Abstract
The prevalence of infection with hepatitis A virus (HAV), HBV, HCV, HDV, and HEV was evaluated in 249 apparently healthy individuals, including 122 inhabitants in Ulaanbaatar, the capital city of Mongolia, and 127 age- and sex-matched members of nomadic tribes who lived around the capital city. Overall, hepatitis B surface antigen (HBsAg) was detected in 24 subjects (10%), of whom 22 (92%) had detectable HBV DNA. Surprisingly, HDV RNA was detectable in 20 (83%) of the 24 HBsAg-positive subjects. HCV-associated antibodies were detected in 41 (16%) and HCV RNA was detected in 36 (14%) subjects, none of whom was coinfected with HBV, indicating that HBV/HCV carriers account for one-fourth of this population. Antibodies to HAV and HEV were detected in 249 (100%) and 28 (11%) subjects, respectively. Of 22 HBV DNA-positive subjects, genotype D was detected in 21 subjects and genotype F was detected in 1 subject. All 20 HDV isolates recovered from HDV RNA-positive subjects segregated into genotype I, but these differed by 2.1 to 11.4% from each other in the 522- to 526-nucleotide sequence. Of 36 HCV RNA-positive samples, 35 (97%) were genotype 1b and 1 was genotype 2a. Reflecting an extremely high prevalence of hepatitis virus infections, there were no appreciable differences in the prevalence of hepatitis virus markers between the two studied populations with distinct living place and lifestyle. A nationwide epidemiological survey of hepatitis viruses should be conducted in an effort to prevent de novo infection with hepatitis viruses in Mongolia.
Mongolia, more commonly known as Outer Mongolia, is located in Northern Asia. With an area of more than 1.57 million square kilometers and a population of 2.46 million as of 2002, Mongolia has a population density of only 1.6 people per square kilometer, one of the lowest in the world (36). Currently, individuals under 35 years of age make up 70% of the population of Mongolia and the average age of the population is 21 years. Geographical conditions and a very low population density make communication, transport, and health service provision difficult. Nearly one-half of the total population lives in cities and towns, and 20% of the population still live a nomadic lifestyle (http://www.un-mongolia.mn/who/review.html). Communicable diseases are still one of the main health problems in Mongolia, and viral hepatitis accounted for 41% of the registered communicable diseases in Mongolia in 1997 (http://www.un-mongolia.mn/who/review.html). However, to date, little is known about the prevalence of infection with hepatitis viruses, including hepatitis A virus (HAV), HBV, HCV, HDV (or hepatitis delta virus), and HEV among healthy individuals in this country.
HAV is an important pathogen which has been responsible for a common form of acute viral hepatitis in many parts of the world where sanitation is suboptimal (10). HEV shares several characteristics with HAV, and these nonenveloped RNA viruses are both transmitted via the fecal-oral route. HEV infection is an important public health concern in much of Asia and Africa, and one epidemic has been documented in Mexico (28). As with HAV, there is no evidence of chronic HEV infection in humans. The extent of enterically transmitted viral hepatitis caused by HAV and HEV has yet to be explored in Mongolia.
HDV is a defective virus requiring helper functions from HBV and is currently classified into three genotypes based on nucleotide sequence comparison: genotype I, which is widely distributed throughout the world; genotype II, which is mainly found in Asia; and genotype III, which is found in countries in South America, including Peru, Colombia, and Venezuela, where several cases of fulminant hepatitis have been documented (29). However, to our knowledge, there has been no report on HDV infection in Mongolia.
Hepatitis B is a major worldwide health problem, with over 350 million chronically infected individuals, some of whom develop chronic hepatitis that progresses to cirrhosis and eventually to hepatocellular carcinoma. The prevalence of HBV infection is generally high in Asia and Africa (4). HCV infects an estimated 170 million persons worldwide, although the prevalence of HCV infection varies by geographic region, with the highest reported prevalence in Egypt (6 to 28%; mean, 22%) (16). HCV infection is characterized by a high rate of progression to chronic infection, and some patients with chronic infections develop cirrhosis and eventually hepatocellular carcinoma (14, 37). High rates of positivity for HBsAg or anti-HCV among blood donors or outpatient volunteers visiting general hospitals in Mongolia have been reported (7, 15, 18; T. Oyunsuren, S. Togos, Z. Odgerel, B. Dashnyam, and T. Delger, Proc. 4th Int. Meet. Hepatitis C Virus Related Viruses Mol. Virol. Pathog., p. 202, 1997). However, the prevalence of infections with the five known hepatitis viruses—HAV, HBV, HCV, HDV, and HEV—among apparently healthy individuals in Mongolia, in relation to living place and lifestyle, is poorly understood in Mongolia. Therefore, the aims of the present study were to investigate the prevalence of HAV, HBV, HCV, HDV, and HEV infections among 249 inhabitants in Mongolia, stratified by age, gender, residence and lifestyle, and to examine the genotypes of HBV, HCV, and HDV in infected individuals, in order to better understand the molecular epidemiology of hepatitis viruses in this country.
MATERIALS AND METHODS
Serum samples.
Serum samples were collected from a total of 249 apparently healthy individuals (126 males and 123 females; age, mean ± standard deviation, 48.4 ± 13.9 years; range, 23 to 86 years) in Mongolia between 25 September and 2 October 2002 after we obtained informed consent. Among the 249 inhabitants, 122 (49%) lived in apartment houses in the central area of Ulaanbaatar, the capital city of Mongolia. The remaining 127 individuals, who were age and sex matched with the 122 inhabitants living in Ulaanbaatar, were members of nomadic tribes who lived in “gers” (movable houses) around the capital city.
Sera from the inhabitants were tested for antibodies against HAV (anti-HAV [total]) by enzyme-linked immunosorbent assay (HAT-EIA; Denka Seiken, Tokyo, Japan). The presence of HBsAg and the corresponding antibodies (anti-HBs) was determined by passive hemagglutination with commercial assay kits (Mycell HBsAg [RPHA] and Mycell anti-HBs [PHA], respectively; Institute of Immunology Co. Ltd., Tokyo, Japan). Antibodies to HCV (anti-HCV) were assayed by the hemagglutination method (Abbott HCV PHA-II; Dainabot, Tokyo, Japan). To detect the immunoglobulin G (IgG) class of antibodies to HEV (anti-HEV IgG), enzyme-linked immunosorbent assay was performed using purified recombinant ORF2 protein of HEV genotype IV that had been expressed in the pupae of silkworms as the antigen probe, as described previously (19). The specificity of the anti-HEV assay was verified by absorption with the same recombinant ORF2 protein that was used as the antigen probe or a mock protein obtained from the pupae of silkworms infected with nonrecombinant baculovirus, as described elsewhere (1).
Detection of HBV DNA and determination of HBsAg subtype and HBV genotype.
The presence of HBV DNA was determined by the method described previously (11), with slight modifications. Briefly, nucleic acids were extracted from 100 μl of serum using a commercially available kit (SMITEST EX-R&D; Genome Science Co. Ltd., Tokyo, Japan) and were tested for HBV DNA by nested PCR using primers derived from the well-conserved areas in the S gene region of the HBV genomes of all eight genotypes (A to H) reported thus far (3, 20, 24, 33) and Perkin-Elmer AmpliTaq DNA polymerase (Roche Molecular Systems, Inc., Branchburg, N.J.). The first-round PCR (94°C for 2 min before the start of cycling: 94°C for 30 s, 55°C for 30 s, and 72°C for 90 s, with an additional 7 min in the last cycle) was performed for 35 cycles with primers HB095 (sense, 5′-GAG TCT AGA CTC GTG GTG GAC-3′) and HB184 (antisense, mixture of two sequences: 5′-CGA ACC ACT GAA CAA ATG GCA CCG C-3′ and 5′-CGC ACC ACT GAA CAA ATT GCA C-3′). The second-round PCR for 25 cycles was carried out under the same conditions as the first-round PCR except for extension for 60 s with primers HB097 (sense, 5′-GAC TCG TGG TGG ACT TCT CTC-3′) and S2-2 (antisense, 5′-GGC ACT AGT AAA CTG AGC CA-3′). The amplification product of the first-round PCR was 461 bp (nucleotides [nt] 244 to 704), and that of the second-round PCR was 437 bp (nt 251 to 687): the nucleotide numbers are in accordance with a genotype C HBV isolate of 3,215 nt (AB033550).
The HBsAg subtype was determined based on the nucleotide sequence of codons 122 and 160 of the S gene (22, 23). The HBV genotype was determined by phylogenetic analysis of the above-mentioned S gene sequence (396 nt; primer sequences at both ends excluded).
Detection of HCV RNA and genotyping of HCV.
Sera from individuals with anti-HCV were assayed for HCV RNA by reverse transcription-PCR using primers derived from well-conserved areas of the 5′ untranslated region of the HCV genome as previously described (26). HCV genotypes 1a, 1b, 2a, 2b, and 3a were determined by the previously described method with a slight modification (25). In brief, the original genotype 1b-specific antisense primer (primer 133) was replaced by another primer, primer 492 (9).
Detection of HDV RNA.
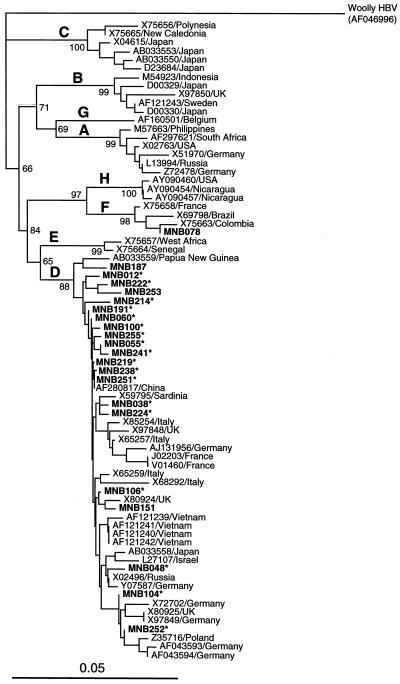
The presence of HDV RNA was determined in RNAs extracted from 100 μl of serum by reverse transcription-PCR with nested primers derived from conserved areas of all reported HDV genomes of genotypes I, II, and III (for accession numbers of reported strains, see Fig. 2). Briefly, the extracted RNAs were heated at 70°C for 3 min, chilled quickly on ice, and subjected to cDNA synthesis with reverse transcriptase (Superscript II; Invitrogen, Tokyo, Japan) and primer D13 (5′-GGA YCA CMG MMG AAG GAA GGC CCT-3′ [where Y is T or C and M is A or C]). The cDNAs were heat denatured at 95°C for 15 min and were subjected to the first-round PCR with Platinum TaqDNA polymerase (Invitrogen) and primers D9 (5′-CTC GCY GGC GCC GGC YGG GCA AC-3′) and D13 for 35 cycles (94°C for 2 min before the start of cycling: 94°C for 30 s, 55°C for 30 s, and 72°C for 75 s [additional 7 min in the last cycle]). The second-round PCR for 25 cycles was carried out under the same conditions as the first-round PCR except for extension for 60 s with primers D11 (5′-GGC YGG GCA ACA TTC CGA RGG-3′ [where R is A or G]) and D14 (5′-GAA GGC CCT SGA GAA CAA GA-3′ [where S is C or G]). The amplification product of the first-round PCR was 592 bp (nt 707 to 1298), and that of the second-round PCR was 565 bp (nt 719 to 1283); nucleotide numbers are in accordance with the prototype HDV isolate (X04451). The PCR product of the second-round PCR was subjected to electrophoresis on an agarose gel, and a sample with a visible band at 565 bp was considered to be positive for HDV RNA. The HDV genotype was determined by phylogenetic analysis of the amplified HDV sequence (522 to 526 nt; primer sequences at both ends excluded).
FIG. 2.
Phylogenetic tree constructed by the neighbor-joining method based on the partial nucleotide sequence (522 to 526 nt) of 51 HDV isolates. In addition to the 20 Mongolian isolates obtained in the present study, which are indicated in boldface type, 31 reported HDV isolates of genotypes I to III whose entire sequence is known were included for comparison. The reported isolates are indicated with the accession number followed by the name of the country of isolation, when available. Bootstrap values are indicated for the major nodes as a percentage of the data obtained from 1,000 resamplings.
Sequence analysis of PCR products.
The amplification products were sequenced directly on both strands using the BigDye Terminator Cycle Sequencing Ready Reaction kit on an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Foster City, Calif.). Sequence analysis was performed using Genetyx-Mac (version 12.0.6; Genetyx Corp., Tokyo, Japan) and ODEN (version 1.1.1) from the DNA Data Bank of Japan (National Institute of Genetics, Mishima, Japan) (12). Sequence alignments were generated by CLUSTAL W (version 1.8) (34). Phylogenetic trees were constructed by the neighbor-joining method (30). Bootstrap values were determined with 1,000 resamplings of the data sets (6). The final tree was obtained using the TreeView program (version 1.6.6) (27).
Statistical analysis.
Statistical analyses were performed using the χ2 test for comparison of proportions between two groups and the Mann-Whitney U test for comparison of continuous variables between two groups. Differences were considered to be statistically significant when P was <0.05.
Nucleotide sequence accession numbers.
The sequences determined in the present study have been deposited in the DDBJ, GenBank, and EMBL nucleotide databases under accession no. AB119010 to AB119051.
RESULTS
Prevalence of hepatitis virus markers stratified by gender and residence.
Overall, HBsAg was detected in 24 (10%) of 249 inhabitants in Mongolia, of whom 22 (92%) had detectable HBV DNA. Surprisingly, HDV RNA was detectable in 20 (83%) of the 24 HBsAg-positive subjects. Among the 24 HBsAg-positive subjects, 18 (75%) were positive for both HBV DNA and HDV RNA, whereas 4 (17%) were positive for HBV DNA but negative for HDV RNA. In the remaining two subjects, only HDV RNA was detectable, although HDV cannot replicate in the absence of HBV. Therefore, it is likely that these two HBsAg-positive subjects had HBV viremia at a level lower than the detection limit of the PCR assay used. Anti-HBs were found in 101 subjects (41%), indicating a high prevalence of HBV infection in the studied population. HCV-associated antibodies were detected in 41 subjects (16%), and HCV RNA was detected in 36 subjects (14%), all of whom had a hemagglutination titer of ≥212. In the five anti-HCV-positive, HCV RNA-negative subjects, the hemagglutination titer of anti-HCV was 210 or lower. Of note, none of the HBsAg-positive subjects had concurrent HCV infection, indicating that the HBV/HCV-viremic subjects, who were possibly chronic hepatitis virus carriers, accounted for approximately one-fourth (24% or 60 of 249) of the study population. In addition, antibodies to HAV were detected in all 249 subjects (100%) and antibodies to HEV were detected in 28 subjects (11%).
Table 1 compares the prevalences of various serological and virological markers of hepatitis viruses among the subjects stratified by gender or residence. The prevalence of HBsAg was significantly higher among males than among females (13 versus 6%; P = 0.0370). Similarly, HBV DNA was detected significantly more frequently among males than among females (13 versus 5%; P = 0.0297). On the contrary, the prevalence of anti-HCV and HCV RNA tended to be higher among females than among males, although the differences were not statistically significant. There was an extremely high prevalence of hepatitis virus infections, and there were no significant differences in the prevalences of various hepatitis virus markers between the population of 122 subjects who lived in apartment houses in the center of Ulaanbaatar and the population of 127 subjects who lived in gers around the capital city, although the prevalence of HBsAg tended to be higher among the subjects living in gers (P = 0.2360).
TABLE 1.
Prevalence of hepatitis virus markers among 249 inhabitants of Mongolia stratified by gender and residenceb
| Inhabitant | Mean age ± SD (yr) | No. (%) of inhabitants positive for:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| HBsAg | Anti-HBs | HBV DNAc | HDV RNAd | Anti-HCV | HCV RNAe | Anti-HAV (total) | Anti-HEV IgG | ||
| Gender | 48.4 ± 13.9 | 24 (10) | 101 (41) | 22 (9) | 20 (8) | 41 (16) | 36 (14) | 249 (100) | 28 (11) |
| Male (n = 126) | 49.6 ± 14.2 | 17 (13) | 56 (44) | 16 (13) | 13 (10) | 16 (13) | 13 (10) | 126 (100) | 14 (11) |
| Female (n = 123) | 47.2 ± 13.6 | 7 (6) | 45 (37) | 6 (5) | 7 (6) | 25 (20) | 23 (19) | 123 (100) | 14 (11) |
| Residence | |||||||||
| Apartment house (n = 122) | 49.0 ± 14.3 | 9 (7) | 53 (43) | 9 (7) | 8 (7) | 21 (17) | 17 (14) | 122 (100) | 15 (12) |
| Gera (n = 127) | 47.9 ± 13.5 | 15 (12) | 48 (38) | 13 (10) | 12 (9) | 20 (16) | 19 (15) | 127 (100) | 13 (10) |
A movable house for nomadic tribes living around Ulaanbaatar.
P values for all subjects were not significant, except with subjects positive for HBsAg and HBV DNA. The prevalence of HBsAg was significantly higher among males than among females (P = 0.0370), as was the prevalence of HBV DNA (P = 0.0297).
Only individuals with HBsAg were tested for HBV DNA.
Only individuals with HBsAg were tested for HDV RNA.
Only individuals with anti-HCV were tested for HCV RNA.
Age-dependent prevalence of hepatitis virus markers.
The age-specific prevalence of various hepatitis virus markers is shown in Table 2. Anti-HCV and HCV RNA were detected significantly more frequently in the age group of 50 to 86 years than in the age group of 23 to 49 years (24% [26 of 110] versus 11% [15 of 139] [P = 0.0066] and 21% [23 of 110] versus 9% [13 of 139], [P = 0.0100], respectively). However, positivity for HBsAg, HBV DNA, anti-HBs, HDV RNA, or anti-HEV IgG was distributed almost equally among the age groups, and there were no significant age-dependent differences in the prevalences of these five hepatitis virus markers. The prevalence of anti-HEV IgG tended to be lower among the subjects in their 20s (4% or 1 of 27) than among the older population (12% or 27 of 222), although the difference fell short of being statistically significant (P = 0.1890).
TABLE 2.
Age-dependent prevalence of hepatitis virus markers among 249 inhabitants of Mongolia
| Age (yr) | No. of inhabitants | No. (%) of individuals positive for:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| HBsAg | Anti-HBs | HBV DNAa | HDV RNAb | Anti-HCV | HCV RNAc | Anti-HAV (total) | Anti-HEV IgG | ||
| 23-29 | 27 | 2 (7) | 9 (33) | 1 (4) | 1 (4) | 2 (7) | 2 (7) | 27 (100) | 1 (4) |
| 30-39 | 47 | 6 (13) | 17 (36) | 6 (13) | 6 (13) | 5 (11) | 5 (11) | 47 (100) | 5 (11) |
| 40-49 | 65 | 7 (11) | 29 (45) | 6 (9) | 6 (9) | 8 (12) | 6 (9) | 65 (100) | 8 (12) |
| 50-59 | 40 | 3 (8) | 15 (38) | 3 (8) | 2 (5) | 10 (25) | 10 (25) | 40 (100) | 4 (10) |
| 60-69 | 51 | 5 (10) | 21 (41) | 5 (10) | 4 (8) | 12 (24) | 10 (20) | 51 (100) | 7 (14) |
| 70-86 | 19 | 1 (5) | 10 (53) | 1 (5) | 1 (5) | 4 (21) | 3 (16) | 19 (100) | 3 (16) |
Only individuals with HBsAg were tested for HBV DNA.
Only individuals with HBsAg were tested for HDV RNA.
Only individuals with anti-HCV were tested for HCV RNA.
Distribution of HBV, HDV, and HCV genotypes.
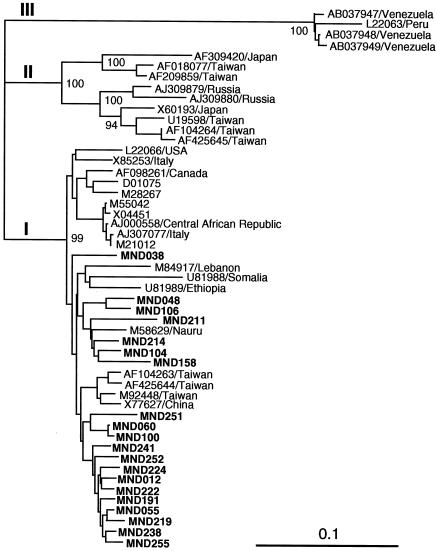
The 22 HBV isolates obtained in the present study were given designations consisting of the prefix MNB followed by the number of the subject (subjects 012 to 106 in Ulaanbaatar and subjects 151 to 255 around the capital city); the letters “MN” stand for Mongolia and the letter “B” stands for HBV. The 22 isolates differed by 0.5 to 7.6% from each other in the partial S gene sequence of 396 nt, indicating the genetic variability of HBV circulating in Mongolia. Figure 1 depicts the phylogenetic tree constructed based on the 396-nt S gene sequence of the 22 HBV isolates obtained in the present study, along with those from 52 representative HBV isolates of genotypes A to H thus far reported, using a woolly HBV as an outgroup. Of the 22 Mongolian HBV isolates, 21 (95%) were most closely related to genotype D isolates and the remaining 1 was closest to genotype F isolates; the serum sample from which the genotype F strain (MNB078) was isolated was negative for HDV RNA. Eight genotype D HBV isolates recovered from inhabitants in Ulaanbaatar were interspersed among isolates from members of Mongolian nomadic tribes living around Ulaanbaatar and from infected individuals in Europe and other Asian countries. All HBsAg-positive samples of genotype D were typed as subtype ayw, and that of genotype F was typed as subtype adw.
FIG. 1.
Phylogenetic tree constructed by the neighbor-joining method based on the partial nucleotide sequence of the S gene (396 nt) of 74 HBV isolates, using a woolly HBV (AF046996) as an outgroup. In addition to the 22 Mongolian isolates found in the present study (which are indicated in boldface type for visual clarity), 52 reported HBV isolates of genotypes A to H whose entire sequence is known were included for comparison. Each reported isolate is indicated with the accession number followed by the name of the country where it was isolated. Asterisks denote Mongolian HBV strains that were isolated from serum samples that were also positive for HDV RNA. Bootstrap values are indicated for the major nodes as a percentage of the data obtained from 1,000 resamplings.
HDV isolates were recovered from the 20 HDV RNA-positive subjects and were given designations consisting of the prefix MND followed by the number of the subject; the letter “D” stands for HDV. When phylogenetic analysis of the 20 HDV isolates obtained in the present study and 31 previously reported HDV isolates was performed, the 20 HDV isolates obtained in the present study were classified exclusively into genotype I (Fig. 2). However, the 20 Mongolian isolates were 2.1 to 11.4% different from each other in the HDV sequence of 522 to 526 nt, indicating dual genetic heterogeneity of the HBV and HDV circulating in Mongolia. Of the 36 HCV RNA-positive samples, 35 (97%) were typeable into genotype 1b and the remaining 1 sample was typeable into genotype 2a. The HCV genotype 2a isolate was obtained from a 57-year-old female (subject 135) who lived in a ger around Ulaanbaatar.
DISCUSSION
In the present study, we conducted an epidemiological survey of infection with five known hepatitis viruses—HAV, HBV, HCV, HDV, and HEV—in 249 23- to 86-year-old apparently healthy individuals living in or around Ulaanbaatar, the capital city of Mongolia. The studied population included two age- and sex-matched groups differing mainly by living place and lifestyle. To our surprise, there were no significant differences in the prevalences of various hepatitis virus markers between the two groups of the 122 subjects who lived in the urban area and the 127 subjects who led a nomadic lifestyle. Symbolizing a high prevalence of hepatitis virus infections, anti-HAV was found in all of the subjects studied, as is the case in many parts of the world where sanitation is suboptimal. Anti-HEV IgG was also prevalent, suggesting the presence of acute hepatitis E in this country.
According to the Mongolia health sector review published by the Government of Mongolia and the World Health Organization in June 1999 (http://www.un-mongolia.mn/who/review.html), communicable diseases are still one of the main health problems in Mongolia, and approximately 25,300 cases of 25 different infectious diseases were registered (111 per 10,000 members of the population) in 1997, the incidence being nearly twice as high as that in 1993. The increase was mainly due to high increases in the incidence of viral hepatitis, tuberculosis, brucellosis, and shigellosis: there is no separate registration yet for viral hepatitis C in Mongolia. In East Asian countries, the prevalence of HBsAg and anti-HCV in the general population or blood donors has been reported to be high, for example, 3.7% in China (Beijing) and 6.3% in Thailand for HBsAg and 3.9% in China (Beijing) and 4.1% in Thailand for anti-HCV (17, 35). Surprisingly, the subjects in our present study showed much higher rates of HBsAg (10%) and anti-HCV (16%) positivity than the population in neighboring countries. However, the prevalence of HBsAg among our subjects was comparable with the reported prevalence of HBsAg in Mongolia: 6.9% (n = 189, general population in 1990) (T. Oyunsuren et al., Proc. 4th Int. Meet. Hepatitis C Virus Related Viruses Mol. Virol. Pathog.), and 9.1% (n = 121, blood donors in 1995) (15). Similarly, the anti-HCV-positive rate among our subjects was comparable with the reported prevalence of anti-HCV among blood donors or the general population in Mongolia (10.7 to 36.3%) (15, 18; T. Oyunsuren et al., Proc. 4th Int. Meet. Hepatitis C Virus Related Viruses Mol. Virol. Pathog.). In addition, higher rates of HBsAg and anti-HCV positivity were reported among 150 outpatient volunteers visiting two general hospitals in Ulaanbaatar (28.7 and 48.0%, respectively), although the underlying disease was not clarified (7). These results indicate that Mongolia is one of the countries with the highest rates of both HBV and HCV carriage in the world.
Although HBV infection is usually minimally symptomatic in early childhood, chronic carriage is likely to occur if the infection is acquired at a young age. Vertical transmission from mother to neonate occurs at disproportionately high rates in Asian populations (21, 31, 32). Therefore, it is likely that mother-to-infant transmission, combined with horizontal transmission in childhood, has historically produced a high prevalence of HBsAg in the Mongolian population. One Mongolian HBV strain was classified into genotype F, and the remaining 21 strains were grouped into genotype D; this corroborates the finding of Alestig et al. (2) that all nine Mongolian HBV strains studied were of genotype D. The distribution of HBV genotypes is geographically confined, with genotype D in the Mediterranean and Middle East regions and genotype F in the Americas (13); genotype F is rare in Asian countries. Fifteen (71%) of the 21 Mongolian HBV strains belonging to genotype D in the present study were most closely related to the Chinese strain (AF280817) that had been isolated in the Ningxia Hui Autonomous Region, which is bordered by the Inner Mongolia Autonomous Region to the north, with 99.0 to 100% nucleotide sequence identity; MNB251 was 100% identical to the Chinese strain. However, the Mongolian genotype D HBV strains were not clearly separate from the European and other Asian genotype D sequences, from which they differed by only 0.5 to 3.8%. Of note, our Mongolian strains of genotype D were 97.1 to 100% similar to the seven Mongolian strains reported by Alestig et al. (2), in the common 384-nt sequence within the S gene. The finding that genotype D HBV strains predominantly circulate in Mongolia may reflect the close economic and cultural contact with Eastern Europe and the Mediterranean area where genotype D prevails, as in Ningxia and Hami in the northern region of China, an area where HBV of genotype D or subtype ayw is prevalent and which had historical contact with Mediterranean people through the Silk Road (35).
Epidemiological studies of HDV infection in HBsAg-positive individuals have shown a worldwide, but nonuniform, distribution; areas of high prevalence include the Mediterranean basin (8). However, no data of HDV infection have been available for Mongolia. In the present study, 20 (83%) of 24 HBsAg-positive Mongolians were found to be coinfected with HDV, although they were symptom free. As pockets of serious liver disease leading to fulminant hepatitis are frequently recorded in South America (29), it has to be clarified in future studies whether frequent dual infection of HBV and HDV is associated with severe liver diseases in Mongolia.
In the present study, the prevalence of anti-HCV and HCV RNA tended to increase in older age, similar to that observed in Japan (37). As transfusion of blood or blood products contaminated with HCV, improper disinfection of medical equipment, and tattooing or traditional medicine where the skin is broken (through which blood contaminated with HCV may be introduced) have been suspected to be causes of HCV transmission in Japan in the past (14, 37), it is likely that a similar situation still exists in Mongolia. However, to elucidate the transmission route of HCV in this country, their lifestyle, folk remedies, and customs should be taken into consideration. In Mongolia in 1998, among all newly diagnosed cancers, 36% were in the liver, followed by the stomach (15%) and lung (11%) (http://www.un-mongolia.mn/who/review.html). Therefore, it is beyond doubt that the leading causes of liver cancer are HBV and HCV infections. Since 1991, the hepatitis B vaccine has been included in Mongolia's universal childhood vaccination program (http://www.un-mongolia.mn/who/review.html), and the vaccination program for hepatitis B has successfully reduced the rate of chronic HBV carriage in the immunized generation (5).
In conclusion, the present study found that infection with HAV, HBV, HCV, HDV, and HEV was highly prevalent among adults 23 to 86 years of age in Mongolia, not only among those living in an urban area but also among nomadic tribes who live in gers, suggesting that hepatitis viruses cause acute or chronic liver disease, regardless of living place and lifestyle in Mongolia. The finding that HBV- or HCV-viremic subjects, probably chronic hepatitis virus carriers, accounted for one-fourth of the studied population and the presence of extremely frequent dual infection with HBV and HDV stress the necessity of nationwide epidemiological surveys of hepatitis viruses, particularly HBV and HCV, which may be related to the development of cirrhosis and hepatocellular carcinoma, in order to prevent de novo infection with hepatitis viruses and to suppress the spread and development of liver diseases in this country.
Acknowledgments
We are grateful to Makoto Mayumi for his advice and encouragement during this study.
This work was supported in part by grants from the Ministry of Health, Labor, and Welfare of Japan and a grant, High Technology Center of Kagawa Nutrition University, from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
REFERENCES
- 1.Aikawa, T., M. Kojima, M. Takahashi, T. Nishizawa, and H. Okamoto. 2002. Identification of indigenous hepatitis E virus from a Japanese patient who contracted sporadic acute hepatitis in 1982. J. Infect. Dis. 186:1535-1536. [DOI] [PubMed] [Google Scholar]
- 2.Alestig, E., C. Hannoun, P. Horal, and M. Lindh. 2001. Hepatitis B virus genotypes in Mongols and Australian aborigines. Arch. Virol. 146:2321-2329. [DOI] [PubMed] [Google Scholar]
- 3.Arauz-Ruiz, P., H. Norder, B. H. Robertson, and L. O. Magnius. 2002. Genotype H: a new Amerindian genotype of hepatitis B virus revealed in Central America. J. Gen. Virol. 83:2059-2073. [DOI] [PubMed] [Google Scholar]
- 4.Blum, H. E., S. Wieland, E. Walter, F. von Weizsacker, W.-B. Offensperger, and D. Moradpour. 1998. Natural course of HBV infection, p. 75-92. In R. Koshy and W. H. Caselmann (ed.), Hepatitis B virus. Molecular mechanisms in disease and novel strategies for therapy. Imperial College Press, London, United Kingdom.
- 5.Edstam, J. S., N. Dulmaa, P. Nymadawa, A. Rinchin, J. Khulan, and A. M. Kimball. 2002. Comparison of hepatitis B vaccine coverage and effectiveness among urban and rural Mongolian 2-year-olds. Prev. Med. 34:207-214. [DOI] [PubMed] [Google Scholar]
- 6.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 7.Fujioka, S., H. Shimomuta, Y. Ishii, J. Kondo, K. Fujio, F. Ikeda, M. Miyake, S. Kusachi, and T. Tsuji. 1998. Prevalence of hepatitis B and C virus markers in outpatients of Mongolian general hospitals. Kansenshogaku Zasshi 72:5-11. [DOI] [PubMed] [Google Scholar]
- 8.Gerin, J. L., J. L. Casey, and R. H. Purcell. 2001. Hepatitis delta virus, p. 3037-3050. In D. M. Knipe, P. M. Howley, D. E. Griffin, M. A. Martin, R. A. Lamb, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 9.Holland, P. V., J. M. Barrera, M. G. Ercilla, C. T. Yoshida, Y. Wang, G. A. B. de Olim, B. Betlach, K. Kuramoto, and H. Okamoto. 1996. Genotyping hepatitis C virus isolates from Spain, Brazil, China, and Macau by a simplified PCR method. J. Clin. Microbiol. 34:2372-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollinger, F. B., and S. U. Emerson. 2001. Hepatitis A virus, p. 799-840. In D. M. Knipe, P. M. Howley, D. E. Griffin, M. A. Martin, R. A. Lamb, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 11.Iizuka, H., K. Ohmura, A. Ishijima, K. Satoh, T. Tanaka, F. Tsuda, H. Okamoto, Y. Miyakawa, and M. Mayumi. 1992. Correlation between anti-HBc titers and HBV DNA in blood units without detectable HBsAg. Vox Sang. 63:107-111. [DOI] [PubMed] [Google Scholar]
- 12.Ina, Y. 1994. ODEN: a program package for molecular evolutionary analysis and database search of DNA and amino acid sequences. Comput. Appl. Biosci. 10:11-12. [DOI] [PubMed] [Google Scholar]
- 13.Kidd-Ljunggren, K., Y. Miyakawa, and A. H. Kidd. 2002. Genetic variability in hepatitis B viruses. J. Gen. Virol. 83:1267-1280. [DOI] [PubMed] [Google Scholar]
- 14.Kiyosawa, K., E. Tanaka, T. Sodeyama, K. Yoshizawa, K. Yabu, K. Furuta, H. Imai, Y. Nakano, S. Usuda, K. Uemura, S. Furuta, Y. Watanabe, J. Watanabe, Y. Fukuda, T. Takayama, and the South Kiso Hepatitis Study Group. 1994. Transmission of hepatitis C in an isolated area in Japan: community-acquired infection. Gastroenterology 106:1596-1602. [DOI] [PubMed] [Google Scholar]
- 15.Kondo, Y., M. Mizokami, T. Nakano, T. Kato, R. Ueda, M. Mukaide, K. Hikiji, T. Ishida, D. Dorjsuren, B. Dashnyam, and T. Oyunsuren. 1997. Prevalence and molecular epidemiology of GB virus/hepatitis G virus infection in Mongolia. J. Med. Virol. 52:143-148. [PubMed] [Google Scholar]
- 16.Lauer, G. M., and B. D. Walker. 2001. Hepatitis C virus infection. N. Engl. J. Med. 345:41-52. [DOI] [PubMed] [Google Scholar]
- 17.Luengrojanakul, P., K. Vareesangthip, T. Chainuvati, K. Murata, F. Tsuda, H. Tokita, H. Okamoto, Y, Miyakawa, and M. Mayumi. 1994. Hepatitis C virus infection in patients with chronic liver disease or chronic renal failure and blood donors in Thailand. J. Med. Virol. 44:287-292. [DOI] [PubMed] [Google Scholar]
- 18.Lvov, D. K., E. L. Samokhvalov, F. Tsuda, N. A. Selivanov, H. Okamoto, V. M. Stakhanova, I. V. Stakhgildyabin, N. V. Doroshenko, T. L. Yashina, S. N. Kuzin, I. A. Suetina, P. G. Deryabin, L. A. Ruzaeva, V. N. Bezgodov, L. A. Firsova, S. N. Sorinson, and S. Mishio. 1996. Prevalence of hepatitis C virus and distribution of its genotypes in Northern Eurasia. Arch. Virol. 141:1613-1622. [DOI] [PubMed] [Google Scholar]
- 19.Mizuo, H., K. Suzuki, Y. Takikawa, Y. Sugai, H. Tokita, Y. Akahane, K. Itoh, Y. Gotanda, M. Takahahsi, T. Nishizawa, and H. Okamoto. 2002. Polyphyletic strains of hepatitis E virus are responsible for sporadic cases of acute hepatitis in Japan. J. Clin. Microbiol. 40:3209-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norder, H., A. M. Courouce, and L. O. Magnius. 1994. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of hepatitis B virus, four of which represent two new genotypes. Virology 198:489-503. [DOI] [PubMed] [Google Scholar]
- 21.Okada, K., I. Kamiyama, M. Inomata, M. Imai, Y. Miyakawa, and M. Mayumi. 1976. e antigen and anti-e in the serum of asymptomatic carrier mothers as indicators of positive and negative transmission of hepatitis B virus to their infants. N. Engl. J. Med. 294:746-749. [DOI] [PubMed] [Google Scholar]
- 22.Okamoto, H., M. Imai, Y. Miyakawa, and M. Mayumi. 1987. Site-directed mutagenesis of hepatitis B surface antigen sequence at codon 160 from arginine to lysine for conversion of subtypic determinant from r to w. Biochem. Biophys. Res. Commun. 148:500-504. [DOI] [PubMed] [Google Scholar]
- 23.Okamoto, H., M. Imai, F. Tsuda, T. Tanaka, Y. Miyakawa, and M. Mayumi. 1987. Point mutation in the S gene of hepatitis B virus for a d/y or w/r subtypic change in two blood donors carrying a surface antigen of compound subtype adyr or adwr. J. Virol. 61:3030-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okamoto, H., F. Tsuda, H. Sakugawa, R. I. Sastrosoewignjo, M. Imai, Y. Miyakawa, and M. Mayumi. 1988. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J. Gen. Virol. 69:2575-2583. [DOI] [PubMed] [Google Scholar]
- 25.Okamoto, H., H. Tokita, M. Sakamoto, M. Horikita, M. Kojima, H. Iizuka, and S. Mishiro. 1993. Characterization of the genomic sequence of type V (or 3a) hepatitis C virus isolates and PCR primers for specific detection. J. Gen. Virol. 74:2385-2390. [DOI] [PubMed] [Google Scholar]
- 26.Okamoto, H., S. Mishiro, H. Tokita, F. Tsuda, Y. Miyakawa, and M. Mayumi. 1994. Superinfection of chimpanzees carrying hepatitis C virus of genotype II/1b with that of genotype III/2a or I/1a. Hepatology 20:1131-1136. [PubMed] [Google Scholar]
- 27.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 28.Purcell, R. H., and S. U. Emerson. 2001. Hepatitis E virus, p. 3051-3061. In D. M. Knipe, P. M. Howley, D. E. Griffin, M. A. Martin, R. A. Lamb, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 29.Quintero, A., N. Uzcategui, C. L. Loureiro, L. Villegas, X. Illarramendi, M. E. Guevara, J. E. Ludert, L. Blitz, F. Liprandi, and F. H. Pujol. 2001. Hepatitis delta virus genotypes I and III circulate associated with hepatitis B virus genotype F in Venezuela. J. Med. Virol. 64:356-359. [DOI] [PubMed] [Google Scholar]
- 30.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 31.Shiraki, K., N. Yoshihara, T. Kawana, H. Yasui, and M. Sakurai. 1977. Hepatitis B surface antigen and chronic hepatitis in infants born to asymptomatic carrier mothers. Am. J. Dis. Child. 131:644-647. [DOI] [PubMed] [Google Scholar]
- 32.Stevens, C. E., R. P. Beesley, J. Tsui, and W. C. Lee. 1975. Vertical transmission of hepatitis B antigen in Taiwan. N. Engl. J. Med. 292:771-774. [DOI] [PubMed] [Google Scholar]
- 33.Stuyver, L., S. De Gendt, C. Van Geyt, F. Zoulim, M. Fried, R. F. Schinazi, and R. Roussau. 2000. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J. Gen. Virol. 81:67-74. [DOI] [PubMed] [Google Scholar]
- 34.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, Y., Q.-M. Tao, H.-Y. Zhao, F. Tsuda, R. Nagayama, K. Yamamoto, T. Tanaka, H. Tokita, H. Okamoto, Y. Miyakawa, and M. Mayumi. 1994. Hepatitis C virus RNA and antibodies among blood donors in Beijing. J. Hepatol. 21:634-640. [DOI] [PubMed] [Google Scholar]
- 36.World Health Organization. 2003. Work of WHO in the Western Pacific region. Report of the Regional Director to the Regional Committee for the Western Pacific. Statistical annex, p. 212-227. World Health Organization, Regional Office for the Western Pacific, Manila, Philippines.
- 37.Yoshizawa, H. 2002. Hepatocellular carcinoma associated with hepatitis C virus infection in Japan: projection to other countries in the foreseeable future. Oncology 62(Suppl. 1):8-17. [DOI] [PubMed] [Google Scholar]