Abstract
Diabetes and its associated hyperglycemia induce multiple changes in liver function, yet we know little about the role played by translational control of gene expression in mediating the responses to these conditions. Here, we evaluate the hypothesis that hyperglycemia-induced O-GlcNAcylation of the translational regulatory protein 4E-BP1 alters hepatic gene expression through a process involving the selection of mRNA for translation. In both streptozotocin (STZ)-treated mice and cells in culture exposed to hyperglycemic conditions, expression of 4E-BP1 and its interaction with the mRNA cap-binding protein eIF4E were enhanced in conjunction with downregulation of cap-dependent and concomitant upregulation of cap-independent mRNA translation, as assessed by a bicistronic luciferase reporter assay. Phlorizin treatment of STZ-treated mice lowered blood glucose concentrations and reduced activity of the cap-independent reporter. Notably, the glucose-induced shift from cap-dependent to cap-independent mRNA translation did not occur in cells lacking 4E-BP1. The extensive nature of this shift in translational control of gene expression was revealed using pulsed stable isotope labeling by amino acids in cell culture to identify proteins that undergo altered rates of synthesis in response to hyperglycemia. Taken together, these data provide evidence for a novel mechanism whereby O-GlcNAcylation of 4E-BP1 mediates translational control of hepatic gene expression.
Regulation of gene expression at the level of translation initiation plays a critical role in biological processes such as cellular proliferation, development, and response to biological cues and environmental stresses. Recruitment of ribosomes to mRNA is the rate-limiting step in translation initiation, which in mammalian cells occurs through both cap-dependent and cap-independent mechanisms. According to the traditional model proposed for cap-dependent translation, recruitment of the ribosome onto eukaryotic mRNA occurs upon recognition of the m7GTP cap at the 5′-end of mRNA by eIF4F, followed by binding of the eIF4F-mRNA complex to the 40S ribosomal subunit (1). Alternatively, some cellular mRNAs contain unique RNA elements known as internal ribosomal entry sites (IRES) that enable them to use a cap-independent mechanism of translation initiation (2). An IRES is a highly structured nucleotide sequence located in the 5′-untranslated region (UTR) that promotes binding of 40S ribosomal subunits to a portion of the mRNA at or near the AUG start codon (3). The inclusion of IRES elements in the 5′-UTR of messages allows them to be translated under physiological stress conditions wherein cap-dependent protein synthesis is compromised (4–7).
With the exception of eIF4E, the same canonical initiation factors that facilitate cap-mediated loading of ribosomes onto mRNA are required for cap-independent initiation (8). Further, eIF4E has been shown to function as a negative modulator of IRES-mediated translation by increasing competition from capped mRNAs for initiation factor complexes (9). The interaction of eIF4E and eIF4G is of critical importance for cap-dependent initiation and is principally regulated by eIF4E sequestering proteins, such as 4E-BP1. Binding of 4E-BP1 to eIF4E is mutually exclusive of eIF4E interaction with eIF4G and is controlled by the sequential phosphorylation of serine/threonine residues on 4E-BP1 (10). Hypophosphorylated 4E-BP1 binds eIF4E strongly; however, upon phosphorylation 4E-BP1 releases eIF4E, allowing eIF4E to interact with eIF4G, which promotes ribosome loading onto the mRNA 5′-cap. Thus, conditions that promote 4E-BP1 binding to eIF4E potentially act as a switch between cap-dependent and cap-independent translation. Intriguingly, we have recently showed that under hyperglycemic conditions 4E-BP1 is also modified by addition of N-Acetylglucosamine to Ser or Thr residues (O-GlcNAcylation), which enhanced its interaction with eIF4E independent of phosphorylation status (11).
Hyperglycemia increases the flux of glucose through the hexosamine biosynthetic pathway (HBP) to increase the production of uridine diphosphate (UDP) N-Acetylglucosamine, which promotes protein O-GlcNAcylation (12) and contributes to the pathophysiology of diabetes (13). O-linked GlcNAcylation has been shown to influence protein function by altering subcellular localization, protein-protein interactions, DNA binding, enzyme activity, and turnover rates (14–18). O-GlcNAcylation modification is dynamic, cycling rapidly on and off proteins in a manner that is more reminiscent of protein phosphorylation than other forms of common glycosylation (19). The O-GlcNAcylation cycling reactions are catalyzed by the enzymes O-GlcNAcylation transferase and O-GlcNAcase (20–22), which strongly associate with subpopulations of cytosolic ribosomes, suggesting that they play an important role in regulating mRNA translation (23). In the current study, we evaluated the hypothesis that hyperglycemia causes elevated O-GlcNAcylation of 4E-BP1 and enhanced binding of 4E-BP1 to eIF4E, which in turn causes a shift from cap-dependent to cap-independent mRNA translation. Overall, the results support the conclusion that O-GlcNAcylation of 4E-BP1 expression underlies a hyperglycemia-mediated shift in gene expression.
RESEARCH DESIGN AND METHODS
Male (RFL12xBLK6) F1 mice expressing a bicistronic Renilla luciferase (LucR)–fibroblast growth factor (FGF-2) IRES–firefly luciferase (LucF) mRNA were treated with 50 mg/mL streptozotocin (STZ) for 5 days to induce diabetes. RFL12 mice have previously been described (24). Diabetes phenotype was confirmed by blood glucose concentrations >400 mg/dL. Four weeks after STZ treatment, phlorizin was administered subcutaneously twice daily for 7 days (11). Procedures were approved by the Pennsylvania State University College of Medicine Institutional Animal Care and Use Committee. Analysis of protein phosphorylation in liver homogenates and immunoprecipitations was performed as previously described (11). LucR and LucF activity was measured by Dual-Glo Luciferase Assay (Promega).
Cell culture and transfections.
Primary cultures of hepatocytes were prepared from the liver of RFL12x-BLK6F1 and previously described Eif4ebp1 or Eif4ebp2 knockout mice (25) using the Worthington Hepatocyte Isolation System (Worthington Biochemical). Prior to plating, Eif4ebp1 or Eif4ebp2 knockout hepatocytes were transfected with a bicistronic reporter plasmid containing the vascular endothelial growth factor (VEGF) IRES using an Amaxa Hepatocyte Nucleofector kit (Lonza). Hepatocytes were seeded onto plates coated with rat tail collagen in Williams E Medium (Gibco) supplemented with 5% FBS (Atlas), 1% penicillin/streptomycin, 1 μmol/L DMSO, 4 μg/mL insulin, 2 mmol/L GlutaMAX (Gibco), and 15 mmol/L HEPES, pH 7.4. After 6 h, cells were transferred to Williams E Medium supplemented with 0.5% penicillin/streptomycin, 0.1 μmol/L DMSO, 1% ITS+ (Gibco), 2 mmol/L GlutaMAX, and 15 mmol/L HEPES, pH 7.4, containing either 11 or 33 mmol/L glucose.
Cultures of wild-type and Eif4ebp1;Eif4ebp2 double knockout mouse embryo fibroblasts (MEFs) were maintained in Dulbecco’s modified Eagle’s medium lacking sodium pyruvate and containing either 25 or 5 mmol/L glucose plus 20 mmol/L mannitol as an osmotic control supplemented with 10% FBS and 1% penicillin/streptomycin. Transfections were performed using Xtremegene HP (Roche). Where indicated, cells were treated with 3 mmol/L glucosamine (Sigma) or 50 nmol/L thiamet G (Cayman) to enhance protein O-GlcNAcylation.
Pulsed stable isotope labeling by amino acids in cell culture.
Cultures of wild-type MEFs were maintained in Dulbecco’s modified Eagle’s medium designed for SILAC (Sigma). The medium was supplemented with 105 mg/L l-leucine, 146 mg/L l-lysine–HCl, 84 mg/L l-arginine, 10% dialyzed FBS (Sigma), and 1% penicillin/streptomycin. For the low-glucose condition, MEF cultures were maintained in 5 mmol/L glucose supplemented with 20 mmol/L mannitol and labeled for 6 h in medium supplemented as described above except that lysine was replaced with 4,4,5,5-D4- l-lysine and arginine with l-arginine-13C6. For the high-glucose condition, MEF cultures were maintained in 25 mmol/L glucose supplemented with 3 mmol/L glucosamine and labeled in medium containing l-lysine-13C615N2 and l-arginine-13C615N4. Cells were washed with PBS and harvested in 4% sodium dodecyl sulfate, 0.1 mol/L dithiothreitol, and 0.1 mol/L Tris-HCl pH 7.6. Aliquots of lysates (0.1 mg protein) from low and high glucose–treated cells were combined and prepared using filter-aided sample preparation methods as previously described (26) with the following modifications: washes were with 40% 2,2,2-trifluoroethanol (TFE) and 0.05 mol/L ammonium bicarbonate, pH 8.5; cysteine reduction was with 50 mmol/L tris(2-carboxyethyl)phosphine (TCEP), 40% TFE, and 0.05 mol/L ammonium bicarbonate, pH 8.5; and protein digestion was with endoproteinase LysC, trypsin, and 0.05 mol/L ammonium bicarbonate, pH 8.5.
Mass spectrometry analysis.
The proteolytic digest was subjected to two-dimensional liquid chromatography–tandem mass spectrometry separation and analysis using an ABSciex 5600 TripleTOF mass spectrometer with a Nanoflow source and ProteinPilot 4.2 Beta analysis software. Initial SCX fractionation was performed as previously described (27,28). Each strong cation exchange (SCX) fraction was dried down and resuspended in 2% (v/v) acetonitrile and 0.1% (v/v) formic acid prior to reverse-phase C18 nanoflow–liquid chromatography separation on a cHiPLC Nanoflex system equipped with a trap column (200 µm × 0.5 mm Reprosil-Pur C18-AQ 3 µm 120 Å) and a separation column (75 µm × 15 cm Reprosil-Pur C18-AQ 3 µm 120 Å), using a 185-min gradient of 0.1% formic acid with increasing percentages of acetonitrile containing 0.1% formic acid, delivered into a 5600 TripleTOF mass spectrometer using a NanoSpray III source (ABSciex) with a 10-μm ID nanospray tip (New Objective, Woburn, MA). Combined MS and MS/MS spectra were analyzed by ProteinPilot 4.2 Beta software searched against the complete Mouse RefSeq database from NCBI (27,303 sequences total, from 4 February 2012, concatenated to a reversed sequence Decoy database derived from the same RefSeq database, plus a list of 389 common laboratory contaminants). Protein identifications were accepted if they had an estimated local false discovery rate of <5%, using the Proteomics System Performance Evaluation Pipeline algorithm to calculate the false discovery rates (29).
Ingenuity Pathways Analysis.
The raw data obtained from the pulsed stable isotope labeling by amino acids in cell culture (pSILAC) analysis was uploaded and analyzed using proteome software Ingenuity Pathways Analysis (IPA) (www.ingenuity.com). A medium:high (M:H) cutoff of 0.5–1.5 was set to identify proteins whose synthesis rates were differentially regulated.
Statistical analysis.
The data are expressed as means + SE. Statistical analysis is described in the figure legends.
RESULTS
Diabetes promotes cap-independent translation and enhances 4E-BP1 expression.
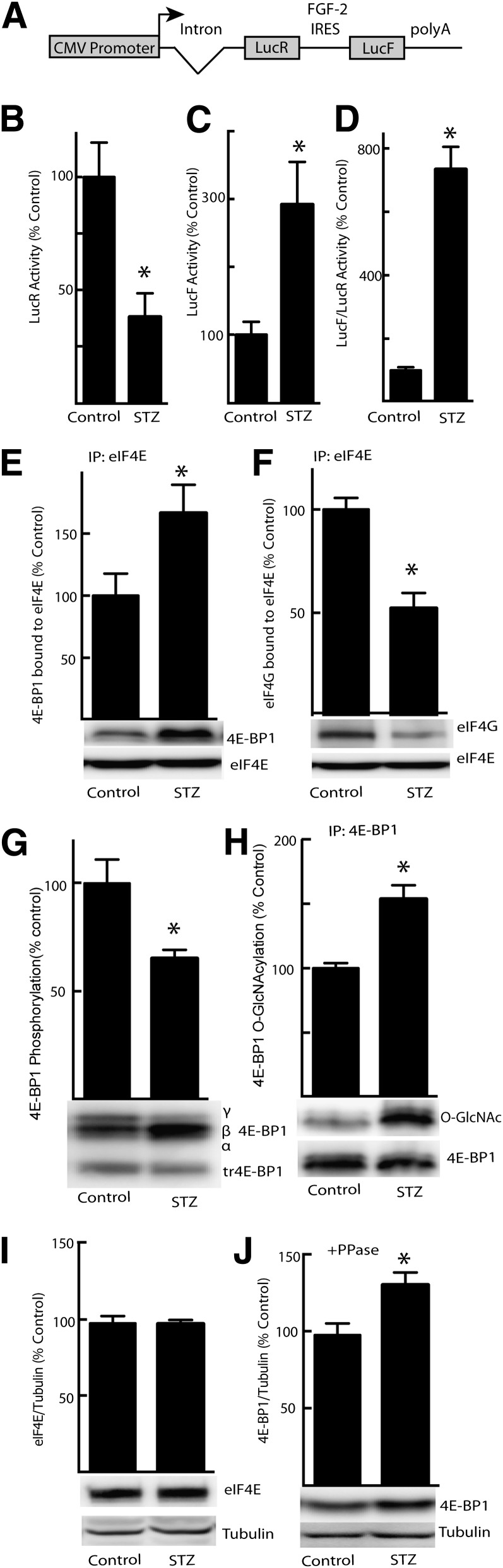
In the current study, we used a transgenic mouse model that expresses a bicistronic mRNA containing two open reading frames (ORFs) encoding two distinct luciferase enzymes: LucR and LucF. Translation of the first ORF (LucR) occurs in a cap-dependent manner, whereas translation of the second ORF (LucF) is driven by the FGF-2 IRES that is located between the two cistrons (Fig. 1A). Similar transgenic mice have been used to demonstrate that diabetes leads to upregulated expression of FGF-2 by increased utilization of the FGF-2 IRES in the aorta (30). Four weeks after the induction of diabetes, blood glucose levels were raised from 189 ± 12 to 540 ± 26 mg/dL and the activity of the LucR reporter was reduced by 62% in the liver of mice with STZ-induced type 1 diabetes compared with nondiabetic littermates (Fig. 1B), whereas LucF activity was elevated by 192% (Fig. 1C). As a result, the relative LucF-to-LucR activity ratio was enhanced by 633% (Fig. 1D). To investigate a potential mechanism for mediating the shift from cap-dependent to cap-independent translation in the liver of STZ-treated mice, the interaction of 4E-BP1 and eIF4G with eIF4E was evaluated. The amount of 4E-BP1 bound to eIF4E was elevated in the liver of diabetic compared with control mice (Fig. 1E), whereas the amount of eIF4G bound to eIF4E was reduced (Fig. 1F). These changes were accompanied by decreased 4E-BP1 phosphorylation (Fig. 1G) and increased O-GlcNAcylation (Fig. 1H). Whereas the expression of eIF4E in the liver of STZ-treated and control mice was not different (Fig. 1I), total hepatic 4E-BP1 expression was increased (Fig. 1J) and likely contributed to increased interaction with eIF4E.
FIG. 1.
STZ-induced diabetes produces a shift from cap-dependent to cap-independent translation. Liver supernatant fractions (1,000g) were prepared from transgenic mice after 4 weeks of STZ-induced diabetes or from control mice treated with vehicle only as described in research design and methods. A: Diagram of bicistronic transgene expressed by transgenic mice. B: Translation of LucR occurs in a cap-dependent manner. C: Translation of LucF is regulated by the FGF-2 IRES in a cap-independent manner. D: Ratio of LucF to LucR activity in liver supernatant fraction represents the relative translation from each reporter. The interaction of 4E-BP1 (E) and eIF4G (F) with eIF4E was examined by immunoprecipitating eIF4E and measuring the amount of 4E-BP1 and eIF4G in the immunoprecipitate (IP) by Western blot analysis. Phosphorylation (G) and O-GlcNAcylation (H) of 4E-BP1 as well as total eIF4E (I) and 4E-BP1 (J) content was measured by treating supernatant fractions with λ-phosphatase (PPase) followed by Western blot analysis as previously described (11). Values are means ± SE for a single experiment (n = 8). Statistical significance of the differences between means was assessed by Student t test and is denoted by *P < 0.05.
Role of hyperglycemia in promoting cap-independent translation.
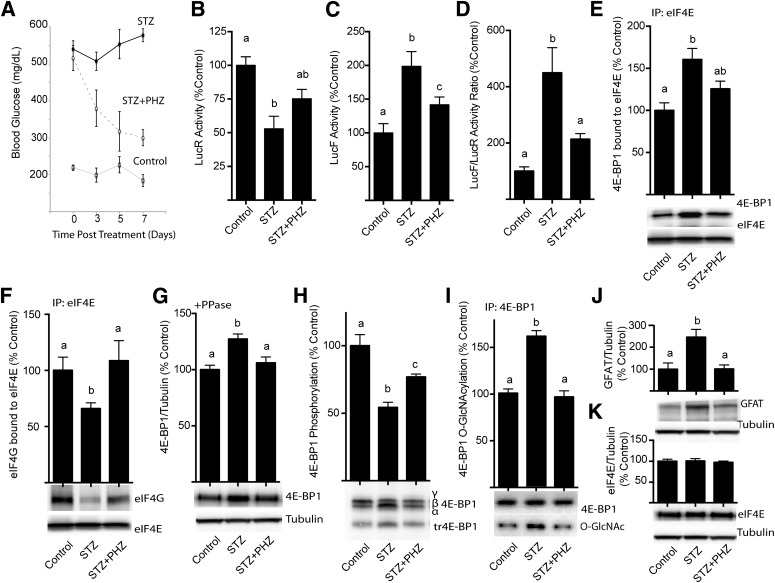
The role of hyperglycemia in mediating the diabetes-induced shift toward cap-independent translation was assessed using phlorizin treatment, which rapidly lowers blood glucose concentrations by blocking intestinal glucose absorption and producing renal glucosuria (31). Phlorizin treatment reduced the nonfasting blood glucose of mice with STZ-induced diabetes from a concentration of 539 ± 22 to 300 ± 16 mg/dL (Fig. 2A). LucR activity was repressed in the liver of diabetic compared with control mice, and upon phlorizin treatment its activity was elevated by 47% (Fig. 2A). Whereas LucF activity was elevated by 99% in diabetic compared with control mice, after treatment with phlorizin LucF activity was elevated by only 42% (Fig. 2C). Lowering blood glucose levels reduced the relative LucF-to-LucR ratio observed in the liver of diabetic mice by 52% (Fig. 2D). Phlorizin treatment of diabetic mice also reduced the amount of 4E-BP1 in eIF4E immunoprecipitates (Fig. 2E) and concomitantly elevated the interaction of eIF4E with eIF4G (Fig. 2F). The reduced interaction of 4E-BP1 with eIF4E upon phlorizin treatment was likely in part due to lower 4E-BP1 expression (Fig. 2G) as well as increased phosphorylation (Fig. 2H) and decreased O-GlcNAcylation (Fig. 2I) of 4E-BP1 compared with untreated diabetic mice. Similar to previous findings with Ins2Akita/+ diabetic mice (11), phlorizin treatment attenuated the elevated hepatic expression of glutamine-fructose-6-phosphate amidotransferase (GFAT) (Fig. 2J) but did not alter total eIF4E expression (Fig. 2K).
FIG. 2.
Phlorizin lowers blood glucose concentrations in STZ-treated mice and represses the diabetes-induced shift from cap-dependent to cap-independent translation in liver. Diabetes was induced in transgenic mice by STZ injection. Four weeks after the induction of diabetes, control and STZ mice were subcutaneously injected with solvent or phlorizin (PHZ) twice daily for 7 days to lower blood glucose concentrations (A). B and C: Luciferase activity was assessed in 1,000g supernatant fractions from liver extracts. Translation of LucR is under control of the cytomegalovirus (CMV) promoter in a cap-dependent manner, and LucF is regulated by the FGF-2 IRES. D: Ratio of LucF to LucR activity in liver supernatant fraction. The interaction of 4E-BP1 (E) and eIF4G (F) with eIF4E was examined by immunoprecipitating eIF4E and measuring the amount of each protein in the immunoprecipitate (IP) by Western blot analysis. G: 4E-BP1 content was measured by treating the supernatant fraction with λ-phosphatase (PPase) followed by Western blot analysis as previously described (11). H: Phosphorylation of 4E-BP1 was assessed as the ratio of the protein in the hyperphosphorylated γ-isoform to the total amount of 4E-BP1 in all isoforms. I: O-GlcNAcylation of 4E-BP1 was assessed by immunoprecipitating 4E-BP1 and measuring the amount of O-GlcNAcylation (O-GlcNAc) by Western blot relative to the total amount of 4E-BP1. Total content of GFAT (J) and eIF4E (K) was assessed by Western blot analysis. Values are means ± SE for two independent experiments (n = 8). Statistical significance was assessed by ANOVA followed by Holm-Sidak multiple-comparisons test to compare the mean of each group with the mean of every other group. Statistical significance is denoted by the presence of different letters above the bars on the graphs. Bars with different letters are statistically different; P < 0.05.
Hyperglycemia promotes cap-independent translation by increasing flux through the HBP.
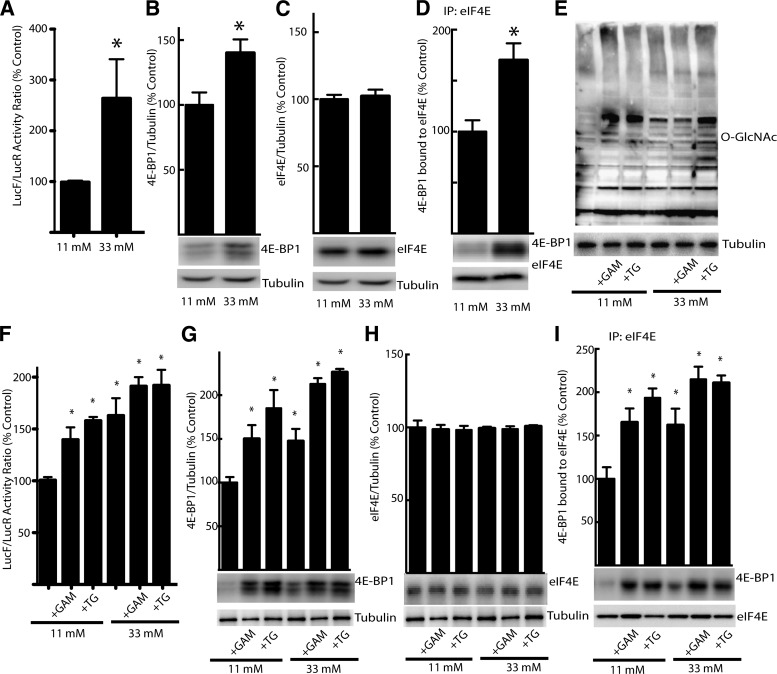
GFAT is the rate-limiting enzyme of the HBP and as such regulates the production of UDP N-Acetylglucosamine, which serves as the substrate for protein O-GlcNAcylation. Changes in GFAT expression suggest that flux through the HBP plays an important role in mediating the effects of hyperglycemia on 4E-BP1 O-GlcNAcylation and expression. For further evaluation of the role of hyperglycemia and the HBP in cap-dependent and cap-independent translation, hepatocytes isolated from the liver of the transgenic bicistronic reporter mice were incubated in medium containing either 11 or 33 mmol/L glucose to match blood glucose concentrations observed in control and STZ-treated mice. Hyperglycemic conditions dramatically enhanced the relative cap-independent translation (Fig. 3A) and 4E-BP1 expression (Fig. 3B) without altering eIF4E expression (Fig. 3C). Immunoprecipitation of eIF4E revealed elevated interaction of 4E-BP1 with eIF4E in hepatocytes maintained in the presence of high compared with low glucose (Fig. 3D). When hepatocytes maintained in low glucose medium were treated with glucosamine to directly stimulate the HBP or thiamet G, a potent and selective inhibitor of O-GlcNAcase that dramatically elevates protein O-GlcNAcylation (32) by bypassing GFAT, total protein O-GlcNAcylation was enhanced (Fig. 3E). Similarly, both glucosamine and thiamet G enhanced the LucF-to-LucR activity ratio in hepatocytes when maintained in low glucose medium (Fig. 3F). Both glucosamine and thiamet G enhanced expression of 4E-BP1 (Fig. 3G) but not eIF4E (Fig. 3H), and immunoprecipitation of eIF4E revealed elevated interaction of 4E-BP1 with eIF4E (Fig. 3I). Overall, these findings support the conclusion that the hyperglycemia-induced shift from cap-dependent to cap-independent luciferase reporter activity was mediated by changes in protein O-GlcNAcylation that coincide with alterations in the expression of 4E-BP1 and its interaction with eIF4E.
FIG. 3.
Hyperglycemic conditions mediate a shift in translation of FGF-2 reporter from cap-dependent to cap-independent translation in isolated hepatocytes and enhance 4E-BP1 expression via the HBP. For evaluation of the role of the HBP, hepatocytes isolated from transgenic mice were maintained in medium containing 11 or 33 mmol/L glucose for 24 h followed by exposure to 3 mmol/L glucosamine (GAM), which feeds directly into the pathway, or thiamet G (TG), an inhibitor of O-GlcNAcase. A and F: The relative translation of LucR and LucF was evaluated. The expression of 4E-BP1 (B and G) as well as eIF4E (C and H) was assessed in 1,000g cell supernatant fractions by Western blot analysis. D and I: The interaction of 4E-BP1 with eIF4E was examined by immunoprecipitating eIF4E and measuring the amount of 4E-BP1 in the immunoprecipitate (IP) by Western blot analysis. E: Total protein O-GlcNAcylation (O-GlcNAc) was assessed in cell supernatant fractions by Western blot. Values are means + SE for two independent experiments (n = 4). Statistical significance of the differences between means was assessed by Student t test and is denoted by *P < 0.05.
Ablation of 4E-BP1/2 prevents hyperglycemia-mediated shift from cap-dependent to cap-independent translation.
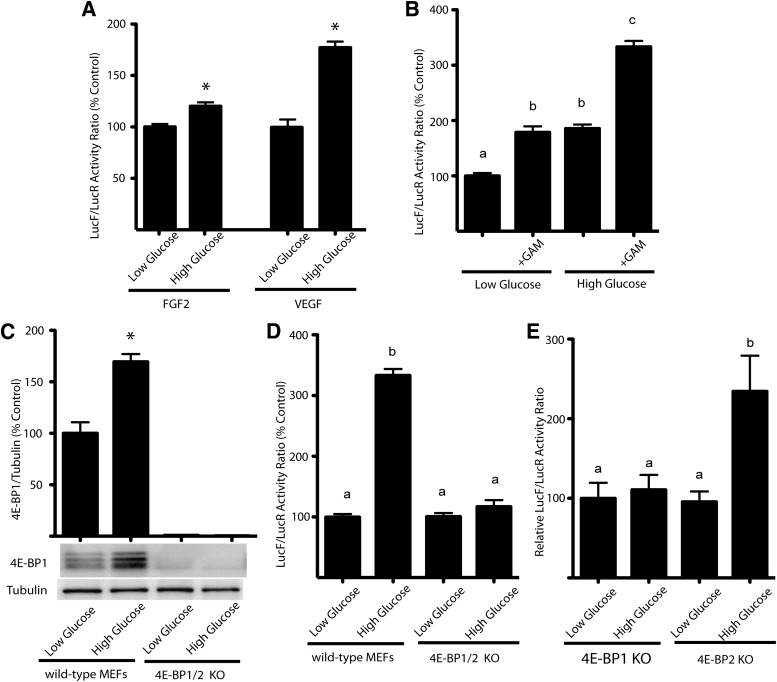
The role of hyperglycemia in modulating the shift from cap-dependent to cap-independent translation was further assessed using MEFs transiently transfected with bicistronic luciferase constructs in which the activity of LucR occurs in a cap-dependent manner and the activity of LucF is regulated by either the FGF-2 or the VEGF. Under high- compared with low-glucose conditions, the ratio of LucF to LucR activity increased by 20% for the FGF-2 IRES and 77% for the VEGF IRES (Fig. 4A). Further, the ratio of LucF to LucR activity with the VEGF IRES also increased by 79% when cells maintained in low-glucose medium were treated with glucosamine (Fig. 4B). When wild-type and 4E-BP1/2 knockout (Eif4ebp1;Eif4ebp2) MEFs were analyzed after transient transfection with the bicistronic luciferase constructs containing the VEGF IRES, the expression of 4E-BP1 was increased by 72% in cells maintained in medium containing high glucose and treated with glucosamine compared with the low-glucose condition (Fig. 4C). There was no detectable 4E-BP1 under either condition in knockout cells. Exposure to high glucose and glucosamine increased the ratio of LucF to LucR activity in wild-type cells to 333% that of the low-glucose condition. In contrast, there was no effect on the ratio of LucF to LucR activity with treatment of 4E-BP1/2 knockout cells (Fig. 4D). For confirmation of the role of 4E-BP1 specifically, hepatocytes from 4E-BP1/2 knockout mice were transfected with a bicistronic plasmid containing the IRES for VEGF and maintained in medium containing either low glucose or high glucose with glucosamine. There was no significant difference in the ratio of LucF to LucR activity under the high- and low-glucose conditions for 4E-BP1 knockout hepatocytes, whereas high glucose increased the ratio of LucF to LucR activity in 4E-BP2 knockout hepatocytes to 234% that of the low-glucose condition (Fig. 4E). These results demonstrate that expression of 4E-BP1 but not 4E-BP2 is necessary for the hyperglycemia-induced shift from cap-dependent to cap-independent translation.
FIG. 4.
Hyperglycemic conditions mediate a shift from cap-dependent to cap-independent translation that is dependent on 4E-BP1. A: MEFs were incubated in medium containing high (25 mmol/L glucose) or low (5 mmol/L glucose, 20 mmol/L mannitol) glucose. Cells were transiently transfected with a bicistronic plasmid containing the IRES for either FGF-2 or VEGF. Whole-cell lysates were harvested, and the relative translation of LucR and LucF was evaluated. B: MEF cells were transiently transfected with bicistronic luciferase plasmid containing the VEGF IRES and were incubated in either 5 or 25 mmol/L glucose in the presence and absence of 3 mmol/L glucosamine. C: Wild-type and Eif4ebp1;Eif4ebp2 double knockout MEFs were incubated in medium containing either low glucose or high glucose with 3 mmol/L glucosamine. The content of 4E-BP1 relative to tubulin was assessed in whole-cell lysates by Western blot analysis. D: Whole-cell lysates were prepared, and the relative translation of LucR and LucF was evaluated. Values are means ± SE for two independent experiments (n = 4). E: Hepatocytes from Eif4ebp1;Eif4ebp2 knockout mice were transfected with a bicistronic plasmid containing the IRES for VEGF and maintained in medium containing either low glucose or high glucose with 3 mmol/L glucosamine. Values are means ± SE for two independent experiments (n = 3). Statistical significance was assessed by either Student t test or ANOVA followed by Holm-Sidak multiple-comparisons test to compare the mean of each group with the mean of every other group. Statistical significance is denoted by * (A and C) or the presence of different letters above the bars on the graphs (B, D, and E). Bars with different letters are statistically different; P < 0.05.
pSILAC identifies proteins with altered synthetic rates under hyperglycemic conditions.
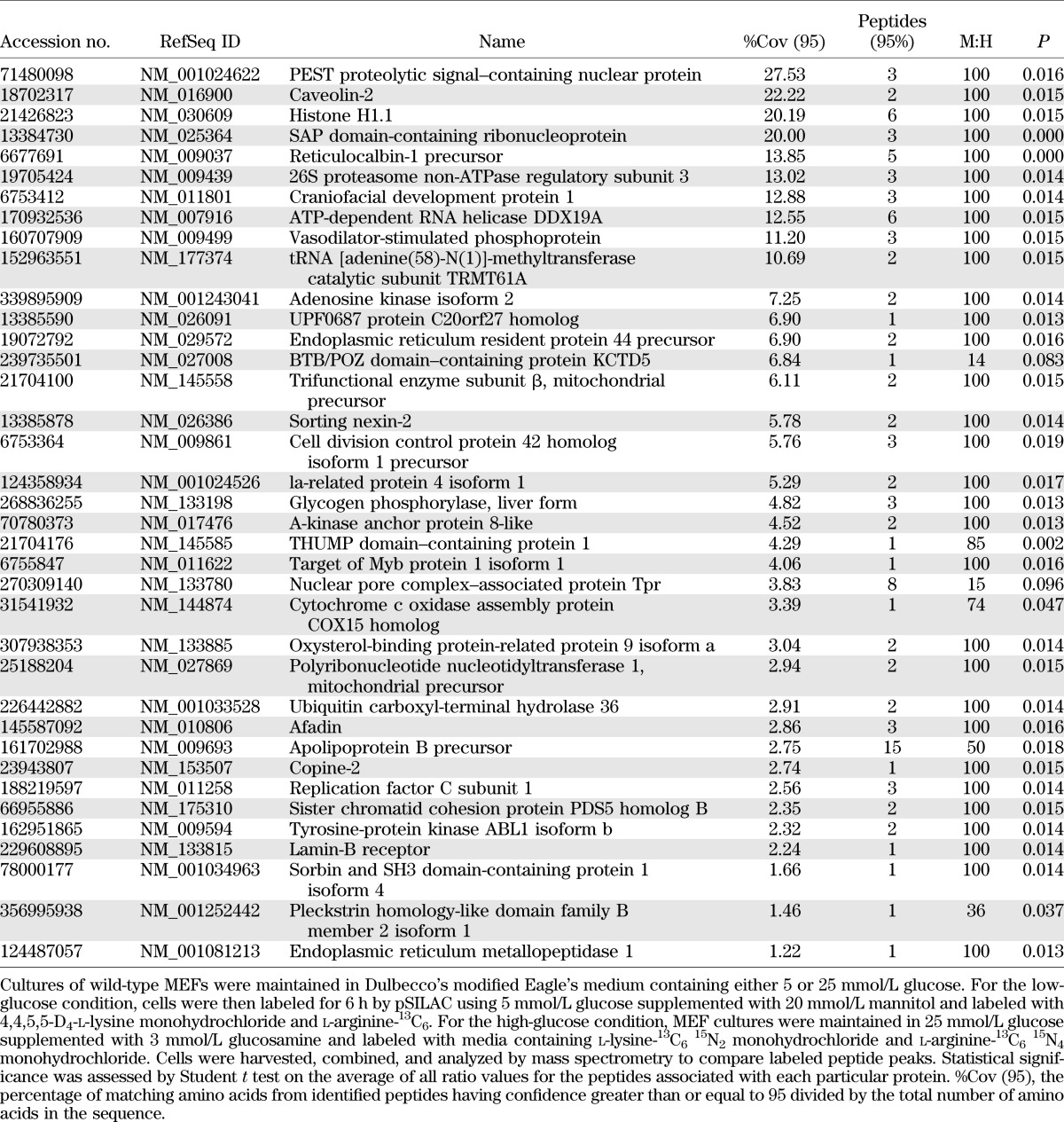
To identify hyperglycemia-induced changes in the translational control of protein expression on a global scale, we used pSILAC to evaluate newly synthesized proteins and quantitate changes in their accumulation rates in MEFs in the presence of either low glucose or high glucose supplemented with glucosamine, as these conditions produced the largest increase in the LucF-to-LucR ratio (Fig. 4B). In total, 1,355 and 2,905 proteins were identified with high confidence in two independent runs. While the majority of proteins had medium-to-heavy ratios of ~1:1, indicating that their translation was not significantly altered by hyperglycemic conditions, the synthesis of a number of proteins was significantly altered between the two conditions. The top-scoring proteins whose synthesis was either upregulated or repressed under high- compared with low-glucose condition are listed in Tables 1 and 2, respectively.
TABLE 1.
Top proteins identified by pSILAC with elevated synthetic rates in cells exposed to hyperglycemic conditions
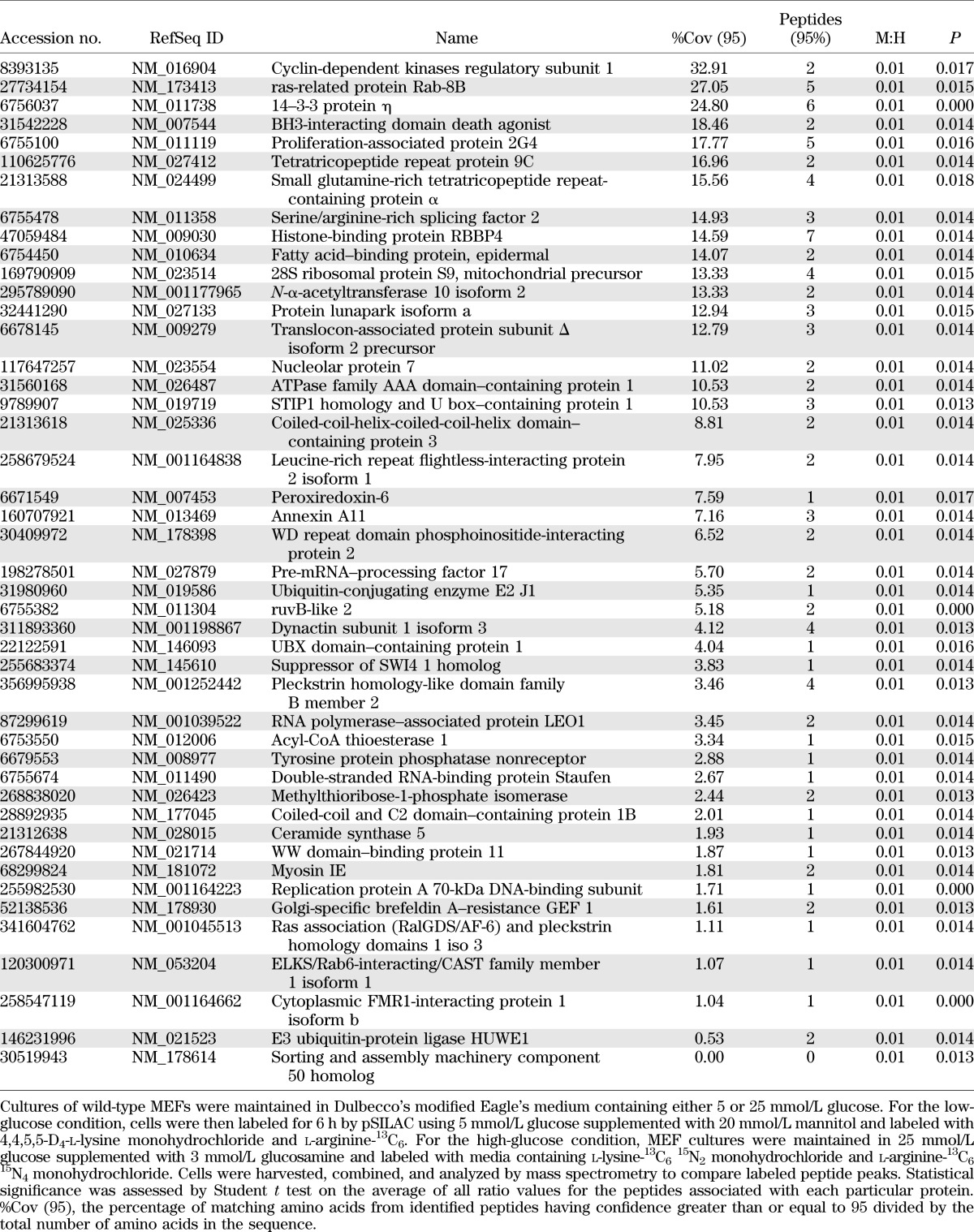
TABLE 2.
Top proteins identified by pSILAC with repressed synthetic rates in cells exposed to hyperglycemic conditions
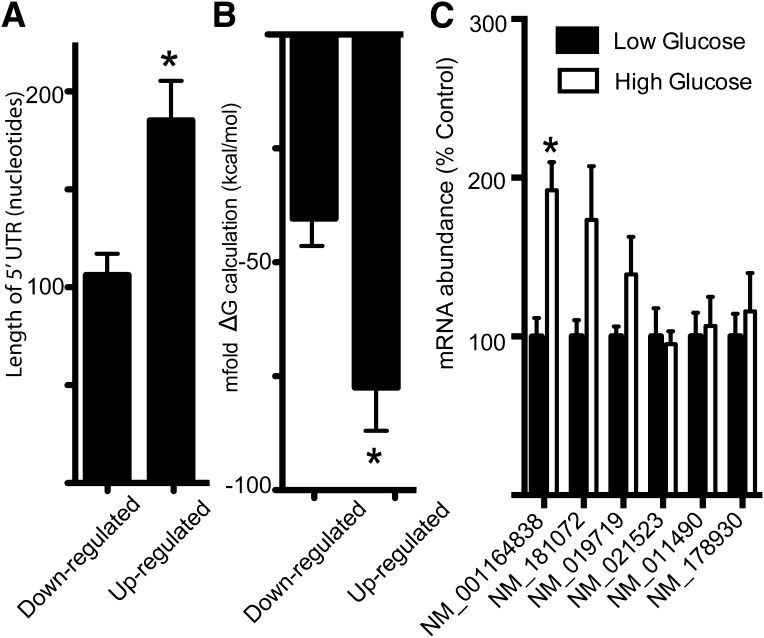
In the classic model of cap-dependent translation initiation, ribosome scanning of the 5′-UTR is impaired by strong secondary structures. Characteristics that decrease the efficiency of cap-dependent translation include the length of the 5′-UTR and the complexity of secondary structure (33). Thus, we predicted that the high-glucose condition would repress the translation of mRNAs with less complex 5′-UTRs in favor of mRNAs with more structured 5′-UTRs. Most cellular IRES elements are 150–300 nucleotides in length, although some are as short as 22 nucleotides (34). The mRNA-encoding proteins whose synthesis was upregulated by high glucose averaged 185.3 nucleotides in length and were 74% longer than the average number of nucleotides in 5′-UTRs of mRNAs encoding proteins whose synthesis was downregulated by high glucose (Fig. 5A). To investigate the complexity of these 5′-UTRs, we used mfold (http://mfold.rna.albany.edu/?q=mfold) to compute secondary structures and calculate a corresponding free energy change for folding (ΔG). The top-scoring proteins whose synthesis was downregulated by high-glucose conditions have an average ΔG of −41.1 kcal/mol, whereas the top-scoring proteins whose synthesis was upregulated by high-glucose conditions have a significantly lower average ΔG value of −77.5 kcal/mol, indicative of more stable secondary structures (Fig. 5B). To verify that the hyperglycemia-induced enhancement in the synthetic rates of proteins with longer 5′-UTRs was not the result of elevated mRNA transcription, we evaluated the abundance of mRNAs corresponding to six proteins from Table 1 with the longest 5′-UTRs (Fig. 5C). Of the six analyzed, only one exhibited increased abundance, and that was 50-fold less than the accumulation of newly synthesized protein. Changes in protein degradation may also impact the M-to-H ratio observed during pSILAC; however, its contribution is relatively small (35). These findings suggest that hyperglycemia favors the translation of messages with longer/more highly structured 5′-UTRs. Although no proteins encoded by mRNAs containing known IRES structures were identified among the top-scoring upregulated proteins, several proteins were identified whose mRNAs are listed on IRESite, the database of experimentally verified IRES structures (36). Among these, cellular inhibitor of apoptosis 1 (c-IAP1) (gi|133922596), heat shock 70-kDa protein 1A (HSPA1A) (gi|40254361), cold shock domain containing E1 (UNR) (gi|240255574), and utrophin A (UTRA) (gi|110431378) were all elevated by nearly twofold in cells exposed to high-glucose conditions in two independent runs, while BCL2 (gi|6753200) increased by nearly 80-fold in the second run but was not detected in the initial run.
FIG. 5.
Hyperglycemic conditions upregulate translation of mRNAs with more complex 5′-UTRs. The database of transcriptional start sites (dbTSS [http://dbtss.hgc.jp/]) was used to identify the 5′-UTRs of the top-scoring proteins identified by pSILAC with altered expression after incubation of MEFs in medium containing either 5 mmol/L glucose supplemented with 20 mmol/L mannitol or with 25 mmol/L glucose supplemented with 3 mmol/L glucosamine. The list of these proteins can be found in Tables 1 and 2. A: The average 5′-UTR length for the mRNAs of the top-scoring proteins was calculated. B: Secondary structures of 5′-UTRs for the mRNAs of the top-scoring proteins were computed with mfold (http://mfold.rna.albany.edu/?q=mfold) to generate a corresponding free energy change for folding (ΔG). For reference, Kozak (45) found that 5′ sequences with predicted ΔG values of −30 kcal/mol had no effect on translation, whereas those with predicted ΔG values of −50 kcal/mol reduced cap-dependent translation by 85–95%, suggesting that the 43S PIC can melt moderately stable structures but stalls at more stable structures. C: RNA was extracted from MEF whole-cell lysates after a standard TRIzol protocol. mRNA expression was normalized to internal controls for glyceraldehyde-3-phosphate dehydrogenase and α-tubulin. All values are means ± SE for two independent experiments (n = 37–45 for A and B; n = 6 for C). Statistical significance of the differences between means was assessed by Student t test and is denoted as *P < 0.05.
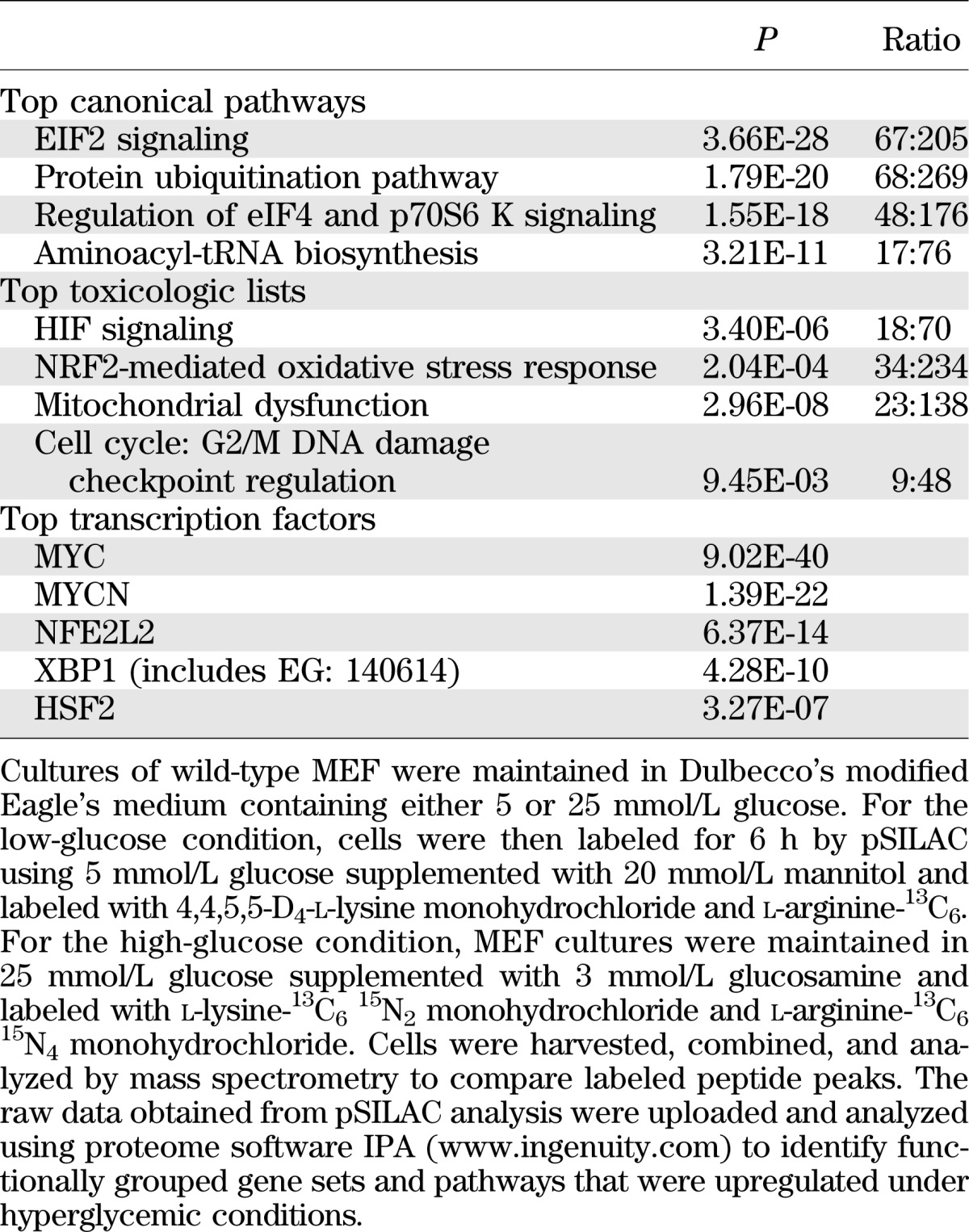
Functional relationships between genes that were differentially expressed in low and high glucose were investigated with IPA. IPA showed that the proteins whose accumulation was upregulated are associated with pathways that regulate eIF2, eIF4F, and S6K1 signaling; protein ubiquitination; and aminoacyl-tRNA biosynthesis (Table 3). Further, the most significant toxicologic list pathways were hypoxia-inducible factor (HIF) signaling, oxidative stress mediated by nuclear factor erythroid 2–related factor 2 (Nrf2), mitochondrial dysfunction, and DNA damage checkpoint regulation of cell cycle phase G2/M (Table 3). Both HIF-1 and Nrf2 contain IRES sequences in their 5′-UTR that allow their translation to be maintained under stress conditions that are inhibitory to cap-dependent translation (6,37). IPA was also used to predict which mRNAs encoding transcription factors were most likely responsible for the altered pattern of expression under low- and high-glucose conditions. The top transcription factor altered by high glucose was the proto-oncogene myc (Table 3), whose 5′-UTR contains an IRES. Activation of myc was predicted under high-glucose conditions based on a broad network of interacting proteins (Supplementary Fig. 1).
TABLE 3.
IPA of protein synthetic rates in cells exposed to hyperglycemic conditions
DISCUSSION
The findings of the current study provide insight into a novel mechanism through which diabetes-induced hyperglycemia alters the selection of mRNA for translation. In the liver of diabetic mice, 4E-BP1 exhibited reduced phosphorylation, elevated O-GlcNAcylation, and enhanced interaction with eIF4E compared with nondiabetic mice. These alterations in 4E-BP1 were associated with a shift from cap-dependent to cap-independent translation using bicistronic luciferase reporter assays. For demonstration of the component of the diabetic state responsible for this shift, diabetic mice were treated with phlorizin to reduce blood glucose concentrations. When the blood glucose concentration was lowered, the interaction of 4E-BP1 with eIF4E returned to nondiabetic levels and the shift from cap-dependent to cap-independent reporter activity was reversed. A similar shift toward cap-independent mRNA translation was observed with cells in culture upon exposure to hyperglycemic conditions or under conditions that promoted protein O-GlcNAcylation. O-GlcNAcylation of 4E-BP1 correlated with the hyperglycemia-induced shift from cap-dependent to cap-independent translation, and expression of 4E-BP1 was necessary for this effect. We extended these findings using pSILAC to identify novel proteins that undergo altered rates of synthesis in high- versus low-glucose conditions. Examination of the expression pattern of these proteins revealed a hyperglycemia-induced shift toward proteins with more complex 5′-UTRs. Overall, the results are consistent with a model wherein the O-GlcNAcylation of 4E-BP1 results in elevated expression and promotes its interaction with eIF4E (11), thereby altering gene expression in response to hyperglycemia and diabetes.
Regulation of the initiation complex eIF4F by posttranslational modification is of critical importance in the selection of mRNAs for cap-dependent translation initiation. Binding of hypophosphorylated 4E-BP1 to eIF4E, which is mutually exclusive of interaction with eIF4G, prevents assembly of functional eIF4F complexes to repress loading of ribosomes onto the mRNA 5′-cap. The interaction of 4E-BP1 with eIF4E is not only regulated by phosphorylation; O-GlcNAcylation of 4E-BP1 also enhances its binding to eIF4E independent of 4E-BP1 phosphorylation status (10). Thus, hyperglycemia-induced sequestration of eIF4E potentially downregulates the synthesis of a broad array of proteins. However, under conditions where eIF4E is limiting, the same canonical initiation factors can facilitate the cap-independent loading of ribosomes onto mRNA that contain IRES elements (8). Therefore, by reducing the competition from capped mRNAs, hyperglycemia-induced sequestration of eIF4E potentially acts as a positive regulator of cap-independent translation in response to conditions of stress.
A critical role for 4E-BP1 in upregulating cap-independent translation has previously been demonstrated (38). Viral IRES-mediated translation is promoted when cellular cap-dependent translation is diminished under conditions of cellular stress (38). However, 4E-BP1 knockdown prevents upregulation of viral cap-independent translation under conditions of amino acid starvation (39). Furthermore, 4E-BP1 plays a crucial role in mediating differential protein expression during hypoxia (40). Overexpressed 4E-BP1 and eIF4G mediate a hypoxia-activated switch from cap-dependent to cap-independent mRNA translation that promotes increased tumor angiogenesis and growth (41). It has been previously demonstrated that the presence of 4E-BP1/2 is also necessary for increased expression of VEGF in both the retina during diabetes and cells maintained under hyperglycemic conditions (42). The results of the current study suggest that increased VEGF expression occurs through increased use of internal ribosome entry sites in response to increased flux through the HBP. This finding likely extends to other mRNAs that are translated through an IRES-dependent mechanism, as overexpression of O-GlcNAcylation transferase produces an accumulation of 80S monosomes, suggesting that excessive O-GlcNAcylation suppresses translation initiation (23).
To explore the effect of hyperglycemia on mRNA translation, we used pSILAC to identify proteins that undergo altered rates of synthesis. Hyperglycemia promoted translation of mRNAs that contained long and structured 5′-UTRs, a characteristic associated with mRNAs containing IRES. This observation does not directly indicate that hyperglycemic conditions favor translation of all mRNAs with IRES domains. Instead, it is more likely that hyperglycemia-induced modification of the translational machinery enhances translation of a specific subset of messages, some of which contain IRES domains. In this study, hyperglycemia specifically enhanced IRES-mediated translation from luciferase reporters driven by either the FGF-2 or VEGF IRES sequence. Using pSILAC, we also observed increased translation of the following cellular mRNAs that have been previously shown to contain IRES structures: B-cell lymphoma 2 (BCL2), 78-kDa glucose-regulated protein (BiP), c-IAP1, HSPA1A, Runt-related transcription factor 1 (RunX1), UNR, and UTRA. IRES containing mRNAs often encode proteins with crucial biological functions, such as the major vascular growth factors VEGF, FGF-2, platelet-derived growth factor, and RunX1. Thus, enhanced hyperglycemia-mediated translation of angiogenic proteins potentially disrupts the relative balance of inducers and inhibitors of angiogenesis to promote the neurovascular complications associated with diabetes. Furthermore, our data support a previous microarray analysis of the retinal transcriptome where normalization of systemic glycemia in diabetic rats primarily restored pathways associated with growth factor signaling (43).
A functional link between the metabolic abnormalities associated with disease and regulation of IRES sequences has previously been proposed (44), yet direct evidence remains limiting. In the current study, we find that hyperglycemia and conditions that promote protein O-GlcNAcylation enhance IRES-mediated mRNA translation through a mechanism that is dependent on 4E-BP1. Thus, regulation of 4E-BP1 expression by O-GlcNAcylation represents a novel mechanism for altering the selection of mRNAs for translation under pathophysiological conditions. Elucidation of molecular mechanisms underlying cap-independent translation initiation will not only enhance our understanding of gene expression but also impact the development of treatment strategies for addressing the pathophysiology of diabetes.
Supplementary Material
ACKNOWLEDGMENTS
The research reported here was supported by National Institutes of Health grants DK088416 (to M.D.D.) and DK13499 (to L.S.J.).
No potential conflicts of interest relevant to this article were reported.
M.D.D. researched data and wrote and edited the manuscript. J.S.S. contributed to discussion and reviewed the manuscript. B.A.S. researched data, wrote the manuscript, contributed to discussion, and reviewed and edited the manuscript. S.R.K. and L.S.J. designed experiments, contributed to discussion, and reviewed and edited the manuscript. M.D.D. is the guarantor of this work and as such had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Parts of this study were presented in abstract form at the 72nd Scientific Sessions of the American Diabetes Association, Philadelphia, Pennsylvania, 8–12 June 2012.
The authors thank Dr. N. Sonenberg (McGill University) for kindly providing 4E-BP1/2 DKO MEFs and Dr. A.-C. Prats (INSERM) for providing the bicistronic reporter containing the FGF-2 IRES. The authors also thank W. Dunton (Pennsylvania State University College of Medicine) and M. Wu (Pennsylvania State University College of Medicine) for assistance with animals.
REFERENCES
- 1.Pestova TV, Kolupaeva VG, Lomakin IB, et al. Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci USA 2001;98:7029–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.López-Lastra M, Rivas A, Barría MI. Protein synthesis in eukaryotes: the growing biological relevance of cap-independent translation initiation. Biol Res 2005;38:121–146 [DOI] [PubMed] [Google Scholar]
- 3.Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev 2001;15:1593–1612 [DOI] [PubMed] [Google Scholar]
- 4.Holcik M, Sonenberg N, Korneluk RG. Internal ribosome initiation of translation and the control of cell death. Trends Genet 2000;16:469–473 [DOI] [PubMed] [Google Scholar]
- 5.Stein I, Itin A, Einat P, Skaliter R, Grossman Z, Keshet E. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol Cell Biol 1998;18:3112–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lang KJ, Kappel A, Goodall GJ. Hypoxia-inducible factor-1alpha mRNA contains an internal ribosome entry site that allows efficient translation during normoxia and hypoxia. Mol Biol Cell 2002;13:1792–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandez J, Yaman I, Mishra R, et al. Internal ribosome entry site-mediated translation of a mammalian mRNA is regulated by amino acid availability. J Biol Chem 2001;276:12285–12291 [DOI] [PubMed] [Google Scholar]
- 8.Pestova TV, Hellen CU, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol 1996;16:6859–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Svitkin YV, Herdy B, Costa-Mattioli M, Gingras AC, Raught B, Sonenberg N. Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol Cell Biol 2005;25:10556–10565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gingras AC, Kennedy SG, O’Leary MA, Sonenberg N, Hay N. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes Dev 1998;12:502–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dennis MD, Schrufer TL, Bronson SK, Kimball SR, Jefferson LS. Hyperglycemia-induced O-GlcNAcylation and truncation of 4E-BP1 protein in liver of a mouse model of type 1 diabetes. J Biol Chem 2011;286:34286–34297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu K, Paterson AJ, Chin E, Kudlow JE. Glucose stimulates protein modification by O-linked GlcNAc in pancreatic beta cells: linkage of O-linked GlcNAc to beta cell death. Proc Natl Acad Sci USA 2000;97:2820–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Issad T, Masson E, Pagesy P. O-GlcNAc modification, insulin signaling and diabetic complications. Diabetes Metab 2010;36:423–435 [DOI] [PubMed] [Google Scholar]
- 14.Duverger E, Roche AC, Monsigny M. N-acetylglucosamine-dependent nuclear import of neoglycoproteins. Glycobiology 1996;6:381–386 [DOI] [PubMed] [Google Scholar]
- 15.Roos MD, Su K, Baker JR, Kudlow JE. O glycosylation of an Sp1-derived peptide blocks known Sp1 protein interactions. Mol Cell Biol 1997;17:6472–6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao Y, Miyazaki J, Hart GW. The transcription factor PDX-1 is post-translationally modified by O-linked N-acetylglucosamine and this modification is correlated with its DNA binding activity and insulin secretion in min6 beta-cells. Arch Biochem Biophys 2003;415:155–163 [DOI] [PubMed] [Google Scholar]
- 17.Soesanto YA, Luo B, Jones D, et al. Regulation of Akt signaling by O-GlcNAc in euglycemia. Am J Physiol Endocrinol Metab 2008;295:E974–E980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han I, Kudlow JE. Reduced O glycosylation of Sp1 is associated with increased proteasome susceptibility. Mol Cell Biol 1997;17:2550–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeidan Q, Hart GW. The intersections between O-GlcNAcylation and phosphorylation: implications for multiple signaling pathways. J Cell Sci 2010;123:13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X, Ongusaha PP, Miles PD, et al. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature 2008;451:964–969 [DOI] [PubMed] [Google Scholar]
- 21.Griffith LS, Schmitz B. O-linked N-acetylglucosamine levels in cerebellar neurons respond reciprocally to pertubations of phosphorylation. Eur J Biochem 1999;262:824–831 [DOI] [PubMed] [Google Scholar]
- 22.Cheng X, Cole RN, Zaia J, Hart GW. Alternative O-glycosylation/O-phosphorylation of the murine estrogen receptor beta. Biochemistry 2000;39:11609–11620 [DOI] [PubMed] [Google Scholar]
- 23.Zeidan Q, Wang Z, De Maio A, Hart GW. O-GlcNAc cycling enzymes associate with the translational machinery and modify core ribosomal proteins. Mol Biol Cell 2010;21:1922–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Créancier L, Morello D, Mercier P, Prats AC. Fibroblast growth factor 2 internal ribosome entry site (IRES) activity ex vivo and in transgenic mice reveals a stringent tissue-specific regulation. J Cell Biol 2000;150:275–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Bacquer O, Petroulakis E, Paglialunga S, et al. Elevated sensitivity to diet-induced obesity and insulin resistance in mice lacking 4E-BP1 and 4E-BP2. J Clin Invest 2007;117:387–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods 2009;6:359–362 [DOI] [PubMed] [Google Scholar]
- 27.Moon MS, McDevitt EI, Zhu J, et al. Elevated hepatic iron activates NF-E2-related factor 2-regulated pathway in a dietary iron overload mouse model. Toxicol Sci 2012;129:74–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giardina BJ, Stanley BA, Chiang HL. Comparative Proteomic Analysis of Transition of Saccharomyces cerevisiae from Glucose-Deficient Medium to Glucose-Rich Medium. Proteome Sci 2012;10:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang WH, Shilov IV, Seymour SL. Nonlinear fitting method for determining local false discovery rates from decoy database searches. J Proteome Res 2008;7:3661–3667 [DOI] [PubMed] [Google Scholar]
- 30.Teshima-Kondo S, Kondo K, Prado-Lourenco L, et al. Hyperglycemia upregulates translation of the fibroblast growth factor 2 mRNA in mouse aorta via internal ribosome entry site. FASEB J 2004;18:1583–1585 [DOI] [PubMed] [Google Scholar]
- 31.Oulianova N, Falk S, Berteloot A. Two-step mechanism of phlorizin binding to the SGLT1 protein in the kidney. J Membr Biol 2001;179:223–242 [DOI] [PubMed] [Google Scholar]
- 32.Yuzwa SA, Macauley MS, Heinonen JE, et al. A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat Chem Biol 2008;4:483–490 [DOI] [PubMed] [Google Scholar]
- 33.Komar AA, Mazumder B, Merrick WC. A new framework for understanding IRES-mediated translation. Gene 2012;502:75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Komar AA, Hatzoglou M. Internal ribosome entry sites in cellular mRNAs: mystery of their existence. J Biol Chem 2005;280:23425–23428 [DOI] [PubMed] [Google Scholar]
- 35.Schwanhäusser B, Busse D, Li N, et al. Global quantification of mammalian gene expression control. Nature 2011;473:337–342 [DOI] [PubMed] [Google Scholar]
- 36.Mokrejs M, Vopálenský V, Kolenaty O, et al. IRESite: the database of experimentally verified IRES structures (www.iresite.org). Nucleic Acids Res 2006;34:D125–D130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li W, Thakor N, Xu EY, et al. An internal ribosomal entry site mediates redox-sensitive translation of Nrf2. Nucleic Acids Res 2010;38:778–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yip CK, Murata K, Walz T, Sabatini DM, Kang SA. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol Cell 2010;38:768–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Licursi M, Komatsu Y, Pongnopparat T, Hirasawa K. Promotion of viral internal ribosomal entry site-mediated translation under amino acid starvation. J Gen Virol 2012;93:951–962 [DOI] [PubMed] [Google Scholar]
- 40.Magagnin MG, van den Beucken T, Sergeant K, et al. The mTOR target 4E-BP1 contributes to differential protein expression during normoxia and hypoxia through changes in mRNA translation efficiency. Proteomics 2008;8:1019–1028 [DOI] [PubMed] [Google Scholar]
- 41.Braunstein S, Karpisheva K, Pola C, et al. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol Cell 2007;28:501–512 [DOI] [PubMed] [Google Scholar]
- 42.Schrufer TL, Antonetti DA, Sonenberg N, Kimball SR, Gardner TW, Jefferson LS. Ablation of 4E-BP1/2 prevents hyperglycemia-mediated induction of VEGF expression in the rodent retina and in Muller cells in culture. Diabetes 2010;59:2107–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fort PE, Losiewicz MK, Reiter CE, et al. Differential roles of hyperglycemia and hypoinsulinemia in diabetes induced retinal cell death: evidence for retinal insulin resistance. PLoS ONE 2011;6:e26498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holcík M. Targeting translation for treatment of cancer—a novel role for IRES? Curr Cancer Drug Targets 2004;4:299–311 [DOI] [PubMed] [Google Scholar]
- 45.Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc Natl Acad Sci USA 1986;83:2850–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.