Abstract
Summary: We present Mutascope, a sequencing analysis pipeline specifically developed for the identification of somatic variants present at low-allelic fraction from high-throughput sequencing of amplicons from matched tumor-normal specimen. Using datasets reproducing tumor genetic heterogeneity, we demonstrate that Mutascope has a higher sensitivity and generates fewer false-positive calls than tools designed for shotgun sequencing or diploid genomes.
Availability: Freely available on the web at http://sourceforge.net/projects/mutascope/.
Contact: oharismendy@ucsd.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
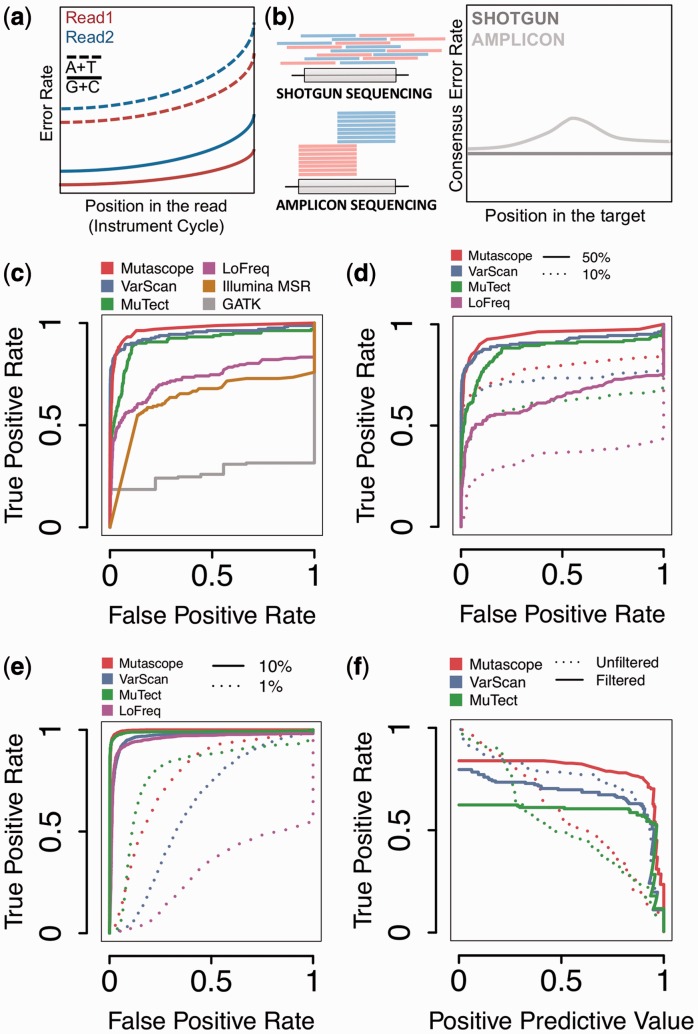
The accurate detection of somatic mutations in tumors is critical for precise diagnostic and selection of targeted therapies (Boyd, 2013), but the low-allelic fraction frequently encountered in heterogeneous or poor cellularity clinical specimens renders this task challenging. In current clinical assays, amplicons covering the exons of 10–100 cancer genes are amplified via polymerase chain reaction-based or analogous approaches and sequenced at high depth to identify mutations present in <5% of a DNA sample (Harismendy et al., 2011). Despite high coverage depth, the error rate resulting from systematic sequencing bias (Harismendy et al., 2009), can hinder the detection of mutations. Although experimental (Hiatt et al., 2013) or analytical (McKenna et al., 2010) methods, or comparison with the normal DNA (Cibulskis et al., 2013; Koboldt et al., 2012) can mitigate this effect, most analysis strategies were developed for sequencing of random shotgun DNA fragments, and thus do not take into account systematic errors specific to amplicon sequencing. In amplicon sequencing, loci are covered by reads with identical genomic starting positions, and because the error rate increases along the length of the read (Fig. 1a), a variable consensus error rate exists over the target (Fig. 1b). Analytical strategies specifically designed for amplicon sequencing have the potential to enhance the mutation detection accuracy of current clinical assays, especially at low-allelic fraction.
Fig. 1.
Mutascope principle and performance. (a) The sequencing error rate varies based on the read type (blue and red), position in the read (x-axis) or reference base sequenced (lines). (b) Paired reads (red and blue) from shotgun and amplicon sequencing distribute differently over the targeted region (gray box) resulting in different consensus error rates (right panel). (c–e) Comparison of 4–6 tools by ROC analysis showing the classification of mutations at low-allelic fraction (1–10%) in the MIX samples (c), after down-sampling reads to 50 or 10% of maximum coverage (d), or using 1 and 10% allelic fraction variants from TNS pairs. (f) Evolution of the true-positive rate and positive predicted value from the MIX sample low-allele frequency variants (1–10%) before (dotted line) and after (continuous line) application of high-confidence filters
Here, we present Mutascope, a software dedicated to the detection of mutations at low-allelic fraction from amplicon sequencing of matched tumor-normal samples pairs. Mutascope determines the amplicon of origin for each read and measures the specific experimental error rate from sequencing the normal DNA. The mutations in the tumor are then identified by comparison with the error rate using a binomial statistics and classified as germ line or somatic by comparison with the normal DNA. A set of filters adapted to amplicon sequencing then eliminates false-positive calls. We used two experimental datasets, a mixture of 8 normal DNA (MIX) and a set of 80 tumor-normal spiked-in (TNS) pairs derived from 38 different normal germ line DNA samples to measure the performance of the approach in comparison with other mutation callers.
2 METHODS
Data generation: The data from the MIX sample or used in the preparation of the TNS pairs was generated using microdroplet polymerase chain reaction amplification (Harismendy et al., 2011) of 1736 amplicons from 47 genes clinically actionable for breast cancer (Supplementary Methods), followed by high-throughput sequencing of 151 nt long paired-end reads on a MiSeq sequencer (Illumina, San Diego, CA, USA) resulting in 981-fold average coverage depth. The data are available through the Short Reads Archive (SRA) at the NCBI (SRA067609 and SRA067610).
Analysis principle: Mutascope aligns the reads to the genome using BWA-SW algorithm (Li and Durbin, 2009). Multi-mapping reads, reads with a low Smith–Watterman score, or not aligning to the specified amplicons are removed. Mutascope then determines the amplicon of origin for each read and measures the error rate using the normal DNA sequencing, stratified by the major drivers of sequencing errors: nucleotide, position in the read and read type (forward or reverse). The allelic fraction of the mutation is compared with this error rate using a binomial test for significance; the mutations are then classified as germ line or somatic using a Fisher exact test. The germ line genotypes are determined using a Bayesian likelihood method. Finally, Mutascope filters out false-positive variants using, for example, read group bias, low-average mutant allele quality or predicted false positive from non-specific amplification.
Benchmarking: The benchmarking was performed using ROCR package (Sing et al., 2005). The prediction score used for the classification corresponded to the binomial P-value for Mutascope, somatic P-value for VarScan (Koboldt et al., 2012), tumor Fstar LOD score for MuTect (Cibulskis et al., 2013) and VCF quality score for LoFreq, GATK and Illumina MiSeq Reporter (McKenna et al., 2010; Wilm et al., 2012). All false-negative prediction scores were set to 0. All benchmarked tools relied on the same alignment performed by Mutascope, except for Illumina MiSeq reporter that performs its own alignment. Whenever allowed, each tool was run without extensive previous filtering to strictly compare the accuracy of the mutation detection. Complete methods are available as Supplementary Methods.
3 RESULTS
We benchmarked Mutascope against other mutation callers using sequencing data generated from a mixture of 8 normal DNA samples with known genotypes (MIX sample) resulting in ‘somatic mutations’ at variable allelic fraction. The classification of the 162 somatic mutations at low-allelic fraction (0.01–0.1) by Mutascope was more accurate than other standard tools (area under the curve: 0.97—Fig. 1c). Not surprisingly, tools designed to identify heterozygotes in diploid genomes were missing most mutations (GATK), whereas tools dedicated to tumor-normal pairs performed better (VarScan and MuTect). To estimate the impact of coverage depth, we selected reads from the MIX sample down to 50 or 10% (490 and 98×, respectively) of the maximum. As expected, the sensitivity decreased equally for all the tools considered (Fig. 1d).
To expand the performance evaluation to additional mutations and experimental conditions, we prepared a set of 80 TNS pairs by mixing reads obtained from sequencing 38 normal DNA. Using these, we interrogated 402 unique ‘somatic mutations’ (between 17 and 55 per pair) at an allelic fraction of 0.01 or 0.1 (40 pairs each). Mutascope was more accurate to detect mutations at an allelic fraction of 0.1 rather than 0.01 (Fig. 1e), and in the former case, its performance was comparable with MuTect and superior to VarScan or LoFreq.
Finally, we tested the effect of the empirical filters applied by each tool after the classification. These filters are important to eliminate false positives resulting from unpredictable sources of error and not accounted for by the statistical model. Although Mutascope’s filters, such as the read group bias and non-specific amplification, are specifically compatible with amplicons sequencing, we adjusted the parameters of the other tools to ensure a fair comparison, such as strand bias and minimum alternate allele frequency filters. When applied to the mutations at low-allelic fraction in the MIX samples, these filters increase the sensitivity and positive predictive value (Fig. 1f and Supplementary Discussion). The set of high-confidence filters from MuTect affects the sensitivity the most. This observation highlights synergies between Mutascope’s two core statistical components: the experimentally driven mutation detection (binomial test) and tumor-normal comparison (Fisher test) resulting in a superior performance.
Therefore, by design, Mutascope specifically optimizes the mutation detection and filtering for deep amplicon sequencing. The resulting higher accurate detection of somatic mutations at low-allelic fraction increases utility in cancer molecular diagnostics.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr P. Carpenter, Dr H. Park and Dr H. Anton-Culver for the collection of samples; Dr Bao and Dr Messer for helpful discussions; RainDance Tech. (Lexington, MA, USA) for technical assistance.
Funding: National Cancer Institute (1R21CA155615-01A1 and 1R21CA152613-01 to O.H. and K.A.F.); NCATS (UL1RR031980 to Dr Firestein); a pilot award from the NIH Center of Excellence Grant to the San Diego Center for Systems Biology (P50 GM085764).
Conflict of Interest: none declared.
REFERENCES
- Boyd SD. Diagnostic applications of high-throughput DNA sequencing. Annu. Rev. Pathol. 2013;8:381–410. doi: 10.1146/annurev-pathol-020712-164026. [DOI] [PubMed] [Google Scholar]
- Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harismendy O, et al. Evaluation of next generation sequencing platforms for population targeted sequencing studies. Genome Biol. 2009;10:R32. doi: 10.1186/gb-2009-10-3-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harismendy O, et al. Detection of low prevalence somatic mutations in solid tumors with ultra-deep targeted sequencing. Genome Biol. 2011;12:R124. doi: 10.1186/gb-2011-12-12-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiatt JB, et al. Single molecule molecular inversion probes for targeted, high accuracy detection of low frequency variation. Genome Res. 2013;23:843–854. doi: 10.1101/gr.147686.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, et al. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing T, et al. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- Wilm A, et al. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012;40:11189–11201. doi: 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.