Abstract
Objective:
Monitoring of influenza virus shedding and optimization of multiplicities of infection (MOI) is important in the investigation of a virus one step growth cycle and for obtaining a high yield of virus in vaccine development and conventional basic diagnostic methods. However, eluted infectious viruses may still be present immediately after virus inoculation and when cells are washed following virus cultivation which may lead to a false positive virus infectivity assay.
Materials and Methods:
In this experimental study, we investigated influenza virus progeny production in Madin-Darby canine kidney (MDCK) cells with five different MOI at determined time points. The results were analyzed by end point titration tests and immunofluorescence assay.
Results:
Higher titers of eluted virus were observed following a high MOI inoculation of virus in cell culture. Most probably, this was the result of sialic acid residues from viral hemagglutin in proteins that were cleaved by neuraminidase glycoproteins on the surface of the influenza virus, which promoted viral spread from the host cell to the culture supernatant or during endocytosis, where viruses recycle to the cell surface by recycling endosomes which culminated in virus shedding without replication.
Conclusion:
We demonstrated that the pattern of influenza virus progeny production was dose-dependent and not uniform. This production was influenced by several factors, particularly MOI. Understanding the exact features of viral particle propagation has a major impact in producing high virus yields in the development of vaccines. Use of lower MOI (0.01) could result in accurate, precise quantitative assays in virus diagnosis and titration methods.
Keywords: Influenza Virus, Virus Shedding, Endosome, MDCK Cells
Introduction
Influenza viruses are major causes of respiratory tract infection, resulting in significant morbidity and mortality. They are now recognized as a major public health concern and have a significant health and economic burden (1).
Knowledge about the details of influenza virus cultivation within a suitable cell culture is of utmost importance for investigation of its replication and vaccine development. Now a days, much should be done to increase our knowledge about the importance of cell culture based techniques for vaccine development and virus multiplication identification. These techniques should be applied by researchers as before, because they seem to have been forgotten(2).
The tissue culture infective dose 50% (TCID50) is one of the basic quantification tests for monitoring in vitro influenza virus replication in both research and development (R&D). Although traditional cell culture-based methods are generally slow, labor intensive and time consuming, they are an important, crucial step in viral seed preparation. In the case of influenza virus, the TCID50 test is confirmed by the hemagglutination assay (HA), which provides greater reliability. These methods are applicable for further evaluations of influenza virus replication and in optimization of multiplicities of infection (MOI) for virus cultivation in large scales such as vaccine production, determining virus shedding at different time points or in vitro evaluation of new antiviral drugs (3-5).
Supernatants are used for quantification of the TCID50 in the cultivation of influenza viruses. However, some of the eluted viruses remain detectable, causing false positive test results.
The more the cells are permissive at the virus attachment level and on cell endocytic capacity for internalization, there will be less numbers of eluted viruses in culture supernatants (6, 7).In this study, we have determined the titers of packaged virus at various time points postinfection with different MOI of 1, 0.1, 0.01, 0.001, and 0.0001 in Madin-Darby canine kidney (MDCK) cells.
Materials and Methods
Quantification of influenza virus by plaque formation on Madin-Darby canine kidney (MDCK) cells
The MDCK cell lines represent one of the most efficient cell systems for the plaque assay of influenza viruses that are currently available. In this experimental study, we inoculated MDCK cells in six-well culture plates with serial dilutions of the A⁄ Puerto Rico ⁄8 ⁄34 (PR8) virus which was adsorbed in one hour. The inoculums were removed and we washed the cells washed three times with phosphate-buffered saline (PBS). The cell monolayers were covered with a first layer that contained 0.8% cell grade agar (Sigma, St.Louis, MI, USA) in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Karlsruhe, Germany), antibiotics (100 IU ⁄ml penicillin and 100 μg ⁄ml streptomycin) without serum, and 2 μg ⁄ml L-1-tosylamido- 2-phenylethyl chloromethyl ketone (TPCK)- treated trypsin (Sigma, St.Louis, MI, USA). Plates were incubated for 72 hours and cells overlaid with 1:1000 neutral red (Sigma, St.Louis, MI, USA), 0.8% agar and DMEM for plaque visualization. All culture incubations were performed in a 37˚C, 5% CO2 humidified incubator (8, 9).
Inoculation of cells with multiplicities of infection (MOI) of viruses
MDCK cells were cultured in DMEM that contained 10% fetal calf serum (FCS, Gibco, Karlsruhe, Germany), 100 IU/ml penicillin and 100 mg/ml streptomycin in six-well plates for 24 hours. Subsequently, cells were washed twice with PBS buffer and inoculated with a ten-fold serial dilution of PR8 virus stock, which resulted in an MOI of 1.0, 0.1, 0.01, 0.001 and 0.0001. After one hour at 37˚C, the cells were washed three times with PBS and supplemented with 3 ml of DMEM that contained 100 IU/ml penicillin and 100 mg/ml streptomycin, without FCS. Finally, 2 μg/ml TPCK-treated trypsin was added to each well (3).
Time point measurement of virus infectivity titers in Madin-Darby canine kidney (MDCK) cells by 50% tissue culture infective dose (TCID50)
We harvested and analyzed culture supernatants for TCID50at the following time points (t) post-infection: 1 (immediately after adsorption), 2, 3, 4, 5, 6, 7, 8, 12, 24 and 48 hours. The virus sample was diluted in a 96-well tissue culture plate that contained MDCK. The titration was performed in quadruplicate. In order to confirm TCID50 results, the spot HA assay was performed. Virus titers were calculated according to the method of Spearman- Karber (10).
Hemagglutination assay (HA)
At each time point, 50 μl volumes of the culture supernatants were harvested and diluted in a twofold dilution with PBS.A total of 50 μl of a 0.5% suspension of fowl red blood cells were added to each dilution in a V-shaped microtiter plate. Following gentle agitation, the plates were left undisturbed for 30 minutes at room temperature (RT). The last dilution that showed complete hemagglutination was considered the end point and expressed as hemagglutination units (HAU) per test volume (11, 12).
Using indirect immunofluorescence for detecting influenza nucleoprotein (NP)
MDCK cells were cultured in 24-well culture plates. Virus inoculation culture supernatants were harvested and monolayer cellswashed three times with PBS, then fixed in 4% formaldehyde for 15 minutes. After washing, 0.02% Triton X-100 (Sigma-Aldrich, Steinheim, Germany) was added and MDCK Cells were incubated for 10 minutes atRT. Antiinfluenza nucleoprotein (anti-NP) monoclonal antibody (Abcam, USA) that contained 1% bovine serum albumin (BSA) was added, and wells were agitated for one hour at RT, washed three times with PBS, then incubated with fluorescein isothiocyanate (FITC)-conjugated secondary antibody anti-mouse IgGthat contained 1% BSA for 1 hour. After washing and drying, cells were observed by inverted immunofluorescence microscopy (Jenus, China) (13, 14).
Results
Plaque formation on Madin-Darby canine kidney (MDCK) cells
Influenza PR8 initiated plaque formation after 48 hours. This resulted in large, flat, indented plaques in the agar (Fig 1). We counted the well that had 15-100 plaques and the viral titer was calculated according to a current microbiology protocol. After adding the second layer of agar that contained 1:1000 neutral red, plates were wrapped in foil to prevent any photodynamic reaction by neutral red (15, 16)
Fig 1.

Plaques formed by A⁄Puerto Rico⁄8⁄34 (PR8) viruses on monolayer Madin-Darby canine kidney (MDCK) cells.
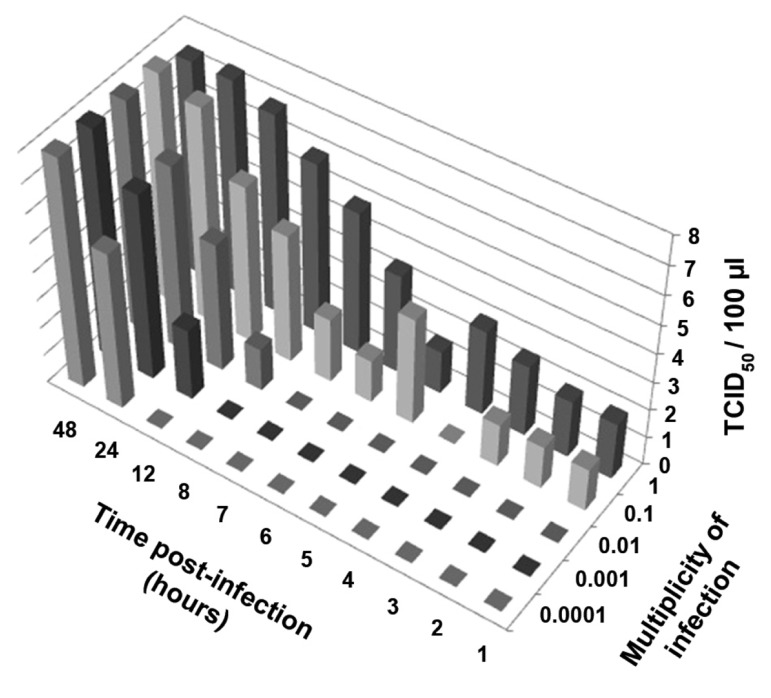
Determining the highest dilution of viral suspension infectious units using the 50% tissue culture infectious dose (TCID50 ) assay
As can be seen in figure 2, at the MOI of 1 and 0.1, the virus infectivity assay was immediately positive (t1) post-infection. However traces of eluted viruses can create false positives in TCID50 analyses. At lower MOI, because the lower titers of viruses have been used for infectivity, eluted viruses are not present. In the current study, virus progeny production was initiated at the following MOI: 0.01 (t8), 0.001 (t12) and 0.0001 (t24).
Fig 2.
Growth chart of A⁄Puerto Rico⁄8⁄34 (PR8) according to the time point virus infectivity assay for supernatant from Madin-Darby canine kidney (MDCK) cells inoculated at different MOI. The results were expressed asTCID50 /100μl.
Quantification of influenza viruses by hemagglutination assay (HA)
As shown in figure 3, after 12 hours post-infection, there were 512 HA/50 μl in the MDCK cell supernatant infected at an MOI of 1 and 32 HA/50 μl in the culture supernatant of MDCK cells infected at an MOI of 0.1. At MOI of 0.01, the HA result was 8, at 0.001 it was 128and for 0.0001, the HA test result was 256 units at 24 hours post-infection. Approximately 48 hours post-infection, all MOI reached the same level of HA units; at 72 hours, they reached a peak.
Fig 3.
Kinetics of PR8 strain of Influenza A virus replication based on the hemagglutination assay (HA) of Madin-Darby canine kidney (MDCK) cell supernatant, inoculated with different MOI. The results were expressed as HA/50μl.
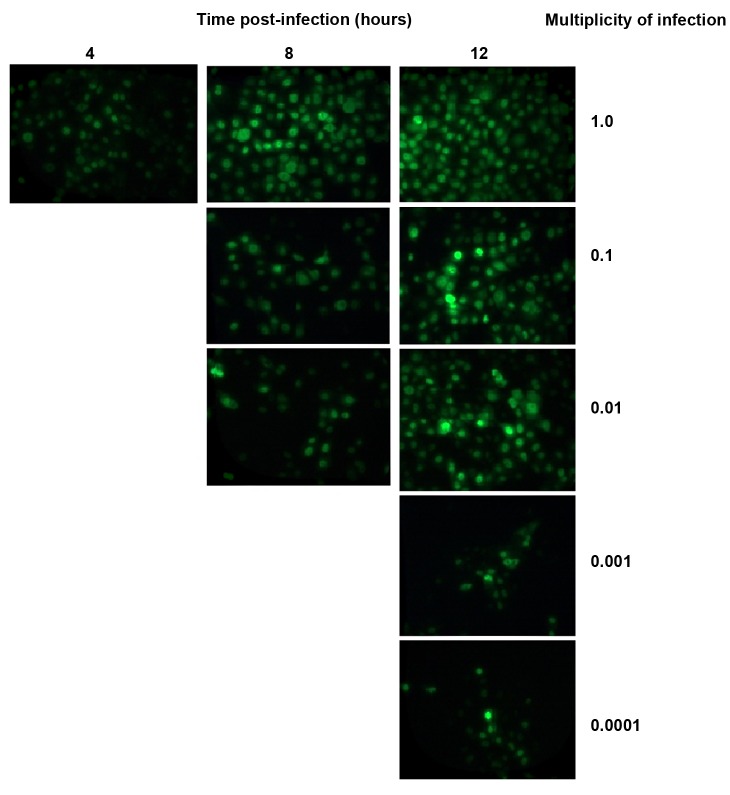
Immunofluorescence assay
At the MOI of 1, it tookfour hours for a positive result according to the immunofluorescence assay of the influenza virus NP protein (Fig 4). The numbers of NP-expressed cells increased with time elapses, such that after12 hours post-infection; the majority of cells were florescence positive at the MOI of 1. Infected cells with MOI of 0.1 and 0.01 were positive in 8 hours. When MDCK cells were infected at lower MOI (0.001 and 0.0001), the cells were florescence-positive at 12 hours post-infection. The NP protein is an indicator for switching from transcription to replication.
Fig 4.
Kinetics of PR8 strain of Influenza A virus replication based on immunofluorescence assay. MDCK cells were inoculated with different MOI and fixed in different times. Then, infected cells observed after incubation with fluorescein labeled antibody specific for the NP protein of influenza viruses (Magnification ×400).
Discussion
MOI optimization and monitoring of influenza virus shedding has a major impact on high virus yield and outcome of virus seed selection for vaccines. Influenza viruses are quantified either by a unit of hemagglutination or by determining infectious units using the TCID50 or plaque assays. The interval between inoculation and appearance of detectable virus hemagglutination or TCID50 bears an inverse relationship to amount of the inoculated virus (15).
The TCID50 assay is a method to gauge the infectious virus in a sample by determining the highest dilution of the sample that can infect 50% of the cells in culture (15). According to TCID50 results, at higher MOI (1 and 0.1) eluted viruses remain in the supernatant and do not complete a single virus synthesis cycle, most likely because neuraminidase glycoproteins separate sialic acid residues from viral hemagglutinin proteins. Subsequently,it promotes the spread of the virus from the host cell to culture supernatant(17,18) or the virus returns to the cell surface by recycling endosome which results in virus shedding without progeny production (19).
We detected progeny virus release at 5 and 6 hours post-infection for MOI 1 and 0.1 whereas for MOI of 0.01, 0.001 and 0.0001 they were detectable at 8, 12 and 24 hours after infection, respectively. Henle and Liuhad suggested that the duration of reproductive cycle of influenza virus is 5 to 6 hours based on the length of the interval between the virus inoculation and increasing viral infective titer. Apparently, the elution of some adsorbed virus lead in a substantial amount of asynchronous of infection which confuses the actual virus one-step growth curve (20). Gaush and Smith have shown that a single virus progeny production cycle of influenza virus requires 8 to 10 hours in MDCK cells compared to 20 hours in Chang’s conjunctival cells (21). Since the HA assay is dependent on the amount of hemagglutinin on the surface of influenza viruses and not on the capability of the virus to replicate, this assay quantifies viral particles apart from their infectivity. Quantitatively, one HAU is equivalent to approximately to 2×106 infectious or noninfectious virus particles/50 μl. At an MOI of 1, 8 hours post-infection is necessary to obtain a positive HA result, whereas for an MOI of 0.1, at 12 hours a positive result is observed (22).
At lower MOI, although HA activity is positive up to 24 hours post-infection, they reach the same HAU within 72 hours. Freymann et al. have demonstrated the appearance of hemagglutin in averaged 8.4, 12.5, 16.3 and 23 hours post-infection when they inoculated 10-day old eggs at MOI of 106.5, 105.5, 104.5 and 101.5 EID50, respectively (6).
Portela and Digard (23) in addition to Rimmelzwaan et al. (3) have shown that NP mRNA and NP protein are synthesized during the early infectious phase, which is controlled by the conversion of cRNA into new vRNA. However, Kawakami et al. suggest that both replication and transcription occur simultaneously in the early phase of infection (24). In the current study, we have shown the switch from transcription to translation. We reported that the time to trigger shifting from transcription to replication was around 4 hours post-infection based on NP protein production with an MOI of 1.Replicative intermediate cRNA, stabilized by the newly synthesized NP and viral polymerase, regulates replication (25). Viral matrixM1 protein and NEP/NS2 protein, which is responsible for vRNP nuclear export, inhibit viral transcription at the late phase of infection. Small RNAs generated by the influenza virus (26) and host factors such as BAT1, heat shock protein 90, the minimal chromosome maintenance, Tat-SF1, and DNA dependent RNA polymerase II are involved in influenza vRNA synthesis (27). Experiments have shown that NP protein expression began 4 hours postinfection, whereas at an MOI of 0.1 and 0.01, the time point for NP protein expression was 8 hours post-infection. At an MOI of 0.001, it took 12 hours for the NP protein to be observed by indirect immunofluorescence (Fig 4).
Conclusion
In this study we compared virus shedding at different time points by determining the TCID5 and A titer. Our data revealed that in higher MOI (1 and 0.1) eluted viruses remained in the supernatant. It took approximately 5 hours for the influenza virus to complete multiplication and produce new progeny at an MOI of 1. The best MOI for influenza virus monitoring in cell culture was 0.01 because no traces of eluted virus were detectable by TCID50 and virus progeny production began 8 hours post-infection. At MOI of 0.001 and 0.0001, virus progeny were detectable at 12 and 24 hours after infection, respectively. The optimum time for harvesting the influenza virus has been shown to be 72 hours post-infection due to virus reach the pick of progeny production and the pH changes in cell culture media is not tangible up to 72 hours post-infection.
Acknowledgments
This study was supported by a grant from the Research Deputy of Tarbiat Modares University, Faculty of Medical Sciences and Influenza Research Laboratory, Pasteur Institute of Iran, Tehran, Iran. There is no conflict of interest in this article.
References
- 1.Medina RA, García-Sastre A. Influenza A viruses: new research developments. Nat Rev Microbiol. 2011;9(8):590–603. doi: 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Monto AS. Global burden of influenza: what we know and what we need to know. Int Congr Ser. 2004;1263:3–11. [Google Scholar]
- 3.Rimmelzwaan GF, Baars M, Claas EC, Osterhaus AD. Comparison of RNA hybridization, hemagglutination assay, titration of infectious virus and immunofluorescence as methods for monitoring influenza virus replication in vitro. J Virol Methods. 1998;74(1):57–66. doi: 10.1016/s0166-0934(98)00071-8. [DOI] [PubMed] [Google Scholar]
- 4.Genzel Y, Behrendt I, König S, Sann H, Reichl U. Metabolism of MDCK cells during cell growth and influenza virus production in large-scale microcarrier culture. Vaccine. 2004;22(17-18):2202–2208. doi: 10.1016/j.vaccine.2003.11.041. [DOI] [PubMed] [Google Scholar]
- 5.Kaverin NV, Webster RG. Impairment of multicycle influenza virus growth in Vero (WHO) cells by loss of trypsin activity. J Virol. 1995;69(4):2700–2703. doi: 10.1128/jvi.69.4.2700-2703.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freymann M, Tamm I, Green RH. Growth curves of influenza virus based on hemagglutination titers in individual embryonated eggs. Yale J Biol Med. 1951;23(4):269–276. [PMC free article] [PubMed] [Google Scholar]
- 7.Hahon N, Booth JA, Eckert HL. Cell attachment and penetration by influenza virus. Infect Immun. 1973;7(3):341–351. doi: 10.1128/iai.7.3.341-351.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Appleyard G, Maber HB. Plaque formation by influenza viruses in the presence of trypsin. J Gen Virol. 1974;25(3):351–357. doi: 10.1099/0022-1317-25-3-351. [DOI] [PubMed] [Google Scholar]
- 9.Dong J, Matsuoka Y, Maines TR, Swayne DE, O'Neill E, Davis CT, et al. Development of a new candidate H5N1 avian influenza virus for pre-pandemic vaccine production. Influenza Other Respi Viruses. 2009;3(6):287–295. doi: 10.1111/j.1750-2659.2009.00104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hubert J. Spearman-Karber method, Bioassay. 2nd ed. Dubuque (Iowa): Hunt Publishing; 1984:. 65-66 [Google Scholar]
- 11.Kistner O, Barrett PN, Mundt W, Reiter M, Schober- S, Dorner F. Development of a mammalian cell (Vero) derived candidate influenza virus vaccine. Vaccine. 1998;16(9-10):960–968. doi: 10.1016/s0264-410x(97)00301-0. [DOI] [PubMed] [Google Scholar]
- 12.Othman F, Ideris A, Motalleb G, Eshak ZB, Rahmat A. Oncolytic effect of Newcastle disease virus AF2240 strain on the MCF-7 breast cancer cell line. Yakhteh. 2010;12(1):17–24. [Google Scholar]
- 13.Jamali A, Sabahi F, Bamdad T, Hashemi H, Mahboudi F, Kheiri MT. A DNA vaccine-encoded nucleoprotein of influenza virus fails to induce cellular immune responses in a diabetic mouse model. Clin Vaccine Immunol. 2010;17(4):683–687. doi: 10.1128/CVI.00445-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin BC, Lai CJ. The influenza virus nucleoprotein synthesized from cloned DNA in a simian virus 40 vector is detected in the nucleus. J Virol. 1983;45(1):434–438. doi: 10.1128/jvi.45.1.434-438.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szretter KJ, Balish AL, Katz JM. Influenza: propagation, quantification, and storage. Curr Protoc Microbiol. 2006. Chapter 15. Unit 15G.1. [DOI] [PubMed] [Google Scholar]
- 16.Tobita K, Sugiura A, Enomoto C, Furuyama M. Plaque assay and primary isolation of influenza A viruses in an established line of canine kidney cells (MDCK) in the presence of trypsin. Med Microbiol Immunol. 1975;162(1):9–14. doi: 10.1007/BF02123572. [DOI] [PubMed] [Google Scholar]
- 17.Mitnaul LJ, Matrosovich MN, Castrucci MR, Tuzikov AB, Bovin NV, Kobasa D, et al. Balanced hemagglutinin and neuraminidase activities are critical for efficient replication of influenza A virus. J Virol. 2000;74(13):6015–6020. doi: 10.1128/jvi.74.13.6015-6020.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagner R, Matrosovich M, Klenk HD. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev Med Virol. 2002;12(3):159–166. doi: 10.1002/rmv.352. [DOI] [PubMed] [Google Scholar]
- 19.Sieczkarski SB, Whittaker GR. Dissecting virus entry via endocytosis. J Gen Virol. 2002;83(7):1535–1545. doi: 10.1099/0022-1317-83-7-1535. [DOI] [PubMed] [Google Scholar]
- 20.Henle W, Liu OC. Studies on host-virus interactions in the chick embryo-influenza virus system VI.Evidence for multiplicity reactivation of inactivated virus. J Exp Med. 1951;94(4):305–322. doi: 10.1084/jem.94.4.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaush CR, Smith TF. Replication and plaque assay of influenza virus in an established line of canine kidney cells. Appl Microbiol. 1968;16(4):588–594. doi: 10.1128/am.16.4.588-594.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hierholzer J, Killington R. Virus isolation and quantitation. In: Mahy BW, Kangro HO, editors. Virology methods manual. London: Academic Press Limited; 1996. pp. 25–46. [Google Scholar]
- 23.Portela A, Digard P. The influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication. J Gen Virol. 2002;83(4):723–734. doi: 10.1099/0022-1317-83-4-723. [DOI] [PubMed] [Google Scholar]
- 24.Kawakami E, Watanabe T, Fujii K, Goto H, Watanabe S, Noda T, et al. Strand-specific real-time RT-PCR for distinguishing influenza vRNA, cRNA, and mRNA. J Virol Methods. 2011;173(1):1–6. doi: 10.1016/j.jviromet.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vreede FT, Jung TE, Brownlee GG. Model suggesting that replication of influenza virus is regulated by stabilization of replicative intermediates. J Virol. 2004;78(17):9568–9572. doi: 10.1128/JVI.78.17.9568-9572.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez JT, Varble A, Sachidanandam R, Zlatev I, Manoharan M, García-Sastre A, et al. Influenza A virus-generated small RNAs regulate the switch from transcription to replication. Proc Natl Acad Sci USA. 2010;107(25):11525–11530. doi: 10.1073/pnas.1001984107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe T, Watanabe S, Kawaoka Y. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe. 2010;7(6):427–439. doi: 10.1016/j.chom.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]