Abstract
Postmortem studies of synapses in human brain are problematic due to the axial resolution limit of light microscopy and the difficulty preserving and analyzing ultrastructure with electron microscopy. Array tomography overcomes these problems by embedding autopsy tissue in resin and cutting ribbons of ultrathin serial sections. Ribbons are imaged with immunofluorescence, allowing high-throughput imaging of tens of thousands of synapses to assess synapse density and protein composition. The protocol takes approximately 3 days per case, excluding image analysis, which is done at the end of the study. Parallel processing for transmission electron microscopy (TEM) using a protocol modified to preserve structure in human samples allows complimentary ultrastructural studies. Incorporation of array tomography and TEM into brain banking is a potent way of phenotyping synapses in well-characterized clinical cohorts to develop clinico-pathological correlations at the synapse level. This will be important for research in neurodegenerative disease, developmental diseases, and psychiatric illness.
INTRODUCTION
During ageing and neurodegenerative diseases, synapses become dysfunctional and degenerate, contributing to cognitive decline and dementia. Similarly in developmental disorders such as autism and fragile × syndrome, synapses are dysfunctional due to developmental abnormalities. The highly plastic nature of synapses makes them appealing targets for therapeutic interventions1, 2. The study of synapses in the human brain is thus of great interest, however has been hampered in the past by several factors.
Methods to study synapses
Electron microscopy (EM) is the only method for obtaining ultrastructural information about synapses. However, preservation of ultrastructure postmortem in human brain is often poor if the methodology used has not been optimized for human postmortem material. In addition the technique can be prohibitively time-consuming and technically difficult. Moreover, serial section stereology is required to generate accurate EM synapse counts3, and immuno-EM techniques for labeling synaptic proteins are often hampered by poor antibody penetration and high levels of non-specific staining4.
Using fluorescent immunostaining methods to label pre- and postsynaptic structures for counting and determining size is desirable, but synapses are too small to be effectively imaged using traditional light microscopy (including confocal and multiphoton imaging). In human neocortex, synapse apposition length is approximately 350nm5. While the lateral (x–y direction) resolution of light microscopes is adequate for imaging synapses (~250nm), the axial, or z-resolution is too poor (~1000nm)6. For structural or calcium imaging studies of postsynaptic dendritic spines in animal models, this confound can be overcome by labeling a subset of synapses or dendritic spines with a fluorescent marker and imaging only the spines that protrude laterally from the dendrite in the imaging field7, 8. However, this technique is not applicable to human material. Golgi silver staining fills a subset of neurons and is feasible to perform on human tissue allowing similar spine density analyses, but it does not allow for immuno-labeling to investigate synaptic proteins. Synaptic protein intensity levels in thick sections labeled with immunohistochemistry9, 10, or measured in western blots or ELISA of homogenized tissue11–13, have been used as an approximation of synapse density in humans, but these do not reveal information about changes in synapse size, composition, or spatial relationships to pathological lesions. Advances in biochemical isolation of synapses and their analysis have made examining protein composition of synapses more feasible in human samples but these preparations still do not allow investigation of the other relevant parameters14.
Development of the protocol
Recently, Micheva and Smith developed a technique called array tomography which they applied to study synapses in rodent brain15, 16. Array tomography overcomes the axial resolution obstacle by physically cutting fixed brain specimens into ribbons of 70nm serial sections (Fig. 1). These ribbons are then stained with standard immunofluorescence techniques and images taken of serial sections to allow reconstruction of three-dimensional volumes of tissue in great detail. Importantly, this technique allows stripping off antibodies and re-staining with multiple markers to determine the protein composition present at each synapse.16 Along with protein signatures, synapse density and size can be calculated from the three dimensional datasets. Array tomography has resolution in the z-direction equivalent to electron microscopy and allows high throughput imaging of thousands of synapses with many more markers than was possible previously. Using this technique in a mouse model of Alzheimer disease, we were able to quantify synapse loss around senile plaques and to determine that oligomeric amyloid beta is present at postsynaptic densities and correlates with synapse shrinkage and loss17.
Fig. 1.
Principles of array tomography. The resolution of light and confocal microscopes is limited in the z plane to ~1000nm (a) which is too large to accurately image synapses. The volume imaged in a single optical section with confocal microscopy (b) can contain multiple objects (numbered 1–2) at the same x–y position, which are seen as a single puncta on the output of the confocal image (c). Array tomography overcomes this z-resolution problem by physically sectioning the tissue at a resolution smaller than a single synapse (d). An example of a 10 µm thick cryostat section stained with PSD95 (c) shows the haze of staining and lack of discrete synapses, while a single array tomography section stained with PSD95 (d) shows clear individual puncta. Scale bar represents 5 µm.
Because array tomography is applied to postmortem rodent tissue, we adapted it for use in human brain18, 19. To preserve antigenicity of proteins and prevent freezing-induced destruction of membranes, best results are obtained when fresh tissue is prepared at autopsy. We have therefore inserted preparation of array tomography tissue into the stream of collecting tissue in the Massachusetts Alzheimer Disease Research Center brain bank and the Medical Research Council UK (MRC) funded University of Edinburgh brain bank. Collecting array tomography tissue allows detailed investigation of synapse number and protein composition in human samples. Parallel processing for electron microscopy (EM), which uses much of the same equipment, allows complimentary investigation of synaptic ultrastructure when tissue preservation is adequate. Here we detail protocols of brain banking and analysis of samples for AT and EM that have been modified specifically for human brain tissue.
During the process of brain banking, frozen specimens and fixed specimens for standard histology are collected from the same regions of the same cases as the array tomography and EM blocks. Processing the tissue in this manner allows for investigation of correlations between synapse data and biochemical, molecular, and pathological markers from the same brain. Therefore, the incorporation of array tomography and EM preparations into brain banking has the potential to allow clinico-pathological correlations to extend to the level of the synapse, which may contribute particularly useful information in the current setting of other forms of deep phenotyping of well-characterized clinical cohorts. We believe array tomography and EM are ideally incorporated whenever possible into fresh brain tissue collection, since preparation of tissue for these techniques is relatively inexpensive. Furthermore, these techniques provide opportunity for effective collaboration between multiple research groups, since dozens of blocks can be made from each region of interest and prepared blocks of brain are stable indefinitely, allowing them to be easily shared.
Applications of the Protocol
Array tomography was originally developed to study synapses in rodent brain, where it has been used to characterize the protein composition of synapses in healthy brain16, 20, 21 and to demonstrate synaptic loss and proteins involved in synaptic degeneration in Alzheimer’s disease mouse models17, 22, 23. In stargazer mice, a model of epilepsy, array tomography revealed higher perisynaptic and lower synaptic levels of AMPARs (2-amino-3-(3-hydroxy-5-methyl-isoxazol-4-yl)propanoic acid receptor)24. The technique has been extended to the study of synaptic connectivity in songbird25, 26 and zebrafish27.
In human brain, we have used array tomography to investigate synapse degeneration in Alzheimer’s disease and improved our understanding of how apolipoprotein E4 contributes to Alzheimer’s disease risk18. From the three-dimensional image stacks obtained with array tomography, synapse density (number of synapses/volume of tissue), synaptic puncta size, and colocalization of proteins of interest with pre or postsynaptic markers can be determined. Unlike biochemical isolation of synapses and immunoprecipitation experiments which require homogenization of the tissue, array tomography allows investigation of the protein composition of individual synapses in the context of the surrounding brain tissue; for example, using array tomography allows for quantification of the relationship between pathological lesions and synapse size, number, and/or composition.
Beyond synapses, array tomography is useful for examining the morphology, localization, and protein composition of many other small biological structures. It has been used to examine the microstructural changes in blood vessel walls during abdominal aortic aneurisms in mice28, the collagenous matrix of human optic nerve head29, and mitochondrial size and distribution in neurons of a tauopathy mouse model and human Alzheimer patients, where we found an association between tau pathology with and mitochondrial transport19.
Experimental Design
Array tomography (AT) experiments can be broken down into several independent stages: tissue collection, sectioning tissue, optimization of staining, staining and imaging, and image analysis (Figure 2).
Fig. 2.
Flowchart showing the timing of AT and EM experiments.
Tissue collection
For tissue preparation, we have incorporated array tomography and EM preparations into routine brain bank collection. We have managed to conduct very effective EM studies on tissues retrieved from donors with long post mortem intervals, up to 100 hours. In our experience a key element in tissue preservation for ultrastructure analysis is post mortem cold storage of the cadaver, with cold storage in a mortuary of around 4–6°C significantly reducing structural degradation. Since we focus on neurodegenerative diseases, we collect 10–12 regions per brain relevant to degeneration as shown in Figure 3. We collect approximately 10 small samples (1mm×1mm×5mm) from each region in each case. Orientation of the samples is important particularly in laminar structures such as neocortex, to ensure that all layers are present in the block facing the cutting surface. In parallel, adjacent blocks are prepared for EM to allow for ultrastuctural studies. Samples are fixed, dehydrated, and embedded then stored at room temperature (20–25 °C) until needed for studies (Fig. 4). The rest of the hemisphere from which AT and EM samples are taken is frozen in slices between aluminum plates and samples from the same regions can be used for biochemical and molecular studies30. The contralateral hemisphere is fixed and used for pathological diagnoses and disease quantification by standard neuropathologic methods e.g. Braak staging and CERAD (Consortium to Establish a Registry for Alzheimer Disease) scoring for Alzheimer pathology. EM blocks are processed for analysis of synaptic ultrastructure using modified methods (see ref31), requiring changes to the protocol with respect to fixation (see below).
Fig. 3.
Regions collected for neurodegenerative disease studies. a– superior frontal cortex, b – anterior cingulate cortex, c- superior temporal cortex, d– entorhinal cortex, e – amygdala, f - posterior superior putamen, g – dentate gyrus granular cell layer, h – CA1, i– superior cerebellar vermis, j – substantia nigra. Scale bar represents 1cm. Inset 2 cm wide.
Fig. 4.
Small 1 cm3 samples from multiple brain regions are bisected and one half dropped in AT fixative (a). After fixing for 20–30 minutes, samples are further divided into 1mm × 1mm × 5 mm blocks (b), taking care to maintain all six layers in cortical blocks with the pial surface (arrowhead) on one end and the white matter (arrow) on the other end of the long face. Samples are dehydrated and embedded in LR White resin then stored at room temperature (c). A diamond facing knife is used to make the top and bottom edges of the trapezoid surrounding the tissue block parallel. A block from substantia nigra of a Parkinsons disease patient is shown with neuromelanin-rich dopaminergic neurons visible (d). Parallel edges are essential to cutting straight ribbons of 70nm serial sections, which are silver-gold in color (e). Ribbons are mounted on a coverslip by guiding them with an eyelash to the interface of the water and coverslip, and the position of the ribbon marked on the reverse side of the coverslip with permanent marker (f).
Sectioning tissue
From each tissue block needed for an AT study, ribbons of 50–100nm thick serial sections are cut using a histo jumbo diamond knife on an ultracut microtome. Ribbons are dried onto cover slips with the thickness required for the objective used for synapse quantification (in our case 0.16–0.19 mm). For testing tissue quality and optimizing staining, short ribbons of 5–10 sections can be used. For quantifying synapse numbers and proteins present at synapses, longer ribbons of 20–50 sections are used. Even longer ribbons can be used to reconstruct larger structures such as entire neurons or volumes of cortex15 or pathological lesions such as senile plaques17 and tangles19. The maximum length of ribbon we have used is approximately 200 sections, allowing reconstruction of ~15 µm depth. Ribbons can be stained immediately after sectioning provided they are dried on the cover slip before staining begins. Alternatively, ribbons can be cut in advance, dried, and stored in dust-free boxes. The longest we have stored ribbons before staining is 6 months, but longer periods are likely to be acceptable.
Before tissue samples (blocks) are used for AT experiments, a ribbon from each block is stained with the nuclear marker DAPI (4',6-diamidino-2-phenylindole) and a synaptic marker to assess quality of the tissue. In addition, we confirm that all 6 cortical layers are identifiable in each cortical block and we stain for markers of relevant pathology and note the types of lesions present in each block (for example plaques and tangles in Alzheimer’s disease, Lewy bodies and neurites in Parkinson’s disease and Dementia with Lewy Bodies, etc). For each new experiment, staining of proteins of interest is optimized by testing antibodies on array tissue using standard immunofluorescence controls. A negative control with no primary antibody is particularly important in aged human brain samples, which may contain large numbers of autofluorescent puncta.
Optimisation of staining
Each primary antibody should be optimised for use with array tomography. Several concentrations can be initially tried along with a negative control with no primary antibody. If available, positive control tissue should be used to confirm staining. For example, to test Aβ antibodies, we use tissue from aged APP/PS1 overexpressing mice, which have accumulations of Aβ in plaques and we co-stain with a different Aβ antibody that has already been tested for array tomography. Ideal negative controls include tissue from knockout animals. For example, we validated the use of an apolipoprotein E antibody by testing it on apoE knockout mice and observed no staining.18 Several different primary antibodies can be used simultaneously if host species are different and enough fluorescence channels are available on the imaging microscope. Fluorophores on secondary antibodies should be chosen based on microscope filter sets and not overlap with fluorescence of the nuclear label (DAPI – 358 nm in our case), which is necessary to align the images. Other immunostains can also be used for alignment if stained objects are large and sparse enough to be followed across multiple sections like NeuN neuronal staining or staining of neurofibrillary tangles. We have used combinations of secondary antibodies labeled with fluorophores with 488nm (FITC), 550nm (Rhodamine/Cy3), and 650nm (Cy5/far red) absorption wavelengths to visualize 3 separate proteins on the same ribbon when the primary antibodies are raised in different species. Care should be taken when using red and far red secondary antibodies in the same session as some overlap between both emission spectra can be observed with very strong stains. Using different combinations of secondary antibodies during optimization of staining allows determination of whether there is bleed-through between these channels. More stains can also be achieved by stripping and re-probing array ribbons after imaging, as was elegantly shown by Micheva, Smith, et al. in mice16. This is also possible in human sections as demonstrated in Supplemental Fig. 1, but the quality of reimaging is likely both tissue and antibody dependent, and so we in practice reserve this technique for detailed studies of individual samples rather than for high throughput studies of dozens of cases with multiple sampling sites per case.
Staining and Imaging
Once the staining protocol is optimized for an experiment, ribbons are immunostained and image stacks acquired (Fig. 4). Staining fades quickly so ribbons are imaged within 24 hours of staining. To avoid sampling biases as much as possible, multiple blocks from each region in each case are used (2–3 per case), and multiple image stacks are acquired from each ribbon (2–3). At least 2 ribbons from different cases are stained and imaged on the same day to control for any variability in staining quality from day to day. In experiments comparing different groups (Alzheimer’s disease and non-demented elderly control cases for example), at least one ribbon from each group is included in each batch of staining and imaging. The number of cases needed varies depending on the effect size. We find an n=5–6 per group is usually sufficient to detect >20% differences in synapse density. There is a large amount of variability in synapse density both between individual cases and between sampling sites within each case. In non-demented subjects, we observe approximately 20% variability (coefficient of variance 0.2) in synapse density in layers II–III of superior temporal gyrus between cases and similarly ~20% variability between sampled regions within this cortical area within the same case, which is why we recommend extensive sampling (multiple blocks, ribbons and stacks from each case).
Image analysis
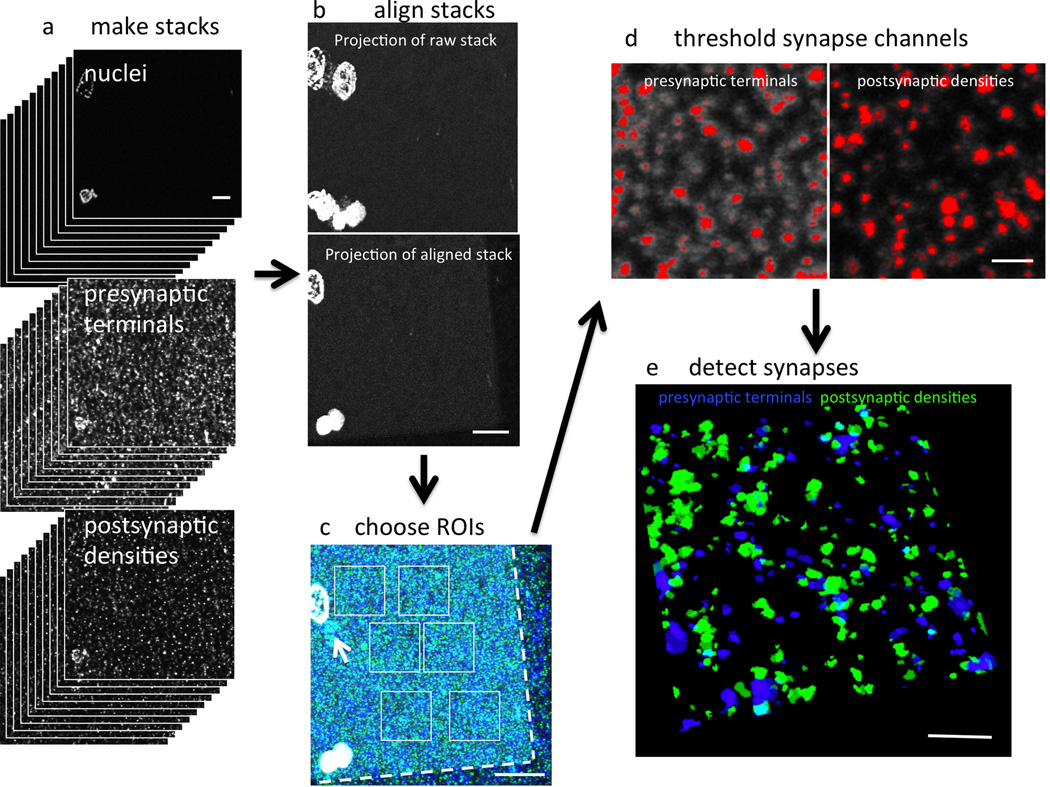
Image processing includes making stacks from the images on each section, aligning the images using nuclei (or amyloid plaques in Alzheimer’s disease samples) as fiduciary markers and applying that alignment to all other channels (Fig. 5). Regions of interest on the stack (“crops”) are then chosen for analysis. In most experiments, we choose regions of neuropil that do not contain any nuclei or capillaries. In disease cases, we also note the position of each crop relative to any pathological lesions (e.g. distance from the nearest amyloid plaque). Before image analysis, we collect all data for the experiment from the 4–5 cases per group.
Fig. 5.
Imaging array tomography ribbons. A tile scan of the entire ribbon is collected at low magnification (a). A region of interest is selected from the same place on two consecutive sections (b) using nuclei (grey) or amyloid plaques (stained green with AW7 antibody) as fiduciary markers. Then high-resolution images are taken on each section of the ribbon at the region of interest (c). Note the punctate staining of presynaptic terminals labeled red with synaptophysin immunostaining. Scale bars represent 2 mm (a), 100 µm (b), and 10 µm (c).
For image analysis, the experimenter is blind to experimental group or clinical condition (i.e. diseased versus healthy). Being blind to condition is important for our current method of analysis which requires the experimenter to choose a thresholding algorithm (in the Fiji program http://fiji.sc) in order to convert the image stacks to binary. These stacks are then analyzed with the Watershed program developed by Brad Busse and Stephen Smith15. Where possible, the same thresholding algorithm in Fiji is applied to all images in the study. However, if there is significant variability in the staining between days, different thresholding paradigms may need to be used for different days, highlighting the need to have cases from each experimental or clinical condition run each day. The Watershed program introduces a size threshold and only counts objects as synapses if they are present in more than one consecutive section. These definitions of synaptic puncta are necessary to remove non-specific staining (likely from non-specific secondary antibody binding). Output image stacks obtained from this program are then used for colocalization studies in MATLAB to find puncta from different channels that share voxels or to determine the distance between objects in different channels. This is useful for defining potential synaptic partners that contain a presynaptic element and a postsynaptic density within 0.5 µm of each other18. We have validated automated detection of synapses using a manual physical dissector stereological technique on the same raw image stacks used for threshold-based analysis22. However, future improvements to image analysis should explore non-threshold based methods of analysis that maintain the ability to restrict the characteristics of synaptic puncta and allow precise localization of the edges of objects for colocalization studies.
Image processing and data analysis are largely automated with ImageJ (Fiji) macros and MATLAB programs but data require quality checking. Statistical considerations are typical to most biological experiments, including checking the normality of data to determine whether parametric or non-parametric statistics are necessary, taking the mean or median of each case then comparing groups as appropriate. When the number of cases available is limited as often occurs in human studies, more statistical power can be gained by using all data points from each subject (instead of only the mean or median value from each person) and comparing the study groups with a cluster analysis that avoids pseudoreplicating the data32 by controlling for the data points coming from different subjects (courtesy Dr R Betensky33).
Limitations
Analysis of human brain tissue by array tomography has strengths and limitations. The strengths of this technique are the ability to examine multiple protein markers at small structures with excellent resolution and the ability to obtain high-throughput quantitative measures of synapse density, size, and protein composition. However, array tomography only allows observation of immunolabeling. Antibody penetration of the LR White resin is not always efficient, and epitopes are not always preserved, limiting proteins of interest that can be studied. Further, immunolabeling does not give ultrastructural anatomical data, which must be acquired using the more difficult and time-consuming electron microscopy methods. We advocate brain banking storage of samples suitable for array tomography and TEM (transmission electron microscopy), along with formalin fixed paraffin embedded brain tissue for conventional histology, and frozen tissue for biochemical and molecular analyses. Array tomography promises to provide an additional powerful approach to understanding synaptic composition and structure in a wide group of human diseases.
MATERIALS
REAGENTS
Human tissue. We have used tissue from the Massachusetts Alzheimer Disease Research Center and Edinburgh Brain Bank following institutional review board approval. CRITICAL Tissue must be obtained in accordance with national and local regulations.
-
-
Paraformaldehyde – EM grade 16% aqueaous (Electron Microscopy Sciences; cat. no. 15710). CAUTION Toxic, use in hood and handle with gloves.
-
-
Glutaraldehyde – EM grade 50% aqueaous (Electron Microscopy Sciences; cat. no. 16310). CAUTION Toxic, use in hood and handle with gloves.
-
-
Durcupan resin, Sigma (Cat. No. 44610-1EA)
-
-
Propylene Oxide, Sigma (Cat. No. 56671-5ML-F)
-
-
Osmium Tetroxide, Sigma (Cat. No. 201030-500MG). CAUTION Fumes are toxic, open only in hood.
-
-
Uranyl Acetate, Sigma (Cat. No. 73943-5G) CAUTION Toxic and slightly radioactive, use in hood and handle with gloves.
-
-
10× (0.1mM) PBS, Gibco (Cat. No. 71011-044)
-
-
DAPI, 1000× Invitrogen (Cat. No. D3571)
-
-
LR White, Electron Microscopy Sciences (Cat. No. 14381-CA) CAUTION Toxic before polymerization. Use in hood and handle with gloves.
-
-
Ethanol, ThermoFisher (Cat. No. 04-355-223) CAUTION Flammable.
-
-
PAP pen, Fisher (Cat. No. 237-69-300)
-
-
Glycine Sigma (Cat. No. G8898)
-
-
TBS: Tris Base, Fisher (Cat. No. BP125); Sodium Chloride, Sigma (Cat. No. S3014); Hydrochloric Acid, Sigma (Cat. No. 77-86-1).
-
-
Sucrose, Sigma (S8501)
-
-
Tween 20, Fisher (Cat. No. BP 337-500)
-
-
Xylenes, Fisher (Cat. No. X5-1) CAUTION Toxic, use in hood and handle with gloves.
-
-
Weldwood contact cement (DAP products)
-
-
Immumount, ThermoFisher (Cat. No. 99-904-02)
-
-
Gelatin powder, J.T. Baker (Cat. No. 2124-01)
-
-
Sodium Dodecyl Sulfate, Fisher (Cat. No. BP166-500)
-
-
Sodium Hydroxide solution 2N, Fisher (Cat. No. SS269-1)
-
-
Appropriate antibodies (see Table 1 for a list of antibodies we use)
Table 1.
Antibodies tested on human AT sections. N/A not applicable
| Antibody | target | Manufacturer | Catalogue Number |
Conc. | Host Species |
|---|---|---|---|---|---|
| PSD95 | excitatory postsynaptic densities | Abcam | ab12093 | 1:50 | Goat |
| PSD95 | excitatory postsynaptic densities | Cell Signaling | D27E11 | 1:50 | Rabbit |
| Synapsin I | presynaptic terminals | Millipore | AB1543 | 1:50 | Rabbit |
| Synaptophysin | presynaptic terminals | Abcam | ab8049 | 1:50 | Mouse |
| ApoE | ApoE | Abcam | ab7620 | 1:25 | Goat |
| LB509 | α-synuclein | Abcam | Ab27766 | 1:50 | Mouse |
| GAD65 | Glutamate decarboxylase (inhibitory) | Millipore | AB5082 | 1:50 | Rabbit |
| NAB61 | Oligomeric Aβ | Virginia Lee | N/A | 1:50 | Mouse |
| R1282 | Aβ | Dennis Selkoe | N/A | 1:50 | Rabbit |
| AW7 | Aβ | Domonic Walsh | N/A | 1:50 | Rabbit |
| Tau13 | Total tau | Peter Davies | N/A | 1:50 | Mouse |
| pan Tau | Total tau | Dako | A002401-2 | 1:50 | Rabbit |
| VDAC1/Porin | Mitochondria | Abcam | ab34726 | 1:50 | Rabbit |
| Alz50 | Misfolded tau | Peter Davies | N/A | 1:50 | Mouse IgM |
| EAAT1 | Astrocytic excitatory aminoacid transporter | Abcam | ab416 | 1:50 | Rabbit |
| VGLUT1 | Excitatory presynaptic terminals | Millipore | AB5905 | 1:50 | Guinea Pig |
| GFAP | Astrocyte cytoskeleton | Sigma | G9269 | 1:50 | Rabbit |
| PHF1 | Phospho-tau (ser396/404) | Peter Davies | N/A | 1:25 | Mouse |
| Tubulin | Cytoskeleton | Sigma | T67983 | 1:50 | Mouse |
| Syn1 | α-synuclein | BD | 610787 | 1:50 | Mouse |
EQUIPMENT
-
-
0.45 micron syringe filters, Fisher (Cat. No. 09-7193)
-
-
20 mL syringe, BD (Cat. No. 309661)
-
-
18G needle 1.5 inch, BD, (Cat. No. 305196)
-
-
Histo Jumbo diamond knife, (Diatome; Cat. No. HI 6792)
-
-
diamond facing knife, (Diatome; Cat. No. CT966)
-
-
ultramicrotome, (Leica Ultracut UCT)
-
-
gelatin capsules, (Electron Microscopy Science; Cat. No. 70100)
-
-
capsule holders, (Electron Microscopy Science; Cat. No. 70022)
-
-
Oven, Quincy Lab Inc, (Model 20 GC Lab Oven)
-
-
wec prep blades, (Electron Microscopy Science; Cat. No. 71933)
-
-
coverslips, (Fisher ; cat. no. 12-544-E;No 1.5;0.16–0.19 mm thick)
-
-
fluorescence microscope – for example a Zeiss AxioImager Z2 equipped with automated stage, digital camera, 10× objective, 63× 1.2 NA plan apochromat oil objective, and array tomography automated imaging plugins in AxioVision software. Note that any fluorescence microscope with high-resolution objective can be used for manual image acquisition.
-
-
NIH FIJI/Image J software (freeware http://fiji.sc) with plugins multistackreg (developed by Brad Busse and available at http://bradbusse.net/downloads.html), TurboReg, and StackReg34
-
-
Matlab software (Mathworks)
-
-
Philips CM12 transmission electron microscope equipped with a Gatan digital camera. Note any transmission electron microscope can be used.
-
-
Vibratome Series 1000 Sectioning System
REAGENT SETUP
AT fixative Prepare 4% paraformaldehyde, 2.5% sucrose in 0.01M PBS. For 40 mL combine 10mL 16% PFA, 1 g sucrose, 4mL of 0.1M PBS, fill up to 40mL with dH20. CRITICAL Fixative should be prepared fresh for every experiment.
EM fixative Mix paraformaldehyde, glutaraldehyde, 0.2M EM-grade phosphate buffer and ddH2O for final concentrations of 4% Paraformaldehyde, 2.5% gluteraldhyde in 0.1M EM-grade phosphate buffer. Adjust pH to 7.4. CRITICAL Fixative should be prepared fresh for every experiment.
Uranyl Acetate Solution Mix 20 mL 70% alcohol with 0.2 g uranyl acetate powder to produce a 1% solution. This solution must be made up fresh for each experimental run and cannot be stored or it will develop crystals that prevent clear imaging. CAUTION Toxic and slightly radioactive, use in hood and handle with gloves.
AT wash buffer Combine 1.75 g sucrose and 187.5 mg glycine, fill up to 50 mL PBS for a final solution of 3.5% glucose, 50mM glycine in 0.01mM PBS. Filter with a 0.45 micron syringe filter. This solution can be stored at 4 °C for up to 3 months.
AT glycine solution Dissolve 187.5 mg glycine in 50mL of 0.01mM PBS for a 50 mM glycine solution. Filter with a 0.45 micron syringe filter. This solution can be stored at 4 °C for up to 3 months.
AT block buffer Combine 1.25 mL of 1% Tween-20 stock (100 µl in 10 ml of H2O) and 250 µl of 10% BSA solution in Tris buffer up to 50 mL for a final solution of 0.05% Tween and 0.1% BSA in TrisBase Solution. Filter with a 0.45 micron syringe filter. This solution can be stored at 4 °C for up to 3 months.
AT stripping buffer Combine 2 mg SDS and 1 mL 2N sodium hydroxide solution into a total volume of 10 mL (with dH2O) for a final concentration of 0.02% SDS, 0.2N NaOH. Mix well and store at room temperature for up to 3 months.
Antibody Solutions
In AT block buffer, add antibodies at optimized concentrations (typically 1:10–1:100). In secondary antibody solution, add 1:1000 of 10 mg/mL DAPI solution for a final concentration of 0.01mg/mL DAPI. Centrifugation of antibodies for 2 min at 13,000×g helps reduce background signal.
Tris Buffered Saaline Solution (TBS) Combine 60g Tris Base and 89.5 g sodium chloride in 10 L dH2O, add 12 N hydrochloric acid until pH is 7.4 (approximately 35mL). Store at room temperature for up to 7 days.
Cement mixture Combine ~5mL Weldwood contact cement with an equal volume of xylenes and stir well with a toothpick. !CAUTION Xylenes are toxic. They should be handled in a fume hood using gloves. Store at room temperature until the mixture is too thick to use.
Gelatin subbing coverslips for AT stripping experiments Dissolve 1.5g gelatin in 750 mL dH2O on a stirring hotplate for a 0.2% solution. Heat and stir until dissolved (do not boil). Place Fisher #1.5 coverslips into slide holders. Pour gelatin solution into slide holder for 1 minute, drain gelatin solution away, and dry coverslips for 1hour in a 37 °C incubator or overnight at room temperature. Repeat coating in gelatin solution and dry. Store coverslips long-term (months) at room temperature in original boxes when dry.
PROCEDURE
Tissue Collection and Fixation for AT and EM – TIMING 3–4 hours
-
1
At autopsy, remove the brain (and spinal cord when available). CRITICAL STEP Ensure proper consent and approvals are obtained for post-mortem examination, including retention and use of tissues for research. The tissue removal should be performed by an appropriately qualified and trained pathologist. PAUSEPOINT While a short post mortem interval (PMI) is recommended, if there is a longer PMI cold storage in a mortuary facility, should be used to limit structural degradation.
-
2
Sample brain pH, weigh brain and collect CSF for future analyses.
-
3
Bisect brain along the midline into 2 hemispheres. Drop one hemisphere into 10% formalin fixative for future pathological and histochemical studies.
-
4
Starting with a section through the mammillary bodies, section the contralateral fresh hemisphere into 1 cm coronal sections. From the coronal sections take regions of approximately 1 cm3 from the regions of interest (Fig. 2). For neurodegenerative diseases, we collect tissue from the following regions: superior frontal cortex, anterior cingulate cortex, superior temporal cortex, entorhinal cortex, amygdala, posterior superior putamen, dentate gyrus granular cell layer, cornu ammonis 1 (CA1) of the hippocampus, superior cerebellar vermis, substantia nigra, and spinal cord (anterior horn gray matter) where available, particularly in Amyotrophic Lateral Sclerosis cases. For cortical regions, ensure the entire cortical mantle from pia to white matter is contained in the sample. The rest of the coronal section can be snap frozen between aluminum plates for separate biochemical and molecular studies.
-
5Bisect the 1cm3 samples (maintaining all of the cortical layers in each cortex sample).
- drop one half of the sample in AT fixative 20–30 min at room temperature then move to step 6.
- Drop the other half of the sample into EM fixative for 48hr at 4 °C. Then follow EM processing step 15).
-
6
Further dissect the AT tissue samples into approximately 1mm × 1mm × 5mm blocks taking care to maintain the orientation of cortical samples such that all 6 layers are included in the longest dimension (Fig. 3, Supplemental Video 1). Make approximately 10 AT blocks per region. ?TROUBLESHOOTING
-
7
Place small blocks back in AT fixative. Fix AT samples for a further 2 hours. CRITICAL STEP. Do not fix AT samples for more than a total of 3 hours to avoid marked decreases in antibody binding.
-
8
Remove fixative from AT samples and wash samples at least 5 min in AT wash buffer. PAUSE POINT. Samples can be left overnight in a fresh change of wash buffer at 4 °C or you can proceed directly to dehydration.
AT tissue dehydration and resin embedding – TIMING 2 hours plus 2 overnight steps
-
9
Dehydrate AT samples by placing them in solutions of graded ethanols for 5 minutes each: 50% ethanol, 70% ethanol, 95% ethanol, 100% ethanol, repeat 100% ethanol.
-
10
Incubate 5 minutes in each of the following 50:50 mixture of 100% ethanol and LR White, LR White alone, and then repeating the LR White. Leave AT samples in LR White overnight at 4 °C. CAUTION LR White is toxic before polymerization. Gloves should be worn.
-
11
Fill gelatin capsules with cold LR White. CRITICAL STEP. Be sure to fill the capsules completely as LR White will not polymerize if there is too much air in the capsule.
-
12
Place one 1mm × 1mm × 5mm tissue sample into each capsule with a printed label in the capsule containing the case number, region, and sample number and place the cap on the capsules. CRITICAL POINT. Do not use pencil or permanent marker; it will dissolve in the resin and the blocks will be indistinguishable.
-
13
Place AT capsules into an oven set at 52 °C overnight. CRITICAL STEP. If the temperature drops below 50 °C or capsules are not left long enough the resin will not harden properly and the blocks will not section properly.
-
14
Remove capsules from oven. When ready these capsules can be processed as described in step 25. AT and EM resins are non-toxic once polymerized. PAUSE POINT. Capsules can be stored at room temperature indefinitely.
EM tissue dehydration and resin embedding – TIMING 4 hours spread over 4 days.
-
15
Transfer EM samples from step 5 to 0.1M Phosphate buffer and store at 4 °C for at least 24hrs.
-
16
Cut 70µm sections on a vibratome and float in PB.
-
17
Incubate vibratome sections for EM in 1% Osmium Tetroxide for 30mins at room temperature (protected from the light)
-
18
Wash EM sections in 3 changes of 0.1M Phosphate buffer and then ddH2O.
-
19
Dehydrate the EM sections by placing them in solutions of graded ethanols: 10 mins in 50%, 40 mins in 70% containing 1% Uranyl Acetate (protected from the light), 15 mins in 95%, 10 mins in 100%, 10 mins in 100% (dried and stored over molecular sieves), followed by 2×10 mins in 100% Propylene Oxide.
-
20
Incubate EM sections in Durcupan resin for 24hrs at room temperature
-
21
Mount sections on microscope slides and cover-slip in Durcupan.
-
22
Transfer slides to an oven at 50 °C for 48hrs to allow the resin to polymerise.
-
23
Smear the slide and coverslip with a thin film of barrier cream to allow the removal of the coverslip with a thin blade.
-
24
Cut out region of interest from EM sections (~1mm×1mm) and superglue to the tip of a pre-set Durcupan resin blocks for subsequent sectioning.
PAUSEPOINT Store slides or prepared blocks until needed at room temperature
Tissue Sectioning for AT and EM – TIMING ~1 hour per sample
-
25
Remove gelatin capsule from polymerized AT resin block to be cut
-
26
Insert an AT or EM block into the chuck of the ultramicrotome and trim excess resin from around the tissue block with a Wec prep blade, to make a trapezoid around the tissue (Fig. 4d). Ensure the blade is sharp otherwise the block will be difficult to cut and chunks of the tissue may accidentally break off.
-
27
Place the chuck in the cutting position and turn the tissue block such that the trapezoid is on its side (long side vertical). Using the diamond facing knife, cut the face of the block until it is even and the tissue has been reached. The tissue can only be as wide as the width of the facing knife’s blade. Otherwise it is not possible to obtain an even face. If the tissue is too wide, repeat step 26 until it is not wider than the facing knife’s blade.
-
28
Move the facing knife to the edges of the trapezoid and cut the edges of the block until the long sides of the trapezoid are exactly parallel. CRITICAL POINT. In order to obtain long ribbons, it is essential that the top and bottom edge of the trapezoid are parallel. A narrow block face (< 1mm in the direction of cutting) also helps produce straight ribbons, which are important for collecting good quality image stacks. ?TROUBLESHOOTING
-
29
Apply a mixture of cement and xylene (roughly 50:50) to the parallel sides of the block so that sections adhere together during cutting and form a contiguous ribbon. This step is not necessary for EM blocks.
-
30
Fill the boat of a histo jumbo diamond knife with filtered dH2O and place a microscope slide in the boat. For AT sections, Label a coverslip with the block to be cut and either place the coverslip onto the microscope slide partially submerged in the water or wait and insert after cutting the ribbon in step 33. CRITICAL STEP. The coverslip should be of the thickness needed for the microscope objective to be used for high-resolution imaging (in our case 0.16–0.19 mm). If stripping and re-probing of sections is desired, use gelatin-subbed coverslips.
-
31
Section the tissue into ribbons of 70nm serial sections (30–40 sections per ribbon for AT, 3–5 sections for EM) using a histo jumbo diamond knife. CRITICAL POINT. Because of the glue, the sections should be sticking together to form a ribbon. They should be the same thickness and the ribbon should be as straight as possible. The tissue should be distinguishable from the resin. When beginning to section, start with the tissue at a greater thickness (e.g. 500 nm) and then decrease the thickness in a stepwise fashion until 70 nm is reached. 70 nm sections are silver-gold in color when observed through the ultramicrotome oculars floating on water. Fluctuations in temperature often make the readout on the ultracut inaccurate so thickness should be judged by section color (Fig 4). ?TROUBLESHOOTING
-
32
Detach the ribbon from the blade by gently running the eyelash along the surface of the blade behind the ribbon. For EM samples, mount short ribbons with an eyelash (glued to a stick) onto formvar-coated EM grids. For AT samples, place the first section of the ribbon on the coverslip (inserted into the boat in step 30) at the interface of the glass and water in the knife boat and gently remove water from the knife boat with a syringe until the ribbon is adhered to the coverslip. Allow the section to dry. To aid finding dried ribbons for staining, outline the ribbon with permanent marker on the reverse side of the coverslip while it is still moist. An alternative method for mounting AT ribbons on a coverslip is to gently slide a coverslip under the ribbon after it is cut and gently move the ribbon to the coverslip and draw back gently. CRITICAL POINT. If the coverslip is brought back too fast the ribbon may not dry well. If the ribbon is not straight imaging will be more difficult. It is also important to pull back and not up. Pulling up will cause the water in the boat to flow out.CRITICAL POINT. Mount the ribbon as straight as possible, it will make imaging and analysis easier. ?TROUBLESHOOTING PAUSE POINT Dried ribbons can be stored protected from dust indefinitely at room temperature.
-
33
Perform standard TEM imaging and analysis as previously published35, 36 on EM samples. Proceed to step 34 for AT samples.
Immunohistochemistry for AT – TIMING 3 hours with one optional overnight step
-
34
Outline the dried ribbon with a hydrophobic PAP pen.
-
35
Incubate the ribbon in AT glycine solution (~100 µL, or enough to cover the entire ribbon) for 5 min at room temperature to remove residual aldehydes from fixation. CRITICAL POINT From this step, keep ribbon wet at all times.
-
36
Gently remove glycine solution with a pipette then immediately cover the ribbon with blocking solution. Incubate in AT block buffer for 10–30 min at room temperature. CRITICAL POINT While staining the ribbon it is easiest to keep the pipettes near the ribbon; however, take care not to touch the ribbon as this can damage it and potentially ruin a region of interest.
-
37
Remove blocking buffer and immediately apply primary antibody solution (diluted in blocking buffer at concentrations of 1:10–1:1000 optimized for each primary, see table 1). Leave 2 hours at room temperature or overnight at 4 °C. Note for each experiment, identical parameters should be maintained for the staining procedure. PAUSE POINT Can be left at 4 °C overnight.
-
38
Gently wash the ribbon 3× with TBS by removing liquid with a pipette from one side of the ribbon and simultaneously adding TBS to the other side for 10–15 seconds each wash.
-
39
Apply fluorescently labeled secondary antibodies diluted in blocking buffer with DAPI (0.01mg/mL) to stain nuclei. Incubate 30 min at room temperature. Alternatively, DAPI can be applied for 20 minutes after washing off secondary antibody solution.
-
40
Wash with TBS as in step 38.
-
41
Invert coverslip and place on microscope slide with a mounting medium that prevents fluorescence bleaching (Immumount).
-
42
Image immediately if possible or within 24 hours as fluorescence fades quickly.
Imaging AT – TIMING ~2 hours per ribbon
-
43
On a fluorescence microscope, locate the ribbon with a low magnification objective (10× or 20×). Using a 63× objective, check the quality of immunostaining. CRITICAL POINT If the staining has high background, is present in structures where there should not be staining (e.g. synaptic proteins in nuclei or capillaries), or if staining is present in all wavelengths (indicative of autofluorescence), STOP and go back to step 26. Cut a new ribbon and try different staining conditions. ?TROUBLESHOOTING.
-
44If you have automated software for array tomography image acquisition (Zeiss AxioVision) follow option A, otherwise follow option B.
- Option A) Automated software (more details regarding using the automated method can be found in the supplemental information).
- Acquire a tilescan image of the entire ribbon at low magnification in the DAPI channel (Fig. 5). Choose a region of interest (ROI) to image at higher resolution. If examining cortex, be sure to image the same cortical layer in all cases to reduce variability. Use the tilescan or manually examine the ribbon to make sure the region of interest is present on every section of the ribbon and is not obscured in any section by artifacts or rips in the tissue.
- Use a marker to select the same nucleus in the ROI on the first two sections of the tile scan and use the AT macros to generate a position list of the ROIs on all of the sections in the ribbon (see supplemental data for precise instructions on the AxioImager’s AT macros).
- Change to high resolution objective (63× plan apochromat oil 1.2 NA) and focus on the first section in the ribbon.
- Adjust the exposure time to optimize for each channel avoiding saturation.
- B) Manual imaging.
- For manual imaging, find the ROI on the first section and take an image of all channels. Manually move to the next section, find the ROI, and take an image, repeating on all sections. CRITICAL POINT. Make sure that the same cell bodies are clearly present in every image and further that they are close to the same location in every image..
-
45
Save all images in the stack with the section number in the filename.
-
46
Collect data from 2–3 blocks per case (for each region) making 2–3 stacks per ribbon.
-
47
If stripping and re-probing is desired, remove coverslip immediately after imaging by gently placing a razor blade under the edge. Rinse off mounting medium thoroughly with distilled water (running for 30 seconds then soaking for 5 minutes). Apply stripping buffer for 15 minutes then wash well in water as above. Dry thoroughly before re-staining from step 35. As a control for antibody elution, a small ribbon or small section of the original ribbon should be treated identically for the first round of staining but have no primary antibodies applied during the second round of staining. This ribbon should be re-incubated with secondary antibodies and DAPI and checked on a microscope to be sure there is no residual staining from the first set of primary antibodies (Supp. Fig. 1).
AT Image Analysis – TIMING varies depending on the number of stacks in the study.
-
48
Convert the individual images from each section into a stack (with a separate stack for each channel). CRITICAL POINT. Custom ImageJ macros make image analysis steps much faster. Macros that we have developed are provided in Supplementary Data 1 for individuals to modify for different systems/experiments.
-
49
Use the multistackreg ImageJ plugin (developed by Brad Busse and available at http://bradbusse.net/downloads.html, which requires TurboReg and StackReg macros34) to align the stack of the nuclear staining (DAPI channel) with the rigid body algorithm and save the transformation file. Check that the DAPI alignment worked by scrolling through the stack then to apply to other channels. CRITICAL POINT Ensure alignment files are saved or it will be impossible to get the other channels similarly aligned. After aligning the DAPI channel, check by eye that the alignment worked well then apply the transformation file to all other channels from the same stack (Fig. 6). Save the aligned stacks. ?TROUBLESHOOTING
-
50
If multiple days of imaging are used after stripping, align the subsequent days using the mutlistackreg plugin as above. Then align the second day to the first using the multistackreg option “align second stack to first” to align the DAPI from day 2 to the DAPI stack from day 1. Save the transformation file. Merge DAPI stacks from Day 1 and Day 2 as different colors to ensure proper alignment. If alignment was good, apply the saved transformation file to the other channels from day 2 with the “align second stack to first” option.
-
51
Choose ROIs on the stack for automated analysis (counting stained objects and determining their sizes). For synapse analysis, choose the ROIs in the neuropil in areas without any nuclei or capillaries. Be sure that the areas chosen for density analysis are present throughout the entire stack and not interrupted as a result of the alignment. A standard ROI size makes analysis simpler, for example a 20 µm × 20 µm box with depth defined as the number of sections in the stack times the thickness (usually 70 nm). Crop multiple ROIs per stack (“crops”) and save each crop with a coded number. CRITICAL POINT. This is the step where we introduce the blind coding. At this stage, a spreadsheet should be started recording for each crop the case number, relevant case information (diagnosis, genotype, gender, etc) block number, stains used, image date, crop volume, and number of sections in the crop. After all crops are generated for the study, the identifying data is hidden in the spreadsheet and analyses performed by an experimenter blind to these data. Maintaining a detailed spreadsheet with all of the information about crop is paramount to keeping track of the data. CRITICAL STEP. Before proceeding, it is best to collect all image stacks and crops needed for the study.
-
52For automated analysis of object density, size, and colocalization, each crop must be assigned a threshold to convert the images to a binary format. Thresholds should be chosen to most accurately reflect the staining of synaptic puncta observed by eye. The threshold chosen should not cause clusters of small puncta to be melded into one object in the binary image while still maintaining most of the smallest, lowest intensity puncta. This can be done manually (option B) or with automated algorithms (option A).
- Automated thresholding
- On representative crops from each staining day, choose a manual threshold that best captures the staining (Fig. 5).
- Run all of the automated thresholding paradigms offered by FIJI on a representative sample of the data. Be sure to select “stack.” Choose the paradigm that is closest to the one you would choose manually.
- Apply this paradigm to all crops and save the thresholded images (using a macro to batch the thresholding saves time, see supplemental data).
- Manual thresholding.
- Choose each threshold for every crop. This should be done by a single experimenter blinded to condition, ideally all on the same computer in one session to minimize variability.
-
53
Process each image with Watershed, a program written by Brad Busse and Stephen Smith which defines objects larger than a given threshold (10 voxels for synapses) that are present in more than one consecutive section. This program generates output images containing positive staining and an objects file that lists the number of objects and the size of each object. Critical step: the Watershed program should be installed in the same folder where all the crops were saved.
-
54
Import the Watershed Output files into the spreadsheet. The program will give number and size of stained objects.
-
55
Calculate the density of synapses in each crop (density = number of synaptic puncta divided by the volume of the crop). Note crop volume = crop box size * number of sections in stack * thickness of sections (70nm). ?TROUBLESHOOTING
-
56
For colocalization or distance between objects analysis run the output files through a MATLAB script that calculates the nearest object between channels (see Supplementary Data 2 for an example script). ImageJ can also be used to determine the colocalization between objects.
Fig. 6.
Image analysis. Stacks are made for each stain from the images of the ribbon (a). The image stack containing large objects such as nuclei is aligned (b) and the alignment applied to all other channels. Regions of interest (ROIs) are chosen on a projected image stack to ensure that they do not contain cell bodies, staining artifacts (arrow) or blank spaces resulting from alignment (outside dotted lines, c). Thresholds are applied to the ROIs to convert them to binary images. Red in (d) shows pixels on a single slice of the ROI detected with thresholding. The ROIs are then run through two analysis programs, one that detects synapses and another to determine the distance between objects in different channels. A 3D reconstruction of 22 serial sections of the pre and postsynapses detected in the ROI from d are shown in e. Presynaptic terminals were labeled with synaptophysin and postsynaptic densities with PSD95. Scale bars represent 10 µm (a–c) and 2 µm (d–e).
TIMING
To acquire an image stack from one tissue sample using array tomography can be as fast as 3 days from initial tissue collection. Similarly, EM images can be acquired within 6 days of tissue collection. However, multiple blocks from multiple cases are needed for each study. Experiments are broken down into three largely independent phases as shown in the flow chart in Figure 2: 1. Tissue collection, which occurs on an ongoing basis as brains are donated and autopsies performed (blue); 2. Sectioning, staining, and imaging cases (green) until staining is optimized and all data have been collected for the experiment; and 3. Analysis of images. Once the staining procedure is optimized for each experiment, data collection is straightforward. Image processing (aligning, stacking, cropping images) can be done after each imaging session, but it is best to conduct image analysis (thresholding, counting and measuring objects, and colocalization studies) after all data has been collected for the study. An AT experiment generates large amounts of data: approximately 10 cases, 30 ribbons, 90 stacks, 500 crops, 4 channels per crop, and tens of thousands of synapses to analyze. Thus the image analysis steps take several weeks to complete, depending on the size of the study. We find that mistakes are often generated in these large datasets either in the automated macros or in typing or copying errors, so extensive quality checking and control of the data are necessary.
TROUBLESHOOTING
Troubleshooting advice can be found in table 2.
Table 2.
Troubleshooting
| Step | Problem | Possible Reason | Solution |
|---|---|---|---|
| 6 | The 1 cm3 piece of tissue breaks apart while trying to dice it into 1mm×1mm×5mm blocks | Dull razor blade, slicing too thin | Start over with a new section taken from the same region. Because of how small the blocks are, there should be ample tissue to get 10 blocks even if a mistake is made on a few. Wait until fixative has hardened a bit more the tissue before starting. |
| 29 | Block face not cutting full sections (only cutting one side or one corner) | Not aligned in the chuck properly | It can be very difficult to determine if the block is straight in the chuck. The easiest way to tell is to try to line up one side of the faced block with the vertex of the knife between the front and side faces. This makes it easier to observe if the blade is actually running straight through the tissue or not. |
| 32 | Sections not coming off straight | Tissue not aligned properly in the chuck | Use the reflection of the blade on the tissue to make sure that the blade is hitting the tissue at a perfectly parallel angle at both the top and bottom. Use the same reflection to make sure the blade is equidistant from the left and right sides. |
| Sections not equal thickness | humidity imbalance, temperature imbalance | Keep hands away from the blade itself as much as possible. Keep movement in and out of the room to a minimum. Leave it and try again in an hour. | |
| Tissue not visible | Tissue is not being cut | Using the facing knife, cut deeper into the block until the tissue is visible as bumpy sections when cutting ~500nm comoared to the absolutely smooth sections cut when only resin is present. | |
| 34 | Ribbon not straight on coverslip | Not moved on to the coverslip straight | If the ribbon is not as straight as desired on the coverslip two things can be done. First, by resubmerging the coverslip in water, one can often get the ribbon free and try again. However, this sometimes results in the ribbon falling apart. Further, once the first section of the ribbon has attached to the coverslip, either remove the ribbon from the boat or resubmerge it but do not try to adjust its orientation once it is attached. Second, a new ribbon can always be cut. |
| 44 | Poor staining | Primary antibody concentrations not optimized | Test varying concentrations of primary antibody (from 1:10–1:1000). Try overnight incubation at 4 °C vs 2 hours room temperature. Increase blocking time (to 30 min or 1 hour). If this does not work, try a different antibody that has worked for immunoEM or IHC on paraffin embedded tissue. |
| 53 | Stacks not aligning | Curved ribbon or change in location of high resolution image on sections. | Try starting alignment on a different section in the stack. Discard sections at the beginning or end of the stack that will not align (as long as all remaning sections are contiguous, e.g. cannot delete one section from the middle of a stack). |
| 58 | Synapse density numbers unexpectedly much higher or lower than expected values | Thresholding algorithm inaccurate | If synaptic densities calculated by AT are far from reported values from previous studies, check the thresholding algorithm to make sure it is accurately detecting synapses. |
ANTICIPATED RESULTS
At low magnification, (10× or 20×) areas of interest can be identified on the ribbon, for example finding the cortical layer to be examined using the pattern of DAPI staining and finding pathological lesions (Fig. 4). Any imperfections in the ribbon can be seen at this magnification and avoided when choosing fields for high-resolution imaging.
Pre and postsynapses can be labeled in human tissue with several markers (Fig. 7, table 1). We have used array tomography to examine synapse loss and the role of amyloid beta, apolipoprotein E and tau in synaptic degeneration18, 22, 23. Examples of amyloid beta plaques, apolipoprotein E, tau, and glial markers in AT sections from Alzheimer disease patients are shown in figure 7. Other types of pathological lesions and their synaptic effects can be examined including alpha synuclein in Lewy bodies (Fig. 7) Array tomography can also be used to examine glial processes and their interactions with synapses (Fig. 7). Mitochondrial localization in tubulin-filled processes and cell bodies can be observed (Fig 7).
Fig. 7.
Examples of AT imaging. A 3D reconstruction of a 47.6 µm × 47.6 µm × 6.3 µm volume of temporal cortex from an Alzheimer patient shows in great detail the association of astrocytic processes labeled with GFAP with senile plaques labeled with the NAB61 antibody specific for oligomeric Aβ (o Aβ, a). An 8 µm × 6 µm × 0.56 µm volume reconstruction from watershed output files shows synapses with apposed presynaptic (blue, VGLUT1) and postsynaptic (green, PSD95) elements (arrows, b). The arrowhead indicates a tripartite synapse with an astrocytic endfoot (red, EAAT1, b). Mitochondrial distribution (c, blue VDAC1/porin) within tubulin-labeled cell bodies (green) in an AD case shows reduction of mitochondria in neurons with accumulations of misfolded Alz50 positive tau (red). Staining Aβ with R1282 antbody and misfolded tau with Alz50 antibody in AD temporal cortex shows a neurofibrillary tangle (arrow, d) and plaque-associated dystrophic neurites (arrowhead, d) in a single section. Apolipoprotein E (ApoE) is associated with plaques (arrowhead, e) and synapses (arrow, e) in a single section from an AD brain. Lewy bodies (arrow, f) and Lewy neurites (arrowheads, f) stained with the LB509 antibody in a maximum projection of 21 sections from a case of dementia with Lewy bodies (DLB). Scale bars represent 10 µm in (a) and (c–f); 1 µm in (b).
Although ultrastructure is often compromised in human tissue postmortem, the quality of tissue obtained using the current modified protocol is usually more than adequate for undertaking quantitative assessment of synaptic integrity and ultrastructural features (e.g. synaptic vesicle densities and localization). At low magnification (Fig. 8), it is possible to identify numerous synaptic profiles and undertake synaptic density measurements. At higher magnification (>5,000×) it is possible to identify individual synaptic profiles and their organelles (particularly synaptic vesicles and mitochondria; Fig. 8). EM analysis can also be used alongside AT to uncover pathological changes at the synapse. An example of a degenerating cortical synapse from a patient with ALS is shown in Fig. 8.
Fig. 8.
Examples of EM imaging of healthy and degenerating human synapses. Electron micrograph of grey matter from Brodmann’s area 19 of human brain shows several synapses in a single field of view (a). A higher magnification image of synapses in Brodmann’s area 46 from a human brain shows retained ultrastructure of post-synaptic spines (black arrow) and pre-synaptic boutons containing numerous 50nm synaptic vesicles (white arrow) (b). Degenerating synapses, revealed by electron dense presynaptic terminals (white arrow) and degenerating synaptic mitochondria (black arrow), can be readily identified in tissue from an ALS patient (Brodmann’s area 19) (c). Scale bars represent 2 µm in (a) and 0.5 µm in (b) and (c).
In conclusion, the data acquired with AT and EM in concert with all of the other clinical and pathological data from the same cases have the potential to enhance our knowledge about synapses and other small structures in human brain both in health and disease.
Supplementary Material
Acknowledgements
We thank the patients and their families for their generous donations. We also thank Kristina Micheva and Stephen Smith for training us in the array tomography technique; Dennis Selkoe, Virginia Lee, and Dominic Walsh for antibodies; and Kishore Kuchibhotla and Scott Raymond for MATLAB programming. Harvard catalyst supplied biostatistical support. This work was supported by NIH grants R00AG033670 (TLS, KRK), T32AG000277 (KJK, BTH), P50AG05134 (BTH, ASP, MPF); Alzheimer Research UK (AMP), the Sylvia Aitken Charitable Trust (CS, AKW, MEB, THB, SA, THG), and Medical Research Council UK G110616 (CS).
Footnotes
Author contribution statement:
K.R.K, T.L.S.-J., A.S.-P., A.M.P., R.K, and K.J.K performed array tomography experiments. C.S. and M.F. consulted on neuropathology. K.R.K. C.S., D.M., A.S.-P., R.K., and T.L.S.-J collected human brain tissue. A.K.W., M.E.B., T.H.B., S.A. and T.H.G. designed and performed electron microscopy experiments. K.R.K., A.S.-P., A.M.P., K.J.K, C.S., T.H.G., B.T.H., and T.L.S.-J. discussed the data and prepared the manuscript.
The authors declare that they have no competing financial interests.
List of supplementary Information
Supplementary Figure 1. Stripping and re-probing array ribbons. A ribbon from the putamen of a DLB patient was stained on day 1 with anti α–synuclein antibody LB509 (mouse monoclonal) and DAPI to label nuclei. After imaging, the antibodies were stripped in SDS-containing buffer, rinsed, and re-probed with the α–synuclein antibody syn1 (mouse monoclonal) and DAPI. The aligned and merged images from days 1 and 2 show overlap of the synuclein stain in aggregates in the neuropil (arrows). An adjacent ribbon was re-probed with only secondary antibodies and no antibody staining was observe, confirming the efficacy of antibody elution. Scale bar represents 10 µm.
Supplementary Table 1. Characteristics of human subjects used.
Supplementary Video 1. Demonstration of making AT blocks from fresh tissue.
Summpementary Methods. Instructions for automated imaging of AT ribbons with AxioVision macros.
Supplementary Data 1. Image J macro examples for AT analysis.
Supplementary Data 2. MATLAB script for colocalization analysis of AT images.
References
- 1.Morrison JH, Baxter MG. The ageing cortical synapse: hallmarks and implications for cognitive decline. Nat Rev Neurosci. 2012;13:240–250. doi: 10.1038/nrn3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koffie RM, Hyman BT, Spires-Jones TL. Alzheimer's disease: synapses gone cold. Mol Neurodegener. 2011;6:63. doi: 10.1186/1750-1326-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mayhew TM. How to count synapses unbiasedly and efficiently at the ultrastructural level: proposal for a standard sampling and counting protocol. J Neurocytol. 1996;25:793–804. doi: 10.1007/BF02284842. [DOI] [PubMed] [Google Scholar]
- 4.Mayhew TM, Muhlfeld C, Vanhecke D, Ochs M. A review of recent methods for efficiently quantifying immunogold and other nanoparticles using TEM sections through cells, tissues and organs. Ann Anat. 2009;191:153–170. doi: 10.1016/j.aanat.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Scheff SW, Price DA, Sparks DL. Quantitative assessment of possible age-related change in synaptic numbers in the human frontal cortex. Neurobiol Aging. 2001;22:355–365. doi: 10.1016/s0197-4580(01)00222-6. [DOI] [PubMed] [Google Scholar]
- 6.Pawley JB. Handbook of biological confocal microscopy. New York, NY: Springer; 2006. [Google Scholar]
- 7.Holtmaat A, et al. Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat Protoc. 2009;4:1128–1144. doi: 10.1038/nprot.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuchibhotla KV, et al. Abeta plaques lead to aberrant regulation of calcium homeostasis in vivo resulting in structural and functional disruption of neuronal networks. Neuron. 2008;59:214–225. doi: 10.1016/j.neuron.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wakabayashi K, Honer W, Masliah E. Synapse alterations in the hippocampal-entorhinal formation in Alzheimer's disease with and without Lewy body disease. Brain Research. 1994;667:24–32. doi: 10.1016/0006-8993(94)91709-4. [DOI] [PubMed] [Google Scholar]
- 10.Moore D, et al. Cortical and subcortical neurodegeneration is associated with HIV neurocognitive impairment. AIDS (London, England) 2006;20:879–887. doi: 10.1097/01.aids.0000218552.69834.00. [DOI] [PubMed] [Google Scholar]
- 11.DeKosky S, Scheff S, Styren S. Structural correlates of cognition in dementia: quantification and assessment of synapse change. Neurodegeneration : a journal for neurodegenerative disorders, neuroprotection, and neuroregeneration. 1996;5:417–421. doi: 10.1006/neur.1996.0056. [DOI] [PubMed] [Google Scholar]
- 12.Masliah E, Terry R, Alford M, DeTeresa R, Hansen L. Cortical and subcortical patterns of synaptophysinlike immunoreactivity in Alzheimer's disease. The American journal of pathology. 1991;138:235–246. [PMC free article] [PubMed] [Google Scholar]
- 13.Clare R, King V, Wirenfeldt M, Vinters H. Synapse loss in dementias. Journal of Neuroscience Research. 2010;88:2083–2090. doi: 10.1002/jnr.22392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tai H-C, et al. The synaptic accumulation of hyperphosphorylated tau oligomers in Alzheimer disease is associated with dysfunction of the ubiquitin-proteasome system. The American journal of pathology. 2012;181:1426–1435. doi: 10.1016/j.ajpath.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Micheva KD, Smith SJ. Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron. 2007;55:25–36. doi: 10.1016/j.neuron.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Micheva KD, Busse B, Weiler NC, O'Rourke N, Smith SJ. Single-Synapse Analysis of a Diverse Synapse Population: Proteomic Imaging Methods and Markers. Neuron. 2010;68:639–653. doi: 10.1016/j.neuron.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koffie RM, et al. Oligomeric amyloid beta associates with postsynaptic densities and correlates with excitatory synapse loss near senile plaques. Proc Natl Acad Sci U S A. 2009;106:4012–4017. doi: 10.1073/pnas.0811698106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koffie RM, et al. Apolipoprotein E4 effects in Alzheimer's disease are mediated by synaptotoxic oligomeric amyloid-beta. Brain. 2012;135:2155–2168. doi: 10.1093/brain/aws127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kopeikina KJ, et al. Tau Accumulation Causes Mitochondrial Distribution Deficits in Neurons in a Mouse Model of Tauopathy and in Human AD Brain. Am J Pathol. 2011;179:2071–2082. doi: 10.1016/j.ajpath.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soiza-Reilly M, Commons KG. Quantitative analysis of glutamatergic innervation of the mouse dorsal raphe nucleus using array tomography. J Comp Neurol. 2011;519:3802–3814. doi: 10.1002/cne.22734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nanguneri S, Flottmann B, Horstmann H, Heilemann M, Kuner T. Three-dimensional, tomographic super-resolution fluorescence imaging of serially sectioned thick samples. PLoS One. 2012;7:e38098. doi: 10.1371/journal.pone.0038098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kopeikina KJ, et al. Synaptic alterations in the rTg4510 mouse model of tauopathy. J Comp Neurol. 2013;521:1334–1353. doi: 10.1002/cne.23234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Calignon A, et al. Propagation of tau pathology in a model of early Alzheimer's disease. Neuron. 2012;73:685–697. doi: 10.1016/j.neuron.2011.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lacey CJ, Bryant A, Brill J, Huguenard JR. Enhanced NMDA receptor-dependent thalamic excitation and network oscillations in stargazer mice. J Neurosci. 2012;32:11067–11081. doi: 10.1523/JNEUROSCI.5604-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oberti D, Kirschmann MA, Hahnloser RH. Projection neuron circuits resolved using correlative array tomography. Front Neurosci. 2011;5:50. doi: 10.3389/fnins.2011.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oberti D, Kirschmann MA, Hahnloser RH. Correlative microscopy of densely labeled projection neurons using neural tracers. Front Neuroanat. 2010;4:24. doi: 10.3389/fnana.2010.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robles E, Smith SJ, Baier H. Characterization of genetically targeted neuron types in the zebrafish optic tectum. Front Neural Circuits. 2011;5:1. doi: 10.3389/fncir.2011.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saatchi S, et al. Three-dimensional microstructural changes in murine abdominal aortic aneurysms quantified using immunofluorescent array tomography. J Histochem Cytochem. 2012;60:97–109. doi: 10.1369/0022155411433066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winkler M, et al. High resolution three-dimensional reconstruction of the collagenous matrix of the human optic nerve head. Brain Res Bull. 2010;81:339–348. doi: 10.1016/j.brainresbull.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vonsattel J, et al. An improved approach to prepare human brains for research. Journal of neuropathology and experimental neurology. 1995;54:42–56. doi: 10.1097/00005072-199501000-00006. [DOI] [PubMed] [Google Scholar]
- 31.Wright AK, Wishart TM, Ingham CA, Gillingwater TH. Synaptic protection in the brain of WldS mice occurs independently of age but is sensitive to gene-dose. PLoS One. 2010;5:e15108. doi: 10.1371/journal.pone.0015108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freeberg TM, Lucas JR. Pseudoreplication is (still) a problem. J Comp Psychol. 2009;123:450–451. doi: 10.1037/a0017031. [DOI] [PubMed] [Google Scholar]
- 33.Rudinskiy N, et al. Orchestrated experience-driven Arc responses are disrupted in a mouse model of Alzheimer's disease. Nat Neurosci. 2012;15:1422–1429. doi: 10.1038/nn.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thevenaz P, Ruttimann UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- 35.Koch S, et al. Morphologic and functional correlates of synaptic pathology in the cathepsin D knockout mouse model of congenital neuronal ceroid lipofuscinosis. Journal of neuropathology and experimental neurology. 2011;70:1089–1096. doi: 10.1097/NEN.0b013e318238fc28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gillingwater T, et al. Delayed synaptic degeneration in the CNS of Wlds mice after cortical lesion. Brain : a journal of neurology. 2006;129:1546–1556. doi: 10.1093/brain/awl101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.