Abstract
Salivary gland tumors constitute a heterogeneous group of uncommon diseases that pose significant diagnostic and therapeutic challenges. However, the recent discovery of a translocation-generated gene fusion network in salivary gland carcinomas as well in benign salivary gland tumors opens up new avenues for improved diagnosis, prognostication, and development of specific targeted therapies. The gene fusions encode novel fusion oncoproteins or ectopically expressed normal or truncated oncoproteins. The major targets of the translocations are transcriptional coactivators, tyrosine kinase receptors, and transcription factors involved in growth factor signaling and cell cycle regulation. Notably, several of these targets or pathways activated by these targets are druggable. Examples of clinically significant gene fusions in salivary gland cancers are the MYB–NFIB fusion specific for adenoid cystic carcinoma, the CRTC1–MAML2 fusion typical of low/intermediate-grade mucoepidermoid carcinoma, and the recently identified ETV6–NTRK3 fusion in mammary analogue secretory carcinoma. Similarly, gene fusions involving the PLAG1 and HMGA2 oncogenes are specific for benign pleomorphic adenomas. Continued studies of the molecular consequences of these fusion oncoproteins and their down-stream targets will ultimately lead to the identification of novel driver genes in salivary gland neoplasms and will also form the basis for the development of new therapeutic strategies for salivary gland cancers and, perhaps, other neoplasms.
Keywords: Fusion oncogenes, Salivary gland neoplasms, Adenoid cystic carcinoma, Mucoepidermoid carcinoma, Biomarker, Targeted therapy, MYB–NFIB, CRTC1–MAML2, ETV6–NTRK3
Introduction
Chromosome rearrangements, in particular translocations, may result in fusion oncogenes encoding oncoproteins with transforming properties. More than 800 fusion oncogenes have thus far been described in a variety of human neoplasms and many of these are now being recognized as important diagnostic and prognostic biomarkers and as novel targets for therapy [1, 2]. Recent studies have shown that gene fusions may account for at least 20 % of human cancer morbidity. The majority of gene fusions are found in leukemias and sarcomas and comparatively few in carcinomas [1]. The reason for this difference is not fully known but may at least partly be due to the fact that recurrent balanced chromosome rearrangements are rare in carcinomas whereas they are common in leukemias and sarcomas [2]. In contrast, several carcinomas have recently been shown to express fusion oncogenes resulting from submicroscopic, often intrachromosomal, rearrangements [3–7]. The most prominent of these are the ETS (ERG or ETV1) and TMPRRS2 fusions found in a high frequency of prostate cancer [4]. Several intrachromosomally generated gene fusions have also been identified in salivary gland neoplasms [3, 6–8]. Future studies using next generation sequencing strategies are expected to lead to the identification of new fusions oncogene in epithelial malignancies, including those derived from salivary glands [9].
Fusion genes are clinically important, potent oncogenes as demonstrated by their ability to induce tumors in various transgenic mouse tumor models. For example, the ETV6–NTRK3 fusion, typical of secretory carcinoma of the breast and mammary analogue secretory carcinoma of the salivary glands, can induce breast cancer in mice through transformation of committed alveolar bipotent or CD61+ luminal progenitor cells [10]. Many fusion oncogenes have also been shown to be tumor-type specific and are therefore useful as diagnostic biomarkers [8, 11, 12]. The majority of fusions identified in solid tumors encode aberrant transcription factors while a minority express chimeric proteins that deregulate growth factor signaling [13]. Taken together, these and other studies (references in Stenman et al. [8]) clearly demonstrate that fusion genes and their down-stream targets are pathogenetically and clinically significant oncogenes and as such also key targets for the development of new cancer therapies. The perhaps most well-known example of this is the tyrosine kinase inhibitor Imatinib mesylate that effectively inhibits the BCR–ABL fusion oncoprotein in chronic myeloid leukemia [14].
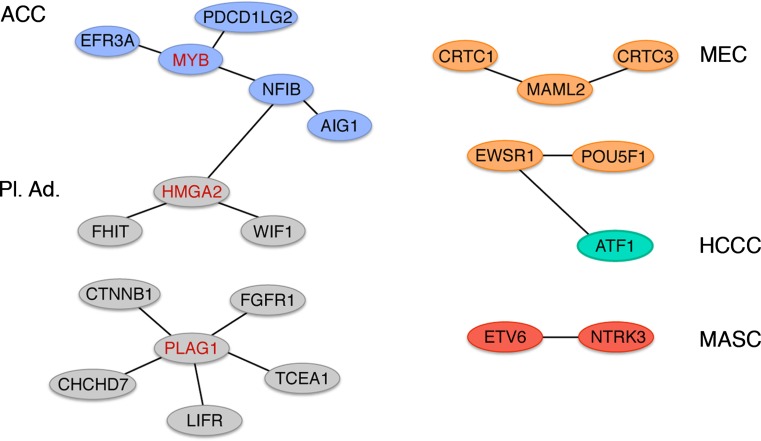
Several subtypes of salivary gland tumors are characterized by recurrent chromosome translocations which recently have been shown to result in a network of oncogenic gene fusions (Fig. 1) [8, 11]. The fusions encode novel fusion proteins as well as ectopically expressed normal or truncated proteins, and are found in both benign and malignant tumors. The major targets of the translocations are transcriptional coactivators, tyrosine kinase receptors, and transcription factors involved in growth factor signaling and cell cycle regulation. The aim of this paper is to review the current literature on fusion oncogenes in benign and malignant salivary gland tumors and discuss their molecular, clinical, and therapeutic consequences.
Fig. 1.
A translocation-generated network of oncogenic gene fusions in salivary gland tumors. The multiple translocation target genes MYB, HMGA2, and PLAG1 are indicated in red. ACC adenoid cystic carcinoma, MEC mucoepidermoid carcinoma, HCCC hyalinizing clear cell carcinoma, MASC mammary analogue secretory carcinoma, Pl. Ad. pleomorphic adenoma
MYB-NFIB Gene Fusion in Adenoid Cystic Carcinoma
Adenoid cystic carcinoma (ACC) is the second most common salivary gland malignancy but may also occur in exocrine glands in several other anatomical locations, including breast, sinonasal tract, tracheobronchial tree, cervix, and vulva [15, references in 8, 9]. It is an aggressive, but slowly growing cancer with an often fatal outcome. More than 80 % of patients with head and neck ACC die in 10–15 years after diagnosis. Until recently, little was known about the molecular pathogenesis of ACC. However, we recently showed that a recurrent t(6;9)(q22–23;p23–24) translocation in ACC [16] consistently results in a fusion of the MYB oncogene to the transcription factor gene NFIB (Fig. 1) [17]. MYB belongs to a family of proteins that functions as transcriptional regulators. The MYB protein contains three functional key domains, an N-terminal DNA-binding domain, a centrally located transcription activation domain, and a C-terminal negative regulatory domain involved in transcriptional repression [reviewed in 18, 19]. MYB plays an important role in the control of cell proliferation, apoptosis, and differentiation and is highly expressed in immature, proliferating cells and is down-regulated as cells become differentiated [19].
In the MYB-NFIB fusion oncogene, which is highly overexpressed in ACC, the 3′-part of MYB, including target sites for negatively regulating microRNAs, is replaced by one or more of the last coding exons of NFIB (Fig. 2a). The predicted MYB-NFIB fusion proteins retain the DNA-binding and transactivation domains of MYB, and is therefore expected to activate MYB target genes. Indeed, several MYB targets, including BCL2, KIT, CD34, BIRC3, MYC, and MAD1L1, were shown to be overexpressed in ACC compared to normal salivary gland and breast tissue [8, 17].
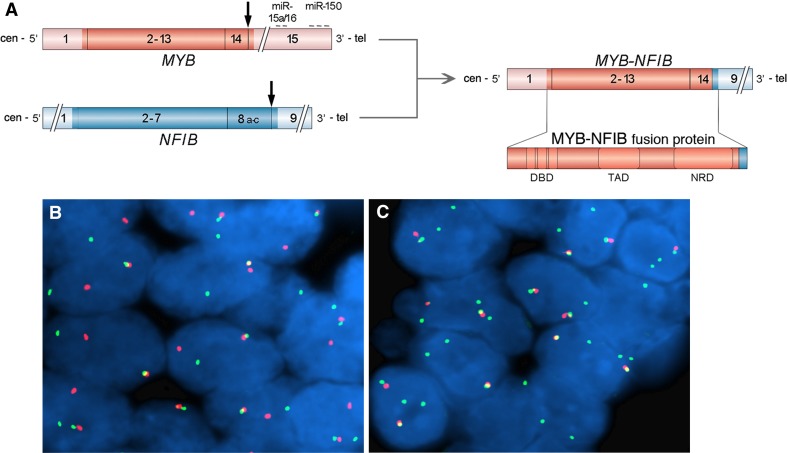
Fig. 2.
a Schematic illustration of the MYB and NFIB genes as well as the MYB–NFIB fusion oncogene (coding exons are shown i darker red and bluecolors) and the resulting fusion oncoprotein. Translocation breakpoints are shown by vertical arrows and binding sites for negatively regulating miRNAs in the 3′-UTR of MYB are indicated. DBD DNA binding domain, TAD transactivation domain, NRD negative regulatory domain. b and c FISH analysis of MYB rearrangements in adenoid cystic carcinoma using a dual-color MYB break-apart probe consisting of a centromeric (green) and a telomeric (red) probe covering the MYB locus and its flanking sequences. Interphase nuclei from a MYB–NFIB fusion-positive tumor (b), showing an intact signal (fused red/green signals) and a split signal (separated red and green signals), indicating a breakpoint within the MYB gene. Panel c shows interphase nuclei from a tumor with 1–2 intact red/green signals as well as 1–2 green signals indicating a selective gain of the MYB gene
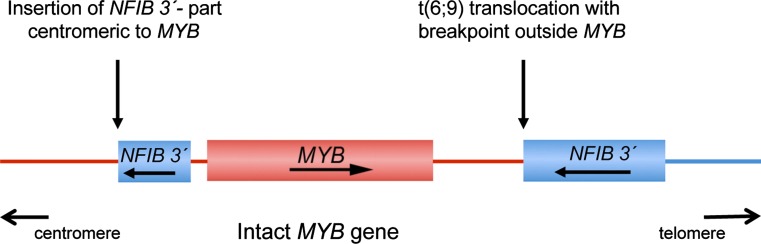
Previous studies have identified a subset of ACC that are MYB–NFIB fusion-negative but still overexpress MYB mRNA and protein. Detailed genomic characterization (including next generation sequencing) of several such cases have revealed insertions of a segment from 9p23–p22.3, including the 3′-part of NFIB, immediately centromeric to the MYB locus (Fig. 3) [9, 20, 21]. In these cases we can only speculate that enhancer elements upstream of MYB perhaps in combination with other regulatory elements in the 3′-part of NFIB and its flanking sequences may contribute to the activation of MYB. There are also cases reported with MYB activation and breakpoints distal to MYB (Fig. 3) [9, 21]. These cases may be similar to the t(6;7) translocations found in a subset of T-ALL in which the TCRB Cβ enhancer is juxtaposed 10–100 kb distal to MYB, leading to transcriptional deregulation of MYB [22]. Taken together, available data indicate that at least 80–90 % of ACC have MYB activation by gene fusion or other mechanisms leading to overexpression of MYB–NFIB fusion proteins or an apparently normal MYB oncoprotein. In contrast, the MYB–NFIB fusion has not been found in any non-ACC carcinomas of the head and neck, confirming the high specificity of the MYB–NFIB fusion for ACC. The fact that MYB activation is found in such a high frequency of ACCs regardless of the site of tumor origin, indicate that the MYB–NFIB fusion is a key oncogenic event and hallmark of ACC [9, 17, 21, 23, 24]. From a diagnostic point of view, the MYB–NFIB fusion and/or MYB activation may be detected by RT-PCR analysis of fusion transcripts, FISH analysis using probes for MYB, and/or NFIB (Fig. 2b, c), or by immunohistochemical staining of MYB-proteins. In addition to being a diagnostic biomarker for ACC, MYB and its downstream targets are also potential therapeutic targets. Continued studies aiming at identifying the transcriptional targets of the fusion will therefore be crucial to develop new therapies that may improve the survival of patients affected by this aggressive and often fatal disease.
Fig. 3.
Schematic illustration of two alternative mechanisms of activation of MYB in fusion-negative adenoid cystic carcinomas, that is insertions of a segment from 9p23–p22. 3, including the 3′-part of NFIB, immediately centromeric to the MYB locus or t(6;9) translocations with breakpoints 10–100 kb distal to MYB. In both cases, the proximity of the 3′-part of NFIB and its flanking sequences to MYB leads to transcriptional activation of an apparently intact MYB gene
Similar to the CRTC1–MAML2 and EWSR1–POU5F1fusions (see below), the MYB–NFIB fusion has also been encountered in sporadic dermal cylindromas [25]. These are benign tumors often located in the head and neck region with certain histopathologic similarities to ACC. Two-thirds (67 %) of cylindromas either express MYB–NFIB fusion transcripts and/or stain positive for MYB proteins. These results together with our previous observations [26–29] further strengthens the evidence for common molecular pathways of importance for the development of both benign and malignant, breast, salivary, and skin adnexal tumors.
CRTC1–MAML2 Gene Fusion in Mucoepidermoid Carcinoma
Mucoepidermoid carcinoma (MEC), the most common salivary gland carcinoma, is characterized by a unique and recurrent t(11;19)(q21–22;p13) translocation that occur in a high frequency of MECs [8, 11, 30]. The translocation results in a fusion of the two transcriptional coactivators MAML2 and CRTC1 (also known as MECT1, TORC1 or WAMTP1) and is a characteristic feature of MECs of salivary, bronchial, and thyroid glands [31–33]. The CRTC1–MAML2 fusion gene is composed of exon 1 of CRTC1 linked to exons 2–5 of MAML2. CRTC1 belongs to a family of highly conserved CREB (cAMP response element-binding protein) coactivators [34, 35] whereas MAML2 belongs to a family of Mastermind-like, nuclear proteins that functions as coactivators for Notch receptors [36, 37]. The fusion encodes a chimeric protein in which the Notch-binding domain of MAML2 is replaced by the CREB-binding, coiled-coil domain of CRTC1 fused to the transactivation domain of MAML2 [31, 32]. The molecular consequences of the fusion is not yet fully understood. However, functional studies have shown that the N-terminal part of the fusion protein, including the CREB-binding domain, is crucial for transforming activity [38] and that the fusion protein activates transcription of cAMP/CREB target genes, including PEPCK1, AREG, MMP10, IL6, NR4A2, and NR4A3 [39, Enlund et al. unpublished data]. Preliminary studies using different small molecule inhibitors of the EGFR (AREG-amphiregulin) or PKA (cAMP-dependent kinase) pathways have shown that they can inhibit the proliferation of MEC-derived cell lines in vitro, suggesting that targeting these pathways may offer a new approach to systemic treatment of CRTC1–MAML2 positive MECs [39].
Several independent and well-documented studies have shown that the CRTC1–MAML2 fusion preferentially occurs in low/intermediate-grade MECs with favorable prognosis [29, 40 and refs. therein]. Based on a recent arrayCGH study of genomic imbalances in fusion-positive and fusion-negative MECs, we proposed that MEC may be subdivided in (a) low-grade, fusion-positive tumors with no or few genomic imbalances and favorable prognosis, (b) high-grade, fusion-positive tumors with multiple genomic imbalances (including deletions of the tumor suppressor gene CDKN2A) and unfavorable prognosis, and (c) a heterogeneous group of high-grade, fusion-negative non-MEC adenocarcinomas with multiple genomic imbalances and unfavorable outcome [40]. There is sufficient evidence at hand indicating that the CRTC1–MAML2 fusion is a clinically useful biomarker that distinguishes true MECs, most of which have an excellent prognosis, from fusion-negative MEC-like tumors with a more unfavorable prognosis.
Is the CRTC1–MAML2 Gene Fusion a Recurrent Feature in Warthin Tumor?
Previous cytogenetic studies have shown that a subset of Warthin tumors, the second most common subtype of benign salivary gland tumor, have an apparently identical t(11;19)(q21–22;p13) translocation as the one found in MEC [32 and refs. therein]. RT-PCR analysis of a few such cases have shown that this translocation also results in a CRTC1–MAML2 gene fusion [32, 33]. Subsequent studies have suggested that these tumors may be described as metaplastic variants of Warthin tumors, representing early signs of MEC developing within Warthin tumors or mis-diagnosed mucoepidermoid carcinomas [41, 42]. Interestingly, an identical CRTC1-MAML2 fusion has also been identified in clear cell hidradenoma of the skin and breast [26, 28]. This is a benign skin adnexal tumor with certain morphologic similarities to MEC. Taken together, these observations indicate that the CRTC1–MAML2 fusion is etiologically linked to benign and low-grade malignant, histogenetically similar tumor types originating from exocrine glands in different anatomical locations.
EWSR1–POU5F1 Gene Fusion in High-Grade MAML2-Negative Mucoepidermoid Carcinoma
As discussed above, there is a subgroup of high-grade CRTC1–MAML2 fusion-negative tumors with a partly MEC-like morphology. In an effort to further characterize these tumors we have identified cases with a t(6;22)(p21;q12) translocation resulting in an EWSR1-POU5F1 gene fusion (Fig. 1) [27]. Moreover, we also identified an identical gene fusion in a subset of less well-differentiated cutaneous hidradenomas (in contrast to the clear cell variants) [27]. The chimeric EWSR1–POU5F1 protein consists of the N-terminal domain of EWSR1 linked to the DNA-binding domain of the transcription factor POU5F1. POU5F1 is important during the early stages of development to maintain the pluripotent status of embryonic stem cells. Notably, the morphology of the EWSR1–POU5F1 positive tumors were more immature compared to the CRTC1–MAML2 positive tumors, in line with the known consequences of overexpression of POU5F1.
A similar EWSR1–POU5F1 fusion has also been found in a case of an undifferentiated bone tumor of the pelvis [43] as well as in a subset of deep-seated benign and malignant soft tissue myoepithelial tumors of children and young adults [44]. In contrast, none of five salivary myoepithelial carcinoma ex pleomorphic adenomas analyzed in that study had rearrangements of EWSR1, suggesting that at least a subset of myoepithelial tumors of the salivary glands may not be related to their soft tissue counterparts.
EWSR1–ATF1 Gene Fusion in Low-Grade Hyalinizing Clear Cell Carcinoma
Recently, a rare salivary gland carcinoma, hyalinizing clear cell carcinoma (HCCC), was shown to have t(12;22)(q13;q12) translocations resulting in EWSR1–ATF1 gene fusions [45, Fehr et al. unpublished data]. HCCC is a low-grade carcinoma with distinctive clear-cell morphology and pattern of hyalinization often in combination with focal mucinous differentiation. The EWSR1–ATF1 fusion has been found in >80 % of HNCCC. By contrast, the fusion is not detected in any of the morphological mimics: epithelial-myoepithelial carcinoma, myoepithelial carcinoma or MEC, demonstrating its usefulness as a diagnostic biomarker for HCCC.
Interestingly, EWSR1–ATF1 fusions were originally described in conventional clear cell sarcomas (of tendons and aponeurosis) [46] and have recently also been encountered in angiomatoid fibrous histiocytomas [47] as well as in a few cases of soft tissue myoepithelial tumors [48]. Taken together, these observations provide evidence for a unifying concept of salivary gland and soft tissue tumors with clear cell morphology.
ETV6–NTRK3 Gene Fusion in Mammary Analogue Secretory Carcinoma of Salivary Glands (MASC)
Mammary analogue secretory carcinomas of the salivary glands (MASC) is a recently described subtype of salivary gland carcinoma, with strong histologic and immunohistochemical resemblance to secretory carcinoma (SC) of breast [49, 50]. In addition to the morphologic similarities, MASC and SC of the breast also have important genetic similarities since they both share a t(12;15)(p13;q25) chromosomal translocation [51], resulting in an identical ETV6–NTRK3 gene fusion (Fig. 1). The fusion is found in the majority of MASCs (>90 %) and is an important biomarker that may help distinguish MASC from acinic cell carcinoma and low-grade cystadenocarcinoma. The ETV6–NTRK3 fusion gene, encodes a chimeric tyrosine kinase that activates two major effector pathways, i.e., the Ras-MAP kinase (MAPK) mitogenic pathway and the phosphatidyl inositol-3-kinase (PI3K)-AKT pathway both of which seem to be required for ETV6–NTRK3 transformation [51–53]. It should be noted that ETV6–NTRK3 gene fusions are found in several other tumor types, including congenital mesoblastic nephroma, congenital fibrosarcoma, and acute myeloid leukemia [52], indicating that the fusion oncoprotein has transforming activity in a variety of cell types. Interestingly, a recent study of mammary-type secretory carcinoma of the skin suggests these tumors are negative for the ETV6–NTRK3 gene fusions [54]. The finding of the ETV6–NTRK3 fusion in SC of the breast and salivary glands is of particular interest because of the recently described MYB–NFIB fusion in ACCs of both glands [17]. These observations further strengthens the evidence for common molecular pathways of importance for development of breast and salivary gland neoplasms.
PLAG1 and HMGA2 Gene Fusions in Pleomorphic Adenoma
Pleomorphic adenoma is the most common histologic subtype of salivary gland tumor. It is a benign tumor with a highly variable morphology that sometimes may cause diagnostic problems. Extensive cytogenetic studies of pleomorphic adenomas have shown that they are characterized by recurrent translocations or intrachromosomal rearrangements with breakpoints preferentially affecting 8q12 (>50 % of the cases) and 12q14–15 (10–15 % of the cases) [reviewed in 8, 11]. The translocations/rearrangements invariably result in gene fusions involving the transcription factor genes PLAG1 and HMGA2 [8, 11, 55, 56]. PLAG1 encodes a developmentally regulated DNA-binding zinc finger protein that is part of a family of cell cycle progression-related proteins. Ectopic overexpression of PLAG1 due to promoter swapping with at least one of five other genes (CTNNB1, FGFR1, LIFR, CHCHD7, TCEA1)(Fig. 1), cause deregulation of PLAG1 target genes and activation of the IGF-II signaling pathway [57, reviewed in 8, 11]. Previous studies have demonstrated that PLAG1 is also involved in gene fusions with two other genes (HAS2 and COL1A2) in benign lipoblastomas [reviewed in 8, 11].
HMGA2 belongs to the high mobility group (HMG) protein gene family which encodes proteins that are heterogeneous, nonhistone components of chromatin [reviewed in 8, 11]. HMGA2 functions as an architectural transcription factor which regulates transcription through its binding to the minor groove of AT-rich DNA and is also involved in the regulation of recombination and chromatin structure. The protein contains three DNA-binding domains, a spacer domain, and a highly acidic C-terminal domain. Genomic rearrangements of the 3′-part of HMGA2 due to fusions with the 3’-parts of NFIB, WIF1 or FHIT (Fig. 1) result in activation of the expression of HMGA2 and its target genes, including the cell cycle regulators CCNA1 and CCNB2 [58, 59]. The molecular mechanism leading to activation of HMGA2 is still partly unknown. Recent studies have indicated that a major mechanism may include loss of target-sites for negatively regulating Let-7 microRNAs in the 3′-UTR as a consequence of gene fusion [60]. Interestingly, HMGA2 is also involved in gene fusion in a variety of benign mesenchymal tumors with 12q14–15 rearrangements, including lipoma, uterine leiomyoma, hamartomas of the breast and lung, fibroadenoma of the breast, angiomyxoma, endometrial polyps, and bone and soft tissue chondroma [reviewed in 8, 11].
The PLAG1 and HMGA2 fusions in pleomorphic adenoma have not been encountered in any other histopathologic subtypes of salivary gland neoplasms and may therefore be useful as biomarkers in diagnostically challenging cases with morphologies partly overlapping with adenoid cystic carcinoma, polymorphous low-grade adenocarcinoma and other salivary gland carcinomas.
Gene Fusion in Carcinoma-Ex-Pleomorphic Adenoma
Our knowledge about the molecular abnormalities involved in the transformation of a benign pleomorphic adenoma into a carcinoma-ex-pleomorphic adenoma (Ca-ex-PA) is still limited. The malignant component is frequently a poorly differentiated adenocarcinoma or undifferentiated carcinoma but may also be virtually any other subtype of salivary gland carcinoma, such as MEC, salivary duct carcinoma or ACC [15]. Molecular studies of small series and single cases of Ca-ex-PA have shown that they express pleomorphic adenoma specific gene fusions involving PLAG1 and HMGA2 [7, 61]. In addition, amplification of multiple genes within 12q13–15 (in particular MDM2 and HMGA2-WIF1 gene fusions, TP53 mutation, deletions of 5q23.2–q31.2, gains of 8q12.1 (PLAG1) and 8q22.1–q24.1 (MYC), and amplification of HER2 have been identified as genetic events of importance for malignant transformation [7].
Conclusions
Salivary gland tumors constitute a heterogeneous group of uncommon diseases that pose significant diagnostic and therapeutic challenges. The primary treatment of salivary gland neoplasms is surgical resection with or without post-operative radiotherapy. For patients presenting with locally advanced, recurrent or metastatic disease the treatment options are currently limited and mainly palliative. However, the recent discovery of a comprehensive translocation-generated gene fusion network in salivary gland carcinomas and benign salivary gland neoplasms opens up new avenues for improved diagnosis and development of specific targeted therapies. The fusions encode novel fusion proteins or ectopically expressed normal or truncated proteins. The major targets of the translocations are transcriptional coactivators, tyrosine kinase receptors, and transcription factors involved in growth factor signaling and cell cycle regulation. Continued molecular characterization of these fusion oncoproteins and their down-stream targets will ultimately lead to the identification of novel driver genes in salivary gland neoplasms and will also form the basis for the development of new therapeutic strategies for salivary gland cancers and, perhaps, other neoplasms.
Acknowledgments
I thank Marta Persson for excellent help with the illustrations. Work presented in this review was supported by the Swedish Cancer Society, IngaBritt and Arne Lundberg Research Foundation, the Adenoid Cystic Carcinoma Research Foundation, and BioCARE—a National Strategic Cancer Research program at University of Gothenburg.
References
- 1.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 2.Mitelman F, Johansson B, Mertens F, editors. Mitelman database of chromosome aberrations and gene fusions in cancer: http://cgap.nci.nih.gov/Chromosomes/Mitelman, 2013.
- 3.Asp J, Persson F, Kost-Alimova M, Stenman G. CHCHD7-PLAG1 and TCEA1-PLAG1 gene fusions resulting from cryptic, intrachromosomal 8q rearrangements in pleomorphic salivary gland adenomas. Genes Chromosomes Cancer. 2006;45:820–828. doi: 10.1002/gcc.20346. [DOI] [PubMed] [Google Scholar]
- 4.Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, Yu J, Wang L, Montie JE, Rubin MA, Pienta KJ, Roulston D, Shah RB, Varambally S, Mehra R, Chinnaiyan AM. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 5.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, Bando M, Ohno S, Ishikawa Y, Aburatani H, Niki T, Sohara Y, Sugiyama Y, Mano H. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 6.Persson F, Winnes M, Andrén Y, Wedell B, Dahlenfors R, Asp J, Mark J, Enlund F, Stenman G. High-resolution array CGH analysis of salivary gland tumors reveals fusion and amplification of the FGFR1 and PLAG1 genes in ring chromosomes. Oncogene. 2008;27:3072–3080. doi: 10.1038/sj.onc.1210961. [DOI] [PubMed] [Google Scholar]
- 7.Persson F, Andrén Y, Winnes M, Wedell B, Nordkvist A, Gudnadottir G, Dahlenfors R, Sjögren H, Mark J, Stenman G. High-resolution genomic profiling of adenomas and carcinomas of the salivary glands reveals amplification, rearrangement, and fusion of HMGA2. Genes Chromosomes Cancer. 2009;48:69–82. doi: 10.1002/gcc.20619. [DOI] [PubMed] [Google Scholar]
- 8.Stenman G, Andersson MK, Andrén Y. New tricks from an old oncogene: gene fusions and copy number alterations of MYB in human cancer. Cell Cycle. 2010;9:2986–2995. doi: 10.4161/cc.9.15.12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Persson M, Andrén Y, Moskaluk CA, Frierson HF, Jr, Cooke SL, Futreal PA, Kling T, Nelander S, Nordkvist A, Persson F, Stenman G. Clinically significant copy number alterations and complex rearrangements of MYB and NFIB in head and neck adenoid cystic carcinoma. Genes Chromosomes Cancer. 2012;51:805–817. doi: 10.1002/gcc.21965. [DOI] [PubMed] [Google Scholar]
- 10.Li Z, Tognon CE, Godinho FJ, Yasaitis L, Hock H, Herschkowitz JI, Lannon CL, Cho E, Kim SJ, Bronson RT, Perou CM, Sorensen PH, Orkin SH. ETV6-NTRK3 fusion oncogene initiates breast cancer from committed mammary progenitors via activation of AP1 complex. Cancer Cell. 2007;12:542–558. doi: 10.1016/j.ccr.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stenman G. Fusion oncogenes and tumor type specificity—insights from salivary gland tumors. Semin Cancer Biol. 2005;15:224–235. doi: 10.1016/j.semcancer.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Åman P. Fusion oncogenes in tumor development. Semin Cancer Biol. 2005;15:236–243. doi: 10.1016/j.semcancer.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 13.Helman LJ, Meltzer P. Mechanisms of sarcoma development. Nat Rev Cancer. 2003;3:685–694. doi: 10.1038/nrc1168. [DOI] [PubMed] [Google Scholar]
- 14.Druker BJ. Translation of the Philadelphia chromosome into therapy for CML. Blood. 2008;112:4808–4817. doi: 10.1182/blood-2008-07-077958. [DOI] [PubMed] [Google Scholar]
- 15.Barnes L, Eveson JW, Reichart P, Sidransky D, editors. World health organization classification of tumours. Pathology and genetics of head and neck tumours. Lyon: IARC Press; 2005. Pp. 209–74.
- 16.Nordkvist A, Mark J, Gustafsson H, Bang G, Stenman G. Non-random chromosome rearrangements in adenoid cystic carcinoma of the salivary glands. Genes Chromosomes Cancer. 1994;1994(10):115–121. doi: 10.1002/gcc.2870100206. [DOI] [PubMed] [Google Scholar]
- 17.Persson M, Andrén Y, Mark J, Horlings HM, Persson F, Stenman G. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci USA. 2009;106:18740–18744. doi: 10.1073/pnas.0909114106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh IH, Reddy EP. The myb gene family in cell growth, differentiation and apoptosis. Oncogene. 1999;18:3017–3033. doi: 10.1038/sj.onc.1202839. [DOI] [PubMed] [Google Scholar]
- 19.Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat Rev Cancer. 2008;8:523–534. doi: 10.1038/nrc2439. [DOI] [PubMed] [Google Scholar]
- 20.Mitani Y, Li J, Rao PH, Zhao YJ, Bell D, Lippman SM, Weber RS, Caulin C, El-Naggar AK. Comprehensive analysis of the MYB-NFIB gene fusion in salivary adenoid cystic carcinoma: incidence, variability, and clinicopathologic significance. Clin Cancer Res. 2010;16:4722–4731. doi: 10.1158/1078-0432.CCR-10-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitani Y, Rao PH, Futreal PA, Roberts DB, Stephens PI, Zhao YI, Zhang L, Mitani M, Weber RS, Lippman SM, Caulin C, El-Naggar AK. Novel chromosomal rearrangements and break points at the t(6;9) in salivary adenoid cystic carcinoma: association with MYB-NFIB chimeric fusion, MYB expression, and clinical outcome. Clin Cancer Res. 2011;17:7003–7014. doi: 10.1158/1078-0432.CCR-11-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clappier E, Cuccuini W, Kalota A, Crinquette A, Cayuela JM, Dik WA, Langerak AW, Montpellier B, Nadel B, Walrafen P, Delattre O, Aurias A, Leblanc T, Dombret H, Gewirtz AM, Baruchel A, Sigaux F, Soulier J. The C-MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (T-ALL), the translocation defining a new T-ALL subtype in very young children. Blood. 2007;110:1251–1261. doi: 10.1182/blood-2006-12-064683. [DOI] [PubMed] [Google Scholar]
- 23.Brill LB, Kanner WA, Fehr A, Andrén Y, Moskaluk CA, Löning T, Stenman G, Frierson HF., Jr Analysis of MYB Expression and MYB-NFIB gene fusions in adenoid cystic carcinoma and other salivary neoplasms. Mod Pathol. 2011;24:1169–1176. doi: 10.1038/modpathol.2011.86. [DOI] [PubMed] [Google Scholar]
- 24.West RB, Kong C, Clarke N, Gilks T, Lipsick JS, Cao H, Kwok S, Montgomery KD, Varma S, Le QT. MYB expression and translocation in adenoid cystic carcinoma and other salivary gland tumors with clinicopathological correlation. Am J Surg Pathol. 2011;35:92–99. doi: 10.1097/PAS.0b013e3182002777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fehr A, Kovács A, Löning T, Frierson H, Jr, van den Oord J, Stenman G. The MYB-NFIB gene fusion—a novel genetic link between adenoid cystic carcinoma and dermal cylindroma. J Pathol. 2011;224:322–327. doi: 10.1002/path.2909. [DOI] [PubMed] [Google Scholar]
- 26.Behboudi A, Winnes M, Gorunova L, van den Oord JJ, Mertens F, Enlund F, Stenman G. Clear cell hidradenoma of the skin-a third tumor type with a t(11;19)-associated TORC1-MAML2 gene fusion. Genes Chromosomes Cancer. 2005;43:202–205. doi: 10.1002/gcc.20168. [DOI] [PubMed] [Google Scholar]
- 27.Möller E, Stenman G, Mandahl N, Hamberg H, Mölne L, van den Oord JJ, Brosjö O, Mertens F, Panagopoulos I. POU5F1, encoding a key regulator of stem cell pluripotency, is fused to EWSR1 in hidradenoma of the skin and mucoepidermoid carcinoma of the salivary glands. J Pathol. 2008;215:78–86. doi: 10.1002/path.2327. [DOI] [PubMed] [Google Scholar]
- 28.Winnes M, Mölne L, Suurküla M, Andrén Y, Persson F, Enlund F, Stenman G. Frequent fusion of the CRTC1 and MAML2 genes in clear cell variants of cutaneous hidradenomas. Genes Chromosomes Cancer. 2007;46:559–563. doi: 10.1002/gcc.20440. [DOI] [PubMed] [Google Scholar]
- 29.Behboudi A, Enlund F, Winnes M, Andrén Y, Nordkvist A, Leivo I, Flaberg E, Szekely L, Mäkitie A, Grenman R, Mark J, Stenman G. Molecular classification of mucoepidermoid carcinomas-prognostic significance of the MECT1-MAML2 fusion oncogene. Genes Chromosomes Cancer. 2006;45:470–481. doi: 10.1002/gcc.20306. [DOI] [PubMed] [Google Scholar]
- 30.Nordkvist A, Gustafsson H, Juberg-Ode M, Stenman G. Recurrent rearrangements of 11q14-22 in mucoepidermoid carcinoma. Cancer Genet Cytogenet. 1994;74:77–83. doi: 10.1016/0165-4608(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 31.Tonon G, Modi S, Wu L, Kubo A, Coxon AB, Komiya T, O’Neil K, Stover K, El-Naggar A, Griffin JD, Kirsch IR, Kaye FJ. t(11;19)(q21;p13) translocation in mucoepidermoid carcinoma creates a novel fusion product that disrupts a Notch signaling pathway. Nat Genet. 2003;33:208–213. doi: 10.1038/ng1083. [DOI] [PubMed] [Google Scholar]
- 32.Enlund F, Behboudi A, Andren Y, Öberg C, Lendahl U, Mark J, Stenman G. Altered Notch signaling resulting from expression of a WAMTP1-MAML2 gene fusion in mucoepidermoid carcinomas and benign Warthin’s tumors. Exp Cell Res. 2004;292:21–28. doi: 10.1016/j.yexcr.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 33.Tirado Y, Williams MD, Hanna EY, Kaye FJ, Batsakis JG, El-Naggar AK. CRTC1/MAML2 fusion transcript in high-grade mucoepidermoid carcinomas of salivary and thyroid glands and Warthin′s tumors: implications for histogenesis and biologic behavior. Genes Chromosomes Cancer. 2007;46:708–715. doi: 10.1002/gcc.20458. [DOI] [PubMed] [Google Scholar]
- 34.Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, Montminy M. TORCs: transducers of regulated CREB activity. Mol Cell. 2003;12:413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 35.Iourgenko V, Zhang W, Mickanin C, Daly I, Jiang C, Hexham JM, Orth AP, Miraglia L, Meltzer J, Garza D, Chirn GW, McWhinnie E, Cohen D, Skelton J, Terry R, Yu Y, Bodian D, Buxton FP, Zhu J, Song C, Labow MA. Identification of a family of cAMP response element-binding protein coactivators by genome-scale functional analysis in mammalian cells. Proc Natl Acad Sci USA. 2003;100:12147–12152. doi: 10.1073/pnas.1932773100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin SE, Oyama T, Nagase T, Harigaya K, Kitagawa M. Identification of new human mastermind proteins defines a family that consists of positive regulators for notch signaling. J Biol Chem. 2002;277:50612–50620. doi: 10.1074/jbc.M209529200. [DOI] [PubMed] [Google Scholar]
- 37.Wu L, Sun T, Kobayashi K, Gao P, Griffin JD. Identification of a family of mastermind-like transcriptional coactivators for mammalian notch receptors. Mol Cell Biol. 2002;22:7688–7700. doi: 10.1128/MCB.22.21.7688-7700.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu L, Liu J, Gao P, Nakamura M, Cao Y, Shen H, Griffin JD. Transforming activity of MECT1-MAML2 fusion oncoprotein is mediated by constitutive CREB activation. EMBO J. 2005;24:2391–2402. doi: 10.1038/sj.emboj.7600719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coxon A, Rozenblum E, Park YS, Joshi N, Tsurutani J, Dennis PA, Kirsch IR, Kaye FJ. Mect1-Maml2 fusion oncogene linked to the aberrant activation of cyclic AMP/CREB regulated genes. Cancer Res. 2005;65:7137–7144. doi: 10.1158/0008-5472.CAN-05-1125. [DOI] [PubMed] [Google Scholar]
- 40.Jee KJ, Persson M, Heikinheimo K, Passador-Santos F, Aro K, Knuutila S, Odell EW, Mäkitie A, Sundelin K, Stenman G, Leivo I. Genomic profiles and CRTC1-MAML2 fusion distinguish different subtypes of mucoepidermoid carcinoma. Mod Pathol. 2012; Sep 28:154. doi:10.1038/modpathol. [DOI] [PubMed]
- 41.Winnes M, Enlund F, Mark J, Stenman G. The MECT1-MAML2 gene fusion and benign Warthin′s tumour: is the MECT1-MAML2 gene fusion specific to mucoepidermoid carcinoma? J Mol Diagn. 2006;8:394–395. doi: 10.2353/jmoldx.2006.060020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fehr A, Röser K, Belge G, Löning T, Bullerdiek J. A closer look at Warthin tumors and the t(11;19) Cancer Genet Cytogenet. 2008;180:135–139. doi: 10.1016/j.cancergencyto.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 43.Yamaguchi S, Yamazaki Y, Ishikawa Y, Kawaguchi N, Mukai H, Nakamura T. EWSR1 is fused to POU5F1 in a bone tumour with translocation t(6;22)(p21;q12) Genes Chromosomes Cancer. 2005;43:217–222. doi: 10.1002/gcc.20171. [DOI] [PubMed] [Google Scholar]
- 44.Antonescu CR, Zhang L, Chang NE, Pawel BR, Travis W, Katabi N, Edelman M, Rosenberg AE, Nielsen GP, Dal Cin P, Fletcher CD. EWSR1-ATF1 fusion in soft tissue myoepithelial tumors. A molecular analysis of sixty-six cases, including soft tissue, bone, and visceral lesions, showing common involvement of the EWSR1 gene. Genes Chromosomes Cancer. 2010;49:1114–1124. doi: 10.1002/gcc.20819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Antonescu CR, Katabi N, Zhang L, Sung YS, Seethala RR, Jordan RC, Perez-Ordoñez B, Have C, Asa SL, Leong IT, Bradley G, Klieb H, Weinreb I. EWSR1-ATF1 fusion is a novel and consistent finding in hyalinizing cler-cell sarcoma of salivary gland. Genes Chromosomes Cancer. 2011;50:559–570. doi: 10.1002/gcc.20881. [DOI] [PubMed] [Google Scholar]
- 46.Zucman J, Delattre O, Desmaze C, Epstein AL, Stenman G, Speleman F, Fletchers CD, Aurias A, Thomas G. EWS and ATF-1 gene fusion induced by t(12;22) translocation in malignant melanoma of soft parts. Nat Genet. 1993;4:341–345. doi: 10.1038/ng0893-341. [DOI] [PubMed] [Google Scholar]
- 47.Rossi S, Szuhai K, Ijszenga M, Tanke HJ, Zanatta L, Sciot R, Fletcher CD. Dei Tos AP, Hogendoorn PC. EWSR1-CREB1 and EWSR1-ATF1 fusion genes in angiomatoid fibrous histiocytoma. Clin Cancer Res. 2007;13:7322–7328. doi: 10.1158/1078-0432.CCR-07-1744. [DOI] [PubMed] [Google Scholar]
- 48.Flucke U, Mentzel T, Verdijk MA, Slootweg PJ, Creytens DH, Suurmeijer AJ, Tops BB. EWSR1-ATF1 chimeric transcript in a myoepithelial tumor of soft tissue: a case report. Hum Pathol. 2012;43:764–768. doi: 10.1016/j.humpath.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 49.Skálová A, Vanecek T, Sima R, Laco J, Weinreb I, Perez-Ordonez B, Starek I, Geierova M, Simpson RH, Passador-Santos F, Ryska A, Leivo I, Kinkor Z, Michal M. Mammary analogue secretory carcinoma of salivary glands, containing the ETV6-NTRK3 fusion gene: a hitherto undescribed salivary gland tumor entity. Am J Surg Pathol. 2010;34:599–608. doi: 10.1097/PAS.0b013e3181d9efcc. [DOI] [PubMed] [Google Scholar]
- 50.Fehr A, Löning T, Stenman G. Mammary analogue secretory carcinoma of the salivary glands with ETV6-NTRK3 gene fusion. Am J Surg Pathol. 2011;35:1600–1602. doi: 10.1097/PAS.0b013e31822832c7. [DOI] [PubMed] [Google Scholar]
- 51.Tognon C, Knezevich SR, Huntsman D, Roskelley CD, Melnyk N, Mathers JA, Becker L, Carneiro F, MacPherson N, Horsman D, Poremba C, Sorensen PH. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2:367–376. doi: 10.1016/S1535-6108(02)00180-0. [DOI] [PubMed] [Google Scholar]
- 52.Lannon CL, Sorensen PH. ETV6-NTRK3: a chimeric protein tyrosine kinase with transformation activity in multiple cell lineages. Semin Cancer Biol. 2005;15:215–223. doi: 10.1016/j.semcancer.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 53.Wai DH, Knezevich SR, Lucas T, Jansen B, Kay RJ, Sorensen PH. The ETV6-NTRK3 gene fusion encodes a chimeric protein tyrosine kinase that transforms NIH3T3 cells. Oncogene. 2000;19:906–915. doi: 10.1038/sj.onc.1203396. [DOI] [PubMed] [Google Scholar]
- 54.Kazakov DV, Hantschke M, Vanecek T, Kacerovska D, Michal M. Mammary-type secretory carcinoma of the skin. Am J Surg Pathol. 2010;34:1226–1227. doi: 10.1097/PAS.0b013e3181e4f49d. [DOI] [PubMed] [Google Scholar]
- 55.Kas K, Voz ML, Röijer E, Åström AK, Meyen E, Stenman G, Van de Ven WJ. Promoter swapping between the genes for a novel zinc finger protein and beta-catenin in pleiomorphic adenomas with t(3;8)(p21;q12) translocations. Nat Genet. 1997;15:170–174. doi: 10.1038/ng0297-170. [DOI] [PubMed] [Google Scholar]
- 56.Geurts JM, Schoenmakers EF, Röijer E, Åström AK, Stenman G, van de Ven WJ. Identification of NFIB as recurrent translocation partner gene of HMGIC in pleomorphic adenomas. Oncogene. 1998;16:865–872. doi: 10.1038/sj.onc.1201609. [DOI] [PubMed] [Google Scholar]
- 57.Voz ML, Mathys J, Hensen K, Pendeville H, Van Valckenborgh I, Van Huffel C, Chavez M, Van Damme B, De Moor B, Moreau Y, Van de Ven WJ. Microarray screening for target genes of the proto-oncogene PLAG1. Oncogene. 2004;23:179–191. doi: 10.1038/sj.onc.1207013. [DOI] [PubMed] [Google Scholar]
- 58.Tessari MA, Gostissa M, Altamura S, Sgarra R, Rustighi A, Salvagno C, Caretti G, Imbriano C, Mantovani R, Del Sal G, Giancotti V, Manfioletti G. Transcriptional activation of the cyclin A gene by the architectural transcription factor HMGA2. Mol Cell Biol. 2003;23:9104–9116. doi: 10.1128/MCB.23.24.9104-9116.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Martino I, Visone R, Wierinckx A, Palmieri D, Ferraro A, Cappabianca P, Chiappetta G, Forzati F, Lombardi G, Colao A, Trouillas J, Fedele M, Fusco A. HMGA proteins up-regulate CCNB2 gene in mouse and human pituitary adenomas. Cancer Res. 2009;69:1844–1850. doi: 10.1158/0008-5472.CAN-08-4133. [DOI] [PubMed] [Google Scholar]
- 60.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bell D, N Myers J, Rao PH, El-Naggar AK. t(3;8) as the sole chromosomal abnormality in a myoepithelial carcinoma ex pleomorphic adenoma: a putative progression event. Head Neck. 2012; Jan 27. doi:10.1002/hed.22926. . [DOI] [PubMed]