Abstract
Drosophila dMyc (dMyc) is known for its role in cell-autonomous regulation of growth. Here we address its role in the fat body (FB), a metabolic tissue that functions as a sensor of circulating nutrients to control the release of Drosophila Insulin-like peptides (Dilps) from the brain influencing growth and development. Our results show that expression of dMyc in the FB affects development and animal size. Expression of dMyc, but not of CycD/cdk4 or Rheb, in the FB diminishes the ability to retain Drosophila Insulin-like peptide-2 (DILP2) in the brain during starvation, suggesting that expression of dMyc mimics the signal that remotely controls the release of Dilps into the hemolymph. dMyc also affects glucose metabolism and increases the transcription of Glucose-transporter-1 mRNA, and of Hexokinase and Pyruvate-Kinase mRNAs, key regulators of glycolysis. These animals are able to counteract the increased levels of circulating trehalose induced by a high sugar diet leading to the conclusion that dMyc activity in the FB promotes glucose disposal. dMyc expression induces cell autonomous accumulation of triglycerides, which correlates with increased levels of Fatty Acid Synthase and Acetyl CoA Carboxylase mRNAs, enzymes responsible for lipid synthesis. We also found the expression of Stearoyl-CoA desaturase, Desat1 mRNA significantly higher in FB overexpressing dMyc. Desat1 is an enzyme that is necessary for monosaturation and production of fatty acids, and its reduction affects dMyc’s ability to induce fat storage and resistance to animal survival. In conclusion, here we present novel evidences for dMyc function in the Drosophila FB in controlling systemic growth. We discovered that dMyc expression triggers cell autonomous mechanisms that control glucose and lipid metabolism to favor the storage of nutrients (lipids and sugars). In addition, the regulation of Desat1 controls the synthesis of triglycerides in FB and this may affect the humoral signal that controls DILP2 release in the brain.
Keywords: Myc, Drosophila, DILP2, Glycolysis, Trehalose, Fatty Acid Synthesis, Desat1
INTRODUCTION
Myc activity controls fundamental processes including animal development, cell proliferation, and tissue growth (Bellosta and Gallant, 2010; Dang, 2012). In Drosophila mutations in the sole dmyc gene reduce body size (hence the name diminutive) (Johnston et al., 1999) and analysis of dmyc’s target genes reveals a high prevalence for genes regulating ribosome biogenesis and protein synthesis (Grewal et al., 2005; Hulf et al., 2005; Orian et al., 2003). Myc activity lies downstream of numerous growth factors and nutrient signaling pathways including the SWH (Salvador-Warts-Hippo) pathway, to control growth in epithelial cells (Huang et al., 2005; Neto-Silva et al., 2010; Ziosi et al., 2010), and of the insulin/TOR signaling pathway, that regulates dMyc protein stability through the ubiquitin degradation pathway (Parisi et al., 2011; Zhang et al., 2006). Myc function is known to control metabolic pathways upstream of glycolysis and glutaminolysis (Osthus et al., 2000; Wise et al., 2008), particularly in cancer cells where c-Myc activity couples glucose and glutamine metabolism with growth and biomass (Dang, 2012; Yuneva et al., 2012). Much less is known about the role of Myc in the regulation of lipid metabolism. A few studies pinpoint at the ability of adipocytes and liver cells to increase c-myc mRNA in response to caloric restriction, suggesting that Myc may be necessary to maintain the basal metabolic rate of these cells during low nutrient conditions (Horikawa et al., 1986; Kim et al., 1991). Previous studies demonstrated that animals expressing c-myc in the liver showed increase blood glucose disposal and resistance to streptozotocian-induced diabetes. These animals favor glycolysis and lipogenesis (Valera et al., 1995), suggesting a function for Myc in the regulation of carbohydrate metabolism in non-cancerous conditions (Riu et al., 1996).
In Drosophila, the fat body (herein referred as FB) is a metabolic tissue with similar physiological functions as the mammalian adipose tissue and liver. The FB acts as storage for sugars and fats, which are mobilized to sustain basal cellular energy supplies when nutrients are scarce (Liu et al., 2009), and regulates systemic growth in response to nutrients and hormonal signals (Tennessen and Thummel, 2011). The FB functions as a sensor for amino-acids in the hemolymph (Britton and Edgar, 1998; Davis and Shearn, 1977), and when their concentration is abundant, releases humoral factors to remotely control the release of Drosophila insulin like peptide 2 (DILP2) from the Insulin Producing Cells (IPCs) (Geminard et al., 2009; Ikeya et al., 2002). The signals that control the production of these factors in the FB and their identity have not been elucidated yet. Recently, the expression of Dilp6, a member of the Drosophila Insulin like peptide produced by the FB, was shown to negatively regulate DILP2 release in the brain of adult flies (Bai et al., 2012). In addition, Unpaired (Upd2), a cytokine member of the unpaired family secreted from the FB, was also shown to control DILP2 release and to reduce systemic growth (Rajan and Perrimon, 2012). dMyc expression in the FB, was reduced at late third instar by ecdysone signaling to reduce growth allowing the onset of metamorphosis to proceed (Delanoue et al., 2010), while the opposite was shown in animals during starvation where dmyc mRNA in FB was sustained by dFOXO transcriptional activity (Teleman et al., 2008) probably to maintain the basal metabolic rate necessary for the animal to survive.
We decided to use the Drosophila FB to identify the key effectors that couple Myc activity to changes in metabolic pathways essential for animal growth and survival. With this study, we were able to demonstrate that expression of dMyc in the FB induces systemic growth and confers resistance to starvation. Animals expressing dMyc in the FB have acquired the ability to facilitate DILP2 release from the IPCs, suggesting that dMyc activity affects the humoral signals in the FB that control DILP2. cg-dMyc animals have increased levels of sugars in the FB and transcription of Glucose-transporter-1 mRNA, Hexokinase-A and -C and Pyruvate-Kinase mRNAs. These animals are able to counteract trehalosemia induced by high sugar diet. dMyc also increases the level of triglycerides, which correlates with increased levels in the FB of Fatty Acid Synthase, Acetyl CoA Carboxylase mRNAs and of Stearoyl-CoA desaturase, Desat1 mRNA. Desat1 is the Drosophila orthologue of the vertebrate Stearoyl-CoA desaturase-1/SCD1, a key enzyme necessary for the formation of monosaturated fatty acids and lipid biosynthesis (Mauvoisin and Mounier, 2011). Desat1 function is conserved from Drosophila to vertebrates (Brock et al., 2007; Kohler et al., 2009), where mutations in SCD1 result in a dramatic reduction of body fat, insulin sensitivity and reduced overall animal size (Ntambi et al., 2002; Zheng et al., 1999). Our data show that expression of Desat1 in the FB is necessary for dMyc-induced regulation of systemic growth, fat storage and resistance to starvation, placing this enzyme as a novel component in dMyc’s function in controlling lipid metabolism and systemic growth.
MATERIALS AND METHODS
Fly husbandry
cg-GAL4 (Bloomington stock 7011), UAS-GS1 RNAi (Bloomington stock 40836), UAS-dMyc RNAi (VDRC stock 2947 and Bloomington stock 25784), UAS-Pyruvate Kinase RNAi (VDRC stock 49533), UAS-Hexokinase-C RNAi (VDRC stock 35337) and UAS-Desat1 RNAi (VDRC stock 33338). UAS-HA-dMyc (Bellosta et al., 2005), yw hs-Flp122; Act5C>FRT-CD2-FRT>Gal4; UAS-GFP/TM6b (Saucedo et al., 2003), hemolectin-GAL4 (Goto et al., 2001), pumpless-GAL4 (Zinke et al., 1999). Animals were raised at low density, at 25°C on a standard corn meal containing 6 g/L agar, 75 g/L corn flour, 100 g/L white sugar, 50 g/L fresh yeast, and 10 g/L inactivated yeast powder, along with nipagin and propionic acid (Acros Organic).
Nile-Red and Oil-Red O tissue staining
For Nile-Red: Larvae were bisected in PBS. Fat bodies were isolated from carcasses, fixed for 20 min in 4% paraformaldehyde (Electron Microscopy Science) in PBS, washed in 0.3% PBS triton X-100 and incubated for 30 min in a 0.01mM Nile Red Solution (SIGMA), then photographed under a Zeiss LSM510 confocal microscope. Oil Red O staining of larval tissues was performed as described previously (Gutierrez et al., 2007).
Quantitative Real Time PCR
Total RNA was isolated using RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. Extracted RNAs were quantified using a UV spectrophotometer and RNA integrity was confirmed with ethidium bromide staining. The purified RNA (1 μg) was used as template for cDNA synthesis using SuperScript II (Invitrogen). The SYBR Green PCR Kit (Qiagen) was used, and products were quantified using the ABI Prism 7300 system. The relative level for each gene was calculated using the 2-DDCt method (Hulf et al., 2005) and reported as arbitrary units compared to actin5C as a control. At least three separate experiments were performed in duplicate. Primers were designed using Primer3 (Rozen and Skaletsky, 2000). Primers list in available in the Supplementary Materials.
Starvation assay
At least 30 staged late L2 larvae from each genotype for each replica were collected and transferred in single well plastic plates containing paper soaked with PBS; survival rate was determined every 12 hrs. These experiments were repeated three times. Statistical analysis was performed using a z-test (Supplementary Appendix 1).
Larval development
At least 30 staged L1 larvae (24 hrs AED) of each genotype were collected in grape agar and transferred into regular corn meal food. The developmental stage was scored every 12 hrs and identified according to the mouth hook/spiracle morphology (Britton and Edgar, 1998). To quantify growth differences, age-matched larvae were collected from the culture medium, washed and killed by microwave pulse. Larval volume was measured using Photoshop software and the formula 4/3π (L/2)*(l/2)2 (L= length l= width) was used.
Glucose and triglyceride measurement
Trehalose and TAG levels were measured in both fed and starved larvae. 72 hr larvae were either kept in normal fly food or starved for 24 hrs in petri dishes containing filter paper moistened with PBS. Five to ten larvae were homogenized on ice in 200 μl of PBS for glucose assays, or in 0.1% PBS Tween for triglyceride assays. Homogenates were centrifuged for 3 min at 5,000 rpm and the supernatants were incubated at 70°C for 5 min to inactivate endogenous enzymes. For glucose assays, trehalose was converted to glucose by adding Trehalase (Sigma) at a final concentration of 0.025 U/ml and incubating for 15 min at 37°C. The samples were mixed with 500 μl of Glucose Reagent HK (Sigma) for 15 min at room temperature and absorbance measured at 340 nm according to the manufacturer’s protocol. Glucose measurements from undigested samples were subtracted to give the amount of glucose obtained from trehalose. For hemolymph assays, hemolymph was pooled from 5 larvae and 1ul collected, diluted 1:20 in PBS, and heat inactivated at 70°C for 5 min. 2ul of diluted hemolymph was used for each glucose assay. Triglyceride measurements were performed by incubating 20 μl of sample with 400 μl of Free Glycerol Reagent (Sigma) for 15 min at RT, followed by 100 μl Triglyceride Reagent (Sigma) for 15 min, and absorbance read at 540nm according to the manufacturer’s protocol. Glucose and triglyceride concentrations were normalized against total protein concentration in each sample as measured by a BCA assay (Pierce). Assays were repeated a minimum of three times. Triglycerides were also measured in fat bodies collected from ten larvae per genotype.
Western blot
Tissues from staged third instar larvae were collected in lysing buffer (50 mM Hepes pH 7.4, 150 mM NaCl, 1% Triton, 1mM EDTA, with phosphatase and protease inhibitors (Roche). Protein concentrations were quantified using Bradford reagent. Equal amounts were resolved on 8% SDS-PAGE gels and transferred to nitrocellulose membranes. Western blotting was performed using mouse monoclonal anti dMyc antibody (Prober and Edgar, 2002), rat monoclonal anti epitope tag HA antibody (Roche) and mouse monoclonal anti-actin antibody (Millipore). Enhanced chemiluminescence (ECL-GE Healthcare) was used for detection. Quantification of dMyc proteins was performed from photographs using ImageJ. The intensity of each band (pixels) was compared to the intensity of the same band in cg- control from the same blot.
anti-DILP2 and anti dFOXO immunofluorescence
Brains from larva at 96 hrs AEL were dissected in PBS, fixed in PBS/4% formaldehyde for 20 min, and washed in PBS-0.3% Triton X-100 (PBT). Tissues were blocked in PBT/5% BSA. Primary rat-anti-DILP2 was used overnight at 4°C (Geminard et al., 2009) followed by Alexa488 anti rat-secondary antibodies (Invitrogen). Tissues were mounted in Vectashield with DAPI (Vectorlabs) and fluorescence images were acquired using a Zeiss LSM 510 confocal microscope. The quantification of the mean pixels corresponding to the intensity of fluorescence in the IPCs was determined using the histogram tool in ImageJ, using similar parameters as described previously (Geminard et al., 2009). Data are expressed in arbitrary units, and represent the average fluorescence intensity in the IPCs and relative standard deviations were calculated using four independent experiments, each including at least ten animals of each genotype and conditions. dFOXO staining FBs from larva at 96 hrs AEL or starved for 24 hrs in PBS, were dissected in PBS, fixed in PBS/4% formaldehyde for 20 min, and washed in PBS-0.3% Triton X-100 (PBT). Tissues were blocked in PBT/5% BSA. Primary rat-anti-dFOXO was used overnight at 4°C followed by Alexa-488 anti rat-secondary antibodies (Invitrogen). Tissues were mounted in Vectashield with DAPI (Vectorlabs) and fluorescence images were acquired using a Zeiss LSM 510 confocal microscope. The anti dFOXO antibody was developed during this study using the region CASMETSRYEKRRGRAKKRVEALR from the Drosophila peptide sequence as an antigen. The peptide was linked to a protein carrier and injected into rabbit. The serum was affinity purified using the peptide used as antigen and AP anti-dFOXO used 1:200 (Prosci-inc, CA).
Proteomics
Three independent collections of FB from third instar larvae of the different genotypes were harvested in 0.66% Rapigest (Waters Corp.) with dithiothreitol (6 mM), sonicated, boiled and digested to peptides with trypsin. Analysis of peptides was performed at the Quantitative Proteomics Center at Columbia University using an ultrahigh-pressure liquid chromatograph and a quadrupole-time-of-flight mass spectrometer using methods described previously (Oswald et al., 2011). This system uses a shotgun proteomics method called MSE in which mass spectra are recorded at alternate low (precursor) and high (product) fragmentation voltages (Silva et al., 2006), resulting in a more complete representation of the intensity of a peptide in a chromatographic peak. Peptides were analyzed in a LC/MS run on a 0.75 μm ID × 25 cm reverse phase 1.7-μm particle diameter C18 column at a flow rate of 300 nL/min with an acetonitrile/formic acid gradient on a NanoAcquity UPLC (Waters Corp.). Spectra were recorded with a 0.6 s scan time with a Synapt mass spectrometer (Waters Corp). MS and MSE spectra were analyzed with ProteinLynx Global Server (PLGS) software (Waters Corp.). In addition to PLGS software, we used the Elucidator Protein Expression Data Analysis System software (Rosetta Biosoftware). This program provided multivariate statistical analysis tools, principal components analysis, ANOVA, and hierarchical cluster analysis for analyzing these data.
RESULTS
dMyc expression in the fat body affects animal size
To understand whether Myc activity in adipocytes regulates metabolic pathways that affect growth and survival, we modulated dMyc expression in the larval fat body (FB), using the UAS-Gal4 binary system (Brand et al., 1994). The cg (collagen type IV) promoter (Asha et al., 2003) fused with Gal4, was used to drive expression of UAS-dMyc or UAS-dMyc-RNAi in the FB. dMyc protein expression was analyzed in FB from yw1118; cg-Gal4; UAS-HA-dMyc (hereinafter called cg-dMyc), yw-1118; cg-Gal4; UAS-dMyc-RNAi (hereinafter called cg-dMyc-RNAi), and yw1118; cg-Gal4 (hereinafter called cg-) third instar larvae, using anti dMyc antibodies (Prober and Edgar, 2002). These data showed that in the FB anti dMyc antibodies recognize two major bands of 100 and 130KDa bands that were significantly reduced in extracts from cg-dMyc RNAi FBs (Figure 1A, left panel). However, since dMyc was expressed with an HA tag, we were able to use anti-HA antibody to detect one band at 100 KDa that was present only in the extracts of FB from cg-HA-dMyc larvae (Figure 1A, right panel). Analysis of organismal size and developmental delay revealed that cg-dMyc animals develop with a similar rate as cg larvae (Figure 1C and Table 1), but they reach pupation at a significantly bigger size than control animals. Similar results were obtained using two independent insertions of UAS-dMyc (t-test P-values<0.05 for both lines). In contrast, RNAi-mediated reduction of dMyc in the FB resulted in animals that reached pupation with a delay of 48 hours at their third instar transition (Figure 1C and Table 1); these animals were smaller throughout development (P<0.0001 in a t-test when compared to cg- larvae) and eclosed as small adults (Figure 1B-D and E). Two independent chromosomal insertions were used for these experiments, and the dMyc-RNAi line on Chromosome III resulted in larval lethality, a defect that was rescued by coexpression of UAS-dMyc (Table 1, and Supplementary Figure 1). Phenotypic analysis of the adult animals showed that cg-dMyc flies were 17% heavier than control cg- animals (Figure1D-E), on the contrary, RNAi-mediated reduction of dMyc resulted in flies that eclosed with and with significantly reduced body size and weight (Figure 1D-E and Table 1) and two days of delay (Supplementary Figure 1B). To assess whether the differences in body weight measured in cg-dMyc and cg-dMyc-RNAi adults animals reflected changes in the size of the peripheral organs, we measured the cell size and number in the wing and the eye of adult animals. In the wing blade each cell is accessorized with a single bristle called trichome. We measured the number of trichomes in a defined area of the wing blade in animals of the three different genotypes. This analysis revealed that flies expressing dMyc in the FB had an increased size and number of the cells in their wings (Figure 1E, P<0.0001 in a t-test) indicating that expression of dMyc in the FB increases the size of the peripheral organs. On the contrary, cg-dMyc-RNAi animals displayed an opposite phenotype with wings that were small due to a reduced number and size of the their cells (Figure 1E, P<0.0001 in a t-test). Similar conclusions were obtained using the ommatidia that constitute the compound eye (Figure 1E). We tested whether the effect on systemic growth was specific for dMyc activity in the FB using pumpless-Gal4 (Zinke et al., 1999) another driver for the FB. This analysis gave us similar results as those obtained with cg-Gal4 (Supplementary Figure 2). Since the cg promoter is active in the FB and in the hemocytes (Asha et al., 2003), we used the hemolectin-Gal4 (Goto et al., 2003) and Serpent-hemocytes-Gal4 (Bruckner et al., 2004) and showed that expression of dMyc in hemocytes was not associated with any alteration in developmental timing or body size (Supplementary Figure 3). Finally, in order to understand whether the systemic growth induced by dMyc was associated with its role in inducing cell size in FB, we expressed the growth inducers cyclin D/Cdk4 (Prober and Edgar, 2002) and Rheb (Patel et al., 2003; Saucedo et al., 2003; Stocker et al., 2003) in the FB using the cg-Gal4 driver, and cell size and organismal size were measured. This analysis showed that even though expression of dMyc, CycD/Cdk4 or Rheb induced cell-autonomous growth (Supplementary Figure 4), only expression of dMyc was able to induce growth at the organismal level (Figure 1E). Taken together these data suggest that increasing dMyc activity in the FB specifically induces systemic growth.
Figure 1. Expression of dMyc in the FB non-autonomously affects animal size.
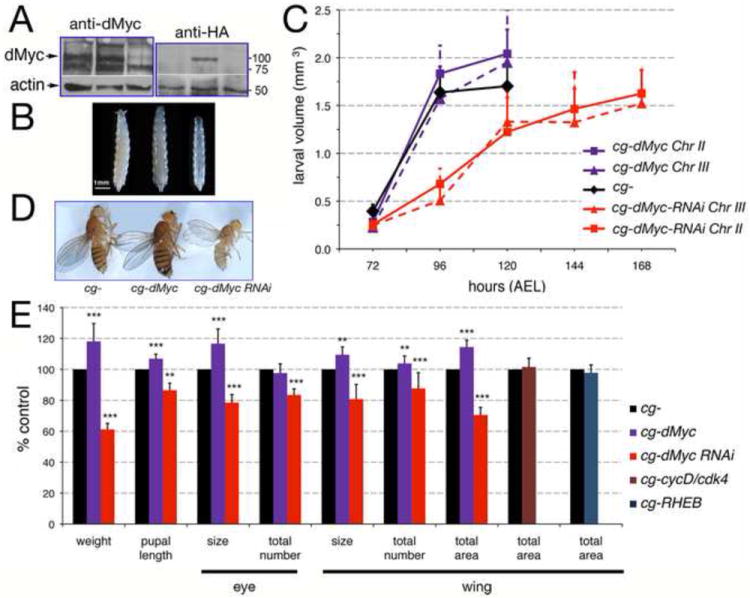
(A) Western blot showing dMyc protein expression in lysates from FB of third instar larvae. dMyc proteins are visible as 100 and 130 KDa bands that are recognized by anti-dMyc antiserum (Prober and Edgar, 2002), and as an 100KDa band that is recognized by anti HA tag antibodies only present in the extract from cg-HA-dMyc larvae. Actin was used as a control for loading. (B) Photographs of third instar larvae. (C) Measurement of larval growth; values are expressed in larval volume (mm3) and hours after egg laying (AEL) at 25°C. Two lines for each UAS-dMyc or UAS-dMyc-RNAi were used. The P-values from a Student t-test at 120 hrs of development are P= 0.008 for cg-dMyc on Chr II and P= 0.01 for cg-dMyc on Chr III. In cg-UAS-dMyc-RNAi animals the total duration of larval development was altered, with pupation reached at 168 hours AEL compared to 120 hours in cg-control or cg-dMyc animals, L1/L2 and L2/L3 transitions were unaffected; a t-test of P<0.001 was calculated for both cg-dMyc-RNAi lines in all the time point of development. Error bars indicate the standard deviation from four independent experiments; approximately 50 larvae for each time point were used. (D) Adult females. (E) Body weight, pupal length and measurement of growth in the eye was calculated by measuring the size of single ommatidia as a measurement of cell size and their total number in each eye as measurement for proliferation. Growth in the wing was calculated by measuring the size of a trichome for cell size, and their total number for proliferation. The total wing area was calculated as in the Material and Methods. Data are expressed as % of cg- control flies (n=20) for female flies. *P<0.05, **P <0.01, ***P <0.001; P-values were calculated from a Student t-test. Off note: all the lines used were isogenic for the yw-1118 chromosome. No significant differences in the timing of development and body size were observed in the parental lines used in those crosses (Supplementary Figure 13 A and B).
Table 1.
The indicated UAS-transgenes were expressed in the fat body using the cg-Gal4 promoter. Two different lines were used for UAS-dMyc and UAS-dMyc-RNAi with different chromosomal insertions.
| Genotype | pupal length (mm) | a days of development | b % of adults at eclosion | c area wing (mm2) | c size of thricome (μm2) | c total number of thricomes/wing |
|---|---|---|---|---|---|---|
| cg-Gal4/+; +/+ | 3.05 ± 0.07 (63) | 10.72 ± 0.31 (443) | 099.8 (443) | 1.96 ± 0.06 (16) | 133 ± 9.5 (16) | 14713 ± 1094 (16) |
| cg-Gal4/UAS-dMyc (II); +/+ | 3.24 ± 0.07 (52) *** | 11.27 ± 0.07 (287) | 100.0 (287) | 2.24 ± 0.06 (16) *** | 149 ± 6.1 (16) *** | 15060 ± 1094 (16) |
| cg-Gal4/+; UAS-dMyc(III)/+ | 3.18 ± 0.09 (46) *** | 11.50 ± 0.22 (356) | 099.2 (356) | nd | nd | nd |
| cg-Gal4/UAS-dMyc-RNAi (II); +/+ | 2.80 ± 0.14 (48) *** | 12.90 ± 0.37 (304) | 054.8 (304) | 1.40 ± 0.10 (16) *** | 113 ± 6.1 (16) *** | 12404 ± 0980 (16) *** |
| cg-Gal4/+; UAS-dMyc-RNAi (III)/+ | 2.35 ± 0.17 (20) *** | PL | PL | PL | PL | PL |
| cg-Gal4/UAS-dMyc (II); UAS-dMyc-RNAi (III)/+ | 3.04 ± 0.14 (55) | 11.30 ± 0.14 (225) | 067.7 (225) | 1.66 ± 0.11 (16) | 125 ± 9.2 (16) | 13259 ± 1310 (16) |
From egg-deposition until adults eclosion.
Data were calculated as % of the total animals of the expected genotype.
see Materials & Methods for calculation.
PL: pupal lethal. nd: not determined.
P-value from t Student test is P<0.001 compared to cg-Gal4. In parenthesis is the total number of animals in the experiments.
dMyc expression in the FB affects DILP2 release from the Insulin Producing Cells
During feeding, the FB remotely controls DILP2 release from the brain by secreting factors in the hemolymph to induce the release of DILP2 from the insulin producing cells (IPCs) (Geminard et al., 2009). Given the systemic effect on growth by dMyc expression in the FB we analyzed if misexpression of dMyc in the FB affects DILP2 protein localization within the IPCs. Animals were grown in corn medium then starved in PBS, and DILP2 protein in the IPCs was quantified by immunofluorescence. This analysis showed that while DILP2 protein accumulates in the IPCs of cg-control larvae after starvation (P<0.001, in a t-test), in cg-dMyc animals this regulation was absent and the intensity of fluorescence in the IPCs was similar in animals in feeding or starving conditions (P= 0.957) (Figure 2A, Supplementary Figure 5). On the contrary, animals with reduced dMyc levels in the FB showed an accumulation of DILP2 in the IPCs when in feeding conditions (Figure 2A, P=0.004, in a t-test and Supplementary Figure 5). The lack of DILP2 protein accumulation after starvation in cg-dMyc animals was attributable mainly to changes in DILP2 protein since dilp2 mRNA levels did not change in these animals (Supplementary Figure 6A-D). To analyze if DILP2 activity was solely responsible for mediating the systemic growth induced by dMyc, we expressed dMyc in the FB of dilp2,3,5 mutant animals. dilp2,3,5 homozygous mutant flies are viable and hatch as small adults with a delay of about 14 days (Gronke et al., 2010). These experiments, showed that reduction of dilp2,3, 5 expression blocked the effect of dMyc on systemic growth (Figure 2B, P=0.114) suggesting that dMyc activity in the FB required functional dilps signaling to induce systemic growth. The use of a triple mutant background was necessary to avoid compensatory effects between the different dilps (Broughton et al., 2008; Ikeya et al., 2002). dilp2, 3 and 5-mRNAs levels in the IPCs did not vary among the three different genotypes (Supplementary Figure 6A-B), however we measured a significant decrease in the level of dilp2, 3 and 5-mRNAs levels in cg-control animals upon starvation, which was not visible in brain from cg-dMyc larvae (Supplementary Figure 6C-D).
Figure 2. dMyc expression in the FB remotely affects DILP2 release from the Insulin Producing Cells (IPCs) and reduces the level of trehalose in the hemolymph.
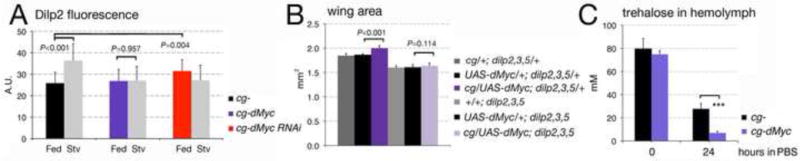
(A) Third instar larvae were raised in corn meal food then starved for 24 hours in PBS. Anti-DILP2 fluorescence was quantified as described previously (Geminard et al., 2009). P values were calculated from a Student t-test from n=10 larvae for each time point and genotype and four independent experiments. (B) dMyc systemic growth is abolished in dilp2, 3,5 mutant background. Growth was measured in the wing of adult females flies of the indicated genotype and expressed in mm2. P-values were calculated from Student t-test from n=15 adults for each genotype, from two independent experiments. (C) Quantification of trehalose (mM) in the hemolymph of cg- control and cg-dMyc larvae in feeding and after starvation. ***P<0.001 value was calculated from three independent experiments. Error bars represent the standard deviations.
DILPs stimulate the uptake of circulating sugars from the hemolymph (Rulifson et al., 2002; Tatar et al., 2003), and their levels in the IPCs are inversely proportional to the amount of circulating trehalose (larval sugar) (Geminard et al., 2009). Trehalose is a dimer of two molecules of glucose and is transported as a monomer through the hemolymph to the tissues where it is uptaken by a class of glucose transporters (Escher and Rasmuson-Lestander, 1999). During starvation, the concentration of trehalose decreases over time, as a direct result of DILP2 retention in the IPCs (Dus et al., 2011; Luo et al., 2012) hence we decided to measure the level of trehalose in the hemolymph as an indirect readout for DILP2 activity. This analysis showed that in starvation the levels of circulating trehalose in cg-dMyc animals decreased more rapidly then in cg-control larvae (Figure 2C, and Supplementary Figure 7), indirectly supporting the hypothesis that cg-dMyc larvae have higher concentration of circulating DILP2 than cg- control animals.
dMyc expression in the FB increases sugar storage, glycolytic enzymes and counteracts increased glycemia
We then analyzed whether the expression of dMyc in the FB affects the storage and mobilization of trehalose in the whole animal. cg-dMyc larvae raised on regular food, displayed a significant increased in trehalose concentration in the whole body, whereas cg-dMyc-RNAi animals, of same developmental stage, showed the opposite effect (Figure 3A). After 24 hrs of starvation, trehalose levels were reduced with a similar ratio in all three genotypes indicating that dMyc in the FB did not influence sugar mobilization. Analysis of enzymes responsible for glucose metabolism, showed an increase in the level of the glucose transporter Glut1-mRNA, and of the glycolysis enzymes Hexokinase-C and Hexokinase-A-mRNAs and of Pyruvate Kinase-mRNAs (Figure 3B). In addition, we found that expression of dMyc correlated with the increased level of dilp6-mRNA. DILP6 is expressed in the FB and produced during the non-feeding stage (Okamoto et al., 2009; Slaidina et al., 2009; Zhang et al., 2009). In adult fly, dilp6 transcription is regulated by dFOXO and correlates with a decrease in DILP2 release, resulting in reduced insulin signaling (Bai et al., 2012).
Figure 3. Expression of dMyc in the FB affects glucose metabolism.
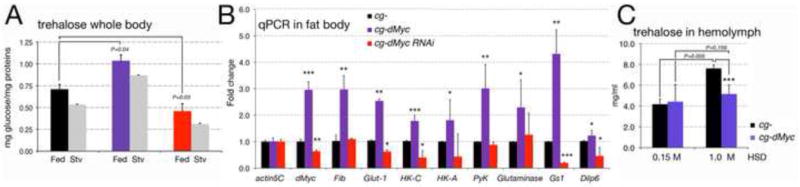
(A) Trehalose in the whole larvae was measured from animals in fed and starving conditions. P-values were calculated from a Student t-test using 10 larvae per genotype in each time point from four independent experiments. Trehalose was normalized to protein concentration. (B) Quantitative RT-PCR in FB from third instar larvae showing the indicated transcript levels in cg-control, cg-Myc and cg-dMyc-RNAi animals; actin5C was used as the internal control. *P <0.05, **P <0.01, ***P <0.001; P-values were calculated from a Student t-test from at least four independent experiments. (C) Quantification of trehalose (mM) in the hemolymph of cg-control and cg-dMyc larvae in medium containing 0.15M or 1M sucrose (Musselman et al., 2011). P-values were calculated from a Student t-test from n=15 cg- and cg-dMyc larvae in different sucrose concentrations. Error bars represent the standard deviations.
Transgenic mice expressing c-myc in the liver maintain lower glycemia after treatment with streptozotocin, a drug that causes a predisposition to diabetes (Riu et al., 1996). We analyzed whether a similar mechanism was also present in flies. Feeding larvae a high-sugar diet (HSD) increases sugars in the hemolymph (Musselman et al., 2011), and while cg- animals fed a HSD exhibited a rise in circulating trehalose (Figure 3C, P<0.05), the cg-dMyc counterparts maintained an invariant trehalosemia (Figure 3C, P=0.159). These data support the idea that dMyc in the FB is able to reduce the level of sugars in the hemolymph via a remote control of dilps and through a tissue autonomous regulation of glucose uptake in FB. Increase in glucose metabolism is associated to glutamine uptake to promote cell survival (Wellen et al., 2010), in addition, c-myc induces the consumption of glutamine to favor growth in cancer cells (Wise et al., 2008). In cg-dMyc FBs, we found increased expression of CG42708 mRNA, encoding for Glutaminase and of Glutamine synthetase1 (GS1) mRNA, suggesting that the events that regulate glutamine metabolism by Myc are present also in the Drosophila FB.
dMyc expression in the FB affects storage of TAG and survival in low nutrient conditions
Morphological analysis of FBs, using the lipid-specific dye Nile Red, revealed that fat cells from cg-dMyc animals contained cytoplasmic lipid droplets smaller in size compared to that in cg- controls (Figure 4A-B and E). This effect was more exacerbated when dMyc was expressed in flip out clones using the actin-promoter (Supplementary Figure 8A). On the contrary, the lipid droplets from cg-dMyc-RNAi FB cells were abnormally enlarged (Figure 4C and E, and supplementary 8B) resembling vesicles from starved animals (Figure 4D). Analysis of the cell size and triglyceride (TAG) content showed that fat cells from cg-dMyc animals were larger (Figure 4F) and contained more TAG per protein compared to cg-control (Figure 4G). The opposite effect was observed in FB from cg-dMyc-RNAi animals. Increased levels of TAG result from increased synthesis or reduced degradation of fatty acids. Thus we analyzed if the expression of specific enzymes involved in fatty acid metabolism were affected by dMyc expression. This analysis revealed that FBs from cg-dMyc animals have increased levels of Perilipin2-mRNA, the orthologue of the vertebrate lipid storage droplet gene-2 (LSD-2) (Bickel et al., 2009), a protein that functions to regulate lipid storage and favors lipogenesis (Gronke et al., 2003; Okumura, 2011; Tansey et al., 2001). In addition, we found increased expression of CG11198 and CG3524 (Palanker et al., 2009), encoding for putative orthologues of the vertebrate Acetyl-Co Carboxylase (ACC) and Fatty Acid Synthase (FAS) respectively. dMyc also upregulates the expression of CG5887-Desat1 mRNA (Wicker-Thomas et al., 1997), the orthologue of the vertebrate Stearoyl CoA Desaturase (SDC1) an enzyme that has been associated together with ACC and FAS with the control of de-novo synthesis of lipids in the liver (Paton and Ntambi, 2009). On the contrary, the expression of CG2107-mRNA (Palanker et al., 2009), encoding for a putative orthologue of a carnitine transporter, a key regulator of beta-fatty acid oxidation was reduced in cg-dMyc FB (Figure 4H), indicating that the fat cells in these animals have increased fatty synthesis and reduced oxidation.
Figure 4. Altered dMyc expression in the FB increases triglycerides (TAG) and confers resistance to starvation.
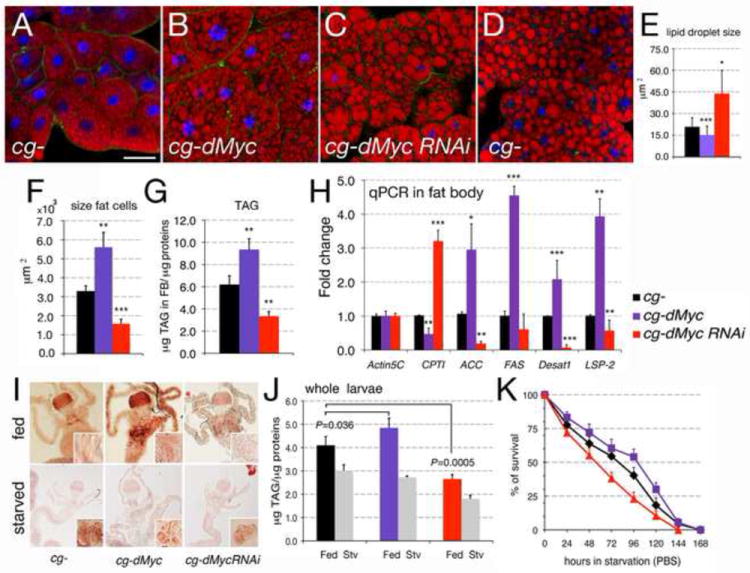
(A-D) Photographs of FB from third instar larvae of the indicated genotypes. Lipid vesicles are stained with Nile Red, nuclei with DAPI and Alexa-488-Phalloidin to visualize the cell membrane. (E) Size of the vesicles in fat cells was analyzed from photographs (A-C). (F) size of cells in the FB (G) triglyceride (TAG) content in cells from the FB; TAG concentration was normalized to the amount of protein; P-values were calculated from a Student t-test from four independent experiments, 10 larvae in each experiment were used; (H) Quantitative RT-PCR from FB from third instar larvae from cg-control, cg-Myc and cg-dMyc-RNAi animals; actin5C was used as control. *P <0.05, **P <0.01, ***P <0.001; P-values were calculated from Student t-test from at least four independent experiments. (I) Oil red O staining of midgut, caeca, and oenocytes (insets) dissected from larvae of the indicated genotype fed (upper panel) or starved for 24 hrs in PBS (lower panel). Insets show oenocytes stained with Nile Red. (J) Content of triglycerides (TAG) was normalized per protein concentration in whole larvae in feeding and after starvation in PBS. P-values were calculated from Student t-test from at three independent experiments. (K) Age-matched late L2 larvae were transferred in PBS, and larval viability was calculated every 12 hours. cg-dMyc larvae show resistance to starvation when compared to cg-control animals, while cg-dMyc-RNAi die early. P<0.001 in a z-test for the survival rate between the three genotypes (see Appendix-1 for calculation). Error bars represent the standard deviations.
Lipids stored principally in the mid-gut and gastric caeca, are cleared from those tissues during starvation and accumulate in the oenocytes, a cluster of cells with functions similar to the vertebrate liver (Gutierrez et al., 2007). To analyze whether dMyc expression in the FB affects lipid mobilization, third instar larvae were starved, and lipid content was visualized in feeding and after starvation using Oil Red O (Palanker et al., 2009). In feeding conditions lipids were present more abundantly in the mid-gut and gastric caeca of cg-dMyc animals (Figure 4I middle panel). These animals also showed the presence of lipid vesicles in their oenocytes, which during feeding are normally emptied of lipids (compare insets in the left and middle panel). After 24h of starvation, lipids were equally cleared from the mid-gut and caeca from all three genotypes and accumulate in the oenocytes with no difference between the three genotypes (Figure 4I, lower panels). Measurement of TAG from whole animals in feeding conditions confirmed that cg-dMyc larvae have a small but significant increase in the level of TAG compared to cg- controls (P<0.05), while cg-dMyc-RNAi displayed a dramatic reduction in their lipid content (P<0.001) (Figure 4J). Upon starvation the TAG stores were reduced at a similar rate in all three genotypes, suggesting that expression of dMyc in FB does not affect lipid mobilization.
Animals store triglycerides and sugars to survive periods of scarce nutrient availability. We analyzed whether the differences in the content of sugars (Figure 3A) and TAG (Figure 4J) between the animals of the three genotypes could affect their ability to survive starvation. Animals were raised in normal food until they reached the mid-second larval instar, at which point they were transferred to PBS and their viability scored every 12 hours. These experiments showed that whereas only 50% of cg- control survived after 80 hours of starvation, the half-life of cg-dMyc animals during starvation reached 100 hours (P<0.0001, in a z-test analysis, see Appendix 1 for statistic analysis). Conversely, cg-dMyc-RNAi larvae reached their half-life at about 56 hours of starvation (Figure 4K) (P<0.0001, in a z-test).
From these data, we conclude that expression of dMyc in FB locally and remotely increases lipid concentration and storage, and correlates with a better survival during nutrient starvation.
Reduction of Desat1 in FB, but not of PyK and HK-C affects the ability of dMyc to induce systemic growth
The increase of dMyc expression in the FB during starvation (Figure 5A) is induced by the transcription factor FOXO that translocates to the nucleus (Figure 5B) to directly induce dmyc-mRNA (Teleman et al., 2008). In order to assess which pathways were sustained by dMyc in the FB during starvation, we undertook a proteomic approach to compare the proteomes of FBs from animals with different dMyc levels in feeding and starving conditions. To confirm that a more sustained expression effectively correlates with an increase in protein abundance, we focused our attention on the genes that were previously identified by quantitative RT-PCR analysis (Figure 3B and 4H). This analysis revealed that FBs expressing dMyc have significantly increased Desat1, Pyruvate Kinase, Hexokinase-C and Glutamine Synthetase1 protein levels (Table 2) and the expression of these enzymes was further increased upon starvation (Table 2, P<0.0001, using a Student’s t test). Next we used the cg-Gal4 driver in combination with UAS-RNAi lines to analyze if the silencing of these genes in the FB interferes with the ability of dMyc to induce systemic growth. This analysis revealed that reduction of PyK or HK-C expression did not affect the ability of dMyc to induce organismal growth, as dMyc expression was able to significantly induce growth (***P<0.001) as measured in adults (Figure 5C) and in pupae (Supplementary Figure 9), even when PyK or HK-C expression was reduced. On the contrary, reduction of Desat1 completely blunted the ability of dMyc to influence body size (Figure 5C). Reduction of GS1 in the FB with the available RNAi line resulted in pupal lethality, preventing us from further performing any genetic analysis (Supplementary Figure 9). Efficiency of the RNAi line in FB was tested (Supplementary Figure 10).
Figure 5. Reduction of desaturase Desat1 affects dMyc systemic growth.
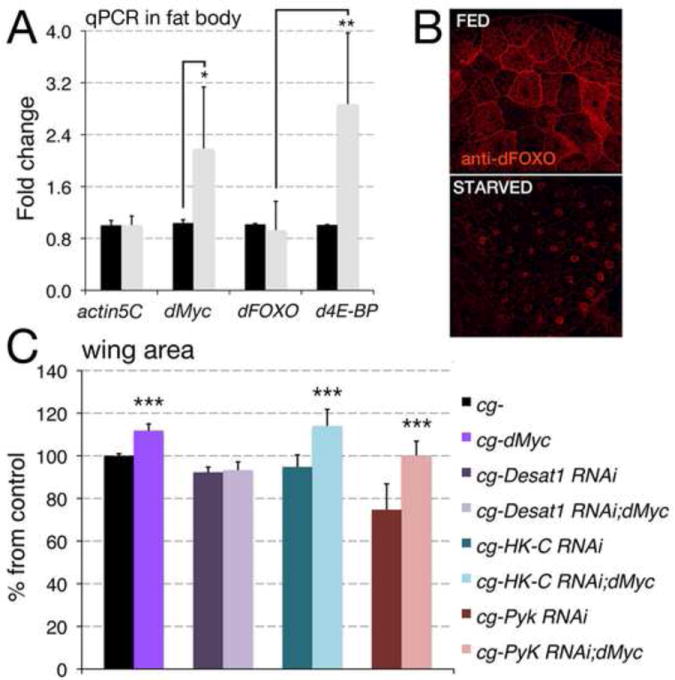
(A) Quantitative RT-PCR from the FB of cg-control third instar larvae in feeding or starved for 24 hrs; actin5C was used as a control. *P <0.05, **P <0.01, ***P <0.001; P-values were calculated from a Student t-test from at least four independent experiments. (B) Photographs of FB from animals in feeding or starved conditions stained for dFOXO protein using anti dFOXO antibodies (Material and Methods). In feeding condition dFOXO is located in the cytoplasm, starvation reduces systemic insulin signaling allowing dFOXO to translocate into the nucleus. (C) Measurement of growth in the wing was calculated by measuring the total wing area, calculated as in Material and Methods. Data are expressed as % of cg- control flies using at least n=20 female for each genotype. *P<0.05, **P <0.01, ***P <0.001; P-values were calculated from a Student t-test. Error bars represent the standard deviations.
Table 2.
Quantitation and identification for digest of proteins in FB from cg-dMyc and cg- control larvae.
| Protein Name | UniProt ID | Ratio FED | Ratio STARVED | P-value FED | P-value STARVED |
|---|---|---|---|---|---|
| Desat1 | Q7K4Y0 | 1.18 | 2.64 | 0.0012 | 8.86E-43 |
| Pyruvate kinase | KPYK | 1.78 | 2.28 | 6.42E-07 | 2.72E-32 |
| Hexokinase C | Q7JYW9 | 1.41 | 1.98 | 0.003 | 3.82E-28 |
| Glutamine synthetase | GLNA1 | 1.38 | 1.43 | 1.52E-24 | 2.02E-21 |
The data are expressed as ratio from FB from cg-dMyc/cg-control animal in feeding or after 24 hours of starvation in PBS. P-values were calculated using ANOVA from three independent experiments.
Reduction of Desat1 in the FB affects accumulation of TAGs and dMyc resistance to starvation
In mice, mutation of scd1 reduces the level of TAG in the adipose tissue resulting in leaner animals (Ntambi et al., 2002). We found that reduction of Desat1 in the FB (cg-Desat1 RNAi) results in smaller pupae (Supplementary Figure 9) and adults with reduced body size (Figure 5C). The larval FB of cg-Desat1 RNAi animals was composed of smaller cells (not shown) containing less TAG (Figure 6A, P=4.5E-06; using a Student’s t test). Expression of dMyc increases the levels of TAG/protein in the larval FB of about 20% in both cg- control and in cg-Desat1 RNAi; dMyc larvae (Figure 6A). However, the overall content of TAG/protein in cg-Desat RNAi; dMyc animals was still lower then that of cg-control animals (Figure 6A, P=0.003; using a Student’s t test). cg-dMyc animals accumulate TAGs and are resistant to nutrient starvation (Figure 4M). We analyzed whether reduction of Desat1 could play a role in the ability of cg-dMyc animals to resist starvation. Animals were raised in normal food until they reached the mid-second larval instar, then they were transferred to PBS and their viability scored every 12 hours. This analysis showed that while cg-dMyc larvae reached their half-life after 100 hrs of starvation, cg-Desat1-RNAi; dMyc larvae reached the same point only after 48 hrs, with a similar rate as cg-Desat1-RNAi animals (Figure 6B), thus suggesting that reduction of Desat1 compromises the ability of cg-dMyc larvae to survive on a starvation regimen. cg-Desat1 RNAi animals are smaller, and their level of TAG in the FB is reduced compared to cg-control larvae (Figure 6A). We then analyzed if reduction of Desat1 affected the size of the fat cells. We use the UAS/Gal4 technique to induce flip-out clones expressing UAS-Desat1-RNAi under the actin promoter. Clones were marked by co-expression of UAS-GFP and scored in the FB. The size of Desat1-RNAi GFP-positive cells was compared to that of wt GFP-negative cells surrounding the clones (Figure 6 C). This analysis revealed that reduction of Desat1 decreased the cell-size to 74% compared to 100% in control wt cells (P<0.0001, using a t test). As expected, expression of dMyc increased the size of the cells in the FB to 179%, but when dMyc was expressed in combination with Desat1 RNAi, its ability to induce growth was reduced to 139% (Figure 6D). However, the overall ability of dMyc to induce growth in a Desat1 RNAi background (23+39%) was comparable to that in cg-control fat cells (79%), leading to the conclusion that depletion of Desat1 did not significantly affect dMyc’s ability to promote cell growth. However, cg-Desat1; dMyc animals are smaller, have a lower content of TAG/protein but have a similar resistance to starvation as cg-Desat1 RNAi larvae, suggesting that Desat1 activity could potentially mediate dMyc’s effect on resistance to starvation.
Figure 6. Reduction of Desat1 affects dMyc’s ability to accumulate TAG and to increase the cell size in the FB.
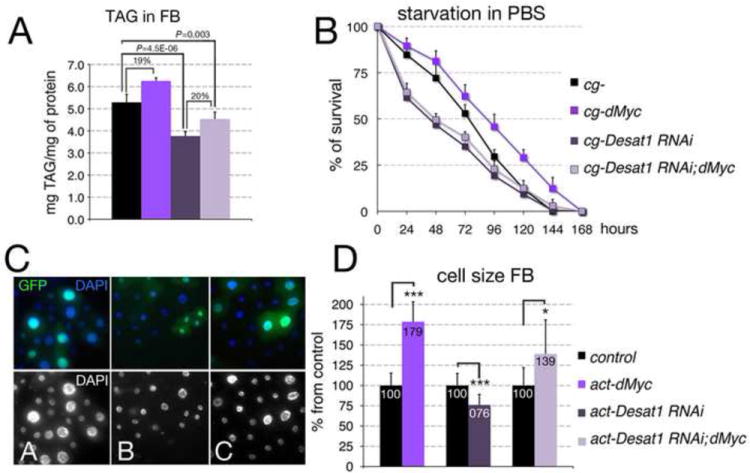
(A) Triglyceride (TAG) content in cells from FBs of the indicated genotype; TAG concentration was normalized to the amount of protein; P-values were calculated from a Student t-test from three independent experiments, 10 larvae per genotype were used in each experiment. (B) Age-matched late L2 larvae were transferred in PBS, and larval viability was calculated every 12 hours. P-values from a Student t-test from three independent experiments P<0.001 was calculated for cg-control and cg-dMyc at all time points. cg-Desat1 RNAi and cg-Desat1 RNAi; dMyc animals are significantly smaller then cg-control animals (P<0.001), while no significant difference was found between cg-Desat1 RNAi and cg-Desat1 RNAi; dMyc (P>0.05). Error bars represent the standard deviations. (C) Actin flip-out clones in FB expressing dMyc (A), Desat1RNAi (B) or Desat1-RNAi; dMyc are marked with GFP, nuclei are stained with DAPI, lower panel is only DAPI. (D) Numbers represent the size of cells from flip-out clones in FB; size is expressed as % of the control wild-type GFP negative cells surrounding the clones (100). *P <0.05, **P <0.01, ***P <0.001; P-values were calculated from Student t-test from at least 40 cells. Error bars represent the standard deviations.
DISCUSSION
Here we show that modulation of dMyc activity in the fat body (FB) controls animal size, recapitulating the phenotype of ubiquitous dMyc expression (de la Cova et al., 2004; Johnston et al., 1999; Pierce et al., 2004). This function is specific for dMyc activity since other known growth inducers like CycD/Cdk4 or Rheb, do not produce the same effect (Figure 1E). The Rheb/TOR signaling pathway couples nutrient availability with growth (Arsham and Neufeld, 2006; Wullschleger et al., 2006). Recent data showed that expression of Rheb in Drosophila S2 cells stimulates ribosomal biogenesis and cell size but did not promote glucose import (Hall et al., 2007). In our experiments we found that dMyc regulates the expression of Glucose transporter-1 (Figure 7). Moreover, cells of the FB expressing dMyc have increased intracellular concentration of glucose (not shown) suggesting that dMyc actively promotes the uptake of glucose in these cells. Decreasing ribosomal biogenesis, using a mutation in the ribosomal-protein RpL14 Minute (3) 66D (Saeboe-Larssen et al., 1997) did not blunt dMyc’s ability to induce organismal growth (Supplementary Figure 11), supporting the idea that other signals in addition to protein synthesis are induced by dMyc in FB and lead to an increase in body size.
Figure 7. MODEL of pathways induced by dMyc in the FB.
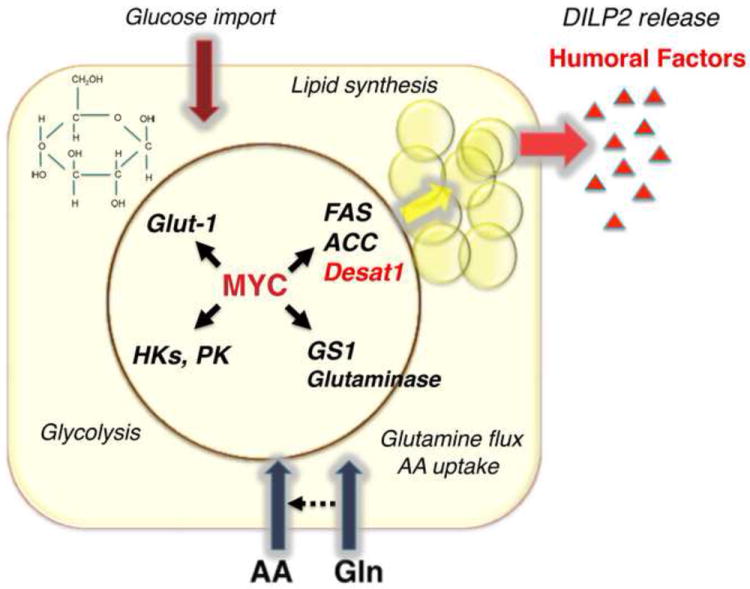
Expression of dMyc in FB favors the secretion of humoral factors responsible for the release of DILP2 from the brain. We have identified potential pathways that are induced by dMyc in the FB. dMyc increases the expression of Fatty Acid Synthase (FAS), Acetyl CoA (ACC) and of Desat1, responsible for the synthesis and modification of fatty acids. Hexokinases (HK-A and C) and Pyruvate Kinases (PyK), glycolytic enzymes, and of Glucose Transporter-1 (Glut-1). Glutamate Synthase (GS1) and Glutaminase, key enzymes in the synthesis of glutamine and glutaminolysis. Evidence also indicates that dMyc increases the expression of SLC38-11 and SLC1A5 amino acid/glutamine transporters suggesting a role in controlling glutamine flux in the FB.
The FB functions as a sensor for the amino acid concentration in the hemolymph and in response to a diet rich in proteins, produces humoral factors that control the release of DILP2 from the brain (Geminard et al., 2009). Our data show that dMyc expression in the FB inhibits the retention of DILP2 in the IPCs during starvation (Figure 2A), suggesting that an enhanced dMyc activity in the FB mimics a fed state or is sufficient to induce the secretion of factors responsible for DILP2 release from the IPCs. Accumulation of DILP2 in the IPCs leads to decreased trehalosemia (level of circulating trehalose in the hemolymph) (Geminard et al., 2009). The ability of cg-dMyc animals to decrease trehalosemia and to affect the amount of DILP2 released during starvation, together with the finding that DILP2 is required for the dMyc-dependent systemic increase in size, led us to conclude that dMyc control of systemic growth depends on the presence of circulating DILPs. However the nature of the humoral factors governing DILP2 release and the mechanism that triggers the secretion of these factors are not clear yet. To attempt at the identification of the pathways involved in their production, we analyzed by quantitative RT-PCR the expression levels of key enzymes of glycolysis, glutamine and lipid metabolic pathways upon dMyc induction in FB.
In the FB dMyc increases cell autonomously the level of Hexokinase-A, the putative homologue of human Hexokinase-IV or glucokinase, Hexokinase-C, and of Pyruvate Kinase-mRNAs (Figure 3B) key enzymes that control glycolysis. Similarly, expression of c-Myc in the liver induced the transcription of glycolytic enzymes and of the glucose transporter GLUT2, resulting in decreased glycemia and counteracting streptozotocin (STZ) induced diabetes (Riu et al., 1996). Subsequently, we found that cg-dMyc larvae, grown on a high sugar diet (1M sucrose), are able to reduce their level of trehalosemia (Figure 3C), suggesting that a similar mechanism is conserved also in flies. Our results could be explained by the ability of dMyc to increase Glut-1 mRNA and favor glucose uptake, which may reduce the concentration of sugar in the hemolymph, and by dMyc’s ability to increase the rate of glucose consumption via glycolysis.
Quantitative PCR analysis also revealed that expression of dMyc in the FB upregulates Glutaminase and Glutamine Synthetase (GS1) mRNA, key regulators in glutamine metabolism (Figure 3B). In addition, we found an increase in the expression of the Drosophila orthologue of the vertebrate LAT1/SLC1A5 also called minidiscs (mnd) (Martin et al., 2000) and of SLC38/11, two members of the L-and N-type family of amino acid transporter-1 (Mackenzie and Erickson, 2004) (Supplementary Figure 12). mnd which is highly expressed in the FB, was shown to function non autonomously to control organismal size (Martin et al., 2000), suggesting a function in controlling organismal size. In vertebrates, LAT1/SLC1A5 controls the efflux of L-glutamine that precedes the import of essential amino acids to induce the activation of mTOR signaling (Nicklin et al., 2009). Similarly, we could hypothesize that dMyc in the FB controls glutamine flux to favor the uptake of essential amino acids resulting in activation of glutamine metabolism. Glutamine flux is also controlled by Glutamine Synthetase, and very recently GS1 in vertebrates was identified as a direct target of FOXO-3 to maintain glutamine homeostasis and promote autophagy and cell survival (van der Vos et al., 2012). In the Drosophila FB, dFOXO is necessary to sustain dmyc-mRNA expression during starvation (Teleman et al., 2008). We can speculate that the dFOXO-dMyc axis functions in the FB to control GS1 activity to regulate glutamine-dependent pathways to sustain animal survival.
dMyc expressing animals have an increased content of TAG and glucose in the whole body that correlates with increased transcriptional levels of Fatty Acid Synthase (FAS) and Acetyl CoA (ACC). In the vertebrate liver and adipocytes FAS and ACC are transcriptionally regulated in response to glucose concentration by the Carbohydrate Response-Element Binding Protein, Max-Like protein (Mlx) and MondoA (Uyeda and Repa, 2006). Mlx and MondoA form heterodimers that bind to the E-box sequences in the ACC and FAS promoter to induce their expression, and to reduce glucose uptake (Billin and Ayer, 2006; Billin et al., 2000). In the presence of high levels of glutamine, the activity of Mlx/Mondo heterodimers decreases and their negative effect on glucose uptake is relieved, resulting in increased glucose concentration (Kaadige et al., 2009). In addition, Drosophila Mondo interacts genetically with dMyc (Billin and Ayer, 2006) raising the possibility that misexpression of dMyc may change the stoichiometry of Mondo/Mlx heterodimers to control ACC and FAS and glucose uptake in the FB.
The proteomes of fat bodies from animals with different dMyc levels revealed a significant increase of the glycolytic enzymes (PyK and HK-C), Glutamine Synthetase (GS1) and Desat1 particularly during starvation (Table 2). To our surprise, we found that only reducing Desat1 blunted the ability of dMyc to induce organismal growth (Figure 5C). Desat1, or SCD1 in vertebrates, is the rate limiting enzyme that catalyzes the biosynthesis of monosaturated fatty acids, components of triglycerides and phospholipids (Mauvoisin and Mounier, 2011). In Drosophila Desat1 was increased in proteomes from starved animals and is necessary in the FB to induce autophagy (Kohler et al., 2009). Desat1 activity is required for the ability of dMyc to induce fat synthesis, and to increase cell size (Figure 6). In our model, we place Desat1 function downstream of dMyc activity to regulate systemic growth (Figure 7), but since we did not find dMyc binding sites in its promoter we concluded that dMyc does not directly activate the expression of this enzyme. Reduction of Desat1 reduces the ability of cg-dMyc larvae to resist starvation, probably because Desat1-RNAi animals have decreased cell-size and accumulate less TAG in their fat cells, suggesting that they are unable to activate survival pathways. In support of this hypothesis, mutant desat1 animals have been found defective in autophagy, one of the survival mechanisms that animals activate in the FB during starvation (Kohler et al., 2009). Human SCD1/Desat1 plays a relevant role in carcinogenesis to control de-novo synthesis of lipids and its level is upregulated in different tumors (Igal, 2010; Scaglia et al., 2009). FAS and ACC expression also increase in cancer cells, where their ability to promote de novo synthesis of lipids provides a growth advantage to the tumor cells (Menendez and Lupu, 2007). Myc’s ability to control FAS and ACC and SCD1/Desat1 expression suggest a novel function for Myc to control lipid metabolism and survival that may be relevant in normal and pathological conditions.
Our studies are centered in the regulation of metabolic pathways by dMyc in FB. However, dMyc expression in this organ is also developmentally regulated by ecdysone, which reduces dmyc mRNA in late third instar to constrain growth (Delanoue et al., 2010). At the same time reduction of insulin signaling activates dFOXO and results in dMyc expression (Figure 5 A) (Teleman et al., 2008). Our idea is that these opposite mechanisms that regulate dMyc expression may coexist in the FB to ensure proper function of specific metabolic processes induced by dMyc and are necessary for larval survival when nutrients are reduced.
In conclusion, our findings establish a novel link between dMyc expression in the fat body and the non-cell-autonomous control of growth, metabolism, and survival of the organism. Our data identify two potential signals that are influenced by dMyc expression in FB (Figure 7). The first mimics the non-autonomous control of DILP2 release from the brain induced by humoral factors produced by the FB during feeding. The second identifies Desat1 as a novel component for dMyc activity, that may be relevant for its function in controlling fatty acid metabolism and the process of de-novo synthesis of lipids as a mechanism of cell survival in cancer cell metabolism. These observations may have significant implications for our understanding not only of the systemic control of growth and metabolism but also create the need to explore Myc function at the physiological level in controlling lipid homeostasis and metabolism in diseases such as diabetes and cancer.
Supplementary Material
Acknowledgments
We thank Esteban Tabak (CIMS-NYU) for performing statistical analysis and Pierre Leopold (University of Nice-France) for anti-DILP2 antibodies. We thank the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) for providing transgenic RNAi fly stocks used in this study. This work was supported by a Public Health Service grant from the NIH-SC1DK085047 and the Stewart Trust to P.B. NIH-GM078464 to L. A. Johnston and the Foundation “CaRisBo” to F.P.
Footnotes
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arsham AM, Neufeld TP. Thinking globally and acting locally with TOR. Curr Opin Cell Biol. 2006;18:589–597. doi: 10.1016/j.ceb.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Asha H, Nagy I, Kovacs G, Stetson D, Ando I, Dearolf CR. Analysis of Rasinduced overproliferation in Drosophila hemocytes. Genetics. 2003;163:203–215. doi: 10.1093/genetics/163.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai H, Kang P, Tatar M. Drosophila insulin-like peptide-6 (dilp6) expression from fat body extends lifespan and represses secretion of Drosophila insulin-like peptide-2 from the brain. Aging Cell. 2012;11:978–985. doi: 10.1111/acel.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellosta P, Gallant P. Myc Function in Drosophila. Genes Cancer. 2010;1:542–546. doi: 10.1177/1947601910377490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellosta P, Hulf T, Balla Diop S, Usseglio F, Pradel J, Aragnol D, Gallant P. Myc interacts genetically with Tip48/Reptin and Tip49/Pontin to control growth and proliferation during Drosophila development. Proc Natl Acad Sci U S A. 2005;102:11799–11804. doi: 10.1073/pnas.0408945102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel PE, Tansey JT, Welte MA. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim Biophys Acta. 2009;1791:419–440. doi: 10.1016/j.bbalip.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billin AN, Ayer DE. The Mlx network: evidence for a parallel Max-like transcriptional network that regulates energy metabolism. Curr Top Microbiol Immunol. 2006;302:255–278. doi: 10.1007/3-540-32952-8_10. [DOI] [PubMed] [Google Scholar]
- Billin AN, Eilers AL, Coulter KL, Logan JS, Ayer DE. MondoA, a novel basic helix-loop-helix-leucine zipper transcriptional activator that constitutes a positive branch of a max-like network. Mol Cell Biol. 2000;20:8845–8854. doi: 10.1128/mcb.20.23.8845-8854.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Manoukian AS, Perrimon N. Ectopic expression in Drosophila. Methods Cell Biol. 1994;44:635–654. doi: 10.1016/s0091-679x(08)60936-x. [DOI] [PubMed] [Google Scholar]
- Britton JS, Edgar BA. Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development. 1998;125:2149–2158. doi: 10.1242/dev.125.11.2149. [DOI] [PubMed] [Google Scholar]
- Brock TJ, Browse J, Watts JL. Fatty acid desaturation and the regulation of adiposity in Caenorhabditis elegans. Genetics. 2007;176:865–875. doi: 10.1534/genetics.107.071860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton S, Alic N, Slack C, Bass T, Ikeya T, Vinti G, Tommasi AM, Driege Y, Hafen E, Partridge L. Reduction of DILP2 in Drosophila triages a metabolic phenotype from lifespan revealing redundancy and compensation among DILPs. PLoS One. 2008;3:e3721. doi: 10.1371/journal.pone.0003721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckner K, Kockel L, Duchek P, Luque CM, Rorth P, Perrimon N. The PDGF/VEGF receptor controls blood cell survival in Drosophila. Dev Cell. 2004;7:73–84. doi: 10.1016/j.devcel.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Dang CV. MYC on the Path to Cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis KT, Shearn A. In vitro growth of imaginal disks from Drosophila melanogaster. Science. 1977;196:438–440. doi: 10.1126/science.403606. [DOI] [PubMed] [Google Scholar]
- de la Cova C, Abril M, Bellosta P, Gallant P, Johnston LA. Drosophila myc regulates organ size by inducing cell competition. Cell. 2004;117:107–116. doi: 10.1016/s0092-8674(04)00214-4. [DOI] [PubMed] [Google Scholar]
- Delanoue R, Slaidina M, Leopold P. The steroid hormone ecdysone controls systemic growth by repressing dMyc function in Drosophila fat cells. Dev Cell. 2010;18:1012–1021. doi: 10.1016/j.devcel.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Dus M, Min S, Keene AC, Lee GY, Suh GS. Taste-independent detection of the caloric content of sugar in Drosophila. Proc Natl Acad Sci U S A. 2011;108:11644–11649. doi: 10.1073/pnas.1017096108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escher SA, Rasmuson-Lestander A. The Drosophila glucose transporter gene: cDNA sequence, phylogenetic comparisons, analysis of functional sites and secondary structures. Hereditas. 1999;130:95–103. doi: 10.1111/j.1601-5223.1999.00095.x. [DOI] [PubMed] [Google Scholar]
- Geminard C, Rulifson EJ, Leopold P. Remote control of insulin secretion by fat cells in Drosophila. Cell Metab. 2009;10:199–207. doi: 10.1016/j.cmet.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Goto A, Kadowaki T, Kitagawa Y. Drosophila hemolectin gene is expressed in embryonic and larval hemocytes and its knock down causes bleeding defects. Dev Biol. 2003;264:582–591. doi: 10.1016/j.ydbio.2003.06.001. [DOI] [PubMed] [Google Scholar]
- Goto A, Kumagai T, Kumagai C, Hirose J, Narita H, Mori H, Kadowaki T, Beck K, Kitagawa Y. A Drosophila haemocyte-specific protein, hemolectin, similar to human von Willebrand factor. Biochem J. 2001;359:99–108. doi: 10.1042/0264-6021:3590099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SS, Li L, Orian A, Eisenman RN, Edgar BA. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat Cell Biol. 2005;7:295–302. doi: 10.1038/ncb1223. [DOI] [PubMed] [Google Scholar]
- Gronke S, Beller M, Fellert S, Ramakrishnan H, Jackle H, Kuhnlein RP. Control of fat storage by a Drosophila PAT domain protein. Curr Biol. 2003;13:603–606. doi: 10.1016/s0960-9822(03)00175-1. [DOI] [PubMed] [Google Scholar]
- Gronke S, Clarke DF, Broughton S, Andrews TD, Partridge L. Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS Genet. 2010;6:e1000857. doi: 10.1371/journal.pgen.1000857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez E, Wiggins D, Fielding B, Gould AP. Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature. 2007;445:275–280. doi: 10.1038/nature05382. [DOI] [PubMed] [Google Scholar]
- Hall DJ, Grewal SS, de la Cruz AF, Edgar BA. Rheb-TOR signaling promotes protein synthesis, but not glucose or amino acid import, in Drosophila. BMC Biol. 2007;5:10. doi: 10.1186/1741-7007-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa S, Sakata K, Hatanaka M, Tsukada K. Expression of c-myc oncogene in rat liver by a dietary manipulation. Biochem Biophys Res Commun. 1986;140:574–580. doi: 10.1016/0006-291x(86)90770-9. [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Hulf T, Bellosta P, Furrer M, Steiger D, Svensson D, Barbour A, Gallant P. Whole-genome analysis reveals a strong positional bias of conserved dMyc-dependent E-boxes. Mol Cell Biol. 2005;25:3401–3410. doi: 10.1128/MCB.25.9.3401-3410.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igal RA. Stearoyl-CoA desaturase-1: a novel key player in the mechanisms of cell proliferation, programmed cell death and transformation to cancer. Carcinogenesis. 2010;31:1509–1515. doi: 10.1093/carcin/bgq131. [DOI] [PubMed] [Google Scholar]
- Ikeya T, Galic M, Belawat P, Nairz K, Hafen E. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr Biol. 2002;12:1293–1300. doi: 10.1016/s0960-9822(02)01043-6. [DOI] [PubMed] [Google Scholar]
- Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P. Drosophila myc regulates cellular growth during development. Cell. 1999;98:779–790. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaadige MR, Looper RE, Kamalanaadhan S, Ayer DE. Glutamine-dependent anapleurosis dictates glucose uptake and cell growth by regulating MondoA transcriptional activity. Proc Natl Acad Sci U S A. 2009;106:14878–14883. doi: 10.1073/pnas.0901221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Fletcher DL, Campion DR, Gaskins HR, Dean R. Effect of dietary manipulation on c-myc RNA expression in adipose tissue, muscle and liver of broiler chickens. Biochem Biophys Res Commun. 1991;180:1–7. doi: 10.1016/s0006-291x(05)81246-x. [DOI] [PubMed] [Google Scholar]
- Kohler K, Brunner E, Guan XL, Boucke K, Greber UF, Mohanty S, Barth JM, Wenk MR, Hafen E. A combined proteomic and genetic analysis identifies a role for the lipid desaturase Desat1 in starvation-induced autophagy in Drosophila. Autophagy. 2009;5 doi: 10.4161/auto.5.7.9325. [DOI] [PubMed] [Google Scholar]
- Liu Y, Liu H, Liu S, Wang S, Jiang RJ, Li S. Hormonal and nutritional regulation of insect fat body development and function. Arch Insect Biochem Physiol. 2009;71:16–30. doi: 10.1002/arch.20290. [DOI] [PubMed] [Google Scholar]
- Luo J, Becnel J, Nichols CD, Nassel DR. Insulin-producing cells in the brain of adult Drosophila are regulated by the serotonin 5-HT(1A) receptor. Cell Mol Life Sci. 2012;69:471–484. doi: 10.1007/s00018-011-0789-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie B, Erickson JD. Sodium-coupled neutral amino acid (System N/A) transporters of the SLC38 gene family. Pflugers Arch. 2004;447:784–795. doi: 10.1007/s00424-003-1117-9. [DOI] [PubMed] [Google Scholar]
- Martin JF, Hersperger E, Simcox A, Shearn A. minidiscs encodes a putative amino acid transporter subunit required non-autonomously for imaginal cell proliferation. Mech Dev. 2000;92:155–167. doi: 10.1016/s0925-4773(99)00338-x. [DOI] [PubMed] [Google Scholar]
- Mauvoisin D, Mounier C. Hormonal and nutritional regulation of SCD1 gene expression. Biochimie. 2011;93:78–86. doi: 10.1016/j.biochi.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- Musselman LP, Fink JL, Narzinski K, Ramachandran PV, Hathiramani SS, Cagan RL, Baranski TJ. A high-sugar diet produces obesity and insulin resistance in wild-type Drosophila. Dis Model Mech. 2011;4:842–849. doi: 10.1242/dmm.007948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto-Silva RM, de Beco S, Johnston LA. Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap. Dev Cell. 2010;19:507–520. doi: 10.1016/j.devcel.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, Myer VE, MacKeigan JP, Porter JA, Wang YK, Cantley LC, Finan PM, Murphy LO. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntambi JM, Miyazaki M, Stoehr JP, Lan H, Kendziorski CM, Yandell BS, Song Y, Cohen P, Friedman JM, Attie AD. Loss of stearoyl-CoA desaturase-1 function protects mice against adiposity. Proc Natl Acad Sci U S A. 2002;99:11482–11486. doi: 10.1073/pnas.132384699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto N, Yamanaka N, Yagi Y, Nishida Y, Kataoka H, O’Connor MB, Mizoguchi A. A fat body-derived IGF-like peptide regulates postfeeding growth in Drosophila. Dev Cell. 2009;17:885–891. doi: 10.1016/j.devcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura T. Role of lipid droplet proteins in liver steatosis. J Physiol Biochem. 2011;67:629–636. doi: 10.1007/s13105-011-0110-6. [DOI] [PubMed] [Google Scholar]
- Orian A, van Steensel B, Delrow J, Bussemaker HJ, Li L, Sawado T, Williams E, Loo LW, Cowley SM, Yost C, Pierce S, Edgar BA, Parkhurst SM, Eisenman RN. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 2003;17:1101–1114. doi: 10.1101/gad.1066903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, Xu Y, Wonsey D, Lee LA, Dang CV. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 2000;275:21797–21800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- Oswald ES, Brown LM, Bulinski JC, Hung CT. Label-free protein profiling of adipose-derived human stem cells under hyperosmotic treatment. J Proteome Res. 2011;10:3050–3059. doi: 10.1021/pr200030v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanker L, Tennessen JM, Lam G, Thummel CS. Drosophila HNF4 regulates lipid mobilization and beta-oxidation. Cell Metab. 2009;9:228–239. doi: 10.1016/j.cmet.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi F, Riccardo S, Daniel M, Saqcena M, Kundu N, Pession A, Grifoni D, Stocker H, Tabak E, Bellosta P. Drosophila insulin and target of rapamycin (TOR) pathways regulate GSK3 beta activity to control Myc stability and determine Myc expression in vivo. BMC Biol. 2011;9:65. doi: 10.1186/1741-7007-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel PH, Thapar N, Guo L, Martinez M, Maris J, Gau CL, Lengyel JA, Tamanoi F. Drosophila Rheb GTPase is required for cell cycle progression and cell growth. J Cell Sci. 2003;116:3601–3610. doi: 10.1242/jcs.00661. [DOI] [PubMed] [Google Scholar]
- Paton CM, Ntambi JM. Biochemical and physiological function of stearoyl-CoA desaturase. Am J Physiol Endocrinol Metab. 2009;297:E28–37. doi: 10.1152/ajpendo.90897.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce SB, Yost C, Britton JS, Loo LW, Flynn EM, Edgar BA, Eisenman RN. dMyc is required for larval growth and endoreplication in Drosophila. Development. 2004;131:2317–2327. doi: 10.1242/dev.01108. [DOI] [PubMed] [Google Scholar]
- Prober DA, Edgar BA. Interactions between Ras1, dMyc, and dPI3K signaling in the developing Drosophila wing. Genes Dev. 2002;16:2286–2299. doi: 10.1101/gad.991102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan A, Perrimon N. Drosophila cytokine unpaired 2 regulates physiological homeostasis by remotely controlling insulin secretion. Cell. 2012;151:123–137. doi: 10.1016/j.cell.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riu E, Bosch F, Valera A. Prevention of diabetic alterations in transgenic mice overexpressing Myc in the liver. Proc Natl Acad Sci U S A. 1996;93:2198–2202. doi: 10.1073/pnas.93.5.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- Saeboe-Larssen S, Urbanczyk Mohebi B, Lambertsson A. The Drosophila ribosomal protein L14-encoding gene, identified by a novel Minute mutation in a dense cluster of previously undescribed genes in cytogenetic region 66D. Mol Gen Genet. 1997;255:141–151. doi: 10.1007/s004380050482. [DOI] [PubMed] [Google Scholar]
- Saucedo LJ, Gao X, Chiarelli DA, Li L, Pan D, Edgar BA. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol. 2003;5:566–571. doi: 10.1038/ncb996. [DOI] [PubMed] [Google Scholar]
- Scaglia N, Chisholm JW, Igal RA. Inhibition of stearoylCoA desaturase-1 inactivates acetyl-CoA carboxylase and impairs proliferation in cancer cells: role of AMPK. PLoS One. 2009;4:e6812. doi: 10.1371/journal.pone.0006812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva EM, Serra VV, Ribeiro AO, Tome JP, Domingues P, Faustino MA, Neves MG, Tome AC, Cavaleiro JA, Ferrer-Correia AJ, Iamamoto Y, Domingues MR. Characterization of cationic glycoporphyrins by electrospray tandem mass spectrometry. Rapid Commun Mass Spectrom. 2006;20:3605–3611. doi: 10.1002/rcm.2766. [DOI] [PubMed] [Google Scholar]
- Slaidina M, Delanoue R, Gronke S, Partridge L, Leopold P. A Drosophila insulin-like peptide promotes growth during nonfeeding states. Dev Cell. 2009;17:874–884. doi: 10.1016/j.devcel.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker H, Radimerski T, Schindelholz B, Wittwer F, Belawat P, Daram P, Breuer S, Thomas G, Hafen E. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat Cell Biol. 2003;5:559–565. doi: 10.1038/ncb995. [DOI] [PubMed] [Google Scholar]
- Tansey JT, Sztalryd C, Gruia-Gray J, Roush DL, Zee JV, Gavrilova O, Reitman ML, Deng CX, Li C, Kimmel AR, Londos C. Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity. Proc Natl Acad Sci U S A. 2001;98:6494–6499. doi: 10.1073/pnas.101042998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M, Bartke A, Antebi A. The endocrine regulation of aging by insulin-like signals. Science. 2003;299:1346–1351. doi: 10.1126/science.1081447. [DOI] [PubMed] [Google Scholar]
- Teleman AA, Hietakangas V, Sayadian AC, Cohen SM. Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab. 2008;7:21–32. doi: 10.1016/j.cmet.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Tennessen JM, Thummel CS. Coordinating growth and maturation - insights from Drosophila. Curr Biol. 2011;21:R750–757. doi: 10.1016/j.cub.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeda K, Repa JJ. Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab. 2006;4:107–110. doi: 10.1016/j.cmet.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Valera A, Pujol A, Gregori X, Riu E, Visa J, Bosch F. Evidence from transgenic mice that myc regulates hepatic glycolysis. FASEB J. 1995;9:1067–1078. doi: 10.1096/fasebj.9.11.7649406. [DOI] [PubMed] [Google Scholar]
- van der Vos KE, Eliasson P, Proikas-Cezanne T, Vervoort SJ, van Boxtel R, Putker M, van Zutphen IJ, Mauthe M, Zellmer S, Pals C, Verhagen LP, Groot Koerkamp MJ, Braat AK, Dansen TB, Holstege FC, Gebhardt R, Burgering BM, Coffer PJ. Modulation of glutamine metabolism by the PI(3)K-PKB-FOXO network regulates autophagy. Nat Cell Biol. 2012;14:829–837. doi: 10.1038/ncb2536. [DOI] [PubMed] [Google Scholar]
- Wellen KE, Lu C, Mancuso A, Lemons JM, Ryczko M, Dennis JW, Rabinowitz JD, Coller HA, Thompson CB. The hexosamine biosynthetic pathway couples growth factor-induced glutamine uptake to glucose metabolism. Genes Dev. 2010;24:2784–2799. doi: 10.1101/gad.1985910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker-Thomas C, Henriet C, Dallerac R. Partial characterization of a fatty acid desaturase gene in Drosophila melanogaster. Insect Biochem Mol Biol. 1997;27:963–972. doi: 10.1016/s0965-1748(97)00077-5. [DOI] [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Yuneva MO, Fan TW, Allen TD, Higashi RM, Ferraris DV, Tsukamoto T, Mates JM, Alonso FJ, Wang C, Seo Y, Chen X, Bishop JM. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab. 2012;15:157–170. doi: 10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu J, Li CR, Momen B, Kohanski RA, Pick L. Deletion of Drosophila insulin-like peptides causes growth defects and metabolic abnormalities. Proc Natl Acad Sci U S A. 2009;106:19617–19622. doi: 10.1073/pnas.0905083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HH, Lipovsky AI, Dibble CC, Sahin M, Manning BD. S6K1 regulates GSK3 under conditions of mTOR-dependent feedback inhibition of Akt. Mol Cell. 2006;24:185–197. doi: 10.1016/j.molcel.2006.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Eilertsen KJ, Ge L, Zhang L, Sundberg JP, Prouty SM, Stenn KS, Parimoo S. Scd1 is expressed in sebaceous glands and is disrupted in the asebia mouse. Nat Genet. 1999;23:268–270. doi: 10.1038/15446. [DOI] [PubMed] [Google Scholar]
- Zinke I, Kirchner C, Chao LC, Tetzlaff MT, Pankratz MJ. Suppression of food intake and growth by amino acids in Drosophila: the role of pumpless, a fat body expressed gene with homology to vertebrate glycine cleavage system. Development. 1999;126:5275–5284. doi: 10.1242/dev.126.23.5275. [DOI] [PubMed] [Google Scholar]
- Ziosi M, Baena-Lopez LA, Grifoni D, Froldi F, Pession A, Garoia F, Trotta V, Bellosta P, Cavicchi S. dMyc functions downstream of Yorkie to promote the supercompetitive behavior of hippo pathway mutant cells. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


