Abstract
Pathogens exhibit remarkable abilities to flout therapeutic intervention. This outcome is driven by evolution, either as a direct response to intervention (e.g. the evolution of antibiotic resistance), or through long-term coevolution generating host or parasite traits that interact with therapy in undesirable or unpredicted ways. To make progress, the concepts and techniques of evolutionary biology must be deeply integrated with traditional approaches to immunology and pathogen biology. An interdisciplinary approach can inform control strategies, or even patient treatment, positioning us to meet the current and future challenges of controlling infectious diseases.
The last century has seen considerable progress in controlling infectious diseases, but success is far from universal. Pathogens remain a major health burden, with over 9.8 million people per year dying from infections (over 16.5% of annual deaths), half of them children1. Thus, despite intense investment in the development of vaccines and therapeutics in the 20th century, old (existing and resurgent) and new (emerging) pathogens remain a significant threat. Some treatments or control efforts begin effectively, and then lose ground. Other disease problems appear intractable, and control programs fail at an early stage. What causes failure? What underlies variability in the short and longer-term success of control measures? A major problem is evolution. And evolution, aside from being pervasive, often works in complex ways with outcomes that are challenging to predict.
This is the age of biology, but to fully exploit the moment, we need to recognise the scale and complexity of the problems posed by evolution, and then find a way to productively integrate evolutionary biology with traditional biomedical research. The fact that these fields typically approach problems in markedly disparate ways is a challenge that needs to be overcome if we are to understand, control or even manipulate evolutionary processes to our benefit.
Evolution: the scale of the problem
The threat posed by evolution is vast and falls into two broad categories. First, intervention drives evolution. The evolution of bacterial resistance to antibiotics and vector resistance to pesticides are the classic (and ongoing) examples of intervention-driven evolution2-4, though many other interventions may provoke undesirable evolution5-8. It is crucial to note that, in the majority of these cases, resistance did not evolve as expected, and intuition stemming from an overly simplistic view of the evolutionary process has proven a poor guide to identifying threats and exploiting new opportunities for control (Box 1).
Second, past evolution confounds intervention. Complex traits that have been fine-tuned by aeons of natural selection and coevolution can incidentally thwart control efforts. These include the classic examples of pathogen adaptations: hiding with an non-antigenic cloak9, antigen switching10, or directly manipulating the immune system11-15, each of which defies our efforts to identify targets for vaccination. Evolved traits can also become intervention-driven threats. For example helminth parasites appear to have the capacity to monitor host immune status and shift reproduction into high gear, thereby enhancing transmission7. Such plasticity in reproductive schedules, which the parasite has the capacity for due to a long evolutionary interaction with immune systems, can also be direct responses to immune effectors that are boosted by vaccination7. Host adaptations can also confound treatment. For example, it was thought that anaemia was a pathological consequence of bacterial infection, but patients died when iron was given because anaemia was actually an adaptation by the host to remove the iron upon which bacteria rely. The role of iron remains a current treatment issue for a variety of parasites and pathogens16,17. To get treatment right, we must understand why an adaptation such as anaemia evolved. A similar reasoning applies to fever. Should we always treat mild fever during an infection? This depends on whether the fever, or any pathology, is an evolutionary adaptation, and if so for whom – the host or the parasite? (Box 1).
Productively integrating evolutionary biology
Creating novel, long-lasting, therapies against historically elusive pathogens (e.g. Plasmodium, helminths), or sustaining the control initiative against resurgent and new pathogens (e.g. tuberculosis, influenza, or opportunistic bacteria), will require the integration of pathogen biology and immunology with evolutionary biology. From the outset, integration will involve overcoming language barriers-evolutionary biologists and traditional biomedical researchers often speak very different languages, and we should begin preparing young scientists to become fluent in diverse terminologies. These disciplines also differ substantially in their respective approaches. Infectious disease biologists have traditionally interrogated systems at the molecular and cellular levels, while evolutionary biologists more often consider whole organism fitness, polymorphism and changes in populations through time (Figure 1). For instance, immunologists strive to reduce variation (e.g. environmental, genetic) to elucidate mechanistic pathways, but for ecologists and evolutionary biologists, variation is the subject, as they analyse changes in fitness in relation to genetic and environmental heterogeneity. There is room for the biomedical sciences to shift away from a focus on inbred model organisms in the laboratory to emphasise the responses of real hosts in the wild18,19. Equally, there is room for evolutionary biology and ecology to strengthen their appreciation of the mechanistic underpinnings of traits, and transform this knowledge into predictive models. We may require more study systems, as humans and mice may not be ideal for evolutionary studies, but the role of traditional model systems could also be expanded to test whether patterns observed in the laboratory can be generalised to other environments (e.g. 19,20,21).
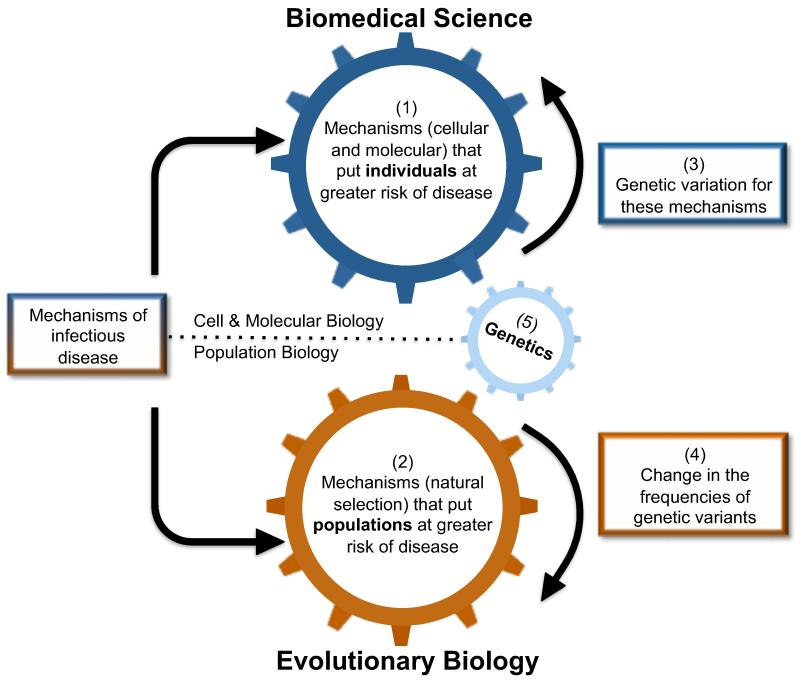
Figure 1.
A central aim of all biomedical research is to elucidate mechanisms associated with infectious disease. Much basic biomedical research, for example immunology or pathogen biology, has its roots in cellular and molecular biology, and as such, has sought to (1) identify cellular or molecular mechanisms (e.g. pathogen virulence factors or host immune deficiencies) that place an individual at risk of contracting or expressing disease. The study of evolution (2), by contrast, is principally a form of population biology, and in the context of infectious disease, evolutionary biology seeks to identify mechanisms (prominently, natural selection) that change whole populations towards either a greater or lower average risk of infection. There has been substantial progress towards identifying (3) genetic variation for the cellular/molecular mechanisms that put individuals at greater risk of disease; evolutionary biology then studies (4) how the frequencies of these genetic variants may change over time due to natural selection. Thus, genetics (5), either molecular or quantitative, provides an important link between traditional biomedical science and evolutionary biology.
We do not expect integration to be easy. Disentangling the complex molecular biology of host-pathogen interactions is not straightforward, and although the elegant simplicity of Darwin’s theory of natural selection can tempt us into thinking the evolutionary outcomes will be easy to predict, they are not (Box 1). Resistance to even a single drug may involve multiple mechanisms and mutational targets, and predicting the spread of resistance in real populations will require complex, parameter-rich models where estimating each parameter will represent a significant challenge. This complexity means we cannot use a cursory understanding of evolutionary biology to guess at evolutionary outcomes. Technological transformations, particularly in genomics, bioinformatics and computing power, are already aiding integration by creating new opportunities to map genotype and molecular mechanism onto phenotype22 in models and non-models alike. We will require scientists with mastery of the new technologies, but perhaps the most important skill will be the ability to contextualise the ever-expanding quantities of data.
With the right data and outlook, we can begin to ask biomedically relevant questions at multiple levels, creating a synergy that allows biological insights to be translated into control strategies. For example, an important question is why hasn’t natural selection purged susceptible (to infection) genotypes from natural populations? Genetic work has revealed possible answers by showing that there are evolutionary trade-offs, where parasite-resistant individuals suffer chronic disease. A recent example, obtained by combining data from association studies and characterising signatures of natural selection, is a polymorphism in the human Apo-L1 protein that confers trypanosome resistance, but also kidney disease23. Sickle-cell anemia and malaria is the entrenched example of such a trade-off, but even this is being reassessed, as data point to new understanding of why sickle cells are protective. In particular, rather than reducing parasite load, it is now surmised that sickle cells release more heme, inducing the production of heme-neutralising systems that assist with both the sickle cell pathology and cerebral malaria24. This form of protection, where pathology is reduced without reducing parasite numbers, puts fundamentally different selective pressures on parasite populations25. Ultimately, analysis of selection and the resulting polymorphism can generate predictive biomarkers, which assist analyses of disease spread. Whilst genotyping and phenotyping in the field remain challenging in areas with poor health infrastructure, as genome sequencing studies continue to analyse more dispersed human populations, it might be possible to use immune polymorphisms to predict disease susceptibility and, with understanding of local pathogen diversity, therapies could even be tailored to specific geographical groups.
In addition to these human studies, multi-generational data sets on wild mammals are now allowing us to associate immunological data with comprehensive measures of Darwinian fitness in the context of optimal immunity. For example, the novel application of immunological tools has shown that highly immune-responsive feral sheep suffer autoimmunity, which reduces reproductive success but enhances survival due to their lower parasite burdens26. The balance of the immune system is also influenced by coinfection: removing one set of pathogens in a co-infection or altering commensal populations can increase susceptibility to other pathogens27, leading to an imbalanced immune response28-30, with potential for autoimmunity, allergy and asthma. Hence, pathogen exposure, and long-term coevolutionary interactions, may not always select striking and discrete polymorphisms such as those at Apo-L1, and instead may select for complex and graduated genetic responses (best studied with the tools of quantitative genetics) that respect the demands of polyparasitism. New pathogen control strategies must address optimal immunity in the real world, where multiple infections are the norm, to avoid adverse immune consequences or the emergence of new, or previously rare, pathogens.
Thus, analyses of how pathogens, and pathogen diversity, evolve in response to control measures are crucial. New approaches are making it possible to rapidly identify medically relevant features of pathogens and the timescales on which they arise. For example, population genomic analysis of ongoing epidemics are elucidating the timing of the emergence of drug resistance, changes in host range and pathogenesis, and identifying source populations and species likely to seed the next epidemic31-33. The pace at which we can now sequence genomes is key here, and, ultimately, such phylodynamic approaches may provide the early-warning signals for disease emergence or drug resistance, allowing us to help predict, and limit, their spread34-39. Similarly, laboratory selection of parasite resistance to new drugs, coupled with genome sequencing and other molecular techniques, is proving useful in identifying the mechanisms of drug action, cross resistance to other drugs and the potential for control failure in both Mycobacterium tuberculosis40 and Plasmodium41, amongst other top killers. Biomedical researchers can and should follow up the identification key traits with mathematical models that can generate predictions for the impact of control strategies. If we could travel back in time to the beginning of the antibiotic era and use evolutionary biology to apply these drugs in a manner less likely to provoke undesired evolution, we would. The lessons learned through integrative strategies can and must be applied to future pathogen control because the cost of developing and testing new therapeutics is so high that any strategy to prolong their efficacy, however slightly, is both morally and economically compelling.
Box 1. Threats and Opportunities.
Evolutionary outcomes can be challenging to predict. This has generated many unforeseen dangers, but an appreciation of the evolutionary strategies used by pathogens can also foster intriguing solutions to therapy and controlling transmission.
Resistance at a bargain. The ability of microbes to resist antibiotics should negatively impact their fitness in the absence of treatment, as resistance traits are likely to be costly. However, mutations that confer resistance are often quickly followed by additional mutations, elsewhere in the genome, that compensate the costs of resistance. Compensatory mutations can even increase the fitness of the resistant genotype above that of the original sensitive genotype, creating ‘superbugs’ which outcompete other bacteria both in the presence and absence of antibiotics.
Old enemies. A standard view has been that long-term host-pathogen associations inevitably co-evolve to become less harmful. However, parasites actually face an evolutionary dilemma as reproducing too quickly may kill the host before transmission takes place, but sometimes there are advantages to reproducing rapidly (even when this harms the host), such as upon co-infection with a competitor. These counteracting demands lead to the evolution of an intermediate level of virulence which maximises parasite transmission2. There is a clear potential for treatments that reduce virulence without directly harming pathogens to favour fast-growing parasites5,6.
Family matters. Many aspects of pathogen biology only make sense in light of the evolutionary theory of kin selection. For example, some Salmonella bacteria induce an immune response that effectively empties the host gut of competing pathogens. However, this immune response will also harm Salmonella, but this suicide strategy makes sense if it helps nearby kin who share genes with the original Salmonella strain42.
Hit ’em late, hit ’em softly. Evolutionary theory has suggested a blueprint for “evolution proof” insecticides: target old rather than young individuals43,44. At first sight, this counters the intuition that the best way to control a pest is to hit them quickly and hard, but an insecticide that kills young mosquitoes maximises selection for resistance. However, evolutionary biology predicts that an insecticide that kills later, once most reproduction has occurred, will minimise selection for resistance, as the strength of natural selection decreases with age. In parasites where transmission generally occurs from older vectors, e.g. Plasmodium, there may be an age window that could be targeted when selection for insecticide resistance is weak and before transmission.
RAMPs. Ribosomally-synthesised antimicrobial peptides(RAMPs) are part of the innate immune system of all multicellular organisms, and bacteria do not appear to have resistance to them, raising hope that these peptides might be used therapeutically. However, resistance easily evolves45 when RAMPs are studied under the conditions that evolutionary theory predicts will provoke the evolution of resistance. Coincidentally, these conditions are similar to the those bacteria would experience if RAMPs were used therapeutically.
Deceptive epitopes. Immunodominance is the preference of the immune system towards a limited set of epitopes. By strategically positioning immunodominant epitopes, the influenza virus can “lock” the immune system into a relatively ineffective and strain-specific response. Thus, evolution suggests that attempting to mimic the natural immune response may play into the pathogen’s hands. Instead, vaccine constructs that mask the immunodominant deceptive epitopes may allow the immune response to be refocused toward less antigenic but more conserved epitopes, generating more effective, wide ranging immunity46,47.
Refernces
- 1. http://www.who.int/infectious-disease-report/pages/grfindx.html.
- 2.McGreer A, Low D. Is resistance futile. Nature Medicine. 2003;9:390–392. doi: 10.1038/nm0403-390. [DOI] [PubMed] [Google Scholar]
- 3.MacLean CR, Hall AR, Perron GG, Buckling A. The population genetics of antibiotic resistance: integrating molecular mechanisms and treatment contexts. Nature Reviews Genetics. 2010;11:405–414. doi: 10.1038/nrg2778. [DOI] [PubMed] [Google Scholar]
- 4.McCarroll L, et al. Insecticides and mosquito-borne disease. Nature. 2000;407:961–962. doi: 10.1038/35039671. [DOI] [PubMed] [Google Scholar]
- 5.Gandon S, Day T. Evidences of parasite evolution after vaccination. Vaccine. 2008;26:C4–C7. doi: 10.1016/j.vaccine.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 6.Gandon S, Mackinnon MJ, Nee S, Read AF. Imperfect vaccines and the evolution of pathogen virulence. Nature. 2001;414:751–755. doi: 10.1038/414751a. [DOI] [PubMed] [Google Scholar]
- 7.Babayan S, Read A, Lawrence R, Bain O, Allen J. Filarial parasites develop faster and reproduce earlier in response to host immune effectors that determine filarial life expectancy. PLoS. Biol. 2010;8:e1000525. doi: 10.1371/journal.pbio.1000525. doi:10.1371/journal.pbio.1000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneider P, Bell A, Read A, Reece S. Antimalarial drugs: unexpected evolutionary consequences. Malar. J. 2010;9:P45. [Google Scholar]
- 9.Conradt U, Schmidt J. Double surface-membrane in Plecocercoids of Ligula intestinalis (Cestoda: Pseudophyllidea) 1992;78:123–129. doi: 10.1007/BF00931653. 0044-3255. [DOI] [PubMed] [Google Scholar]
- 10.Deitsch KW, Lukehart SA, Stringer JR. Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nat Rev Micro. 2009;7:493–503. doi: 10.1038/nrmicro2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grainger J, et al. Helminth secretions induce de novo T cell Foxp3 expression and regulatory function through the TGF-{beta} pathway. J Exp Med. 2010 doi: 10.1084/jem.20101074. doi:10.1084/jem.20101074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schroeder H, Skelly P, Zipfel P, Losson B, Vanderplasschen A. Subversion of complement by hematophagous parasites. Dev. Comp. Immunol. 2009;33:5–13. doi: 10.1016/j.dci.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hastie K, Kimberlin C, Zandonatti M, Macrae I, Saphire E. Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3′ to 5′ exonuclease activity essential for immune suppression. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1016404108. doi:10.1073/pnas.1016404108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maizels R, et al. Helminth parasites--masters of regulation. Immunological Reviews. 2004;201:89–116. doi: 10.1111/j.0105-2896.2004.00191.x. [DOI] [PubMed] [Google Scholar]
- 15.Harnett W, Harnett M. Molecular basis of worm-induced immunomodulation. Parasite Immunol. 2006;28:535–543. doi: 10.1111/j.1365-3024.2006.00893.x. doi:10.1111/j.1365-3024.2006.00893.x. [DOI] [PubMed] [Google Scholar]
- 16.Bullen JJ, Rogers HJ, Spalding PB, Ward CG. Natural resistance, iron and infection: a challenge for clinical medicine. Journal of Medical Microbiology. 2006;55:251–258. doi: 10.1099/jmm.0.46386-0. [DOI] [PubMed] [Google Scholar]
- 17.Portugal S, et al. Host-mediated regulation of superinfection in malaria. Nat Med. 2011;17:732–737. doi: 10.1038/nm.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kafatos FC, Eisner T. Unification in the century of biology. Science. 2004;303:1257. doi: 10.1126/science.303.5662.1257. [DOI] [PubMed] [Google Scholar]
- 19.Pedersen A, Babayan S. Wild Immunology. 2010;20:643–650. doi: 10.1111/j.1365-294X.2010.04938.x. 0962-1083. [DOI] [PubMed] [Google Scholar]
- 20.Scott ME. Heligmosomoides polygrus (Nematoda): susceptible and resistant strains are indistinguishable following natural infection. Parasitology. 1991;103:429–438. doi: 10.1017/s0031182000059953. [DOI] [PubMed] [Google Scholar]
- 21.Leslie M. Immunology Uncaged. Science. 2010;327:1573–1573. doi: 10.1126/science.327.5973.1573. [DOI] [PubMed] [Google Scholar]
- 22.Paterson S, et al. Antagonistic coevolution accelerates molecular evolution. Nature. 2010;464:275–278. doi: 10.1038/nature08798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Genovese G, et al. Association of Trypanolytic ApoL1 Variants with Kidney Disease in African Americans. Science. 329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferreira A, et al. Sickle Hemoglobin Confers Tolerance to Plasmodium Infection. Cell. 2011;145:398–409. doi: 10.1016/j.cell.2011.03.049. [DOI] [PubMed] [Google Scholar]
- 25.Roy BA, Kirchner JW. EVOLUTIONARY DYNAMICS OF PATHOGEN RESISTANCE AND TOLERANCE. Evolution. 2000;54:51–63. doi: 10.1111/j.0014-3820.2000.tb00007.x. [DOI] [PubMed] [Google Scholar]
- 26.Graham A, et al. Fitness Correlates of Heritable Variation in Antibody Responsiveness in a Wild Mammal. Science. 2010;330:662–665. doi: 10.1126/science.1194878. [DOI] [PubMed] [Google Scholar]
- 27.Telfer S, et al. Species Interactions in a Parasite Community Drive Infection Risk in a Wildlife Population. Science. 2010;330:243–246. doi: 10.1126/science.1190333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graham AL, Allen JE, Read AF. Evolutionary causes and consequences of immunopathology. Annual Review of Ecology Evolution and Systematics. 2005;36:373–397. [Google Scholar]
- 29.Maizels RM. Infections and allergy - helminths, hygiene and host immune regulation. Curr Opin Immunol. 2005;17:656–661. doi: 10.1016/j.coi.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 30.Wilson MS, et al. Suppression of allergic airway inflammation by helminthinduced regulatory T cells. J Exp Med. 2005;202:1199–1212. doi: 10.1084/jem.20042572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rambaut A, et al. The genomic and epidemiological dynamics of human influenza A virus. Nature. 2008;453:615–619. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu W, et al. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature. 2010;467:420–425. doi: 10.1038/nature09442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pybus OG, Rambaut A. Evolutionary analysis of the dynamics of viral infectious disease. Nat Rev Genet. 2009;10:540–550. doi: 10.1038/nrg2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharp PM, Robertson DL, Hahn BH. Cross-species transmission and recombination of AIDS viruses. Phil. Trans. R. Soc. B. 1995;349:41–47. doi: 10.1098/rstb.1995.0089. [DOI] [PubMed] [Google Scholar]
- 35.Hughes GJ, et al. Molecular phylodynamics of the heterosexual HIV epidemic in the United Kingdom. PLoS Pathog. 2009;5:e1000590. doi: 10.1371/journal.ppat.1000590. doi:10.1371/journal.ppat.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McWilliam, et al. Transmission networks and population turnover of echovirus 30. J Virol. 2009;83:2109–2118. doi: 10.1128/JVI.02109-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matthews L, et al. Heterogeneous shedding of Escherichia coli O157 in cattle and its implications for control. Proc Natl Acad Sci U S A. 2006;103:547–552. doi: 10.1073/pnas.0503776103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chase-Topping M, Gally D, Low C, Matthews L, Woolhouse M. Supershedding and the link between human infection and livestock carriage of Escherichia coli O157. Nat Rev Microbiol. 2008;6:904–912. doi: 10.1038/nrmicro2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raynor J, Liu W, Peeters M, Sharp P, Hahn BH. A plethora of Plasmodium species in wild apes: a source of human infection. Trends Parasitol. 2011;27:222–229. doi: 10.1016/j.pt.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andries K, et al. A Diarylquinoline Drug Active on the ATP Synthase of Mycobacterium tuberculosis. Science. 2005;307:223–227. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 41.Rottmann M, et al. Spiroindolones, a Potent Compound Class for the Treatment of Malaria. Science. 2010;329:1175–1180. doi: 10.1126/science.1193225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown S, Le Chat L, Taddei F. Evolution of virulence: triggering host inflammation allows invading pathogens to exclude competitors. Ecol. Lett. 2008;11:44–51. doi: 10.1111/j.1461-0248.2007.01125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Read AF, Lynch PA, Thomas M. How to Make Evolution-Proof Insecticides for Malaria Control. PLoS Biol. 2009;7:e1000058. doi: 10.1371/journal.pbio.1000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koella JC, Lorenz L, Bargielowskia I. Microsporidians as Evolution-Proof Agents of Malaria Control? Advances in Parasitology. 2009;68:315–327. doi: 10.1016/S0065-308X(08)00612-X. [DOI] [PubMed] [Google Scholar]
- 45.Perron GG, Zasloff M, Bell G. Experimental evolution of resistance to an antimicrobial peptide. Proceedings of the Royal Society B: Biological Sciences. 2006;273:251–256. doi: 10.1098/rspb.2005.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nara PL, et al. How Can Vaccines Against Influenza and Other Viral Diseases Be Made More Effective? PLoS Biol. 2010;8:e1000571. doi: 10.1371/journal.pbio.1000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Im E-J, et al. Protective Efficacy of Serially Up-Ranked Subdominant CD8+ T Cell Epitopes against Virus Challenges. PLoS Pathog. 2011;7:e1002041. doi: 10.1371/journal.ppat.1002041. [DOI] [PMC free article] [PubMed] [Google Scholar]