Abstract
Genome-wide association studies have proven to be highly effective at defining relationships between single nucleotide polymorphisms (SNPs) and clinical phenotypes in complex diseases. Establishing a mechanistic link between a noncoding SNP and the clinical outcome is a significant hurdle in translating associations into biological insight. We demonstrate an approach to assess the functional context of a diabetic nephropathy (DN)-associated SNP located in the promoter region of the gene FRMD3. The approach integrates pathway analyses with transcriptional regulatory pattern-based promoter modeling and allows the identification of a transcriptional framework affected by the DN-associated SNP in the FRMD3 promoter. This framework provides a testable hypothesis for mechanisms of genomic variation and transcriptional regulation in the context of DN. Our model proposes a possible transcriptional link through which the polymorphism in the FRMD3 promoter could influence transcriptional regulation within the bone morphogenetic protein (BMP)-signaling pathway. These findings provide the rationale to interrogate the biological link between FRMD3 and the BMP pathway and serve as an example of functional genomics-based hypothesis generation.
While genome-wide association studies (GWASs) are effective at projecting genetic variants to complex disease phenotype, establishing the corresponding mechanistic link remains difficult. This is especially true for single nucleotide polymorphisms (SNPs) in non–protein coding regions of the genome that may affect regulatory function in a manner that is only evident in a particular functional context (1). One such context may be a biological process determined by genes whose transcription is synchronized by common regulatory elements within their promoters (2,3). A SNP located in one of these regulatory elements may alter or disrupt this coordinated regulation, leading to a change in gene expression and subsequently phenotype. It may be possible to identify such a mechanism via a change to a transcription factor binding site (TFBS) by a candidate SNP; we demonstrate this strategy for a SNP affecting the diabetic nephropathy (DN)-associated bone morphogenetic protein (BMP)-signaling pathway. The approach allows us to generate testable hypotheses from GWAS candidates falling in promoter regions and has the potential to help understand the functional impact of genetic variants in DN and other complex genetic diseases.
DN is the leading cause of end-stage renal disease in the U.S. (4), and ~20–40% of all patients with either type 1 diabetes (T1D) or type 2 diabetes (T2D) develop DN (5–7). DN has a significant heritability (8), providing the rationale for performing GWASs to discover genetic loci implicated in DN (9). Initial DN GWASs discovered candidate genetic loci for predisposition to DN for both T1D and T2D (8,10). However, these associations of a locus with DN do not explain how associated alleles affect the mechanism of disease. Unfortunately, this situation is typical of most GWAS of complex genetic disorders, while loci whose effects have been functionally confirmed are generally associated with Mendelian disorders. An example is the autosomal dominant disorder multiple osteochondromas, for which a SNP located in the EXT1 promoter eliminates a TFBS and increases promoter activity (11). For complex diseases, any large-scale analysis involving luciferase assays, electrophoretic mobility shift assays (EMSAs), and ELISAs are simply not feasible for hundreds of disease-associated SNPs. Data-driven approaches including the one outlined in this manuscript are necessary to prioritize the number of testable hypotheses for further experimental validation.
Establishing the functional context of a SNP is important in defining such hypotheses. Our group has previously used a functional context approach to identify proteins associated with the glomerular slit diaphragm in DN (12). In that work, a regulatory module detected in the promoters of a few known slit diaphragm genes predicted other slit diaphragm molecules after a genome-wide promoter search. Here, our integrative approach combines regulatory SNP prediction, transcriptional promoter modeling, and pathway analysis capable of decoding putative transcriptional pathomechanisms of DN (Fig. 1). We focus on the candidate gene FERM domain containing 3 (FRMD3) identified by a GWAS of the Genetics of Kidneys in Diabetes (GoKinD) study collection (13). In that study, the SNP rs1888747 showed the strongest risk association (P = 4.7 × 10−7; OR = 1.45) with DN within T1D subjects. Despite different study designs, this SNP also reached statistical significance level in a replication study of 1,305 participants of the Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications (EDIC) study, as well as in a subcohort of Japanese subjects with T2D (14). This polymorphism remained significantly associated with DN in a random-effects meta-analysis of genetic variants reproducibly associated with DN (15). Additionally, we have recently shown that rs1888747 is significantly associated with DN among 66 large T2D families from the Joslin T2D family collection (16). The SNP rs1888747 is located on chromosome 9q in the extended promoter region of FRMD3. FRMD3 has not previously been implicated in the pathogenesis of DN, T1D, or T2D.
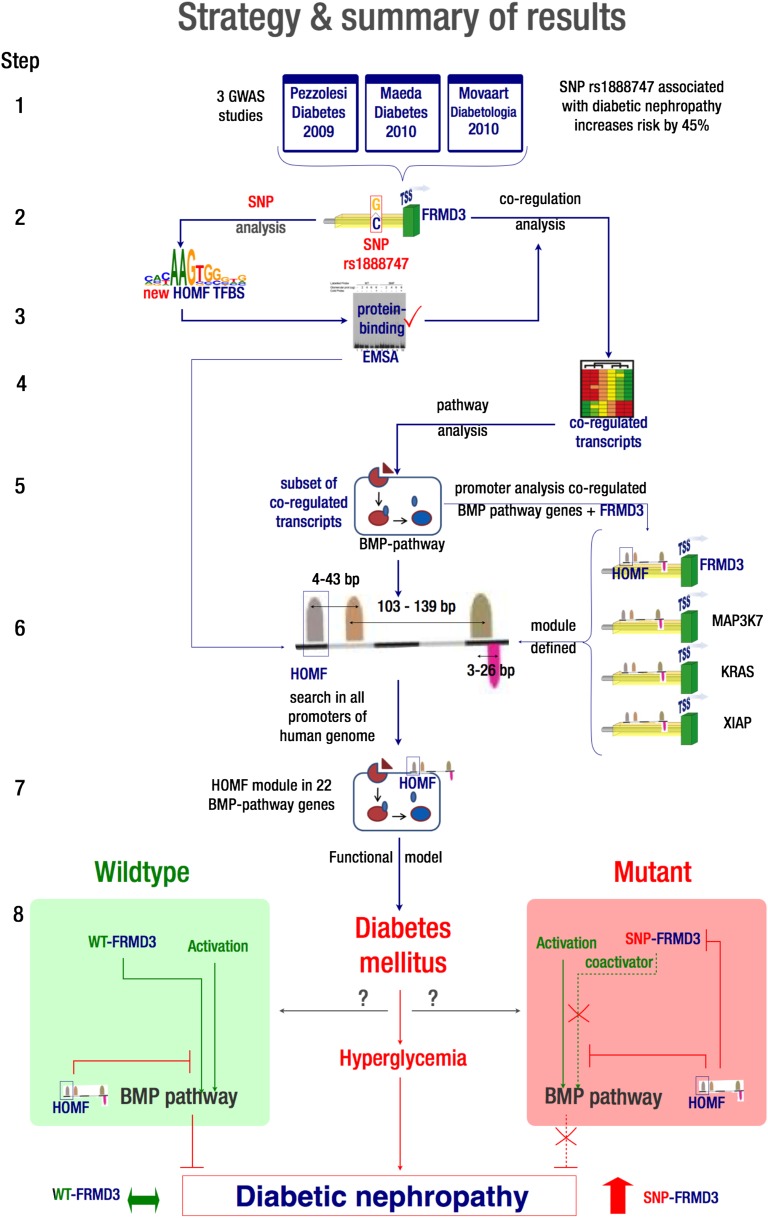
FIG. 1.
Overview of the analysis strategy (2–7) to identify the putative regulatory effect of GWAS candidate (1) on FRMD3 regulation (2), linking the gene to transcriptional regulation of the BMP pathway (4–7) and DN and suggesting a hypothetical regulatory model (8).
Here, we describe both our in silico approach and its use to derive the hypothesis that a DN risk allele brings FRMD3 under the control of a proposed transcriptional regulatory module and inhibits renal expression of FRMD3. The approach not only detects a transcriptional regulatory pattern affected by the candidate SNP but also connects known DN-associated pathways to the GWAS-derived candidate gene, providing the testable model system for further insight into the pathophysiology of DN.
RESEARCH DESIGN AND METHODS
Strategy.
We hypothesized that the SNP rs1888747, reported to be associated with DN by Pezzolesi et al. (13) and located in the proximity of FRMD3, is a regulatory SNP that alters the transcription factor binding capabilities of the FRMD3 proximal promoter region. We assumed that the binding site putatively affected by the SNP is part of a molecular TFBS framework involved in this transcriptional change, which should also be conserved in promoters of functionally connected (i.e., covarying) transcripts. Finding those transcripts might enable us to detect the framework by including the polymorphism in the FRMD3 promoter and thus place the SNP into a DN-relevant functional context.
We used comparative promoter analysis to determine common regulatory elements of FRMD3 and its coexpressed transcripts. Promoters of functionally linked transcripts are likely to contain conserved (nonrandom associated) TFBS frameworks. SNP-related TFBS alterations have the potential to integrate genomic features with transcriptional regulatory functions. A detailed overview of our study and strategy can be found in Fig. 1.
Human renal biopsies.
Renal biopsy samples were procured from 22 participants in a clinical trial (17) with an extended follow-up that provides an opportunity to examine the etiology of DN in T2D as well as the effect of treatment with losartan on the onset and progression of diabetic kidney disease. Renal biopsy specimens were processed and analyzed as previously described (12,18,19). Subjects' aggregate clinical and histological characteristics are summarized in Supplementary Table 2.
FRMD3 expression in subjects with DN and either normal or decreased glomerular filtration rate.
We compared glomerular FRMD3 expression levels as well as individual estimated glomerular filtration rate measurements of 22 Pima Indians with normal GFR with a cohort of seven T2D subjects with chronic kidney disease (CKD) stage 3 to assess whether FRMD3 gene expression would correlate with renal function. Statistical analysis comparing the two groups was done using GraphPad Prism 5 with a two-tailed t test (Mann-Whitney U test, 95% CI). P < 0.05 was considered statistically significant.
Pathway analysis of FRMD3 coexpressed transcripts.
When genes are coregulated under various biological conditions, their corresponding -expression profiles may show relative similarity or coexpression (20). We identified FRMD3 coexpressed transcripts by calculating Pearson r correlation between the expression profiles of FRMD3 and all other genes expressed above background. These coexpressed, potentially coregulated transcripts were then analyzed to identify transcripts known to be functionally related using Ingenuity Pathway Analysis software (version 8.5; Ingenuity Systems, Redwood City, CA [http://www.ingenuity.com]). The software detects enriched canonical pathways in a given gene set. Default settings were applied.
Renal function associated with FRMD3 coexpressed transcripts.
An unsupervised hierarchical clustering analysis of the 22 Pima Indians (T2D DN) using the expression levels of 581 FRMD3 coexpressed genes (including FRMD3) was performed (MeV, version 4.5.1, Euclidean distance, average linkage method). The two main branches in the dendrogram showed 100% support (bootstrap, n = 1,000). They were further analyzed for differences in their FRMD3 expression and their ability to associate with clinical and histologic subgroups, as this would link FRMD3 coexpressed transcripts with a disease-associated phenotype. Renal function measures, iothalamate GFR (iGFR) (in milliliters per minute) measured by a urinary clearance method that used cold iothalamate (21), the albumin-to-creatinine ratio (ACR), and the fractional mesangial area were compared between the two clusters. ΔACR/year and ΔiGFR/year were calculated by subtracting the corresponding value from the time of enrollment into the study from the latest available value divided by the number of years of follow-up. Fractional mesangial area was determined as previously described (22). Statistical analysis comparing the two major cluster branches was done using GraphPad Prism 5 with a two-tailed t test (Mann-Whitney U test [95% CI]). P < 0.05 was considered statistically significant.
Computational promoter analysis and evaluation.
Promoter regions for the eight FRMD3 coexpressed BMP pathway members were extracted (version 07/2009; ElDorado, Genomatix), and promoter modeling was performed to detect common transcriptional regulatory elements potentially influenced by the SNP of interest. For the FRMD3 promoter, we extracted a sequence of ±320 nucleotides (nt) around the SNP of interest, rs1888747. A sequence length of 320 nt was chosen to allow the detection of a four-element promoter module starting at the SNP position with an estimated average distance of 80 nt between the centers of two consecutive elements. The SNP rs1888747 is located at position 85345371 on chromosome 9 (Genome Build 36.3) in the extended promoter sequence of FRMD3 (1904 nt proximal to the first transcription start region). We determined potential TFBS generated or lost by the SNP rs1888747 (MatInspector, Genomatix) as described by Cartharius et al. (23). The FRMD3 promoter sequence was analyzed both with and without the risk allele. A promoter module is defined as a set of two or more TFBS of a defined order, orientation, and distance range acting together in a certain functional context (see Fessele et al. [2]).
We searched for a common module among promoter sequences of a subset of the eight FRMD3 coexpressed BMP pathway members and the SNP-altered sequence of the FRMD3 promoter (FrameWorker, Genomatix). Variance and distance between the individual promoter elements were altered until a module with more than two elements was discovered. We required more than two elements to be identified in our search, since more complex modules have been shown to be associated with more specific biological function (24). In addition, the promoter module was required to occur in at least two of the eight FRMD3 coexpressed BMP pathway members as well as in the FRMD3 promoter sequence at the position of rs1888747.
We evaluated the significance of the promoter module by searching a genome-wide human promoter database for additional genes whose promoters would also contain potential binding capabilities for the defined framework identified in the previous step (ModelInspector, Genomatix). For achievement of comparable preconditions, this search was conducted after adjustment for the promoter sequence of all genes in the promoter database (version 7/2009; ElDorado, Genomatix) (93,372 promoters) to the same sequence length where rs1888747 was found in the promoter of FRMD3. Additional BMP pathway members identified by this approach were evaluated for their enrichment in comparison with the total number of additionally detected genes.
EMSA.
EMSA was conducted to evaluate protein-binding differences of the FRMD3 wild-type (WT) and SNP-altered sequence. While this method does not allow conclusions about the actual binding protein itself, it is an effective way for an initial assessment of regulatory capabilities of an SNP in a noncoding region. The following steps were taken:
Glomerular isolation: glomeruli from five 3-month-old C57BL/6J mouse kidneys were isolated (25) with modifications in the nylon membranes used (100-µm nylon sieve; Sefar, Briarcliff Manor, NY).
Nuclear extracts: nuclear protein extracts from adult mouse kidneys and livers, glomeruli isolated from adult murine kidneys, and 293 cells were prepared as previously described (2).
EMSA analysis: oligonucleotides corresponding to the WT DNA sequence 5′-ACAAGGCTCTGGGAAACCAACTGGCCATTGTCAACAATAATA-3′ or to the SNP sequence 5′-ACAAGGCTCTGGGAAACCAAGTGGCCATTGTCAACAATAATA-3′ and complimentary strands were annealed and end-labeled with 32P-dCTP (26). Nuclear protein extracts were incubated in buffer with poly dIdC or poly dAdT and 10,000 cpm end-labeled oligonucleotide as previously described (26). For competition experiments, unlabeled DNA was added to the binding reactions at a 100-fold excess of the radiolabeled oligonucleotide. The DNA-protein complexes were resolved on 6% nondenaturing polyacrylamide gels in Tris-Borate-EDTA buffer at 120 V for 2.5 h. Gels were dried and exposed to XOMAT film (Eastman Kodak) overnight. The intensity of the DNA-protein complex was measured using the software ImageJ 1.44p (NIH, http://imagej.nih.gov/ij/). A paired t test (GraphPad Prism 5) was used to assess the significance of the mean intensity in the SNP sequences compared with the WT sequence.
RESULTS
Defining clinical and functional association of FRMD3.
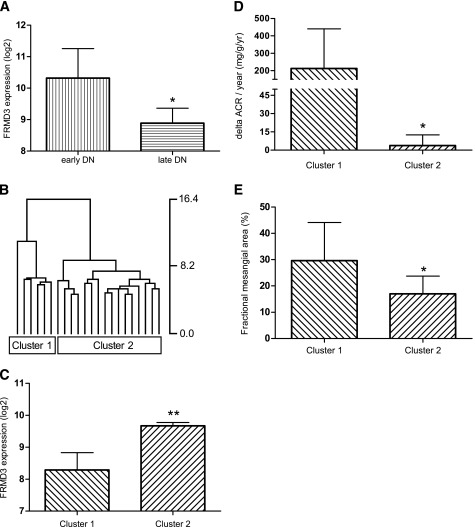
To assess the functional relationship between FRMD3 and DN, we related steady state mRNA levels to the available clinical outcome parameters. We found FRMD3 transcript levels decreased significantly with progression of DN (mean ± SD 8.9 ± 1.2 in DN with CKD stage 3 compared with 10.3 ± 0.9 in DN with normal GFR [P < 0.02]) (Fig. 2A).
FIG. 2.
A: FRMD3 is repressed with progression of DN. FRMD3 gene expression comparing 22 Pima Indians with T2D and normal GFR with a cohort of 7 T2D with CKD stage 3. Data are displayed as means ± SD. Glomerular FRMD3 expression in early DN (Pima) 10.3 ± 0.9 and in CKD3 DN 8.9 ± 1.2. estimated glomerular filtration rate in early DN 104 ± 19 mL/min/1.73 m2 and in CKD stage 3 DN 53 ± 33 mL/min/1.73 m2 (P < 0.002, Mann-Whitney U test, 95% CI). B: FRMD3 coregulated genes segregate DN patients in defined subgroups. Cluster dendrogram of 581 FRMD3-correlated genes (including FRMD3) in a cohort of 22 Pima Indians with T2D DN. The two main branches (cluster 1 and cluster 2) of the dendrogram show 100% support and reflect distinct clinical groups (see D). C: FRMD3 and coregulated BMP pathway members are repressed in cluster 1. FRMD3 and BMPR2, CREB1, KRAS, MAP3K7, PRKAR2B, SMAD5, and XIAP (7 of 8 BMP pathway members) are significantly (**P < 0.008) downregulated in cluster 1 compared with cluster 2 (Mann-Whitney U test, two-tailed, 95% CI). Expression data are displayed as means ± SD. Glomerular FRMD3 expression cluster 1, 8.29 ± 0.54; cluster 2, 9.67 ± 0.41. D: FRMD3/BMP repression is associated with increase of albuminuria. Clinical measures of ΔACR/year comparing the two main cluster branches from B. Data are displayed as means ± SD. ΔACR/year cluster 1, 212.4 ± 227.9, is significantly (*P = 0.017) increased compared with ΔACR/year in cluster 2, 3.7 ± 8.7. (Mann-Whitney U test, two-tailed, 95% CI). E: FRMD3/BMP repression is associated with increase of fractional mesangial area. Histologic measures of fractional mesangial area (%) comparing the two main cluster branches from B. Data are displayed as means ± SD. Mesangial expansion was significantly (*P = 0.04) increased in cluster 1 (30 ± 14%) compared with cluster 2 (17 ± 7%) (Mann-Whitney U test, two-tailed, 95% CI).
As FRMD3 had no prior link to DN, we used a data-driven approach to establish a putative clinical and functional context for FRMD3 in DN. Starting from a list of 17,589 transcripts expressed on the Affymetrix microarray chip, 16,956 passed the cutoff filter (median + 2 × SD of the 27 Poly-A Affymetrix negative controls’ expression baseline [27]) and were tested for correlation with FRMD3. Transcriptional coregulation orchestrated by common upstream transcriptional regulatory elements (2) provided the rationale that FRMD3-correlated transcripts (similar mRNA expression patterns) might be linked to regulatory pathways in DN, which in turn may help establish the link between FRMD3 and the disease.
We identified 581 FRMD3 coexpressed transcripts (|r| ≥ 0.65, FDR ≤ 0.02; for top 10 transcripts with the highest |r| value, see Supplementary Table 1). The majority (518) of the 581 FRMD3 coexpressed transcripts were concordantly regulated with FRMD3, as were the top 10 (sorted by |r| value) FRMD3 coexpressed transcripts. For 5 of those top 10 transcripts or close variants, an association with diabetes or cardiovascular or inflammatory diseases has been published (Supplementary Table 1), consistent with the relevance of this gene set to the pathophysiology of DN.
Expression of FRMD3 and its correlated transcripts is linked to early progression in DN.
Hierarchical clustering using the expression signatures of FRMD3 coexpressed transcripts detected two distinct clusters (Fig. 2B and C). Patients contained in cluster 1 had a significantly (P = 0.017) higher ΔACR/year of 212.4 ± 227.9 compared with cluster 2 (ΔACR/year of 3.7 ± 8.7 [Fig. 2D]). Mesangial expansion, a key histologic feature of DN (22), was significantly (P = 0.04) increased in cluster 1 (30 ± 14%) compared with cluster 2 (17 ± 7%) (Fig. 2E). ΔGFR showed a similar trend but missed statistical significance. Observation times were similar in both patient groups (cluster 1, 9.0 ± 2.2 years; cluster 2, 9.5 ± 0.9 years; P = 0.91). In cluster 1, with higher ΔACR/year, the gene expression of seven out of the eight BMP pathway genes (BMPR2, CREB1, KRAS, MAP3K7, PRKAR2B, SMAD5, and XIAP) was lower than in cluster 2. This concordance of transcriptional regulation of FRMD3 and BMP pathway members with renal outcome measures points toward a common molecular mechanism responsible for the coregulation of FRMD3 and several BMP pathway members.
Pathway analysis of FRMD3 coexpressed transcripts.
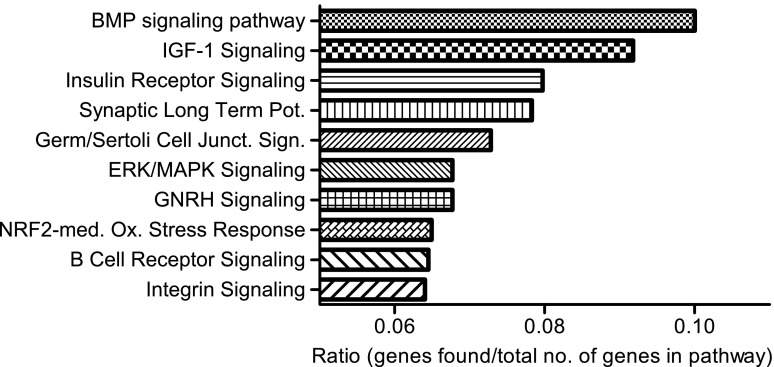
We determined the functional context of FRMD3 and its 581 coexpressed transcripts by mapping them to known canonical pathways. Among them, the BMP signaling pathway was found to be the pathway with the strongest enrichment with eight BMP pathway members coexpressed with FRMD3 (BMPR2, CREB1, KRAS, MAP3K7, PRKAR1B, PRKAR2B, SMAD5, and XIAP) (Fig. 3). This finding is consistent with previous publications attributing DN-protective properties to the BMP pathway (rev. in 28,29) and indicates that the biological context defined for FRMD3 and its coexpressed transcripts might indeed be relevant for DN.
FIG. 3.
Functional association of FRMD3-correlated genes. Top 10 pathways (Ingenuity Pathways Analysis; Ingenuity Systems) of 581 FRMD3-correlated genes sorted by the ratio of members of the pathway among FRMD3-correlated genes vs. total number of members of that pathway. P ≤ 0.001 for all pathways. Junct., junction; med., mediated; Ox., oxidative; Pot., potentiation; Sign., signaling.
Defining putative SNP function.
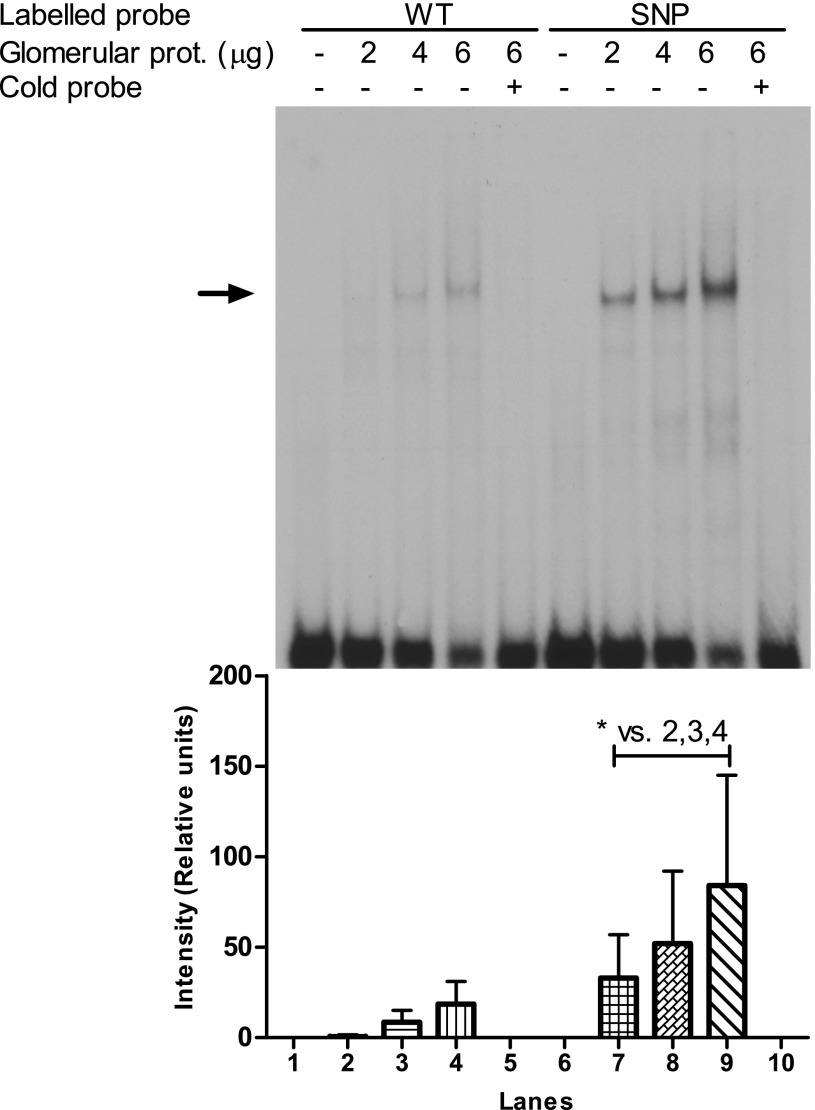
In silico comparison of sequence variants with and without the risk allele identified a potential homeodomain factor (HOMF) TFBS covering the SNP position. This TFBS was not detected in the presence of the nonrisk allele in the FRMD3 promoter (Fig. 1, step 2). An EMSA of oligonucleotides corresponding to the WT and SNP-altered sequence of glomerular extracts from C57Black6 mice supports these predictions: the sequence with the disease-associated SNP shows a >4.7 times relative increase (intensity WT vs. SNP: 1 vs. 57, 15 vs. 92, and 31 vs. 145, respectively) in protein binding compared with the WT DNA sequence (Fig. 4). These results show that rs1888747 affects protein binding, suggesting the generation of a putative TFBS by that particular SNP.
FIG. 4.
Increased binding of glomerular nuclear extracts to DN-associated genomic region. EMSA from oligonucleotides corresponding to the WT DNA sequence and SNP-altered sequence (SNP) of glomerular extracts from C57Black6 mice. The nonspecific competitor poly(dIdC) was used. Arrow indicates position of protein-bound oligos. With increasing amounts of protein used, a distinct binding signal can be detected in the SNP sequence but to a lesser amount in the WT-sequence as displayed in the Intensity Blot. Intensity of the DNA-protein complex in lane 2 was set to 1.0. A paired t test showed that the mean intensity was significantly higher in the SNP sequences compared with the WT sequence (*P = 0.04). prot., protein.
Putative transcriptional mechanism for coregulation of FRMD3 and BMP pathway members.
After extraction of the proximal promoter sequences of the eight BMP genes coexpressed with FRMD3, we identified promoter frameworks shared among BMP genes as well as the FRMD3 promoter sequence with the risk allele. For FRMD3 and four of the eight FRMD3 coexpressed BMP pathway members (XIAP, KRAS, PRKAR2B, and MAP3K7), we found a module with four TFBS (HOMF, BRNF [Brn POU domain factors], BRN5 [Brn-5 POU domain factors], and GATA [GATA binding factors]) where the SNP rs1888747 occurs in the first (HOMF) TFBS of FRMD3 (for details of the framework, see Fig. 1, step 6). This framework provides the molecular basis for a proposed coregulatory pattern of FRMD3 and BMP pathway members. A genome-wide search in a human promoter database (ModelInspector/ElDorado, Genomatix) identified an additional set of 18 BMP pathway members containing the four TFBS modules in their promoters. An enrichment analysis showed that detecting the promoter module in 22 (18 newly identified plus 4 original BMP pathway members) of the total 72 BMP pathway genes as annotated by Ingenuity Pathway Analysis software achieved an enrichment score of 4.2 and a significant z score of 7.6. These findings suggest that the four TFBS promoter modules could mediate the transcriptional coregulation of BMP pathway members and FRMD3 in the functional context of DN. Our results provide a rationale and an experimental framework to define a regulatory link between FRMD3 and the BMP pathway in DN.
DISCUSSION
With the emerging capabilities to capture the genetic and molecular underpinnings of diabetes complications, molecular-based disease definition can lead to individual risk assessments and selection of targeted therapies (30). Describing gene-environment interactions will be a critical step toward molecular disease definition. A series of studies currently aims to link genetic variation to diabetes complications (13,31–33). Genetic variants can affect the phenotype by directly altering the coding sequence of a gene, resulting in a qualitative change in the encoded protein. Alternatively, variants can alter regulatory regions in the genome, resulting in quantitative changes of the transcript. Research in monogenetic diseases has established a clear path forward to define the consequences of protein coding variants. Defining the consequences of regulatory variants on gene expression, particularly in complex diseases, is still in its infancy. The current study aims to provide one possible way forward to identify potential regulatory effects of DN-associated noncoding variants and their link to complex regulatory networks in DN.
Regulatory network analysis starting from a putative causal SNP needs to be embedded in an in-depth analysis of the functional context of the affected gene. This context is required to reveal regulatory mechanisms represented by TFBS frameworks active in regulatory regions of the genes of interest. In general, regulatory SNPs can be inferred if a known or potential TFBS is directly affected by the polymorphism (34). However, since individual TFBS are often not sufficient for regulatory functions, their functional contributions can only be assessed in the appropriate regulatory context, i.e., the interaction with other TFBS (35). Disease-relevant pathways and transcriptional covariance can serve as selection criteria for genes belonging to that functional context. Regulatory links identified by this approach allow prediction of transcriptional alterations, which can be tested in the context of disease.
This strategy presented in our study is applicable whenever a transcriptional change of the GWAS gene is observed and coregulated transcriptional networks can be identified. However, although this implies finding a group of coexpressed genes, the pathway association might not always be as clear-cut as in our case, which might result in testing multiple associated pathways with the strategy presented above. A direct hit of the SNP in a TFBS is an advantage, but proximity to a potential TFBS framework most likely would suffice to alter TFBS function. In case no such framework can be found with any associated pathway, alternative bioinformatics methods for the selection of genes of a similar functional context can be tested, including protein-protein interaction networks (36), phylogenetic conservation (12), or epigenetic/epigenomic approaches (37). With the increasing availability of genetic mapping of expression quantitative trait loci studies in DN cohorts, expression quantitative trait loci will be linked directly to the physical location of transcripts differentially expressed in DN and thereby support promoter modeling approaches as described by our example (38,39).
The study presented here started from a worst-case scenario, as a testable hypothesis had to be developed for the role of a noncoding SNP in a gene without known function in DN. We followed a sequential strategy integrating multiple lines of genetic and genomic evidence for hypothesis generation (see Fig. 1 for overview). First, the candidate SNP rs1888747 in the proximal promoter region of FRMD3 prompted us to search for the functional context of the TFBS framework covering the candidate SNP. Pathway analysis of coexpressed transcripts revealed a significant enrichment for the BMP pathway (40). BMPs are part of the transforming growth factor-β superfamily (41) and have a well-established role in kidney development, cell growth, cell differentiation, chemotaxis, and apoptosis of various cell types (42). An imbalance of BMP7 agonists like kielin/chordin-like protein and BMP7 antagonists like gremlin has been described in DN (29). Decreased expression of BMP7 and its agonists has been associated with increased profibrotic activity in animal models of DN (43), consistent with a protective effect of BMP activation in DN. Promoter modeling for the FRMD3 promoter sequence as well as for eight coexpressed transcripts led to the discovery of a BMP pathway–specific TFBS framework that identified a total of 22 BMP pathway members in a genome-wide promoter sequence search.
Our results support the hypothesis of a functional connection of the SNP with reduced FRMD3 expression, as the SNP-created binding site is located in a likely repressive promoter module. Since this module is shared between regulatory regions of 22 genes of the BMP pathway, BMP genes could be suppressed by the same mechanism using the shared module. The risk allele generates the necessary binding sites of the BMP module in the FRMD3 promoter and, as for BMPs, represses FRMD3 with deleterious impact on DN including inhibition of the protective effects of the BMP pathway. Interestingly, a BMP-focused candidate gene study by the GoKinD Study Group was not able to identify statistically significant DN-associated SNPs in the genes BMP2, BMP4, and BMP7 (44). The above hypothesis establishes a trans-association of the DN-associated SNP linking BMP genes to the risk of DN via FRMD3.
Proposed model connecting FRMD3 and BMP pathway.
Based on our findings, we developed a testable hypothesis for the functional impact of the SNP rs1888747 in DN. We suggest that our proposed TFBS framework is generally inhibitory in the context of renal gene expression and may act as a negative regulatory feedback loop to balance BMP pathway action. A maximum parsimony of all known facts is consistent with the idea that one FRMD3 function is to aid in the activation of BMP pathway gene expression, providing some counterbalance to the inhibitory effect of the TFBS framework defined for BMPs above. This is consistent with the observed higher expression of BMP genes in the absence of the risk allele. However, the risk allele brings FRMD3 under the control of the same negative BMP feedback loop, effectively abolishing the positive impact of FRMD3 on BMP expression. As a result, BMP-mediated protective effects on renal tissue, and thus renal protection, are reduced in individuals with the polymorphism, which is consistent with the observed DN phenotype associated with the polymorphism. FRMD3 and BMP pathway gene repression is correlated to the severity of the renal phenotype. Recent GWASs of T2D subjects also detected SNPs in the FRMD3 gene region to be associated with diabetic retinopathy, possibly relating to a uniform connection of FRMD3 and BMP pathway members in diabetes end organ damage (45).
The strength of this approach is its ability to predict functional connections based solely on regulatory networks as exemplified by significantly enriched transcriptional TFBS frameworks in the absence of direct protein-based evidence. We currently do not know how the connection between FRMD3 and BMP pathway members is mediated. We found no evidence at the protein, RNA, or microRNA level. Therefore, FRMD3 is thought to influence currently unknown regulatory intermediates. Even in this case, the model provides an explanation of how this SNP could bring the transcriptional regulation of FRMD3 under the same control as the coregulated BMP genes via the four TFBS regulatory module. While beyond the scope of our manuscript, functionality can now be established experimentally in vivo. The model approach introduced here provides insight into genomic variation and the mechanisms of transcriptional regulation and provides the basis for targeted experimental design. FRMD3 appears to be a promising target for these experiments, as comparative genome mapping data also confirmed FRMD3 as a nephropathy candidate gene in mice (46). The functional context proposed in this study could be experimentally validated by several approaches. Luciferase promoter reporter assays corresponding to WT and disease-associated alleles could be used to determine the functional impact of the rs1888747 SNP on FRMD3 expression, and functional consequences of FRMD3 gene silencing/overexpression on the expression of BMP pathway members can be tested in vitro. The impact of the polymorphism in DN in vivo can be evaluated using mice transgenic for the FRMD3 locus with and without the disease-associated polymorphism. As our data provide a functional link of BMP signaling pathway members to other potentially DN-associated pathways such as the IGF-1 and insulin receptor signaling pathway, results from these functional assays can be interpreted with regard to all pathways shown to be enriched among FRMD3-correlated transcripts.
Our work provides a paradigm of how functional genomics–based hypothesis generation can be implemented by a stepwise integration of regulatory SNP prediction, transcriptional promoter modeling, and pathway analysis. Our model approach provides a novel strategy to extend insight into the mechanisms of genomic variation and transcriptional regulation to regulatory networks informing subsequent experimental design. The general approach can be applied for different questions in the field of GWAS and transcriptomic data integration. The method is also suitable for the analysis of experimentally derived TFBS datasets, such as ChIP-Seq data or panels of in vivo protein-bound DNA elements, generated by genomic footprinting (47). Furthermore, information from chromatin histone modifications, potentially regulatory sequences, or phylogenetic footprinting studies can be linked to regulatory networks. In the context of DN, our work presents a novel starting point for hypothesis generation in molecular medicine in DN.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported in part by National Institutes of Health grants R01-DK058549 to A.S.K. and K01-DK090125 to M.G.P., by the O’Brien Renal Center (P30-DK081943) and the National Center for Integrative Bioinformatics (U54-DA021519) to M.K., and from the Federal Ministry of Education and Research, Germany (01EX1021L) to T.W.
No potential conflicts of interest relevant to this article were reported.
S.M. and V.N. conceived the project, designed the experiments, performed transcriptomic data analysis, and prepared the manuscript. S.R.P. conducted the EMSA assays and prepared the manuscript. F.E. performed the transcriptomic data analysis. R.G.N. phenotyped the Pima Indian participants, provided data and tissue samples for gene expression studies, and reviewed and edited the manuscript. E.J.W. phenotyped the Pima Indian participants, provided data and tissue samples for gene expression studies, performed the morphometric characterization of the kidney tissue, and reviewed and edited the manuscript. M.G.P. prepared the manuscript. A.S.K. supervised the study. A.R. performed transcriptomic data analysis. B.J.K. and T.W. prepared the manuscript. M.K. conceived the project, designed the experiments, reviewed and edited the manuscript, and supervised the study. S.M. is the guarantor of this work and as such had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Parts of this study were presented in abstract form at the Nexus Symposium of the International Society of Nephrology, Geneva, Switzerland, 30 June–2 July 2010.
The authors thank C.V. Komorowsky, Internal Medicine and Nephrology, University of Michigan, for helpful discussion of the renal morphometric data, as well as the authors’ collaborators from University College Dublin (E.P. Brennan, C. Godson, and F. Martin) for discussion on functional relationships between FRMD3 and BMP pathway members.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db12-1416/-/DC1.
REFERENCES
- 1.Freedman ML, Monteiro AN, Gayther SA, et al. Principles for the post-GWAS functional characterization of cancer risk loci. Nat Genet 2011;43:513–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fessele S, Maier H, Zischek C, Nelson PJ, Werner T. Regulatory context is a crucial part of gene function. Trends Genet 2002;18:60–63 [DOI] [PubMed] [Google Scholar]
- 3.Liu R, McEachin RC, States DJ. Computationally identifying novel NF-kappa B-regulated immune genes in the human genome. Genome Res 2003;13:654–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.USRDS 2010 Annual Data Report: Atlas of Chronic Kidney Disease and End-Stage Renal Disease in the United States. Bethesda, MD, U.S. Renal Data System, 2010 [Google Scholar]
- 5.Hasslacher C, Ritz E, Wahl P, Michael C. Similar risks of nephropathy in patients with type I or type II diabetes mellitus. Nephrol Dial Transplant 1989;4:859–863 [DOI] [PubMed] [Google Scholar]
- 6.Gross JL, de Azevedo MJ, Silveiro SP, Canani LH, Caramori ML, Zelmanovitz T. Diabetic nephropathy: diagnosis, prevention, and treatment. Diabetes Care 2005;28:164–176 [DOI] [PubMed] [Google Scholar]
- 7.Bloomgarden ZT. Diabetic nephropathy. Diabetes Care 2005;28:745–751 [DOI] [PubMed] [Google Scholar]
- 8.Freedman BI, Bostrom M, Daeihagh P, Bowden DW. Genetic factors in diabetic nephropathy. Clin J Am Soc Nephrol 2007;2:1306–1316 [DOI] [PubMed] [Google Scholar]
- 9.Conway BR, Maxwell AP. Genetics of diabetic nephropathy: are there clues to the understanding of common kidney diseases? Nephron Clin Pract 2009;112:c213–c221 [DOI] [PubMed] [Google Scholar]
- 10.Ichinose K, Kawasaki E, Eguchi K. Recent advancement of understanding pathogenesis of type 1 diabetes and potential relevance to diabetic nephropathy. Am J Nephrol 2007;27:554–564 [DOI] [PubMed] [Google Scholar]
- 11.Jennes I, Zuntini M, Mees K, et al. Identification and functional characterization of the human EXT1 promoter region. Gene 2012;492:148–159 [DOI] [PubMed] [Google Scholar]
- 12.Cohen CD, Klingenhoff A, Boucherot A, et al. Comparative promoter analysis allows de novo identification of specialized cell junction-associated proteins. Proc Natl Acad Sci USA 2006;103:5682–5687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pezzolesi MG, Poznik GD, Mychaleckyj JC, et al. DCCT/EDIC Research Group Genome-wide association scan for diabetic nephropathy susceptibility genes in type 1 diabetes. Diabetes 2009;58:1403–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maeda S, Araki S, Babazono T, et al. Replication study for the association between four Loci identified by a genome-wide association study on European American subjects with type 1 diabetes and susceptibility to diabetic nephropathy in Japanese subjects with type 2 diabetes. Diabetes 2010;59:2075–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mooyaart AL, Valk EJ, van Es LA, et al. Genetic associations in diabetic nephropathy: a meta-analysis. Diabetologia 2011;54:544–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pezzolesi MG, Jeong J, Smiles A, et al. Family-based association analysis confirms the role of the chromosome 9q21.32 locus in the susceptibility of diabetic nephropathy. PLoS ONE. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weil EJ, Fufaa G, Jones LI, et al. Effect of losartan on prevention and progression of early diabetic nephropathy in American Indians with type 2 diabetes. Diabetes. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen CD, Frach K, Schlöndorff D, Kretzler M. Quantitative gene expression analysis in renal biopsies: a novel protocol for a high-throughput multicenter application. Kidney Int 2002;61:133–140 [DOI] [PubMed] [Google Scholar]
- 19.Thacker SG, Berthier CC, Mattinzoli D, Rastaldi MP, Kretzler M, Kaplan MJ. The detrimental effects of IFN-α on vasculogenesis in lupus are mediated by repression of IL-1 pathways: potential role in atherogenesis and renal vascular rarefaction. J Immunol 2010;185:4457–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Do JH, Choi DK. Clustering approaches to identifying gene expression patterns from DNA microarray data. Mol Cells 2008;25:279–288 [PubMed] [Google Scholar]
- 21.Myers BD, Nelson RG, Tan M, et al. Progression of overt nephropathy in non-insulin-dependent diabetes. Kidney Int 1995;47:1781–1789 [DOI] [PubMed] [Google Scholar]
- 22.Weil EJ, Lemley KV, Mason CC, et al. Podocyte detachment and reduced glomerular capillary endothelial fenestration promote kidney disease in type 2 diabetic nephropathy. Kidney Int 2012;82:1010–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cartharius K, Frech K, Grote K, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 2005;21:2933–2942 [DOI] [PubMed] [Google Scholar]
- 24.Werner T. Target gene identification from expression array data by promoter analysis. Biomol Eng 2001;17:87–94 [DOI] [PubMed] [Google Scholar]
- 25.Wiggins JE, Goyal M, Sanden SK, et al. Podocyte hypertrophy, “adaptation,” and “decompensation” associated with glomerular enlargement and glomerulosclerosis in the aging rat: prevention by calorie restriction. J Am Soc Nephrol 2005;16:2953–2966 [DOI] [PubMed] [Google Scholar]
- 26.Patel SR, Dressler GR. Expression of Pax2 in the intermediate mesoderm is regulated by YY1. Dev Biol 2004;267:505–516 [DOI] [PubMed] [Google Scholar]
- 27.Berthier CC, Bethunaickan R, Gonzalez-Rivera T, et al. Cross-species transcriptional network analysis defines shared inflammatory responses in murine and human lupus nephritis. J Immunol 2012;189:988–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maciel TT, Kempf H, Campos AH. Targeting bone morphogenetic protein signaling on renal and vascular diseases. Curr Opin Nephrol Hypertens 2010;19:26–31 [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Zhang Q. Bone morphogenetic protein-7 and Gremlin: New emerging therapeutic targets for diabetic nephropathy. Biochem Biophys Res Commun 2009;383:1–3 [DOI] [PubMed] [Google Scholar]
- 30.Offit K. Personalized medicine: new genomics, old lessons. Hum Genet 2011;130:3–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jurado J, Ybarra J, Romeo JH, Garcia M, Zabaleta-Del-Olmo E. Angiotensin-converting enzyme gene single polymorphism as a genetic biomarker of diabetic peripheral neuropathy: longitudinal prospective study. J Diabetes Complications 2012;26:77–82 [DOI] [PubMed] [Google Scholar]
- 32.Kang P, Tian C, Jia C. Association of RAGE gene polymorphisms with type 2 diabetes mellitus, diabetic retinopathy and diabetic nephropathy. Gene 2012;500:1–9 [DOI] [PubMed] [Google Scholar]
- 33.Sandholm N, Salem RM, McKnight AJ, et al. DCCT/EDIC Research Group New susceptibility loci associated with kidney disease in type 1 diabetes. PLoS Genet 2012;8:e1002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benson CC, Zhou Q, Long X, Miano JM. Identifying functional single nucleotide polymorphisms in the human CArGome. Physiol Genomics 2011;43:1038–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson PJ, Werner T. Pathways and promoter networks analysis provides systems topology for systems biology approaches. Semin Nephrol 2010;30:477–486 [DOI] [PubMed] [Google Scholar]
- 36.Jayapandian M, Chapman A, Tarcea VG, et al. Michigan Molecular Interactions (MiMI): putting the jigsaw puzzle together. Nucleic Acids Res 2007;35:D566–D571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feinberg AP. Epigenomics reveals a functional genome anatomy and a new approach to common disease. Nat Biotechnol 2010;28:1049–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keller B, Martini S, Sedor J, Kretzler M. Linking variants from genome-wide association analysis to function via transcriptional network analysis. Semin Nephrol 2010;30:177–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schadt EE, Monks SA, Drake TA, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature 2003;422:297–302 [DOI] [PubMed] [Google Scholar]
- 40.Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors 2004;22:233–241 [DOI] [PubMed] [Google Scholar]
- 41.Ducy P, Karsenty G. The family of bone morphogenetic proteins. Kidney Int 2000;57:2207–2214 [DOI] [PubMed] [Google Scholar]
- 42.Zeisberg M, Kalluri R. Reversal of experimental renal fibrosis by BMP7 provides insights into novel therapeutic strategies for chronic kidney disease. Pediatr Nephrol 2008;23:1395–1398 [DOI] [PubMed] [Google Scholar]
- 43.Wang S, de Caestecker M, Kopp J, Mitu G, Lapage J, Hirschberg R. Renal bone morphogenetic protein-7 protects against diabetic nephropathy. J Am Soc Nephrol 2006;17:2504–2512 [DOI] [PubMed] [Google Scholar]
- 44.McKnight AJ, Pettigrew KA, Patterson CC, Kilner J, Sadlier DM, Maxwell AP, Warren 3/UK GoKinD Study Group Investigation of the association of BMP gene variants with nephropathy in Type 1 diabetes mellitus. Diabet Med 2010;27:624–630 [DOI] [PubMed] [Google Scholar]
- 45.Hu C, Zhang R, Yu W, et al. CPVL/CHN2 genetic variant is associated with diabetic retinopathy in Chinese type 2 diabetic patients. Diabetes 2011;60:3085–3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garrett MR, Pezzolesi MG, Korstanje R. Integrating human and rodent data to identify the genetic factors involved in chronic kidney disease. J Am Soc Nephrol 2010;21:398–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hesselberth JR, Chen X, Zhang Z, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods 2009;6:283–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.