also enriched with tetraspanins.17,18 In addition, the formation of SAMs was dependent on actomyosin activity and dynamin-mediated endocytosis, as is the case with rear-end retraction in migrating cells.19,20 Our findings revealed that SAMs are closely correlated with rear-end retraction in migrating cells.
Results
Proteomic analysis of SAMs
To comprehensively identify the protein components of SAMs, A549 cells that had been cultured on laminin-511 in serum-free conditions were treated with EGTA to prepare SAMs. Scanning electron microscopy showed that EGTA treatment evoked the retraction and rounding of cells, leaving long, thin and branched protrusions (SAMs) firmly bound to the substrates (Fig. 1A). These results are consistent with a previous report,16 in which SAMs were prepared from rat hepatoma CMH5123 cells and examined by scanning electron microscopy. To perform proteomic analysis, SAMs were harvested from the dishes with SDS after the detachment of cells by treatment with EGTA. Determination of the protein content in SAMs and detached cells indicated that only 0.88 ± 0.03% (n = 3) of the total cellular protein was recovered in SAMs. Separation of the SAM proteins by SDS-PAGE showed that their banding pattern was obviously different from that of proteins in detached cells (Fig. 1B).
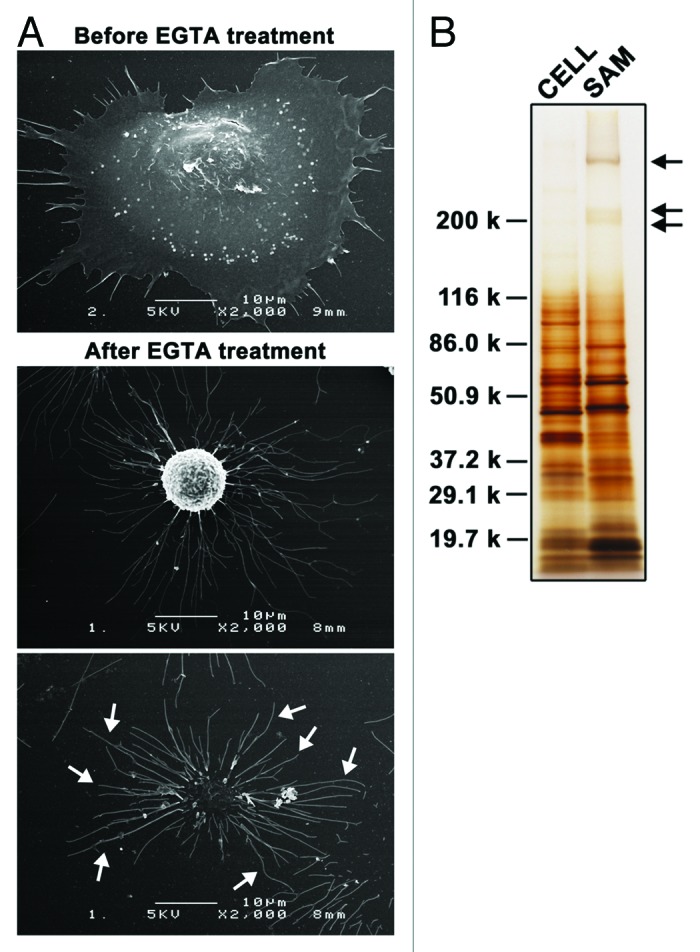
Figure 1. Substrate-attached materials on laminin-511. (A) A549 cells were cultured on laminin-511-coated dishes for 2h30min. Cells were then treated with EGTA for 15 min and fixed. Scanning electron micrographs were obtained as described in Materials and Methods. Arrows indicate SAMs. (B) SAMs were prepared after detaching the cells by treatment with EGTA as described in Materials and Methods, following which they were separated by SDS-PAGE under reducing conditions and silver-stained. Lysates were also prepared from detached cells (CELL) and analyzed by SDS-PAGE. The positions of molecular weight markers are shown on the left. Arrows indicate laminin-511, which was used to coat dishes.
The SAM proteins separated by SDS-PAGE were subjected to in-gel digestion with trypsin, and the resulting peptides were extracted from the gels and analyzed by LC-MS/MS. LC-MS/MS analyses of three independent SAM preparations resulted in the detection of 1971, 3018, and 2691 proteins per analysis (Tables S1–3), 1739 proteins of which were reproducibly detected (Table S4). In the present study, we focused on plasma transmembrane proteins, because they should include cell adhesive molecules and regulators, which are important in initiating cellular responses at the interface of cell–ECM interactions. We found that the plasma transmembrane proteins thus detected in SAMs include tetraspanins (CD9 and CD81) and integrins, CD44 and Lu/BCAM, which also have been detected in tetraspanin-enriched microdomains (also referred to as the tetraspanin web) (Table 1; Table S4).21-23 Notably, protein components of focal adhesions were not detected in SAMs, except for α-parvin (Table S4).
Table 1. Plasma transmembrane proteins detected by LC-MS/MS analysis of SAMs.
| Category | Protein | Gene | Association with TSPANs† |
|---|---|---|---|
| Tetraspanins |
CD9 antigen |
(CD9) |
|
| CD81 antigen |
(CD81) |
|
|
| Tetraspanin-4 |
(TSPAN4) |
|
|
| Tetraspanin-14 |
(TSPAN14) |
|
|
| Integrins |
Integrin α3 |
(ITGA3) |
+ |
| Integrin α6 |
(ITGA6) |
+ |
|
| Integrin αv |
(ITGAV) |
+ |
|
| Integrin β1 |
(ITGB1) |
+ |
|
| Integrin β5 |
(ITGB5) |
+ |
|
| Ig superfamily |
Ig superfamily member 8/EWI2 |
(IGFSF8) |
+ |
| Lutheran blood group glycoprotein |
(BCAM) |
+ |
|
| CD166 antigen |
(ALCAM) |
+ |
|
| Basigin/CD147‡ |
(BSG) |
NR |
|
| Poliovirus receptor/NECL-5 |
(PVR) |
NR |
|
| Cell adhesion molecules |
CD44 antigen |
(CD44) |
+ |
| Cadherin-1 |
(CDH1) |
+ |
|
| Claudin-1 |
(CLDN1) |
+ |
|
| Receptors |
Transferrin receptor protein 1 |
(TFRC) |
NR |
| Renin receptor |
(ATP6AP2) |
NR |
|
| Ephrin type-A receptor 2 |
(EPHA2) |
NR |
|
| Neuropilin-1 |
(NRP1) |
NR |
|
| Receptor-type Tyr-protein phosphatase F |
(PTPRF) |
NR |
|
| Scavenger receptor class B member 1 |
(SCARB1) |
NR |
|
| Plasminogen receptor (KT) |
(C9orf46) |
NR |
|
| Retinoic acid-induced protein 3 |
(GPRC5A) |
NR |
|
| Endothelial protein C receptor |
(PROCR) |
NR |
|
| Proteases |
Disintegrin and metalloproteinase domain-containing protein 10 |
(ADAM10) |
+ |
| Matrix metalloproteinase-14/MT1-MMP |
(MMP14) |
+ |
|
| Nicastrin |
(NICA) |
+ |
|
| Transporters |
4F2 cell-surface antigen heavy chain¶ |
(SLC3A2) |
+ |
| Large neutral amino acids transporter small subunit 1 |
(SLC7A5) |
NR |
|
| Na+-coupled neutral amino acid transporter 2 |
(SLC38A2) |
NR |
|
| Na+/K+-transporting ATPase subunit β -1 |
(ATP1B1) |
NR |
|
| Others | Myoferlin |
(MYOF) |
NR |
| Protein LYRIC |
(MTDH) |
NR |
|
| Extended synaptotagmin-2 |
(ESYT2) |
NR |
|
| Proteolipid protein 2 |
(PLP2) |
NR |
|
| Amphiregulin |
(AREG) |
NR |
|
| Amyloid β A4 protein |
(APP) |
NR |
|
| Kunitz-type protease inhibitor 2 | (SPINT2) | NR |
The listed plasma transmembrane proteins were reproducibly detected in three independent LC-MS/MS analyses of SAMs prepared from A549 cells cultured on laminin-511. †+ indicates the proteins that have been shown to interact with tetraspanins.49-60 NR, not reported. ‡Protein reported to interact with integrin α3β1 and α6β1.61 ¶Protein reported to interact with integrin β1.62,63 See also Table S4.
Enrichment of tetraspanins in SAMs
To further examine the occurrence of proteins that have been shown to associate with focal adhesions and tetraspanin-enriched microdomains in SAMs, we performed immunoblot analysis (Fig. 2A). Talin, α-actinin, vinculin, paxillin, and α-parvin, which are components of focal adhesions, were scarcely detected in SAMs, as was the case with actin. In contrast, the two tetraspanins CD9 and CD81 were significantly enriched in SAMs when compared with lysates prepared from detached cells, although another tetraspanin, CD151, was not concentrated in SAMs but was detected at a relatively high level. The lower abundance of CD151 may explain why CD151 was detected in only one of three proteomic analyses (Table S2). The failure of LC-MS/MS analysis may be due to the difficulty in the detection of tetraspanins by LC-MS/MS, possibly because a major part of their sequence is transmembranous.24 Integrin β1, integrin α3, CD44, and ADAM10, which are known to be associated with tetraspanins, were also detected in SAMs at relatively high levels compared with focal adhesion proteins. Similar results, including the detection of large quantities of tetraspanins, were also obtained with HT-1080 cells (Fig. S1). Consistent with these results, SAMs remaining on laminin-coated surfaces after EGTA treatment were positively immunostained with anti-CD9, anti-CD81, and anti-CD151 antibodies (Fig. 2B). The signals for CD81 were less pronounced than those for CD9 and CD151, possibly due to the reduced reactivity of the anti-CD81 antibody toward formaldehyde-fixed SAMs. These results indicate that SAMs contain tetraspanins and their associated proteins, but not focal adhesion proteins.
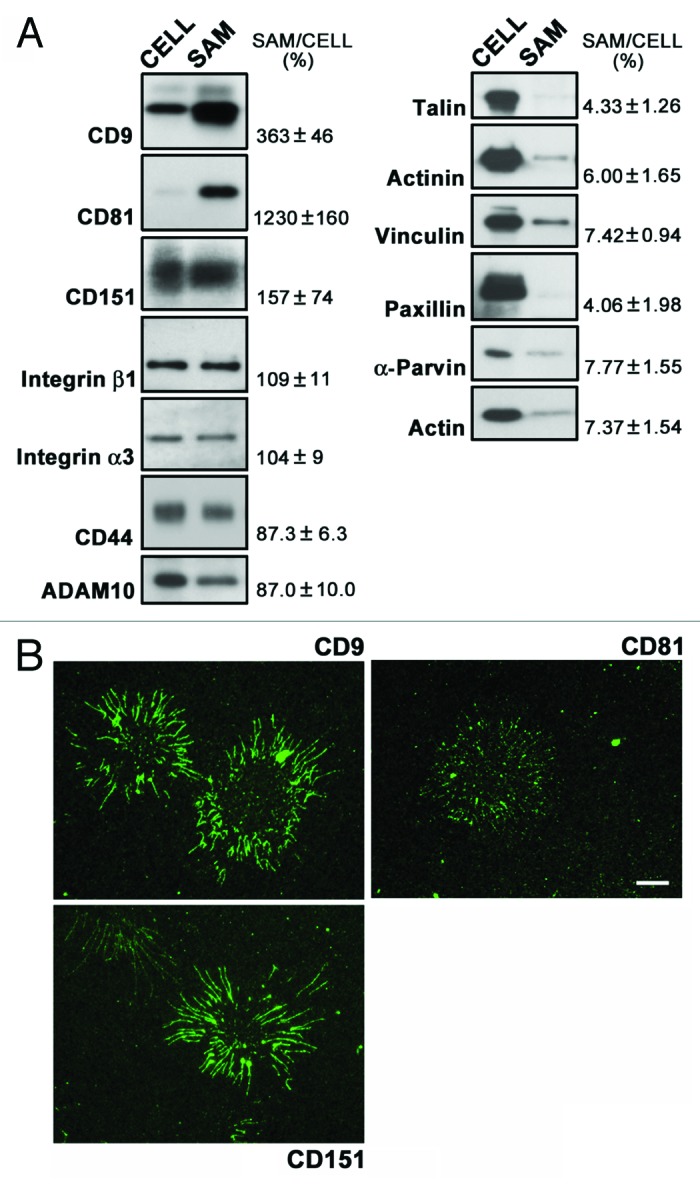
Figure 2. Detection of tetraspanins and their associated proteins in SAMs. (A) SAMs were prepared following the treatment of A549 cells cultured on laminin-511 with EGTA as described in Materials and Methods. Lysates (CELL) were also prepared from the cells detached by the EGTA treatment. SAMs and lysates were separated by SDS-PAGE, and subsequently immunoblotted with the antibodies indicated on the left of the blots. Data are representative of three separate experiments. The relative intensities of bands detected in SAMs to those in cell lysates are shown on the right. Values represent means ± SD from three independent experiments. (B) A549 cells cultured on laminin-511 were treated with EGTA. SAMs remaining on the substrates were immunostained with anti-CD9, anti-CD81, and anti-CD151 antibodies as described in Materials and Methods. Bar represents 10 μm.
It has been reported that migrating cells exhibit retraction fibers on their tails and leave behind “footprints” or “migration tracks” that contain integrins and tetraspanins, but not focal adhesion components.17,25-27 Immunofluorescence staining of A549 cells migrating on laminin-511 showed that CD9, CD81, and CD151 were detected discontinuously but throughout retraction fibers (Fig. 3), overlapping with F-actin. It should be noted that F-actin signals retracted more readily than tetraspanin signals, leaving behind the tetraspanin+/F-actin− regions. These findings raise the possibility that SAMs are closely related to footprints and retraction fibers based on not only their morphology but also their protein composition.
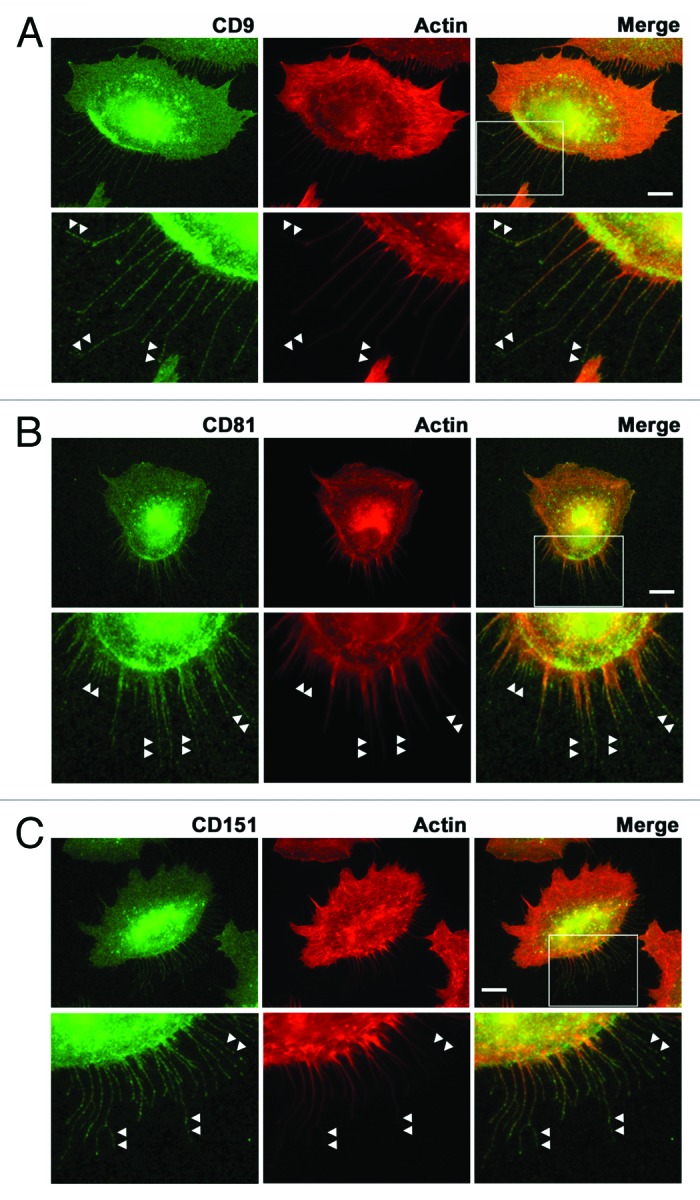
Figure 3. Localization of tetraspanins in retraction fibers in migrating cells. A549 cells migrating on laminin-511 were stained with anti-CD9 (A), anti-CD81 (B), and anti-CD151 (C) antibodies (green) and rhodamine-phalloidin (red) as described in Materials and Methods. The lower panels show high magnification views of the boxed areas. Arrowheads indicate tetraspanin-positive but F-actin-negative regions in retraction fibers. Bars represent 10 μm.
Involvement of actomyosin activity and dynamin-dependent endocytosis in the formation of SAMs
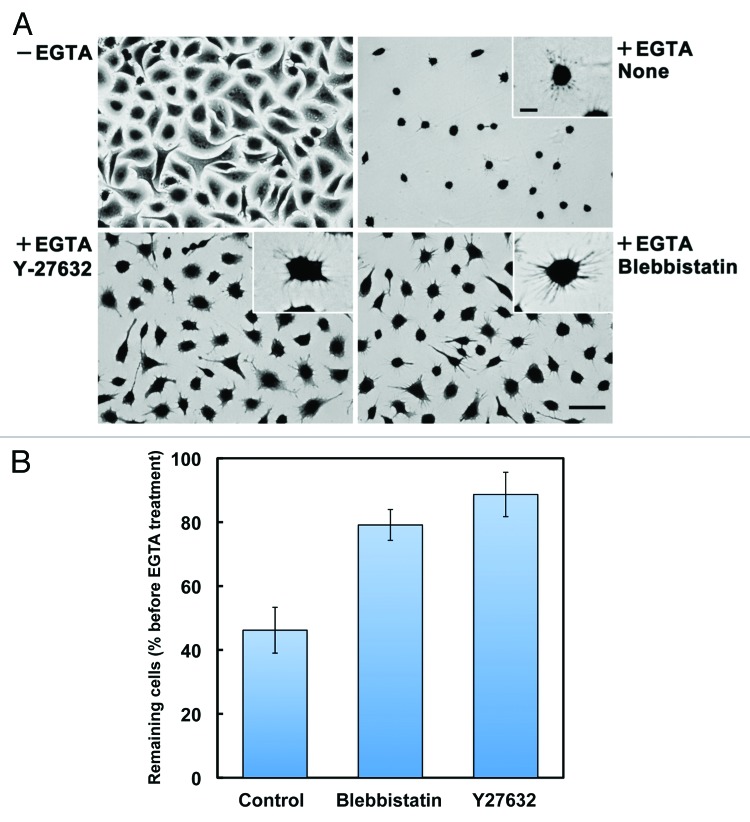
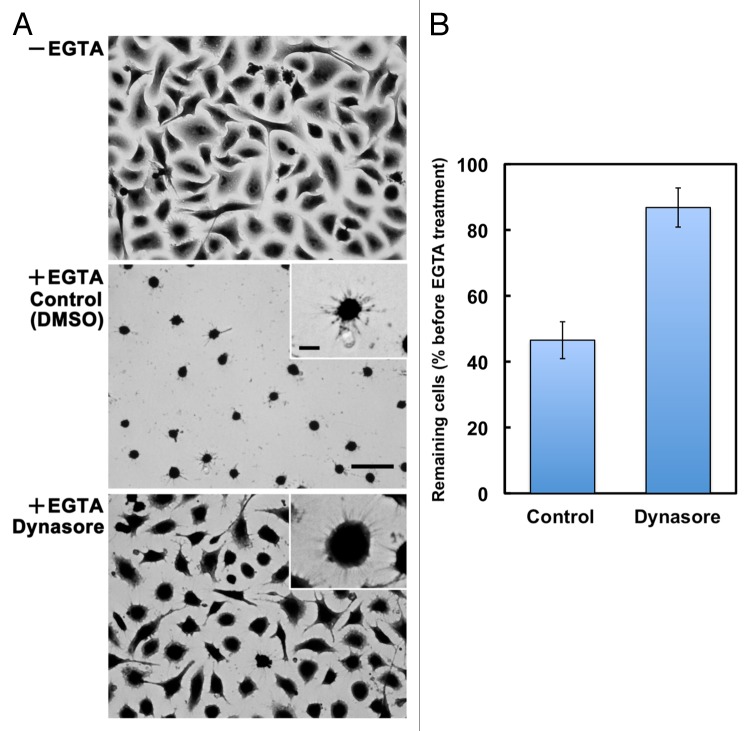
To assess whether the process of SAM formation resembles that of rear-end retraction in migrating cells, we examined the effects of inhibiting actomyosin contractility, which is critically involved in rear-end retraction in migrating cells.19 When cells cultured on laminin-511 were treated with EGTA in the presence of Y-27632 and blebbistatin, inhibitors of ROCK and myosin II, respectively, cell retraction and detachment were significantly attenuated, leaving the cells spread on the substrates (Fig. 4). However, even in the presence of the inhibitors, EGTA-treated cells extended many thin protrusions at their peripheries, indicating that these inhibitors delay, rather than completely block, the formation of SAMs. In A549 cells migrating on laminin-511, these inhibitors also disturbed the detachment of retraction tails, thereby stimulating the elongation of the retraction tails (Vids. S1–S3). We also investigated whether clathrin-dependent endocytosis, which is also involved in rear-end retraction in migrating cells,28,29 participates in cell retraction following EGTA treatment. As shown in Figure 5, the cell retraction and detachment were blocked by the addition of dynasore, a dynamin inhibitor, which inhibits clathrin-dependent endocytosis.30 As with Y-27632 and blebbistatin, dynasore did not completely prevent, but rather retarded, the formation of SAMs. The migration of A549 cells on laminin-511 was also suppressed by dynasore with an apparent loss of front-rear polarity and retraction tails (Vid. S4). These results support the possibility that SAMs are analogous to retraction fibers and footprints, and that they are formed and left on the substrates through similar molecular processes.
Figure 4. Involvement of actomyosin contractility in the formation of SAMs. (A) A549 cells were allowed to adhere to laminin-511 for 2h30min (−EGTA). Then, cells were treated with EGTA (+EGTA) for 15 min in the absence (None) or presence of 10 μM Y-27632 or 20 μM (±)-blebbistatin, following which they were stained with toluidine blue. Bar represents 50 μm. High-magnification images are shown in the insets (bar indicates 10 μm). (B) Cells remaining on the substrates were quantified as described in Materials and Methods. Values represent the mean ± SD (n = 3).
Figure 5. Involvement of dynamin activity in the formation of SAMs. (A) A549 cells were allowed to adhere to laminin-511 for 2h30min (−EGTA). Then, cells were treated with EGTA (+EGTA) for 15 min in the presence of 0.1% DMSO (Control) or 100 μM dynasore, following which they were stained with toluidine blue. Bar represents 50 μm. High-magnification images are shown in the insets (bar indicates 10 μm). (B) Cells remaining on the substrates were quantified as described in Materials and Methods. Values represent the mean ± SD (n = 3).
Morphological similarities between SAMs and retraction fibers on different ECM proteins
To further address the similarity between SAMs and retraction fibers and footprints, we compared cell migration on laminin-511 and type I collagen by time-lapse phase-contrast microscopy. Cells on laminin-511 showed thick and stable retraction fibers on their tails, although the retraction fibers of cells on type I collagen were thin and fragile (Fig. 6; Vids. S5 and S6). The diameters of the retraction fibers were 0.357 μm ± 0.058 (n = 133) on laminin-511 and 0.232 μm ± 0.035 on type I collagen (n = 105). Consistent with these results, scanning electron microscopy of EGTA-treated cells on laminin-511 and type I collagen demonstrated that the SAMs on laminin-511 were greater in number and thicker than those on type I collagen (Fig. 7A and B). Immunoblot analysis showed that greater amounts of integrins β1 and α3, CD9, CD81, CD151, ADAM10, and CD44 were detected in SAMs on laminin-511 than in SAMs on type I collagen (Fig. 7C). These results are in agreement with the morphological distinctions between the SAMs on laminin-511 and type I collagen. In contrast, the cytoskeletal protein, keratin 18, the RNA-binding protein, G3BP, the nuclear protein, histone H3, and the mitochondrial protein, prohibitin, were detected at almost the same levels in SAMs remaining on laminin-511 and type I collagen. It seems likely, therefore, that integrins β1 and α3, CD9, CD81, CD151, ADAM10, and CD44 are intrinsic components of SAMs, while keratin 18, G3BP, histone H3, and prohibitin, which were detected in large amounts by mass spectrometric analysis, are entrapped in SAMs independently of the mechanism operating in cell retraction.
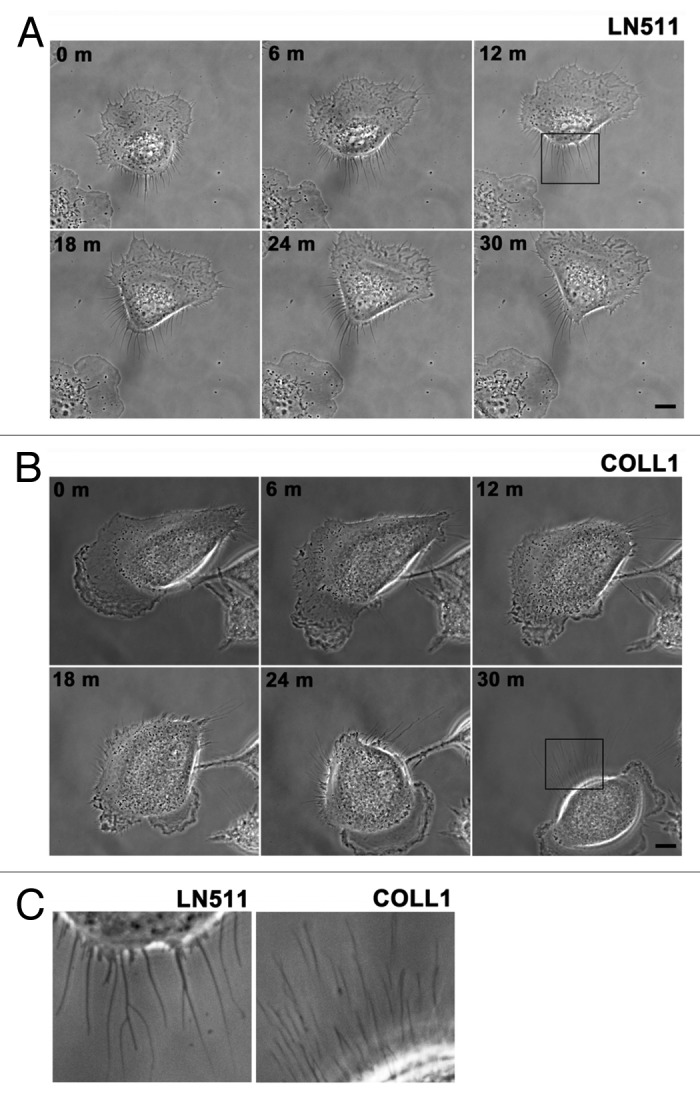
Figure 6. Morphological differences in retraction fibers in cells migrating on laminin-511 or type I collagen. (A and B) Migration of A549 cells on laminin-511 (LN511, [A]) or type I collagen (COLL1, [B]) was recorded by time-lapse microscopy as described in Materials and Methods. Bars represent 10 μm. See also Vids. S1 and S2. (C) The left and right panels show high magnification views of the areas boxed in (Aand B), respectively. The data are representative of 43 and 21 cells migrating on laminin-511 or type I collagen, respectively.
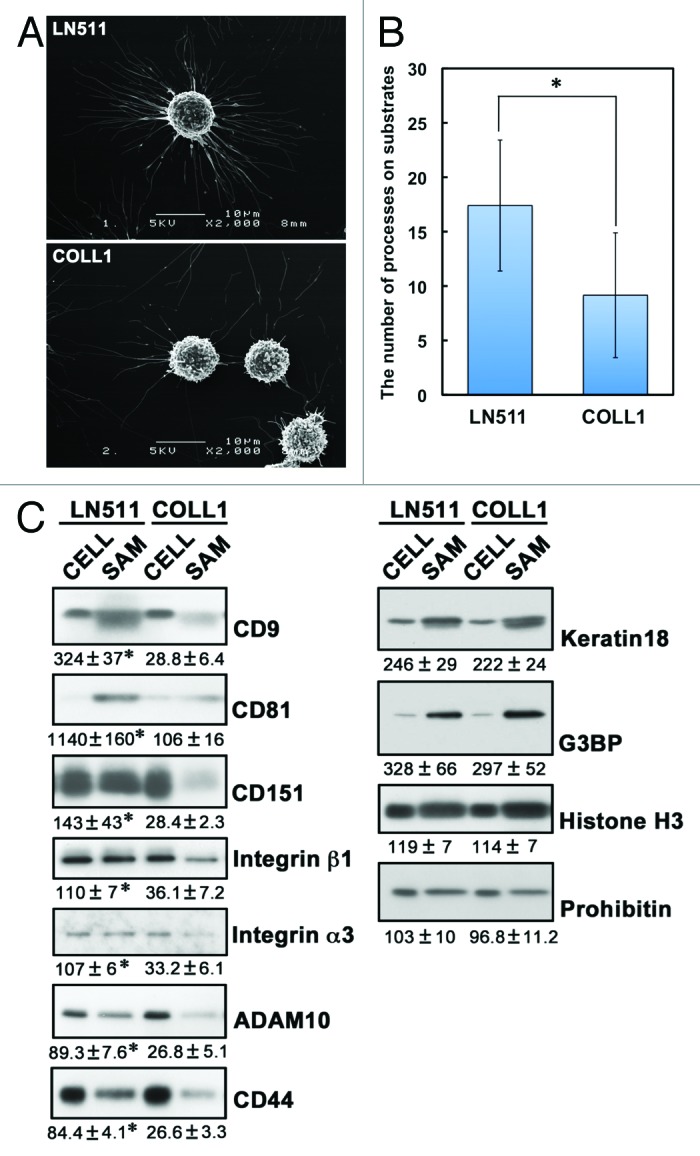
Figure 7. Differences in SAMs remaining on laminin-511 and type I collagen. (A) A549 cells were cultured on laminin-511 (LN511)- or type I collagen (COLL1)-coated dishes for 2h30min. The cells were then treated with EGTA for 15 min and fixed. Scanning electron micrographs were obtained as described in Materials and Methods. (B) The number of cell processes per cell was counted in the electron micrographs. Values represent the mean ± SD (n = 27 for laminin-511 and n = 28 for type I collagen; *p < 0.01, the Student t test). (C) SAMs were prepared following EGTA treatment of A549 cells cultured on laminin-511 (LN511) or type I collagen (COLL1) as described in Materials and Methods. Lysates (CELL) were also prepared from the cells detached by the EGTA treatment. These proteins were then immunoblotted with the antibodies indicated on the right of the blots. Data are representative of three separate experiments. The relative intensities of bands detected in SAMs to those in cell lysates are shown below the blots. Values represent means ± SD from three independent experiments. Statistical analysis was performed using the Student t test, *p < 0.05.
Discussion
SAMs were discovered in the 1970s as cellular feet that remained on substrates after detachment of cells with EGTA, although there have been few reports on SAMs in the past two decades. In the present study, we focused on SAMs as cell–ECM adhesion machineries, and performed the comprehensive determination of their protein components to uncover the molecular mechanisms underlying cellular events regulated by cell–ECM adhesions. Our data indicate that SAMs are closely related to the retraction fibers and footprints that appear during rear-end retraction in migrating cells, thus revealing new aspects of SAMs as cell adhesion structures.
At the rear of migrating cells, retraction fibers are observed during rear-end detachment. At the tips of these fibers, “membrane ripping” occurs, thereby leaving footprints on substrates.25,31 We found that CD9, CD81, and CD151 were enriched in SAMs. Integrins were also recovered in SAMs, but focal adhesion components, including talin, α-actinin, vinculin, paxillin, and α-parvin, were scant. These properties of SAMs are reminiscent of those of footprints and retraction fibers. It has been reported that footprints contain high amounts of integrins in various types of cells, including keratinocytes and fibroblasts.25-27 Focal adhesion constituents such as talin and vinculin are scarcely present in footprints. Peñas et al.17 reported that tetraspanins such as CD9 and CD81 exist at high levels in the footprints of keratinocytes. In human prostate cancer Du145 cells, CD81 was shown to be present in footprints and retraction fibers in which F-actin was scarcely detected.18 Consistent with these reports, we observed that in A549 cells migrating on laminin-511, CD9, CD81, and CD151 were detected throughout retraction fibers, and even in the regions near their tips where F-actin was scant. It is conceivable, therefore, that SAMs are structurally analogous to the footprints and retraction fibers of migrating cells.
Inhibition of ROCK and myosin II by Y-27632 and blebbistatin attenuated the formation of SAMs by EGTA treatment. It is known that Rho/ROCK signaling mediates the retraction of the trailing edges of cells and is implicated in adhesion disassembly during cell detachment.19 Inhibition of Rho kinase induces an elongated morphology with impaired rear-end detachment.32 In addition, fibroblasts deficient in myosin IIA show impaired adhesion disassembly and rear-end detachment.33 We also observed similar phenomena in A549 cells migrating on laminin-511 in the presence of Y-27632 and blebbistatin by time-lapse microscopy. Therefore, rear-end retraction in migrating cells and SAM formation by EGTA treatment may be driven by similar machineries that involve actomyosin activity.
We also found that the formation of SAMs was suppressed by dynasore, an inhibitor of dynamin, which also participates in rear-end retraction by mediating clathrin-dependent endocytosis.8,20,28 By time-lapse microscopy, we observed that dynasore effectively inhibited the migration of A549 cells on laminin-511. It has been reported that the endocytosis and recycling of integrin αvβ3 contributes to adhesion release in neutrophils migrating on vitronectin.34 In the migration of mouse fibroblast NIH3T3 and human fibrosarcoma HT1080 cells, clathrin-mediated endocytosis is involved in focal adhesion disassembly.20,28,29 Taken together, the formation of SAMs and rear-end retraction in migrating cells seem to share similar mechanisms involving actomyosin activity and clathrin-dependent endocytosis.
Scanning electron microscopic observations showed that SAMs on type I collagen-coated substrates were fewer and thinner than those on laminin-511-coated substrates. Consistent with these results, tetraspanins and their associated proteins were detected at significantly reduced levels in SAMs on type I collagen compared with SAMs on laminin-511. In contrast, the cytoskeletal protein, keratin 18, the nuclear protein, histone H3, and the mitochondrial protein, prohibitin, were detected in large amounts in SAMs irrespective of the type of substrate. These observations make it likely that the nuclear, mitochondrial, and cytoskeletal proteins frequently detected in our LC-MS/MS analyses are not intrinsic components of SAMs, but this possibility needs to be confirmed extensively by immunoblot analysis of individual proteins in SAMs prepared on laminin-511 and type I collagen.
Tetraspanins, which possess four transmembrane domains, are present in different combinations in almost all types of cells and tissues, and have been implicated in diverse cellular functions involving cell–cell and cell–substratum interactions.21-23 Tetraspanins associate with each other and with other transmembrane proteins, e.g., integrins and immunoglobulin superfamily proteins, thereby forming multimolecular membrane microdomains, often referred to as a tetraspanin-enriched microdomain or the tetraspanin web. Many reports have suggested that tetraspanins are involved in the regulation of cell migration,21,35 although the mechanisms involved remain largely unknown. Several lines of evidence indicate that the deregulation of tetraspanin expression is associated with cancer metastasis.36-38 Zijlstra et al.39 have reported that an anti-CD151 mAb, which blocks metastasis, prevents rear-end detachment during the migration of human epidermoid carcinoma cells, suggesting the involvement of tetraspanins in rear-end retraction in migrating cells. Tetraspanins, which are major components of SAMs, may also participate in SAM formation upon EGTA treatment. Further studies on the roles of tetraspanins in SAM formation will provide a better understanding of the mechanisms underlying rear-end retraction in migrating cells.
It has been reported that GD2/GD3 gangliosides are required for the detection of CD151 in cellular microprocesses left on substrates after the detachment of COS-7 cells by treatment with EDTA.40 Gangliosides have been shown to associate with several tetraspanins, i.e., CD9 and CD82, and thereby regulate protein–protein interactions in tetraspanin-enriched microdomains.41-44 In addition, SAMs contain various kinds of gangliosides, including GD2 and GD3.15,16 Thus, localization of tetraspanins may, at least in part, be because of their association with gangliosides. Given that gangliosides are known to participate in the regulation of cell migration through integrins,45 tetraspanins and gangliosides may cooperatively participate in rear-end retraction in migrating cells.
In summary, our results raise the possibility that the molecular mechanism of SAM formation mimics that of rear-end retraction in migrating cells. Given that the regulation of cell adhesion at the trailing edges is less well defined than that at the leading edges, the analysis of SAMs will provide novel insights into the mechanisms of cell migration. In addition, it has been proposed that the trailing edge of a cell drives cell migration by forming a defined rear prior to the formation of polarized cell protrusions.46 Further analysis of SAMs could enable the molecular mechanisms underlying the formation of front-rear polarization, as well as rear-end retraction, in migrating cells to be elucidated.
Materials and Methods
Cell culture, ECM proteins, antibodies, and reagents
A549 human lung adenocarcinoma and HT-1080 human fibrosarcoma cells were maintained in 10-cm dishes in Dulbecco’s modified Eagle’s medium (DMEM, Sigma) supplemented with heat-inactivated 10% (v/v) fetal bovine serum (FBS; JRH Biosciences). The cells were cultured at 37 °C in a humidified atmosphere containing 5% CO2.
Laminin-511 was purified from the conditioned medium of human choriocarcinoma JAR cells as described previously.47 Type I collagen was purchased from Nitta Gelatin (Cellmatrix Type I-C).
A mouse monoclonal antibody (mAb) against human CD151 (8C3) was produced as described previously.48 Anti-CD9 mouse mAb (MM2/57) was purchased from Chemicon; anti-integrin α3 goat polyclonal antibody (pAb), anti-CD81 mouse mAb (5A6), and anti-CD44 rat mAb (IM7) were purchased from Santa Cruz; anti-paxillin mouse mAb, anti-α-actinin mouse mAb, and anti-integrin β1 mouse mAb were from BD Transduction Lab; α-parvin rabbit pAb was from Cell Signaling; anti-actin rabbit pAb, anti-talin mouse mAb (8D4), and anti-vinculin mouse mAb (hVIN-1) were from Sigma; anti-ADAM10 rabbit pAb was from Millipore; peroxidase-conjugated AffiniPure anti-mouse IgG, anti-rabbit IgG, anti-rat IgG, and anti-goat IgG antibodies were from Jackson Immuno Res; Alexa 488-conjugated anti-mouse IgG antibody and rhodamine-labeled phalloidin were from Molecular Probes. Y-27632, (±)-blebbistatin, and dynasore were purchased from Calbiochem.
Preparation of SAMs
Cells were detached from dishes with PBS containing 0.025% trypsin and 1 mM EDTA. For LC-MS/MS, detached cells were washed 3 times with serum-free DMEM containing 10 mM HEPES-NaOH, pH 7.5, resuspended in the same medium, and then plated on dishes that had been coated with 5 nM laminin-511 and blocked with Protein-free Blocking Reagent (Pierce). For immunoblot analysis, cells were prepared and seeded in the same way as for LC-MS/MS, except that medium containing 10 mM HEPES-NaOH, pH 7.5, and 0.5% (w/v) bovine serum albumin (BSA) was used for cell suspension and blocking of the coated dishes. After culture for 2h30 min, cells were washed twice with PBS and then once with 1 mM EGTA, 1 mg/ml glucose in PBS, and treated with 1 mM EGTA, 1 mg/ml glucose in PBS at 37 °C for 20 min. Cells were completely detached by washing 5 times with PBS containing 1 mM EGTA and then 3 times with PBS. Dishes were washed twice with ice-cold PBS containing 0.2% Brij 97 at 4 °C, and further washed 3 times with ice-cold PBS at 4 °C. SAM proteins on dishes were extracted with 0.25% SDS at 37 °C, and then concentrated with centrifugal filter devices (Amicon Ultra-4, Millipore) for analysis by LC-MS/MS and immunoblotting. Cells detached by the EGTA treatment were lysed with 0.25% SDS and used in immunoblot analysis for comparison with SAMs.
Liquid chromatography coupled with tandem mass spectrometry
Protein samples were separated by SDS-PAGE (SDS-PAGE) and then fixed in the gels with 30% methanol, 10% acetic acid. After washing with deionized water, the gels were cut into >20 rectangular pieces and then washed sequentially with 25 mM ammonium bicarbonate containing 50% acetonitrile, 100% acetonitrile, 100 mM ammonium bicarbonate, and 100% acetonitrile. The gel pieces were dried and then incubated with 100 mM ammonium bicarbonate and 10 mM DTT at 56 °C for 45 min. After cooling to room temperature, the DTT solution was replaced with 100 mM ammonium bicarbonate, 55 mM iodoacetamide and the gel pieces were incubated at room temperature for 30 min in the dark. The gel pieces were washed sequentially with deionized water, 25 mM ammonium bicarbonate containing 50% acetonitrile, 100% acetonitrile, 100 mM ammonium bicarbonate, and 100% acetonitrile. After being dried, the gel pieces were subjected to trypsin digestion at 35 °C overnight with XL-TrypKit (Apro Sci.). Following enzymatic digestion, the resulting peptides were extracted sequentially with 5% trifluoroacetic acid containing 50% acetonitrile, and 100% acetonitrile. The peptides were dried and cleaned up with PepClean C-18 Spin Columns (Pierce). The peptides eluted from the columns with 70% acetonitrile were dried and dissolved in 0.1% trifluoroacetic acid.
LC-MS/MS analyses were performed on a LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific) equipped with a nano-ESI source (AMR) and coupled to a Paradigm MG2 pump (Michrom Bioresources) and an autosampler (HTC PAL, CTC Analytics). A spray voltage of 2200 V was applied. Peptide mixtures were separated on a MagicC18AQ column (100 μm × 150 mm, 3.0 μm particle size, 300 Å, Michrom Bioresources) with a flow rate of 500 nl/min. A linear gradient of 5–30% buffer B in buffer A for 80 min, 30–95% buffer B in buffer A for 10 min, 95% buffer B and 5% buffer A for 4 min, and finally decreasing to 5% buffer B in buffer A, was employed (buffer A = 0.1% formic acid in 2% acetonitrile, buffer B = 0.1% formic acid in 90% acetonitrile). Intact peptides were detected in the Orbitrap at 60 000 resolution. For LC-MS/MS analysis, 10 precursor ions were selected for subsequent MS/MS scans in a data-dependent acquisition mode following each full scan (m/z, 450–1800). A lock mass function was used for the LTQ-Orbitrap to obtain constant mass accuracy during gradient analysis.
Peptides and proteins were identified by means of an automated database search using Proteome Discoverer v.1.3 (Thermo Fisher Scientific) with the MASCOT algorithm against the SwissProt protein database (version 2012_06, 536489 sequences). Taxonomy was set to Homo sapiens (20,312 entries). Search parameters for peptide and MS/MS mass tolerance were 10 ppm and 0.8 Da, respectively, with allowance for two missed cleavages in the trypsin digest. Carbamidomethylation of cysteine was set as a fixed modification and oxidation of methionines was allowed as a variable modification. MASCOT results were filtered with the integrated Percolator based filter using a false discovery rate of <1% (based on PSMs).
Immunoblotting
The protein concentrations of samples were determined using BCA Protein Assay Reagent (Pierce). Samples containing equal amounts of protein were separated by SDS-PAGE and transferred onto polyvinylidene fluoride membranes (Millipore) in 0.1 M Tris base, 0.192 M glycine, and 20% (v/v) methanol using a semi-dry electrophoretic transfer cell (Bio-Rad). The membranes were blocked with 5% (w/v) nonfat dried milk in 0.1% (w/v) Tween 20/TBS (T-TBS) for at least 1 h at room temperature, and then incubated with antibodies in the blocking buffer at 4 °C overnight. After washing three times with T-TBS, the membranes were incubated with peroxidase-coupled secondary antibodies in the blocking buffer at room temperature for 1 h. The membranes were then washed four times with T-TBS and visualized using the ECL chemiluminescence system (GE Healthcare).
Assay for cell retraction and detachment following EGTA treatment
Cells were detached from dishes with PBS containing 0.025% trypsin and 1 mM EDTA, washed with DMEM, 0.5% BSA, 10 mM HEPES-NaOH, pH 7.5, and resuspended in the same medium. Cells were plated on 12-well plates coated with 5 nM laminin-511 and cultured for 2h30min. After washing twice with PBS and then once with PBS, 1 mM EGTA, 1 mg/ml glucose containing an inhibitor (Y-27632, blebbistatin or dynasore), cells were treated with the same saline containing the inhibitor at 37 °C for 15 min. Cells were fixed with 3.7% formaldehyde in PBS for 20 min at room temperature and then stained with 0.1% toluidine blue. Stained cells were photographed under a bright-field microscope (Olympus). To quantify the amounts of cells remaining on substrates, the dye in the stained cells was extracted with 0.5% (w/v) SDS and then subjected to colorimetric measurement at 595 nm.
Immunofluorescence staining
A549 cells suspended in DMEM, 0.5% BSA, 10 mM HEPES-NaOH, pH 7.4 were plated on glass coverslips coated with 10 nM laminin-511. For immunostaining of SAMs, cells were cultured for 2h30min after plating. After washing twice with PBS and then once with PBS, 1 mM EGTA, 1 mg/ml glucose, cells were treated with the same saline containing EGTA at 37 °C for 15 min, and then fixed with 3.7% formaldehyde in PBS. For immunostaining of migrating cells, cells were cultured for 1 h after plating and the medium was then changed to DMEM, 1% FBS, 10 mM HEPES-NaOH, pH 7.4. Two hours after the medium change, cells were fixed with 3.7% formaldehyde in PBS. After fixation, cells were permeabilized with 0.2% Brij 97 for 3 min and blocked with PBS containing 2.5% (w/v) BSA. Cells were then incubated with anti-CD9, anti-CD81, or anti-CD151 mouse mAb at 4°C overnight, followed by washing 3 times with PBS and then incubation with Alexa 488-conjugated anti-mouse IgG antibody at room temperature for 1 h. Actin filaments were stained with rhodamine-conjugated phalloidin. Stained cells were observed under a fluorescent microscope (Axiovert 200, Zeiss).
Time-lapse microscopy
A549 cells suspended in DMEM, 0.5% BSA, 10 mM HEPES-NaOH, pH 7.4 were replated on 35-mm glass-bottom dishes coated with either laminin-511 (10 nM) or type I collagen (300 μg/ml). One hour after plating, the medium was changed to DMEM, 1% FBS, 10 mM HEPES-NaOH, pH 7.4. Two hours after the medium change, cell migration was monitored using an Axiovert 200 inverted microscope (Zeiss). Video images were collected with a CoolSNAP HQ CCD camera (Photometrics) at 30 sec intervals for 30 min using SlideBook software (Intelligent Imaging Innovations).
Scanning electron microscopy
A549 cells suspended in DMEM, 0.5% BSA, 10 mM HEPES-NaOH, pH 7.4 were replated on 35-mm dishes coated with either laminin-511 (5 nM) or type I collagen (300 μg/ml), and cultured for 2h30min. After washing twice with PBS and then once with PBS, 1 mM EGTA, 1 mg/ml glucose, cells were treated with the same saline containing EGTA at 37 °C for 15 min, and then fixed with 2% glutaraldehyde in 0.1 M sodium phosphate buffer (pH 7.4) at 4 °C. The cells were postfixed in 2% osmium tetroxide solution at 4 °C. The samples were dehydrated in a series of ethanol, immersed in tert-butyl alcohol, freeze-dried, and coated with osmium. The samples were examined with a scanning electron microscope (JSM-6320F, JEOL).
Supplementary Material
Acknowledgments
We thank Noriko Sanzen for establishing hybridoma clones for laminins and CD151 and purifying the mAbs, and Keita Kawamura for assistance in the proteomic analyses. This work was supported by Grants-in-Aid for Scientific Research from the Japan Society for Promotion of Science (to M.Y. and K.S.) and for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to K.S.).
Glossary
Abbreviations:
- BSA
bovine serum albumin
- DMEM
Dulbecco’s modified Eagle’s medium
- ECM
extracellular matrix
- FBS
fetal bovine serum
- LC-MS/MS
liquid chromatography coupled with tandem mass spectrometry
- mAb
monoclonal antibody
- pAb
polyclonal antibody
- PBS
phosphate-buffered saline
- SAM
substrate-attached material
- TBS
Tris-buffered saline
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/celladhesion/article/25041
References
- 1.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–87. doi: 10.1016/S0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 2.Harburger DS, Calderwood DA. Integrin signalling at a glance. J Cell Sci. 2009;122:159–63. doi: 10.1242/jcs.018093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geiger B, Yamada KM. Molecular architecture and function of matrix adhesions. Cold Spring Harb Perspect Biol. 2011;3:a005033. doi: 10.1101/cshperspect.a005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo W, Giancotti FG. Integrin signalling during tumour progression. Nat Rev Mol Cell Biol. 2004;5:816–26. doi: 10.1038/nrm1490. [DOI] [PubMed] [Google Scholar]
- 5.Evans R, Patzak I, Svensson L, De Filippo K, Jones K, McDowall A, et al. Integrins in immunity. J Cell Sci. 2009;122:215–25. doi: 10.1242/jcs.019117. [DOI] [PubMed] [Google Scholar]
- 6.Wickström SA, Radovanac K, Fässler R. Genetic analyses of integrin signaling. Cold Spring Harb Perspect Biol. 2011;3:a005116. doi: 10.1101/cshperspect.a005116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huttenlocher A, Horwitz AR. Integrins in cell migration. Cold Spring Harb Perspect Biol. 2011;3:a005074. doi: 10.1101/cshperspect.a005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broussard JA, Webb DJ, Kaverina I. Asymmetric focal adhesion disassembly in motile cells. Curr Opin Cell Biol. 2008;20:85–90. doi: 10.1016/j.ceb.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 9.Terry AH, Culp LA. Substrate-attached glycoproteins from normal and virus-transformed cells. Biochemistry. 1974;13:414–25. doi: 10.1021/bi00700a004. [DOI] [PubMed] [Google Scholar]
- 10.Rosen JJ, Culp LA. Morphology and cellular origins of substrate-attached material from mouse fibroblasts. Exp Cell Res. 1977;107:139–49. doi: 10.1016/0014-4827(77)90395-0. [DOI] [PubMed] [Google Scholar]
- 11.Domen C, Culp LA. Adhesion sites of neural tumor cells. Morphogenesis of substratum-attached material. Exp Cell Res. 1981;134:329–38. doi: 10.1016/0014-4827(81)90433-X. [DOI] [PubMed] [Google Scholar]
- 12.Murray BA, Culp LA. Multiple and masked pools of fibronectin in murine fibroblast cell--substratum adhesion sites. Exp Cell Res. 1981;131:237–49. doi: 10.1016/0014-4827(81)90229-9. [DOI] [PubMed] [Google Scholar]
- 13.Culp LA, Murray BA, Rollins BJ. Fibronectin and proteoglycans as determinants of cell-substratum adhesion. J Supramol Struct. 1979;11:401–27. doi: 10.1002/jss.400110314. [DOI] [PubMed] [Google Scholar]
- 14.Rollins BJ, Culp LA. Glycosaminoglycans in the substrate adhesion sites of normal and virus-transformed murine cells. Biochemistry. 1979;18:141–8. doi: 10.1021/bi00568a022. [DOI] [PubMed] [Google Scholar]
- 15.Mugnai G, Tombaccini D, Ruggieri S. Ganglioside composition of substrate-adhesion sites of normal and virally-transformed Balb/c 3T3 cells. Biochem Biophys Res Commun. 1984;125:142–8. doi: 10.1016/S0006-291X(84)80346-0. [DOI] [PubMed] [Google Scholar]
- 16.Barletta E, Mugnai G, Ruggieri S. Morphological characteristics and ganglioside composition of substratum adhesion sites in a hepatoma cell line (CMH5123 cells) during different phases of growth. Exp Cell Res. 1989;182:394–402. doi: 10.1016/0014-4827(89)90244-9. [DOI] [PubMed] [Google Scholar]
- 17.Peñas PF, García-Díez A, Sánchez-Madrid F, Yáñez-Mó M. Tetraspanins are localized at motility-related structures and involved in normal human keratinocyte wound healing migration. J Invest Dermatol. 2000;114:1126–35. doi: 10.1046/j.1523-1747.2000.00998.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhang XA, Huang C. Tetraspanins and cell membrane tubular structures. Cell Mol Life Sci. 2012;69:2843–52. doi: 10.1007/s00018-012-0954-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009;10:778–90. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ezratty EJ, Partridge MA, Gundersen GG. Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat Cell Biol. 2005;7:581–90. doi: 10.1038/ncb1262. [DOI] [PubMed] [Google Scholar]
- 21.Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005;6:801–11. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 22.Charrin S, le Naour F, Silvie O, Milhiet PE, Boucheix C, Rubinstein E. Lateral organization of membrane proteins: tetraspanins spin their web. Biochem J. 2009;420:133–54. doi: 10.1042/BJ20082422. [DOI] [PubMed] [Google Scholar]
- 23.Yáñez-Mó M, Barreiro O, Gordon-Alonso M, Sala-Valdés M, Sánchez-Madrid F. Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol. 2009;19:434–46. doi: 10.1016/j.tcb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 24.André M, Le Caer JP, Greco C, Planchon S, El Nemer W, Boucheix C, et al. Proteomic analysis of the tetraspanin web using LC-ESI-MS/MS and MALDI-FTICR-MS. Proteomics. 2006;6:1437–49. doi: 10.1002/pmic.200500180. [DOI] [PubMed] [Google Scholar]
- 25.Palecek SP, Schmidt CE, Lauffenburger DA, Horwitz AF. Integrin dynamics on the tail region of migrating fibroblasts. J Cell Sci. 1996;109:941–52. doi: 10.1242/jcs.109.5.941. [DOI] [PubMed] [Google Scholar]
- 26.Rigort A, Grünewald J, Herzog V, Kirfel G. Release of integrin macroaggregates as a mechanism of rear detachment during keratinocyte migration. Eur J Cell Biol. 2004;83:725–33. doi: 10.1078/0171-9335-00431. [DOI] [PubMed] [Google Scholar]
- 27.Kirfel G, Rigort A, Borm B, Schulte C, Herzog V. Structural and compositional analysis of the keratinocyte migration track. Cell Motil Cytoskeleton. 2003;55:1–13. doi: 10.1002/cm.10106. [DOI] [PubMed] [Google Scholar]
- 28.Chao WT, Kunz J. Focal adhesion disassembly requires clathrin-dependent endocytosis of integrins. FEBS Lett. 2009;583:1337–43. doi: 10.1016/j.febslet.2009.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ezratty EJ, Bertaux C, Marcantonio EE, Gundersen GG. Clathrin mediates integrin endocytosis for focal adhesion disassembly in migrating cells. J Cell Biol. 2009;187:733–47. doi: 10.1083/jcb.200904054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. Dynasore, a cell-permeable inhibitor of dynamin. Dev Cell. 2006;10:839–50. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 31.Kirfel G, Rigort A, Borm B, Herzog V. Cell migration: mechanisms of rear detachment and the formation of migration tracks. Eur J Cell Biol. 2004;83:717–24. doi: 10.1078/0171-9335-00421. [DOI] [PubMed] [Google Scholar]
- 32.Worthylake RA, Lemoine S, Watson JM, Burridge K. RhoA is required for monocyte tail retraction during transendothelial migration. J Cell Biol. 2001;154:147–60. doi: 10.1083/jcb.200103048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vicente-Manzanares M, Zareno J, Whitmore L, Choi CK, Horwitz AF. Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J Cell Biol. 2007;176:573–80. doi: 10.1083/jcb.200612043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawson MA, Maxfield FR. Ca(2+)- and calcineurin-dependent recycling of an integrin to the front of migrating neutrophils. Nature. 1995;377:75–9. doi: 10.1038/377075a0. [DOI] [PubMed] [Google Scholar]
- 35.Stipp CS. Laminin-binding integrins and their tetraspanin partners as potential antimetastatic targets. Expert Rev Mol Med. 2010;12:e3. doi: 10.1017/S1462399409001355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richardson MM, Jennings LK, Zhang XA. Tetraspanins and tumor progression. Clin Exp Metastasis. 2011;28:261–70. doi: 10.1007/s10585-010-9365-5. [DOI] [PubMed] [Google Scholar]
- 37.Romanska HM, Berditchevski F. Tetraspanins in human epithelial malignancies. J Pathol. 2011;223:4–14. doi: 10.1002/path.2779. [DOI] [PubMed] [Google Scholar]
- 38.Zöller M. Tetraspanins: push and pull in suppressing and promoting metastasis. Nat Rev Cancer. 2009;9:40–55. doi: 10.1038/nrc2543. [DOI] [PubMed] [Google Scholar]
- 39.Zijlstra A, Lewis J, Degryse B, Stuhlmann H, Quigley JP. The inhibition of tumor cell intravasation and subsequent metastasis via regulation of in vivo tumor cell motility by the tetraspanin CD151. Cancer Cell. 2008;13:221–34. doi: 10.1016/j.ccr.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thorne RF, Mhaidat NM, Ralston KJ, Burns GF. Shed gangliosides provide detergent-independent evidence for type-3 glycosynapses. Biochem Biophys Res Commun. 2007;356:306–11. doi: 10.1016/j.bbrc.2007.02.139. [DOI] [PubMed] [Google Scholar]
- 41.Todeschini AR, Dos Santos JN, Handa K, Hakomori SI. Ganglioside GM2-tetraspanin CD82 complex inhibits met and its cross-talk with integrins, providing a basis for control of cell motility through glycosynapse. J Biol Chem. 2007;282:8123–33. doi: 10.1074/jbc.M611407200. [DOI] [PubMed] [Google Scholar]
- 42.Odintsova E, Butters TD, Monti E, Sprong H, van Meer G, Berditchevski F. Gangliosides play an important role in the organization of CD82-enriched microdomains. Biochem J. 2006;400:315–25. doi: 10.1042/BJ20060259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ono M, Handa K, Sonnino S, Withers DA, Nagai H, Hakomori S. GM3 ganglioside inhibits CD9-facilitated haptotactic cell motility: coexpression of GM3 and CD9 is essential in the downregulation of tumor cell motility and malignancy. Biochemistry. 2001;40:6414–21. doi: 10.1021/bi0101998. [DOI] [PubMed] [Google Scholar]
- 44.Mitsuzuka K, Handa K, Satoh M, Arai Y, Hakomori S. A specific microdomain (“glycosynapse 3”) controls phenotypic conversion and reversion of bladder cancer cells through GM3-mediated interaction of α3β1 integrin with CD9. J Biol Chem. 2005;280:35545–53. doi: 10.1074/jbc.M505630200. [DOI] [PubMed] [Google Scholar]
- 45.Hakomori SI. Glycosynaptic microdomains controlling tumor cell phenotype through alteration of cell growth, adhesion, and motility. FEBS Lett. 2010;584:1901–6. doi: 10.1016/j.febslet.2009.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cramer LP. Forming the cell rear first: breaking cell symmetry to trigger directed cell migration. Nat Cell Biol. 2010;12:628–32. doi: 10.1038/ncb0710-628. [DOI] [PubMed] [Google Scholar]
- 47.Yamada M, Sumida Y, Fujibayashi A, Fukaguchi K, Sanzen N, Nishiuchi R, et al. The tetraspanin CD151 regulates cell morphology and intracellular signaling on laminin-511. FEBS J. 2008;275:3335–51. doi: 10.1111/j.1742-4658.2008.06481.x. [DOI] [PubMed] [Google Scholar]
- 48.Nishiuchi R, Sanzen N, Nada S, Sumida Y, Wada Y, Okada M, et al. Potentiation of the ligand-binding activity of integrin α3β1 via association with tetraspanin CD151. Proc Natl Acad Sci U S A. 2005;102:1939–44. doi: 10.1073/pnas.0409493102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boucheix C, Rubinstein E. Tetraspanins. Cell Mol Life Sci. 2001;58:1189–205. doi: 10.1007/PL00000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berditchevski F. Complexes of tetraspanins with integrins: more than meets the eye. J Cell Sci. 2001;114:4143–51. doi: 10.1242/jcs.114.23.4143. [DOI] [PubMed] [Google Scholar]
- 51.Le Naour F, André M, Boucheix C, Rubinstein E. Membrane microdomains and proteomics: lessons from tetraspanin microdomains and comparison with lipid rafts. Proteomics. 2006;6:6447–54. doi: 10.1002/pmic.200600282. [DOI] [PubMed] [Google Scholar]
- 52.Tarrant JM, Robb L, van Spriel AB, Wright MD. Tetraspanins: molecular organisers of the leukocyte surface. Trends Immunol. 2003;24:610–7. doi: 10.1016/j.it.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 53.Kovalenko OV, Yang XH, Hemler ME. A novel cysteine cross-linking method reveals a direct association between claudin-1 and tetraspanin CD9. Mol Cell Proteomics. 2007;6:1855–67. doi: 10.1074/mcp.M700183-MCP200. [DOI] [PubMed] [Google Scholar]
- 54.Chattopadhyay N, Wang Z, Ashman LK, Brady-Kalnay SM, Kreidberg JA. α3β1 integrin-CD151, a component of the cadherin-catenin complex, regulates PTPmu expression and cell-cell adhesion. J Cell Biol. 2003;163:1351–62. doi: 10.1083/jcb.200306067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yañez-Mó M, Barreiro O, Gonzalo P, Batista A, Megías D, Genís L, et al. MT1-MMP collagenolytic activity is regulated through association with tetraspanin CD151 in primary endothelial cells. Blood. 2008;112:3217–26. doi: 10.1182/blood-2008-02-139394. [DOI] [PubMed] [Google Scholar]
- 56.Lafleur MA, Xu D, Hemler ME. Tetraspanin proteins regulate membrane type-1 matrix metalloproteinase-dependent pericellular proteolysis. Mol Biol Cell. 2009;20:2030–40. doi: 10.1091/mbc.E08-11-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wakabayashi T, Craessaerts K, Bammens L, Bentahir M, Borgions F, Herdewijn P, et al. Analysis of the gamma-secretase interactome and validation of its association with tetraspanin-enriched microdomains. Nat Cell Biol. 2009;11:1340–6. doi: 10.1038/ncb1978. [DOI] [PubMed] [Google Scholar]
- 58.Domínguez F, Simón C, Quiñonero A, Ramírez MA, González-Muñoz E, Burghardt H, et al. Human endometrial CD98 is essential for blastocyst adhesion. PLoS One. 2010;5:e13380. doi: 10.1371/journal.pone.0013380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dornier E, Coumailleau F, Ottavi JF, Moretti J, Boucheix C, Mauduit P, et al. TspanC8 tetraspanins regulate ADAM10/Kuzbanian trafficking and promote Notch activation in flies and mammals. J Cell Biol. 2012;199:481–96. doi: 10.1083/jcb.201201133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gilsanz A, Sánchez-Martín L, Gutiérrez-López MD, Ovalle S, Machado-Pineda Y, Reyes R, et al. ALCAM/CD166 adhesive function is regulated by the tetraspanin CD9. Cell Mol Life Sci. 2013;70:475–93. doi: 10.1007/s00018-012-1132-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berditchevski F, Chang S, Bodorova J, Hemler ME. Generation of monoclonal antibodies to integrin-associated proteins. Evidence that α3β1 complexes with EMMPRIN/basigin/OX47/M6. J Biol Chem. 1997;272:29174–80. doi: 10.1074/jbc.272.46.29174. [DOI] [PubMed] [Google Scholar]
- 62.Merlin D, Sitaraman S, Liu X, Eastburn K, Sun J, Kucharzik T, et al. CD98-mediated links between amino acid transport and beta 1 integrin distribution in polarized columnar epithelia. J Biol Chem. 2001;276:39282–9. doi: 10.1074/jbc.M105077200. [DOI] [PubMed] [Google Scholar]
- 63.Rintoul RC, Buttery RC, Mackinnon AC, Wong WS, Mosher D, Haslett C, et al. Cross-linking CD98 promotes integrin-like signaling and anchorage-independent growth. Mol Biol Cell. 2002;13:2841–52. doi: 10.1091/mbc.01-11-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.