Abstract
The way in which cells adopt different morphologies is not fully understood. Cell shape could be a continuous variable or restricted to a set of discrete forms. We developed quantitative methods to describe cell shape and show that Drosophila hemocytes in culture are a heterogeneous mixture of five discrete morphologies. In an RNAi screen of genes affecting the morphological complexity of heterogeneous populations, we found that most genes regulate the transition between discrete shapes rather than generating new morphologies. In particular, we identified a subset of genes, including the tumour suppressor PTEN, that decrease the heterogeneity of the population leading to populations enriched in rounded or elongated forms. We show that these genes have a highly conserved function as regulators of cell shape in both mouse and human metastatic melanoma cells.
Introduction
Morphological plasticity is critical to organism development – as exemplified by the reversible conversion of embryonic non-migratory epithelial cells to motile mesenchymal cells required for tissue positioning and organisation1. The size of the shape space a cell has the potential to explore reflects its morphological plasticity2. Highly plastic cells explore large regions of shape space compared to cells with stable morphologies. In adult organisms, the shape space available to most differentiated cells is relatively limited, serving to enforce tissue architecture and function. However, during the pathogenesis of diseases such as metastatic cancers, cells can re-acquire the ability to explore shape space and thus “find” a shape that is suitable for migration and invasion2-6. Currently there is little understanding of how the size and topology of cellular shape space is determined by genetic and environmental factors.
To identify how genes contribute to the size and topology of shape space we developed high throughput imaging and computational methods to describe the morphological complexity of cellular populations and applied them to datasets generated by systematic RNA interference (RNAi) screens in Drosophila Kc cells. We first determined whether cells have discrete shapes or whether shape is a continuous variable. Subsequently we identified genes that contribute to the exploration of shape space in Kc cells, as well as those that regulate the topology of shape space itself. Finally we isolated a conserved gene network that regulates contractility and protrusion in Drosophila as well as mouse and human melanoma cells. This demonstrates that the analysis of morphological complexity provides new insights into the signalling networks regulating cell shape.
Results
RNAi screening indicates that Kc cells exist in discrete shapes
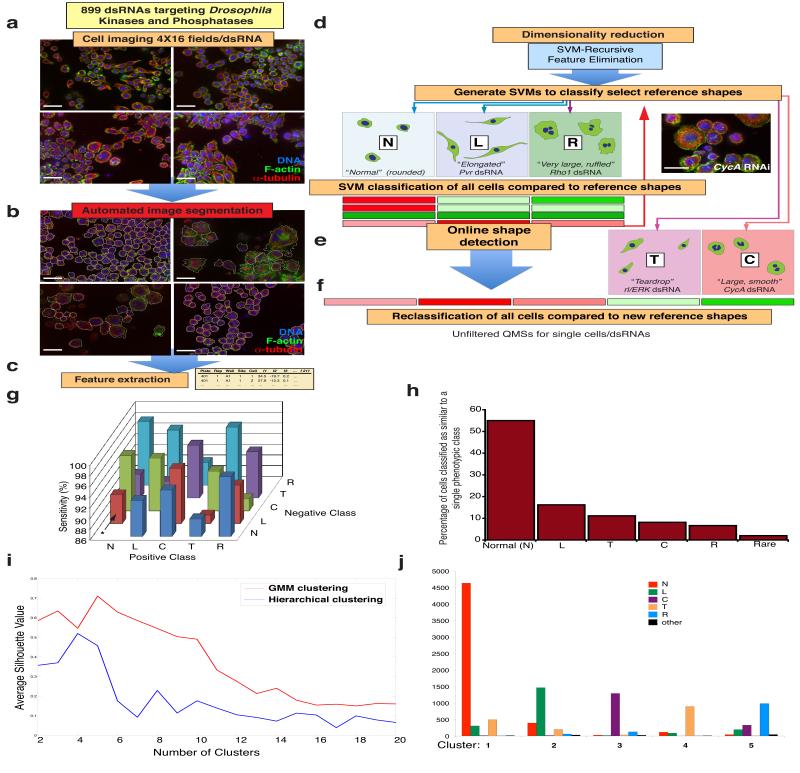
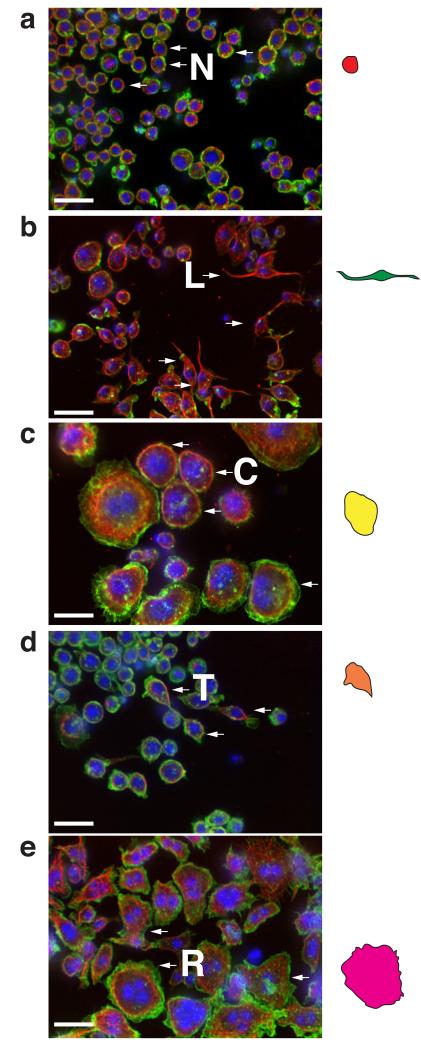
We used RNAi screening in Drosophila Kc167 cells (Kc cells) to explore the contribution of genes to morphological complexity (Fig. 1a-f). We use the term “experimental condition” (EC) for cells treated with dsRNA. Following image processing (see Methods, Supplementary Note, Supplementary Figure 1, and Supplementary Tables S1-S4), we scored cells in each EC on the basis of their similarity to reference shapes7, 8. Briefly, we used human observers (Fig. 1d) and online discovery algorithms (Fig. 1e)8 to identify as many distinct cellular shapes (reference shapes) as possible in the dataset. The majority of cells in the dataset could be characterised as normal (“N”) cells, which are rounded cells with smooth borders of cortical actin (Fig. 2a) together with an additional 4 reference shapes. We labelled these reference shapes as “L”, “C”, “T”, or “R”, which correspond to: elongated, bipolar, spindle shaped cells (L) (Fig. 2b); large cells with smooth edges (C) (Fig. 2c); small, partially polarised “teardrop” shaped cells (T) (Fig.2d); and very large flat cells with ruffled edges (R) (Fig. 2e). Five different support vector machine (SVM)-based classifiers were generated that could distinguish these morphological classes. We also derived specific and sensitive Gaussian SVM classifiers that distinguish between pairs of shapes versus simply one shape from all others (Fig. 1g). Every cell in the dataset was then scored using each classifier to generate a multi-dimensional vector, or a Quantitative Morphological Signature (QMS) that describes the similarity of that cell to each reference shape (Fig. 1f). Thus, unlike the use of absolute measures (e.g., area, size), a QMS is a measure that describes shape relative to other reference shapes. Each cell in the dataset is assigned a QMS, and a mean QMS can be calculated for any given population.
Figure 1. Automated morphological profiling.
(a) Kc167 cells were incubated with 899 single dsRNAs targeting the majority of Drosophila kinases and phosphatases. Experiments were performed in triplicate or quadruplicate in 384-well plates. Following fixation and staining using DAPI, phalloidin, and an anti-α-tubulin antibody, each well was imaged at 16 sites by confocal microscopy. Scale bars, 20 μm. (b) Automated image segmentation and feature extraction were performed to generate feature information for 2,038,641 cell segments. Scale bars, 20 μm. (c) SVM-recursive feature elimination was used to reduce the dimensionality of the data, and SVM-based classifiers were generated for three initial reference shapes (N”, “L”, and “R”). (d) Individual cell segments were initially classified by assigning raw Quantitative Morphological Signatures (QMSs) based on the similarity of the segments to N, L, and R shapes. Scale bar, 20 μm. (e) Subsequently, online phenotype-detection methods8 were implemented to detect the presence of two additional shapes, “T” and “C”. (f) All cells were then re-assigned QMSs based on the similarity of each cell to all five reference shapes, but the comparison to “N” cells is done by calculating a penetrance Z-score of mutant shapes prior to filtering (see Fig. 2). (g) Specificity as determined by cross-validation analysis to determine whether two exemplar shapes are quantitatively different from one another using a new SVM classifier. Each test was performed on 200 test cells from training classes comprised as follows; “N” 2,185 cells; “L” 2,053 cells, “C” 2,041 cells; “T” 2,002 cells; “R” 2,028 cells. For example, if N is considered the positive class, and L is negative class, the N vs L classifier correctly identifies N cells in 91% of tests (asterisk). (h) Cells are classified as most similar to a particular single shape. Cells not assigned to one particular shape are assigned to the rare class. (i) Silhouette index for different cluster numbers using hierarchical clustering (red) or Gaussian Mixture Models (blue) of PC data. (J) Number of cells with a particular morphology (N, L, C, T, R, or other) that are part of a particular cluster (1-6) using hierarchical clustering of principal components.
Figure 2. N, L, C, T, R cells.
(a) Wild-type cells, (b) Pvr-, (c) CycA-, (d) rl/ERK-, or (e) Rho1-deficient cells were fixed, labelled with DAPI, phalloidin, and anti-α-tubulin antibody, and imaged. Arrows denote cells with representative shapes. Coloured cells on right are traces of representative shapes. Scale bars, 20 μm.
To gain a sense of whether our classification systems capture the majority of the morphological variance present in the dataset we asked if most cells in the dataset could be considered as similar to one of the reference shapes. When all cells in the dataset are classified with respect to their similarity to a single phenotypic group, versus determining their similarity to multiple classes simultaneously, we observed that the vast majority of cells could be grouped into the N, L, C, T, or R classes, and that only 2.15% could be classified as other/rare shapes (Fig. 1h). We confirmed this finding using alternative unsupervised classification methods such as Principal Component Analysis (PCA) followed by hierarchical clustering, or Gaussian Mixture Modelling (GMM) to segregate the data into distinct morphological clusters (Fig. 1i). Each of the 5 major morphological classes is populated predominantly by one of N, L, C, T, or R-type cells (Fig. 1j). Thus, perhaps surprisingly, the number of different shapes present in the entire dataset is low, and is well described by 5 different shapes.
As a first step towards understanding the role of different genes in the control of cell shape, we classified the effects of RNAi on the basis of the population mean of single-cell QMS scores, following the filtering out of normal cells and consolidation of replicable phenotypes. Here, the QMS is a 5-dimensional vector that describes the mean similarity of cells to “L”, “C”, “T”, “R” shapes, and a “PZ” score, which the penetrance of all non-normal shapes in the population prior to filtering (Supplementary Table S5). Gene QMSs were organised using average linkage hierarchical clustering to describe phenoclusters (Fig. 3). However, while this analysis reveals how different genes broadly affect the morphology of different populations, it does not account for population heterogeneity.
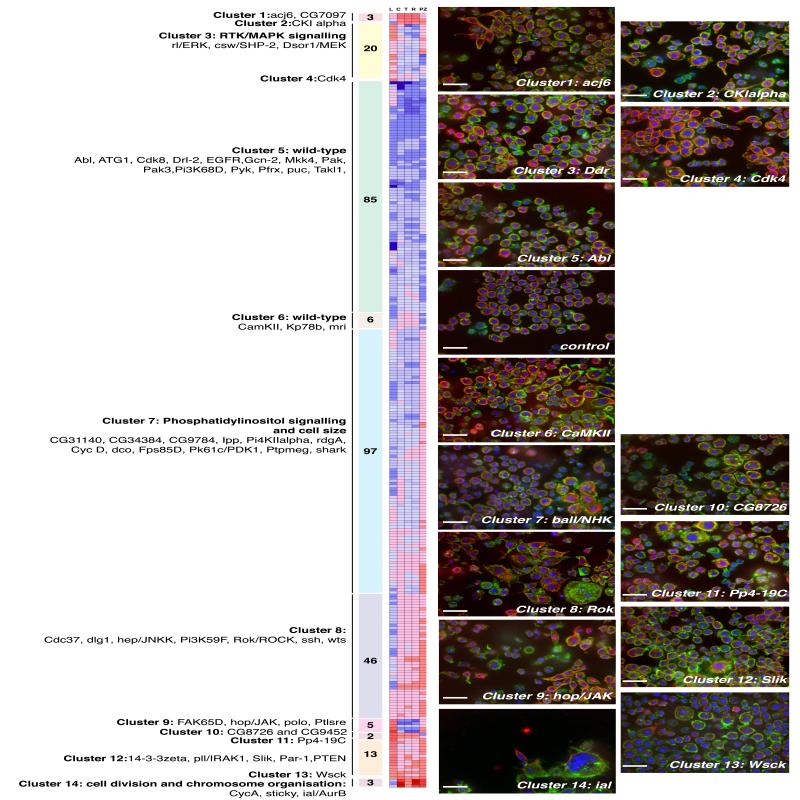
Figure 3. Hierarchical clustering of QMSs.
Average linkage clustering of 284 5-feature QMSs comprising “L”, “C”, “T”, and “R” SVM Z-scores as well as “PZ” (Penetrance Z-scores). Included are treatment conditions following the RNAi-mediated depletion of 282 genes, RNAi-mediated targeting of lacZ, and a signature for control wells. Genes are in the same phenocluster when clustered together at a cut-off distance cut-off (CDC) (an average of uncentered Pearson correlation coefficients) greater than 0.90. At this threshold, we identified 10 different phenoclusters and 4 QMSs (CkIalpha, Cdk4, Pp4-19C, Wsck) that did not cluster with any other gene. The number of genes that comprise each phenocluster are shown in shaded boxes. Some genes that are members of each phenocluster are listed. The largest phenocluster, cluster 5, is composed of 85 ECs that have QMSs that are not significantly different from wild-type cells, even when RNAi penetrance is taken into account (Fig. S1 and Supplementary Methods). The mean QMS of 6 ECs in cluster 6 is also essentially wild-type. Cluster 3 is significantly enriched19 in canonical sevenless Receptor Tyrosine Kinase (RTK) components (p = 6.21e−4), and cluster 7 is significantly enriched for genes involved in phosphatidylinositol signalling (p = 1.19e−4) and cell size (p = 1.07e−3). A complete list of genes in each phenocluster is included in the Supplementary Table 5. Representative image fields from particular phenoclusters are shown, and in some cases a zoomed-in image from a field is displayed on the right or embedded within the image. Nuclei are labelled with Hoechst (blue), polymerized actin is labelled with phalloidin (green), and microtubules are labelled with anti-tubulin antibody (red). Scale bars, 20 μm.
Population of wild-type Kc cells are comprised of 5 shapes
We next sought to leverage single-cell data to determine how genes contribute to the regulation of morphological complexity. We prefer the term complexity to heterogeneity as it better describes the number of shapes that could be considered distinct, versus the total number of shapes in a population – which may represent variations on the same shape. For example, if cells in a population are mostly a single highly variable shape, the heterogeneity of the population is high but the complexity is low. After accounting for differential penetrance of different dsRNAs and identifying dsRNAs with reproducible phenotypes (Supplementary Figure S2), we calculated matrices describing the similarity of the space sampled by an EC to that sampled by all other ECs (Q(1) score, Fig. 4a), as well as to the space sampled by ECs in the same phenocluster (Q(2) score) (Fig. 4a). From these scores, we generated a Q(4) by subtracting the Q(1) score from the Q(2) score (Fig. 4a). A Q(4) score describes the complexity of a population. Populations that sample the same morphological space, and thus have the same complexity as other populations, have low Q(4) scores, while homogeneous populations with low complexity have high Q(4) scores (Fig. 4b, and Supplementary Table S5). By plotting the Q(1) of each EC against all others, we observed that ECs from clusters 5 and 6, or wild-type cells, are very similar to themselves, and to almost all other ECs in the dataset (Fig. 4c). Moreover, control (mock-treated) and lacZ RNAi ECs have the 14th and 18th highest Q(1) scores in the dataset, respectively, and ECs from clusters 5 and 6 are 40/50 highest ranking ECs in terms of Q(1) scores. Only 3.9% of ECs have a Q(1) score significantly different than wild-type (p < 0.05). These data show that wild-type Kc cells have limited morphological complexity that is nearly equivalent to that of the entire dataset of RNAi treatments, and are comprised of different shapes that are well-represented by the 5 reference shapes. Given that the entire dataset is well described by 5 shapes (Fig. 1j), this suggests that gene knockdown most often enriches for shapes that are already present at low levels in populations of wild-type cells.
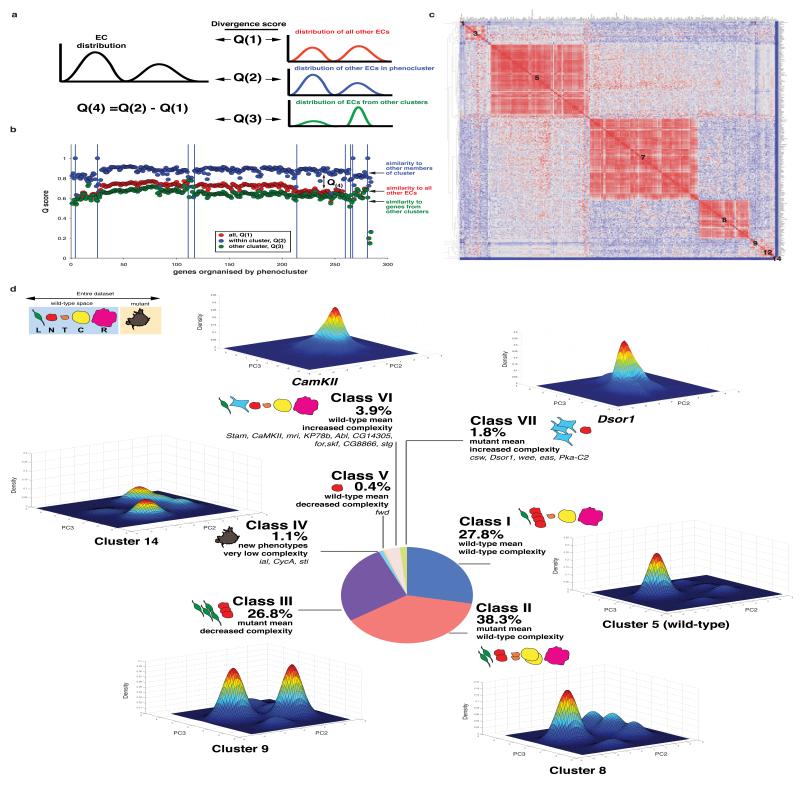
Figure 4. Morphological complexity is a phenotype that can be altered by RNAi.
(a) Diagram explaining the generation of Q(4) scores. For any given EC, the distribution of cells in morphological space is compared to the distribution of all other ECs, all other ECs in a phenocluster (Figure 3), and distribution of ECs from all other clusters to generate Q(1), Q(2), and Q(3) scores respectively. A Q(4) score is the difference between a Q(2) and Q(1) score, while the Q(3) score describes the uniqueness of the population. (b) The average similarity scores (y-axis) between a single EC and all ECs in the dataset (red), all genes in the same phenocluster (blue), or all ECs in different phenoclusters (green) are shown. Genes are organized by phenocluster (left to right) as in Figure 3. (c) Q(1) similarity scores comparing all ECs to each other. Similarity scores are normalized to range between 0-1. Highly similar populations are coloured in red, and dissimilar populations are coloured in blue. Genes are arranged according to their phenocluster membership as described in Figure 3 (denoted by the numbers on the graph). (d) We compared the Q(4) and mean QMSs of different ECs to the scores in wild-type/control EC (e.g. lacZ RNAi) to describe how genetic inhibition can affect the exploration of cells in the pre-defined shape space. A population can belong to one of seven different categories depending on its mean QMS score and Q(4) scores. For 6 different classes we estimated the Gaussian kernel density for PC2 and PC3 of populations sampled from different clusters or ECs that are representative of different classes. Each plot represents the probabilities for the occurrence of different shapes and thus describes the morphological space explored by the population. For each graph the cell numbers are as follows: Cluster 5, 247,341; Cluster 8, 72,196; Cluster 9, 7,358; Cluster 14, 4,144; CamKII, 3,008; Dsor1, 3,603.
RNAi most often decreases the number of shapes present in the wild-type population
To describe phenotypes on the basis of the morphological complexity of cellular populations, we classified genes by their Q(4) and mean QMS (Fig. 3) to generate seven different classes (Fig. 4d): Class (i) is comprised of ECs with a wild-type mean and wild-type complexity (unaffected cells, 27.8% of all ECs). Notably, by plotting density estimations of shape frequency in two principal components, we observe that the five different sub-populations in wild-type cells appear to exist as discrete sub-populations (Fig. 4d). Class (ii) consists of ECs that have an abnormal mean but wild-type complexity (38.3%). In this class RNAi has altered the distribution of cells within sub-populations, but each sub-population remains represented in the population. For example, in Par1-, Rok-, Slik-, SAK-, or trc-deficient populations there is an enrichment of elongated shapes, resulting in a mean score that is different from wild-type cells, but the complexity of this population is the same as wild-type and the population is also enriched in other shapes. In class (iii) are ECs with decreased morphological complexity where one or more sub-populations has been enriched at significant expense of others (Q(4) Z-score > 1.0), (26.8%). Examples of these include 14-3-3zeta-, Pp2B-14D-, Pp2A-29B-, Dgk-, hop/JAK- or PTEN-deficient populations where there is an enrichment of “L” elongated cells but a marked decrease in other shapes. Class (iv) is a small fraction (1.1%) of ECs with an abnormal mean, significantly decreased heterogeneity and morphologies that are different from those sampled by wild-type populations. Here new shapes have been generated, but the overall complexity (number of total shapes) is less than wild-type. For example, ial/AurB- or sticky/CitronK-depleted populations are very homogenous and explore a small region of shape space not sampled by wild-type cells. Class (v) is a single EC, fwd RNAi, which has a wild-type mean and decreased complexity. Class (vi) (3.9% of ECs) have a wild-type mean, but are significantly more complex than the wild-type cells. Class (vii) (1.4% of ECs) have an abnormal mean and significantly increased complexity compared with wild-type cells. Classes (vi) and (vii) include genes such as Stam, CamKII, and Abl, that are of particular interest as morphological complexity is increased but these populations are sampling space within that explored by wild-type cells.
Thus, RNAi does not typically lead to the generation of new shapes, but rather alters the distribution of pre-existing sub-populations that exist in wild-type cells. We propose that cellular morphogenesis of Kc cells is a “canalised processes”9, where cells can only transition between a limited number of stable shapes, and changes in the distribution occur following RNAi because inhibition of different signalling events prevents the ability of cells to transition from one shape to another, effectively trapping them in one or more stable shapes found in wild-type cells.
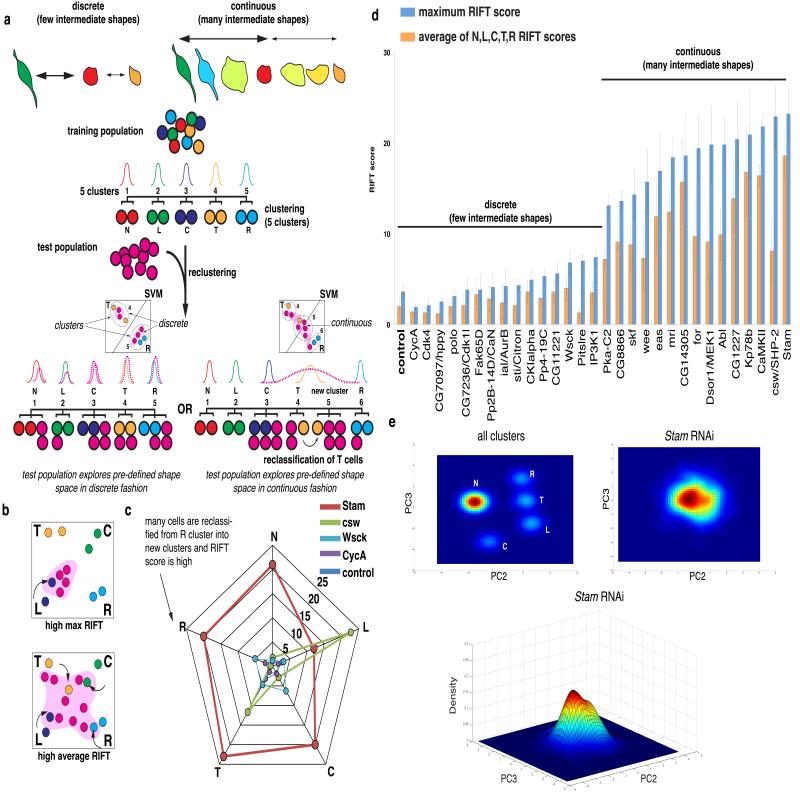
Kc cells make switch-like transitions between discrete shapes
We reasoned that cells could transition between stable shapes in one of two ways. Cells could transition between discrete shapes in a “switch-like” manner, where intermediate forms are highly transient and therefore rarely observed. Alternatively, cells could make continuous transitions where there are a diverse number of morphologies that appear as stable intermediates between shapes. To discriminate between these two possibilities, we calculated a RIFT score (“Rate of Intermediate Forms or Transitions”) for different ECs. The RIFT score quantifies the extent of misclassification by clustering that occurs when populations of cells comprised of 5 shapes from a training pool are mixed in silico with an equal number of cells from an EC (Fig. 5a). A high RIFT score indicates the presence of intermediate shapes in the EC, whereas a low RIFT score suggest that there are very few intermediate shapes in the EC. Deficiency of some genes results in high RIFT scores for all shape classes, and thus accumulation of intermediate forms between all shapes (Fig. 5b,c). In other cases the RIFT score is high only for a particular class, meaning there is an accumulation of forms near a particular shape (Fig. 5b, c). We calculated the maximum RIFT score of populations (Fig. 5d, blue bars) and the average RIFT score (Fig.5, orange bars), although there is typically high correlation between these values (Fig. 5d). For example, we determined the RIFT scores for ten populations with low complexity (high Q(4) score) and ten, including wild-type cells, with high complexity (low Q(4) score), and find that there are few intermediate shapes in wild-type cells. Moreover, RNAi rarely results in an increase in RIFT scores. Thus the morphogenesis of wild-type Kc cells is both discrete and switch-like in nature. However, RNAi-mediated knockdown of Stam (Fig. 5e) and CamKII leads to high average RIFT scores (Fig. 5d), and these ECs have many shapes that can be considered continuous. This suggests the function of these genes is essential for switch-like morphogenesis of Kc cells.
Figure 5. Kc cells exist as discrete sub-populations.
(a) Methodology for generating the Rate of Intermediates or Transitions (RIFT) score. A training class of 500 cells from 5 reference classes (N, L, C, T, R) is clustered, resulting in five different clusters. Test populations are then added to the set, and the entire population is reclustered. If the reclustering results in 5 clusters, the test population is comprised of largely discrete sub-populations and a low RIFT score. However, if after reclustering cells from the training pool are misclassified into new clusters, this indicates the test population has shapes that can be considered intermediate between the reference class resulting in a high RIFT score. (b) ECs can have a high maximum RIFT score (e.g. where a high-percentage of L cells are assigned into new clusters) and also a high average RIFT score. (c) Radar-gram of RIFT scores for Stam, csw/SHP-2, Wsck, CycA, and control ECs. For ECs such as Stam, many cells do not fall into N, L, C, T, R phenoclusters, whereas in csw/SHP-2 populations, many cells of L or T classes are specifically reclassified. (d) The maximum and average RIFT score were calculated for ECs with the 10 highest and 10 lowest Q(4) scores, as well as for 10 normal populations. Error bars represent Standard Deviation (S.D.) following 10 recalculations (using new populations) of the RIFT score. (e) Top panels are a top-down view of the density estimates of randomly sampled cells from all clusters (584,452 cells), or of Stam-deficient cells (1,392 cells). Bottom panel is same density estimate of Stam-deficient cells. RIFT scores for all ECs are listed in Supplementary Table S5.
Melanoma cells exhibit discrete, switch-like morphogenesis
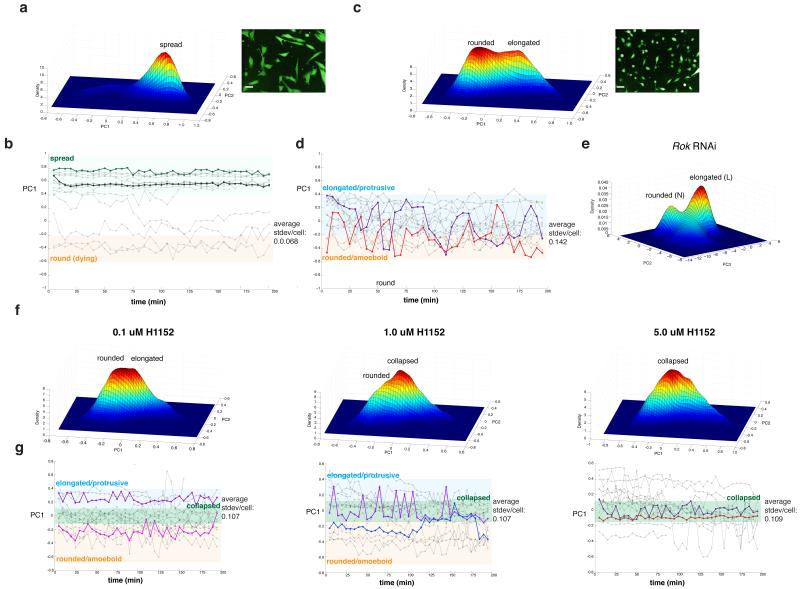
We next sought to determine if our model of discrete, switch-like morphogenesis can be applied to mammalian cells. When cultured on the artificial substrate of rigid tissue culture plastic, metastatic melanoma cells, such as human WM266.4 cells, do not explore shape space in a discrete, switch-like manner akin to Kc cells, as their morphology varies continuously around a single spread morphology (Fig. 6a). We extended these observations by quantifying the morphology of WM266.4 cells plated on plastic over time (Fig. 6b). However when cultured on deformable Collagen-I (Col-I) matrices5, 10-12 that have a stiffness comparable to the epidermis, the morphogenesis of WM266.4 cells becomes discrete. On Col-I WM266.4 cells assume only a rounded (similar to “N” shapes) or an elongated form (similar to “L” shape) (Fig. 6c). Within minutes WM266.4 cells plated on Col-I make rapid switch-like conversions between the shapes (Fig. 6d). This reveals that WM266.4 melanoma cells can explore shape space in a manner similar to Kc cells when plated on substrates that closely resemble their in vivo environment. Kc cells presumably can assume discrete shapes on plastic as they are only weakly adherent. We reasoned that the ability of cells to make switch-like conversions between rounded and elongated shapes could be due to dynamic regulation of protrusive and contractile forces. In support of this notion, knockdown in Kc cells of the Rho-kinase Rok13, a key regulator of cellular contractility leads to an accumulation of elongated “L”, as well as large, flat, and presumably poorly contractile “C” and “R” cells (Fig. 6e); thus morphological transitions are inhibited in Rok-deficient cells. To test the role of contractility in the discrete switch-like morphogenesis of melanoma cells we incubated WM266.4 cells plated on Col-I in increasing doses of the ROCK inhibitor H1152 and tracked their morphology over time. Inhibition of ROCK led to the accumulation of cells with a collapsed morphology that differs from both elongated forms and rounded forms (Fig. 6f) and makes only small continuous variations in shape (Fig. 6g); switch-like transitions do not occur. Thus substrate stiffness and cellular contractility are important factors determining the extent to which cells explore shape space in a discrete versus continuous fashion.
Figure 6. Melanoma cells make switch-like transitions between discrete morphologies on Collagen-I.
WM266.4 cells were plated either on (a, b) plastic or (c, d, f, g) Collagen-I. The Gaussian kernel density estimate of single-cell morphology in 2D PC space are shown in (a), (c), and (f). In (b), (d), and (g) the y-axis corresponds to the PC1 scores of single cells. Elongated cells have high PC1 scores, rounded cells have low PC1 scores and are shaded in orange. Time is described in the x-axes. (e) The Gaussian kernel density estimate of Rok deficient Kc cells (1,808 cells). In (f) and (g) WM266.4 cells were plated on Collagen-I, exposed to increasing doses of ROCK inhibitor H1152, and morphology was quantified 6 hrs later at both a single time point or (f) over time (g). We calculated the magnitude of morphology fluctuations for individual cells by calculating the S.D. in PC1 scores per cell over time. (a,b) 29,476 cells (c,d) 21,061 cells (f, g) 0.1 μM H11552, 36,665 cells; 1.0 μM 12,605 cells; 5. 0μM, 66,374 cells. Scale bars, 20 μm.
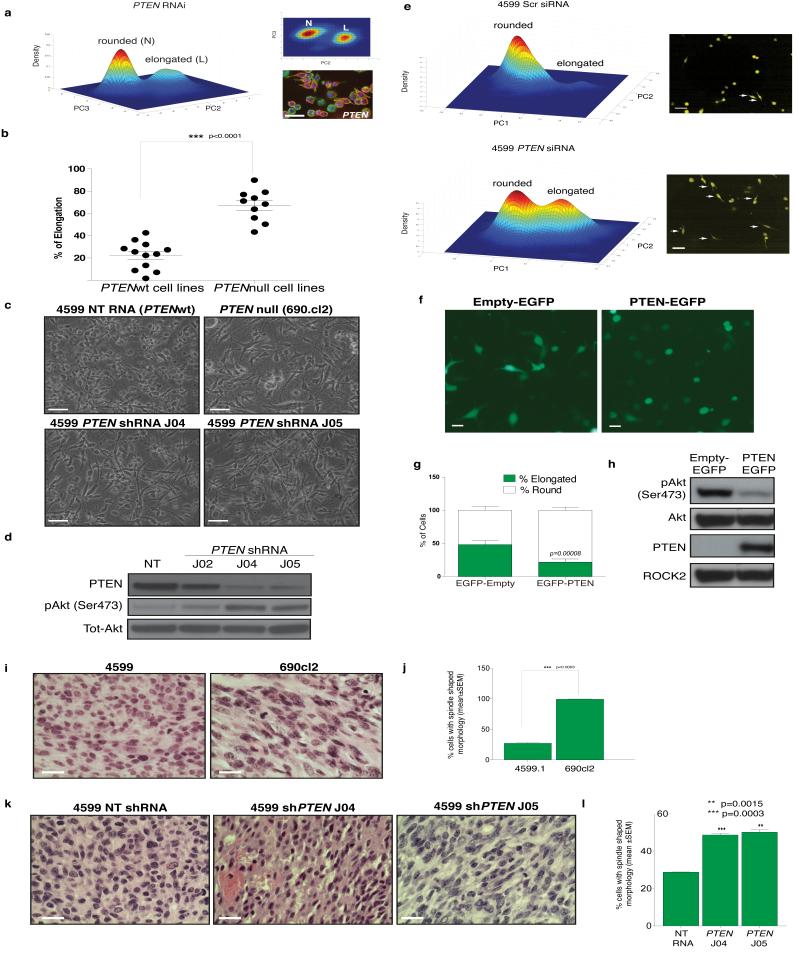
PTEN deficiency promotes bistable populations of rounded and elongated cells
The ratio of rounded to elongated melanoma cells is highly dependent on both environment and genetic background. For example, whereas the ratio of elongated to rounded cells can be as high as 50:50 in the case of WM266.4 cells on Col-I (Fig. 6b), melanoma cells such as A375M2 are mostly rounded5,11. However, the specifc genes that determine the rounded:elongated ratio are largely unknown. We reasoned that we could leverage the results of our morphological screen to gain insight into the factors regulating the conversion between rounded and elongated shape of melanoma cells based on two striking observations. The first is that the shape of WM266.4 cells, which do not express PTEN, phenocopies that of PTEN-deficiency in Drosophila (high ratio of elongated to rounded) (Fig. 7a). Secondly, hop/JAK-deficient Kc populations are also heavily enriched in elongated cells at the expense of other shapes, which is consistent with our recent finding that JAK1 promotes contractility in melanoma cells12. In fact, PTEN and hop/JAK RNAi results in the seventh and eigth highest L-scores respectively in the entire Kc dataset, and both ECs have high Q(4) scores demonstrating that they explore only limited regions of shape space compared to wild-type Kc cells.
Figure 7. Loss of PTEN alters the exploration of shape space.
(a) Density estimation of PTEN deficient Kc cells (1,831 cells). Upper right panel is side view of density estimate shown and lower right panel shows Kc cells stained with DAPI (blue), phalloidin (green), and anti-tubulin antibody (red) following PTEN RNAi. Scale bar, 20 μm. (b) Percentage of elongated cells on the top of thick Col-I (Mean±S.E.M); 250 cells over 5 fields of view per cell line; n = 12 PTENwt and 10 PTENnull cell lines (Supplementary Table S6); Student’s t-test was used to generate p-value (c) Images of 4599.1 cells on thick Col-I, PTEN was stably depleted by two different shRNAs (J04 and J05). NT is a non-targeting shRNA. 690.cl2 cells are shown for comparison. Scale bars, 50 μm. (d) Representative immuno-blot of pSer473 AKT, PTEN and tot-AKT in NT and PTEN shRNA-expressing 4599.1 cells. (e) Density estimation of 4599.1 cells treated with Scr (scrambled) siRNA (upper panel; 1,326 cells) or PTEN RNAi (lower panel; 305 cells) cultured on thick Col-I. Right panels show images of live cells, arrows denote elongated cells. Scale bars, 20 μm. (f) Representative images of WM266.4 cells transfected with Empty-EGFP construct or EGFP-PTEN construct on the top of thick Col-I. Scale bars, 50 μm. (g) Proportion of elongation/rounding following expression of Empty-EGFP or PTEN-EGFP expressing cells (Mean±SD); 200 cells per experiment, n=3 experiments; Student’s t-test was used to generate p-value. (h) Levels of PTEN, pAKT, total (Tot) AKT and ROCK2 (loading control) in Empty-EGFP and EGFP-PTEN transfected WM266.4 cells. (i) Representative images of 690cl2 and 4599.1 tumour sections. Scale bars, 20 μm. (j) The number of elongated cells in body of either 4599.1 or 690cl2 tumours is expressed as a percentage of the total number of cells counted per tumour (Mean±SEM); 200 cells/field assessed in 5 fields view per tumour; n=4 4599.1 and 4 690cl2 tumours; statistical analysis was done using unpaired students t test. (k) Representative images of tumour sections derived from NT or PTEN shRNAs-expressing 4599.1 cells. Scale bars, 100 μm. (l) Percentage of elongated cells in the body of the tumour following control (NT) or PTEN RNAi. (Mean±SEM); 200 cells/field assessed in 5 fields of view over n= 4 NT RNAi and 4 PTEN RNAi tumours.
To determine if PTEN status correlates with the ratio of round:elongated shapes in cell populations, we plated 22 melanoma cell lines (10 PTENnull and 12 PTENwt) on Col-I gel and assessed the ratio of rounded:elongated cells. PTEN loss strongly correlates with an increase in the proportion of elongated to rounded cells (Fig. 7b and Supplementary Table S6). Furthermore depletion of PTEN expression in PTEN wild type mouse 4599.1 or human A375p melanoma cells (Fig. 7c, and Supplementary Figure S3a,b) by independent shRNAs increases the number of elongated cells and increases phosphorylated Akt levels (Fig. 7d and Supplementary Figure S3a,b). We confirmed the effect of PTEN shRNA using quantiative readouts of morphology (Fig. 7e), and show that PTEN depletion increases the number of elongated cells but does not generate any additional shapes. Importantly, re-expression of PTEN in the WM266.4 PTEN null melanoma cells increases the number of rounded cells at the expense of elongated cells (Fig. 7f, g, h). To determine whether PTEN regulates the exploration of shape space in vivo we used orthotopic implantation of melanoma cells from 4599.1 (PTENwt), 4599.1 cell populations stably expressing 2 different PTEN shRNAs or 690cl2 (PTENnull) into the dermis of NOD SCID mice and assessed the shape in haematoxylin and eosin stained sections5, 14. Tumour cells arising from injection of 4599.1 cells are predominantly rounded (Fig. 6i,j) while tumour cells arising from injection of 690cl2 cells (Fig. 7i,j), or 4599.1 cells in which PTEN had been knocked down are markedly elongated (Fig. 7k,l and Supplementary Figure S4). Thus PTEN loss induces elongation cells in tissue culture and in vivo.
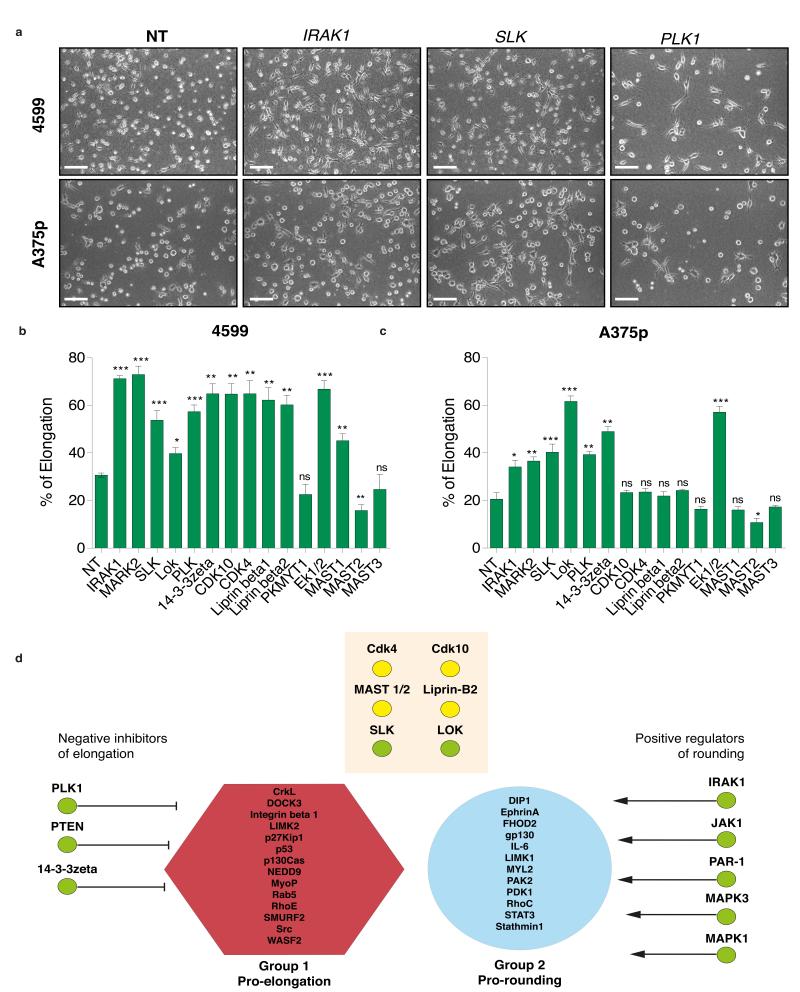
A conserved class of genes that promote rounding
Given that depletion of PTEN and JAK results in bistable populations of elongated and rounded cells in Drosophila, mouse and human cells, we sought to determine if we other genes identified in the Drosophila screen are conserved regulators of morphogenesis. We selected genes whose depletion in Drosophila cells results in a significant increase in the number of elongated cells, or the magnitude of their “L” scores (Supplmentary Table S7). Genes were further prioritised if their inhibition resulted in low complexity (high Q(4) score), and thus were enriched in “L” cells at the expense of other shapes. For example, whereas PLK1- and 14-3-3zeta-deficient Drosophila cell populations are comprised almost exclusively of rounded and elongated shapes, Slik-deficient populations have a high (L) score, but are also enriched in other subpopulations. We only tested genes where we could identify human homologues; in some cases this required targeting of multiple genes (e.g. MAST1, MAST2, MAST3 are homologues of Drosophila CG6498) (Supplementary Table S7). Using siRNA pools (Supplementary Tables S8,S9) we depleted 15 different homologs of 11 different Drosophila genes in 4599.1 mouse and A375p human melanoma cells. When cultured in starving conditions on a thick Col-I matrix, both 4599.1 and A375p convert to elongated cells at a low frequency;we scored populations on the basis of whether siRNA knockdown increases the frequency of elongation (Fig. 8a). RNAi-mediated knockdown of 12/15 genes in mouse (Fig. 8b) and 7/15 genes human cells results in significant increases in elongation that phenocopies their depletion in Drosophila (Supplementary Table S7). For example, depletion of mouse and human IRAK1, PLK1, PTEN, ERK1, and ERK2, led to marked increases in the numbers of elongated cells (Fig. 8b). That 10/11 Drosophila genes whose depletion results in a high L score can be validated as regulators of cell rounding in at least one mammalian metatstatic melanoma tumour line, in addition to PTEN and JAK, strongly highlights the ability of our RNAi screen to identify novel genes that have relevance to disease progression.
Figure 8. A conserved set of genes promotes rounding in Drosophila, mouse, and human cells.
(a) Mouse 4599.1 (upper panels), and human A375p (lower panels) metastatic melanoma cells plated on Col-I following RNAi mediated gene knockdown of IRAK1, SLK, and PLK1. NT is non-targetted/scrambled RNAi. Scale bars, 50 μm. (b) Percentage of elongated cells following knockdown of 15 mouse genes in 4599.1 cells plated on Col-I (Mean±SEM). (c) Percentage of elongated cells following knockdown of 15 human genes in A375p cells plated on Col-I (Mean±SEM). In (b) and (c) 250 cells over n=3 experiments. Student’s t-test was used to generate p-values. Asterisk denotes level of significance where * = p < 0.05, ** = p <0.001, *** = p < 0.0001. (d) Network analysis. We calculated the proximity of different proteins identified in our screen to either “Pro-elongation” proteins, or “Pro-rounding” proteins in protein-protein interaction space (PPI). Length of arrow is scaled to a Z-score that describes the significance of this proximity compared to random proteins, where longer arrows are less significant and thus further away in PPI space. Because inhibition of all proteins here results in elongated shapes, we could classify different proteins as negative regulators of elongation or positive regulators of rounding. Proteins in the orange box are not significantly close to either group. Green circles indicate gene depletion increases percentage of elongation in mouse and human cells, yellow circles indicate gene depletion increases percentage of elongation in mouse cells.
Classifying genes as protrusion antagonists or contractility agonists
Towards gaining systems-level mechanistic insights into how different validated genes identified in our screen regulate cell shape, we performed a network analysis to determine the proximity of different proteins in network space to regulators of protrusiveness or contractility. Proteins previously implicated in controlling cell shape were classified into either “pro-elongation” or “pro-contractility” groups15. We then calculated the average number of edges that separated proteins identified in our screen from proteins in either previously assigned group in protein-protein interaction networks, and judged the signficance of this distance compared to that between other random proteins. Proteins such as JAK1 and IRAK1 are signficantly closer in protein-protein interaction space to the pro-contractility group, whereas PTEN and 14-3-3 zeta are closer to the pro-elongation group (Fig. 8d). Given that depletion of all these genes results in similar elongated shapes, we conclude that JAK1 and IRAK1 promote rounding, but 14-3-3zeta and PTEN negatively inhibit protrusion. This unbiased network is consistent with our previous observation that JAK1 upregulates contractility by activation of the STAT3 transcription factor12, and that PTEN is a negative regulator of PI3K which acts to promote protrusion in multiple other cell-types16. Interestingly, proteins such as ERK1/2 have not been previously associated with an upregulation of contractility. Thus this analysis provides hypotheses for other poorly characterised genes.
Discussion
By implementing methods to quantify mean morphology, complexity, and presence of intermediate forms in cell populations in an RNAi screen of Drosophila Kc cells, we propose that cells can explore shape space in a discrete, switch-like manner. Using live-cell imaging methods in combination with morphological quantification, we demonstrate that this type of morphogenesis is not limited to Drosophila hemocytes, and that metastatic melanoma cells explore shape space in a similar fashion when plated on substrates that mimic their in vivo environment. We propose that many cell types will also exhibit discrete switch-like morphogenesis in vivo, and that it has been the long-standing use of rigid tissue culture plastic which has obscured this aspect of cell shape control. While the model that cells can be constrained to specific regions of shape space is potentially counterintuitive given the highly plastic nature of cell shape and the ability of cells to adopt radically diverse shapes, discrete morphogenesis or morphological canalisation of single cells17 is consistent with the model of signalling networks as dynamical systems that can exist in a limited number of stable states, or attractors18.
That systematic gene inhibition by RNAi can alter shape space and/or alter the mode of morphogenesis from switch-like to continuous (e.g. Stam RNAi), suggests that signalling networks have evolved to couple the topology of their shape space, and how they explore it, to environmental conditions. Metastatic cancer cells may have re-engineered regulatory networks that uncouple the control of morphogenesis from environmental cues, which would otherwise dictate the number of shapes they can assume and how they convert between these shapes. In the case of PTEN it is tempting to speculate that loss of PTEN may promote the adoption of a bistable state where rounded and elongated forms are present in high numbers. By increasing the frequency of rounded and elongated cells this would provide metastatic cells with a survival advantage that is otherwise not gained by adopting only a single shape, or being highly plastic.
Supplementary Material
Figure S1 Workflow for image analysis of RNAi screening data. Nuclei were first segmented from the DAPI channel, and these segments were later integrated with a “cell body image” combined from the α-tubulin and F-actin channels. Scale bars, 20 μm
Figure S2 Accounting for differential penetrance and diverse shapes in RNAi replicates. (a) Representative image of cells treated with sti/Citron (DRSC09739) or Pvr (DRSC03080) dsRNAs. Scale bars, 20 μm. (b) The percentage of normal cells that exists in each population following treatment with a single dsRNA (x-axis) is plotted against the number of cells that were initially analysed (y-axis). ECs with >75% are considered normal. (c) Cells treated with thread/DIAP1 RNAi (DRSC11404), show a 96.7% reduction in viability (n=4 experiments). Scale bars, 20 μm. (d) Representative images of mock-treated cells or cells treated with rl/ERK (DRSC07833) and stained with anti-ERK antibody (green) and DAPI (red). Scale bars, 20 μm. (e) Mean ERK intensity (normalized to DAPI intensity) from 195 individual cells randomly selected from mock-treated populations or populations treated with two different dsRNAs targetting rl/ERK. (f) Mean AKT intensity (normalized to DAPI intensity) from 170 individual cells that were randomly selected from mock-treated populations or populations treated with three different dsRNAs targetting Akt.
Figure S3 Repeatability analysis. (a) Example of the effects of normal cell filtering on Pp2-14D deficient cells. Whereas Pp2-14D dsRNA is 37% penetrant pre-filtering, there are almost no normal cells in the cell population post-filtering. (b) The upper panels show the similarity of the 4-dimensional QMSs (comparison to L, C, T, R shapes) generated by 3 different dsRNAs targeting the same gene. Line colour indicates dsRNAs targeting the same gene. Each point represents the mean normalised Z-score of the cell population (y-axis) describing the similarity to 4 reference shapes (x-axis). The left upper panel shows cases where dsRNAs give dissimilar QMSs, whereas the right upper panel shows cases where dsRNAs give similar QMSs. The lower panels show the similarity of the 4-dimensional QMSs generated by different 4 dsRNAs targeting the same gene. The left lower panel shows cases where dsRNAs give dissimilar QMSs, whereas the right lower panel shows cases where dsRNAs give similar QMSs. (c) The y-axis describes the number of replicable dsRNAs (blue) or non-replicable dsRNAs (red) distributed on the basis of the number of dsRNAs used to target an individual gene in the screen (x-axis). (d) Similarity matrix for dsRNAs targeting 4 genes from Clusters 1 and 2. The colour of each square represents the repeatability of each dsRNA compared with all others in the matrix. A colour towards the red end of the visible spectrum indicates increasing levels of repeatability. Squares below the diagonal depict repeatability analyses performed prior to normal cell filtering. Squares above the diagonal are analyses performed after normal cell filtering. White boxes indicate cases where normal cell filtering decreases the repeatability, meaning that the remaining shapes are dissimilar.
Figure S4 PTEN depletion by RNAi leads to increased numbers of elongated cells. 4599.1 melanoma cells (a) and A375p melanoma cells (b) were transfected with non-targetting (NT) or PTEN RNAi(s) and seeded on a thick layer of Col-I. After 5-16 hrs of serum starvation, cells were photographed under phase contrast. Scale bars, 50 μm. Histograms show quantification of the proportion of elongated cells (Mean±S.D.) in 4599.1 melanoma cells (a) and A375p melanoma cells (b) upon PTEN knockdown; 300 cells per n=3 experiments; Student’s t-test was used to generate p-value. Immuno-blots show the level PTEN and total (Tot) AKT in NT- and PTEN RNAi(s)-transfected 4599.1 (upper panel) and A375p (lower panel).
Figure S5 High magnification images of tumour sections following PTEN RNAi. Representative images of low magnification tumour sections derived from either non-targetting (NT), or PTEN shRNAs-expressing 4599.1 melanoma cells. Scale bars, 100 μm.
Figure S6 Levels of mRNA following siRNA-mediated knockdown in mouse and human melanoma cells.
Figure S7 Uncropped Western blots.
Table S1. Summary of whole-cell geometry features. Each one of 11 whole-cell geometry features is defined by a feature ID among the 211 morphology features. A brief description and the data source from where the specific feature is extracted.
Table S2. Summary of Haralick texture features extracted from the spatial-dependence matrix of each cell segment. The 14 Haralick features are divided into three groups, and the feature IDs among the 211 morphology features, as well as the feature names as defined in the original reference, are listed.
Table S3. Summary of regional geometric features. The 54 regional geometric features are divided into two groups, namely length ratios and area ratios. For each group, the feature IDs among the 211 morphology features are listed; a brief description for feature extraction are supplied; and an simple illustration for feature extraction process is shown.
Table S4. Summary of the four groups within the initial population of each GA run. The 200 individuals in each initial populations for a GA run is divided into four groups. Each group is defined based on the results of the previous SVM-RFE process, and the relationships between individuals in each group and the SVM-RFE results are defined.
Table S5. Quantitative Morphological Signature (QMS), Q(4), max RIFT, and mean RIFT scores for 287 ECs. In the first sheet we describe the number of repeatable cells that comprise each EC (column C), the number of cells from individual populations targetted by individual amplicons that contribute to total cell number. Each ECs QMS is comprised of L, T, C, and R scores and a PZ score. Cluster number is determined by hierarchical clustering (Fig. 3). In the second sheet the raw amplicon data for all tested amplicons is listed.
Table S6. Morphological comparison of PTEN wild-type and PTEN-deficient cells. Detail of data that is summarised in Fig. 7b.
Table S7. Drosophila genes chose for validation in mouse and human melanoma cells.
Table S8. Sequences for siRNAs and shRNAs used in mouse and human knockdown experiments.
References
- 1.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Keren K, et al. Mechanism of shape determination in motile cells. Nature. 2008;453:475–480. doi: 10.1038/nature06952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mogilner A, Keren K. The shape of motile cells. Curr Biol. 2009;19:R762–771. doi: 10.1016/j.cub.2009.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guarino M, Rubino B, Ballabio G. The role of epithelial-mesenchymal transition in cancer pathology. Pathology. 2007;39:305–318. doi: 10.1080/00313020701329914. [DOI] [PubMed] [Google Scholar]
- 5.Sanz-Moreno V, et al. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135:510–523. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 6.Wolf K, et al. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J Cell Biol. 2003;160:267–277. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bakal C, Aach J, Church G, Perrimon N. Quantitative morphological signatures define local signaling networks regulating cell morphology. Science. 2007;316:1753–1756. doi: 10.1126/science.1140324. [DOI] [PubMed] [Google Scholar]
- 8.Yin Z, et al. Using iterative cluster merging with improved gap statistics to perform online phenotype discovery in the context of high-throughput RNAi screens. BMC Bioinformatics. 2008;9:264. doi: 10.1186/1471-2105-9-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waddington CH. The Strategy of Genes. Allen & Unwin; 1957. [Google Scholar]
- 10.Gadea G, Sanz-Moreno V, Self A, Godi A, Marshall CJ. DOCK10-mediated Cdc42 activation is necessary for amoeboid invasion of melanoma cells. Curr Biol. 2008;18:1456–1465. doi: 10.1016/j.cub.2008.08.053. [DOI] [PubMed] [Google Scholar]
- 11.Sahai E, Marshall CJ. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat Cell Biol. 2003;5:711–719. doi: 10.1038/ncb1019. [DOI] [PubMed] [Google Scholar]
- 12.Sanz-Moreno V, et al. ROCK and JAK1 signaling cooperate to control actomyosin contractility in tumor cells and stroma. Cancer Cell. 2011;20:229–245. doi: 10.1016/j.ccr.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 13.Tan C, Stronach B, Perrimon N. Roles of myosin phosphatase during Drosophila development. Development. 2003;130:671–681. doi: 10.1242/dev.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Viros A, et al. Improving melanoma classification by integrating genetic and morphologic features. PLoS Med. 2008;5:e120. doi: 10.1371/journal.pmed.0050120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanz-Moreno V, Marshall CJ. The plasticity of cytoskeletal dynamics underlying neoplastic cell migration. Curr Opin Cell Biol. 2010;22:690–696. doi: 10.1016/j.ceb.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 16.Ridley AJ, et al. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 17.Waddington CH. Canalization of development and genetic assimilation of acquired characters. Nature. 1959;183:1654–1655. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- 18.Kauffman SA. The Origins of Order. Self-Organization and Selection in Evolution. Oxford University Press; New York, Oxford: 1993. [Google Scholar]
- 19.Dhomen N, et al. Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell. 2009;15:294–303. doi: 10.1016/j.ccr.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 20.Dankort D, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Workflow for image analysis of RNAi screening data. Nuclei were first segmented from the DAPI channel, and these segments were later integrated with a “cell body image” combined from the α-tubulin and F-actin channels. Scale bars, 20 μm
Figure S2 Accounting for differential penetrance and diverse shapes in RNAi replicates. (a) Representative image of cells treated with sti/Citron (DRSC09739) or Pvr (DRSC03080) dsRNAs. Scale bars, 20 μm. (b) The percentage of normal cells that exists in each population following treatment with a single dsRNA (x-axis) is plotted against the number of cells that were initially analysed (y-axis). ECs with >75% are considered normal. (c) Cells treated with thread/DIAP1 RNAi (DRSC11404), show a 96.7% reduction in viability (n=4 experiments). Scale bars, 20 μm. (d) Representative images of mock-treated cells or cells treated with rl/ERK (DRSC07833) and stained with anti-ERK antibody (green) and DAPI (red). Scale bars, 20 μm. (e) Mean ERK intensity (normalized to DAPI intensity) from 195 individual cells randomly selected from mock-treated populations or populations treated with two different dsRNAs targetting rl/ERK. (f) Mean AKT intensity (normalized to DAPI intensity) from 170 individual cells that were randomly selected from mock-treated populations or populations treated with three different dsRNAs targetting Akt.
Figure S3 Repeatability analysis. (a) Example of the effects of normal cell filtering on Pp2-14D deficient cells. Whereas Pp2-14D dsRNA is 37% penetrant pre-filtering, there are almost no normal cells in the cell population post-filtering. (b) The upper panels show the similarity of the 4-dimensional QMSs (comparison to L, C, T, R shapes) generated by 3 different dsRNAs targeting the same gene. Line colour indicates dsRNAs targeting the same gene. Each point represents the mean normalised Z-score of the cell population (y-axis) describing the similarity to 4 reference shapes (x-axis). The left upper panel shows cases where dsRNAs give dissimilar QMSs, whereas the right upper panel shows cases where dsRNAs give similar QMSs. The lower panels show the similarity of the 4-dimensional QMSs generated by different 4 dsRNAs targeting the same gene. The left lower panel shows cases where dsRNAs give dissimilar QMSs, whereas the right lower panel shows cases where dsRNAs give similar QMSs. (c) The y-axis describes the number of replicable dsRNAs (blue) or non-replicable dsRNAs (red) distributed on the basis of the number of dsRNAs used to target an individual gene in the screen (x-axis). (d) Similarity matrix for dsRNAs targeting 4 genes from Clusters 1 and 2. The colour of each square represents the repeatability of each dsRNA compared with all others in the matrix. A colour towards the red end of the visible spectrum indicates increasing levels of repeatability. Squares below the diagonal depict repeatability analyses performed prior to normal cell filtering. Squares above the diagonal are analyses performed after normal cell filtering. White boxes indicate cases where normal cell filtering decreases the repeatability, meaning that the remaining shapes are dissimilar.
Figure S4 PTEN depletion by RNAi leads to increased numbers of elongated cells. 4599.1 melanoma cells (a) and A375p melanoma cells (b) were transfected with non-targetting (NT) or PTEN RNAi(s) and seeded on a thick layer of Col-I. After 5-16 hrs of serum starvation, cells were photographed under phase contrast. Scale bars, 50 μm. Histograms show quantification of the proportion of elongated cells (Mean±S.D.) in 4599.1 melanoma cells (a) and A375p melanoma cells (b) upon PTEN knockdown; 300 cells per n=3 experiments; Student’s t-test was used to generate p-value. Immuno-blots show the level PTEN and total (Tot) AKT in NT- and PTEN RNAi(s)-transfected 4599.1 (upper panel) and A375p (lower panel).
Figure S5 High magnification images of tumour sections following PTEN RNAi. Representative images of low magnification tumour sections derived from either non-targetting (NT), or PTEN shRNAs-expressing 4599.1 melanoma cells. Scale bars, 100 μm.
Figure S6 Levels of mRNA following siRNA-mediated knockdown in mouse and human melanoma cells.
Figure S7 Uncropped Western blots.
Table S1. Summary of whole-cell geometry features. Each one of 11 whole-cell geometry features is defined by a feature ID among the 211 morphology features. A brief description and the data source from where the specific feature is extracted.
Table S2. Summary of Haralick texture features extracted from the spatial-dependence matrix of each cell segment. The 14 Haralick features are divided into three groups, and the feature IDs among the 211 morphology features, as well as the feature names as defined in the original reference, are listed.
Table S3. Summary of regional geometric features. The 54 regional geometric features are divided into two groups, namely length ratios and area ratios. For each group, the feature IDs among the 211 morphology features are listed; a brief description for feature extraction are supplied; and an simple illustration for feature extraction process is shown.
Table S4. Summary of the four groups within the initial population of each GA run. The 200 individuals in each initial populations for a GA run is divided into four groups. Each group is defined based on the results of the previous SVM-RFE process, and the relationships between individuals in each group and the SVM-RFE results are defined.
Table S5. Quantitative Morphological Signature (QMS), Q(4), max RIFT, and mean RIFT scores for 287 ECs. In the first sheet we describe the number of repeatable cells that comprise each EC (column C), the number of cells from individual populations targetted by individual amplicons that contribute to total cell number. Each ECs QMS is comprised of L, T, C, and R scores and a PZ score. Cluster number is determined by hierarchical clustering (Fig. 3). In the second sheet the raw amplicon data for all tested amplicons is listed.
Table S6. Morphological comparison of PTEN wild-type and PTEN-deficient cells. Detail of data that is summarised in Fig. 7b.
Table S7. Drosophila genes chose for validation in mouse and human melanoma cells.
Table S8. Sequences for siRNAs and shRNAs used in mouse and human knockdown experiments.