Abstract
A series of flexible carbocyclic pyrimidine nucleosides has been designed and synthesized. In contrast to previously reported “fleximers” from our laboratory, these analogues have the connectivity of the heterocyclic base system “reversed”, where the pyrimidine ring is attached to the sugar moiety, rather than the five membered imidazole ring. As was previously seen with the ribose fleximers, their inherent flexibility should allow them to adjust to enzyme binding site mutations, as well as increase the affinity for atypical enzymes. Preliminary biological screening has revealed surprising inhibition of adenosine deaminase, despite their lack of resemblance to adenosine.
Keywords: Fleximers, carbocyclic nucleosides, adenosine deaminase, 5-substituted uracils, pyrimidine based inhibitors
INTRODUCTION
The development of drug resistance to currently used therapeutics is one of the major challenges medicinal chemists are facing today. One common resistance mechanism arises when mutations occur within the active site of a target enzyme. A single point mutation of an amino acid residue within the binding pocket can result in an unfavorable steric or electronic clash and prevent an inhibitor from binding properly.[1] Recent reports have shown that flexible inhibitors can overcome resistance mechanisms and retain activity by utilizing secondary residues within the binding site not previously involved in the enzyme’s mechanism of action.[2–5] Additionally, the flexibility of the inhibitor may allow it to “masquerade” as a different compound, which would slow the onset of resistance that can arise from repeated exposure to a particular chemotherapeutic agent.[2,3]
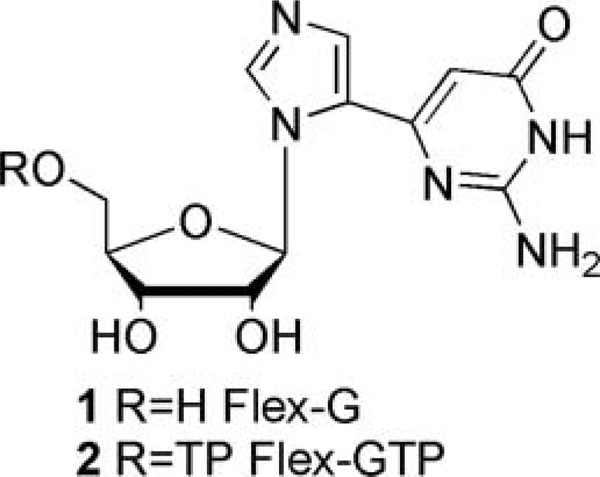
One focus of our research has been to impart flexibility to the nucleobase scaffold to potentially increase inhibitor function and recognition. The purine scaffold of the “fleximer” has been split into the individual imidazole and pyrimidine rings that remain attached via a single carbon–carbon bond.[6–9] As a result, the nucleobase retains the requisite hydrogen bonding and aromatic features necessary for recognition, while gaining the flexibility to adapt to an enzyme’s binding site and potential mutations. In that regard, we have previously reported that a flexible guanosine nucleoside (1, Figure 1), served as an inhibitor of S-adenosylhomocysteine hydrolase (SAHase), an adenosine-metabolizing enzyme.[10] The flexibility of the base unit yielded rotational and conformational changes that allowed the guanosine analogue (Flex-G) to mimic adenosine.[10] In addition to this, our investigations with GTP fucose pyrophosphorylase (GFPP) have shown that the triphosphate of Flex-G (Flex-GTP, 2) was preferred over the natural substrate, guanosine triphosphate (GTP).[11] The ability of Flex-GTP to interact with secondary amino acids within the binding site led to more favorable binding interactions, thereby increasing the affinity in GFPP when compared to GTP.[11,12] In addition to our findings, groups such as Hudson et al. have pursued a series of analogues using click chemistry in which they have found interesting fluorescent properties.[13] It is important to note that, to date, no toxicity has been observed with any of the members of this interesting class of nucleosides.
FIGURE 1.
Flex-G and Flex-GTP.
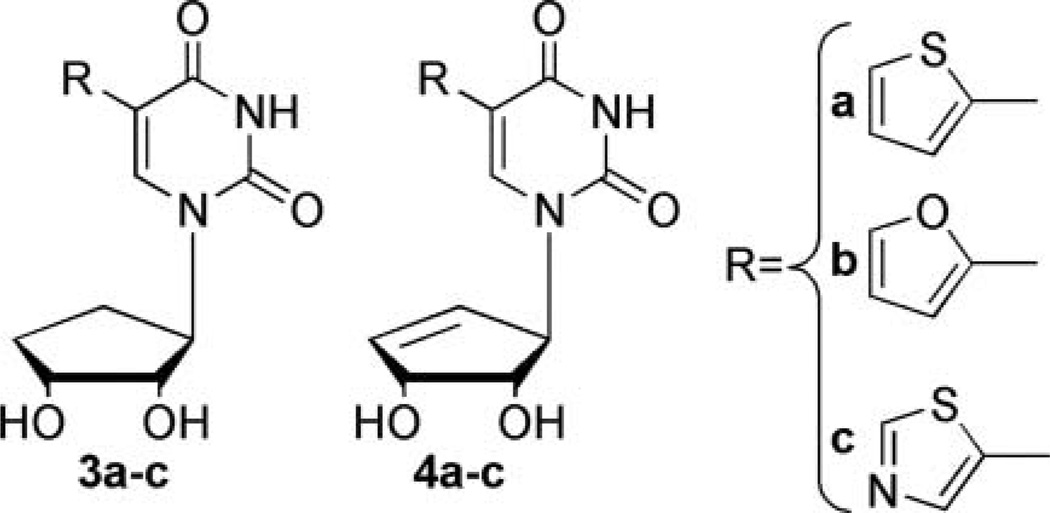
As an extension of our initial studies with the ribose and 2′-deoxyribose fleximers,[14] we have designed a series of “reverse” fleximers,[15] where the purine base scaffold is connected to the sugar moiety at the N-3 of the pyrimidine ring rather than the N-9 of the imidazole ring, as shown in Figure 2 (3a–c and 4a–c). This reverses the conformation of the split purine base, which can also be viewed as a C5-substituted pyrimidine. This type of connectivity is also found in Isoadenosine (IsoA)[16,17] and while IsoA exhibited interesting biological properties, it was not extensively pursued because of instability to both acidic and basic conditions, resulting in a 1,3-migration to afford adenosine.[17]
FIGURE 2.
“Reverse” carbocyclic targets.
Earlier efforts in our laboratory to address the stability issues of IsoA prompted us to the employ the more stable carbocyclic sugar scaffold.[18] This structural change imparts increased stability by rendering the labile glycosidic hemiaminal bond a tertiary amine.[19,20] In addition, carbocyclic nucleosides such as aristeromycin (Ari), 5′-norAri, Neplanocin A (NpcA), and their truncated derivatives are potent inhibitors of SAHase; thus, the carbocyclic modification was considered important for the design of the reverse fleximers.[20–22]
Several ribose compounds structurally similar to our target compounds have been reported. Tor et al. utilized substituted pyrimidine analogues as a fluorescent bioprobe to study DNA helical structure.[23] Herdewijn et al. developed a series of C5 substituted pyrimidine analogues that proved active against HSV-1 due to phosphorylation by the viral thymidine kinase (TK).[24–26] In contrast, our analogues would not be expected to be phosphorylated by TK, since they lack the requisite 5′-hydroxyl that would undergo phosphorylation, however, could inhibit other nucleoside-metabolizing enzymes such as the aforementioned SAHase. Moreover, it was speculated that they may be able to serve as inhibitors of both purine metabolizing enzymes, such as SAHase, but might also prove to be inhibitors of pyrimidine metabolizing enzymes as was noted with Herdewijn’s analogues.
As described herein, these novel pyrimidine nucleosides were not recognized by SAHase as anticipated, but rather, by adenosine deaminase (ADA), something we did not anticipate given their lack of resemblance to adenosine. Although the exact mechanism of action has yet to be elucidated, speculation is that the compounds have a similar effect as exhibited by inosine and xanthosine, which act as product inhibitors. Moreover, given the reports that nucleoside ADA inhibitors can exhibit activity against lymphomas and leukemias due to elevated levels of ADA present in malignant human lymphocytes, this finding may prove significant.[27–29]
RESULTS AND DISCUSSION
Chemistry
In considering the design of the target compounds, the truncated systems were initially chosen due to the toxicity exhibited by Ari and NpcA as a result of phosphorylation at the 5′-hydroxyl.[30] Additionally, the thiophene, furan, and thiazole substituents were chosen instead of the more traditional imidazole due to the higher activity exhibited by Herdewijn’s analogues, as well as the reported inability to remove the N-methyl group from the imidazole analogue.[31]
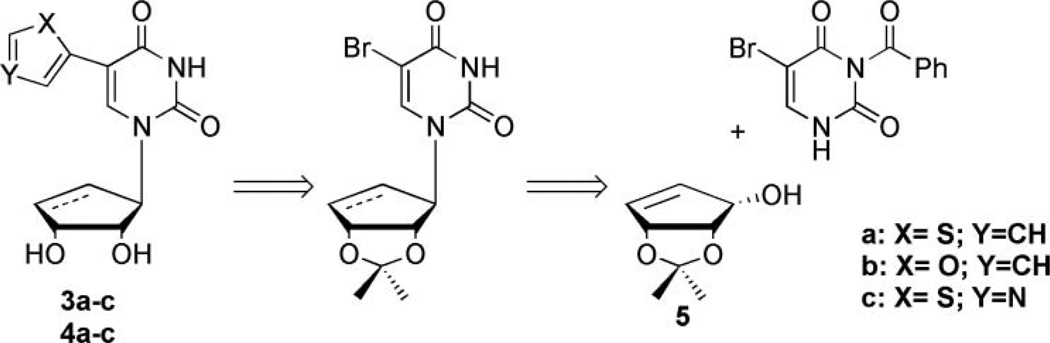
The synthesis of 3a–c and 4a–c was envisioned starting from a well-known enone intermediate,[32] which can undergo a Luche reduction[33] to form the allylic alcohol (5, Figure 3) that can then be coupled using Mitsunobu conditions[34] to attach the pyrimidine. Subsequent organometallic coupling, to the various 5-membered ring systems, would then produce the desired targets.[26,31]
FIGURE 3.
Retrosynthetic approach to targeted fleximers.
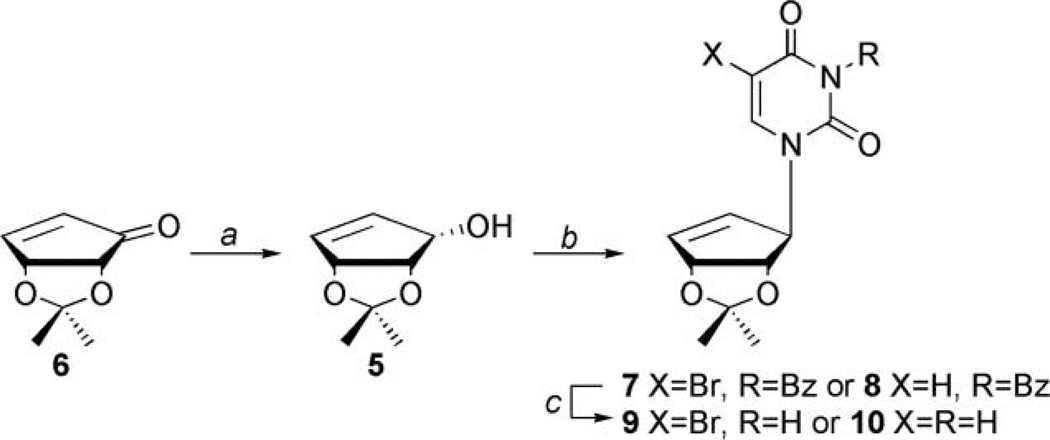
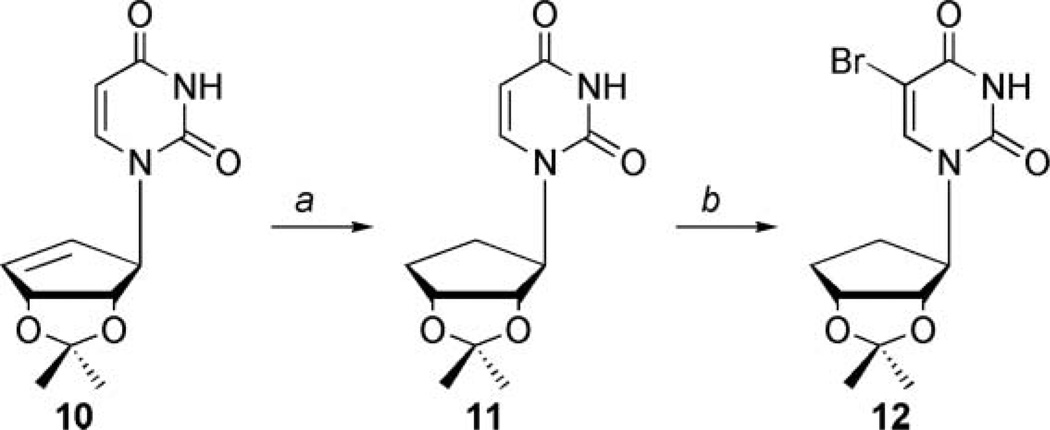
As outlined in Scheme 1, enone 6, which is synthesized from d-ribose,[35] was stereospecifically reduced down to allylic alcohol 5, using Luche reduction conditions[36] and then coupled to either N3-benzoyl-5-bromouracil or N3-benzoyluracil[37] through the use of the Mitsunobu[34] reaction to afford 7 and 8, respectively.
SCHEME 1.
Synthesis of truncated substituted uracil carbocycles. Conditions: (a) NaBH4, MeOH, CeCl3·7H2O, r.t., 1 hour, yield: 93%; (b) N3-benzoyluracil or N3-benzoyl-5-bromouracil, DPPE, DIAD, acetonitrile, r.t., 18 hours, for 7 yield: 78%; for 8 yield: 70%; and (c) methanolic ammonia, r.t., 12 hours, for 9 yield: 62%; for 10 yield: 99%.
It should be noted that the most obvious route to the saturated carbocycles 3a–c involved reducing the double bond in the cyclopentenyl ring of products 4a–c; however, the presence of the C5 substituents prohibited this approach, due to unwanted side reactions that would result from a palladium catalyzed hydrogenation. In addition, reduction prior to that point would have removed the halide on the pyrimidine ring. Once the benzoyl protected products, 7 and 8 were obtained, they were deblocked using mildly basic conditions to yield 9 and 10.
Synthetic efforts then shifted to the saturated carbocyclic series (3a–c) shown in Scheme 2, which could be obtained from intermediate 10. To accomplish this, two additional steps were required to install the necessary bromine functionality. Compound 10 was first reduced using catalytic hydrogenation conditions with Pd/C to afford 11 in quantitative yield, which was subsequently brominated at the C5 position of the uracil ring using NBS in dimethoxyethane and water to afford 12.[38]
SCHEME 2.
Synthesis of saturated carbocyclic C5-bromouracil. Conditions: (a) Pd/C, MeOH, H2 25 psi, 5 hours, quantitative and (b) NBS, NaN3, DME, H2O, r.t., 24 hours, yield: 48%.
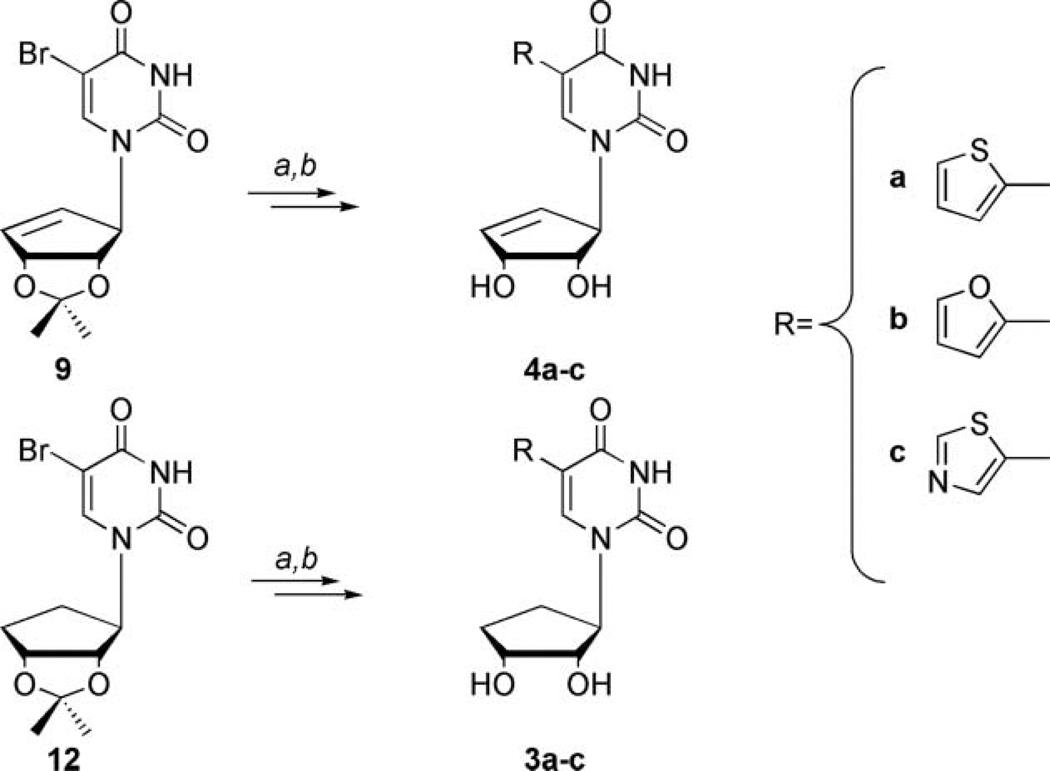
As shown below in Scheme 3, intermediates 9 and 12 were then subjected to Stille coupling methodology using the corresponding tin reagents of the thiazole, thiophene, and furan. These are commercially available or readily constructed using literature procedures.[39,40] These tin reagents can be coupled using PdCl2(PPh3)2 or Pd(PPh3)4,[24,41,42] then subsequent deblocking of the 2′,3′-diol of each of the compounds was readily achieved through the use of mildly acidic conditions employing trifluoroacetic acid and water to provide 3a–c or 4a–c in good yields.
SCHEME 3.
Synthesis of reverse fleximer truncated carbocycles. Conditions: (a) for 3a and 4a, 12 or 9 respectively, tributylstannyl thiophene, dioxane, PdCl2(PPh3)2, reflux; for 3b and 4b, 12 or 9, respectively, tributylstannyl furan, dioxane, PdCl2(PPh3)2, reflux for 3c and 4c, 12 or 9, respectively, tributylstannyl thiazole, dioxane, Pd(PPh3)4, reflux; (b) TFA:H2O (2:1) in THF, r.t., 6 hours. Yield over two steps: for 3a yield: 90%; for 3b yield: 90%; for 3c yield: 89%; for 4a yield: 55%; for 4b yield: 58%; for 4c yield: 42%.
Assays
Given our initial goal of developing SAHase inhibitors, we then screened the reverse fleximers against SAHase using a standard assay previously reported by us and others.[43,44] The SAHase assay is a dual enzyme assay that monitors the rate of hydrolysis of SAH by SAHase to homocysteine and adenosine. This reaction utilizes adenosine deaminase (ADA) to monitor the conversion of adenosine to inosine, which results in a decrease of absorbance at 265 nm. Initially the compounds appeared to inhibit SAHase, however, upon further investigation it was revealed the compounds were not inhibiting SAHase, but instead ADA (Table 1). ADA converts adenosine to inosine, and is linked to the deactivation of various nucleoside drugs such as AraA through deamination via ADA.[45–47]
TABLE 1.
Percent inhibition and fitness of analogues in the active site of ADA
| Compounda | Percent inhibitionb | Fitness scorec |
|---|---|---|
| Inosine | 31% | 50.00 |
| 3a | 36% | 50.90 |
| 3b | 35% | 48.70 |
| 3c | 28% | 47.29 |
| 4a | 48% | 54.78 |
| 4b | 55% | 53.62 |
| 4c | 49% | 51.90 |
Compounds were dissolved in DMSO and tested at 100 µM for analysis.
Calculated as: [1-(reaction rate of inhibited ADA/reaction rate of uninhibited ADA)]*100%. Rates were determined by monitoring the decrease in absorbance at 265 nm from the conversion of adenosine to inosine.
Analogues were built in Chem3D then transferred to Sybyl before analysis. Fitness score is the relative affinity of the ligand to the binding site.
Due to compounds 3a–c and 4a–c structural similarity to inosine, the deaminated product of adenosine following metabolism by ADA, computational studies were conducted to uncover possible reasons for this unusual activity. The structures were docked using Gold 4.1.2[48] in bovine ADA [pdb file: 1KRM] that had been crystallized with the transition state analogue, 6-hydroxy-1,6-dihydropurine ribose.[49] Each molecule was docked to the binding site and scored. The top three results of each docked inhibitor were selected for analysis. Of the six inhibitors, four scored higher than inosine (Table 1).
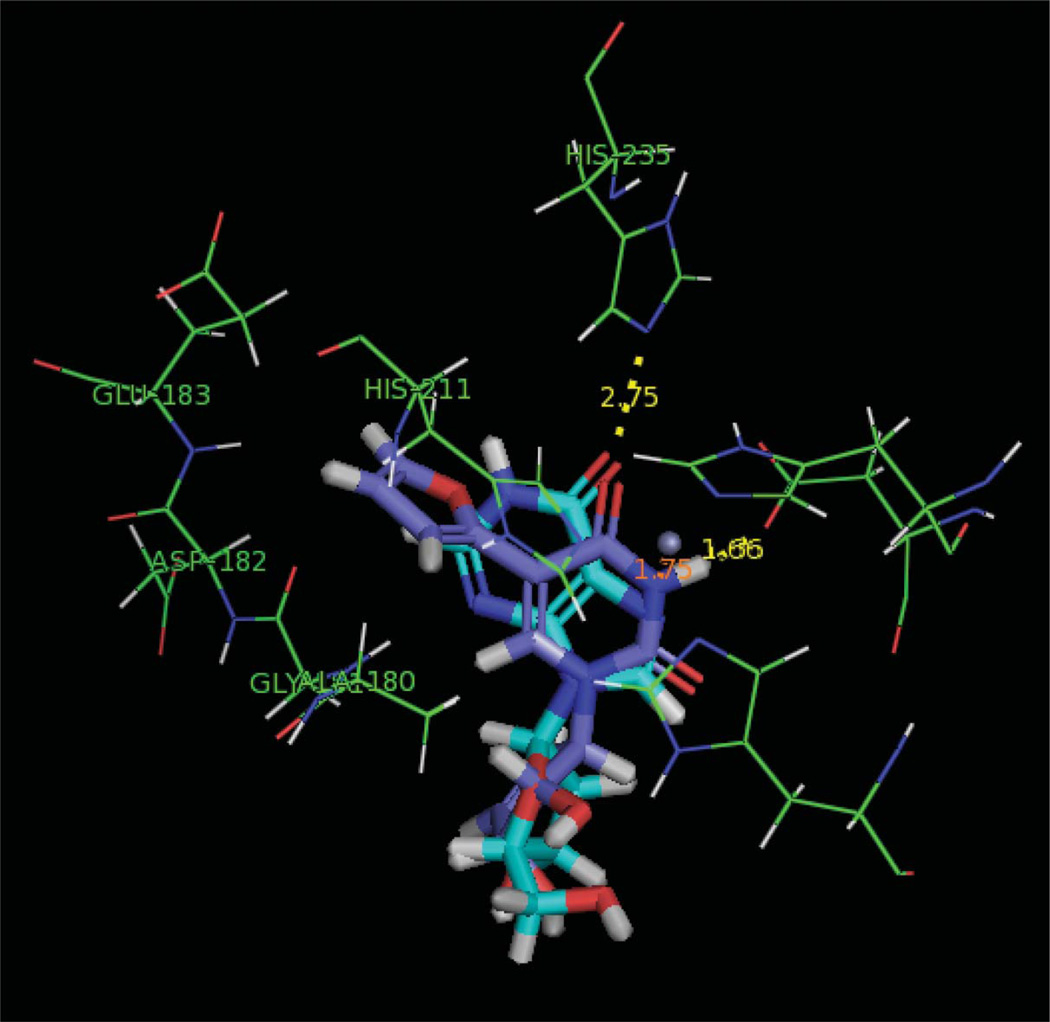
Upon further inspection of the ADA binding site, a large binding pocket adjacent to the adenosine binding site was discovered. This adjacent pocket can accommodate the pendant heteroaromatic rings as depicted in Figure 4 with inosine and 4b. Furan analogue 4b exhibited the best inhibition as well as fitness score as shown in Table 1. Thus, it appears that the new bonding interactions provided by the added flexibility and length of the C5 substitute pendant ring are likely assisting in the recognition and binding. Interestingly, the more potent inhibitors exhibited a distinct perpendicular conformation for the pendant and pyrimidine rings, while the remainder of the nucleoside scaffold overlaps with inosine.
FIGURE 4.
Reverse fleximer 4b (purple) and inosine (cyan) in the active site of Bovine ADA (Color figure available online).
In comparing the computational results shown in Table 1 for the six inhibitors, several conclusions can be drawn. One, the unsaturated analogues seem to be better inhibitors than the saturated series, which coincides with the results observed for the inhibition. Also in agreement is that the thiophene and furan pendant systems have similar activities that are expected to be potentially better than the thiazole pendant system due to their computational fitness. We are now focused on exploring additional heterocyclic and aromatic C5 substituents and exploring the incorporation of linkers between the pyrimidine and C5 substituent to lengthen the “reach” of the substituent into the adjacent pocket, in hopes of increasing the affinity of the enzyme for these nucleoside analogues.
The compounds were also subjected to broad screen antiviral testing by Professor Jan Balzarini at the Katholieke Universiteit (Leuven, Belgium) against viruses such as HSV-1, HSV-2, vaccinia and influenza types A and B. Disappointingly, none of the compounds showed activity comparable to Ribavirin.
CONCLUSION
In summary, a novel series of carbocyclic “reverse fleximer” nucleosides was successfully synthesized and resulted in a new type of adenosine deaminase inhibitor. These findings serve as further evidence that flexible inhibitors can be recognized in atypical enzymatic systems, thus conformational adaptability may prove to be beneficial for future therapeutics. Current efforts are underway to synthesize the corresponding Ari and NpcA analogues to see if the addition of the 4′-hydroxymethyl imparts recognition by viral TK as was observed with Herdewijn’s series, as well as possible recognition by other enzymes that require the 5′-hydroxyl. The results of those studies will be reported elsewhere as they become available.
EXPERIMENTAL SECTION
General
All chemicals were obtained from commercial sources and used without further purification unless otherwise noted. Anhydrous DMF, MeOH, DMSO, and toluene were purchased from Fisher Scientific (Pittsburgh, PA, USA). Anhydrous THF, acetone, CH2Cl2, CH3CN and ether were obtained using a solvent purification system (mBraun Labmaster 130, Stratham, NH, USA). Melting points are uncorrected. NMR solvents were purchased from Cambridge Isotope Laboratories (Andover, MA, USA). All 1H and 13C NMR spectra were obtained on a JEOL ECX 400 MHz NMR, operated at 400 and 100 MHz and referenced to internal tetramethylsilane (TMS) at 0.0 ppm. The spin multiplicities are indicated by the symbols s (singlet), d (doublet), dd (doublet of doublets), t (triplet), q (quartet), m (multiplet), and b (broad). Reactions were monitored by thin-layer chromatography (TLC) using 0.25 mm Whatman Diamond silica gel 60-F254 precoated plates. Column chromatography was performed using silica gel (63–200 μ) from Dynamic Adsorptions Inc. (Norcross, GA, USA), and eluted with the indicated solvent system. Yields refer to chromatographically and spectroscopically (1H and 13C NMR) homogeneous materials. Mass spectra were recorded at the Johns Hopkins Mass Spectrometry Facility (Baltimore, MD, USA).
General Procedure for Mitsunobu Coupling of Protected Uracils to Cyclopentenol
To a stirred suspension of 5 (1.06 g, 6.80 mmol), protected uracil (10.2 mmol), and 1,2-bis(diphenylphosphino)ethane (1.49 g, 3.37 mmol) in acetonitrile (75 mL) was added dropwise DIAD (2.0 mL, 10.2 mmol) at 0°C. The reaction mixture was allowed to stir at room temperature (r.t.) for 18 hours. The solvent was then removed under reduced pressure and the resulting residue purified using silica gel chromatography eluting with hexanes:EtOAc (1:2).
1-[(2′,3′-O-Isopropylidene)-4′-cyclopenten-1′-yl]-3-benzoyl-5-bromouracil (7)
Off-white crystalline solid (4.02 g, 9.28 mmol, 78%). 1H NMR (CDCl3): δ 1.34 (s, 3H), 1.41 (s, 3H), 4.62 (d, 1H, J = 14.9 Hz), 5.34 (dd, 1H, J = 14.8, 3.5 Hz), 5.40 (d, 1H, J = 3.5 Hz), 5.78 (dt, 1H, J = 13.8 Hz), 6.32 (dt, 1H, J = 3.4, 13.8 Hz), 7.41 (s, 1H), 7.48 (t, 2H, J = 18.3 Hz), 7.64 (t, 1H, J = 18.3 Hz), 7.90 (d, 2H, J = 17.2 Hz). 13C NMR (CDCl3): δ 22.7, 25.6, 70.0, 83.5, 84.4, 96.8, 112.7, 128.8, 129.4, 130.7, 131.0, 135.5, 140.1, 140.7, 149.1, 156.5, 158.1, 167.5. HRMS calculated for C19H17BrN2O5 [M+H]+ 432.0320; found, 432.0315.
1-[(2′,3′-O-Isopropylidene)-4′-cyclopenten-1′-yl]-3-benzoyl-uracil (8)
Clear colorless foam (1.69 g, 4.77 mmol, 70%). 1H NMR (DMSO-d6): δ 1.33 (s, 3H), 1.41 (s, 3H), 4.71 (d, 1H, J = 13.7Hz), 5.27 (s, 1H), 5.29 (d, 1H, J = 11.5 Hz), 5.81 (d, 1H, J = 19.4 Hz), 5.84 (dd, 1H, J = 5.7, 13.7 Hz), 6.23 (dt, 1H, J = 4.6, 13.8 Hz), 7.51 (d, 1H, J = 19.5 Hz), 7.57 (t, 2H, J = 19.5 Hz), 7.78 (t, 1H, J = 19.5 Hz), 7.97 (d, 2H, J = 21.7 Hz). 13C NMR (DMSO-d6): δ 24.4, 26.2, 69.1, 83.2, 84.6, 101.1, 111.8, 128.9, 129.0, 130.1, 131.6, 135.0, 139.0, 143.0, 149.9, 163.0, 168.8. HRMS calculated for C19H18N2O5 [M+H]+ 355.1294; found, 355.1282.
General Procedure for the Removal of the Benzoyl Protecting Group from Uracil Intermediates
To a stirred solution of benzoyl protected carbocyclic uracil (7.08 mmol) in MeOH (10 mL) was addedmethanolic ammonia (100 mL) and allowed to react for 18 hours. The reaction mixture was condensed under reduced pressure and then re-evaporated from EtOH (3 × 100 mL). The crude residue was purified by silica gel chromatography eluting with hexanes:EtOAc (1:4).
1-[(2′,3′-O-Isopropylidene)-4′-cyclopenten-1′-yl]-5-bromouracil (9)
Off-white solid (1.98 g, 5.75 mmol, 62%). 1H NMR (CDCl3) δ 1.35 (s, 3H), 1.45 (s, 3H), 4.56 (d, 1H, J = 14.9 Hz), 5.35 (dd, 1H, J = 3.5, 14.9 Hz), 5.46 (s, 1H), 5.76 (dd, 1H, J = 3.4, 13.8 Hz), 6.34 (dt, 1H, J = 4.6, 14.9 Hz), 7.28 (s, 1H), 8.67 (s, 1H). 13C NMR (CDCl3) δ 25.6, 27.3, 69.2, 83.7, 84.3, 97.0, 112.7, 129.0, 140.0, 140.5, 149.8, 158.7. HRMS calculated for C12H13BrN2O4 [M+H 79Br]+ 329.0137, [M+H 81Br]+ 331.0117; found, 329.0122, 331.0110.
1-[(2′,3′-O-Isopropylidene)-4′-cyclopenten-1′-yl]-uracil (10)
Off-white solid (1.77 g, 7.07 mmol, 99%). 1H NMR (CDCl3): δ 1.31 (s, 3H), 1.41 (s, 3H), 4.56 (d, 1H, J = 14.9 Hz), 5.31 (dd, 1H, J = 2.3, 14.9), 5.41 (d, 1H, J = 4.6), 5.66 (d, 1H, J = 20.6 Hz), 5.72 (dd, 1H, J = 3.5, 13.7 Hz), 6.23 (d, 1H, J = 14.9 Hz), 6.98 (d, 1H, J = 20.6 Hz), 9.90 (s, 1H). 13C NMR (CD3OD): δ 24.4, 26.2, 68.4, 83.4, 84.6, 101.2, 111.7, 129.3, 138.5, 142.5, 151.3, 165.0. HRMS calculated for C12H14N2O4 [M+H]+ 251.1032; found, 251.1026.
Preparation of 1-[(2′,3′-O-Isopropylidene)-cyclopentan-1′-yl]-uracil (11)
A solution of 10 (1.77 g, 7.07 mmol) and Pd/C (150 mg) in MeOH (100 mL) was hydrogenated for 3 hours at 25 psi. The reaction mixture was filtered through a pad of celite and the solvent removed under vacuum to give 11 as an off-white foam (1.54 g, 6.11 mmol, 86%). 1H NMR (CDCl3): δ 1.27 (s, 3H), 1.46 (s, 3H), 1.88–1.93 (m, 2H), 2.11–2.15 (m, 1H), 2.29–2.34 (m, 1H), 4.37–4.39 (m, 1H), 4.76 (dd, 1H, J = 6.9, 15.5 Hz), 4.80 (td, 1H, J = 5.7, 14.9 Hz), 5.66 (d, 1H, J = 20.6 Hz), 7.07 (d, 1H, J = 20.7 Hz), 8.24 (bs, 1H). 13C NMR (DMSO-d6): 24.4, 27.3, 29.1, 31.2, 63.9, 80.3, 84.4, 101.8, 110.0, 143.9, 152.6, 163.8. HRMS calculated for C12H16N2O4 [M+H]+ 253.1183; found, 253.1189.
Preparation of 1-[(2′,3′-O-Isopropylidene)-cyclopentan-1′-yl]-5-bromouracil (12)
To a stirred solution of 9 (1.19 g, 4.72 mmol) in 1,2-dimethoxyethane (200 mL) was added a solution of sodium azide (920 mg, 14.15 mmol) and NBS (1.01 g, 5.66 mmol) in water (5 mL) and stirred for 24 hours at r.t. The reaction mixture was then concentrated in vacuo and the resulting residue purified by silica gel chromatography eluting with hexanes:EtOAc (1:2) to afford 12 as a white solid (750 mg, 2.27 mmol, 48%). 1H NMR (CDCl3): δ 1.30 (s, 3H), 1.49 (s, 3H), 1.92–1.97 (m, 2H), 2.14–2.19 (m, 1H), 2.32–2.36 (m, 1H), 4.41 (m, 1H), 4.77 (dd, 1H, J = 6.9, 15.5 Hz), 4.82 (td, 1H, J = 5.7, 14.9 Hz), 7.42 (s, 1H), 9.33 (s, 1H). 13C NMR (CDCl3): δ 15.2, 24.5, 28.7, 29.2, 65.1, 81.3, 85.2, 89.8, 110.9, 150.5, 165.2, 165.3. HRMS calculated for C12H15BrN2O4 [M+H 79Br]+ 331.0293, [M+H 81Br]+ 333.0243; found, 331.0287, 333.0259.
General Procedure for the Stille Coupling with Stannyl Thiophene and Stannyl Furan Reagents
To a stirred solution of the desired 5-bromo carbocyclic nucleoside (1.74 mmol) and aryl tin (8.72 mmol) in dioxane (50 mL) was added PdCl2(PPh3)2 (122 mg, 0.17 mmol) and the temperature held at 90°C for 18 hours. The reaction mixture was then evaporated to dryness and the residue purified by silica gel chromatography eluting with hexanes:EtOAc (1:1).
1-[(2′,3′-O-Isopropylidene)-4′-cyclopenten-1′-yl]-5-(thiophene-2-yl)-uracil
Off-white solid (343 mg, 1.03 mmol, 59%). 1H NMR (CDCl3) δ 1.36 (s, 3H), 1.46 (s, 3H), 4.63 (d, 1H, J = 14.9 Hz), 5.37 (dd, 1H, J = 2.3, 11.4 Hz), 5.50 (d, 1H, J = 4.6 Hz), 5.82 (dd, 1H, J = 3.4, 13.8 Hz), 6.34 (dt, 1H, J = 4.5, 14.9 Hz), 7.04 (dd, 1H, J = 9.2, 12.6 Hz), 7.29 (d, 1H, J = 3.4 Hz), 7.31 (s, 1H), 7.35 (dd, 1H, J = 3.4, 9.1 Hz), 9.18 (s, 1H). 13C NMR (CDCl3) δ 25.6, 27.3, 69.3, 83.7, 84.5, 112.6, 124.8, 125.8, 127.3, 129.3, 133.1, 136.2, 139.5, 141.1, 160.7, 172.1. HRMS calculated for C16H16N2O4S [M+H]+ 332.0830; found, 332.0840.
1-[(2′,3′-O-Isopropylidene)-4′-cyclopenten-1′-yl]-5-(furan-2-yl)-uracil
Off-white solid (520 mg, 1.64 mmol, 63%). 1H NMR (CDCl3) δ 1.35 (s, 3H), 1.46 (s, 3H), 4.62 (d, 1H, J = 13.7 Hz), 5.41 (dd, 1H, J = 2.6, 12.6 Hz), 5.55 (d, 1H, J = 3.5 Hz), 5.81 (dt, 1H, J = 3.5, 13.8 Hz), 6.35 (dt, 1H, J = 3.4, 14.9 Hz), 6.44 (dd, 1H, J = 4.6, 8.0 Hz), 7.03 (d, 1H, J = 8.0 Hz), 7.34 (d, 1H, J = 4.5 Hz), 7.44 (s, 1H), 8.93 (s, 1H). 13C NMR (CDCl3) δ 22.0, 29.8, 67.1, 69.0, 83.8, 84.5, 109.7, 112.1, 128.5, 129.5, 134.7, 136.7, 139.4, 141.3, 149.8, 160.1. HRMS calculated for C16H16N2O5 [M]+ 316.1059; found, 316.1053.
1-[(2′,3′-O-Isopropylidene)-cyclopentan-1′-yl]-5-(thiophene-2-yl)uracil
Off-white solid (111 mg, 0.31 mmol, 99%).1H NMR (CDCl3): δ 1.32 (s, 3H), 1.51 (s, 3H), 1.95–2.05 (m, 2H), 2.21–2.25 (m, 1H), 2.39–2.41 (m, 1H), 4.44–4.49 (m, 1H), 4.83–4.85 (m, 1H), 4.88 (dd, 1H, J = 6.9, 14.8 Hz), 7.04 (dd, 1H, J = 9.2, 12.6), 7.29 (dd, 1H, J = 2.3, 12.6), 7.39 (dd, 1H, J = 2.3, 9.1 Hz), 7.43 (s, 1H), 8.49 (s, 1H). 13C NMR (CD3OD): δ 12.6, 23.4, 26.0, 26.6, 27.9, 28.9, 31.0, 66.7, 80.9, 85.5, 109.7, 111.4, 123.6, 125.3, 126.2, 139.3. HRMS calculated for C16H18N2O4S [M]+ 334.0987; found, 334.0979.
1-[(2′,3′-O-Isopropylidene)-cyclopentan-1′-yl]-5-(furan-2-yl)-uracil
Off-white solid (100 mg, 0.31 mmol, 99%). 1H NMR (CDCl3) δ 1.32 (s, 3H), 1.51 (s, 3H), 1.97–2.01 (m, 2H), 2.19–2.24 (m, 1H), 2.38–2.42 (m, 1H), 4.55–4.59 (m, 1H), 4.81 (dd, 1H, J = 5.7, 14.9 Hz), 4.87 (td, 1H, J = 5.7, 14.8 Hz), 6.46 (dd, 1H, J = 4.6, 9.2 Hz), 7.02 (d, 1H, J = 8.0 Hz), 7.36 (d, 1H, J = 4.5 Hz), 7.57 (s, 1H), 8.17 (s, 1H). 13C NMR (CD3OD): δ 13.7, 17.4, 24.5, 27.0, 28.4, 29.2, 31.6, 66.9, 80.7, 84.7, 109.5, 112.1, 136.5, 141.3, 149.5, 159.6. HRMS calculated for C16H18N2O5 [M]+ 318.1216; found, 318.1219.
General Procedure for the Stille Coupling with Stannyl Thiazole Reagents
To a stirred solution of the desired 5-bromo carbocyclic nucleoside (1.10 mmol) and 5-(tributylstannyl)thiazole (900 mg, 2.41 mmol) in dry THF (50 mL) was added Pd(PPh3)4 (50 mg) and refluxed under nitrogen for 72 hours. The reaction mixture was then evaporated to dryness and the resulting residue purified by silica gel chromatography eluting with EtOH:CH2Cl2 (5:95).
1-[(2′,3′-O-Isopropylidene)-4′-cyclopenten-1′-yl]-5-(thiazol-5-yl)-uracil
Off-white solid (100 mg, 0.30 mmol, 52%). 1H NMR (CD3OD) δ 1.36 (s, 3H), 1.42 (s, 3H), 4.72 (d, 1H, J = 13.7 Hz), 5.38 (dd, 1H, J = 2.3, 14.9 Hz), 5.42 (s, 1H), 5.85 (dd, 1H, J = 4.6, 14.9 Hz), 6.26 (dt, 1H, J = 4.6, 13.7 Hz), 7.83 (s, 1H), 8.17 (s, 1H), 8.90 (s, 1H). 13C NMR (CD3OD) δ 24.5, 26.3, 70.02, 83.3, 84.9, 106.1, 111.7, 129.1, 129.5, 138.4, 138.7, 139.2, 150.3, 153.9, 162.1. HRMS calculated for C15H15N3O4S [M+H]+ 334.0862; found, 334.0855.
1-[(2′,3′-O-Isopropylidene)-cyclopentan-1′-yl]-5-(thiazol-5-yl)-uracil
Off-white solid (96.2 mg, 0.29 mmol, 95%). 1H NMR (DMSO-d6): δ 1.21 (s, 3H), 1.39 (s, 3H), 1.67–1.74 (m, 1H), 1.95–1.99 (m, 1H), 2.10–2.19 (m, 2H), 4.56–4.60 (m, 1H), 4.73–4.77 (m, 1H), 4.81 (dd, 1H, J = 0.8, 14.9 Hz), 8.22 (s, 1H), 8.28 (s, 1H), 8.96 (s, 1H), 11.78 (bs, 1H). 13C NMR (CD3OD): δ 12.7, 23.5, 26.1, 29.0, 29.5, 30.9, 31.8, 67.0, 80.8, 84.4, 111.4, 128.7, 130.8, 132.5, 140.7. HRMS calculated for C15H17N3O4S [M+H]+ 336.1018; found, 336.1010.
General Procedure for the Removal of the 2′,3′-isopropylidene Protecting Group
A solution of 2′,3′-protected carbocyclic nucleoside (0.22 mmol) in TFA:H2O (9 mL, 2:1) was stirred at r.t. for 18 hours. The reaction mixture was evaporated under reduced pressure and the resulting residue was dissolved in EtOH (3 × 10 mL) and concentrated three times under vacuum. The crude product was purified over silica gel eluting with EtOAc:acetone:MeOH:H2O (8:1:1:0.5).
1-[(2′,3′-Dihydroxy)-4′-cyclopenten-1′-yl]-5-(thiophene-2-yl)-uracil (4a)
Yellow solid (60.3 mg, 0.21 mmol, 94%). λmax = 259 nm. 1H NMR (DMSO-d6): δ 4.07 (q, 1H), 4.42 (bs, 1H), 4.81 (d, 1H, J = 13.8 Hz), 4.99 (d, 1H, J = 17.2 Hz), 5.36 (d, 1H, J = 14.9 Hz), 5.89 (dd, 1H, J = 4.5, 14.9 Hz), 6.08 (dt, 1H, J = 5.8, 16.0 Hz), 7.01 (dd, 1H, J = 9.1, 12.6 Hz), 7.42 (d, 1H, J = 12.6 Hz), 7.44 (dd, 1H, J = 2.3, 9.1 Hz), 7.74 (s, 1H), 11.94 (s, 1H). 13C NMR (DMSO-d6): δ 19.1, 31.5, 67.3, 72.9, 76.2, 119.1, 123.5, 126.3, 126.9, 133.1, 137.2, 162.0, 181.0. HRMS calculated for C13H12N2O4S [M]+ 292.0518; found, 292.0515.
1-[(2′,3′-Dihydroxy)-4′-cyclopenten-1′-yl]-5-(furan-2-yl)-uracil (4b)
Off-white foam (20.0 mg, 0.07 mmol, 92%). λmax = 256 nm. 1H NMR (DMSO-d6): δ 3.93 (q, 1H, J = 13.7 Hz), 4.40 (m, 1H), 4.85 (d, 1H, J = 14.9 Hz), 5.00 (d, 1H, J = 16.1 Hz), 5.39 (dt, 1H, J = 3.4, 10.3 Hz), 5.89 (dd, 1H, J = 4.6, 14.9 Hz), 6.13 (dt, 1H, J = 5.8, 16.0 Hz), 6.49 (dd, 1H, J = 4.6, 9.2 Hz), 6.83 (d, 1H, J = 6.9 Hz), 7.52 (s, 1H), 7.60 (dd, 1H, J = 2.3, 4.6 Hz), 11.60 (s, 1H). 13C NMR (acetone-d6): 67.5, 73.4, 76.4, 106.2, 108.2, 111.5, 132.2, 135.2, 137.9, 141.2, 146.8, 150.3, 160.1 HRMS calculated for C13H12N2O5 [M+H]+ 277.0825; found, 277.0821.
1-[(2′,3′-Dihydroxy)-4′-cyclopenten-1′-yl]-5-(thiazol-5-yl)-uracil (4c)
Off-white foam (71.7 mg, 0.22 mmol, 81%). λmax = 260 nm. 1H NMR (acetone-d6): δ 4.19 (q, 1h, J = 13.7 Hz), 4.25–4.27 (m, 1H), 4.68–4.69 (m, 1H), 5.52–5.54 (m, 1H), 6.02 (dd, 1H, J = 5.7, 16.0 Hz), 6.21 (dt, 1H, J = 5.7, 10.3 Hz), 7.00 (dd, 1H, J = 9.2, 12.6 Hz), 7.36 (dd, 1H, J = 3.4, 13.7 Hz), 7.41 (dd, 1H, J =2.3, 9.2Hz), 7.73 (s, 1H), 10.32 (bs, 1H). 13C NMR (acetone-d6): δ 13.5, 24.5, 64.0, 71.0, 75.5, 106.0, 129.2, 139.1, 140.1, 159.3, 152.9. HRMS calculated for C12H11N3O4S [M+H]+ 294.0549; found 294.0547.
1-[(2′,3′-Dihydroxy)-cyclopentan-1′-yl]-5-(thiophene-2-yl)-uracil (3a)
Off-white foam (5.7 mg, 0.02 mmol, 91%). λmax = 260 nm. 1H NMR (acetone-d6): δ 1.65–1.68 (m, 1H), 1.86–1.89 (m, 1H), 2.09–2.16 (m, 2H), 3.83 (bs, 1H), 4.09–4.11 (m, 1H), 4.25 (bs, 1H), 4.42 (q, 1H, J = 11.5 Hz), 4.70–4.79 (m, 1H), 7.01 (dd, 1H, J = 9.1, 12.6 Hz), 7.35 (dd, 1H, J = 2.3, 12.6 Hz), 7.47 (dd, 1H, J = 2.3, 9.1 Hz), 8.01 (s, 1H), 10.28 (s, 1H). 13C NMR (acetone-d6): δ 12.7, 23.8, 64.1, 70.7, 74.3, 107.1, 128.7, 131.0, 138.6, 141.1, 158.9, 160.9, 161.2. HRMS calculated for C13H14N2O4S [M+H]+ 295.0753; found, 295.0747.
1-[(2′,3′-Dihydroxy)-cyclopentan-1′-yl]-5-(furan-2-yl)-uracil (3b)
White solid (20.0 mg, 0.07 mmol, 91%). λmax = 255 nm. 1H NMR (acetone-d6): δ 1.63–1.66 (m, 1H), 1.87–1.90 (m, 1H), 2.11–2.17 (m, 2H), 3.83 (bs, 1H), 4.08–4.11 (m, 1H), 4.25 (s, 1H), 4.42 (q, 1H, J = 11.4 Hz), 4.72–4.78 (m, 1H), 6.47 (dd, 1H, J = 3.5, 8.1 Hz), 6.93 (d, 1H, J = 6.8 Hz), 7.50 (d, 1H, J = 3.4 Hz), 7.95 (s, 1H), 10.24 (bs, 1H). 13C NMR (acetoned6): δ 12.5, 23.68, 64.3, 71.0, 74.6, 106.9, 127.6, 131.3, 137.9, 141.4, 158.6, 161.1, 161.7. HRMS calculated for C13H14N2O5 [M+H]+ 279.0981; found, 279.0952.
1-[(2′,3′-Dihydroxy)-cyclopentan-1′-yl]-5-(thiazol-5-yl)-uracil (3c)
White solid (25.2 mg, 0.09 mmol, 94%). λmax = 260 nm. 1H NMR (DMSO-d6): δ 1.51–1.54 (m, 1H), 1.67–1.70 (m, 1H), 1.95–2.21 (m, 2H), 3.90 (bs, 1H), 4.20–4.24 (m, 1H), 4.59 (d, 1H, J = 6.9 Hz), 4.68 (q, 1H, J = 22.9 Hz), 4.96 (d, 1H, J = 16.1 Hz), 8.33 (s, 1H), 8.34 (s, 1H), 8.95 (s, 1H), 11.72 (s, 1H). 13C NMR (DMSO-d6): δ 24.8, 29.0, 62.5, 70.7, 75.3, 105.8, 129.3, 139.3, 140.5, 150.8, 154.1, 161.8. HRMS calculated for C12H13N3O4S [M+H]+ 296.0705; found, 296.0703.
SAHase Assay
The enzyme-coupled continuous assay in the hydrolysis direction was performed as previously reported.[43] Briefly, SAH is hydrolyzed to homocysteine and adenosine, which is subsequently converted by adenosine deaminase into ammonia and inosine, a process associated with a decrease of absorbance at 265 nm. Each assay was conducted in thermostatted 1 cm quartz cuvettes at 37°C maintained by a Peltier unit on a Cary 100 ultraviolet-visible spectrophotometer. Enzyme assay solution typically contained 50 mM potassium phosphate at pH 7.4, 0.39 units of adenosine deaminase (Worthington Biochemical Corp., Lakewood, NJ, USA), 0.132 nM of SAHase (provided by Dr. Lynne Howell), 10 μM of SAH and various concentrations of inhibitors in a total volume of 1000 μL. The reactions were initiated by the addition of SAH. In all cases, we ascertained that SAH hydrolysis catalyzed by SAHase was rate limiting under testing conditions (data not shown). The kinetic data were analyzed using KaleidaGraph 4.0 (Synergy Inc., Reading, PA, USA). Based on a competitive inhibition mechanism, the Ki value was determined using the equation, v = kcat × [S] × [E]/{Km × (1 + [I]/Ki) + [S]}; where v, kcat, [S], [E], Km, [I], and Ki stand for the initial reaction rates, rate constant, substrate concentration, enzyme concentration, Michaelis–Menten constant, inhibitor concentration, and dissociation constant of the enzyme-inhibitor complex, respectively. A previously reported Km value of 7.9 μM for SAH in the hydrolysis direction was used for all calculations.[50]
ADA Assay
The ADA assay was performed in a similar manner to the SAHase assay conducted in thermostatted 1 cm quartz cuvettes at 37°C maintained by a Peltier unit on a Cary 100 ultraviolet-visible spectrophotometer. ADA mitigates the reaction of adenosine to inosine, which can be monitored by measuring the decrease in absorbance at 265 nm. Assay solutions contained 50 mM potassium phosphate at pH 7.4, 0.001 units of adenosine deaminase (Worthington Biochemical Corp.), 3 μM of adenosine at a total volume of 1000 μL. Inosine, 3a–c and 4a–c (100 μM) were monitored for the consumption of substrate. All reactions were performed in triplicate, and then the rates were averaged. Relative Inhibition was determined by dividing the rate of the inhibited reaction by the rate of the reaction with no inhibitor present. Bovine adenosine deaminase was used due to similarity to human adenosine deaminase and its availability.
Docking
Six compounds were devised for binding to Bovine ADA (Protein Data Bank ID: 1KRM).[49] Each derivative was built in Chem3D and then transferred to Sybyl. The derivatives were assigned with Gasteiger–Huckel charges and were energy minimized in Sybyl using the Powell method with 1000 iterations. The Tripos Mol2 files generated in Sybyl were merged into one file and transferred to Gold 4.1.2.[48] Each derivative was then docked to Bovine ADA as the receptor.
The crystal structure of Bovine ADA included a bound inosine, zinc, carbon dioxide, and an amine. The inosine, carbon dioxide and the amine were removed electronically. After protonation of the N-terminus and deprotonation of the C-terminus, the charges of the protein functionalities were determined as described for the above derivatives. The inosine was used for the receptor-grid generation and the final grid volume corresponding to the active site of the protein was 20 × 20 × 20 Å. Standard precision docking was applied with flexibility of ring flips, twists, and internal hydrogen bonding in the ligands and no positional constraints were applied to the ligands.
The structures were docked using Gold 4.1.2[48] in Bovine ADA (Protein Data Bank ID: 1KRM).[49] Each molecule was docked to the binding site and scored. For each docked inhibitor, the top three results were selected for analysis. From the analysis, one structure for each inhibitor was selected and of the six inhibitors, four scored higher than the natural substrate.
Acknowledgments
We thank Professors Sunny Zhou (Northeastern University) for assistance with the SAHase and ADA assay. This work was supported by the National Institutes of Health (NIH) (R01 CA97634 to KSR, NIH T32 GM066706 CBI Fellowship (SCZ). We are also grateful to Dr. Phil Mortimer (Johns Hopkins Mass Spectrometry Facility) for his invaluable assistance with the HRMS analysis.
Footnotes
Publisher's Disclaimer: Full terms and conditions of use: http://www.tandfonline.com/page/terms-and-conditions
This article may be used for research, teaching, and private study purposes. Any substantial or systematic reproduction, redistribution, reselling, loan, sub-licensing, systematic supply, or distribution in any form to anyone is expressly forbidden.
The publisher does not give any warranty express or implied or make any representation that the contents will be complete or accurate or up to date. The accuracy of any instructions, formulae, and drug doses should be independently verified with primary sources. The publisher shall not be liable for any loss, actions, claims, proceedings, demand, or costs or damages whatsoever or howsoever caused arising directly or indirectly in connection with or arising out of the use of this material.
REFERENCES
- 1.Silverman RB. The Organic Chemistry of Drug Design and Drug Action. 2nd ed. San Diego: Elsevier; 2004. [Google Scholar]
- 2.Das K, Bauman JD, Clark AD, Jr, Frenkel YV, Lewi PJ, Shatkin AJ, Hughes SH, Arnold E. High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: strategic flexibility explains potency against resistance mutations. Proc. Natl. Acad. Sci. USA. 2008;105:1466–1471. doi: 10.1073/pnas.0711209105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Das K, Lewi PJ, Hughes SH, Arnold E. Crystallography and the design of anti-AIDS drugs: conformational flexibility and positional adaptability are important in the design of non-nucleoside HIV-1 reverse transcriptase inhibitors. Prog. Biophys. Mol. Biol. 2005;88:209–231. doi: 10.1016/j.pbiomolbio.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Lewi PJ, deJonge M, Daeyaert F, Koymans L, Vinkers M, Heeres J, Janssen PA, Arnold E, Das K, Clark AD, Jr, Hughes SH, Boyer PL, deBethune MP, Pauwels R, Andries K, Kukla M, Ludovici D, DeCorte B, Kavash R, Ho C. On the detection of multiple-binding modes of ligands to proteins, from biological, structural, and modeling data. J. Comput. Aided Mol. Des. 2003;17:129–134. doi: 10.1023/a:1025313705564. [DOI] [PubMed] [Google Scholar]
- 5.Tuske S, Sarafianos SG, Clark AD, Jr, Ding J, Naeger LK, White KL, Miller MD, Gibbs CS, Boyer PL, Clark P, Wang G, Gaffney BL, Jones RA, Jerina DM, Hughes SH, Arnold E. Structures of HIV-1 RT-DNA complexes before and after incorporation of the anti-AIDS drug tenofovir. Nat. Struct. Mol. Biol. 2004;11:469–474. doi: 10.1038/nsmb760. [DOI] [PubMed] [Google Scholar]
- 6.Seley KL, Zhang L, Hagos A, Quirk S. “Fleximers”. Design and synthesis of a new class of novel shape-modified nucleosides. J. Org. Chem. 2002;67:3365–3373. doi: 10.1021/jo0255476. [DOI] [PubMed] [Google Scholar]
- 7.Seley KL, Zhang L, Hagos A. “Fleximers”. Design and synthesis of two novel split nucleosides. Org. Lett. 2001;3:3209–3210. doi: 10.1021/ol0165443. [DOI] [PubMed] [Google Scholar]
- 8.Seley KL, Salim S, Zhang L. “Molecular chameleons”. Design and synthesis of C-4-substituted imidazole fleximers. Org. Lett. 2005;7:63–66. doi: 10.1021/ol047895v. [DOI] [PubMed] [Google Scholar]
- 9.Seley KL, Salim S, Zhang L, O’Daniel PI. “Molecular chameleons”. Design and synthesis of a second series of flexible nucleosides. J. Org. Chem. 2005;70:1612–1619. doi: 10.1021/jo048218h. [DOI] [PubMed] [Google Scholar]
- 10.Seley KL, Quirk S, Salim S, Zhang L, Hagos A. Unexpected inhibition of S-adenosyl-L-homocysteine hydrolase by a guanosine nucleoside. Bioorg. Med. Chem. Lett. 2003;13:1985–1988. doi: 10.1016/s0960-894x(03)00331-7. [DOI] [PubMed] [Google Scholar]
- 11.Quirk S, Seley KL. Substrate discrimination by the human GTP fucose pyrophosphorylase. Biochemistry. 2005;44:10854–10863. doi: 10.1021/bi0503605. [DOI] [PubMed] [Google Scholar]
- 12.Quirk S, Seley KL. Identification of catalytic amino acids in the human GTP fucose pyrophosphorylase active site. Biochemistry. 2005;44:13172–13178. doi: 10.1021/bi051288d. [DOI] [PubMed] [Google Scholar]
- 13.St Amant AH, Bean LA, Guthrie JP, Hudson RH. Click fleximers: a modular approach to purine base-expanded ribonucleoside analogues. Org. Biomol. Chem. 2012;10:6521–6525. doi: 10.1039/c2ob25678a. [DOI] [PubMed] [Google Scholar]
- 14.Wauchope OR, Velasquez M, Seley-Radtke KL. Synthetic routes to a series of proximal and distal 2′-deoxy fleximers. Synthesis. 2012;44:3496–3504. doi: 10.1055/s-0032-1316791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zimmermann SC, Sadler JM, Andrei G, Snoeck R, Balzarini J, Seley-Radtke KL. Carbocyclic 5′-nor “reverse” fleximers. Design, synthesis, and preliminary biological activity. MedChemComm. 2011;2:650–654. doi: 10.1039/C1MD00094B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar S, Wilson SR, Leonard NJ. Structure of 3-isoadenosine. Acta Crystallogr. C. 1988;44(Pt 3):508–510. doi: 10.1107/s0108270187011144. [DOI] [PubMed] [Google Scholar]
- 17.Gerzon K, Johnson IS, Boder GB, Cline JC, Simpson PJ, Speth C, Leonard NJ, Laursen RA. Biological activities of 3-isoadenosine. Biochim. Biophys. Acta. 1966;119:445–461. doi: 10.1016/0005-2787(66)90120-1. [DOI] [PubMed] [Google Scholar]
- 18.Seley KL, Mosley SL, Zeng FX. Carbocyclic lsoadenosine analogues of neplanocin A. Org. Lett. 2003;5:4401–4403. doi: 10.1021/ol035696q. [DOI] [PubMed] [Google Scholar]
- 19.Tseng CK, Marquez VE, Fuller RW, Goldstein BM, Haines DR, McPherson H, Parsons JL, Shannon WM, Arnett G, Hollingshead M, Driscoll JS. Synthesis of 3-deazaneplanocin A, a powerful inhibitor of S-adenosylhomocysteine hydrolase with potent and selective in vitro and in vivo antiviral activities. J. Med. Chem. 1989;32:1442–1446. doi: 10.1021/jm00127a007. [DOI] [PubMed] [Google Scholar]
- 20.Marquez VE. Carbocyclic nucleosides. Adv. Antivir. Drug Des. 1996;2:89–146. [Google Scholar]
- 21.De Clercq E. S-adenosylhomocysteine hydrolase inhibitors as broad spectrum antiviral agents. Biochem. Pharmacol. 1987;36:2567–2575. doi: 10.1016/0006-2952(87)90533-8. [DOI] [PubMed] [Google Scholar]
- 22.De Clercq E. Advances in Antiviral Drug Design. Amsterdam, The Netherlands: Elsevier Science; 1996. [Google Scholar]
- 23.Greco NJ, Tor Y. Simple fluorescent pyrimidine analogues detect the presence of DNA abasic sites. J. Am. Chem. Soc. 2005;127:10784–10785. doi: 10.1021/ja052000a. [DOI] [PubMed] [Google Scholar]
- 24.Herdewijn PAMM. 5-Substituted-2′-deoxyuridines as anti-hsv-1 agents—synthesis and structure-activity relationship. Antiviral Chem. Chemother. 1994;5:131–146. [Google Scholar]
- 25.De Winter H, Herdewijn P. Understanding the binding of 5-substituted 2′-deoxyuridine substrates to thymidine kinase of herpes simplex virus type-1. J. Med. Chem. 1996;39:4727–4737. doi: 10.1021/jm960278v. [DOI] [PubMed] [Google Scholar]
- 26.Wigerinck P, Snoeck R, Claes P, De Clercq E, Herdewijn P. Synthesis and antiviral activity of 5-heteroaryl-substituted 2′-deoxyuridines. J. Med. Chem. 1991;34:1767–1772. doi: 10.1021/jm00110a003. [DOI] [PubMed] [Google Scholar]
- 27.Yoshikawa N, Nakamura K, Yamaguchi Y, Kagota S, Shinozuka K, Kunitomo M. Reinforcement of antitumor effect of Cordyceps sinensis by 2′-deoxycoformycin, an adenosine deaminase inhibitor. In Vivo. 2007;21:291–295. [PubMed] [Google Scholar]
- 28.Lowe JK, Gowans B, Brox L. Deoxyadenosine metabolism and toxicity in cultured L5178Y cells. Cancer Res. 1977;37:3013–3017. [PubMed] [Google Scholar]
- 29.Wilson JM, Mitchell BS, Daddona PE, Kelley WN. Purinogenic immunodeficiency diseases.Differential effects of deoxyadenosine and deoxyguanosine on DNA synthesis in human T lymphoblasts. J. Clin. Invest. 1979;64:1475–1484. doi: 10.1172/JCI109606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan C-S, Liu S, Wnuk SF, Robins MJ, Borchardt RT. Design and synthesis of S-adenosylhomocysteine hydrolase inhibitors as broad-spectrum antiviral agents. In: DE Clercq E, editor. Advances in Antiviral Drug Design. Vol. 2. Amsterdam, The Netherlands: Elsevier; 1996. pp. 41–88. [Google Scholar]
- 31.Wigerinck P, Pannecouque C, Snoeck R, Claes P, De Clercq E, Herdewijn P. 5-(5-Bromothien-2-yl)-2′-deoxyuridine and 5-(5-chlorothien-2-yl)-2′-deoxyuridine are equipotent to (E)-5-(2-bromovinyl)-2′-deoxyuridine in the inhibition of herpes simplex virus type I replication. J. Med. Chem. 1991;34:2383–2389. doi: 10.1021/jm00112a011. [DOI] [PubMed] [Google Scholar]
- 32.Yang MM, Ye W, Schneller SW. Preparation of carbocyclic S-adenosylazamethionine accompanied by a practical synthesis of (-)-aristeromycin. J. Org. Chem. 2004;69:3993–3996. doi: 10.1021/jo040119g. [DOI] [PubMed] [Google Scholar]
- 33.Luche J-L. Lanthanides in organic chemistry. 1. Selective 1,2 reductions of conjugated ketones. J. Am. Chem. Soc. 1978;100:2226–2227. [Google Scholar]
- 34.Mitsunobu O. The use of diethyl azodicarboxylate and triphenylphosphine in synthesis and transformation of natural products. Synthesis. 1981;1981:1–28. [Google Scholar]
- 35.Moon HR, Choi WJ, Kim HO, Jeong LS. Improved and alternative synthesis of D- and L-cyclopentenone derivatives, the versatile intermediates for the synthesis of carbocyclic nucleosides. Tetrahedron-Asymmetr. 2002;13:1189–1193. [Google Scholar]
- 36.Luche J-L, Rodriguez-Hahn L, Crabbe P. Reduction of natural enones in the presence of cerium trichloride. J. Chem. Soc. Chem. Commun. 1978:601–602. [Google Scholar]
- 37.Russ P, Schelling P, Scapozza L, Folkers G, Clercq ED, Marquez VE. Synthesis and biological evaluation of 5-substituted derivatives of the potent antiherpes agent (north)-methanocarbathymine. J. Med. Chem. 2003;46:5045–5054. doi: 10.1021/jm030241s. [DOI] [PubMed] [Google Scholar]
- 38.Obika S, Uneda T, Sugimoto T, Nanbu D, Minami T, Doi T, Imanishi T. 2′-O,4′-C-methylene bridged nucleic acid (2′,4′-BNA): synthesis and triplex-forming properties. Bioorgan. Med. Chem. 2001;9:1001–1011. doi: 10.1016/s0968-0896(00)00325-4. [DOI] [PubMed] [Google Scholar]
- 39.Pinhey JT, Roche EG. Chemistry of organolead(IV) tricarboxylates. Synthesis and electrophilic heteroarylation reactions of 2- and 3-thienyl-, and 2- and 3-furyllead tricarboxylates. J. Chem. Soc. Perkin. Trans. 1988;1(1972–1999):2415–2421. [Google Scholar]
- 40.Zambon A, Borsato G, Brussolo S, Frascella P, Lucchini V. Efficient access to 5-substituted thiazoles by a novel metallotropic rearrangement. Tetrahedron Lett. 2007;49:66–69. [Google Scholar]
- 41.Wigerinck P, Kerremans L, Claes P, Snoeck R, Maudgal P, Declercq E, Herdewijn P. Synthesis and antiviral activity of 5-thien-2-Yl-2′-deoxyuridine analogs. J. Med. Chem. 1993;36:538–543. doi: 10.1021/jm00057a003. [DOI] [PubMed] [Google Scholar]
- 42.Arnau N, Cortes J, Moreno-Manas M, Pleixats R, Villarroya M. Palladium(0)-catalyzed allylation of heterocycles with cyclopentene derivatives. J. Heterocycl. Chem. 1997;34:233–239. [Google Scholar]
- 43.Mosley SL, Bakke BA, Sadler JM, Sunkara NK, Dorgan KM, Zhou ZS, Seley-Radtke KL. Carbocyclic pyrimidine nucleosides as inhibitors of S-adenosylhomocysteine hydrolase. Bioorgan. Med. Chem. 2006;14:7967–7971. doi: 10.1016/j.bmc.2006.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dorgan KM, Wooderchak WL, Wynn DP, Karschner EL, Alfaro JF, Cui Y, Zhou ZS, Hevel JM. An enzyme-coupled continuous spectrophotometric assay for S-adenosylmethionine-dependent methyltransferases. Anal. Biochem. 2006;350:249–255. doi: 10.1016/j.ab.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 45.Bloch A, Robins MJ, McCarthy JR., Jr The role of the 5′-hydroxyl group of adenosine in determining substrate specificity for adenosine deaminase. J. Med. Chem. 1967;10:908–912. doi: 10.1021/jm00317a034. [DOI] [PubMed] [Google Scholar]
- 46.Brink JJ, Lepage GA. 9-Beta-D-arabinofuranosyladenine as an inhibitor of metabolism in normal and neoplastic cells. Can J. Biochem. Physiol. 1965;43:1–15. doi: 10.1139/o65-001. [DOI] [PubMed] [Google Scholar]
- 47.Lepage GA, Junga IG. Metabolism of purine nucleoside analogs. Cancer Res. 1965;25:46–52. [PubMed] [Google Scholar]
- 48.Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved protein-ligand docking using GOLD. Proteins: Struct. Funct. Bioinf. 2003;52:609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 49.Kinoshita T, Nishio N, Nakanishi I, Sato A, Fujii T. Structure of bovine adenosine deaminase complexed with 6-hydroxy-1,6-dihydropurine riboside. Acta Crystallogr. D. 2003;59:299–303. doi: 10.1107/s090744490202190x. [DOI] [PubMed] [Google Scholar]
- 50.Elrod P, Zhang J, Yang X, Yin D, Hu Y, Borchardt RT, Schowen RL. Contributions of active site residues to the partial and overall catalytic activities of human S-adenosylhomocysteine hydrolase. Biochemistry. 2002;41:8134–8142. doi: 10.1021/bi025771p. [DOI] [PubMed] [Google Scholar]