Abstract
The beet cyst nematode Heterodera schachtii induces syncytia in the roots of Arabidopsis thaliana, which are its only nutrient source. One gene, At1g64110, that is strongly up-regulated in syncytia as shown by RT-PCR, quantitative RT-PCR, in situ RT-PCR and promoter::GUS lines, encodes an AAA+-type ATPase. Expression of two related genes in syncytia, At4g28000 and At5g52882, was not detected or not different from control root segments. Using amiRNA lines and T-DNA mutants, we show that At1g64110 is important for syncytium and nematode development. At1g64110 was also inducible by wounding, jasmonic acid, salicylic acid, heat and cold, as well as drought, sodium chloride, abscisic acid and mannitol, indicating involvement of this gene in abiotic stress responses. We confirmed this using two T-DNA mutants that were more sensitive to abscisic acid and sodium chloride during seed germination and root growth. These mutants also developed significantly smaller roots in response to abscisic acid and sodium chloride. An in silico analysis showed that ATPase At1g64110 (and also At4g28000 and At5g52882) belong to the ‘meiotic clade’ of AAA proteins that includes proteins such as Vps4, katanin, spastin and MSP1.
Keywords: Arabidopsis thaliana, Heterodera schachtii, syncytium, AAA+ ATPase, GUS, amiRNA, abiotic stress, DAA1
Introduction
Nematodes are a large group of animals that include free-living and parasitic species of animals, humans and plants. Plant pathogenic nematodes parasitize a large variety of plant species, especially the roots, and cause serious damage to crop plants. Among the economically important pathogens are two main groups with a sedentary lifestyle. They induce a feeding site within the plant root that consists of several giant cells in case of root-knot nematodes (genus Meloidogyne) or a syncytium in case of cyst nematodes (genera Heterodera and Globodera). These feeding sites are the sole source of nutrients throughout the whole life of these sedentary nematodes (Gheysen and Mitchum, 2011).
Cyst nematodes enter the plant roots as second-stage juveniles (J2). They select a single root cell (initial syncytial cell) within the central cylinder, and induce a syncytium that expands by incorporating up to a few hundred neighbouring cells by partial cell-wall dissolution. Adult male cyst nematodes only feed from the syncytium until their third moult, and then leave their feeding site to mate with females. Female cyst nematodes never leave their feeding site and continue to feed after fertilization. They produce several hundred eggs that remain within their enlarged bodies, which subsequently harden to form cysts, which protect the eggs until infective J2 larvae hatch again under favourable conditions (Sobczak and Golinowski, 2011).
Development of the syncytium from the initial syncytial cell requires partial cell-wall dissolutions to neighbouring cells, and is probably initiated by nematode secretions that are delivered into the plant through the nematode stylet. The development of syncytia requires coordinated expression of a variety of plant genes, including expansins, cellulases and pectinases that are important for the degradation of cell walls, leading to incorporation of new cells into the growing syncytium (Goellner et al., 2001; Wieczorek et al., 2006, 2008). Syncytial cell walls are not only partially degraded but also undergo modifications that require the synthesis of new cell-wall polysaccharides. Cell-wall ingrowths that are thought to be important for the transport of water and solutes are produced at the interface between syncytia and xylem vessels in syncytia associated with female nematodes (Jones and Northcote, 1972; Siddique et al., 2012), and the outer cell walls of the syncytium are strengthened. The cells of the syncytium undergo drastic changes in ultrastructure and activity. The large central vacuole is fragmented into many small vacuoles, and the nuclei and nucleoli are enlarged due to endo-reduplication of DNA (Niebel et al., 1996). Syncytia show high metabolic activity, evident from proliferation of the endoplasmic reticulum and accumulation of ribosomes and mitochondria in a dense granular cytoplasm (Golinowski et al., 1996; Sobczak et al., 1997). At the transcriptome level, preferential up-regulation of GO categories related to metabolic activity was found (Szakasits et al., 2009). The nematodes withdraw solutes and nutrients from the syncytium through a feeding tube formed at the tip of their stylet, thus creating a strong sink within the plant. The feeding tube also acts as a molecular sieve, such that only molecules up to 20–30 kDa are taken up (Böckenhoff and Grundler, 1994; Urwin et al., 1998).
The sugar beet cyst nematode Heterodera schachtii completes its lifecycle on Arabidopsis thaliana roots in vitro within 6 weeks (Sijmons et al., 1991). This interaction has been established as a model system due to the fact that the translucent Arabidopsis roots facilitate microscopic study of the development of this and other nematode species inside living roots (Wyss and Grundler, 1992). We have recently performed a transcriptome analysis of syncytia induced by H. schachtii in Arabidopsis roots that revealed that, of 21 138 genes represented on the Affymetrix GeneChip, 18.4% had a higher expression level and 15.8% had a lower expression level in syncytia compared with control roots (Szakasits et al., 2009).
One of the most strongly up-regulated genes (At1g64110, Table S1) was found to encode an AAA+-type ATPase. This gene was also found to be regulated by the R2R3 MYB transcription factor DUO1 and induced in sperm cells (Borg et al., 2011). It was therefore named DAA1 (DUO1-activated ATPase1). AAA+-type ATPases form a large superfamily of proteins that contain a P-loop NTPase domain. They have a diverse range of functions, for example as subunits of proteases or as molecular chaperones involved in the unfolding and disaggregation of macromolecules (Iyer et al., 2004; Ammelburg et al., 2006). Considering the rearrangements that take place in the cells that are incorporated into the syncytium, it is clear that such activities are needed for the development of syncytia. We therefore studied expression of the At1g64110/DAA1 gene in detail, and show its importance for the biotic interaction with the beet cyst nematode H. schachtii and also for abiotic stress responses.
Results
Expression of ORTHO000440 genes in Arabidopsis
The At1g64110/DAA1 gene belongs to a small sub-family of three very similar genes in A. thaliana and Arabidopsis lyrata, designated ORTHO000440 by PLAZA (http://bioinformatics.psb.ugent.be/plaza/, Proost et al., 2009) within the gene family HOM000025. The two other genes in this sub-family are At4g28000, which is expressed at very low levels in roots and syncytia (Szakasits et al., 2009), and At5g52882, which is not represented on the Affymetrix Arabidopsis GeneChip (Table S1). We performed a semiquantitative RT-PCR using RNA isolated from syncytia cut from infected roots at 5 and 15 dpi (days post-inoculation). Corresponding root segments without nematode infection were used as a control (Figure S1). Expression of At1g64110 and At5g52882 was clearly detected. Expression of At1g64110 was stronger in syncytia than in control root segments, but there was not much difference between syncytia and control root segments in terms of expression of At5g52882, confirming transcriptome data (Szakasits et al., 2009). We then used quantitative RT-PCR (Table 1) to compare expression in 15 dpi syncytia cut from infected roots with that in control root segments without root tips (as used by Szakasits et al., 2009). Expression of At1g64110 was strongly up-regulated in syncytia, with a fold change of 97.2, but expression of At5g52882 was not different in syncytia compared to control root segments (fold change of 0.95). In order to localize expression of At1g64110 in and around the syncytium, we used in situ RT-PCR on sections from 15 dpi syncytia (Figure 1). A high level of At1g64110 transcripts was clearly detected in syncytia. In uninfected root sections, expression was detected in pericycle cells but not in cells of the stele. No products were observed in control root sections without Taq polymerase.
Table 1.
ORTHO000440 gene expression in syncytia (quantitative RT-PCR)
| ΔΔCt (log2) | Fold change | |
|---|---|---|
| At1g64110 | 7.255 | 97.2 |
| At5g52882 | 0.08 | 0.95 |
Expression in 15 dpi syncytia (cut from the roots) compared to control root segments comprising the elongation zone without root tips. Values are the means of three technical and three biological replicates.
Figure 1.
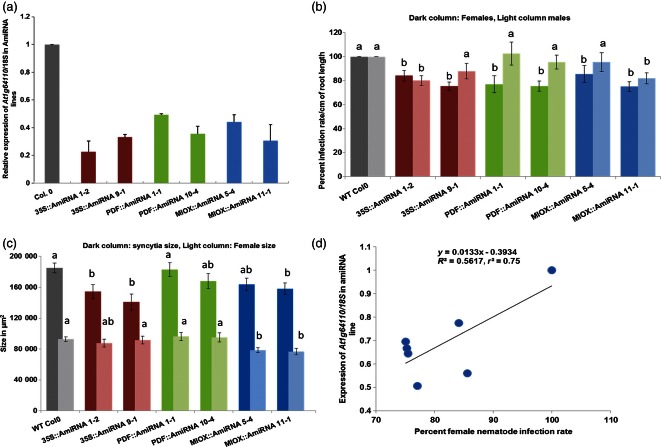
In situ RT-PCR of At1g64110 expression in 15 dpi syncytia and uninfected roots. (a) Strong signal for At1g64110 mRNA in the syncytium. In addition, staining is visible in pericycle cells. (b) Control reaction performed without Taq polymerase on a section of 15 dpi syncytium. Staining is not visible in either the feeding site or surrounding cells. (c) Control reaction on a root section above the syncytium showing signals for At1g64110 in pericycle cells. S, syncytium; Ep, epidermis; En, endodermis; P, pericycle. Scale bars = 50 μm.
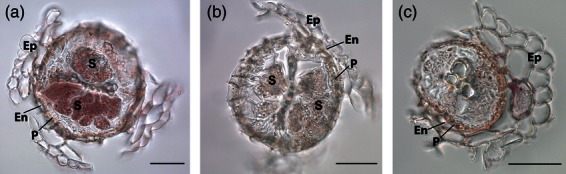
We performed RT-PCR for all three genes using RNA from various organs and growth stages (Figure 2). Expression of At1g64110 and At5g52882 was detected in most organs. At1g64110 and At5g52882 were expressed in the roots and shoots of 5- and 14-day-old seedlings. At1g64110 was expressed in stems and flowers but not in siliques, whereas At5g52882 was expressed in stems and flowers and also in siliques. Expression of At4g28000 was only detected in siliques and weakly in flowers. The results for At1g64110 and At4g28000 are in line with GeneChip data available at Genevestigator (https://www.genevestigator.com/, Zimmermann et al., 2004) (Figure S2).
Figure 2.
RT-PCR using RNA isolated from seedlings grown on MS medium (5S, 5-day-old shoots; 14S, 14-dayold shoots; 5R, 5-day-old roots; 14R, 14-day-old roots) or on soil (5WL, 5-week-old leaves; CL, cauline leaves, ST, stems; FL, flowers; SIL, siliques). Primers for the 18S gene were used for control reactions.
GUS expression analysis of At1g64110
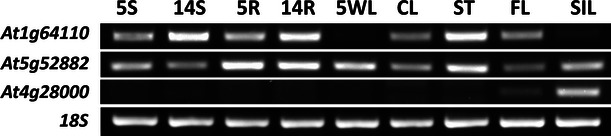
Of the three genes of this sub-family, only At1g64110 expression was strongly induced in syncytia. We therefore created promoter::GUS lines to further study its expression. A representative line was selected, and the general GUS expression pattern was assessed (Figure 3). Seedlings showed staining in the roots and the shoots. Older rosette leaves were not stained except for trichomes, with some staining at the edge of the leaves. We also noted staining at the petiole where the leaves were cut from the plant, suggesting induction of the gene by wounding. We confirmed this by cutting the leaves, which led to staining at the cut sites. Cauline leaves also showed some staining at the edges, and also strong staining in the trichomes. Stems were also stained, and we observed staining of the filaments of the anthers and the veins of the sepals in flowers. Some staining was also observed at the base of the stigma, and also in young siliques but not old siliques. Young siliques were also stained at the abscission zone, and seeds were only stained after imbibition. Lastly, GUS expression was observed in sperm cells after artificial germination of pollen grains.
Figure 3.
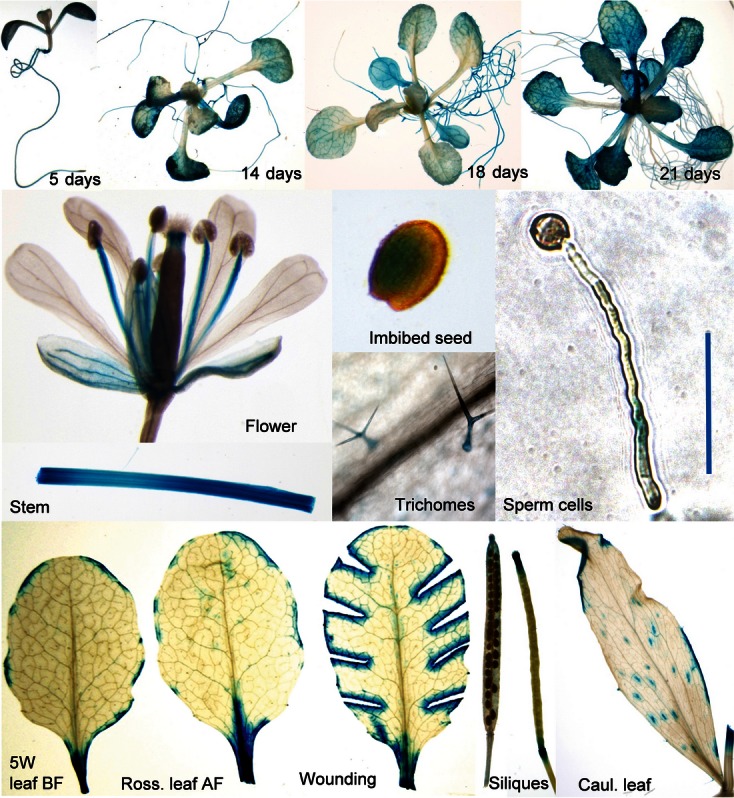
GUS expression at various growth stages (5-, 14-, 18- and 21-day-old seedlings) and in various organs of a representative At1g64110 promoter::GUS line. BF, before flowering; AF, after flowering. Scale bar = 100 μm.
The promoter::GUS line was used to determine the expression of At1g64110 during syncytium development (Figure 4). Even at 12 hpi (hours post-inoculation), faint GUS staining was visible that appeared to be associated with cells that are then incorporated into the developing syncytium. Clear GUS staining of the syncytium was detected at 24 hpi, and at all time points thereafter (48 hpi, 3 dpi, 4 dpi, 5 dpi, 9 dpi and 12 dpi), up to 15 dpi. At 20 dpi, the GUS staining was much weaker. Uninfected seedlings showed GUS staining in roots and shoots at all time points (Figure S3) that decreased in older seedlings, and there was usually no staining in roots by 22 days.
Figure 4.

GUS expression in syncytia of a representative At1g64110 promoter::GUS line at various times after inoculation. There was strong expression of GUS throughout the development of nematode-induced syncytia, starting from cell differentiation for syncytia formation (12 hpi) up to mature syncytia (15 dpi), and decreasing at 20 dpi. Asterisks indicate cells differentiating into syncytia; N, nematode; S, syncytium. Scale bar = 100 μm. The last image shows the root elongation zone and root tip from a non-infected plant.
Down-regulation of At1g64110 enhances resistance against H. schachtii
The strong up-regulation of At1g64110 in syncytia indicated an important function of this gene for syncytium development. We tested this by producing artificial micro RNA (amiRNA) lines to down-regulate the gene using three promoters: the CaMV 35S promoter, the Pdf2.1 promoter (Siddique et al., 2011), and the Miox5 promoter (Siddique et al., 2009). The 35S promoter has been reported to be only active in younger syncytia and down-regulated in old syncytia (Urwin et al., 1997; Goverse et al., 1998), whereas both Pdf2.1 and Miox5 have been shown to be strongly up-regulated in syncytia. For each promoter, we selected two independent homozygous lines with at least 50% down-regulation of the At1g64110 gene (Figure 5a). We performed infection assays for all lines, and found that the number of female nematodes was significantly reduced in each line but the number of males was not significantly different (Figure 5b). Contrasting results were found for the size of syncytia and females in these lines (Figure 5c). In the 35S promoter lines, the size of the syncytia was significantly reduced but not the size of females. In the Miox5 promoter lines, the sizes of both females and syncytia were reduced. No differences were found for the Pdf2.1 promoter miRNA lines. A strong correlation (r2 = 0.82) was found between the nematode infection rate and silencing of At1g64110 in 11 tested amiRNA lines with the various promoters (Figure 5d). A low positive correlation was found between syncytium size and female nematode size (r2 = 0.73 and r2 = 0.32, respectively) and silencing of At1g64110 in amiRNA lines (Figure S4)
Figure 5.
Down-regulation of At1g46110 expression enhances resistance against H. schachtii. Three promoters (35S, Pdf2.1 and Miox5) were used to drive expression of an artificial At1g46110 miRNA gene. (a) At1g46110 expression in miRNA lines measured by quantitative RT-PCR relative to expression of the 18S gene in syncytia cut from infected roots at 15 dpi. Values are means ± SEM for three biological and three technical replicates. (b) Infection rate per cm of root length calculated at 15 dpi for male and female nematodes. (c) Size of female syncytia and female nematodes at 14 dpi (n = 24). In (b) and (c), data were analysed for significant differences using ANOVA (P < 0.05) and LSD. Values are means ± SE. Different letters indicate differences that are significant at P < 0.05. (d) Correlation of infection rate with expression of At1g64110 in miRNA lines with various promoters.
At1g64110 and At5g52882 are important for syncytium and nematode development
We isolated homozygous T-DNA mutants for the At1g64110 gene (two mutants with insertions in exons) and also for the At5g52882 gene (one mutant with an insertion in an exon and one mutant with an insertion in the promoter), which was also found to be expressed in syncytia (Figure 6). In addition to these single mutants, we constructed a double mutant by crossing the lines SAIL_804_F09 (At1g64110) and SALK_150112 (At5g52882). PCR was used to confirm homozygosity of all mutants. We excluded the At4g28000 gene from this analysis because it is not expressed in syncytia. Except for the SALK_146218 mutation, which is located in the promoter, the other three T-DNA lines were knockout mutants, as shown by quantitative RT-PCR. No expression was detected for At1g64110 in either mutant line (SALK_025850 and SAIL_804_F09). For At5g52882, no expression was detected in SALK_150112; however, normal expression was detected in SALK_146218 (Table 2). We analysed the seedling root phenotype of these mutants. There was a significant decrease in root area and length for SALK_146218 (At5g52882) compared to wild-type seedlings. For all other mutants, including the double mutant, no differences were found (Figure S5). We tested the susceptibility of the mutants (single and double) and compared them with Col wild-type plants (Figure 7). Single mutants of gene At5g52882 had the least effect. The number of female and male nematodes was not significantly different from wild-type, and also the size of syncytia and of female nematodes was the same. However, both single mutants of gene At1g64110 were clearly more resistant than wild-type. They supported fewer females and males, and also syncytia and females were smaller than those from wild-type plants. The double mutant was not significantly different from the At1g64110 single mutants. We also analysed expression of the defence marker genes Pdf1.2 (Thomma et al., 1998) and PR1 (Uknes et al., 1992) in the mutants (Figure 8). We found no difference in expression in seedlings of any of the mutants compared to wild-type seedlings.
Figure 6.
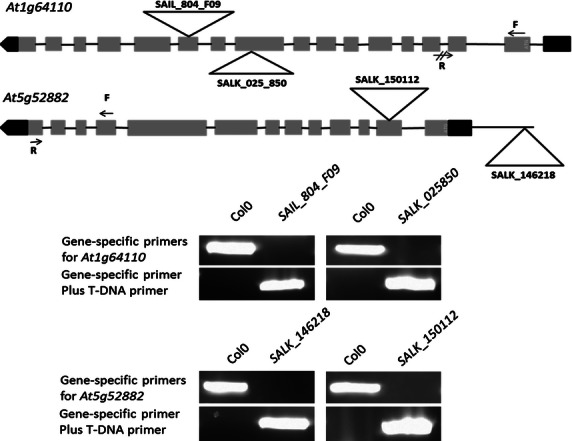
T-DNA mutants for At1g46110 and At5g52882. The intron/exon structure and the site of the T-DNA insertion are shown for both genes. PCR using gene-specific primers and the T-DNA primer confirmed homozygous mutants.
Table 2.
Expression of At1g64110 and At5g52882 in T-DNA mutants
| Locus | T-DNA mutant lines | Expression |
|---|---|---|
| At1g64110 | SAIL_804_F09 | ND |
| SALK_025850 | ND | |
| At5g52882 | SALK_150112 | ND |
| SALK_146218 | + |
ND, not detected; +, normal expression.
RNA was extracted from 7-day-old seedlings to detect expression of At1g64110 and At5g52882 in corresponding T-DNA mutant lines by quantitative RT-PCR.
Figure 7.
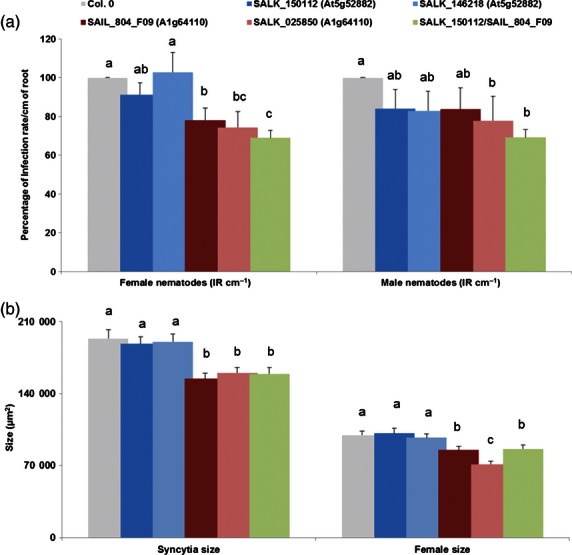
Infection assays for knockout mutants of At1g64110 (SAIL_804_F09 and SALK_025850), At5g52882 (SALK_150112 and SALK_146218) and a double mutant (SALK_150112/SAIL_804_F09) compared with Col wild-type plants. (a) Infection rate per cm root length calculated at 15 dpi, with wild-type set as 100%, for female and male nematodes. (b) Size of female syncytia and female nematodes at 14 dpi (n = 30). Different letters indicate differences that are significant at P < 0.05 (anova and LSD). Values are means ± SE.
Figure 8.
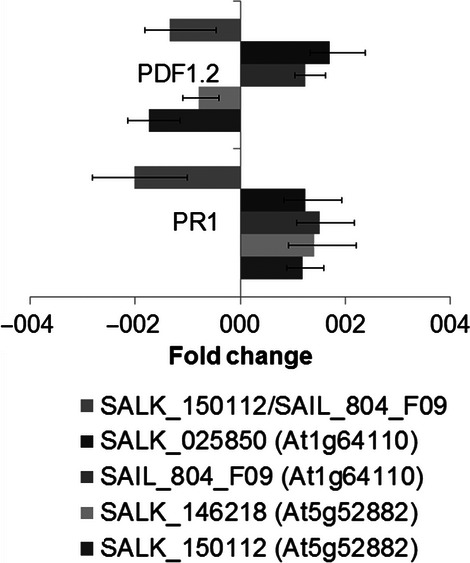
Expression of defence marker genes in At1g46110 and At5g52882 T-DNA mutants. Relative expression of Pdf1.2 and PR1 measured by quantitative RT-PCR in seedlings of knockout mutants compared with Col wild-type plants. There was no change in expression of Pdf1.2 and PR1 in any of the mutants. Values are means ± SEM for three technical and three biological replicates.
At1g64110 is involved in the response to abiotic stress
The finding that At1g64110 was induced by wounding prompted us to test its induction by jasmonic acid (JA). We also included salicylic acid (SA) as it is commonly involved in resistance responses to biotrophic pathogens. Treatment with methyl jasmonic acid (MeJA) and SA was performed by infiltrating seedlings growing on MS medium. As shown in Figure 9, At1g64110 expression was induced by JA, especially after 3 and 12 h. The strongest response to SA was found at 3 h, after which it declined, but increased again at 48 h.
Figure 9.
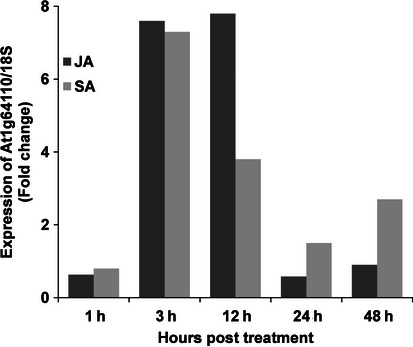
Relative expression of the At1g64110 gene in response to MeJA (100 μM) and SA (1 mm). The graph shows the results for one of two experiments with similar results.
We further performed quantitative RT-PCR to study expression of the ATPase gene At1g64110 in response to various abiotic stress conditions, including wounding, abscisic acid (ABA), cold, heat, drought, salt and osmotic stresses at the 14-day seedling stage and 5-week rosette stage. The At1g64110 gene was highly induced in response to these various abiotic stresses (Figure 10). However, the induction was more profound in response to ABA, mannitol and NaCl at the early seedling stage. To test the importance of this gene in the response to these physiological stresses, we produced synchronized seed populations harvested from plants grown at the same time. These seeds of the same age from control (Col) and two T-DNA insertion knockout mutants of At1g64110 were plated on Murashige and Skoog (MS) medium in the presence of various concentrations of ABA, NaCl or mannitol, and stratified for 3 days at 4°C. The germination percentage was scored, and seedling root lengths were measured. The results revealed that knockout mutants showed slow germination under control conditions (at day 1 but not day 4), and also had increased sensitivity to ABA and NaCl during seed germination and root growth (Figure 11a,b). The mutants also developed significantly smaller roots in response to ABA, NaCl and mannitol (Figure 11c). Together, these experiments indicate that At1g64110 knockout mutants are impaired in germination and root growth in response to ABA, NaCl and mannitol, which is indicative of stress sensitivity and the role of this gene in stress tolerance.
Figure 10.
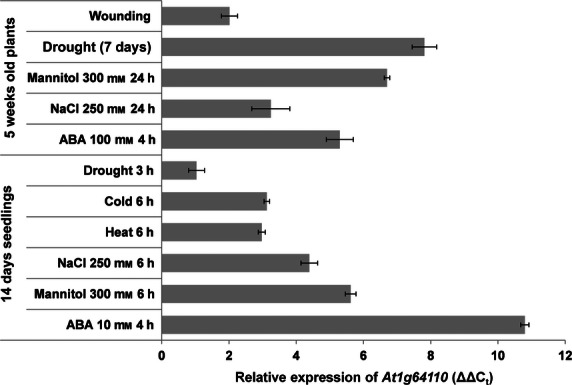
Relative expression of At1g64110 in wild-type seedlings subjected to various stresses at early (14-day-old seedlings on MS medium) and late (5-week-old rosette leaves on soil) developmental stages compared to control plants as measured by quantitative RT-PCR. The experiment was repeated twice with three technical replicates, and the data are shown as 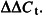 The values are means ± SE.
The values are means ± SE.
Figure 11.
Physiological responses of At1g64110 mutants (SAIL_804_F09 and SALK_025850). (a) Germination of wild-type and two mutants grown for 10 days on MS agar plates containing various ABA concentrations. (b) Effects of ABA and NaCl on germination at day 1 and day 4. (c) Effects of NaCl and mannitol on the root length of seedlings grown on MS agar plates. Data are means ± SE of three experiments with n = 20 (germination) and n = 6 (root length).
At1g64110 is not regulated by DUO1 in syncytia
The induction of At1g64110/DAA1 in sperm cells is regulated by the R2R3 MYB transcription factor DUO1 (Borg et al., 2011). To test whether DUO1, which is not included on the Affymetrix GeneChip, may also be responsible for the strong induction of At1g64110/DAA1 in syncytia, we performed a quantitative RT-PCR experiment. Syncytia were cut out from infected roots and compared to uninfected root segments. No difference was found for the expression of DUO1 in 5 and 15 dpi syncytia compared to roots, indicating that this transcription factor is probably not responsible for the strong expression of At1g64110/DAA1 in syncytia (Table 3).
Table 3.
Expression of DUO1 in 5 and 15 dpi syncytia (quantitative RT-PCR)
| Time point | ΔΔCt (log2) | Fold change |
|---|---|---|
| 5 dpi | −0.75 | −1.68 |
| 15 dpi | 0.24 | 1.18 |
Syncytia were cut from the roots, and expression of DUO1 was compared to that in control root segments. Values are the means of three technical and three biological replicates.
Sequence analysis of ORTHO000440 proteins
The AAA+ ATPase proteins studied here are very similar at the protein level as shown by a sequence alignment (Figure S6). They do not contain a signal sequence (Petersen et al., 2011) but do contain a transmembrane helix at the N-terminus (Figure S7). They contain two AAA domains, but only the second one appears to be functional as it comprises the catalytical Walker A and B sequences (Walker et al., 1982), which are missing from the first domain. Furthermore, a C-terminal extension, similar to the one found in Vps4 (Babst et al., 1998) and presumably important for oligomerization, was detected. Sequence comparison with other ATPases positioned the ORTHO000440 proteins within the central cluster of the AAA family, within the AAA+ superfamily (Ammelburg et al., 2006). Further sub-typing allowed assignment of At1g64110 to the ‘meiotic clade’ of AAA proteins (Frickey and Lupas, 2004b), containing AAA proteins such as Vps4, katanin, spastin and MSP1 (Figure 12). The most pronounced similarity detected is to the membrane-bound MSP1 (mitochondrial sorting of proteins 1) proteins of fungi and metazoa, with BLAST P values as low as 1.0e-70.
Figure 12.
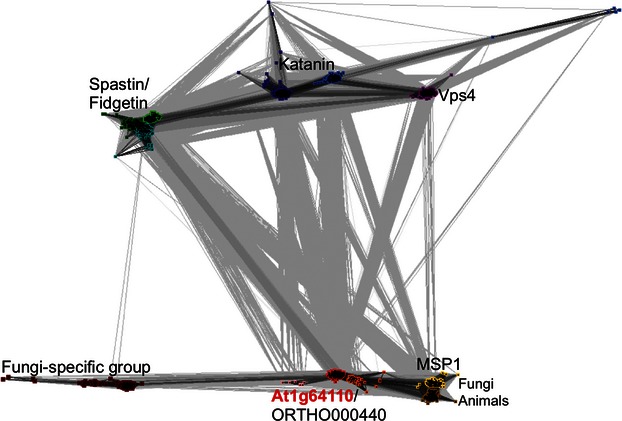
Cluster analysis of the ‘meiotic clade’ of AAA proteins. The cluster map illustrates the relationships of proteins belonging to the meiotic clade of AAA proteins (Frickey and Lupas, 2004b). The map contains 1734 sequences, each of which is represented by a dot. Pairwise similarities of the conserved AAA domain of all sequences were calculated using BLAST (Altschul et al., 1997). Sequences were clustered using CLANS (Frickey and Lupas, 2004a) using similarities with P values < 1.0e-50. Darker lines between two dots indicate lower BLAST P values. The At1g64110/ORTHO000440 cluster (red dots) shows the tightest connections to MSP1 proteins from fungi and metazoa (yellow dots). However, the Spastin (green), Katanin (blue) and Vps4 (purple) clusters also show strong similarities to the At1g64110/ORTHO000440 (red) group.
Discussion
Expression of ATPase genes in response to nematode infection
The ATPase gene At1g64110 was among the most strongly up-regulated genes found in a transcriptome analysis of syncytia induced by the beet cyst nematode H. schachtii in Arabidopsis roots (Szakasits et al., 2009). Here we confirmed the expression in syncytia using RT-PCR, in situ RT-PCR and GUS analysis. Arabidopsis contains two other genes that are related to At1g64110: At4g28000 and At5g52882. At4g28000 was expressed at a very low level in syncytia and control root segments (Szakasits et al., 2009), and was only detected in the present study in flowers and siliques using RT-PCR; thus we have excluded this gene from further experiments. At5g52882 is not represented on the Affymetrix Arabidopsis GeneChip, and therefore no information was available about its expression. We therefore tested the expression of this gene in syncytia using RT-PCR and quantitative RT-PCR. Expression was clearly detected in syncytia and roots, but was not up-regulated in syncytia, in contrast to At1g64110 expression.
At1g64110 is expressed in sperm cells
It has been shown that At1g64110 is expressed in sperm cells, and that this expression is dependent on the R2R3 MYB transcription factor DUO1 (Borg et al., 2011). We confirmed the expression of At1g64110 in sperm cells using our promoter::GUS line. Unfortunately, DUO1 (At3g60460) is not represented on the Arabidopsis GeneChip, and its expression in syncytia was therefore not known. We therefore used quantitative RT-PCR to show that expression of DUO1 was not induced in syncytia compared to uninfected control root segments. Furthermore, examination of the expression level in syncytia (Szakasits et al., 2009) of DUO1 target genes in sperm cells (Borg et al., 2011) revealed that, other than At1g64110, only one gene (DAN1) was up-regulated in syncytia compared to control root segments and DAW1 was down-regulated. All other DUO1 target genes were expressed at a very low level in roots and syncytia (Table S3). Thus, it is unlikely that DUO1 is responsible for the strong up-regulation of At1g64110 in syncytia.
ATPase genes are important for syncytium and nematode development
The strong up-regulation of At1g64110 in syncytia suggested an important function of this gene for syncytium and nematode development. We confirmed this assumption using amiRNA lines and T-DNA knockout mutants. Expression of the amiRNA gene was driven by one of three promoters, and all lines achieved at least 50% reduction of the transcript level in syncytia cut from infected Arabidopsis roots. All lines showed an effect, and especially the number of females developing on these roots was reduced. The reduction in syncytium size in some amiRNA lines and in the At1g64110 T-DNA mutants showed that At1g64110 is important for syncytium development. The smaller syncytia provide fewer nutrients for the nematodes, and thus influence the development of nematodes. The observed effects were stronger with the two T-DNA mutants that were used, probably due to the fact that the miRNA lines still had a significant level of At1g64110 transcripts while both At1g64110 T-DNA mutants were knockout mutants. The effect of the At5g52882 mutants was less pronounced. An At1g64110 and At5g52882 double mutant showed an additive effect, especially on the number of female nematodes developing on the roots. These data indicate that both proteins (and perhaps also At4g28000) have the same function, and that a high level of these proteins is needed for fully functional syncytia.
The ATPase genes discussed here may also be important for the development of clubroot disease. The pathogen Plasmodiophora brassiceae induces formation of galls in the roots of Brassicaceae, including Arabidopsis, which are a severe nutrient sink for the plant, similar to the syncytia induced by cyst nematodes. In a transcriptome analysis of clubroots of Arabidopsis roots, it was found that expression of At1g64110 was almost 54-fold up-regulated (Siemens et al., 2006). It may be interesting to determine whether down-regulation of this gene led to enhanced resistance against Plasmodiophora brassiceae. Furthermore, induction of the gene by SA indicates that it may also be involved in resistance responses to other pathogens.
The ATPase gene At1g64110 is involved in abiotic stress responses
We found that At1g64110 was inducible by MeJA and wounding, heat, cold and drought, as well as NaCl, ABA and mannitol. Involvement of the ATPase gene At1g64110 in abiotic stress has been reported previously. Analysis of drought responses of Boechera holboelli, a relative of Arabidopsis, found that the B. holboelli homologue of At1g64110 was up-regulated during drought responses (Knight et al., 2006). Drought responses in Arabidopsis are regulated by the plasma membrane histidine kinase ATHK1, and At1g64110 is one of the genes that was induced in ATHK1 over-expression lines and down-regulated in an athk1 mutant (Wohlbach et al., 2008). However, expression of ATHK1 in syncytia was rather low, and was not different from that in control root segments (Szakasits et al., 2009), which indicates that the expression of At1g64110 in syncytia may not be regulated by ATHK1. Furthermore, among the genes reported by Wohlbach et al. (2008) as up-regulated by ATHK1, only 11 were up-regulated in syncytia compared to control root segments (Szakasits et al., 2009), while three were down-regulated and the majority (24) showed the same expression in syncytia and roots (Table S4; data from Szakasits et al., 2009). Another transcription factor that was shown to regulate the expression of At1g64110 was AREB1, a leucine zipper protein encoded by At1g45249 that binds to the ABRE motif in the promoter region of ABA-inducible genes (Fujita et al., 2005). Again, most of the genes that were found to be up-regulated in transgenic lines over-expressing a constitutive active form of AREB1 (AREB1ΔQT) were not up-regulated in syncytia (Szakasits et al., 2009), indicating that AREB1 is also probably not involved in the strong up-regulation of At1g64110 in syncytia (Table S5).
A proteomic study of brassinosteroid signal transduction in Arabidopsis using pre-fractionation and two-dimensional difference gel electrophoresis showed that expression of the ATPase-encoding gene At1g64110 is induced in response to the brassinosteroid treatment (Tang et al., 2008). However, it is currently unknown whether brassinosteroids are involved in induction of At1g64110 expression in syncytia.
The induction in response to various abiotic stresses indicated that At1g64110 may be involved in the plant response to abiotic stresses. We tested this using two independent T-DNA mutants, and found that the gene plays an important role in germination and seedling root growth in response to ABA and NaCl. It may be interesting to extend these investigations to other stresses and to test whether the two other genes in this group (At4g28000 and At5g52882) have the same effect.
Function of ORTHO000440 ATPase genes
The three genes studied here belong to the sub-family designated ORTHO000440 by PLAZA (http://bioinformatics.psb.ugent.be/plaza/, Proost et al., 2009) within the gene family HOM000025, and encode typical ATPase proteins. Sequence comparison indicated that they belong to the AAA group within the AAA+ superfamily (Ammelburg et al., 2006).
Proteins of the meiotic clade of AAA proteins, such as Vps4, katanin and spastin, fulfil functions in membrane deformation and microtubule severing (Hill and Babst, 2012; Lumb et al., 2012). Furthermore, the yeast orthologue from the most similar sub-group, MSP1, has been shown to play a role in insertion of proteins into the inner mitochondrial membrane (Nakai et al., 1993). The rather close relationship of At1g64110 to these proteins implicates this plant-specific sub-group in the remodelling of membranes and shaping of their protein content. Such activities are relevant for processes such as formation of a syncytium or the response to stress, and provide a plausible explanation for the effects observed. To further explore the specific function of At1g64110 and related ATPases in Arabidopsis, it will be important to identify their subcellular location and to identify their interacting proteins, for example by using yeast two-hybrid screens or related methods.
Experimental Procedures
Plant cultivation and treatment
Arabidopsis seeds were surface-sterilized for 20 min in 6% w/v sodium hypochlorite, and subsequently washed three times with sterile water for 1 min at room temperature. Seedlings were grown in 9 cm Petri dishes on a modified Knop medium (Sijmons et al., 1991) supplemented with 2% sucrose under sterile conditions or on MS medium (Epple et al., 1995) in a growth chamber at 25°C under a 16 h light/8 h dark cycle.
Germination and growth assays of mutants were performed using age-matched seed populations of wild-type and mutants grown at the same time. For seed germination and root growth assays, seeds were sterilized and plated on 0.8% agar plates containing half-strength MS medium supplemented with various concentrations of ABA, NaCl or d-mannitol (all from Sigma-Aldrich; http://www.sigmaaldrich.com). A seed was considered as germinated when the radical protruded from the seed coat. Three plates each of 20 seeds per line were scored in germination assays. Three plates with six seeds per line were scored for root growth.
The expression of At1g64110 in response to MeJA, SA, ABA, mannitol and NaCl was studied by infiltrating the solutions under vacuum into seedlings grown for 14 days on MS medium containing 3% sucrose (Epple et al., 1995). After infiltration, the seedlings were returned to the growth chamber, and samples were taken at various time points (1, 3, 12, 24 and 48 h). The fold change was determined by quantitative RT-PCR according to the 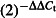 method (Livak and Schmittgen, 2001). The data were corrected by mock infiltration with tap water. Values are the means of two independent experiments (n = 2) and three technical replicates.
method (Livak and Schmittgen, 2001). The data were corrected by mock infiltration with tap water. Values are the means of two independent experiments (n = 2) and three technical replicates.
For treatment of older plants, we used 5-week-old rosette leaves from plants grown on soil. Wounding was performed using forceps, heat treatment was at 37°C, and cold treatment was at 4°C. For drought treatment, soil-grown plants were not watered for 7 days, while seedlings growing in Petri dishes on MS medium were treated by opening the plates and incubating them on a sterile bench. Untreated plants were used as a control for these treatments.
Cloning of an amiRNA construct for At1g64110
Engineering of the precursor containing the artificial microRNA (amiRNA) was performed as described by Schwab et al. (2006). The primer sequences (Table S2) used to create the precursor of the artificial miRNA (amiRNA sequence: 5′-TAAACGTTTATGAAACTCGCC-3′) were generated using the web-based tool ‘Web microRNA Designer’ at http://wmd.weigelworld.org/cgi-bin/mirnatools.pl (Schwab et al., 2006).
The amiRNA-containing precursor was generated by overlapping PCR as described by Schwab et al. (2006). The first round amplified fragments (a) to (c). These were subsequently fused by PCR (d). Oligonucleotide primers I–IV were used to replace the miRNA319a region with the artificial sequence in plasmid pUCmi319a (see Methods S1). Primers A and B were based on the template plasmid sequence. Regeneration of functional miRNA precursors was achieved by combining PCR products A-IV, II-III and I-B in a single reaction with primers A and B.
Arabidopsis transformation
Binary vectors were introduced into Agrobacterium tumefaciens GV3101 for transformation of Arabidopsis Col-0 plants by the floral-dip method as modified by Logemann et al. (2006). Transgenic plants were selected on MS medium containing 50 μg ml−1 kanamycin and 250 μg ml−1 timentin. Dry seeds were sprinkled onto the selection plates at approximately 25–35 seeds per cm2. The plates were incubated at 22°C with a photoperiod of 16 h light/8 h dark for the selection of kanamycin-resistant plants. Arabidopsis plants resistant to kanamycin were transferred to soil in a growth chamber to produce seeds.
RNA isolation
Root segments containing syncytia were excised at 15 dpi and immediately frozen in liquid nitrogen. Control root segments were collected and frozen as described previously (Szakasits et al., 2009). Total RNA was isolated using an RNeasy plant mini kit (Qiagen; http://www.qiagen.com) according to the manufacturer's instructions, including DNase I digestion. The quality and quantity of the RNA was assessed using an Agilent 2100 bioanalyzer (Agilent Technologies; http://www.home.agilent.com). Reverse transcription was performed using SuperScript III reverse transcriptase (Invitrogen; http://www.invitrogen.com) and random primers [oligo(dN6)] according to the manufacturer's instructions.
Semi-quantitative RT-PCR
RT-PCR was performed using RT-PCR Master Mix (USB; http://www.affymetrix.com/estore/browse/brand/usb/usb.jsp) according to the manufacturer's instructions. The primers used are shown in Table S2.
Quantitative RT-PCR
Real-time RT-PCR was performed using an ABI PRISM 7300 sequence detector (Applied Biosystems; http://www.invitrogen.com/site/us/en/home/brands/Applied-Biosystems.html?CID=fl-AppliedBiosystems) as described previously (Siddique et al., 2009). Each quantitative PCR sample contained 12.5 μl Platinum SYBR Green qPCR SuperMix with UDG (uracil DNA glycosylase) and ROX (5-carboxy-X-rhodamine) reference dye (Invitrogen), 2 mm MgCl2, 0.5 μl forward and reverse primer (10 μm), 2 μl cDNA and sufficient water to obtain a 25 μl total reaction volume. The primers used are shown in Table S2. All samples were diluted 1:3, and were analysed in technical triplicates.
Nematode infection assays
H. schachtii cysts were harvested from in vitro stock cultures on roots of mustard plants (Sinapis alba cv. Albatros) growing on 0.2× Knop medium (Sijmons et al., 1991) supplemented with 2% sucrose. Hatching of J2 from the cysts was stimulated by soaking in 3 mm ZnCl2. Larvae were then washed three times in sterile water, and the J2 were resuspended in 0.5% w/v gelrite (Duchefa; http://www.duchefa-biochemie.nl/). Roots of 12-day-old Arabidopsis seedlings growing on Knop medium were inoculated under sterile conditions with approximately 50-60 J2 per plant. Three independent experiments were performed, each comprising five Petri dishes with a total of approximately ten seedlings. At 14 dpi, the size of syncytia (associated with females) and of females was measured as described by Siddique et al. (2009). Briefly, syncytia associated with females were randomly selected and photographed (longitudinal optical sections) using a Zeiss Axiovert 200M inverted microscope equipped with a Zeiss Axiocam digital camera (Zeiss, http://www.zeiss.com/). The syncytia and females were outlined using the Axiovision Kontour tool, and the area was determined. The mean size of syncytia and females was calculated, and data were further statistically analysed using single-factor anova (P < 0.05) and fisher's least significant difference test. One day later (15 dpi), the number of males and females was counted. The data were expressed as nematodes per cm of root length, and were analysed using single-factor anova (P < 0.05) and Duncan's multiple range test.
GUS reporter analysis
The promoter region of At1g64110 was amplified by PCR using 50 ng Arabidopsis genomic DNA as template. Primers (Table S2) included restriction sites for NcoI and EcoRI (underlined) for subsequent cloning into binary vector pPZP3425 (Szakasits et al., 2007). This plasmid contains a double enhanced 35S promoter and a TMV (tobacco mosaic virus) omega element driving the GUS gene. During the cloning procedure, the double enhanced 35S promoter and TMV omega element were replaced by the promoter fragment. The promoter sequence was confirmed by sequencing. We produced 17 GUS lines and tested seven in detail for GUS expression in seedlings and syncytia. From these, we selected the one line that was used here. Histochemical detection of GUS activity was performed by staining using X-Gluc (Biomol; http://www.biomol.de) in 0.1 m sodium phosphate buffer pH 7.0, 0.1% Triton X-100, 0.5 mm K3[Fe(CN)6], 0.5 mM K4[Fe(CN)6] and 10 mM Na2EDTA. After staining, chlorophyll was removed from photosynthetic tissues using 70% v/v ethanol. Various plant parts were stained after flowering of the plants. For seed imbibition, seeds were imbibed at 4°C on filter papers soaked in sterile water under continuous white light for 4 days before GUS staining for 24 h. For analysis of syncytia, infected roots containing syncytia were stained at various times after inoculation by overnight incubation with X-Gluc. The stained syncytia and uninfected roots were photographed under the Axiovert 200M inverted microscope with an integrated camera (AxioCam MRc5).
In vitro pollen germination and GUS staining
For sperm cell staining, fresh anthers were used for in vitro pollen germination. The medium for in vitro pollen germination contained 18% sucrose, 0.01% boric acid, 1 mm CaCl2, 1 mm Ca(NO3)2, 1 mm MgSo4 and 0.5% agar (Difco; http://www.bd.com) at pH 7.0. Sucrose was dissolved first, and then other ingredients were added. Finally, agar was added and the medium was boiled for 3–4 min to dissolve agar (Li et al., 1999). The medium was poured into small Petri dishes (35 × 10 mm). Pollen was dusted onto the medium and allowed to germinate for 3–5 h or sometimes overnight at room temperature. After pollen germination, histochemical detection of GUS activity was performed by staining using the above-mentioned X-Gluc solution and incubation overnight at 37°C. The pollen tubes containing stained sperm cells were photographed as described above.
In situ RT-PCR
In situ RT-PCR was performed as described by Koltai and Bird (2000) and Urbanczyk-Wochniak et al. (2002). Syncytia at 15 dpi were dissected from roots and immediately placed into cold fixation solution (63% v/v ethanol, 2% v/v formalin). After 24 h, syncytia were embedded in 4% low-melting-point agarose, and 25 μm thick sections were prepared using a vibratome (VT100; Leica, http://www.leica.com/). RT-PCR was then performed using digoxigenin-labelled dUTP and primers (Table S2). After a staining reaction with nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate, cross-sections were photographed under an Axiovert 200M inverted microscope with an integrated camera (AxioCam MRc5). Full details of the method are provided in Wieczorek et al. (2006).
Mutant screening
Seeds of SAIL_804_F09, SALK_023626 (At1g64110), SALK_150112 and SALK_146218 (At5g52882) were obtained from the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info/) (Sessions et al., 2002; Alonso et al., 2003). Genomic DNA from segregating plants was screened by PCR using the primers listed in Table S2. Primer sequences were obtained using the tool supplied at the SIGnAL website (http://signal.salk.edu/tdnaprimers.2.html).
Root length and area measurement
Seeds of Arabidopsis plants (Col-0 and T-DNA mutants) were grown on Knop medium as described above. Twelve-day-old plants (corresponding to the infection time point) were photographed using a DM4000 microscope (Leica; http://www.leica-microsystems.com) and an Olympus digital camera (http://www.olympus-europa.com/). The length and diameter of main and all lateral roots was calculated using the measurement tool of las software version 4.1 (http://www.leica-microsystems.com/products/microscope-software/materials-sciences/materials-imaging/details/product/leica-las-interactive-measurement/). The total root area of a plant was calculated by adding the areas of all individual roots. Five to ten individual measurements were used to calculate the mean length and area per plant.
Bioinformatics
Sequence similarity searches indicated the presence of a conserved AAA+ domain in the C-terminal part of At1g64110. Homologues of this domain were identified by searching the National Center for Biotechnology Information non-redundant database using BLAST (Altschul et al., 1997). The 2000 most similar high-scoring segment pairs were retrieved and clustered using CLANS (Frickey and Lupas, 2004a), with BLAST as a comparison tool. Clustering was performed with default parameters at a P value cut-off of 1.0e-50. Searches using HHpred (Soding et al., 2005), a remote homology detection method based on comparison of profile hidden Markov models, revealed the presence of another but degenerate AAA+ domain in the N-terminal portion of At1g64110. The N-terminal transmembrane helix was assigned using TMHMM (Krogh et al., 2001).
Acknowledgments
We appreciate the excellent technical assistance of Sabine Daxböck-Horvath and Martina Niese. This research was supported by grants P16296-B06, P21067-B12 and P20471-B11 from the Austrian Fonds zur Förderung der Wissenschaftlichen Forschung. M.A.A. and S.M.S. were supported by the Higher Education Commission of Pakistan. M.A. is grateful to Andrei Lupas (Department 1, Protein Evolution, Max Planck Institute for Developmental Biology, Tülbingen, Germany) for continuous support.
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article.
Table S1. Gene expression of ATPase genes in syncytia (from Szakasits et al., 2009).
Table S2. Primers used in this work.
Table S3. Expression of validated DUO1 target genes (Borg et al., 2011) in syncytia induced by H. schachtii in Arabidopsis roots.
Table S4. Expression of genes identified as ‘significantly differentially regulated according to both ATHK1 transcript level and sorbitol stress condition’ (Wohlbach et al., 2008) in syncytia induced by H. schachtii in Arabidopsis roots.
Table S5. Expression of genes up-regulated in plants over-expressing AREB1ΔQT (Fujita et al., 2005) in syncytia induced by H. schachtii in Arabidopsis roots.
Figure S1. Expression of At1g64110 and At1g52882 in syncytia as shown by RT-PCR.
Figure S2. Gene expression of At1g64110 and At4g28000 according to Genevestigator.
Figure S3. GUS expression in uninfected seedlings.
Figure S4. Correlation of syncytia size and female nematode size with the expression of At1g64110 in miRNA lines with various promoters.
Figure S5. Phenotypic analysis of T-DNA mutants.
Figure S6. Sequence alignment of ORTHO000440 ATPases.
Figure S7. Domain composition of At1g64110.
Methods S1. Cloning of pUCmi319a.
References
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden T, Schaffer A, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammelburg M, Frickey T, Lupas AN. Classification of AAA+ proteins. J. Struct. Biol. 2006;156:2–11. doi: 10.1016/j.jsb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böckenhoff A, Grundler FMW. Studies on the nutrient uptake by the beet cyst nematode Heterodera schachtii by in situ microinjection of fluorescent probes into the feeding structures in Arabidopsis thaliana. Parasitology. 1994;109:249–255. [Google Scholar]
- Borg M, Brownfield L, Khatab H, Sidorova A, Lingaya M, Twell D. The R2R3 MYB transcription factor DUO1 activates a male germline-specific regulon essential for sperm cell differentiation in Arabidopsis. Plant Cell. 2011;23:534–549. doi: 10.1105/tpc.110.081059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple P, Apel K, Bohlmann H. An Arabidopsis thaliana thionin gene is inducible via a signal-transduction pathway different from that for pathogenesis-related proteins. Plant Physiol. 1995;109:813–820. doi: 10.1104/pp.109.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frickey T, Lupas AN. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004a;20:3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- Frickey T, Lupas AN. Phylogenetic analysis of AAA proteins. J. Struct. Biol. 2004b;146:2–10. doi: 10.1016/j.jsb.2003.11.020. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell. 2005;17:3470–3488. doi: 10.1105/tpc.105.035659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheysen G, Mitchum MG. How nematodes manipulate plant development pathways for infection. Curr. Opin. Plant Biol. 2011;14:415–421. doi: 10.1016/j.pbi.2011.03.012. [DOI] [PubMed] [Google Scholar]
- Goellner M, Wang XH, Davis EL. Endo-β-1,4-glucanase expression in compatible plant–nematode interactions. Plant Cell. 2001;13:2241–2255. doi: 10.1105/tpc.010219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golinowski W, Grundler FMW, Sobczak M. Changes in the structure of Arabidopsis thaliana during female development of the plant-parasitic nematode Heterodera schachtii. Protoplasma. 1996;194:103–116. [Google Scholar]
- Goverse A, Biesheuvel J, Wijers GJ, Gommers FJ, Bakker J, Schots A, Helder J. In planta monitoring of the activity of two constitutive promoters, CaMV 35S and TR2′, in developing feeding cells induced by Globodera rostochiensis using green fluorescent protein in combination with confocal laser scanning microscopy. Physiol. Mol. Plant Pathol. 1998;52:275–284. [Google Scholar]
- Hill CP, Babst M. Structure and function of the membrane deformation AAA ATPase Vps4. Biochim. Biophys. Acta. 2012;1823:172–181. doi: 10.1016/j.bbamcr.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA plus ATPases. J. Struct. Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Jones MG, Northcote DH. Nematode-induced syncytium – a multinucleate transfer cell. J. Cell Sci. 1972;10:789–809. doi: 10.1242/jcs.10.3.789. [DOI] [PubMed] [Google Scholar]
- Knight CA, Vogel H, Kroymann J, Shumate A, Witsenboer H, Mitchell-Olds T. Expression profiling and local adaptation of Boechera holboellii populations for water use efficiency across a naturally occurring water stress gradient. Mol. Ecol. 2006;15:1229–1237. doi: 10.1111/j.1365-294X.2006.02818.x. [DOI] [PubMed] [Google Scholar]
- Koltai H, Bird DM. High throughput cellular localization of specific plant mRNAs by liquid-phase in situ reverse transcription-polymerase chain reaction of tissue sections. Plant Physiol. 2000;123:1203–1212. doi: 10.1104/pp.123.4.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Li H, Lin Y, Heath RM, Zhu MX, Yang Z. Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell. 1999;11:1731–1742. doi: 10.1105/tpc.11.9.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the
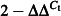 method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar] - Logemann E, Birkenbihl RP, Ulker B, Somssich IE. An improved method for preparing Agrobacterium cells that simplifies the Arabidopsis transformation protocol. Plant Methods. 2006;2:16. doi: 10.1186/1746-4811-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumb JH, Connell JW, Allison R, Reid E. The AAA ATPase spastin links microtubule severing to membrane modelling. Biochim. Biophys. Acta. 2012;1823:192–197. doi: 10.1016/j.bbamcr.2011.08.010. [DOI] [PubMed] [Google Scholar]
- Nakai M, Endo T, Hase T, Matsubara H. Intramitochondrial protein sorting. Isolation and characterization of the yeast MSP1 gene which belongs to a novel family of putative ATPases. J. Biol. Chem. 1993;268:24262–24269. [PubMed] [Google Scholar]
- Niebel A, Engler JD, Hemerly A, Ferreira P, Inze D, VanMontagu M, Gheysen G. Induction of cdc2a and cyc1At expression in Arabidopsis thaliana during early phases of nematode-induced feeding cell formation. Plant J. 1996;10:1037–1043. doi: 10.1046/j.1365-313x.1996.10061037.x. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Proost S, Van Bel M, Sterck L, Billiau K, Van Parys T, Van de Peer Y, Vandepoele K. PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell. 2009;21:3718–3731. doi: 10.1105/tpc.109.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, et al. A high-throughput Arabidopsis reverse genetics system. Plant Cell. 2002;14:2985–2994. doi: 10.1105/tpc.004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique S, Endres S, Atkins JM, et al. Myo-inositol oxygenase genes are involved in the development of syncytia induced by Heterodera schachtii in Arabidopsis roots. New Phytol. 2009;184:457–472. doi: 10.1111/j.1469-8137.2009.02981.x. [DOI] [PubMed] [Google Scholar]
- Siddique S, Wieczorek K, Szakasits D, Kreil DP, Bohlmann H. The promoter of a plant defensin gene directs specific expression in nematode-induced syncytia in Arabidopsis roots. Plant Physiol. Biochem. 2011;49:1100–1107. doi: 10.1016/j.plaphy.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique S, Sobczak M, Tenhaken R, Grundler FM, Bohlmann H. Cell wall ingrowths in nematode induced syncytia require UGD2 and UGD3. PLoS ONE. 2012;7:e41515. doi: 10.1371/journal.pone.0041515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemens J, Keller I, Sarx J, Kunz S, Schuller A, Nagel W, Schmulling T, Parniske M, Ludwig-Muller J. Transcriptome analysis of Arabidopsis clubroots indicate a key role for cytokinins in disease development. Mol. Plant–Microbe Interact. 2006;19:480–494. doi: 10.1094/MPMI-19-0480. [DOI] [PubMed] [Google Scholar]
- Sijmons PC, Grundler FMW, Vonmende N, Burrows PR, Wyss U. Arabidopsis thaliana as a new model host for plant-parasitic nematodes. Plant J. 1991;1:245–254. [Google Scholar]
- Sobczak M, Golinowski W. Cyst nematodes and syncytia. In: Jones J, Gheysen G, Fenoll C, editors. Genomics and Molecular Genetics of Plant–Nematode Interactions. New York: Springer; 2011. pp. 61–82. [Google Scholar]
- Sobczak M, Golinowski W, Grundler FMW. Changes in the structure of Arabidopsis thaliana roots induced during development of males of the plant parasitic nematode Heterodera schachtii. Eur. J. Plant Pathol. 1997;103:113–124. [Google Scholar]
- Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakasits D, Siddique S, Bohlmann H. An improved pPZP vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. Rep. 2007;25:115–120. [Google Scholar]
- Szakasits D, Heinen P, Wieczorek K, Hofmann J, Wagner F, Kreil DP, Sykacek P, Grundler FM, Bohlmann H. The transcriptome of syncytia induced by the cyst nematode Heterodera schachtii in Arabidopsis roots. Plant J. 2009;57:771–784. doi: 10.1111/j.1365-313X.2008.03727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WQ, Deng ZP, Oses-Prieto JA, Suzuki N, Zhu SW, Zhang X, Burlingame AL, Wang ZY. Proteomics studies of brassinosteroid signal transduction using prefractionation and two-dimensional DIGE. Mol. Cell. Proteomics. 2008;7:728–738. doi: 10.1074/mcp.M700358-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BP, Eggermont K, Penninckx IA, Mauch-Mani B, Vogelsang R, Cammue BP, Broekaert WF. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl Acad. Sci. USA. 1998;95:15107–15111. doi: 10.1073/pnas.95.25.15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J. Acquired resistence in Arabidopsis. Plant Cell. 1992;4:645–656. doi: 10.1105/tpc.4.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanczyk-Wochniak E, Filipecki M, Przybecki Z. A useful protocol for in situ RT-PCR on plant tissues. Cell. Mol. Biol. Lett. 2002;7:7–18. [PubMed] [Google Scholar]
- Urwin PE, Moller SG, Lilley CJ, McPherson MJ, Atkinson HJ. Continual green-fluorescent protein monitoring of cauliflower mosaic virus 35S promoter activity in nematode-induced feeding cells in Arabidopsis thaliana. Mol. Plant–Microbe Interact. 1997;10:394–400. doi: 10.1094/MPMI.1997.10.3.394. [DOI] [PubMed] [Google Scholar]
- Urwin PE, McPherson MJ, Atkinson HJ. Enhanced transgenic plant resistance to nematodes by dual proteinase inhibitor constructs. Planta. 1998;204:472–479. doi: 10.1007/s004250050281. [DOI] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek K, Golecki B, Gerdes L, et al. Expansins are involved in the formation of nematode-induced syncytia in roots of Arabidopsis thaliana. Plant J. 2006;48:98–112. doi: 10.1111/j.1365-313X.2006.02856.x. [DOI] [PubMed] [Google Scholar]
- Wieczorek K, Hofmann J, Blochl A, Szakasits D, Bohlmann H, Grundler FMW. Arabidopsis endo-1,4-β-glucanases are involved in the formation of root syncytia induced by Heterodera schachtii. Plant J. 2008;53:336–351. doi: 10.1111/j.1365-313X.2007.03340.x. [DOI] [PubMed] [Google Scholar]
- Wohlbach DJ, Quirino BF, Sussman MR. Analysis of the Arabidopsis histidine kinase ATHK1 reveals a connection between vegetative osmotic stress sensing and seed maturation. Plant Cell. 2008;20:1101–1117. doi: 10.1105/tpc.107.055871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss U, Grundler FMW. Heterodera schachtii and Arabidopsis thaliana, a model host–parasite interaction. Nematologica. 1992;38:488–493. [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.