Abstract
Study Objective
To assess the effects of the cytochrome P450 (CYP) 3A genotype, CYP3A5, on atorvastatin pharmacokinetics and its interaction with clarithromycin.
Design
Prospective, two-phase, randomized-sequence, open-label pharmacokinetic study.
Setting
Clinical research center at a teaching hospital.
Subjects
Twenty-three healthy volunteers who were screened for genotype: 10 subjects carried the CYP3A5*1 allele (expressors) and 13 subjects did not (nonexpressors).
Intervention
In one phase, subjects received a single oral dose of atorvastatin 20 mg. In the other phase, subjects received clarithromycin 500 mg twice/day for 5 days; on day 4 after the morning dose, subjects also received a single oral dose of atorvastatin 20 mg. All subjects participated in both phases of the study, which were separated by at least 14 days.
Measurements and Main Results
Pharmacokinetic parameters of both forms of atorvastatin—atorvastatin acid and atorvastatin lactone—were compared between CYP3A5 expressors and nonexpressors, both in the absence and presence of clarithromycin, a strong CYP3A inhibitor. The acid form is pharmacologically active, and the lactone form has been associated with the atorvastatin’s muscle-related adverse effects. Atorvastatin acid exposure did not differ significantly between CYP3A5 genotype groups. When subjects had not received clarithromycin pretreatment, the area under the concentration-time curve from time zero extrapolated to infinity (AUC0–∞) of atorvastatin lactone was 36% higher in nonexpressors than in expressors (median 47.6 ng•hr/ml [interquartile range (IQR) 37.8–64.3 ng•hr/ml] vs 34.9 ng•hr/ml [IQR 21.6–42.2 ng•hr/ml], p=0.038). After clarithromycin pretreatment, changes in the pharmacokinetic parameters of atorvastatin acid and lactone were not significantly different between the nonexpressors versus the expressors; however, the increase in the AUC0−∞ of atorvastatin lactone was 37% greater in expressors than in nonexpressors (geometric mean ± SD 3.59 ± 0.57 vs 2.62 ± 0.35, p=0.049).
Conclusion
Our data suggest that the CYP3A5 genotype has minimal effects on the pharmacokinetic parameters of atorvastatin and its interaction with clarithromycin; these effects are unlikely to be clinically significant.
Keywords: cytochrome P450, CYP, CYP3A5 genotype, pharmacogenetics, atorvastatin, clarithromycin, drug interaction
Atorvastatin, a 3-hydroxy-3-methylglutaryl coenzyme A reductase inhibitor (statin), is the most widely prescribed drug in the United States. Although it is generally considered a safe drug, 3–8% of patients taking atorvastatin experience muscle-related adverse effects.1 Atorvastatin-induced muscle-related adverse effects may be influenced by the activity of the cytochrome P450 (CYP) 3A enzyme.
Atorvastatin exists in both acid and lactone forms. The acid form is pharmacologically active, and its plasma concentrations are similar to or higher than those of the lactone form.2, 3 The lactone form has been associated with atorvastatin’s muscle-related adverse effects.4 In vitro, it was more toxic to human myocytes than the acid form.5 As atorvastatin lactone has higher affinity for CYP3A than does atorvastatin acid, the activity of CYP3A may play an important role in atorvastatin-induced muscle-related adverse effects.6
The most serious atorvastatin-induced muscle-related adverse effect is rhabdomyolysis, and its risk increases with coadministration of a CYP3A inhibitor.7, 8 The macrolide antibiotic, clarithromycin, is a strong CYP3A inhibitor according to classification by the United States Food and Drug Administration.9 Coadministration of atorvastatin with an inhibitor such as clarithromycin increases the risk of atorvastatin-induced muscle-related adverse effects through inhibition of CYP3A.8
Cytochrome P450 3A is a major drug-metabolizing enzyme subfamily. In adults, it mainly consists of CYP3A4 and CYP3A5.10 The activity of CYP3A is highly variable among individuals; genetic polymorphisms in CYP3A5 (but not CYP3A4) have been shown to contribute to the interindividual variability in CYP3A activity. The CYP3A5 gene contains two common single-nucleotide polymorphisms: CYP3A5*3 and CYP3A5*6.11, 12 The CYP3A5*3 (rs776746: A>G) is more common in Caucasian and Asian individuals, whereas CYP3A5*6 (rs10264272: G>A) is common in African-American individuals.13 Of importance, these single-nucleotide polymorphisms change the enzyme’s function. The CYP3A5*3 allele produces a truncated protein by creating a cryptic splice site in intron 3.11 The CYP3A5*6 allele encodes for an enzyme with very low activity by causing skipping of exon 7 in CYP3A5 transcripts.11 As a result, individuals carrying either CYP3A5*3 or CYP3A5*6, or both alleles, have decreased or absent CYP3A5 activity compared with those with a CYP3A5*1 allele.11, 12, 14 Since both CYP3A4 and CYP3A5 share many substrates and inhibitors,15, 16 the CYP3A5 genotype may influence the pharmacokinetics of CYP3A substrates and the magnitude of CYP3A-mediated drug interactions.
In this study, we tested two hypotheses. First, CYP3A5 genotype influences atorvastatin pharmacokinetics, so CYP3A5*1 carriers would have lower exposure to atorvastatin than non-carriers. Second, CYP3A5 genotype influences the magnitude of atorvastatin interaction with clarithromycin, so CYP3A5*1 carriers would have a greater increase in atorvastatin plasma concentrations after clarithromycin pretreatment. To test the hypotheses, we conducted single-dose atorvastatin pharmacokinetic studies with and without clarithromycin pretreatment in healthy volunteers. Since atorvastatin is a substrate of solute carrier organic anion transporter (SLCO1B1) and its pharmacokinetics are influenced by a polymorphism in the gene (rs4149056: T>C),17–19 we controlled for this polymorphism in our analysis.
Methods
Study Design and Setting
This was a prospective, two-phase, randomized-sequence, open-label, pharmacokinetic study conducted at the University of Florida General Clinical Research Center (Gainesville, FL). The study protocol was approved by the institutional review board at the University of Florida, and all participants provided written informed consent.
Genotype Screening
Healthy subjects aged 18 years or older were screened for CYP3A5 genotype before enrollment. Genomic DNA was isolated from the buccal cells with use of a commercially available kit (Qiagen DNA Blood Isolation Kit; Qiagen, Valencia, CA). Genotyping for CYP3A5*3 (rs776746) and CYP3A5*6 (rs10264272) was performed by using polymerase chain reaction (PCR), followed by pyrosequencing (Pyrosequencing AB, Uppsala, Sweden) with a PSQ HS96A sequencing system (Biotage AB, Uppsala, Sweden), as described previously.20 In addition, genotyping for SLCO1B1 rs4149056: T>C was carried out for all subjects who were enrolled in the study, by using PCR and pyrosequencing. For the SLCO1B1 genotyping, the following PCR and sequencing primers were used, respectively: forward-biotinylated-5′ - AAGGAATCTGGGTCATACATGTGG-3′, reverse-5′-CCCCTATTCCACGAAGCATATT-3′, and reverse sequencing-5′-AAG CATATTACCCATGAAC-3′.
Study Subjects
Subjects were enrolled into the study based on their CYP3A5 genotype such that enrollment was enriched for *1 carriers relative to its presence in the general population. Subjects were ascertained to be healthy by detailed medical history, complete physical examination, and clinical laboratory tests. Subjects were excluded if they had a creatinine clearance value less than 60 ml/minute as estimated by the Cockcroft-Gault equation, an abnormal liver function test result (≥ 2 times the upper limit of normal for alanine aminotransferase or aspartate amino-transferase level), an abnormal serum creatine kinase level (≥ 2 times the upper limit of normal), an allergy to macrolide antibiotics, a previous adverse reaction to clarithromycin, or a body mass index greater than 30 kg/m2. In addition, any subject who was pregnant, smoked tobacco, abused drugs, or consumed more than two alcoholic drinks/day was excluded.
Subjects were grouped based on CYP3A5 genotype status: those who carried a CYP3A5*1 allele were classified as expressors, and the others were classified as nonexpressors.
Study Protocol
The study consisted of two phases: atorvastatin only and atorvastatin plus clarithromycin. Subjects participated in both phases, but the order in which they completed the phases was randomly assigned and stratified by race-ethnicity and sex. The two phases were separated by at least 14 days.
Subjects were asked not to take any prescription or over-the-counter drugs, herbal medicines, or grapefruit products for at least 7 days before and during the study. While taking the study drug(s), subjects were also asked to abstain from alcohol and to limit the intake of caffeine-containing beverage to no more than two drinks/day.
Atorvastatin-Only Phase
Subjects arrived at the clinical research center in the morning after an overnight fast. They were given a single oral dose of atorvastatin 20 mg with 150 ml of water. They were fed 2 hours after atorvastatin administration. All foods were free of caffeine and grapefruit products. At 0, 0.5, 1, 2, 4, 6, 8, 12, 24, 36, and 48 hours after atorvastatin administration, 10 ml of blood was collected from each subject. Plasma was separated within 30 minutes of the blood collection and was stored at −80°C until the sample analysis.
Atorvastatin plus Clarithromycin Phase
Subjects took oral clarithromycin 500 mg twice/day for 5 consecutive days. On the fourth day, they reported to the clinical research center. The morning dose of clarithromycin 500 mg was administered, followed 2 hours later by a single oral dose of atorvastatin 20 mg. Clarithromycin tablets were provided in a blister pack; compliance was assessed by pill count before atorvastatin administration. Those who missed more than one dose were not allowed to continue the study. All the remaining procedures were the same as in the atorvastatin-only phase except that clarithromycin was continued for an additional day.
Determination of Plasma Concentrations of Atorvastatin Acid and Lactone
Plasma concentrations of atorvastatin acid and lactone were quantified according to a published method, with modifications.21 Briefly, 250 µl of plasma samples, which had been kept on ice (4°C) were mixed with 250 µl of chilled sodium acetate buffer (0.1 M, pH 5.0), 10 µl of deuterated internal standards, and 3 ml of methyl tert-butyl ether. The organic layer was separated by centrifugation at 4000 rpm for 10 minutes at 4°C and transferred to a glass tube containing 100 µl of 1% formic acid in acetonitrile. After the organic layer was evaporated under nitrogen, the samples were reconstituted with 60% 5-mM ammonium acetate in acetonitrile (100 µl).
The reconstituted samples (15 µl) were injected onto a Luna Phenyl-Hexyl high-performance liquid chromatography (HPLC) column (100×2 mm, 3 µM; Phenomenex, Torrance, CA). Binary gradient elution was then achieved with HPLC mobile phases composed of 5-mM ammonium acetate with 1% formic acid in water and 5-mM ammonium acetate with 1% formic acid in acetonitrile; the flow rate was 0.2 ml/minute.
The liquid chromatography with tandem mass spectrometry detection (LC–MS-MS) system consisted of a Surveyor HPLC autosampler, Surveyor MS quaternary pump, and a TSQ Quantum Discovery triple quadrupole mass spectrometer (Thermo Scientific, San Jose, CA). The TSQ Quantum mass spectrometer was equipped with an electrospray source and operated in the positive ion mode. The acquisition parameters were spray voltage 5.0 kV, source collision-induced dissociation 10 V, and heated capillary temperature of 300°C. The collision energy was 29 eV for all analytes. The selected reaction monitoring scheme followed transitions of the [M+H]+ precursor to selected product ions with the following values: mass:charge ratio (m/z) 559.2 → 440.2 for atorvastatin acid, 541.2 → 448.2 atorvastatin lactone, 564.2 → 440.2 d5- atorvastatin acid, and 546.2 → 448.2 d5- atorvastatin lactone.
The standard concentrations ranged from 0.1–75 ng/ml. The intra- and interday precision (coefficient of variation) and accuracy (relative error) values were within 11% for the lower limit of quantitation (0.1 ng/ml for both atorvastatin acid and lactone) and low, medium, and high concentrations for atorvastatin acid and atorvastatin lactone.
Determination of Plasma Concentrations of Clarithromycin
Plasma concentrations of clarithromycin were determined by an LC–MS-MS method developed in our laboratory.22 The lower limit of quantitation was 100 ng/ml, and the precision and accuracy (intra- and interday) were within 7.9% and 4.9%, respectively.
Sample Size Calculation
Based on reported variability in atorvastatin area under the plasma concentration–time curve from time zero extrapolated to infinity (AUC0–∞),3, 17, 23 12 subjects in each group were needed to detect an effect size of 0.55 between the groups, with a power of 80% and at an α level of 0.05.
Pharmacokinetics and Statistical Analysis
Maximum plasma concentration observed (Cmax), time at which Cmax occurred, elimination half-life, and AUC0–∞ of atorvastatin acid and lactone in each phase were determined. Apparent oral clearance of atorvastatin acid in each phase was also determined. The partial AUC of clarithromycin was calculated because blood samples were not collected until 2 hours after clarithromycin administration. All of the pharmacokinetic parameters were determined by using WinNonlin, version 3.1 (Pharsight Corp., Mountain View, CA).
Data are presented as median and interquartile range (IQR), unless otherwise specified. Non-normally distributed variables were log transformed where appropriate. For half-life and apparent oral clearance, harmonic means were calculated and their standard deviations were computed with the Norris method.24 Geometric means for ratios of each pharmacokinetic parameter between the two phases were calculated by CYP3A5 genotype status, and standard deviations were computed with Norris’ method to obtain 95% confidence intervals (CIs).24
Wilcoxon rank sum test was used to compare pharmacokinetic parameters of atorvastatin acid and lactone from each phase by CYP3A5 genotype (expressors vs nonexpressors). The test was also used to compare ratios for pharmacokinetic parameters by CYP3A5 genotype.
Multivariable linear regression analysis of the pharmacokinetic parameters of atorvastatin acid and lactone was performed with a normally distributed pharmacokinetic parameter as a dependent variable and potential explanatory variables as independent variables. Potential explanatory variables included CYP3A5 genotype status, sex, race-ethnicity, weight, body mass index, partial AUC of clarithromycin, and SLCO1B1 rs4149056 C carrier status. The SLCO1B1 rs4149056 carrier status and the variables that had a p value less than 0.2 were entered into the multivariable models. A p value less than 0.05 was defined as statistically significant. Statistical analyses were performed with SAS software, version 9.0 (SAS Institute Inc., Cary, NC).
Results
Twenty-three subjects were enrolled into the study: 10 carried the CYP3A5*1 allele (expressors) and 13 did not (nonexpressors). One subject was homozygous for the CYP3A5*1 allele. Baseline characteristics including sex, race-ethnicity, and SLCO1B1 rs4149056 C carrier status were not significantly different between the groups (Table 1).
Table 1.
Baseline Characteristics of the 23 Study Subjects
| Characteristic |
CYP3A5 Expressors (n=10) |
CYP3A5 Nonexpressors (n=13) |
|---|---|---|
| Mean ± SD | ||
| Age (yrs) | 24.2 ± 4.1 | 24.6 ± 4.0 |
| Weight (kg) | 69.6 ± 9.4 | 69.5 ± 9.8 |
| Body mass index (kg/m2) | 23.8 ± 1.8 | 23.3 ± 1.8 |
| No. (%) of Subjects | ||
| Male | 7 (70) | 7 (54) |
| Race-ethnicity | ||
| Caucasian | 6 (60) | 10 (77) |
| Hispanic | 1 (10) | 1 (8) |
| Asian | 3 (30) | 2 (15) |
| SLCO1B1variant carriera | 1 (10) | 4 (31)b |
CYP = cytochrome P450; SLCO = solute carrier organic anion transporter.
p=0.34 (Fisher exact test).
One patient was the SLCO1B1 rs4149056 C/C homozygote.
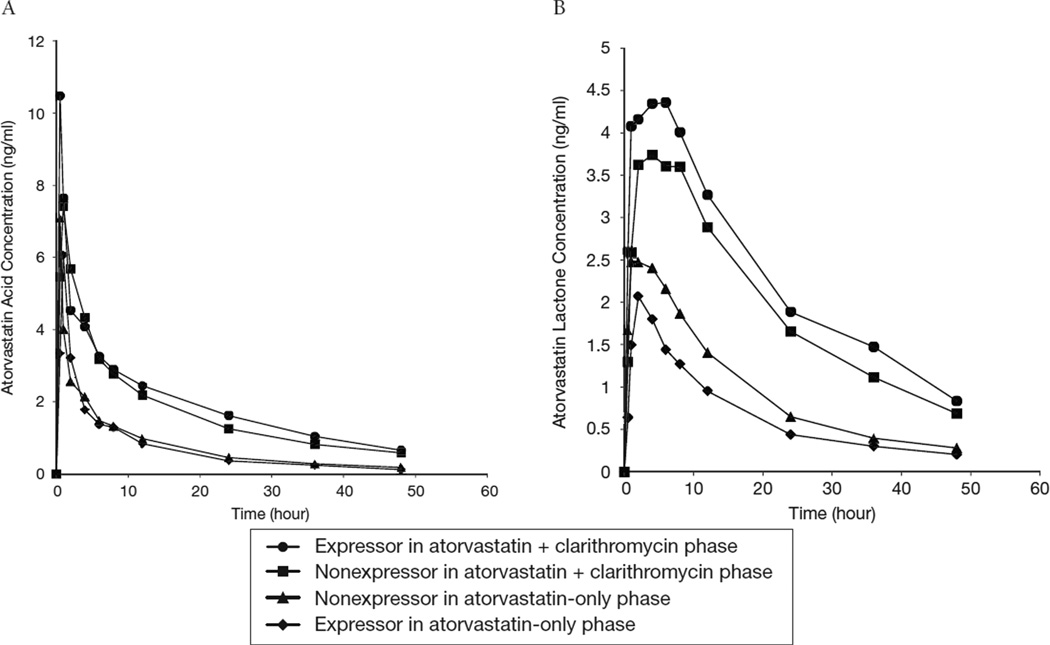
Plasma concentration–time profiles of atorvastatin acid and lactone by CYP3A5 genotype during both phases are shown in Figure 1. Plasma concentrations of atorvastatin acid and lactone were significantly higher in all subjects in the atorvastatin plus clarithromycin phase than in the atorvastatin-only phase.
Figure 1.
Mean plasma concentrations over time of atorvastatin acid (A) and atorvastatin lactone (B) in relation to cytochrome P450 3A5 genotype after 23 healthy volunteers were administered a single dose of atorvastatin either with or without clarithromycin.
The pharmacokinetics of atorvastatin acid and lactone did not differ significantly between expressors and nonexpressors in either phase, with the exception that the AUC0–∞ of atorvastatin lactone was 36% higher (p=0.038) in nonexpressors than in expressors (median 47.6 ng•hr/ml, IQR 37.8–64.3 ng•hr/ml, vs 34.9 ng•hr/ml, IQR 21.6–42.2 ng•hr/ml) in the atorvastatin-only phase (Table 2). However, this was a weak association given the magnitude of the difference. In a multivariable linear regression model, CYP3A5 genotype was weakly associated with the difference in AUC0–∞ of atorvastatin lactone after adjustment for SLCO1B1 rs4149056 C carrier status (p=0.03). There were no significant differences in pharmacokinetic parameters by SLCO1B1 genotype, indicating that the small number of variant carriers did not influence the pharmacokinetic differences observed.
Table 2.
Pharmacokinetic Parameters of and Their Ratios for Atorvastatin Acid and Lactone by CYP3A5 Genotype
| CYP3A5 Expressors (n=10) | |||||
|---|---|---|---|---|---|
| Atorvastatin Analyte by Study Phase |
Cmax (ng/ml) |
Tmax (hrs) |
AUC0–∞ (ng•hr/ml) |
Half-life (hrs) |
Cl/F (L/hr) |
| Median (IQR) |
Harmonic Mean ± SD |
||||
| Atorvastatin only | |||||
| Acid | 6.0 (2.4–8.0) | 1.0 (1.0–2.0) | 40.7 (29.6–55.3) | 13.7 ± 5.4 | 470.9 ± 171.0 |
| Lactone | 2.1 (1.7–2.5) | 2.0 (1.0–2.0) | 34.9 (21.6–42.2)a | 10.9 ± 4.5 | NA |
| Atorvastatin + clarithromycin | |||||
| Acid | 8.2 (6.8–10.1) | 1.0 (0.5–1.0)) | 102.4 (92.1–130.5) | 22.1 ± 9.9 | 180.0 ± 52.2 |
| Lactone | 4.2 (3.1–4.8) | 5.0 (2.0–8.0) | 109.5 (96.7–128.4) | 19.3 ± 5.4 | NA |
| Geometric Mean ± SD (95% CI) | |||||
| Between-phase ratios | |||||
| Acid | 1.62 ± 0.42 | 0.81 ± 0.24 | 2.69 ± 0.36 | 1.55 ± 0.42 | 0.37 ± 0.06 |
| (1.39–1.89) | (0.65–0.97) | (2.45–2.93) | (1.28–1.82) | (0.33–0.41) | |
| Lactone | 1.81 ± 0.27 | 2.32 ± 0.9 | 3.59 ± 0.57b | 1.69 ± 0.27 | NA |
| (1.63–1.99) | (1.73–2.91) | (3.22–3.96) | (1.51–1.87) | ||
| CYP3A5 Nonexpressors (n=13) | |||||
| 5.0 (3.0–13.9) | 1.0 (0.5–1.0) | 38.6 (31.9–51.6) | 13.8 ± 6.6 | 435.7 ± 169.0 | |
| 2.8 (1.6–4.6) | 1.0 (1.0–2.0) | 47.6 (37.8–64.3)a | 13.6 ± 5.2 | NA | |
| 10.8 (7.0–15.2) | 0.5 (0.5–1.0) | 118.7 (93.2–126.3) | 17.2 ± 5.5 | 172.3 ± 42.3 | |
| 4.2 (3.5–5.8) | 4.0 (2.0–6.0) | 127.1 (94.7–178.8) | 19.2 ± 5.2 | NA | |
| Geometric Mean ± SD (95% CI) | |||||
| 1.76 ± 0.48 | 0.77 ± 0.21 | 2.63 ± 0.28 | 1.17 ± 0.28 | 0.38 ± 0.3 | |
| (1.49–2.03) | (0.65–0.89) | (2.47–2.79) | (1.01–1.33) | (0.36–0.40) | |
| 1.73 ± 0.24 | 2.15 ± 0.83 | 2.62 ± 0.35b | 1.36 ± 0.24 | NA | |
| (1.59–1.87) | (1.68–2.62) | (2.40–2.82) | (1.22–1.50) | ||
CYP3A5 = cytochrome P450 3A5 gene; Cmax = maximum plasma concentration; Tmax = time at which maximum plasma concentration was observed; AUC0–∞ = area under the concentration-time curve from time zero extrapolated to infinity; Cl/F = apparent oral clearance, where F is bioavailability; IQR = interquartile range; NA = not applicable; CI = confidence interval; SLCO1B1 = solute carrier organic anion transporter family member 1B1 gene.
p=0.038 (CYP3A5 expressors vs nonexpressors [Wilcoxon rank sum test]); linear regression model: AUC0–∞ = 51.0 – 19.2×CYP3A5 genotype + 9.0×SLCO1B1 genotype (where CYP3A5 expressor = 1, CYP3A5 nonexpressor = 0, SLCO1B1 rs4149056 C carrier = 1, SLCO1B1 rs4149056 C noncarrier = 0); p=0.03 (CYP3A5 genotype); p=0.38 (SLCO1B1 genotype).
p=0.049 (CYP3A5 expressors vs nonexpressors [Wilcoxon rank sum test]); linear regression model (using log transformation): log AUC0–∞ = 0.96 + 0.32×CYP3A5 genotype + 0.01×SLCO1B1 genotype (where CYP3A5 expressor = 1, CYP3A5 nonexpressor = 0, SLCO1B1 rs4149056 C carrier = 1, SLCO1B1 rs4149056 C noncarrier = 0); p=0.047 (CYP3A5 genotype); p=0.96 (SLCO1B1 genotype).
The between-phases ratios for pharmacokinetic parameters of atorvastatin acid were not significantly different between expressors and nonexpressors. Although the ratio for AUC0–∞ of atorvastatin lactone was higher in expressors than in nonexpressors (geometric mean ± SD 3.59 ± 0.57 vs 2.62 ± 0.35, p=0.049), this was a weak association. In a multivariable linear regression model, CYP3A5 genotype was weakly associated with the difference in the ratio for atorvastatin lactone AUC0–∞ after adjustment for SLCO1B1 rs4149056 C carrier status (p=0.047).
The partial AUC of clarithromycin (i.e., hrs 2–10) was not significantly different between CYP3A5 expressors and nonexpressors (mean ± SD 17,719.2 ± 17,391 µg•hr/ml vs 21,894.2 ± 23,690 µg•hr/ml, p>0.15). In addition, the partial AUC of clarithromycin was not a significant variable that predicted AUC0–∞ of atorvastatin lactone and its ratio between the phases.
Discussion
To our knowledge, this is the first study to prospectively evaluate the influence of CYP3A5 genotype on the magnitude of drug interaction between atorvastatin and clarithromycin, a strong CYP3A inhibitor. We have two findings from this study. First, CYP3A5 genotype had minimal influence on atorvastatin pharmacokinetic parameters. In either phase, CYP3A5 genotype was not associated with significant differences in atorvastatin pharmacokinetics. The only exception was that those who carried a functional CYP3A5 allele had a numerically lower AUC0–∞ of atorvastatin lactone than noncarriers in the atorvastatin-only phase, although it was a weak association. Second, CYP3A5 genotype did not influence the magnitude of atorvastatin interaction with clarithromycin. Clarithromycin pretreatment increased the AUC0–∞ of atorvastatin lactone in all of the subjects, and the magnitude of the increase was greater (37%) in those who carried a CYP3A5*1 allele than in those who did not. However, this association was weak, and this difference in magnitude was unlikely to have clinical significance. Overall, our data suggest that CYP3A5 genotype has a very modest effect, at best, on the pharmaco-kinetics of atorvastatin and its drug interaction with clarithromycin.
Our finding that CYP3A5 genotype had minimal influence on atorvastatin pharmacokinetics is consistent with previous studies on atorvastatin metabolism. The in vitro intrinsic clearance of atorvastatin is 2–7-fold higher by CYP3A4 compared with CYP3A5, suggesting a minor role for CYP3A5 in atorvastatin acid metabolism.25 Two studies in humans showed that CYP3A5 genotype was not significantly associated with atorvastatin lactone exposure, which is consistent with the data we report.4, 19 In our study, atorvastatin lactone exposure was modestly higher (36%) in nonexpressors than in expressors, which is unlikely to be clinically relevant because patients with a history of statin-induced myopathy had 2.4-fold higher atorvastatin lactone exposure than those without the history.4 The two previous studies on the effect of CYP3A5 genotype on atorvastatin pharmacokinetics have some important limitations. One study did not evaluate the effect of CYP3A5 genotype on atorvastatin lactone exposure in a prospective manner.4 In addition, this study enrolled few non-Caucasian subjects and had only one of 28 subjects carrying the CYP3A5*1 allele. The other study enrolled only a single ethnic group (Koreans).19 In addition, the study’s main focus was the effect of the SLCO1B1 and ABCB1 polymorphisms on atorvastatin pharmacokinetics. In contrast, our study prospectively enrolled patients based on CYP3A5 genotype, had relatively diverse race-ethnicity, and focused on the effect of CYP3A5 genotype on atorvastatin pharmacokinetics and drug interaction. Of note, to our knowledge, none of the previous studies, examined the effect of CYP3A5 genotype on the magnitude of atorvastatin and clarithromycin interaction.
Although these data suggest a minimal role of CYP3A5 genotype in atorvastatin pharmacokinetics, observational studies have reported a significant association between CYP3A5 genotype and either response to lipid-lowering effect or risk of myopathy or myalgia.26–28 Although some of these studies prospectively followed patients, many studies obtained data retrospectively. In addition, none of these studies accounted for the polymorphisms in the transporter genes that have been associated with atorvastatin pharmacokinetics. 17–19
The metabolism of atorvastatin was significantly decreased by clarithromycin pretreatment as it reduced the apparent oral clearance of atorvastatin acid by about 60% in both expressors and nonexpressors in our study. Clarithromycin pretreatment, however, had a slightly greater magnitude of effect in CYP3A5 expressors. This finding is consistent with a previous study demonstrating that clarithromycin inhibited intestinal CYP3A activity to a greater degree in CYP3A5 expressors than in nonexpressors.29 Given that the AUC0–∞ of atorvastatin lactone did not significantly differ by CYP3A5 genotype after clarithromycin pretreatment, the CYP3A5 genotype is unlikely to influence the risk of atorvastatin-induced muscle-related adverse events when a strong CYP3A inhibitor such as clarithromycin is coadministered with atorvastatin.
Limitations
Our study had several limitations. First, our study did not evaluate the polymorphisms in transporter genes, other than the SLCO1B1 gene, that have been associated with variable atorvastatin pharmacokinetics.2, 17–19 In addition, our study was not prospectively designed to assess effects of the SLCO1B1 polymorphism. Second, because we had only one subject with the CYP3A5*1/*1 genotype, we were not able to evaluate the effect of this genotype on atorvastatin pharmacokinetics and the drug interaction with clarithromycin. The impact of CYP3A5 genotype may only be apparent in homozygous expressors; simvastatin pharmacokinetics are significantly different between the CYP3A5*1/*1 and CYP3A5*3/*3 genotypes.30 Third, the hydroxy metabolites of atorvastatin acid and lactone have been associated with atorvastatin-induced myopathy.4 We measured ortho-hydroxy metabolites but did not measure para-hydroxy metabolites of atorvastatin acid and lactone. Our ortho-hydroxy metabolite data were consistent with those in previous studies (data not shown).2 In addition, pharmacokinetics of atorvastatin acid and lactone in our study were similar to those in other studies.2, 17 Finally, our sample of 23 subjects may be considered relatively small. Based on our post hoc sample size calculation, however, a sample of 10 subjects/group would be enough to detect a 50% difference in exposure, which we consider to be the minimum clinically relevant difference. In addition, our study is an enrichment study whereby the less common genotype is overrepresented in the study population relative to the general population.
Conclusion
The CYP3A5 genotype has minimal effects on the pharmacokinetic parameters of atorvastatin and its interaction with clarithromycin; these effects are not likely to be clinically significant.
Acknowledgments
Supported by National Institutes of Health grant (HL68834) and American Heart Association postdoctoral fellowship grant (0525474B). Dr. Pacanowski is currently employed by the U.S. Food and Drug Administration (FDA), but this work was conducted while he was an employee of the University of Florida. No official FDA endorsement is intended or should be inferred.
References
- 1.Pfizer Inc. Lipitor (atorvastatin) package insert. New York, NY: 2009. [Google Scholar]
- 2.Keskitalo JE, Zolk O, Fromm MF, Kurkinen KJ, Neuvonen PJ, Niemi M. ABCG2 polymorphism markedly affects the pharmacokinetics of atorvastatin and rosuvastatin. Clin Pharmacol Ther. 2009;86:197–203. doi: 10.1038/clpt.2009.79. [DOI] [PubMed] [Google Scholar]
- 3.Backman JT, Luurila H, Neuvonen M, Neuvonen PJ. Rifampin markedly decreases and gemfibrozil increases the plasma concentrations of atorvastatin and its metabolites. Clin Pharmacol Ther. 2005;78:154–167. doi: 10.1016/j.clpt.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Hermann M, Bogsrud MP, Molden E, et al. Exposure of atorvastatin is unchanged but lactone and acid metabolites are increased several-fold in patients with atorvastatin-induced myopathy. Clin Pharmacol Ther. 2006;79:532–539. doi: 10.1016/j.clpt.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 5.Skottheim IB, Gedde-Dahl A, Hejazifar S, Hoel K, Asberg A. Statin-induced myotoxicity: the lactone forms are more potent than the acid forms in human skeletal muscle cells in vitro. Eur J Pharm Sci. 2008;33:317–325. doi: 10.1016/j.ejps.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 6.Jacobsen W, Kuhn B, Soldner A, et al. Lactonization is the critical first step in the disposition of the 3-hydroxy-3-methylglutaryl-CoA reductase inhibitor atorvastatin. Drug Metab Dispos. 2000;28:1369–1378. [PubMed] [Google Scholar]
- 7.Thompson PD, Clarkson P, Karas RH. Statin-associated myopathy. JAMA. 2003;289:1681–1690. doi: 10.1001/jama.289.13.1681. [DOI] [PubMed] [Google Scholar]
- 8.Sipe BE, Jones RJ, Bokhart GH. Rhabdomyolysis causing AV blockade due to possible atorvastatin, esomeprazole, and clarithromycin interaction. Ann Pharmacother. 2003;37:808–811. doi: 10.1345/aph.1C396. [DOI] [PubMed] [Google Scholar]
- 9.U.S. Food and Drug Administration. [Accessed April 1, 2011];Drug development and drug interactions: table of substrates, inhibitors and inducers. Available from www.fda.gov/Drugs/DevelopmentApprovalProcess/DevelopmentResources/DrugInteractionsLabeling/ucm081177.htm.
- 10.Koch I, Weil R, Wolbold R, et al. Interindividual variability and tissue-specificity in the expression of cytochrome P450 3A mRNA. Drug Metab Dispos. 2002;30:1108–1114. doi: 10.1124/dmd.30.10.1108. [DOI] [PubMed] [Google Scholar]
- 11.Kuehl P, Zhang J, Lin Y, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27:383–391. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- 12.Lee SJ, Usmani KA, Chanas B, et al. Genetic findings and functional studies of human CYP3A5 single-nucleotide polymorphisms in different ethnic groups. Pharmacogenetics. 2003;13:461–472. doi: 10.1097/00008571-200308000-00004. [DOI] [PubMed] [Google Scholar]
- 13.Solus JF, Arietta BJ, Harris JR, et al. Genetic variation in eleven phase I drug metabolism genes in an ethnically diverse population. Pharmacogenomics. 2004;5:895–931. doi: 10.1517/14622416.5.7.895. [DOI] [PubMed] [Google Scholar]
- 14.Hustert E, Haberl M, Burk O, et al. The genetic determinants of the CYP3A5 polymorphism. Pharmacogenetics. 2001;11:773–779. doi: 10.1097/00008571-200112000-00005. [DOI] [PubMed] [Google Scholar]
- 15.Huang W, Lin YS, McConn DJ, II, et al. Evidence of significant contribution from CYP3A5 to hepatic drug metabolism. Drug Metab Dispos. 2004;32:1434–1445. doi: 10.1124/dmd.104.001313. [DOI] [PubMed] [Google Scholar]
- 16.Niwa T, Murayama N, Emoto C, Yamazaki H. Comparison of kinetic parameters for drug oxidation rates and substrate inhibition potential mediated by cytochrome P450 3A4 and 3A5. Curr Drug Metab. 2008;9:20–33. doi: 10.2174/138920008783331121. [DOI] [PubMed] [Google Scholar]
- 17.Pasanen MK, Fredrikson H, Neuvonen PJ, Niemi M. Different effects of SLCO1B1 polymorphism on the pharmacokinetics of atorvastatin and rosuvastatin. Clin Pharmacol Ther. 2007;82:726–733. doi: 10.1038/sj.clpt.6100220. [DOI] [PubMed] [Google Scholar]
- 18.He YJ, Zhang W, Chen Y, et al. Rifampicin alters atorvastatin plasma concentration on the basis of SLCO1B1 521T>C polymorphism. Clin Chim Acta. 2009;405:49–52. doi: 10.1016/j.cca.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Lee YJ, Lee MG, Lim LA, Jang SB, Chung JY. Effects of SLCO1B1 and ABCB1 genotypes on the pharmacokinetics of atorvastatin and 2–hydroxyatorvastatin in healthy Korean subjects. Int J Clin Pharmacol Ther. 2010;48:36–45. doi: 10.5414/cpp48036. [DOI] [PubMed] [Google Scholar]
- 20.Langaee TY, Gong Y, Yarandi HN, et al. Association of CYP3A5 polymorphisms with hypertension and antihypertensive response to verapamil. Clin Pharmacol Ther. 2007;81:386–391. doi: 10.1038/sj.clpt.6100090. [DOI] [PubMed] [Google Scholar]
- 21.Jemal M, Ouyang Z, Chen BC, Teitz D. Quantitation of the acid and lactone forms of atorvastatin and its biotransformation products in human serum by high-performance liquid chromatography with electrospray tandem mass spectrometry. Rapid Commun Mass Spectrom. 1999;13:1003–1015. doi: 10.1002/(SICI)1097-0231(19990615)13:11<1003::AID-RCM597>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 22.Shin J, Pauly DF, Johnson JA, Frye RF. Simplified method for determination of clarithromycin in human plasma using protein precipitation in a 96-well format and liquid chromatography-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;871:130–134. doi: 10.1016/j.jchromb.2008.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kantola T, Kivisto KT, Neuvonen PJ. Effect of itraconazole on the pharmacokinetics of atorvastatin. Clin Pharm Ther. 1998;64:58–65. doi: 10.1016/S0009-9236(98)90023-6. [DOI] [PubMed] [Google Scholar]
- 24.Norris N. The standard errors of the geometric and harmonic means and their application to index numbers. Ann Mathematic Stat. 1940;11:445–448. [Google Scholar]
- 25.Park JE, Kim KB, Bae SK, Moon BS, Liu KH, Shin JG. Contribution of cytochrome P450 3A4 and 3A5 to the metabolism of atorvastatin. Xenobiotica. 2008;38:1240–1251. doi: 10.1080/00498250802334391. [DOI] [PubMed] [Google Scholar]
- 26.Kivisto KT, Niemi M, Schaeffeler E, et al. Lipid-lowering response to statins is affected by CYP3A5 polymorphism. Pharmacogenetics. 2004;14:523–525. doi: 10.1097/01.fpc.0000114762.78957.a5. [DOI] [PubMed] [Google Scholar]
- 27.Wilke RA, Moore JH, Burmester JK. Relative impact of CYP3A genotype and concomitant medication on the severity of atorvastatin-induced muscle damage. Pharmacogenet Genomics. 2005;15:415–421. doi: 10.1097/01213011-200506000-00007. [DOI] [PubMed] [Google Scholar]
- 28.Willrich MA, Hirata MH, Genvigir FD, et al. CYP3A53A allele is associated with reduced lowering-lipid response to atorvastatin in individuals with hypercholesterolemia. Clin Chim Acta. 2008;398:15–20. doi: 10.1016/j.cca.2008.07.032. [DOI] [PubMed] [Google Scholar]
- 29.Pinto AG, Wang YH, Chalasani N, et al. Inhibition of human intestinal wall metabolism by macrolide antibiotics: effect of clarithromycin on cytochrome P450 3A4/5 activity and expression. Clin Pharmacol Ther. 2005;77:178–188. doi: 10.1016/j.clpt.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 30.Keskitalo JE, Kurkinen KJ, Neuvoneni PJ, Niemi M. ABCB1 haplotypes differentially affect the pharmacokinetics of the acid and lactone forms of simvastatin and atorvastatin. Clin Pharmacol Ther. 2008;84:457–461. doi: 10.1038/clpt.2008.25. [DOI] [PubMed] [Google Scholar]