Abstract
ACTN4 is an actin-binding protein that participates in cytoskeleton organisation. It resides both in the cytoplasm and nucleus and physically associates with various transcription factors. Here, we describe an effect of ACTN4 expression on transcriptional activity of the RelA/p65 subunit of NF-kB. We demonstrate that ACTN4 enhances RelA/p65-dependant expression of c-fos, MMP-3 and MMP-1 genes, but it does not affect TNC, ICAM1 and FN1 expression. Importantly, actin-binding domains of ACTN4 are not critical for the nuclear translocation and co-activation of RelA/p65-dependent transcription. Collectively, our data suggest that in the nucleus, ACTN4 functions as a selective transcriptional co-activator of RelA/p65.
Keywords: ACTN4, RelA/p65, transcription regulation, MMP
INTRODUCTION
Alpha-actinin 4 (ACTN4) is an actin-binding protein which belongs to the spectrin superfamily. It predominantly localises to the cytoplasm and focal adhesion contacts. Primarily, ACTN4 was found to participate in the cytoskeleton organisation, cytokinesis, regulating cell adhesion and shape [1, 2]. Being a part of cytoskeleton, ACTN4 plays a crucial role in remodelling of actin cytoskeleton and in formation of protrusions, which potentiate migration of normal and cancer cells [3].
According to the domain structure, ACTN4 can be subdivided into three major regions. The N-terminal region includes two calponin homology domains (CH1-2). Due to the actin-binding domain (ABD) located in this region, ACTN4 directly cross-links the actin filaments and drives them to focal adhesion sites via adhesion plaque proteins [4, 5]. The central part of ACTN4 consists of four spectrin repeats (SR1-4), which are responsible for dimerisation in an anti-parallel manner; they also participate to the assembly of multiprotein structures involved both in the cytoskeleton architecture and signal transduction [6]. Finally, together with the C-terminus, spectrin repeats mediate interactions with a wide group of cytoskeleton and non-cytoskeleton proteins such as vinculin, MEKK1 kinase, MAGI, BP-180 [7, 8, 9]. The C-terminus region of ACTN4 includes two EF hand domains (EF1-2) and the calmodulin-homology domain [10].
A nuclear localisation of ACTN4 has been reported [1, 11], as well as its interaction with a number of nuclear proteins like NF-Y [12], DNaseY [13], RelA/p65 subunit of the nuclear factor-κB (NF-κB) [14], heterogeneous nuclear ribonucleoproteins [15] and HDAC7 [16]. ACTN4 may therefore participate in regulation of transcription. Accordingly, ACTN4 modulates transcriptional activity of estrogen receptor, retinoic acid receptor and peroxisome proliferator-activated receptor gamma [17, 18]. Nevertheless, its nuclear functions are not fully understood, and the precise mechanisms of its nuclear translocation have not been revealed yet.
The NF-kB family includes several members, RelA, c-Rel, RelB, p50 and p52, all able to regulate, as transcription factors, a number of molecules involved in cancer [19, 20, 21, 22, 23], cellular senescence [24], hypertonic stress [25], immunity [26] as well as host defence against pathogens [27, 28]. A major function of NF-κB occurs in the innate and adoptive immunity [29] is mediated by its ability to regulate cytokines [30], chemokines [31], and adhesion molecules [32]. However, there are still several open questions related to the selective functions of individual NF-κB family members [33] during a coordinated cellular response to pathogens, and the related molecular mechanisms. The effect on autophagy [34], TNF [35, 36, 37, 38, 39], TRAIL [40, 41] is still under intense investigation [42, 43], and in particular several interactions seems able to fine tuning NF-κB function, including for example regulators of histone acetylation [44], ubiquitin [45] and IAPs [46]. To this end, a potential regulation by ACTN4 could be of particular interest.
To further elucidate ACTN4 role in nuclear processes, we investigated functional significance of its interaction with RelA/p65 subunit of transcriptional factor NF-kB. We have found that ACTN4 can modulate transcriptional activity of RelA/p65. This function does not directly depend on its ability to bind actin or to form dimers. Nevertheless, ACTN4 regions responsible for actin binding and dimerisation are required for its nuclear translocation. Our data indicate that ACTN4 may co-regulate expression of certain genes together with transcription factors such as NF-kB.
RESULTS
ACTN4 does not influence on nuclear accumulation of RelA/p65
The transcription factor NF-kB is sequestered in the cytoplasm of most cells by specific inhibitory protein IkBα [47]. Nuclear translocation of NF-kB is required for its full activation and ability to activate gene expression. In fact, RelA/p65 subunit of NF-kB co-immunoprecipitates with ACTN4 and localises along stress fibres in A431 cells [14, 48]. To study the interaction between ACTN4 and RelA/p65 and their co-localisation in HEK293T cells, we prepared a construct, expressing full-length ACTN4 (ACTN4Fl) with MYC-tag at the N-terminus. Co-immunoprecipitation shows that ACTN4Fl is present in complexes with RelA/p65 in cytoplasm of HEK293T cells (Fig.1 A). Since ACTN4 interacts with RelA/p65 both in the cytoplasm and nucleus, we evaluated the ability of ACTN4 over-expression to affect the RelA/p65 nuclear accumulation. Immunoblotting of nuclear and cytoplasm protein extracts demonstrates that ACTN4Fl over-expression alone does not lead to nuclear accumulation of RelA/p65 in comparison to control cells (Fig.1 B).
Figure 1. Over-expressed ACTN4 co-immunoprecipitates with RelA/p65 in the cytoplasm of HEK293T cells but does not mediate RelA/p65 nuclear translocation.
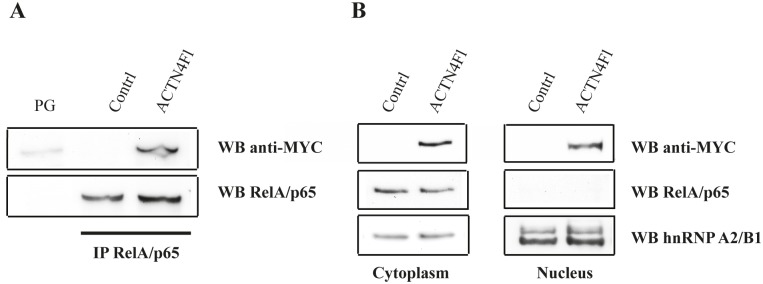
(A) RelA/p65 was immunoprecipitated from cytoplasm protein extracts of HEK293T cells with RelA/p65-specific antibody. Bound proteins were separated by SDS-PAGE followed by immunoblotting with anti-MYC and anti-RelA/p65 antibodies. No RelA/p65 specific signal was detected after incubation of cytoplasm extracts with empty Protein G Sepharose beads (PG lane). (B) Western blot analysis of cytoplasm and nuclear protein extracts using MYC-, RelA/p65- and hnRNPA2/B1-specific antibodies. hnRNPA2/B1 was used as a loading control.
ACTN4 co-activates RelA/p65-dependant expression of c-fos and MMP-3 genes
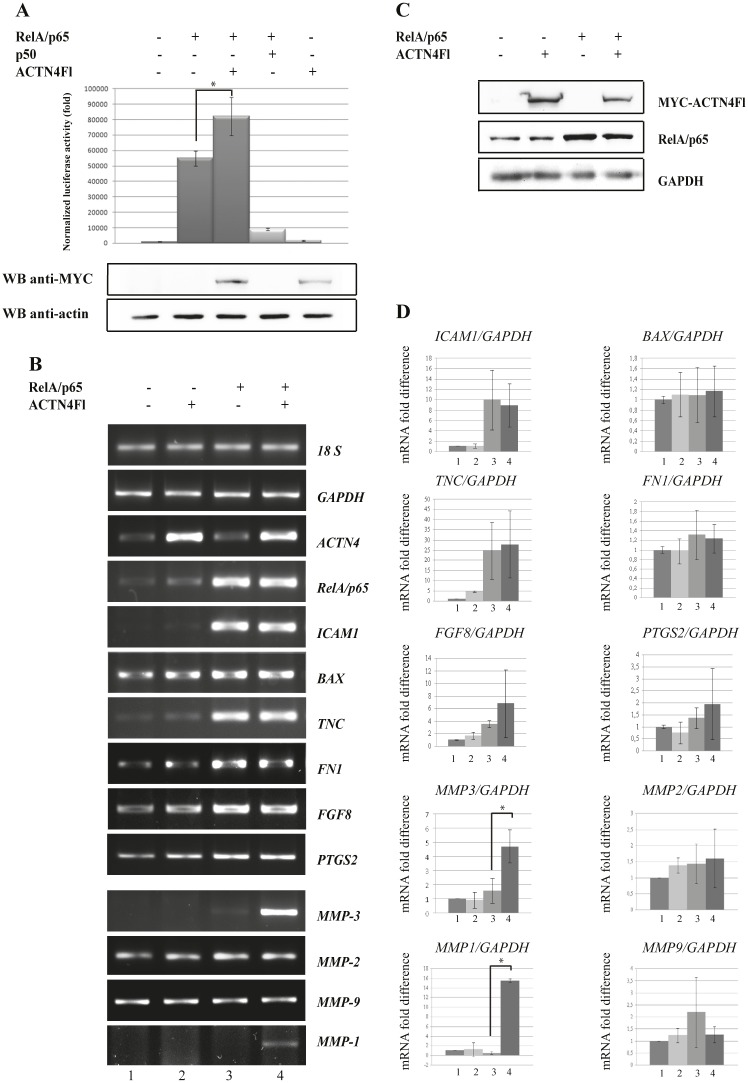
In order to further evaluate a possible influence of ACTN4 on RelA/p65-dependant gene expression, we employed a luciferase transcription assay. The pfLUC reporter construct that contained the luciferase gene under control of the minimal (-59 to +109) c-fos promoter [49] along with ACTN4 and RelA/p65 were transiently transfected into A431 cells. We have found that over-expression of full-length ACTN4 alone does not activate transcription from c-fos promoter (Fig.2 A). However, co-expression of ACTN4Fl with RelA/p65 enhances luciferase activity versus RelA/p65 alone (Fig.2 A). These data indicate that ACTN4Fl specifically co-activates the RelA/p65 subunit of transcription factor NF-kB, and can potentially regulate expression of other RelA/p65-dependent genes. To determine whether this regulation is gene-specific or it is a more general phenomenon, we analysed expression of seven additional genes known to contain RelA/p65 binding site in the promoter. HEK293T cells were transiently transfected with plasmids that express ACTN4Fl and RelA/p65 followed by a semi-quantitative RT-PCR analysis of TNC, ICAM1, FGF8, BAX, PTGS2, FN1 and MMP-3 gene expression (Fig.2 B, D). Over-expression of RelA/p65 alone noticeably induces expression of TNC and ICAM1 genes, but does not effect on expression of BAX, FGF8, PTGS2, and FN1 genes. Moreover, none of these genes is affected by over-expression of ACTN4Fl. However, co-expression of RelA/p65 with ACTN4Fl significantly enhances expression of MMP-3 gene (Fig.2 B lower panel and D). These results indicate that ACTN4Fl can regulate MMP-3 gene expression in a RelA/p65-dependant manner. All together, our data demonstrate that ACTN4 can be a co-activator of RelA/p65, even though not all genes display this effect.
Figure 2. Co-expression of ACTN4Fl and RelA/p65 leads to enhanced expression of RelA/p65-regulated genes.
(A) A431 cells were transfected with luciferase reporter vector containing luciferase gene under c-fos mouse promoter, and expression plasmids with ACTN4Fl, RelA/p65 and p50. The ratios of luciferase-to-β-galactosidase in comparison to an empty luciferase construct are presented. Expression of exogenous ACTN4 (the lower panel) was determined by Western blot analysis using anti-MYC antibody (*P< 0.02 for ACTN4Fl+RelA/p65 versus RelA/p65). (B) ACTN4Fl influence on transcription activity of RelA/p65 subunit of transcription factor NF-kB in regard to MMP-3 gene promoter. HEK293T cells were transfected with plasmids coding ACTN4Fl and RelA/p65. Over-expression of RelA/p65 alone noticeably induces expression of TNC and ICAM1 genes, but does not have significant effect on expression of BAX, FGF8, PTGS2, and FN1 genes. The lower panel represents RT-PCR of MMPs: MMP-2, MMP-9, MMP-1 and MMP-3 genes. (C) Western blot analysis of ACTN4Fl and RelA/p65 expression in HEK293T cells. (D) Quantification data of RelA/p65 regulated genes after transfection of RelA/p65 and ACTN4Fl. The data show fold difference of mRNA level of RelA/p65-regulated genes in regard to GAPDH gene. HEK293T cells were transfected either with ACTN4Fl (lanes 2), or RelA/p65 (lanes 3), or co-transfected with ACTN4 and RelA/p65 (lanes 4). Non-transfected HEK293T cells is shown at lanes 1. Co-expression of RelA/p65 with ACTN4Fl leads to strong increase of MMP-3 gene expression (*P<0.05 - MMP-3 and *P<0.02 - MMP-1 for ACTN4Fl+RelA/p65 versus RelA/p65). The data represent an average of at least three independent experiments, ±SD.
ACTN4 affects matrix metalloproteinases gene expression
Matrix metalloproteinases (MMPs) belong to a large family of endopeptidases that cleave a wide range of substrates including numerous matrix proteins, growth factors and proteases [50]. MMP-3 plays a major role in remodelling of collagens, and shows substrate specificity to several other matrix proteins and proteoglycans [51]. Besides MMP-3, NF-kB can activate other MMP genes, including MMP-1, MMP-9 and MMP-2 [52, 53]. We examined whether ACTN4 might also activate expression of these genes. MMP-2 and MMP-9 genes do not exhibit any changes in their mRNA levels in response to ACTN4Fl and RelA/p65 over-expression (Fig.2 B lower panel and D). However, the mRNA level of the MMP-1 gene is affected by ACTN4Fl and RelA/p65 (Fig.2 B lower panel and D). These data show that other MMPs, such as MMP-1, can be regulated in a similar manner to MMP-3.
Dimerisation and binding to F-actin are not required for the nuclear import and functioning of ACTN4
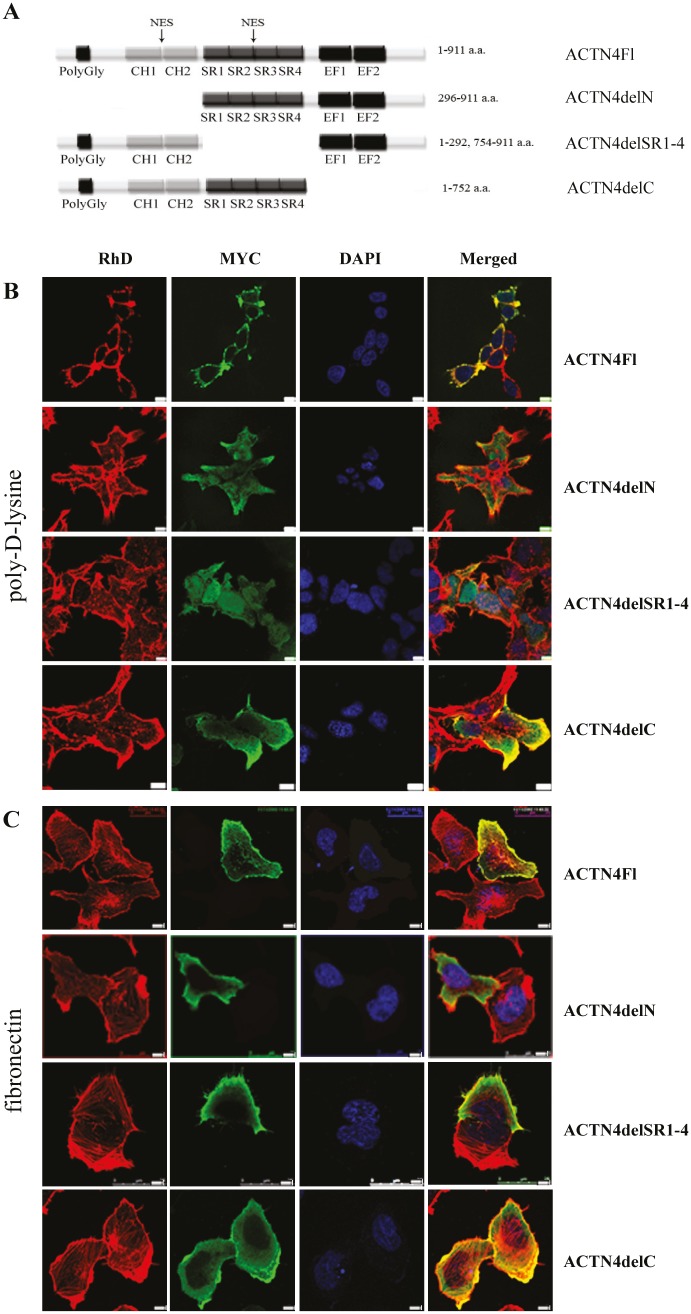
The main functions of ACTN4 in the cytoplasm are mediated by its ability to form homodimers and bind F-actin filaments. Similarly, dimerisation and actin-binding may be critical for translocation of ACTN4 into the nucleus and its function as a co-regulator of RelA/p65. To test this hypothesis, we constructed three expression plasmids, containing ACTN4 with deletions of the N-terminal region with actin-binding domain, the C-terminal region, and SR1-4 domains. All deletion variants contained a MYC-tag at the N-terminus that allowed visualising exogenous protein (Fig.3 A).
Figure 3. Intracellular distribution of the ACTN4 deletion mutants in transfected HEK293T cells.
(A) Scheme of ACTN4 deletion variants with MYC-tag on N-terminus. (B) HEK293T cells were transfected with expression plasmids coding ACTN4Fl, ACTN4delN, ACTN4delSR1-4, ACTN4delC, and cultured on poly-D-lysine coated cover slips. (C) HEK293T cells spread on EMC protein (fibronectin). Bar scale is 5 um.
To determine the intracellular distribution of ACTN4 deletion variants, we carried out transfection of HEK293T cells followed by immunofluorescence using anti-MYC-tag antibodies. Transfected HEK293T cells were spread on cover slips covered with poly-D-lysine for 24 hours. The subcellular distribution of ACTN4 compared to the actin cytoskeleton and the nucleus was obtained by staining of corresponding structures with rodamin-phalloidin (RhD) and DAPI, respectively (Fig.3). Analysis of the localisation of exogenous protein variants shows that the deletion of the N-terminal and SR regions leads to nuclear accumulation of ACTN4 along with disruption of interaction with structures of the actin cytoskeleton (Fig.3 B). The localisation of protein variant with the deletion of C-terminal part of ACTN4 is very similar to the intracellular localisation of ACTN4Fl with the exception of diffused distribution through the cytoplasm along with binding to actin structures (Fig.3 B). These results are in keeping with a previous report, showing the presence of two nuclear export signals in ACTN4 sequence inside calponin-homology and SR domains [11].
Next, we decided to test whether ACTN4 localisation changes in response to stimuli from extracellular matrix (ECM). Thus, we spread transfected HEK293T cells on fibronectin. The cell spreading on fibronectin leads to a predominantly cytoplasmic localisation of the ACTN4delN and ACTN4delSR1-4 deletion mutants (Fig.3 C). This intracellular distribution is opposite to the one observed in cells cultured on poly-D-lysine. These protein variants do not co-localise along the filaments of actin cytoskeleton, but, at the same time, they predominantly localise outside the nucleus. The localisation of ACTN4Fl and ACTN4delC on fibronectin is very similar to that observed on poly-D-lysine. The ACTN4delC protein variant is predominantly localised in cytoplasm except weaker co-localisation with stress-fibres. Taking together, these data demonstrate that the N-terminal and SR domains are critical for the nuclear/cytoplasmic distribution of ACTN4 and F-actin binding in response to ECM cell signalling.
Considering the abovementioned data, we next examined the significance of ACTN4 dimerisation for its ability to activate MMP-3 gene expression. HEK293T cells were transiently transfected with ACTN4delN, ACTN4delSR1-4 and RelA/p65 expression plasmids followed by the analysis of MMP-3 expression level. Western blot analysis of cellular lysates confirms the expression of MYC-tagged ACTN4 deletion variants in HEK293T cells (Fig.4 C). We have found that variants with deletions of the spectrin or N-terminal domains activate MMP-3 gene expression in the presence of RelA/p65 (Fig.4 A, B), however to a slightly less extent than the full-length ACTN4 protein. These results indicate that ACTN4 domains responsible for binding to F-actin and homodimerisation are not critical for co-activation of RelA/65 in respect to MMP-3 gene expression.
Figure 4. Over-expression of ACTN4delN and ACTN4delSR1-4 enhances activation of MMP-3 expression by RelA/p65.
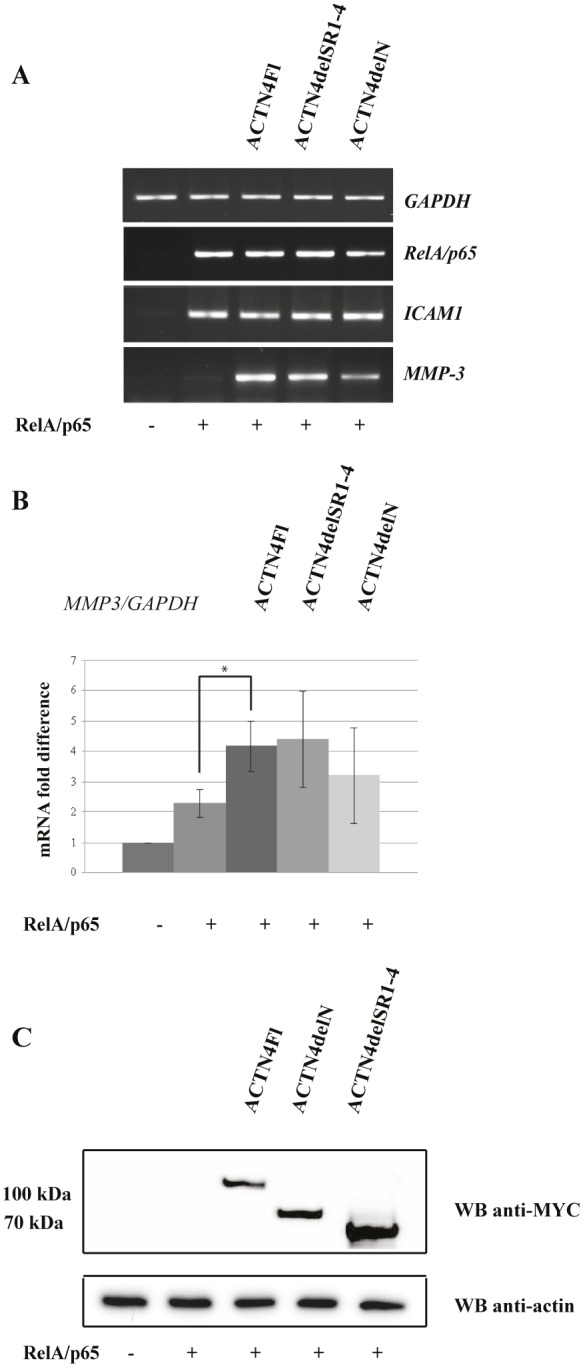
(A) The RT-PCR analysis of GAPDH, RelA/p65, MMP-3, ICAM1 expression level in 67 hours after transfection of HEK293T cells with ACTN4 deletion variants and/or RelAp65. (B) Quantification data of RelA/p65 and ACTN4 deletion variants co-expression. The data represent average fold difference of MMP-3 mRNA level to GAPDH mRNA level of at least three independent experiments, ±SD. (*P<0.01 for MMP-3 for ACTN4Fl+RelA/p65 versus RelA/p65) (C) Western blot analysis of ACTN4 mutants expressed in HEK293T cells using MYC and actin specific antibodies.
DISCUSSION
In the present study, we have demonstrate that ectopic expression of ACTN4 with the RelA/p65 subunit of transcription factor NF-kB, but not ACTN4 alone, potentiates transactivation of the mouse c-fos gene by RelA/p65. In addition, ACTN4 co-activates RelA/p65-mediated gene expression of the matrix metalloproteinases MMP-3 and MMP-1 genes, but does not affect expression of other RelA/p65 target genes, including BAX, TNC, FGF8, PTGS2, ICAM1 and FN1. Moreover, we demonstrate that ACTN4 does not potentiate nuclear accumulation of RelA/p65. Alteration of NF-kB activity via post-translational modifications without inducing nuclear translocation has been demonstrated [54, 55, 56, 57]. Apparently, ACTN4 co-activates RelA/p65-dependent transcription by an unknown mechanism.
It has previously been shown that ACTN4 can be a co-activator of nuclear receptors like ER alpha, RAR, PPAR gamma [17, 18] and transcription factor MEF2 [16]. Along with these observations, our data demonstrate that ACTN4 can be a co-activator/positive regulator for various transcription factors, including RelA/p65. It is quite possible that a common mechanism, which is yet to be uncovered, is involved in all these cases. In addition, we have shown that only MMP-3 and MMP-1, but not MMP-2 and MMP-9 are directly regulated by ACTN4. Similarly, such regulation of different subtypes of MMPs was also observed in recent studies on the impact of oxidative stress and heat shock on cells [51, 58]. The fact that ACTN4 acts as a RelA/p65 co-activator in respect to some genes but not to the others suggests that the requirement in ACTN4 may be promoter-specific.
Nuclear function of ACTN4 as a transcriptional co-activator is likely associated with its cellular distribution. In particular, this effect has been demonstrated for regulation of the Fshb gene via gonadotropin-releasing hormone [59]. Translocation of ACTN4 into the nucleus occurs in spite of an absence of a classic nuclear localisation signal. Recently, two signals of ACTN4 nuclear export were determined in the middle part of calponin-homology domains and spectrin regions. It has been proposed that nuclear import of ACTN4 can be mediated by direct interaction of spectrin region with the nuclear pore complex [11]. Here, we analysed the intracellular distribution of ACTN4 with deletions of the amino-, carboxyl- and SR domains in cells cultured either on poly-D-lysine or on fibronectin. Interestingly, all ACTN4 deletion mutants retained their ability to enter the nucleus in cells on poly-D-lysine. These data indicate that the SR-dependent nuclear import may be not the only mechanism mediating ACTN4 nuclear translocation. Consequently, ACTN4 may contain more than one sequence responsible for nuclear import. Moreover, nuclear accumulation of ACTN4delN and ACTN4delSR1-4 mutants may suggest that these regions contain sequences that keep ACTN4 in the cytoplasm, probably due to their involvement into F-actin binding.
Our immunofluorescence data showed that ACTN4delN and ACTN4delSR1-4 variants displayed nuclear localisation. Therefore, these mutants were selected for analysis of their ability to activate MMP-3 gene expression. According to our results, both protein variants are able to co-activate the MMP-3 gene expression, although to a lesser extent in comparison to the full-length ACTN4 protein. These data also suggest that dimerisation of ACTN4 and its binding to nuclear actin are not critical for RelA/p65-mediated gene expression. Thus, we speculate that the nuclear role of ACTN4 in regulation of transcription factor activity differs from its function in the cytoplasm and does not depend on the actin binding.
MATERIALS AND METHODS
Cell cultures and antibodies
A431 and HEK293T cell lines (Cell Culture Collection, Institute of Cytology, Russian Academy of Sciences) were cultured in complete Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Sigma) in 5% CO2 at 37°C.
The following antibodies were used in this study: anti-MYC-tag (M4439, Sigma), anti-RelA/p65 (sc-372 and sc-8008 AC, Santa Cruz), anti-actin (AC-17, Sigma), anti-hnRNP A2/B1 (R4653, Sigma), anti-GAPDH (2118, Cell Signaling), Alexa 488-conjugated anti-mouse antibodies (A21200, Invitrogen), HRP-conjugated anti-mouse antibodies (A9044, Sigma).
Plasmid constructions
Total RNA was purified from A431 cells using TRIzol Reagent (Invitrogen). The cDNA synthesis was performed by RevertAid – First Strand cDNA Synthesis Kit (Fermentas) according to manufacturer’s protocol. Full length ACTN4 cDNA (ACTN4Fl, 1-2736 nt from start codon) and deletion variants were amplified with primers listed in Table 1 by Pfu DNA-polymerase (Fermentas), and cloned into pCS2MT expression vector. ACTN4 deletion variants were constructed by PCR from ACTN4Fl cDNA: deletion of spectrin-homology domains (ACTN4delSR1-4, 293-753 a.a.); deletion of EF-hand domains (ACTN4delC, 753-911, a.a.); deletion of N-terminus, including ABD (ACTN4delN, 1-295 a.a.).
Table 1. Oligonucleotides used for generation of ACTN4 deletion variants.
| ACTN4 variant | Primer sequence (5’→3’) |
|---|---|
| ACTN4Fl | Forward GGAATTCTATGGTGGACTACCACGCGGCG Reverse GCTCTAGATCACAGGTCGCTCTCGCCATACAAG |
| ACTN4delSR1-4 | Forward CAGATCCTCACCCGCGACGCCAAG Reverse CTCGTTCTCTTGGTTGACAGCCAGCACC |
| ACTN4delN | Forward GGAATTCGATGGAGGACTACGAGAAGCTGGC Reverse GCTCTAGATCACAGGTCGCTCTCGCCATACAAG |
| ACTN4delC | Forward GGAATTCTATGGTGGACTACCACGCGGCG Reverse GCTCTAGATCAGTTCTCCACCTCGTTGATGGTGC |
RT-PCR
Expression of RelA/p65-dependent genes was established by semi-quantitative polymerase chain reaction. cDNA was generated from 2.5 ug of total RNA using RevertAid – First Strand cDNA Synthesis Kit (Fermentas). PCR was performed in 20 ul reaction volume containing 2 ul of cDNA, 1 pmol of forward and reverse primers, 2 mM MgCl2, and 0.2 U Taq-polymerase (Fermentas). The primers used for semi-quantitative PCR are listed in Table 2.
Table 2. Oligonucleotides used for amplification of cDNAs.
| Gene | Primer sequence (5’→3’) |
|---|---|
| Forward Reverse | |
| GAPDH | CCATCTTCCAGGAGCGAGA GGCAGTGATGGCATGGACTGT |
| 18S | AAACGGCTACCACATCCAAG CAATTACAGGGCCTCGAAAG |
| RelA/p65 | CGAATGGCTCGTCTGTAGTG TGGTGGTATCTGTGCTCCTC |
| ACTN4 | GGGCAGAAGAGATTGTGGAC TTGTTCAGGTTGGTGACAGG |
| ICAM1 | CACAGTCACCTATGGCAACG CTGAGACCTCTGGCTTCGTC |
| TNC | CTCTGGTGCTGAACGAACTG GGAAACTGTGAACCCGTAGG |
| FN1 | CTACCAAGGCTGGATGATGG TGTGCCTCTCACACTTCCAC |
| BAX | CATGTTTTCTGACGGCAACT GGAGGAAGTCCAATGTCCAG |
| PTGS2 | TGGCTACAAAAGCTGGGAAG AACTGATGCGTCAAGTGCTG |
| FGF8 | GGACACCTTTGGAAGCAGAG CCCTCGTACTTGGCATTCTG |
| MMP-3 | TGCTTTGTCCTTTGATGCTG CCAGCTCGTACCTCATTTCC |
| MMP-9 | GCCAGTCCACCCTTGTGCTCTT TCGGGCAGGGACAGTTGCTT |
| MMP-1 | TTCGGGGAGAAGTGATGTTC TCCTTGGGGTATCCGTGTAG |
| MMP-2 | GCTCAGATCCGTGGTGAGAT GGTGCTGGCTGAGTAGATCC |
RT-PCR quantification was done by densitometry analysis of Quantity One 1-D software. Statistical comparisons were performed using Student’s t-test of at least three independent experiments.
Immunofluorescence
HEK293T cells were transfected with expression plasmids coding full-length ACTN4 (ACTN4Fl) and deletion variants (ACTN4delSR1-4, ACTN4delN, ACTN4delC) with Turbofect reagent (Fermentas). Cover slips for cell spreading were covered with poly-D-lysine hydro bromide (Sigma) or fibronectin (Fluka) according to manufacturer’s protocol. Following 24 hours after transfection, HEK293T cells were spread on poly-D-lysine for 24 hours or fibronectin for 1 hour. Cells were fixed with 4% paraformaldehyde in PBS at room temperature for 15 minutes, and permeabilized in 0.1% Triton X-100 for 6 minutes. Protein intracellular localisation was detected by specific primary antibodies to MYC-tag and secondary Alexa 488-conjugated anti-mouse antibodies. The cytoskeleton and nucleus were visualised with rhodamine-phalloidine (Invitrogen) and 4,6-diamidino-2-phenylindole (DAPI, Sigma), respectively. Images were acquired using confocal microscope Leica TCS SP5.
Luciferase assay
A431 cells were seed in 6*104 on 24-well plates a day before transfection. Transfection was performed using Lipofectamine 2000 (Invitrogen) according to manufacturer’s instruction. Total amount of DNA was 3 ug/well. The pfLUC reporter construct with the luciferase gene controlled by the minimal (from -56 to +109 nt) c-fos mouse promoter was constructed as previously described [49]. The empty vector pcDNA3.0 was used to maintain an equal amount of DNA in all wells. Thirty hours after transfection, cells were lysed in 1X lysis buffer (6.25 mM Tris-HCl pH7.8, 10 mM DTT, 10 mM EDTA, 50% Glycerol, 5% Trition X-100). Luciferase activity was measured using kit for luciferase assay (BioVision). Activity of β-galactosidase was used to correct differences in transfection efficiency between replicates. The data is representative of at least three independent repeats. Statistical comparisons were performed using Student’s t-test (P < 0.02).
Protein extraction and immunoprecipitation
Nuclear and cytoplasm proteins were extracted as described previously [15]. Briefly, the cells were scrubbed off the culture plates, resuspended in a lysis buffer (0.32 M sucrose, 2 mM MgCl2, 0.1 mM EDTA, 10 mM Tris–HCl, pH 7.9, 1 mM DTT, 0.4 mM PMSF), and incubated until the cells were lysed. The nuclei were purified by centrifugation through 0.5 M sucrose. Anti-RelA/p65 agarose conjugated antibodies (sc-8008 AC) was added to the precleared cytoplasm extracts (1:200 volume to volume ratio). Following overnight incubation, to control extracts without antibodies Protein G Sepharose beads were added for 2 h at +4°C.
SDS-PAGE and Western blotting
SDS-PAGE and Western blotting were performed as described elsewhere [60]. Primary antibodies were diluted in 1X PBS with 0.1% Tween-20 (PBST). Secondary anti-mouse antibodies conjugated to HRP were diluted 1:10000 in PBST. Detection of proteins was performed using Chemidoc (BioRad) with ECL mixture (1.25 mM luminol, 0.2 mM p-coumaric acid, 0.01 % hydrogen peroxide in 150 mM Tris-HCl pH 8.8).
Acknowledgments
This work was supported by FEBS Collaborative Experimental Scholarship for Central and Eastern Europe, grant of RFBR (12-04-32194 mol_a, 10-04-00174, 12-04-32252 mol_a), Russian Government Programme for the Recruitment of the leading scientists into the Russian Institutions of Higher Education (11.G34.31.0069), grant for PhD students supported by Federal Target Program – Scientific and Pedagogical staff of innovative Russia to 2009-2013 years (П174) and Federal Programme for Scientific training in Innovative Russia (№8280).
REFERENCES
- 1.Honda K, Yamada T, Endo R, Ino Y, Gotoh M, Tsuda H, Yamada Y, Chiba H, Hirohashi S. Actinin-4, a novel actin-bundling protein associated with cell motility and cancer invasion. J Cell Biol. 1998;140:1383–1393. doi: 10.1083/jcb.140.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jayadev R, Kuk CY, Low SH, Hori MM. Calcium sensitivity of α-actinin is required for equatorial actin assembly during cytokinesis. Cell Cycle. 2012;11:1929–1937. doi: 10.4161/cc.20277. [DOI] [PubMed] [Google Scholar]
- 3.Gross SR. Actin binding proteins. Their ups and downs in metastatic life. Cell Adhes Migr. 2013:7. doi: 10.4161/cam.23176. dx.doi.org/10.4161/cam.23176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shao H, Wang JHC, Pollak MR, Wells A. a-Actinin-Is Essential for Maintaining the Spreading, Motility and Contractility of Fibroblasts. Plos One. 2010;5:e13921. doi: 10.1371/journal.pone.0013921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eckert MA, Yang J. Targeting invadopodia to block breast cancer metastasis. Oncotarget. 2011;2:562–568. doi: 10.18632/oncotarget.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Djinovic-Carugo K, Gautel M, Ylanne J, Young P. The spectrin repeat: a structural platform for cytoskeletal protein assemblies. FEBS Letters. 2002;513:119–123. doi: 10.1016/s0014-5793(01)03304-x. [DOI] [PubMed] [Google Scholar]
- 7.Christerson LB, Vanderbilt CA, Cobb MH. MEKK1 interacts with alpha-actinin and localizes to stress fibers and focal adhesions. Cell Motil Cytoskeleton. 1999;43:186–198. doi: 10.1002/(SICI)1097-0169(1999)43:3<186::AID-CM2>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez AM, Otey C, Edlund M, Jones JC. Interactions of hemidesmosome component and actinin family members. J Cell Sci. 2001;114:4197–4206. doi: 10.1242/jcs.114.23.4197. [DOI] [PubMed] [Google Scholar]
- 9.Patrie KM, Drescher AJ, Welihinda A, Mundel P, Margolis B. Interaction of two actin binding proteins, synaptopodin and α-actinin-4, with the tight junction protein MAGI-1. J Biol Chem. 2002;277:30183–30190. doi: 10.1074/jbc.M203072200. [DOI] [PubMed] [Google Scholar]
- 10.Broderick MJF, Winder SJ. Spectrin, α-actinin, and dystrophin. Adv Prot Chem. 2005;70:203–233. doi: 10.1016/S0065-3233(05)70007-3. [DOI] [PubMed] [Google Scholar]
- 11.Kumeta M, Yoshimura SH, Harata M, Takeyasu K. Molecular mechanisms underlying nucleocytoplasmic shuttling of actinin-4. J Cell Sci. 2010;123:1020–1030. doi: 10.1242/jcs.059568. [DOI] [PubMed] [Google Scholar]
- 12.Poch MT, Al-Kassim L, Smolinski SM, Hines RN. Two distinct classes of CCAAT box elements that bind nuclear factor-Y/α-actinin-4: potential role in human CYP1A1 regulation. Toxicol App Pharmacol. 2004;199:239–250. doi: 10.1016/j.taap.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 13.Liu QY, Lei JX, Le Blanc J, Sodja C, Ly D, Charlebois C, Walker PR, Yamada T, Hirohashi S, Sikorska M. Regulation of DNaseY activity by actinin-a4 during apoptosis. Cell Death Differ. 2004;11:645–654. doi: 10.1038/sj.cdd.4401401. [DOI] [PubMed] [Google Scholar]
- 14.Babakov VN, Petukhova OA, Turoverova LV, Kropacheva IV, Tentler DG, Bolshakova AV, Podolskaya EP, Magnusson K-E, Pinaev GP. RelA/NF-kB transcription factor associates with α-actinin-4. Exper Cell Res. 2008;314:1030–1038. doi: 10.1016/j.yexcr.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Khotin M, Turoverova L, Aksenova V, Barlev N, Borutinskaite VV, Vener A, Bajenova O, Magnusson K-E, Pinaev G, Tentler D. Proteomic analysis of ACTN4-interacting proteins reveals it’s a putative involvement in mRNA metabolism. Biochem Biophys Res Com. 2010;397:192–196. doi: 10.1016/j.bbrc.2010.05.079. [DOI] [PubMed] [Google Scholar]
- 16.Chakraborty S, Reineke EL, Lam M, Li X, Liu Y, Gao C, Khurana S, Kao HY. Alpha-actinin 4 potentiates myocyte enhancer factor-2 transcription activity by antagonizing histone deacetylase 7. J Biol Chem. 2006;281:35070–35080. doi: 10.1074/jbc.M602474200. [DOI] [PubMed] [Google Scholar]
- 17.Khurana S, Chakraborty S, Lam M, Liu Y, Su YT, Zhao X, Saleem MA, Mathieson PW, Bruggeman LA, Kao HY. Familial focal segmental glomerulosclerosis (FSGS)-linked α-actinin 4 (ACTN4) protein mutants lose ability to activate transcription by nuclear hormone receptors. J Biol Chem. 2012;287:12027–12035. doi: 10.1074/jbc.M112.345421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khurana S, Chakraborty S, Zhao X, Liu Y, Guan D, Lam M, Huang W, Yang S, Kao HY. Identification of a novel LXXLL motif in α-actinin 4-spliced isoform that is critical for its interaction with estrogen receptor α and co-activators. J Biol Chem. 2012;287:35418–35429. doi: 10.1074/jbc.M112.401364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dixit D, Sharma V, Ghosh S, Mehta VS, Sen E. Inhibition of Casein kinase-2 induces p53-dependent cell cycle arrest and sensitizes glioblastoma cells to tumor necrosis factor (TNFα)-induced apoptosis through SIRT1 inhibition. Cell Death Dis. 2012;3:e271. doi: 10.1038/cddis.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walsby E, Pearce L, Burnett AK, Fegan C, Pepper C. The Hsp90 inhibitor NVP-AUY922-AG inhibits NF-κB signaling, overcomes microenvironmental cytoprotection and is highly synergistic with fludarabine in primary CLL cells. Oncotarget. 2012;3:525–34. doi: 10.18632/oncotarget.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chefetz I, Holmberg JC, Alvero AB, Visintin I, Mor G. Inhibition of Aurora-A kinase induces cell cycle arrest in epithelial ovarian cancer stem cells by affecting NFĸB pathway. Cell Cycle. 2011;10:2206–14. doi: 10.4161/cc.10.13.16348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nogueira L, Ruiz-Ontañon P, Vazquez-Barquero A, Moris F, Fernandez-Luna JL. The NFκB pathway: a therapeutic target in glioblastoma. Oncotarget. 2011;2:646–53. doi: 10.18632/oncotarget.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Markert EK, Levine AJ, Vazquez A. Proliferatio and tissue remodeling in cancer: the hallmarks revisited. Cell Death Dis. 2012;3:e397. doi: 10.1038/cddis.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaughan S, Jat PS. Deciphering the role of nuclear factor-kB in cellular senescence. Aging. 2011;3:913–9. doi: 10.18632/aging.100390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hasler U. An example of functional interaction between NFAT5/TonEBP and nuclear factor-kB by hypertonic stress: aquaporin-2 transcription. Cell Cycle. 2011;10:364–5. doi: 10.4161/cc.10.3.14520. [DOI] [PubMed] [Google Scholar]
- 26.Silke J, Vince JE. IAPs, TNF, inflammation and Jürg Tschopp; a personal perspective. Cell Death Differ. 2012;19:1–4. doi: 10.1038/cdd.2011.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim SY, Jeong S, Jung E, Baik KH, Chang MH, Kim SA, Shim JH, Chun E, Lee KY. AMP-activate protein kinase-α1 as an activating kinase of TGF-β-activated kinase 1 has a key role in inflammatory signals. Cell Death Dis. 2012;3:e357. doi: 10.1038/cddis.2012.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papi A, Guarnieri T, Storci G, Santini D, Ceccarelli C, Taffurelli M, De Carolis S, Avenia N, Sanguinetti A, Sidoni A, Orlandi M, Bonafé M. Nuclear receptors agonists exert opposing effects on the inflammation dependent survival of breast cancer stem cells. Cell Death Differ. 2012;7:1208–19. doi: 10.1038/cdd.2011.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mora E, Guglielmotti A, Biondi G, Sassone-Corsi P. Bindarit: an anti-inflammatory small molecule that modulates the NFκB pathway. Cell Cycle. 2012;11:159–69. doi: 10.4161/cc.11.1.18559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bellet MM, Zocchi L, Sassone-Corsi P. The RelB subunit of NFκB acts as a negative regulator of circadian gene expression. Cell Cycle. 2012;11:3304–11. doi: 10.4161/cc.21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandow JJ, Jabbour AM, Condina MR, Daunt CP, Stomski FC, Green BD, Riffkin CD, Hoffmann P, Guthridge MA, Silke J, Lopez AF, Ekert PG. Cytokine receptor signaling activates an IKK-dependent phosphorylation of PUMA to prevent cell death. Cell Death Differ. 2012;19:633–41. doi: 10.1038/cdd.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lippens S, Lefebvre S, Gilbert B, Sze M, Devos M, Verhelst K, Vereecke L, Mc Guire C, Guérin C, Vandenabeele P, Pasparakis M, Mikkola ML, Beyaert R, Declercq W, van Loo G. Keratinocyte-specific ablation of the NF-κB regulatory protein A20 (TNFAIP3) reveals a role in the control of epidermal homeostasis. Cell Death Differ. 2011;18:1845–53. doi: 10.1038/cdd.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saccani S. All p65-containing dimers are not equal. Cell Cycle. 2012;11:646–7. doi: 10.4161/cc.11.4.19279. [DOI] [PubMed] [Google Scholar]
- 34.Criollo A, Chereau F, Malik SA, Niso-Santano M, Mariño G, Galluzzi L, Maiuri MC, Baud V, Kroemer G. Autophagy is required for the activation of NFκB. Cell Cycle. 2012;11:194–9. doi: 10.4161/cc.11.1.18669. [DOI] [PubMed] [Google Scholar]
- 35.Moujalled DM, Cook WD, Lluis JM, Khan NR, Ahmed AU, Callus BA, Vaux DL. In mouse embryonic fibroblasts, neither caspase-8 nor cellular FLICE-inhibitory protein (FLIP) is necessary for TNF to activate NF-κB, but caspase-8 is required for TNF to cause cell death, and induction of FLIP by NF-κB is required to prevent it. Cell Death Differ. 2012;19:808–15. doi: 10.1038/cdd.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rauert H, Stühmer T, Bargou R, Wajant H, Siegmund D. TNFR and TNFR2 regulate the extrinsic apoptotic pathway in myeloma cells by multiple mechanisms. Cell Death Dis. 2011;2:e194. doi: 10.1038/cddis.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan YH, Yu Y, Mao RF, Tan XJ, Xu GF, Zhang H, Lu XB, Fu SB, Yang J. USP4 targets TAK1 to downregulate TNFα-induced NF-κB activation. Cell Death Differ. 2011;18:1547–60. doi: 10.1038/cdd.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vanlangenakker N, Bertrand MJ, Bogaert P, Vandenabeele P, Vanden Berghe T. TNF-induced necroptosis in L929 cells is tightly regulated by multiple TNFR1 complex I and II members. Cell Death Dis. 2011;2:e230. doi: 10.1038/cddis.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song S, Choi K, Ryu SW, Kang SW, Choi C. TRAI promotes caspase-dependent pro-inflammatory responses via PKCδ activation by vascular smooth muscle cells. Cell Death Dis. 2011;2:e223. doi: 10.1038/cddis.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mange LA, May MJ. The NF-kB paradox: RelB induces and inhibits gene expression. Cell Cycle. 2011;10:6–7. doi: 10.4161/cc.10.1.14291. [DOI] [PubMed] [Google Scholar]
- 41.Ge R, Wang Z, Zeng Q, Xu X, Olumi AF. F-box protein 10, an NF-κB-dependent anti-apoptotic protein, regulates TRAIL-induced apoptosis through modulating c-Fos/c-FLIP pathway. Cell Death Differ. 2011;18:1184–95. doi: 10.1038/cdd.2010.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Janssens S, Tinel A. The PIDDosome, DNA-damage-induced apoptosis and beyond. Cell Death Differ. 2012;19:13–20. doi: 10.1038/cdd.2011.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khandelwal N, Simpson J, Taylor G, Rafique S, Whitehouse A, Hiscox J, Stark LA. Nucleolar NF-κB/RelA mediates apoptosis by causing cytoplasmic relocalization of nucleophosmin. Cell Death Differ. 2011;18:1889–903. doi: 10.1038/cdd.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shu G, Tang Y, Zhou Y, Wang C, Song JG. Zac1 is a histone acetylation-regulated NF-κB suppressor that mediates histone deacetylase inhibitor-induced apoptosis. Cell Death Differ. 2011;18:1825–35. doi: 10.1038/cdd.2011.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heinz LX, Rebsamen M, Rossi DC, Staehli F, Schroder K, Quadroni M, Gross O, Schneider P, Tschopp J. The death domain-containing protein Unc5CL is a novel MyD88-independent activator of the pro-inflammatory IRAK signaling cascade. Cell Death Differ. 2012;19:722–31. doi: 10.1038/cdd.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Darding M, Meier P. IAPs: guardians of RIPK1. Cell Death Differ. 2012;19:58–66. doi: 10.1038/cdd.2011.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hayden MS, Goch S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 48.Are AF, Galkin VE, Pospelova TV, Pinaev GP. The p65/RelA subunit of NFkappaB interacts with actin-containing structures. Exp Cell Res. 2000;256:533–544. doi: 10.1006/excr.2000.4830. [DOI] [PubMed] [Google Scholar]
- 49.Saksela K, Baltimore D. Negative Regulation of Immunoglobulin Kappa Light-Chain Gene Transcription by a Short Sequence Homologous to the Murine Bi Repetitive Element. Mol Cell Biol. 1993;13:3698–3705. doi: 10.1128/mcb.13.6.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kohman A, Kammere U, Kapp M, Dietl J, Anacker J. Expression of matrix metalloproteinases (MMPs) in primary human breast cancer and breast cancer cell lines: New findings and review of the literature. BMC Cancer. 2009 doi: 10.1186/1471-2407-9-188. doi: 10.1186/1471-2407-9-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alge-Prigliner CS, Kreutzer T, Obholzer K, Wolf A, Mempl M, Kernt M, Kampik A, Priglinger SG. Oxidative Stress-Mediated Induction of MMP-1 and MMP-3 in Human RPE Cells. Inv Ophtalmol Vis Sci. 2009;50:5495–5503. doi: 10.1167/iovs.08-3193. [DOI] [PubMed] [Google Scholar]
- 52.Bond M, Chase AJ, Baker AH, Newby AC. Inhibition of transcription factor NF-kB reduces matrix metalloproteinase-1, -3, and -9 production by vascular smooth muscle cells. Cardiovasc Res. 2001;50:556–565. doi: 10.1016/s0008-6363(01)00220-6. [DOI] [PubMed] [Google Scholar]
- 53.Chen J-H, Lin H-H, Chiang T-A, Hsu J-D, Ho H-H, Lee Y-C, Wang C-J. Gaseous Nitrogen Oxide Promotes Human Lung Cancer Cell Line A549 Migration, Invasion, and Metastasis via iNOS-Mediated MMP-2 Production. Toxicol Sci. 2008;106:364–375. doi: 10.1093/toxsci/kfn195. [DOI] [PubMed] [Google Scholar]
- 54.Perkins ND, Gilmore TD. Good cop, bad cop: the different faces of NF-kB. Cell Death Differ. 2006;13:759–772. doi: 10.1038/sj.cdd.4401838. [DOI] [PubMed] [Google Scholar]
- 55.Mattson MP, Meffert MK. Roles for NF-kB in nerve cell survival, plasticity, and disease. Cell Death Differ. 2006;13:852–860. doi: 10.1038/sj.cdd.4401837. [DOI] [PubMed] [Google Scholar]
- 56.Lanzillotta A, Sarnico I, Ingrassia R, Boroni F, Branca C, Benarese M, Faraco G, Blasi F, Chiarugi A, Spano P, Pizzi M. The acetylation of RelA in Lys310 dictates the NF-κB-dependent response in post-ischemicinjury. Cell Death Dis. 2010;12:e107. doi: 10.1038/cddis.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Manna SK, Manna P, Sarkar A. Inhibition of RelA phosphorylation sensitizes apoptosis in constitutive NF-kappaB-expressing and chemoresistant cells. Cell Death Differ. 2007;14:158–170. doi: 10.1038/sj.cdd.4401929. [DOI] [PubMed] [Google Scholar]
- 58.Park C-H, Lee MJ, Ahn J, Kim S, Kim HH, Kim KH, Eun HC, Chung JH. Heat Shock-Induced Matrix Metalloproteinase (MMP)-1 and MMP-3 Are Mediated through ERK and JNK Activation and via an Autocrine Interleukin-6 Loop. J Invest Dermatol. 2004;123:1012–1019. doi: 10.1111/j.0022-202X.2004.23487.x. [DOI] [PubMed] [Google Scholar]
- 59.Yu H, Li Z, Ghosh D, Lim TK, He Y, Lin Q. α-Actinin4 nuclear translocation mediates gonadotropin-releasing hormone stimulation of follicle-stimulating hormone β-subunit gene transcription in LβT2 cells. FEBS Letters. 2012;586:1466–71. doi: 10.1016/j.febslet.2012.03.067. [DOI] [PubMed] [Google Scholar]
- 60.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]