Abstract
Phylogenetic information is becoming a recognized basis for evaluating conservation priorities, but associations between extinction risk and properties of a phylogeny such as diversification rates and phylogenetic lineage ages remain unclear. Limited taxon-specific analyses suggest that species in older lineages are at greater risk. We calculate quantitative properties of the mammalian phylogeny and model extinction risk as an ordinal index based on International Union for Conservation of Nature Red List categories. We test for associations between lineage age, clade size, evolutionary distinctiveness and extinction risk for 3308 species of terrestrial mammals. We show no significant global or regional associations, and three significant relationships within taxonomic groups. Extinction risk increases for evolutionarily distinctive primates and decreases with lineage age when lemurs are excluded. Lagomorph species (rabbits, hares and pikas) that have more close relatives are less threatened. We examine the relationship between net diversification rates and extinction risk for 173 genera and find no pattern. We conclude that despite being under-represented in the frequency distribution of lineage ages, species in older, slower evolving and distinct lineages are not more threatened or extinction-prone. Their extinction, however, would represent a disproportionate loss of unique evolutionary history.
Keywords: ordinal index, distinctiveness, phylogenetic generalized linear mixed models, phylogenetic age
1. Introduction
Past extinctions and current extinction risk are not distributed randomly among taxa, and analyses of extant species have identified various factors that explain the selectivity of extinction risk [1,2]. However, these types of studies often overlook the role of evolutionary history as an explicit predictor. Studies of several vertebrate groups [2–9] have found that taxa from older and species-poor lineages are most likely to face extinction. Davies et al. [10] found the opposite pattern in South African plants: fast-evolving and speciose lineages are more prone to extinction. Selectivity in extinction risk could be explained by evolutionary history, represented by a clade's size and age. Older species usually occur in depauperate clades, which result from reduced geographical space, elevated extinction or low speciation [11].
In the early twentieth century, evolutionary biologists proposed that species in long-lived lineages were rare and ultimately fated with extinction [12]. Early analyses of survivorship curves from the fossil record found that the extinction probability of a taxon is independent of its age [13,14]. Reinterpretation of palaeontological studies by evolutionary geneticists briefly suggested that gene pools may introduce inferior morphotypes as they age [15].
Several mechanisms to explain the purported positive association between lineage age and risk of extinction have been proposed. First, extinction probability might stochastically increase through time [4,16]. Second, specialization is known to correlate with extinction risk [17,18]. Older taxa might be more specialized through early occupation of fringe niches, phylogenetic constraint or by having more time to evolve a specialized ecology, behaviour or morphology [19,20]. Specialization to narrow adaptive zones reduces the likelihood of radiation and of per-species background extinction in stable niches. This seems to be the case for relict mammal clades (e.g. monotremes (platypus and echidnas) and xenarthrans (anteaters, armadillos and sloths)) [11]. Third, slow reproductive rates that evolve early in a lineage's history and show a strong phylogenetic signal might limit the capacity for recovery under increased mortality from anthropogenic pressures [3,21].
Conversely, taxon age may have a negative association with extinction proneness. Older lineages might be more robust if greater taxon age reflects better survival ability and resilience [4]. An analysis of multiple animal datasets in the fossil record found that older lineages are closer to an average morphology, ecologically more generalist and able to survive a greater range of environmental changes [22]. Taxon age and extinction risk may be independent. Perhaps those taxa susceptible to extrinsic stresses are already extinct and no pattern is evident. Turvey & Fritz [23] found evidence of an extinction filter operating in the Holocene, and consequently some regional mammal faunas seem less threatened because the intrinsically susceptible species are extinct. If no relationship between lineage age and extinction risk exists, the taxonomic clustering of extinction-biasing traits must not relate to a species’ lineage age. Robust or susceptible species would not be over-represented in any part of the age distribution of extant species.
Mammals have high ecological, economic and social value [24,25]. We focus on mammals because correlates of extinction risk have been investigated extensively using phylogenetic comparative studies. Life-history datasets and phylogenies are available [24], and studies incorporating palaeontological data [23,26] enable interpretation of contemporary patterns on an evolutionary time scale.
If older lineages are intrinsically more extinction-prone, we expect lineage age to correlate positively with extinction risk in extant mammals, given the prevalence of external stresses and high proportion of threatened species. Patterns in this relationship might vary with geographical differences in mammalian diversity and threats. Dubey & Shine [27] found spatial disparity in the mean species ages of reptiles and amphibians. At similar latitudes, species from the Southern Hemisphere are older than species from the Northern Hemisphere. Geographical disparity in mammalian lineage ages also exists in modern mammal assemblages because of historical changes in climate and topography. The prevalence of modern threats varies significantly among mammalian orders [28], as well as the taxonomic clustering of different threat types. Fritz & Purvis [29] found that the phylogenetic pattern of risk caused by harvesting is more strongly clumped than for species threatened by habitat loss or invasive species.
Phylogenies can be analysed to inform macroevolution [11]. The temporal spacing of nodes reveals changes in diversification rates over time [30], and asymmetries in a phylogeny illustrate how clades vary in their underlying probabilities of diversifying [11]. Genetic or morphological differences between species can be used to estimate approximate branching times, constrained by dates from the fossil record. Evolutionary age and clade size can be combined to calculate evolutionary distinctiveness (ED). Evolutionarily distinct species have few living close relatives, slower diversification rates, greater lineage ages and are known to have experienced greater levels of extinction, leading to imbalance in the phylogeny [31]. Phylogenetic age, clade size, diversification and distinctiveness reflect key aspects of a species’ evolutionary history and allow us to analyse modern extinction risk at the species level.
Similar numbers of species extinctions can cause disparate losses of evolutionary history and, potentially, unique phenotypic and functional diversity [9,32]. Measures of taxonomic uniqueness have implications for modern conservation practice. For example, the Zoological Society of London's Evolutionarily Distinct and Globally Endangered (EDGE; [31,33,34]) initiative highlights and protects threatened species that represent the most unique evolutionary history. This system ranks species in terms of ED and global endangerment (GE). So far, no association has been found between ED and GE in analyses of birds and primates [9,33].
This paper investigates the relationship between phylogeny and the selectivity of extinction risk for extant and 14 recently extinct terrestrial non-volant mammal species. We build models at a global scale and for different taxonomic groups and geographical regions. We test for associations between phylogenetic age, genus size and ED, net diversification rate and extinction risk.
2. Material and methods
(a). Data sources
We focused only on terrestrial, non-volant mammals. We followed the International Union for Conservation of Nature (IUCN) Red List [35] nomenclature and excluded data deficient, domestic and taxonomically uncertain species. The final dataset of species traits included 3294 extant species and 14 species that became extinct after 1800, appear in the chosen phylogeny and have information on geographical distribution (see the electronic supplementary material, dataset S1). We collected data on body size as adult mass in grams, following the methods in the PanTHERIA database [36] for calculating measures of central tendency.
Body size is frequently associated with vulnerability to extinction [37], mainly due to its correlation with several other traits that are more directly tied to persistence (e.g. speed of life history, home range size and conflict with humans) [38]. To avoid spurious results, we incorporated body size as a covariate in all of our analyses. We focused on obtaining body size information for all the species, supplementing information in the PanTHERIA [36], MOM v. 4.1 [39] and Morgan [40] datasets with recently published, unpublished, museum and grey literature data (see the electronic supplementary material, dataset S2). Body size data for 202 species could not be located (see the electronic supplementary material, dataset S3), so we imputed the missing values to avoid biases originating from casewise removal of species with missing data [41]. We applied non-parametric missing value imputation using random forests implemented in the R package missForest [42], using a random forest trained on the observed values to predict the missing values. We completed datasets of 17 mammal families that included species with no body size information, and evaluated the statistical deviations of the imputed datasets relative to the complete datasets and to randomly guessed values following Pantanowitz & Marwala [43]. We kept all imputed values, since the missing data imputation did not have a significant negative impact on the statistical properties of the data (mean, first quartile, median, third quartile, standard deviation, variance, combined minimum standard error, mean Mahalanobis distance, linear correlation with target set and maximum percentage deviation).
We obtained phylogenetic age estimates defined as branching times from sister taxa, from an update [44] to a dated and calibrated species-level composite supertree of mammals [30]. We defined genus size as the number of congeners and used the ED metric from the 2011 EDGE list [31]. We collected net diversification rates from Soria-Carrasco & Castresana [45] for mammalian genera identified as monotypic in the same supertree [44], calculated assuming no extinction, or a high extinction fraction.
We used the IUCN Red List status as our response variable of extinction risk. For species-level models, we converted the threat categories to an ordinal index from Least Concern (zero) to Extinct (five). For genus-level analyses, we used the Red List categories to define species as threatened or non-threatened to count the number of threatened species or calculate the proportion threatened per genus (number of threatened species divided by genus size). We counted those species considered vulnerable, endangered, critically endangered or extinct as ‘threatened’, and species classified as least concern or near threatened as ‘non-threatened’.
(b). Statistical analysis
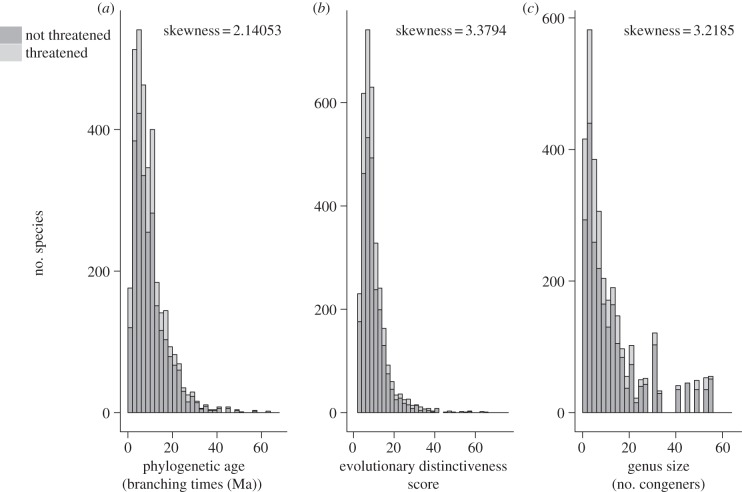
All analyses were carried out in R v. 2.15.2 [46]. An exploratory analysis of the frequency distribution of mammalian species’ lineage ages, ED scores and genus sizes revealed a right skew for all three variables (figure 1). We initially tested for associations at the genus level and then focused on relationships at the species level.
Figure 1.
(a–b) Frequency distributions of the quantitative variables derived from the phylogeny, showing threatened species per bin and skewness values. Ma, millions of years ago.
(i). Genus-level analysis
Past extinctions are less likely to cause misleading branch length values at supraspecific taxonomic levels and at a global scale than at the species level and local scales. Missing taxa lead to overestimated branching times [47] and extinctions of entire genera are less common than species extinctions [21]. However, more higher-order taxon losses than expected by chance are expected under the current extinction regime [2,48]. Initial tests for global patterns followed Johnson et al. [6], including the transformation of lineage ages and genus sizes to base two logarithms. We calculated mean body size, genus age and size, and ED for a global set of 896 genera and quantified extinction risk as the proportion of species threatened. We examined the relationships between extinction risk and the phylogenetic variables using generalized additive models (logit link function, binomial distribution of variance and an extra penalty added to each smooth term so that it can be penalized to zero) using the R package mgvc [49].
This approach treats genera as independent, yet several groups of two or more genera arise from the same nodes in the phylogeny. We repeated the analyses using phylogenetic generalized linear mixed models (PGLMM) to incorporate phylogenetic information as a covariance matrix representing the amount of shared evolutionary history between taxa. We fit the models with the R package MCMCglmm [50].
For all PGLMM analyses, we used an uninformative prior for the random effect [51] and ran each chain for 555 000 iterations with a thinning value of 500 after a burn-in of 50 000, resulting in 1000 samples. All diagnostics of convergence of PGLMM parameters followed Rutkowska et al. [51] using the Gelman–Rubin statistic. Potential scale reduction values were all less than 1.1 among three parallel Markov Chain Monte Carlo (MCMC) chains for models with different starting values [52], and the autocorrelations of posterior samples were all less than 0.1. Effective sample sizes for all fixed effects were all greater than 800. We calculated all parameter estimates after combining the three parallel MCMC chains. We considered fixed effects to be statistically significant when the probabilities in the 99% credible region (based on the highest posterior density interval) did not include zero.
Of the 896 genera with data, only 440 could be clearly identified as nodes in the phylogeny. We modelled the number of species threatened in a genus as a binomial response, treating species classified in the IUCN Red List as extinct, critically endangered, endangered and vulnerable as ‘threatened’, and species listed as near threatened (NT) and least concern as ‘not threatened’.
We analysed the association between diversification rates and threatened species per genus separately, assuming no extinction or high extinction, and including body size as a covariate. We used the values from Soria-Carrasco & Castresana [45] for 173 genera and modelled extinction risk as a binomial response.
In reality, no clear line separates threatened and non-threatened species [53]. Species listed as NT have been considered as both ‘threatened’ and ‘not threatened’ in previous studies [54,55]. To test whether changing the threat threshold influences our results, we repeated all genus-level analyses, considering NT species as not threatened, threatened and excluding NT species altogether.
(ii). Species-level analysis
Threatened species within a genus may differ widely in their risk level and body sizes, so we also performed analyses for species. The amount of difference in a species’ actual extinction risk probably varies between threat categories, which are separated by unequal distances along the underlying continuous variable that they measure [56,57]. We used PGLMM to model extinction risk as an ordered response. Residual variance cannot be identified in ordinal probit models, so it was set at a fixed arbitrary value of one in the prior specification for the variance components of the fixed effects.
We tested for associations at the global scale using the full dataset of 3308 species. To address our biogeographic, taxonomic and ecological hypotheses, we then built models with species subsets defined by global biogeographic regions, orders and for monophyletic groups of two or more orders with similar ecologies [58]. We trimmed the complete phylogenetic tree to match the species subsets used for each model when creating the covariance matrices. We used species distributions from the IUCN spatial dataset [35] and a digitalized map of mammalian zoogeographic regions [59] to divide the global species list into spatial subsets that only included those species that occur exclusively within the region boundary. We divided all mammals into 11 orders (Afrosoricida, Carnivora, Cetartiodactyla, Dasyuromorphia, Didelphimorphia, Diprotodontia, Eulipotyphla, Lagomorpha, Perissodactyla, Primates and Rodentia) and six monophyletic groups: Afrotheria, Euarchonta, Glires, marsupials, ungulates and xenarthrans. We repeated the analysis of our primate dataset excluding lemurs (Lemuroidea, families Daubentoniidae, Indriidae, Lemuridae and Lepilemuridae) to ensure that any result would not be entirely driven by this ancient, distinct and endemic clade with one of the highest levels of threat recorded for any vertebrate group [9,60].
3. Results
(a). Genus-level patterns
Extinction risk increased with body size in the initial genus-level analysis for global data (χ2 = 173.63, estimated d.f. = 8, n = 896, p < 0.0001) and in the PGLMM analysis that accounted for phylogenetically structured data (see the electronic supplementary material, table S3). We found no relationships between genus age or distinctiveness and extinction risk.
We found no significant associations between net diversification rates and the number of threatened species in a genus (see the electronic supplementary material, table S4) for both estimates (with or without extinction). The parameter estimates from both of our models are almost identical. Soria-Carrasco & Castresana [45] found that both rate estimates are highly correlated and suggest that assuming extinction has little impact on comparative analyses.
Different ‘threatened’ versus ‘non-threatened’ thresholds for binomial models did not alter the parameter estimates or nature of the relationships and resulted in reduced sample sizes when excluding NT species. Using the proportion or number of threatened species per genus has the disadvantage of losing the detail that a species-specific assessment provides, especially for smaller genera. This measure is sensitive to varying definitions of genus that may ultimately affect genus sizes in a quantitative analysis. The issue is confounded by the use of arbitrary taxonomic units that are not based on biological principles and might not be monophyletic.
(b). Species-level patterns
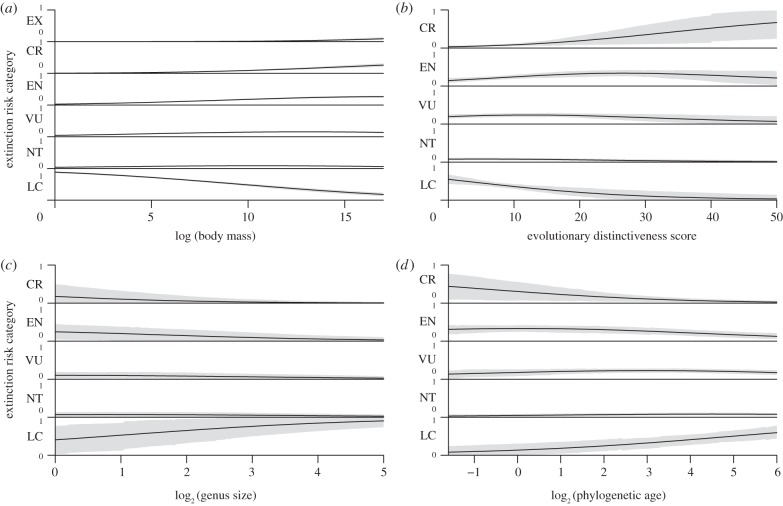
Body size was the only significant predictor of extinction risk in the global model (figure 2a) and in most models for spatial and taxonomic subsets (see the electronic supplementary material, tables S1–S3). We found significant effects for phylogenetic variables in three taxonomic subsets (see the electronic supplementary material, table S1): extinction risk increases with ED for primate species (figure 2b), and lagomorph species (rabbits, hares and pikas) in less speciose genera have higher probabilities of being classified as threatened (figure 2c). Evolutionarily distinct primate species are more likely to be critically endangered than least concern. Lagomorph species in more speciose genera have the highest probability of being listed as least concern, while the probability of being classified into the higher risk categories decreases with increasing number of species in the genus. We found no significant association between evolutionary distinctiveness and extinction risk once we excluded lemurs from the primate data. However, we found a significant negative association between phylogenetic age and extinction risk (see the electronic supplementary material, table S1) in non-lemur primates. The probability of being classified as least concern category is greater with increasing phylogenetic age, and older species have lower probabilities of being classified as endangered or critically endangered (figure 2d).
Figure 2.
Effects of body size, ED and genus size on the probabilities of falling into different extinction risk categories when the effects of other variables are held constant for (a) mammal species globally, (b) primates, (c) lagomorphs and (d) primates, excluding lemurs. Confidence intervals (95%) are shaded in grey. CR, critically endangered; EN, endangered; EX, extinct; LC, least concern; NT, near threatened; VU, vulnerable.
4. Discussion
The phylogenetic traits we chose to reflect a species’ evolutionary history do not generally predict extinction risk in mammals. We suggest that in mammals, there is an overall lack of biologically meaningful associations between evolutionary age, distinctiveness, clade size, diversification and any known extinction-biasing traits, such as body size or ecological versatility, even when these traits have a strong phylogenetic signal [44].
Net diversification rates for genera had no association with extinction risk (see the electronic supplementary material, table S4). Certain clades are characterized by either high diversity and rapid diversification (e.g. carnivores) or low diversity and systemic diversification rate slowing (e.g. Afrotheria and Perissodactyla) [11]. We did not find any general patterns in the species-level analyses of major lineages to suggest that historical differences in diversification influence current extinction risk.
ED predicted extinction risk in primates (figure 2b), a well-studied group with several known predictors of threat [57,61]. Redding et al. [9] found that evolutionarily distinct primate species are ecomorphologically odd and geographically peripheral. The mechanisms that make trait oddness and distance from continental centroid significant drivers of extinction risk for evolutionarily distinct species are unknown, but our results point to lemurs driving the relationship between ED and threat status. We found no significant relationship once we excluded lemurs from the primate analysis (electronic supplementary material, table S1). The pattern of higher risk for younger species of primates (excluding lemurs) is novel for vertebrates (figure 2d).
If younger taxa occupy a smaller geographical range or adaptive space, they may be more sensitive to small-scale environmental perturbations and more susceptible to extinction [14]. Models in Purvis et al. [1] underestimated threat status for primates in tropical countries with especially high levels of deforestation, and concluded that species with lower risk than predicted by intrinsic traits occur in large areas of conserved habitat. Spatially and taxonomically non-random patterns of threat may explain our result of decreasing extinction risk with increasing lineage age for primates (excluding lemurs). For example: bushbabies (family Galagidae) are in an older lineage with no threatened species out of 29. Threat level in the recent and diverse radiation of colobine monkeys (subfamily Colobinae) exceeds 70 per cent (40/53), including several critically endangered island endemics.
Unlike most mammalian lineages, lagomorphs exhibit higher species diversity in the fossil record than in the present, suggesting an ongoing decline in diversity [62]. Eight out of 13 (61%) extant genera of lagomorphs are monotypic and six of these (75%) are threatened. Mooers et al. [26] found that species in depauperate clades experienced disproportionately high extinction during the Holocene, and attribute the disparate loss to the island effect. Except for the extinct Sardinian pika Prolagus sardus [35], no palaeontological data indicate elevated levels of extinction for this group [62]. Lagomorph extinctions were similarly minimal in the Late Pleistocene, when mammal extinctions preferentially affected large-bodied mammals [63]. Our analysis of current extinction risk may have identified the ongoing decline in lagomorph diversity (figure 2c). The decline in perissodactyl diversity evident in the fossil record, and in the clade's downshifted diversification rate that is also predicted by branching models [11,30,48,62], was not evident in our results. A negative association between genus size and extinction risk supports the notion that taxa with more species may have phenotypes or ecologies that cause higher diversification rates [64]. Taxon size could relate to robustness towards external threats, efficient niche partitioning, strong dispersal abilities or wider geographical distributions and niche breadths, while specialization to narrow adaptive zones is perhaps the best predictor of species-poor clades [11].
The frequency distribution of mammalian taxon ages and distinctiveness values are right-skewed (figure 1). We propose that this has led to misleading interpretations of the effect of taxon antiquity on modern extinction risk. Few living species are ancient or extremely distinct, yet most of these are not threatened with extinction. Species at the tail-end of the age distribution are not intrinsically more susceptible to extinction nor more threatened than younger species. The significant negative relationship between phylogenetic age and extinction risk supports our view. We suspect that this right skew in lineage ages reflects the biased ratio between extinction and speciation in deep time, but extinction rates should not be estimated from molecular phylogenies [11]. Branching times from a molecular phylogeny do not reflect species' lifespans or stratigraphic durations and might not represent true node ages times when the phylogeny is built from relationships between extant taxa [6]. We chose the phylogeny with the highest taxonomic coverage and a consistent dating process, based on multi-gene alignment and cladistically robust fossil calibration points [30]. Our results from the genus-level analyses agree with the species-level models, supporting our conclusion that there is a general lack of significant associations between taxon age and extinction risk.
Treating the IUCN Red List categories as an ordered factor in analyses that correct for phylogenetic inertia provides a powerful method for understanding extinction risk. Ordered threat categories help to guide priorities for conservation investment among species and produce a series of recommendations for conservation action for each category [65]. Our approach can identify trends within each threat category, and it avoids losing information by aggregating classifications into dichotomous variables. We avoided elevated type I error rates caused by not preserving the variance structure of the original ordinal ranks [57] when assuming that categories are evenly spaced and continuously varying.
The methods in this study may be applied to investigate the role of phylogenies in the extinction risk patterns of other vertebrates with different evolutionary dynamics. Large databases of life-history traits [24], extinction risk assessments [35] and species-level phylogenies [66] for birds, fish and amphibians are increasingly available for comparative analyses.
Although we conclude that evolutionary history has no consistent association with extinction risk, prioritization methods that combine threat status and evolutionary history are critical, because anthropogenic threats are increasingly pervasive regardless of species’ intrinsic traits [41]. Widespread threats like habitat loss, invasive species and overkill are sampling more of the taxon age distribution, including distinct and ancient species [14].
Acknowledgements
We thank the editor and two anonymous reviewers for comments that improved an earlier version of the manuscript. We are grateful to Nels Johnson for comments on ordinal regression, Jeffrey O. Hanson for assistance with R scripting, Kerrie A. Wilson for comments on manuscript structure and N. Wood, J. Charters and K. Garland for help in error checking the taxonomic and body size databases.
Funding statement
L.D.V.A. was supported by funding from Consejo Nacional de Ciencia y Tecnología (CONACYT) scholarship 308685. D.O.F. was supported by Australian Research Council fellowships DP0773920 and FT110100191.
References
- 1.Purvis A, Gittleman JL, Cowlishaw G, Mace GM. 2000. Predicting extinction risk in declining species. Proc. R. Soc. Lond. B 267, 1947–1952 (doi:10.1098/rspb.2000.1234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Purvis A, Agapow P-M, Gittleman JL, Mace GM. 2000. Non-random extinction and the loss of evolutionary history. Science 288, 328–330 (doi:10.1126/science.288.5464.328) [DOI] [PubMed] [Google Scholar]
- 3.Bennett PM, Owens IPF. 1997. Variation in extinction risk among birds: chance or evolutionary predisposition? Proc. R. Soc. Lond. B 264, 401–408 (doi:10.1098/rspb.1997.0057) [Google Scholar]
- 4.Gaston K, Blackburn T. 1997. Evolutionary age and risk of extinction in the global avifauna. Evol. Ecol. 11, 557–565 (doi:10.1007/s10682-997-1511-4) [Google Scholar]
- 5.Jennings S, Reynolds JD, Polunin NVC. 1999. Predicting the vulnerability of tropical reef fishes to exploitation with phylogenies and life histories. Conserv. Biol. 13, 1466–1475 (doi:10.1046/j.1523-1739.1999.98324.x) [Google Scholar]
- 6.Johnson CN, Delean S, Balmford A. 2002. Phylogeny and the selectivity of extinction in Australian marsupials. Anim. Conserv. 5, 135–142 (doi:10.1017/s1367943002002196) [Google Scholar]
- 7.Meijaard E, Sheil D, Marshall AJ, Nasi R. 2008. Phylogenetic age is positively correlated with sensitivity to timber harvest in Bornean mammals. Biotropica 40, 76–85 (doi:10.1111/j.1744-7429.2007.00340.x) [Google Scholar]
- 8.Russell GJ, Brooks TM, McKinney MM, Anderson CG. 1998. Present and future taxonomic selectivity in bird and mammal extinctions. Conserv. Biol. 12, 1365–1376 (doi:10.1111/j.1523-1739.1998.96332.x) [Google Scholar]
- 9.Redding DW, DeWolff CV, Mooers AØ. 2010. Evolutionary distinctiveness, threat status, and ecological oddity in primates. Conserv. Biol. 24, 1052–1058 (doi:10.1111/j.1523-1739.2010.01532.x) [DOI] [PubMed] [Google Scholar]
- 10.Davies TJ, et al. 2011. Extinction risk and diversification are linked in a plant biodiversity hotspot. PLoS Biol. 9, e1000620 (doi:10.1371/journal.pbio.1000620) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Purvis A, Fritz SA, Rodríguez J, Harvey PH, Grenyer R. 2011. The shape of mammalian phylogeny: patterns, processes and scales. Phil. Trans. R. Soc. B 366, 2462–2477 (doi:10.1098/rstb.2011.0025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiedler PL. 1986. Concepts of rarity in vascular plant species, with special reference to the genus Calochortus Pursh (Liliaceae). Taxon 35, 502–518 (doi:10.2307/1221904) [Google Scholar]
- 13.Van Valen L. 1973. A new evolutionary law. Evol. Theory 1, 1–30 [Google Scholar]
- 14.Boyajian GE. 1991. Taxon age and selectivity of extinction. Paleobiology 17, 49–57 [Google Scholar]
- 15.Ward P. 1992. On Methuselah's trail: living fossils and the great extinctions, p. 212 New York, NY: W H Freeman & Co [Google Scholar]
- 16.Pearson PN. 1998. Speciation and extinction asymmetries in paleontological phylogenies: evidence for evolutionary progress? Paleobiology 24, 305–335 (doi:10.1666/0094-8373-24.3.305) [Google Scholar]
- 17.Arita HT, Robinson JG, Redford KH. 1990. Rarity in Neotropical forest mammals and its ecological correlates. Conserv. Biol. 4, 181–192 (doi:10.1111/j.1523-1739.1990.tb00107.x) [Google Scholar]
- 18.Williams SE, Williams YM, VanDerWal J, Isaac JL, Shoo LP, Johnson CN. 2009. Ecological specialization and population size in a biodiversity hotspot: how rare species avoid extinction. Proc. Natl Acad. Sci. USA 106(Suppl. 2), 19 737–19 741 (doi:10.1073/pnas.0901640106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nosil P. 2002. Transition rates between specialization and generalization in phytophagous insects. Evolution 56, 1701–1706 (doi:10.1111/j.0014-3820.2002.tb01482.x) [DOI] [PubMed] [Google Scholar]
- 20.Nosil P, Mooers AØ. 2005. Testing hypotheses about ecological specialization using phylogenetic trees. Evolution 59, 2256–2263 (doi:10.1111/j.0014-3820.2005.tb00933.x) [PubMed] [Google Scholar]
- 21.McKinney ML. 1997. Extinction vulnerability and selectivity: combining ecological and paleontological views. Annu. Rev. Ecol. Syst. 28, 495–516 (doi:10.1146/annurev.ecolsys.28.1.495) [Google Scholar]
- 22.Liow LH. 2007. Lineages with long durations are old and morphologically average: an analysis using multiple datasets. Evolution 61, 885–901 (doi:10.1111/j.15585646.2007.00077.x) [DOI] [PubMed] [Google Scholar]
- 23.Turvey ST, Fritz SA. 2011. The ghosts of mammals past: biological and geographical patterns of global mammalian extinction across the Holocene. Phil. Trans. R. Soc. B 366, 2564–2576 (doi:10.1098/rstb.2011.0020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.González-Suárez M, Lucas PM, Revilla E. 2012. Biases in comparative analyses of extinction risk: mind the gap. J. Anim. Ecol. 81, 1211–1222 (doi:10.1111/j.1365-2656.2012.01999.x) [DOI] [PubMed] [Google Scholar]
- 25.Fritz SA, Purvis A. 2010. Phylogenetic diversity does not capture body size variation at risk in the world's mammals. Proc. R. Soc. B 277, 2435–2441 (doi:10.1098/rspb.2010.0030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mooers AØ, Goring SJ, Turvey ST, Kuhn TS. 2009. Holocene extinctions and the loss of feature diversity. In Holocene extinctions (ed. Turvey ST.), pp. 263–277 Oxford, UK: Oxford University Press [Google Scholar]
- 27.Dubey S, Shine R. 2010. Geographic variation in the age of temperate-zone reptile and amphibian species: Southern Hemisphere species are older. Biol. Lett. 7, 96–97 (doi:10.1098/rsbl.2010.0557) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mace GM, Balmford A. 2000. Patterns and processes in contemporary mammalian extinction. In Priorities for the conservation of mammalian diversity. Has the panda had its day? (eds Entwistle A, Dunstone N.), pp. 27–52 Cambridge, UK: Cambridge University Press [Google Scholar]
- 29.Fritz SA, Purvis A. 2010. Selectivity in mammalian extinction risk and threat types: a new measure of phylogenetic signal strength in binary traits. Conserv. Biol. 24, 1042–1051 (doi:10.1111/j.1523-1739.2010.01455.x) [DOI] [PubMed] [Google Scholar]
- 30.Bininda-Emonds ORP, et al. 2008. The delayed rise of present-day mammals. Nature 456, 274 (doi:10.1038/nature07347) [DOI] [PubMed] [Google Scholar]
- 31.Collen B, Turvey ST, Waterman C, Meredith HMR, Kuhn TS, Baillie JEM, Isaac NJB. 2011. Investing in evolutionary history: implementing a phylogenetic approach for mammal conservation. Phil. Trans. R. Soc. B 366, 2611–2622 (doi:10.1098/rstb.2011.0109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cadotte MW, Davies JT. 2010. Rarest of the rare: advances in combining evolutionary distinctiveness and scarcity to inform conservation at biogeographical scales. Divers. Distrib. 16, 376–385 (doi:10.1111/j.1472-4642.2010.00650.x) [Google Scholar]
- 33.Redding DW, Mooers AØ. 2006. Incorporating evolutionary measures into conservation prioritization. Conserv. Biol. 20, 1670–1678 (doi:10.1111/j.1523-1739.2006.00555.x) [DOI] [PubMed] [Google Scholar]
- 34.Isaac NJB, Turvey ST, Collen B, Waterman C, Baillie JEM. 2007. Mammals on the EDGE: conservation priorities based on threat and phylogeny. PLoS ONE 2, e296 (doi:10.1371/journal.pone.0000296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.IUCN 2012. IUCN red list of threatened species. Version 2012.1. See http://www.iucnredlist.org. Downloaded on 24 September 2012.
- 36.Jones KE, et al. 2009. PanTHERIA: a species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology 90, 2648 (doi:10.1890/08-1494.1) [Google Scholar]
- 37.Collen B, McRae L, Deinet S, De Palma A, Carranza T, Cooper N, Loh J, Baillie JEM. 2011. Predicting how populations decline to extinction. Phil. Trans. R. Soc. B 366, 2577–2586 (doi:10.1098/rstb.2011.0015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burton AC, Sam MK, Kpelle DG, Balangtaa C, Buedi EB, Brashares JS. 2011. Evaluating persistence and its predictors in a West African carnivore community. Biol. Conserv. 144, 2344–2353 (doi:10.1016/j.biocon.2011.06.014) [Google Scholar]
- 39.Smith FA, Lyons SK, Ernest SKM, Jones KE, Kaufman DM, Dayan T, Marquet PA, Brown JH, Haskell JP. 2003. Body mass of late quaternary mammals. Ecology 84, 3403 (doi:10.1890/02-9003) [Google Scholar]
- 40.Ernest SKM. 2003. Life history characteristics of placental non-volant mammals. Ecology 84, 3402 (doi:10.1890/02-9002) [Google Scholar]
- 41.Fisher DO, Blomberg SP, Owens IPF. 2003. Extrinsic versus intrinsic factors in the decline and extinction of Australian marsupials. Proc. R. Soc. Lond. B 270, 1801–1808 (doi:10.1098/rspb.2003.2447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stekhoven DJ, Bühlmann P. 2011. MissForest— non-parametric missing value imputation for mixed-type data. Bioinformatics 28, 112–118 (doi:10.1093/bioinformatics/btr597) [DOI] [PubMed] [Google Scholar]
- 43.Pantanowitz A, Marwala T. 2009. Evaluating the impact of missing data imputation. In Advanced data mining and applications (eds Huang R, Yang Q, Pei J, Gama J, Meng X, Li X.), pp. 577–586 Berlin, Germany: Springer [Google Scholar]
- 44.Fritz SA, Bininda-Emonds ORP, Purvis A. 2009. Geographical variation in predictors of mammalian extinction risk: big is bad, but only in the tropics. Ecol. Lett. 12, 538–549 (doi:10.1111/j.1461-0248.2009.01307.x) [DOI] [PubMed] [Google Scholar]
- 45.Soria-Carrasco V, Castresana J. 2012. Diversification rates and the latitudinal gradient of diversity in mammals. Proc. R. Soc. B 279, 4148–4155 (doi:10.1098/rspb.2012.1393) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.R Development Core Team 2012. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- 47.Gittleman JL, Jones KE, Price SA. 2004. Using complete phylogenies in comparative biology. In Phylogenetic supertrees: combining information to reveal the tree of life, vol. 4 (ed. Bininda-Emonds ORP.), p. 439 Dordrecht, The Netherlands: Kluwer [Google Scholar]
- 48.McKinney ML. 1998. Branching models predict loss of many bird and mammal orders within centuries. Anim. Conserv. 1, 159–164 (doi:10.1111/j.1469-1795.1998.tb00024.x) [Google Scholar]
- 49.Wood SN. 2011. Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. J. R. Stat. Soc. B 73, 3–36 (doi:10.1111/j.1467-9868.2010.00749.x) [Google Scholar]
- 50.Hadfield JD. 2010. MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J. Stat. Softw. 33, 1–2220808728 [Google Scholar]
- 51.Rutkowska J, Lagisz M, Nakagawa S. 2012. The long and the short of avian W chromosomes: no evidence for gradual W shortening. Biol. Lett. 8, 636–638 (doi:10.1098/rsbl.2012.0083) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gelman A, Rubin DD. 1992. Inference from iterative simulation using multiple sequences. Stat. Sci. 7, 457–472 (doi:10.1214/ss/1177011136) [Google Scholar]
- 53.Robbirt K, Roberts D, Hawkins J. 2006. Comparing IUCN and probabilistic assessments of threat: do IUCN Red List criteria conflate rarity and threat? Biodivers. Conserv. 15, 1903–1912 (doi:10.1007/s10531-005-4307-2) [Google Scholar]
- 54.Agnarsson I, Kuntner M, May-Collado LJ. 2010. Dogs, cats, and kin: a molecular species-level phylogeny of Carnivora. Mol. Phylogenet. Evol. 54, 726–745 (doi:10.1016/j.ympev.2009.10.033) [DOI] [PubMed] [Google Scholar]
- 55.Mooers AØ, Faith DP, Maddison WP. 2008. Converting endangered species categories to probabilities of extinction for phylogenetic conservation prioritization. PLoS ONE 3, e3700 (doi:10.1371/journal.pone.0003700) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Purvis A, Cardillo M, Grenyer R, Collen B. 2005. Correlates of extinction risk: phylogeny, biology, threat and scale. In Phylogeny and conservation (ed. Purvis A.), pp. 295–316 Cambridge, UK: Cambridge University Press [Google Scholar]
- 57.Matthews LJ, Arnold C, Machanda Z, Nunn CL. 2011. Primate extinction risk and historical patterns of speciation and extinction in relation to body mass. Proc. R. Soc. B 278, 1256–1263 (doi:10.1098/rspb.2010.1489) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Valen L. 1971. Adaptive zones and the orders of mammals. Evolution 25, 420–428 (doi:10.2307/2406935) [DOI] [PubMed] [Google Scholar]
- 59.Cox B. 2001. The biogeographic regions reconsidered. J. Biogeogr. 28, 511–523 (doi:10.1046/j.1365-2699.2001.00566.x) [Google Scholar]
- 60.Spathelf M, Waite TA. 2007. Will hotspots conserve extra primate and carnivore evolutionary history? Divers. Distrib. 13, 746–751 (doi:10.1111/j.1472-4642.2007.00386.x) [Google Scholar]
- 61.Cowlishaw G, Pettifor RA, Isaac NJB. 2009. High variability in patterns of population decline: the importance of local processes in species extinctions. Proc. R. Soc. B 276, 63–69 (doi:10.1098/rspb.2008.0767) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lopez-Martinez N. 2008. The lagomorph fossil record and the origin of the European rabbit. In Lagomorph biology (eds Alves P, Ferrand N, Hackländer K.), pp. 27–46 Berlin, Germany: Springer [Google Scholar]
- 63.Cardillo M, Mace GM, Jones KE, Bielby J, Bininda-Emonds ORP, Sechrest W, Orme CDL, Purvis A. 2005. Multiple causes of high extinction risk in large mammal species. Science 309, 1239–1241 (doi:10.1126/science.1116030) [DOI] [PubMed] [Google Scholar]
- 64.Mark AM, Jonathan MB. 2007. Clade age and not diversification rate explains species richness among animal taxa. Am. Nat. 169, E97–E106 (doi:10.1086/512135) [DOI] [PubMed] [Google Scholar]
- 65.Rodrigues ASL, Pilgrim JD, Lamoreux JF, Hoffmann M, Brooks TM. 2006. The value of the IUCN Red List for conservation. Trends Ecol. Evol. 21, 71–76 (doi:10.1016/j.tree.2005.10.010) [DOI] [PubMed] [Google Scholar]
- 66.Fritz SA, Rahbek C. 2012. Global patterns of amphibian phylogenetic diversity. J. Biogeogr. 39, 1373–1382 (doi:10.1111/j.1365-2699.2012.02757.x) [Google Scholar]