Abstract
Circadian (24 h) clocks provide a source of internal timing in most living organisms. These clocks keep time by using complex transcriptional/post-translational feedback loops that are strikingly resilient to changes in environmental conditions. In the last few years, interest has increased in the role of post-transcriptional regulation of circadian clock components. Post-transcriptional control plays a prominent role in modulating rapid responses of the circadian system to environmental changes, including light, temperature and general stress and will be the focus of this review.
Keywords: circadian clock, post-transcriptional regulation, environment
1. Introduction
Circadian (24 h) clocks provide internal timing in most living organisms. In constant darkness and temperature, a circadian rhythm will free-run with its own endogenous periodicity, which is close to, but not exactly 24 h. However, these rhythms can be entrained (or driven) at an exact 24 h periodicity (or even a shorter or longer period) with light and temperature cycles.
Circadian clocks keep time by using complex transcriptional/post-translational feedback loops. One cardinal feature of these clocks is that the period of the oscillation is strikingly resilient to changes in temperature, a property called ‘temperature compensation’ [1]. Yet despite this resilience, other characteristics of the rhythmic phenotype are highly responsive to changes in light and temperature, the two most important environmental variables [2]. Thus, for example, light can reset a clock, or shift its phase and re-entrain it to a new light–dark schedule [3]. High or low temperature can also change the apparent phase of a rhythm but without changing the period [4].
Over the last 30 years, many advances have been made towards understanding the molecular mechanisms of circadian timekeeping. Efforts have mainly focused on transcriptional and post-translational control of circadian components and on the properties of the neuronal networks that drive circadian rhythms in locomotor activity, the most prominent behavioural output of the circadian system in animals. The first evidence showing the importance of post-transcriptional levels of regulation in maintaining circadian rhythmicity was discovered in the single-celled green alga Acetabularia. Interestingly, this unicellular organism can survive for several weeks after the rhizoid containing its nucleus is removed and will continue to photosynthesize rhythmically in constant light, even in the presence of inhibitors of organellar transcription [5,6]. In the last few years, interest has increased in studying the role of post-transcriptional regulation of circadian clock components. Pioneering studies by Carla Green's laboratory demonstrate robust circadian regulation of deadenylation [7–9], which contributes to the control of metabolism by the circadian clock. In addition, many other groups have demonstrated the importance of post-transcriptional control in general clock function [10–13], specific circadian-regulated metabolism [14,15], eclosion rhythms [16,17], locomotor rhythms in flies [18,19], and circadian rhythms in plants and algae [20].
Both post-translational (i.e. protein degradation) and transcriptional controls have been implicated in the response of the molecular clock to changes in environmental cues [21,22]. A growing body of evidence points to post-transcriptional control as an additional key mechanism in mediating this response to the environment. In fact, post-transcriptional control may be the rule rather than the exception, as post-transcriptional control of central clock components plays a key role in adaptation or entrainment in both plants and mammals. In contrast to transcriptional control, post-transcriptional control results in rapid and durable changes in the amounts of circadian components. These changes can be achieved at the level of RNA or protein sequence (through alternative splicing), RNA stability, the ability of a given mRNA to be translated, or transcriptional output (this could be reduced owing to premature 3′ cleavage of the nascent transcript) [23]. In this review, we will focus on two processes that mediate most post-transcriptional regulation in response to changes in the environment: pre-mRNA splicing and post-transcriptional control by microRNAs (miRNAs).
2. Alternative splicing mediates clock responses to the environment
Different steps of gene expression may be regulated to fine-tune the clock in different organisms. One of these steps is pre-mRNA splicing. In pre-mRNA splicing, a ribonucleoprotein complex (the spliceosome) mediates precise cuts at exon–intron boundaries and the assembly of exonic sequences into the mature mRNA [24]. This process is tightly linked functionally, temporally and spatially to transcription, but it is still considered a post-transcriptional event [25]. Splicing involves the recognition of specific target sequences in the intron–exon junctions (5′ and 3′ splice sites) as well as inside the excised intron. When the splicing donor or acceptor sites do not perfectly fit the consensus sequence for the region, alternative versions of a given mRNA can be generated. This process is called alternative splicing and can lead to the generation of many mRNA and protein isoforms from a single genomic sequence [26]. The exact proportion of possible isoforms is achieved through integration of signals in the transcribing pre-mRNA molecule. These signals may involve recruitment of specific RNA-binding proteins (e.g. SR proteins) or changes in the elongation rate of the RNA polymerase II or siRNAs [26]. Interestingly, control of alternative splicing is a prominent way by which the circadian clock adapts to temperature changes in unicellular and pluricellular organisms (see below).
(a). Alternative splicing mediates the response of the circadian clock to temperature in Neurospora
In Neurospora crassa, alternative splicing of frequency, which encodes the central component of the circadian clock, FRQ [27], regulates the response of the circadian clock to temperature changes. The frq gene is rhythmically expressed and contains six open reading frames (uORFs) that encode large (l) and small (s) isoforms of FRQ [28–30]. The expression of the different isoforms is regulated both by alternative promoter usage and by levels of alternative splicing. The abundance and ratio of l-FRQ versus s-FRQ are important for robust free running circadian rhythmicity [28,29].
The amount of l-FRQ increases significantly as temperature rises, whereas s-FRQ levels do not. This leads to a variation in the l-FRQ to s-FRQ ratio as a function of temperature [28,30,31]. The mechanism underlying this variation is alternative splicing: splicing of the sixth intron of frq pre-mRNA (frq-I6), which contains the initiation codon of l-FRQ, is temperature sensitive. The temperature sensitivity of the splicing event is probably due to inefficient recognition of non-canonical splice sites that flank this intron by the splicing machinery at high temperatures, which results in an increasing fraction of l-FRQ [30,31] (figure 1a).
Figure 1.
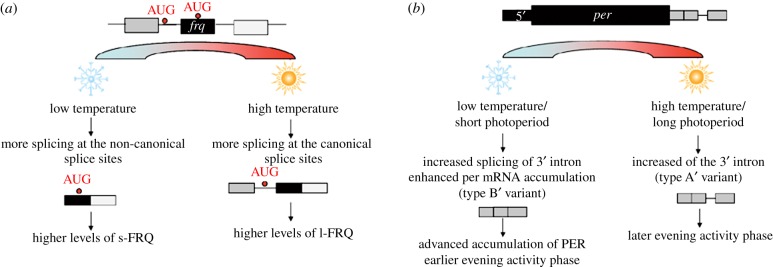
Schematics of how temperature controls alternative splicing of frequency and period. (a) Control of frq alternative splicing at different temperatures (N. crassa). (b) Control of per alternative splicing at different temperatures (Drosophila melanogaster).
FRQ levels are also subject to a temperature-dependent translational control mechanism that is crucial for the temperature-dependent increase in FRQ. The frq gene's uORFs reduce translation initiation of l-FRQ and s-FRQ isoforms. However, as the uORFs do not harbour a strong Kozak consensus sequence, their usage is very limited at higher temperatures. This thermosensitive trapping of scanning ribosomes at uORFs allows adjustment of FRQ levels according to ambient temperature [30].
In summary, two thermosensitive, adaptive mechanisms, splicing of frq-I6 and trapping of scanning ribosomes, regulate abundance of FRQ and the ratio of l-FRQ to s-FRQ. Both processes are based on temperature-influenced recognition of non-consensus splice sites or non-consensus translational initiation sites. These mechanisms are thought to extend the physiological temperature range over which the Neurospora clock functions, keeping FRQ levels in a range compatible with feedback loop function. Indeed, when either of the FRQ isoforms is eliminated, the temperature range permissible for rhythmicity is reduced/narrowed [28].
A later study by Diernfellner et al. [32] shows that the regulation of the l-FRQ to s-FRQ ratio does not seem to be necessary for temperature compensation per se; strains that express only a single FRQ isoform (l-FRQ or s-FRQ) display temperature compensation (at a temperature range between 22°C and 28°C), but their circadian rhythms are either long or short. This suggests that the thermosensitive l-FRQ to s-FRQ ratio provides a molecular means to fine-tune period length, but it may act redundantly with other mechanisms of temperature compensation.
(b). Alternative splicing mediates genome-wide effects of temperature on the circadian clock in Arabidopsis
Alternative splicing is also involved in adaptation of the circadian clock to temperature in Arabidopsis thaliana [33,34]. For example, alternative splicing controls the activity of the gene circadian clock associated 1 (CCA1), which is implicated in temperature compensation [35]. The full-size isoform CCA1α is a key circadian transcriptional activator and acts as a homodimer to activate transcription of target genes. The CCA1β isoform has a protein–protein interaction domain that mediates dimer formation but lacks the N-terminal Myb DNA-binding domain [36]. Hence, CCA1β competitively inhibits CCA1α activity by forming non-functional heterodimers (CCA1α–CCA1β and LHY-CCA1β), which have reduced DNA-binding affinities. Interestingly, low temperatures sway splicing towards the production of the active isoform (CCA1α). Thus, under cold conditions, the Arabidopsis system rebalances the ratio of the inactive form of CCA1 to the active form to produce effective freeze tolerance [36].
Characterization of the Arabidopsis transcriptome through high-throughput sequencing identified a temperature-sensitive alternative splicing event involving intron 4 retention in CCA1 mRNA [37]. Interestingly, this alternative splicing regulation is present not only in A. thaliana, but also in Oryza sativa, Brachypodium distachyon and Populus trichocarpa, mono and dicotyledonous species that diverged from a common ancestor 120–170 Myr ago [38,39].
A recent study used a genome-wide approach to demonstrate the importance of alternative splicing in mediating responses of the Arabidopsis circadian clock to temperature changes [40]. This study evaluated the splicing of all clock components in plants adapted to different temperatures or undergoing temperature transitions. Surprisingly, the analysis showed extensive changes in the splicing of many clock genes. In particular, alternative splicing of the genes late elongated hypocotyl (LHY) and pseudo response regulator 7 (PRR7) led to the production of non-functional transcripts. This regulation is key for modulating smooth transcriptional changes during temperature transitions; regulation at the level of splicing reduces amounts of these particular proteins without affecting the overall transcription rate [40,41]. Interestingly, the authors also found that for clock genes that have been previously implicated in temperature compensation (e.g. CCA1 and LHY [35]) and temperature responsiveness (e.g. PRR9 and PRR7 [42]), the levels of functional transcripts responded in opposite directions to transient cold conditions. For example, CCA1 and PRR9 were upregulated, and LHY and PRR7 downregulated. These changes at low temperature suggest that their effects are balanced or compensated in the clock system.
(c). The effects of temperature on the Drosophila circadian system
Temperature has diverse effects on the Drosophila circadian system probably mediated by different regulatory mechanisms. Temperature-regulated circadian processes include: (i) temperature-dependent changes in the distribution of daily locomotor activity, the so-called ‘temperature seasonal adaptation’ [4,43]; (ii) the capability of clocks to be entrained/synchronized by daily temperature cycles and to be phase-shifted by temperature pulses or steps [3]; (iii) abolition of circadian function at temperatures outside those permissive for rhythm generation; and (iv) temperature compensation. Over the last 15 years, significant advances have been made in our understanding of how Drosophila adapts to changes in temperature. Most of this work has focused on the key role of per mRNA splicing.
The clock is under constant control by external cues, such as light and temperature. Adaptation of Drosophila to different environmental conditions relies on the presence of at least two neuronal circuits, one that controls the morning peak of locomotor activity (M) and another that controls the evening (E) burst of activity [44–46]. The environment fine-tunes the activity pattern by altering the timing of action of these oscillators. This system may help animals to adapt to seasonal changes by altering the phase of these two oscillators in a flexible, presumably adaptive manner [47]. In the laboratory, under hot temperatures and long photoperiods mimicking summer, flies shift their M activity to pre-dawn and their E activity into the early night [4]. By contrast, under shorter day lengths and cooler temperatures mimicking autumn, the M and E activity components fall closer together and occur around the middle of the day, enabling maximal activity at the warmest temperatures. These putative adaptations, which generate the so-called ‘siesta’ at higher temperatures, ensure that the activity of an organism is maximal at a time of day when the temperature is optimal for its activity.
(d). Drosophila temperature adaptation is mediated by a specific splicing event in the period mRNA 3′ untranslated region
In 1999, Edery et al. [4] showed that a thermosensitive splicing event in the per 3′ untranslated region (UTR) is important for the seasonal adaptation or ‘siesta’. The evening advance in the locomotor activity observed at lower temperatures correlates with an advance in the phase of oscillation of tim and per mRNAs. This shift of the molecular clock is driven by the exclusion/splicing of an alternative exon located in per 3′ UTR. At low temperatures, relatively more of the spliced, shorter variant (type B’) of per is present compared with the unspliced variant (type A’). The enhanced efficiency of splicing of per pre-mRNA at low temperatures correlates with an earlier rise in the levels of total per mRNA and protein (figure 1b). By using transgenic flies with mutations that either inhibit per splicing (perA mutant, having mutated 5′ and 3′ splice sites) or those that only express the spliced type B′ transcript (perB′), Majercak et al. [4] observed that both splice-locked variants were unable to phase shift their behaviour at different temperatures. This suggests that it is the act of splicing itself that is important, not the splice products. This could be due to enhancement of 3′ end formation and polyadenylation by the splicing event. Importantly, perA and perB′ mutants can both temperature compensate and entrain to temperature cycles, suggesting that per splicing is not involved in these two cardinal clock features [3]. The mechanism by which the cold-induced splicing of per results in an increase accumulation of per mRNA is still not clear. One possibility is that the assembly of the spliceosomes on the intron increases the production of mature mRNAs. This could be achieved by an increase in transcriptional elongation or by facilitating 3′-end formation.
Further studies [48,49] revealed that the efficiency of per mRNA splicing is regulated not only by temperature but also by the photoperiod. Short photoperiods lead to an increase in the spliced type B′ variant, which accumulates earlier, contributing to increases in the daily upswing and peak levels of per mRNA. On the other hand, long days lead to the production of unspliced per mRNA molecules. Regulation of splicing allows locomotor activity to be fine-tuned to any given set of photoperiodic and temperature conditions. Interestingly, the inhibitory effect of light on per splicing does not require a functional clock. However, the efficiency of per splicing displays low-amplitude oscillation that is dependent on a functional circadian clock. Phospholipase C (encoded by no-receptor-potential-A (norpA)) probably plays a physiological, non-photic role in downregulating the production of spliced transcripts. Briefly, the proportion of the type B′ variant is abnormally high in the norpA mutant and exhibits little daytime decrease, even on warm days. Thus, it appears that irrespective of temperature, the splicing of per mRNA in norpA flies exhibits characteristics that are normally observed only at low temperatures, consistent with the advanced E activity of the mutant even on warm days. This cannot be explained simply by the well-known effect of NorpA on the visual pathway and suggests that norpA is directly involved in thermosensitivity.
In a further study in 2008, the Edery laboratory examined the splicing pattern of per in Drosophila yakuba. Drosophila yakuba is closely related to the cosmopolitan Drosophila melanogaster, but inhabits Afro-equatorial regions, where the species faces no significant seasonal variation in day length or temperature [43,50,51]. Although the per gene of D. yakuba also has a 3′-terminal intron, it is spliced out over a wide range of temperatures and photoperiods, consistent with the marginal effect of temperature on the daily rhythms of per RNA levels and behaviour in this tropical species. This study clearly shows a very tight link between the thermal responsiveness of splicing in the per 3′ UTR and temperature effects on the daily profiles of per mRNA levels and locomotor activity in two different species of Drosophila. Suboptimal splicing signals within the per mRNA underlie the thermosensitivity of splicing efficiency of this mRNA.
A more recent comparative analysis of D. melanogaster sequences in Atlantic coast fly populations from the USA found a number of single nucleotide polymorphisms close to the splicing region [52]. One of these haplotypes had higher splicing levels that correlate with a more cold-adapted behavioural phenotype. One might expect that if these polymorphisms are adaptive, a higher frequency for this haplotype might be found in northern, compared with southern, populations in America but this is evidently not the case [52]. However, as flies have only colonized North America for the past 200–300 years, it may be that over longer evolutionary periods (as in Europe, which flies colonized after the last Ice Age), the polymorphisms may generate a spatial pattern. A relevant example is the European Thr-Gly repeat length polymorphism within the coding region of per, which forms a latitudinal cline on that continent [53].
In a further evolutionary/ecological context, a recent study of fly behaviour in quasi-natural conditions revealed that at European summer temperatures, flies do not show a ‘classic’ siesta, but instead have large mid-day locomotor component, termed A (‘afternoon’) [54]. per splicing has a linear relationship with temperature within the natural 7–30°C range, but the siesta, as measured by the position of the evening (E) locomotor onset, or from the time between morning (M) offset and E onset (ignoring A), is not. Indeed, the phase position of the E onset is quite stable until average temperatures reach 20–22°C, when the E onset then begins to delay, correlating with the further inhibition of per splicing. This would suggest that if the per splicing is relevant to the wild; it is its inhibition at hot temperatures that is more important than its activation at colder temperatures, which makes a certain adaptive sense. However, the ‘fly in the ointment’ is that per01 mutants show essentially the same phenotype as the wild-type in terms of their natural siesta, so this would argue that the mechanism for generating the siesta may be dependent on per splicing (at least at high temperatures), but, oddly, independent of the PER protein. A further relevant laboratory study that introduced natural temperature ranges into the entrainment procedure also observed that behavioural phase changes in colder conditions could be generated as expected, even in the splice-locked per mutants [55]. This would suggest that under more natural thermal ranges, per splicing may not be the only contributor to behavioural phase changes.
It should be noted that per 3′ UTR regions have an important role not only for temperature adaptation but also for normal circadian rhythmicity. Indeed, early experiments performed in the late 1990s showed that flies in which per 3′ UTR has been replaced show long rhythms [13]. The importance of this regulation is evident, but it is not related to temporal control of translation as per mRNA is bound to polysomes, whenever it is expressed [13]. Whether this regulation is related to the splicing changes is not known, but it is certainly an interesting avenue to pursue.
The above-mentioned examples show a common theme of how changes in alternative splicing help balance the levels of key circadian components. In these cases, the alternative splicing events are more prominent at low temperatures and can lead to both the reduction and elevation of key circadian components. There are multiple variations of this theme that involve lower mRNA stability, lower translational efficiency, or the generation of non-productive transcripts. A first glimpse would suggest that the decrease of gene products at lower temperatures is unexpected. While general transcription initiation and elongation should decrease as a consequence of decreased biochemical activity at lower temperatures, protein and mRNA degradation should also decrease. Given that organisms manipulate the mRNA levels of these key regulators, per and frq (often the limiting factor in the circadian molecular cycle), it appears that the proteins alone cannot temperature compensate in most organisms as they do in cyanobacteria [56].
A very important issue that still remains to be examined is the contribution of alternative splicing to regulation of the mammalian circadian clock in response to environmental change (such as temperature, nutrition).
However, regulation has been found to operate in the other direction. A recent study by McGlincy et al. [57] demonstrates the regulatory effect of the murine circadian clock on alternative splicing. The authors found widespread circadian control of alternative splicing, which is tissue-dependent in both phase and amplitude and can be modulated by fasting. This implies the existence of a feedback loop involving alternative splicing and the circadian system. Indeed, it was recently established that the Arabidopsis protein methylase PRMT5, and its Drosophila orthologue dart5, regulate both circadian alternative splicing and period length [58].
3. miRNAs regulate circadian rhythms in response to environmental changes
miRNAs are post-transcriptional regulators of gene expression. These small RNAs recognize their mRNA targets through complete or partial binding with the target 3′ UTR and act mainly by reducing levels of translation and accelerating mRNA turnover. Recently, miRNAs have been implicated in both the core circadian mechanism and in the responses of the clock to the environment. We summarize below only the latter.
(a). miRNAs mediate canonical and stress-related circadian responses in Drosophila
Recently, a handful of papers have demonstrated roles for miRNAs in the Drosophila circadian system [15,19,59]. miRNAs mediate the central clock [18] and circadian output systems such as behaviour [19] and metabolism [15]. Interestingly, miRNAs at least partially regulate the response of the circadian clock to food intake.
In the first study involving a specific miRNA–mRNA pair in the circadian clock, Kadener et al. found that the miRNA bantam modulates the core circadian component clock and that other central clock components such as vri and cwo are also regulated by miRNAs. In addition to controlling the molecular pacemaker itself, miRNAs may be involved in providing robustness to the circadian system. Indeed, the study by Kadener et al. [59] showed that the resistance of the circadian clock to strong transcriptional perturbations, such as a fourfold to fivefold increase in the strength of transcriptional activation by the CLK–CYC dimer, is mediated at least partially by miRNAs. When flies expressing a ‘super-activator’ CYC transgene (CYCVP16) are transferred to an extreme temperature (29°C), their behavioural rhythmicity is dependent on the presence of dicer-1, the last enzyme in the miRNA-processing pathway. This strongly suggests that miRNAs provide robustness to the circadian system by buffering changes in transcriptional components under conditions of genetic, and more commonly, environmental stress.
A recent study by Luo & Sehgal [19] demonstrated that another miRNA, miR-279, regulates behavioural output of the circadian clock through the JAK/STAT pathway. Alterations in miR-279 levels (overexpression or inhibitory mutations) lead to disrupted locomotor activity rhythms. Interestingly, these altered rhythms do not correlate with any abnormality of the molecular clock, suggesting that miR-279 regulates an output pathway rather than the central pacemaker. The authors found that the product of upd, a ligand of JAK/STAT, probably mediates this effect, as knockdown of upd rescues the behavioural phenotype of miR-279 mutants. Moreover, manipulations of the JAK/STAT pathway affect locomotor rhythms, and there seems to be close communication between the clock neurons and some upd-expressing cells. The involvement of JAK/STAT signalling in the circadian clock suggests a mechanism by which this pathway controls behaviour imparted by the circadian clock: as this pathway is typically activated by stress, miRNA regulation could manipulate the output of the circadian neurons without interfering with the molecular clockwork. This is an exciting possibility and work in this direction should be fruitful.
(b). miR-132 mediates entrainment of the circadian clock to light in mammals
In a very elegant study by the Obrietan laboratory [11,60], miR-132 was shown to mediate entrainment of the circadian clock in mice. In brief, the authors identified two miRNAs that are regulated by the circadian and light-induced transcription factor CREB: miR-132 and miR-219. miR-219, which also is regulated directly by the CLK-BMAL1 dimer, affects the circadian pacemaker, as knockdown of this miRNA lengthens the circadian period. In addition, expression from the miR-219 promoter in PC12 cells is significantly increased upon co-expression of CLK and BMAL. Interestingly, the expression of the second CREB-target miRNA, miR-132, is independent of CLK-BMAL1 but displays circadian oscillations in the suprachiasmatic nuclei (SCN) that are dependent on the negative transcriptional regulators CRY1 and CRY2. The authors hypothesized that the circadian expression of miR-132 could be due to activation of CREB in the early-middle day. They showed that light pulses during the subjective night led to a significant increase in the levels of miR-132 in the SCN, suggesting a role in the circadian light input pathway. Downregulation of miR-132 in the SCN using specific antagomirs (cholesterol-conjugated oligonucleotides complementary to miR-132) have little to no effect on the circadian period, but strongly affect circadian locomotor behaviour after a light pulse. Downregulation of miR-132 prior to a light pulse in the subjective night results in a double phase shift in the behavioural rhythms of these mice.
In order to identify potential targets of miR-132, the authors performed a careful bioinformatics analysis. From this analysis, they identified the mRNA encoding the light-induced transcription factor RFX4 as a potential miR-132 target in the SCN. Indeed, and at least in a cell culture system, miR-132 strongly downregulates RFX4 protein expression in a miRNA-site-dependent manner. Lastly, the authors closed the loop and demonstrated that miR-132 can regulate the core circadian molecular oscillator. Thus, miR-132 levels can modulate the negative arm of the circadian clock and the response of the molecular circadian machinery to light. In a follow-up study, Alvarez-Saavedra et al. [61] showed that miR-132 controls the molecular clock in response to light by targeting a specific set of chromatin and translational regulators. The chromatin regulator Mecp2 is involved in the expression of the per1 and per2 promoters, and the proteins Btg2 and Paip2a suppress production of PER1 and PER2 by accelerating mRNA decay. This latter study demonstrates the mechanism by which a specific miRNA regulates entrainment of the circadian clock in the SCN.
4. Conclusions
In summary, the literature reviewed here represents studies that primarily emphasize the central role of post-transcriptional control, mediated largely through miRNAs and alternative splicing. These have been shown to mediate the rapid responses of the circadian clock to environmental changes. Most of this research involves the description of very specific thermosensitive splicing events of equally specific clock genes.
There are several questions that remain to be answered by future research. The work performed in plants suggested that a comprehensive, whole-genome RNA-seq would reveal the scope of environmental-dependent post-transcriptional regulation on clock machinery as well as output genes in other model organisms (e.g. as in [62] or [63]). Additionally, the molecular mechanisms underlying environmental-dependent post-transcriptional control need to be revealed by future research. Lastly, it would be interesting to compare the machineries involved in daily adaptation to routine environmental factors, to those machineries reacting under stress or to extreme environmental factors.
Acknowledgements
S.K. thanks the European Research Council (grant no. 260911), the Human Frontiers Science Program (CDA grant no. 10/2009), the Israeli Science Foundation (grant no. 1015/10) and the Marie Curie Reintegration Grant Program for supporting his research.
Funding statement
O.B. thanks to Israel Cancer Research Laboratory for the support. C.P.K. thanks Biotechnology and Biological Sciences Research Council for their support. J.D.L. receives support as a Canada Research Chair and also from the Natural Sciences and Engineering Research Council of Canada and the Canadian Institutes of Health Research.
References
- 1.Rensing L, Ruoff P. 2002. Temperature effect on entrainment, phase shifting, and amplitude of circadian clocks and its molecular bases. Chronobiol. Int. 19, 807–864 (doi:10.1081/CBI-120014569) [DOI] [PubMed] [Google Scholar]
- 2.Zheng X, Sehgal A. 2012. Speed control: cogs and gears that drive the circadian clock. Trends Neurosci. 35, 574–585 (doi:10.1016/j.tins.2012.05.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glaser FT, Stanewsky R. 2005. Temperature synchronization of the Drosophila circadian clock. Curr. Biol. 15, 1352–1363 (doi:10.1016/j.cub.2005.06.056) [DOI] [PubMed] [Google Scholar]
- 4.Majercak J, Sidote D, Hardin PE, Edery I. 1999. How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron 24, 219–230 (doi:10.1016/S0896-6273(00)80834-X) [DOI] [PubMed] [Google Scholar]
- 5.Mergenhagen D, Schweiger H-G. 1975. The effect of different inhibitors of transcription and translation on the expression and control of circadian rhythm in individual cells of Acetabularia. Exp. Cell Res. 94, 321–326 (doi:10.1016/0014-4827(75)90499-1) [DOI] [PubMed] [Google Scholar]
- 6.Lakin-Thomas PL. 2006. Transcriptional feedback oscillators: maybe, maybe not. J. Biol. Rhyth. 21, 83–92 (doi:10.1177/0748730405286102) [DOI] [PubMed] [Google Scholar]
- 7.Kojima S, Shingle DL, Green CB. 2011. Post-transcriptional control of circadian rhythms. J. Cell Sci. 124, 311–320 (doi:10.1242/jcs.065771) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baggs JE, Green CB. 2003. Nocturnin, a deadenylase in Xenopus laevis retina: a mechanism for posttranscriptional control of circadian-related mRNA. Curr. Biol. 13, 189–198 (doi:10.1016/S0960-9822(03)00014-9) [DOI] [PubMed] [Google Scholar]
- 9.Kojima S, Sher-Chen EL, Green CB. 2012. Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev. 26, 2724–2736 (doi:10.1101/gad.208306.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagel R, Clijsters L, Agami R. 2009. The miRNA-192/194 cluster regulates the Period gene family and the circadian clock. FEBS J. 276, 5447–5455 (doi:10.1111/j.1742-4658.2009.07229.x) [DOI] [PubMed] [Google Scholar]
- 11.Cheng HY, et al. 2007. microRNA modulation of circadian-clock period and entrainment. Neuron 54, 813–829 (doi:10.1016/j.neuron.2007.05.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim C, Lee J, Choi C, Kilman VL, Kim J, Park SM, Jang SK, Allada R, Choe J. 2011. The novel gene twenty-four defines a critical translational step in the Drosophila clock. Nature 470, 399–403 (doi:10.1038/nature09728) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y, Hunter-Ensor M, Schotland P, Sehgal A. 1998. Alterations of per RNA in noncoding regions affect periodicity of circadian behavioral rhythms. J. Biol. Rhythms 13, 364–379 (doi:10.1177/074873098129000192) [DOI] [PubMed] [Google Scholar]
- 14.Gatfield D, Schibler U. 2008. Circadian glucose homeostasis requires compensatory interference between brain and liver clocks. Proc. Natl Acad. Sci. USA 105, 14 753–14 754 (doi:10.1073/pnas.0807861105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vodala S, Pescatore S, Rodriguez J, Buescher M, Chen YW, Weng R, Cohen SM, Rosbash M. 2012. The oscillating miRNA 959–964 cluster impacts Drosophila feeding time and other circadian outputs. Cell Metab. 16, 601–612 (doi:10.1016/j.cmet.2012.10.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sundram V, Ng FS, Roberts MA, Millan C, Ewer J, Jackson FR. 2012. Cellular requirements for LARK in the Drosophila circadian system. J. Biol. Rhyth. 27, 183–195 (doi:10.1177/0748730412440667) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y, Genova G, Roberts M, Jackson FR. 2007. The LARK RNA-binding protein selectively regulates the circadian eclosion rhythm by controlling E74 protein expression. PLoS ONE 2, e1107 (doi:10.1371/journal.pone.0001107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kadener S, et al. 2009. A role for microRNAs in the Drosophila circadian clock. Genes Dev. 23, 2179–2191 (doi:10.1101/gad.1819509) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo W, Sehgal A. 2012. Regulation of circadian behavioral output via a MicroRNA-JAK/STAT circuit. Cell 148, 765–779 (doi:10.1016/j.cell.2011.12.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petrillo E, Sanchez SE, Kornblihtt AR, Yanovsky MJ. 2011. Alternative splicing adds a new loop to the circadian clock. Commun. Integr. Biol. 4, 284–286 (doi:10.4161/cib.4.3.14777) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mehra A, Baker CL, Loros JJ, Dunlap JC. 2009. Post-translational modifications in circadian rhythms. Trends Biochem. Sci. 34, 483–490 (doi:10.1016/j.tibs.2009.06.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reischl S, Kramer A. 2011. Kinases and phosphatases in the mammalian circadian clock. FEBS Lett. 585, 1393–1399 (doi:10.1016/j.febslet.2011.02.038) [DOI] [PubMed] [Google Scholar]
- 23.Keene JD. 2010. Minireview: global regulation and dynamics of ribonucleic acid. Endocrinology 151, 1391–1397 (doi:10.1210/en.2009-1250) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharp PA. 1987. Splicing of messenger RNA precursors. Science 235, 766–771 (doi:10.1126/science.3544217) [DOI] [PubMed] [Google Scholar]
- 25.Munoz MJ, de la Mata M, Kornblihtt AR. 2010. The carboxy terminal domain of RNA polymerase II and alternative splicing. Trends Biochem. Sci. 35, 497–504 (doi:10.1016/j.tibs.2010.03.010) [DOI] [PubMed] [Google Scholar]
- 26.Caceres JF, Kornblihtt AR. 2002. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 18, 186–193 (doi:10.1016/S0168-9525(01)02626-9) [DOI] [PubMed] [Google Scholar]
- 27.Dunlap JC, Loros JJ. 2004. The Neurospora circadian system. J. Biol. Rhythms 19, 414–424 (doi:10.1177/0748730404269116) [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Garceau NY, Loros JJ, Dunlap JC. 1997. Thermally regulated translational control of FRQ mediates aspects of temperature responses in the neurospora circadian clock. Cell 89, 477–486 (doi:10.1016/S0092-8674(00)80228-7) [DOI] [PubMed] [Google Scholar]
- 29.Garceau NY, Liu Y, Loros JJ, Dunlap JC. 1997. Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell 89, 469–476 (doi:10.1016/S0092-8674(00)80227-5) [DOI] [PubMed] [Google Scholar]
- 30.Diernfellner AC, Schafmeier T, Merrow MW, Brunner M. 2005. Molecular mechanism of temperature sensing by the circadian clock of Neurospora crassa. Genes Dev. 19, 1968–1973 (doi:10.1101/gad.345905) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colot HV, Loros JJ, Dunlap JC. 2005. Temperature-modulated alternative splicing and promoter use in the circadian clock gene frequency. Mol. Biol. Cell 16, 5563–5571 (doi:10.1091/mbc.E05-08-0756) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diernfellner A, Colot HV, Dintsis O, Loros JJ, Dunlap JC, Brunner M. 2007. Long and short isoforms of Neurospora clock protein FRQ support temperature-compensated circadian rhythms. FEBS Lett. 581, 5759–5764 (doi:10.1016/j.febslet.2007.11.043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Staiger D, Green R. 2011. RNA-based regulation in the plant circadian clock. Trends Plant Sci. 16, 517–523 (doi:10.1016/j.tplants.2011.06.002) [DOI] [PubMed] [Google Scholar]
- 34.Bieniawska Z, Espinoza C, Schlereth A, Sulpice R, Hincha DK, Hannah MA. 2008. Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol. 147, 263–279 (doi:10.1104/pp.108.118059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gould PD, et al. 2006. The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18, 1177–1187 (doi:10.1105/tpc.105.039990) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seo PJ, Park MJ, Lim MH, Kim SG, Lee M, Baldwin IT, Park CM. 2012. A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell 24, 2427–2442 (doi:10.1105/tpc.112.098723) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong WK, Mockler TC. 2010. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 20, 45–58 (doi:10.1101/gr.093302.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lynch M, Conery JS. 2000. The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155 (doi:10.1126/science.290.5494.1151) [DOI] [PubMed] [Google Scholar]
- 39.Tuskan GA, et al. 2006. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604 (doi:10.1126/science.1128691) [DOI] [PubMed] [Google Scholar]
- 40.James AB, Syed NH, Bordage S, Marshall J, Nimmo GA, Jenkins GI, Herzyk P, Brown JWS, Nimmo HG. 2012. Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell 24, 961–981 (doi:10.1105/tpc.111.093948) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.James AB, Syed NH, Brown JW, Nimmo HG. 2012. Thermoplasticity in the plant circadian clock: how plants tell the time-perature. Plant Signal. Behav. 7, 1219–1223 (doi:10.4161/psb.21491) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salome PA, McClung CR. 2005. PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17, 791–803 (doi:10.1105/tpc.104.029504) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Low KH, Lim C, Ko HW, Edery I. 2008. Natural variation in the splice site strength of a clock gene and species-specific thermal adaptation. Neuron 60, 1054–1067 (doi:10.1016/j.neuron.2008.10.048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stoleru D, Peng Y, Nawathean P, Rosbash M. 2005. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature 438, 238–242 (doi:10.1038/nature04192) [DOI] [PubMed] [Google Scholar]
- 45.Stoleru D, Peng Y, Agosto J, Rosbash M. 2004. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 431, 862–868 (doi:10.1038/nature02926) [DOI] [PubMed] [Google Scholar]
- 46.Grima B, Chelot E, Xia R, Rouyer F. 2004. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature 431, 869–873 (doi:10.1038/nature02935) [DOI] [PubMed] [Google Scholar]
- 47.Stoleru D, Nawathean P, Fernandez MP, Menet JS, Ceriani MF, Rosbash M. 2007. The Drosophila circadian network is a seasonal timer. Cell 129, 207–219 (doi:10.1016/j.cell.2007.02.038) [DOI] [PubMed] [Google Scholar]
- 48.Collins BH, Rosato E, Kyriacou CP. 2004. Seasonal behavior in Drosophila melanogaster requires the photoreceptors, the circadian clock, and phospholipase C. Proc. Natl Acad. Sci. USA 101, 1945–1950 (doi:10.1073/pnas.0308240100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Majercak J, Chen WF, Edery I. 2004. Splicing of the period gene 3′-terminal intron is regulated by light, circadian clock factors, and phospholipase C. Mol. Cell Biol. 24, 3359–3372 (doi:10.1128/MCB.24.8.3359-3372.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ko WY, David RM, Akashi H. 2003. Molecular phylogeny of the Drosophila melanogaster species subgroup. J. Mol. Evol. 57, 562–573 (doi:10.1007/s00239-003-2510-x) [DOI] [PubMed] [Google Scholar]
- 51.Russo CA, Takezaki N, Nei M. 1995. Molecular phylogeny and divergence times of drosophilid species. Mol. Biol. Evol. 12, 391–404 [DOI] [PubMed] [Google Scholar]
- 52.Low KH, Chen WF, Yildirim E, Edery I. 2012. Natural variation in the Drosophila melanogaster clock gene period modulates splicing of its 3′-terminal intron and mid-day siesta. PLoS ONE 7, e49536 (doi:10.1371/journal.pone.0049536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sawyer LA, Hennessy JM, Peixoto AA, Rosato E, Parkinson H, Costa R, Kyriacou CP. 1997. Natural variation in a Drosophila clock gene and temperature compensation. Science 278, 2117–2120 (doi:10.1126/science.278.5346.2117) [DOI] [PubMed] [Google Scholar]
- 54.Vanin S, Bhutani S, Montelli S, Menegazzi P, Green EW, Pegoraro M, Sandrelli F, Costa R, Kyriacou CP. 2012. Unexpected features of Drosophila circadian behavioural rhythms under natural conditions. Nature 484, 371–375 (doi:10.1038/nature10991) [DOI] [PubMed] [Google Scholar]
- 55.Currie J, Goda T, Wijnen H. 2009. Selective entrainment of the Drosophila circadian clock to daily gradients in environmental temperature. BMC Biol. 7, 49 (doi:10.1186/1741-7007-7-49) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tomita J, Nakajima M, Kondo T, Iwasaki H. 2005. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science 307, 251–254 (doi:10.1126/science.1102540) [DOI] [PubMed] [Google Scholar]
- 57.McGlincy NJ, Valomon A, Chesham JE, Maywood ES, Hastings MH, Ule J. 2012. Regulation of alternative splicing by the circadian clock and food related cues. Genome Biol. 13, R54 (doi:10.1186/gb-2012-13-6-r54) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanchez SE, et al. 2010. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 468, 112–116 (doi:10.1038/nature09470) [DOI] [PubMed] [Google Scholar]
- 59.Kadener S, Menet JS, Schoer R, Rosbash M. 2008. Circadian transcription contributes to core period determination in Drosophila. PLoS Biol. 6, e119 (doi:10.1371/journal.pbio.0060119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hansen KF, Sakamoto K, Obrietan K. 2011. MicroRNAs: a potential interface between the circadian clock and human health. Genome Med. 3, 10 (doi:10.1186/gm224) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alvarez-Saavedra M, Antoun G, Yanagiya A, Oliva-Hernandez R, Cornejo-Palma D, Perez-Iratxeta C, Sonenberg N, Cheng HY. 2011. miRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Hum. Mol. Genet. 20, 731–751 (doi:10.1093/hmg/ddq519) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Filichkin SA, Mockler TC. 2012. Unproductive alternative splicing and nonsense mRNAs: a widespread phenomenon among plant circadian clock genes. Biol. Direct. 7, 20 (doi:10.1186/1745-6150-7-20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Voytsekh O, Seitz SB, Iliev D, Mittag M. 2008. Both subunits of the circadian RNA-binding protein CHLAMY1 can integrate temperature information. Plant Physiol. 147, 2179–2193 (doi:10.1104/pp.108.118570) [DOI] [PMC free article] [PubMed] [Google Scholar]


