Abstract
There are no approved therapeutics for the most deadly nonsegmented negative-strand (NNS) RNA viruses, including Ebola (EBOV). To identify new chemical scaffolds for development of broad-spectrum antivirals, we undertook a prototype-based lead identification screen. Using the prototype NNS virus, vesicular stomatitis virus (VSV), multiple inhibitory compounds were identified. Three compounds were investigated for broad-spectrum activity, and inhibited EBOV infection. The most potent, CMLDBU3402, was selected for further study. CMLDBU3402 did not show significant activity against segmented negative-strand RNA viruses suggesting proscribed broad-spectrum activity. Mechanistic analysis indicated that CMLDBU3402 blocked VSV viral RNA synthesis and inhibited EBOV RNA transcription, demonstrating a consistent mechanism of action against genetically distinct viruses. The identification of this chemical backbone as a broad-spectrum inhibitor of viral RNA synthesis offers significant potential for the development of new therapies for highly pathogenic viruses.
Introduction
The nonsegmented, negative sense (NNS) RNA viruses are an order of viruses containing many human diseases. These include long-recognized pathogens such as rabies, mumps, measles and respiratory syncytial virus, as well as more recently identified pathogens such as Nipah, Hendra, and Ebola viruses. For many members of the NNS family, there are no approved therapeutics or vaccines. Furthermore, rapid development of drug resistance to monotherapy has been observed for other RNA viruses, such as influenza (van der Vries et al., 2010; Zhu et al., 2012), suggesting that multiple antivirals will be required for long-term effective treatment of these diseases. Therefore, the development of new therapies is warranted, especially ones that could target multiple members of this human-pathogen laden virus order.
Unlike bacterial diseases, for which many broad-spectrum antibiotics exist, there are no highly effective broad-spectrum small molecules to treat viral diseases. To address this issue, we have sought to identify small chemical probes that show antiviral activity against multiple NNS virus family members. Our hypothesis is that these molecules will target shared steps in virus replication, identifying targets for broad-spectrum antivirals. In this regard, one of the most promising potential targets for therapeutic intervention is the viral RNA dependent RNA polymerase (RdRp).
The viral RdRp is the only protein with enzymatic activity that is produced by all NNS family members. It is necessary for all aspects of viral RNA synthesis, ranging from genome synthesis to mRNA synthesis, capping and polyadenylation (Whelan et al., 2004). The polymerase is a validated antiviral target, as previous studies have shown compounds that directly target the polymerase complex will inhibit virus replication (Li et al., 2007). Additionally, compounds that target cellular proteins important for viral RdRp stability also block viral replication, indicating even indirect targeting of RdRp activity is a valid approach for antagonizing RNA virus replication (Connor et al., 2007).
To identify compounds that might act as broad-spectrum probes of virus function, we used a two-stage screening process. We initially set up a bi-functional screen to identify small molecules that showed strong antiviral activity but little cell toxicity. Our goal was to identify a small pool of lead compounds that could then be tested in a second stage to identify compounds that inhibited multiple NNS family members. Our initial screening assay used the prototypical member of the NNS virus family, vesicular stomatitis virus (VSV). VSV has significant advantages as a virus for tissue culture-based screening, as it is exceptionally well adapted to growth in culture, growing to high titer and causing significant cytopathic effects in a short period of time. Thus, any compounds that show antiviral activity against this virus in cell culture will likely have the capacity to dramatically blunt virus replication.
Once we developed a robust screen, we used the assay to screen a library synthesized by the Center for Chemical Methodology and Library Development at Boston University (CMLD-BU) using Diversity Oriented Synthesis (DOS) principles (Brown et al., 2011; Tan, 2005). This library was a collection of over 2000 diverse, focused sub-libraries averaging 100 compounds each, chosen because it represented a wide range of chemical space with a fairly limited number compounds. This gave us the opportunity to look for novel chemotypes that have not previously been screened for activity against NNS viruses. Our interest in this library was supported by the fact that previous screens using similar composite libraries have identified compounds with antimicrobial activities (Brown et al., 2011; Peng et al., 2007; Thomas et al., 2008).
From our screen, several molecules targeting multiple viruses were identified, underscoring the validity of this approach for discovering broad-spectrum antivirals. From these, we focused on one compound that inhibited the NNS viruses VSV and EBOV. Our analysis showed the compound limited viral mRNA synthesis, suggesting that it was targeting the RdRp. We have thus identified a new probe that shows broad-spectrum inhibition of negative-strand RNA virus infection at transcription, a step for which few chemical probes exist. Further development of this compound could lead to a therapeutic with broad-spectrum activity against NNS RNA viruses.
Results
Identification of antiviral compounds
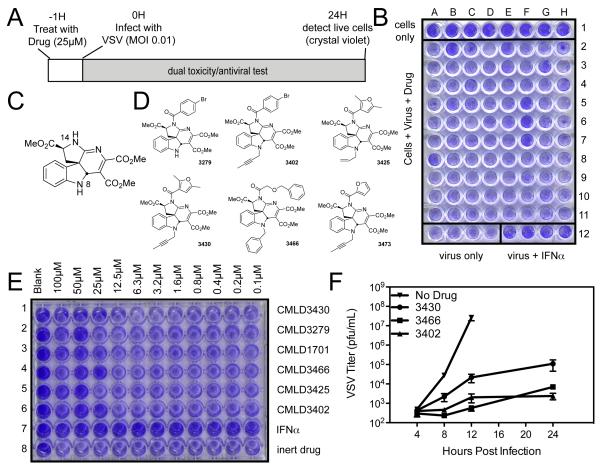
To discover new antiviral probes, we designed a high-throughput screen to identify small molecules that showed antiviral activity with little cellular toxicity. Our screen identified compounds that inhibit infection of VSV, the prototype NNS virus. An example of the overall assay design is shown in Figure 1A, with a sample plate illustrated in Figure 1B. Row 1 contained only cells (with no treatment or virus addition) and served as a negative control for normal cell growth and staining. For rows 2-11, each well was treated with an individual CMLD-BU compound at a final concentration of approximately 25μM. In row 12, 4 wells were treated with interferon (IFN; as a positive control for antiviral activity) and 4 wells were left infected but untreated (as a negative control for VSV infection). One hour after the addition of drugs or IFN, VSV was added to rows 2-12 at an MOI of 0.01 so that approximately 1% of the cells would be initially infected. Productive virus infection would allow spread and killing of all the cells in the well, while inhibition of virus infection at any step in the virus lifecycle would prevent cytopathic effects and cell death. At 24 hours postinfection (hpi), cells were washed, fixed, and stained with crystal violet to determine the abundance of viable cells not affected by virus or drug.
Figure 1.
Screen for antiviral compounds using VSV cytotoxicity (A) Schematic of screen. (B) Sample plate from screen: HeLa cells seeded in all wells and treated with library compounds in rows 2-11 and IFNa control in row 12, columns E-H. Infected with VSV MOI 0.1 in rows 2-12 for 24 hours, then fixed and stained with crystal violet. (C) Basic structure of hit scaffold from preliminary screen. (D) Structures of active, indoline alkaloid-type CMLD-BU compounds identified in the screen. (E) HeLa cells were treated with dilution curve of CMLD-BU compounds for 1 hour before infection with wtVSV, then fixed and stained as in screen. (F) HeLa cells were treated with 50μM CMLD-BU compounds for 1 hour before infection with wtVSV at MOI 0.01. Virus was collected at specified timepoints and titer determined by plaque assay.
In wells where VSV replication had taken place or where the added compounds were cytotoxic, we expected little crystal violet staining due to cell death induced by either phenomenon (see Figure 1B, row 12, columns A-D). In wells not infected with VSV (Figure 1B, row 1) or infected with VSV but treated with IFN (Figure 1B, row 12, columns E-H), we expected a strong purple crystal violet staining. In cells treated with compounds that were not toxic and that blocked the replication of VSV, we also expected strong purple staining, as seen in Figure 1B wells F 5-7. These compounds were scored as positives. The screen was carried out twice, and those compounds that inhibited VSV-mediated cell death in both screens were considered hits and were further investigated.
Our screening of the 2000-compound CMLD-BU library identified a dozen high-confidence hit compounds at a 25μM concentration. Because the screen determined cellular viability as a measure of VSV infection, the hit compounds were not cytotoxic at the concentration screened; thus we were confident that cellular death was not providing us with false positive hits. Included in these hits were several synthetic indoline alkaloid-type compounds that shared a common central chemical scaffold. The scaffold of these active compounds (Figure 1C) is named 1 for the purpose of this paper. In the library that we screened, there were sixteen additional N-14 amide analogues of 1, and forty-eight N-14 sulfonamides (see N-14 labeled in figure 1C). The larger number of the sulfonamides had been part of a separate successful project (Brown et al., 2011)(Brown et al., 2011) studying antimalarial compounds. In our original screen, more than 100 analogs of 1 were screened. Of these, we identified 6 as hits (Figure 1D). Interestingly, these hits were not evenly distributed amongst the amide and sulfonamide compounds. All of the active compounds came from the subset of members diversified at N-14 as amides. The sulfonamides invariably fell below the threshold of activity deemed a hit, with the exception of one compound, which was a very weak hit in the screen.
New lots of the hit compounds were synthesized and tested as serial dilutions in a bioassay similar to that shown in Figure 1A to roughly determine the EC50 of each compound. One column in each row was left untreated with drug or virus (far left). In successive columns, cells were incubated with decreasing dilutions of drug, starting at 100μM. One hour after drug treatment, cells were infected with VSV at an MOI of 0.01, and cell viability was assessed at 24hpi. The results of this test are shown in Figure 1E. The five alkaloid-type compounds are in rows 1, 2, 4, 5, and 6. Row 3 is a non-basic, weakly antiviral compound. Row 7 shows a serial dilution of IFN, as a control for antiviral activity. The complete inhibition of VSV infection by IFN is expected, as the IFN-competent HeLa cells produce more IFN during the infection. Row 8 shows a dilution of a control CMLD-BU compound that was neither cytotoxic nor antiviral in the initial screen. Of the five alkaloid compounds tested, all showed antiviral activity, and the EC50 values ranged from 12.5μM to 50μM.
Confirmation of antiviral activity of hits
We chose three of the five initial indoline alkaloid-type hits to continue a more precise determination of the mechanism of antiviral activity of these compounds. In our initial efforts, we determined how well these compounds blocked the production of new infectious virions following infection. HeLa cells were treated with the compounds at 50μM for 1 hour, then infected with VSV at MOI 0.01. Virus was collected at specified times post infection, and viral titers were determined by plaque assay. In the absence of any CMLD-BU compound, VSV replication was robust, reaching titers of 108 by 12hpi (Figure 1F). However, in cells infected with VSV but treated with CMLDBU3430, there was a 3-log decrease (99.9%) in viral replication, while in the presence of CMLDBU3466 and CMLDBU3402 there was 4-log decrease. This highlighted the strong antiviral activity of these compounds and suggested that CMLDBU3466 and CMLDBU3402 showed the greatest efficacy.
Compounds with activity against VSV have activity against other NNS viruses
In earlier studies, we have noted that compounds with activity against VSV have demonstrated inhibition of other, more pathogenic negative strand RNA viruses (Connor et al., 2007; Smith et al., 2010). To examine whether the compounds identified in this screen showed broad-spectrum antiviral activity, we tested the efficacy against other negative strand RNA viruses. We tested Ebola virus, a member of the Filoviridae family, which is a NNS virus like VSV, as well as Rift Valley fever virus, a member of the Bunyaviridae family, which is a tripartite negative strand virus.
We tested the efficacy of several compounds against Zaire Ebolavirus (EBOV) expressing eGFP (EBOV-eGFP). EBOV-eGFP was used to infect cells in a 96-well plate format, and eGFP expression was read over time with several concentrations of compound. Figure 2A demonstrates that the compounds inhibited EBOV infection when cells were treated with 40μM CMLDBU3402, CMLDBU3279 or CMLDBU3466 and infection levels read over time (data normalized to fluorescent units of DMSO treatment control 5 days post infection). To visualize the inhibition, slides infected with EBOV-eGFP for 3 days in the presence or absence of compound were fixed, stained with DAPI to delineate nuclei and imaged. Figure 2B shows representative images in the absence (No Drug) or presence of 40μM CMLDBU3402 or CMLDBU3466. The compounds inhibited EBOV-eGFP infection, as fewer cells expressed eGFP when treated with CMLD-BU compounds, and cells incubated with either compound showed little cytoxoticity, as evidenced by normal nuclear morphology.
Figure 2.
Effect of CMLDBU3402 on Ebola virus replication. (A) A549 cells were treated with 40μM CMLDBU3402 and infected with EBOV-eGFP at MOI 1. Fluorescence was read at specified timepoints and data normalized to DMSO. Error bars represent standard deviation of triplicates. (B) A549 cells were treated with 40μM CMLDBU3402 and infected with EBOV-eGFP at MOI 1.0 for 65 hours. Cells were fixed and nuclei stained before imaging. (C) A549 cells were treated with 5μM – 100μM CMLDBU3402 and infected with EBOV-eGFP at MOI 1. Fluorescence was read 5 days post infection and data normalized to DMSO. Error bars represent standard deviation of triplicates.
To determine the EC50 for the compounds against EBOV, we treated A549 cells with a dilution curve of the 3 compounds from 100μM to 5μM and read the infection levels by eGFP expression at 5 days post infection. When normalized to infection levels with DMSO alone, CMLDBU3402 and CMLDBU3466 have similar curves, with an EC50 around 20μM, while CMLDBU3279 was less potent, with an EC50 around 40μM (Figure 2C). This pattern corresponded to the inhibition curves seen with VSV, although the compounds were more potent against VSV (Figure 1D and DNS).
Of the structurally similar compounds, CMLDBU3402 showed the greatest inhibitory activity against both VSV and EBOV; therefore, we chose to further investigate its antiviral activity. We tested the efficacy of CMLDBU3402 against Rift Valley fever virus (RVFV), a member of the Bunyaviridae family. Figure 3A shows a micrograph of cells infected with eGFP-expressing RVFV in the absence (top panel) or presence (bottom panel) of CMLDBU3402. These micrographs suggest that there was little effect of CMLDBU3402 on RVFV infection. This conclusion was supported when high MOI (5) and low MOI (0.1) infections with and without drug were quantified by fluorometry (Figure 3B). These data suggested that the compound does not universally inhibit negative-strand RNA virus infection, but does inhibit NNS virus infection.
Figure 3.

Effect of CMLDBU3402 on Bunyavirus gene expression. A549 cells were treated with 20μM CMLDBU3402 and infected with MOI 0.1 or MOI 5.0 of RVFV-GFP for 49 hours. (A) Cells infected with MOI 5.0 were imaged in the absence (top panel) or presence (bottom panel) of CMLDBU3402 at 49 hpi. (B) Fluorescence of GFP expressed by RVFV was measured at 48 hpi and background values subtracted. Error bars represent standard deviation.
Reduction of virus growth by CMLDBU3402 is independent of stereochemistry
The structural similarities of the hit compounds suggest that they inhibit virus replication through a similar mechanism. To begin to determine the mechanism of action, we utilized the most potent compound, CMLDBU3402. To ensure that the compound was not cytotoxic at the concentrations used for treatment, we determined CC50 values for CMLDBU3402. At 6 hours after drug treatment, there was no cytotoxicity seen in HeLa cells up to 100μM (DNS). At 17 hours after drug treatment, there was some cell death seen at drug concentrations over 80μM; however this was not significant compared to DMSO treated cells (p value of 0.0762). These data correlate with the lack of cytotoxicity seen in the original screen (Figure 1), and contend that the concentrations used in this study are not cytotoxic.
Comparison of the structures of the antiviral compounds with structurally similar compounds that lack antiviral activity indicated that small changes in the side chains can have a large effect on antiviral activity (DNS). To determine whether the absolute stereochemistry of CMLDBU3402 also played an important role in its activity, CMLDBU3402 produced from (L)-tryptophan and the enantiomer CMLDBU3402E, prepared from (D)-tryptophan, were confirmed to be enantiomerically pure by chiral HPLC (DNS). Both compounds were tested for activity against a VSV virus that expresses renilla luciferase (VSV-rLuc). A549 cells were treated with dilutions of CMLDBU3402 or CMLDBU3402E for 45 minutes, and then infected with VSV-rLuc at an MOI of 1, which allowed for expression of high levels of luciferase in the absence of antiviral treatment. The infection was allowed to progress for 24 hours before the cells were lysed with Renilla-Glo reagent, read on a luminometer, and normalized to DMSO treatment. Both CMLDBU3402 and CMLDBU3402E inhibited infection to similar degrees (Figure 4A). These data demonstrate that the chirality of the compound did not dramatically alter its antiviral effect. Because the chirality of the compound did not affect the antiviral capacity, the two compounds were used interchangeably.
Figure 4.
Effect of drug treatment on growth of VSV protein expression. (A) A549 cells were treated with CMLDBU3402 or CMLDBU3402E, then infected with VSV-rLuc at MOI 1.0 for 16 hours and lysed and read on luminometer. Normalized to DMSO control; error bars are standard deviation. (B) HeLa cells were treated with 0, 10 or 25μM CMLDBU3402 for 1 hour before infection with wtVSV at MOI 10 for specified times. Immunoblot of VSV matrix protein and actin control. (C) HeLa cells treated with 1, 10 or 25 μM CMLDBU3402 or CMLDBU3402E were infected with wtVSV at MOI 10 for 6 hours. Immunoblot of VSV-G and actin control. (D) A549 cells were treated with 20μM CMLDBU3402 then infected with VSV-eGFP at MOI 5 for 6 hours. Cells were fixed and stained for VSV-G and nuclei. (E) A549 cells infected with VSV-eGFP were counted to determine percent infection in the absence and presence of CMLDBU3402. Error bars are standard deviation.
CMLDBU3402 inhibits viral gene expression
The ability of CMLDBU3402 to block expression of virally encoded rLuc suggested the drug was blocking viral infection at or before protein synthesis. To directly look at the effects of native viral protein accumulation in the presence of drug, we determined the level of the VSV matrix (M) protein in infected cells in the presence or absence of CMLDBU3402. HeLa cells were infected with VSV in the presence 10μM or 25μM CMLDBU3402. Cells were lysed at increasing times postinfection and total protein was separated by SDS-PAGE and transferred to PVDF for immunoblotting with an anti-VSV M antibody. A representative result of these experiments is shown in Figure 4B. In cells that were not treated with drug, the expression of VSV M was readily detected beginning at 4 hours postinfection. The presence of either 10μM or 25μM CMLDBU3402 decreased the expression of M protein at 4 hours by greater than 90%. This same inhibition of M protein accumulation was seen at 6 hours postinfection, with a dose-dependent effect of drug concentration. To determine if expression of other viral proteins was also inhibited, cells treated with either CMLDBU3402 or CMLDBU3402E were lysed at 6 hpi, and the recovered proteins were immunoblotted for the VSV glycoprotein (VSV-G). As shown in Figure 4C, both compounds were highly effective at blocking expression of VSV-G at 25μM.
To determine whether the decrease in viral protein expression was a result of lower protein expression in all infected cells or fewer cells being infected, immunofluorescence was performed to examine the protein expression levels in individual cells. A549 cells were infected with a recombinant VSV that expresses eGFP (VSV-eGFP) at an MOI of 5. At 6 hpi, cells were fixed and stained with an antibody against the VSV glycoprotein (red). Infection of DMSO-treated cells with this virus resulted in the expression of high levels of eGFP and VSV-G (Figure 4D). In cells that were treated with 20μM CMLDBU3402, the expression of both eGFP and the VSV glycoprotein were dramatically decreased. Quantitation of several experiments showed that >60% of DMSO-treated cells infected with VSV-eGFP expressed significant levels of viral proteins, while fewer than 20% of CMLDBU3402-treated cells infected with VSV-eGFP expressed low levels of viral proteins (Figure 4F). These results suggested that CMLDBU3402 decreased both the number of infected cells and the protein expression in each cell, and confirmed that CMLDBU3402 acts to block virus infection at or before the level of viral protein synthesis. We therefore began to test the possible steps of the virus lifecycle, including virus entry, transcription and replication.
CMLDBU3402 does not block viral entry
Recent studies have shown that some compounds targeting VSV replication can inactivate the virus particle directly (Wolf et al., 2010). To determine whether CMLDBU3402 was able to act upon the assembled virus and inactivate it prior to infection, virus and drug were co-incubated for 1 hour at a concentration of 100μM, which is over 5 times the EC50 value (Figure 1D). Following the co-incubation, the virus and drug mixture was diluted past the point of drug efficacy, to 0.25μM, and used to infect untreated cells at MOI 5 (lane 3). As controls, cells were infected with virus alone (lane 1), infected with virus plus 0.25μM CMLDBU3402, a concentration that does not show antiviral activity (lane 5), or infected with virus plus 50μM CMLDBU3402, a concentration that effectively blocks infection (lane 4). Cells were lysed and probed for expression of VSV-G and VSV-M by immunoblot, as above. Pre-incubation of the drug directly with the virus did not inhibit virus infectivity (Figure 5A). Furthermore, pre-treatment of VSV-rLuc virus with CMLDBU3402 in a similar manner did not significantly inhibit rLuc expression as read on a luminometer (p value 0.22; Figure 5B), indicating that the drug was not directly inactivating the virus particle. As a control, an anti-VSV-G inhibitory antibody, 8G5 (Lefrancois and Lyles, 1982a, b), was incubated with the virus or added during infection. In both cases, VSV infection was inhibited, indicating that the time of pre-incubation is sufficient for interaction and inhibition of VSV.
Figure 5.
Effect of pre-treatment of virus with CMLDBU3402. (A) VSV and CMLDBU3402 were incubated for 1 hour before adding to cells at a final concentration of 0.25μM. As controls, 0.25μM or 50μM drug were added to cells before infection with VSV. At 6hpi, cells were lysed and immunoblotted for VSV-G, VSV-M or actin control. (B) VSV-rLuc was incubated with CMLDBU3402 at 100μM or anti-VSV-G antibody 8G5 for 1 hour prior to adding to cells, for a final concentration of 1μM CMLDBU3402. As controls, cells were treated with 1μM or 40μM or 8G5 prior to infection with VSV-rLuc at MOI 1.0. 16 hpi, cells were lysed and read on luminometer. Error bars represent standard deviation of triplicates. (C) HeLa cells were treated with 25μM CMLDBU3402 at specified times and infected with wtVSV at MOI 10. At 4hpi, cells were lysed and immunoblotted for VSV-G, VSV-M and actin as a control.
To determine whether CMLDBU3402 displayed a restricted time of maximum activity, we carried out time-of-addition experiments. Cells were infected with VSV, and drug was added to cells either at the time of virus infection or at 1, 2, 3 or 4 hpi. Accumulation of viral protein was determined at 6 hpi by immunoblotting cell lysates. The results, in Figure 5C, show that concurrent treatment with drug maximally affected virus replication, but CMLDBU3402 was able to diminish the accumulation of viral proteins when added after virus entry had occurred, with a 50% reduction in viral protein production seen when compound was added 2 hours post infection. These data suggest that CMLDBU3402 is able to inhibit the VSV lifecycle at a step after initial entry and release of the virus into the cell but at or before the expression of viral proteins.
CMLDBU3402 inhibits an early stage of viral replication
The finding that CMLDBU3402 is able to inhibit a step post-entry suggested the compound exerts its antiviral effect by inhibiting transcription or translation. To directly test the effects of CMLDBU3402 on viral transcription, a VSV minigenome expression system was used. In this experiment, eGFP expression was directly dependent on the viral polymerase complex without requiring earlier steps in the virus lifecycle. eGFP expression from the minigenome construct was measured in the presence and absence of drug.
Cells were infected with Vaccinia virus expressing the T7 polymerase (VACV-T7) and transfected with a set of plasmids that express the VSV polymerase complex proteins L, N and P and a VSV eGFP-containing minigenome RNA fragment. eGFP would only be produced if the VSV polymerase transcribed eGFP mRNA from the minigenome RNA template. Figure 6A shows that untreated cells expressed eGFP. When CMLDBU3402 was added to the cells, eGFP expression was greatly diminished (Figure 6B). Quantification of eGFP expression showed a decrease in eGFP signal to approximately 33% of signal from minigenome-transfected cells without drug added (DNS), suggesting that CMLDBU3402 was able to directly inhibit VSV transcription.
Figure 6.
Effect of CMLDBU3402 on VSV gene expression. (A-B) BSR-T7 cells were infected with VACV-T7 and transfected with plasmids expressing the VSV polymerase complex, N, P and L along with a mini-genome that expresses eGFP in the absence (A) or presence (B) of CMLDBU3402. (C-D) BSR-T7 cells were infected with VACV-T7 and transfected with a plasmid that expresses GFP under the control of T7 in the absence (C) or presence (D) of CMLDBU3402. (E) Cells infected with VSV at an MOI 10 were pulsed with 3H-uridine to allow incorporation into mRNA. Extracted RNA was run on a gel, and viral mRNA bands for L, G/N and P+M are labeled. (F) 3H-uridine incorporation into viral mRNAs was measured on a scintillation counter. Error bars represent standard deviation of triplicates. (G) Cells infected with VSV at an MOI 10 and treated with DMSO, CMLDBU3402E or Ribavirin concurrent with infection or at 1 or 3 hours post infection. Cells were pulsed with 3H-uridine from 2-4 hours post infection to label viral mRNA. Extracted RNA was run on a gel and viral mRNA bands for G/N and P+M are labeled, with quantitation of P+M normalized to DMSO presented.
To verify that CMLDBU3402 did not inhibit VACV-T7 expression or eGFP translation, cells infected with VACV-T7 were transfected with a control plasmid that expressed eGFP in a T7-dependent context (T7-eGFP). eGFP expression from a T7-driven plasmid was not altered in the presence of CMLDBU3402 (Figure 6C and D). This indicated that the compound does not directly inhibit VACV expression of T7, transcription by the T7 polymerase, or translation of eGFP mRNA.
CMLDBU3402 inhibits viral transcription
To determine more directly if CMLDBU3402 inhibits transcription of viral RNA, BHK cells were infected with VSV in the presence or absence of CMLDBU3402E and pulsed with 3H-uridine from 4-6 hpi in the presence of actinomycin-D to specifically label viral RNA synthesis. Without drug, infected cells showed robust viral mRNA expression, as demonstrated by the detection of four viral mRNA bands, corresponding to L, G, N and P+M, which are similar in size and migrate together (Figure 6F). Addition of CMLDBU3402E blocked the expression of viral mRNAs, indicating that the compound blocks viral mRNA transcription. Figure 6E shows the quantification of viral RNA synthesis from three experiments.
CMLDBU3402 inhibited viral protein expression when added after infection (Figure 5C). To examine if the time of drug addition impacts mRNA synthesis, CMLDBU3402E was added 1 hour before VSV infection, and 1 or 3 hours after VSV infection. Viral transcription was labeled from 4-6 hpi with 3H-uridine in the presence of actinomycin D, as above. As seen in Figure 6G, CMLDBU3402E inhibited viral mRNA accumulation most when added prior to infection, to 54% of DMSO when the P+M band was quantified; however, inhibition of viral mRNA synthesis was seen when drug is added 1 hour after infection as well, to 74% of DMSO. As a control, ribavirin (which is also thought to inhibit RNA synthesis) was added during virus infection, with results similar to CMLDBU3402E. Both drugs inhibited VSV infection most when added prior to infection, but they still inhibit VSV mRNA synthesis when added 1 hour after infection.
CMLDBU3402 inhibits viral transcription in multiple viruses
The ability of CMLDBU3402 to specifically inhibit NNS viruses suggests that the mechanism of action would be similar in VSV- and EBOV-infected cells. To determine if the mechanism of action against EBOV is similar to VSV, we used an ebolavirus minigenome system in the presence of CMLDBU3402. An EBOV minigenome plasmid carrying a renilla luciferase reporter gene was transfected into BHK cells expressing the T7 polymerase along with support plasmids carrying EBOV NP, L, VP30 and VP35 (Muhlberger et al., 1999). The negative-sense copy of the luciferase gene in the minigenome plasmid is made by T7 RNA polymerase and subsequently encapsidated and transcribed by the support EBOV RdRp complex. The resulting rLuc expression was quantified using a luminometer. As shown in Figure 7A, when the minigenome system was transfected into cells, rLuc was expressed. Addition of 40μM CMLDBU3402 blocked rLuc expression, indicating that the mechanism of action against EBOV is similar to that seen with VSV. Therefore, CMLDBU3402 blocks viral transcription of NNS viruses, including VSV and EBOV.
Figure 7.

Effect of CMLDBU3402 on EBOV minigenome-based gene expression. (A) BSR-T7 cells were transfected with the Ebola virus polymerase complex plasmids NP, VP35, L, and VP30 with a minigenome plasmid expressing rLuc in the presence or absence of CMLDBU3402. Cells were then lysed and read on luminometer.
Discussion
Nonsegmented negative-strand RNA viruses include many human diseases, but there are no approved therapeutics for some of the most pathogenic, including Ebola virus. A broad-spectrum antiviral could be used to treat infections with these viruses and as-yet unidentified emerging NNS viruses in the future. We describe here a simple screening approach that identified promising antiviral probes from a diverse small molecule library. Together with other recently-published efforts, this highlights that screening of the library can be fruitful in identifying probe compounds with antimicrobial activities (Benson et al., 2012; Brown et al., 2011; Thomas et al., 2008; Wei et al., 2009).
The identification of five indoline alkaloid-type hits in our screen suggests that this structure (Figure 1C) may be particularly well suited as an antiviral scaffold. In addition to our identification of compounds like CMLDBU3402, other screens have found that related indoline alkaloid-type library members have activity against hepatitis C (HCV) and malaria (Brown et al., 2011; Wei et al., 2009). Of particular interest, related compounds that were originally identified as inhibitors of HCV core assembly with EC50 values in the high micromolar range were readily improved through structural modifications to be effective inhibitors at low nanomolar concentrations (Ni et al., 2011). This suggests that the scaffold in this work, designated 1 in Figure 1C, should be readily improvable through medicinal chemistry derivatization following structure activity relationships.
Though the sub-library of indoline-type compounds contained only a limited number of samples (sixteen N-14 amides, forty-eight N-14 sulfonamides), preliminary SAR trends could be observed. First, N-14 amides were clearly more active than the corresponding N-14 sulfonamides. Second, N-14 amides containing aryl groups were also more active than those with only alkyl amides at N-14, though this latter suggestion needs to be further explored. Finally, N-8 substituents were examined, though only a very limited number of structural variants at this position (N-unsubstituted, methyl, allyl, benzyl, isobutenyl and 2-butynyl) were in the small library initially screened.
Our experiments demonstrate that the lead compounds found to have anti-VSV activity in our screen show antiviral activity against EBOV as well, and thus have the potential to be broad-spectrum antiviral agents. These distantly related viruses both have nonsegmented negative strand RNA genomes, which suggests that the inhibitors may be able to inhibit other NNS viruses as well. While VSV and EBOV encode different genes and have disparate strategies for entry, these viruses show high sequence similarity in the RNA-dependent RNA polymerase and are known to follow the same general mechanism for RNA synthesis (Whelan et al., 2004). This suggested that the RdRp is a potential target of the antiviral activity.
Examination of the mechanism of action of CMLDBU3402 is consistent with this hypothesis. CMLDBU3402 does not interact with the virion directly or inhibit virus entry in studies with VSV (Figure 5). It does, however, block viral gene expression at an early stage of infection and blocks reporter gene expression from a minigenome system for both VSV (Figure 6) and EBOV (Figure 7). Furthermore, VSV viral mRNA synthesis is inhibited by CMLDBU3402 (Figure 6). This is the expected behavior from a chemical probe that inhibits the viral polymerase. Thus, we propose that CMLDBU3402 is inhibiting the RdRp of both VSV and EBOV through a similar mechanism.
There are several possibilities for how CMLDBU3402 might inhibit RNA polymerase activity. The compound is not an obvious nucleoside analog (Figure 1D), which is encouraging in that this decreases the likelihood that there will be nonspecific toxicity effects to host transcription (Moyle, 2000). Studies have shown that pyrimidine synthesis inhibitors can act as broad-spectrum antiviral agents (Hoffmann et al., 2011), but CMLDBU3402 does not appear to be inhibiting pyrimidine or purine synthesis, as supplementation with uridine or adenosine does not restore viral infection (DNS).
How CMDLBU3402 is acting to block viral RNA synthesis is not yet clear. The compound could be interacting with the RdRp complex directly to prevent activity and therefore inhibit the virus. Direct interaction with viral proteins would be expected to have minimal cytotoxicity, but could lead to faster resistance as the viral proteins evolve to avoid the inhibitor (Li et al., 2007). To combat drug resistance, multiple therapeutics may be used concurrently; therefore, the identification of inhibitors at multiple steps in the virus lifecycle is ideal. CMLDBU3402 may also be interacting with host factors that are necessary for viral transcription, therefore preventing virus infection. For example, it has been previously shown that inhibition of HSP90 prevents replication of several NNS viruses (Connor et al., 2007; Smith et al., 2010). It may be more difficult for a virus to evade therapeutics targeting the host, making host-targeted small molecules attractive drug candidates (Schwegmann and Brombacher, 2008).
Although CMLDBU3402 has antiviral activity against disparate viruses, it is important to note that it does not inhibit the replication of all viruses. In our experiments CMLDBU3402 did not inhibit the tripartite negative sense RNA Bunyavirus RVFV, which suggests that the mechanism of action is restricted to the NNS viruses. This may be due to intrinsic differences in the mechanism of action of the polymerase for NNS viruses, or it may be due to host factors that are necessary for NNS virus transcription, but not for other negative strand virus or DNA virus transcription (Whelan et al., 2004). Further understanding of the exact mechanism of action should elucidate the reason for the specificity of this inhibitor.
There have been other broad-spectrum antivirals described in the literature. Ribavirin, which has many proposed mechanisms of action but is thought to target the virus polymerase, inhibits a wide range of virus families, including Arenaviruses, Alphaviruses, and Bunyaviruses (Crotty et al., 2000; Huggins, 1989; Scheidel and Stollar, 1991; Streeter et al., 1973). The exact mechanism of action of ribavirin has been difficult to determine, and there are many negative side effects including hemolytic anemia and possible teratogenicity, limiting its use (Cameron and Castro, 2001). Importantly, it has been shown that ribavirin has very little effect on Ebola virus replication, suggesting that CMLDBU3402 and ribavirin may attack viral polymerase activity in different ways.
Recently, a pyrazine derivative compound, T-705, has been shown to have broad-spectrum antiviral activity against Arenaviruses, Bunyaviruses and Orthomyxoviruses. It appears to act as a nucleoside analog and inhibits the polymerase activity of influenza (Furuta et al., 2009; Gowen et al., 2007; Takahashi et al., 2003). CMLDBU3402 does not appear to act as a nucleoside analog, and does not inhibit Bunyavirus replication; therefore, CMLDBU3402 appears to inhibit the polymerase complex in a manner distinct from T-705.
The fact that several broad-spectrum inhibitors all appear to alter polymerase function indicates how attractive this protein complex is as a target for therapeutic development. Against this group of broad-spectrum antivirals, CMLDBU3402 shows a similar antiviral target, suggesting promise for future development. Further work will be required to determine the precise target of the compound and the extent of its broad-spectrum capacity.
Significance
There are few therapeutics available for many pathogenic RNA viruses, including Ebola virus, a virus with mortality rates up to 90% following infection, and a recently discovered rhabdovirus associated with hemorrhagic fever. Identification of small molecules with broad-spectrum activity against these types of viruses will be important tools in developing successful therapies. Our study has identified a set of indoline alkaloid-like compounds that inhibit the replication of the prototype rhabdovirus VSV and the filovirus EBOV. Analysis of the mechanism of action of one of these compounds, CMLDBU3402, shows that the compound is likely targeting the ability of the virus to synthesize RNA. RNA synthesis is the main catalytic activity of RNA viruses, so these compounds represent probes of a central virus function and a potential drug target for the development of effective broad-spectrum antivirals.
Materials and Methods
Cell Culture and Viruses
A549 (CCL-85), Vero-E6 and HeLa (CCL-2) cells were obtained from the ATCC. Vero-E6 cells were used for growth of Ebola virus. BHK21 cells (ATCC) were used for growth of VSV virus. VSV virus stocks were made by infecting cells with wild-type San Juan at an MOI of 0.01. Supernatant fluids were harvested, clarified and titrated on BHK21 cells.
Plaque Assays
BHK21 cells were infected with serial dilutions of clarified supernatant fluid from infected cells, allowed to sit for one hour, then covered with 1× DMEM-agar and incubated at 37 degrees for 24 hours. Cells were fixed with 4% formaldehyde for one hour. Then, the agar overlay was removed and cells stained with crystal violet.
Library Screening and Compounds
Generation of the screening library and chemical syntheses were performed by the Center for Chemical Methodology and Library Development at Boston University (CMLD-BU), Boston, MA. HeLa cells were seeded at 4,000 cells per well in 96-well plates (Corning) the previous day and infected with VSV at an MOI of 0.01 in the presence of compound. Final compound concentrations of the initial screen were estimated to be approximately 25μM based on an average molecular weight for the library of 400. At 24hpi, cells were washed in PBS, fixed with methanol, stained with crystal violet, and washed. The wells were scored based on color density. Interferon-α was obtained from PBL InterferonSource and used at a concentration of 1-10 IU as a positive control. For all follow-up work, the compounds were resynthesized, validated and used at precise concentrations as referenced in text.
Compound Synthesis
Compounds were synthesized by the Center for Chemical Methodology and Library Development at Boston University. See Supplemental Data for details.
CC50 Assay
HeLa cells were seeded in 96-well plates the previous day and treated with a dilution curve of CMLDBU3402 from 100μM to 2.5μM. After 6 or 17 hours, the cells were lysed and luciferase read according to manufacturer’s instructions using CellTiter-Glo Luminescent Cell Viability Assay system (Promega) on a LUMIstar Omega luminometer (BMG Labtech) for 1 second/well.
VSV Luciferase Assay
A549 cells were seeded in 96-well plates the previous day and infected with VSV-rLuc at an MOI specified in text in DMEM without phenol. Cells were lysed and luciferase read according to manufacturer’s instructions using Renilla-Glo Luciferase Assay system (Promega) on a LUMIstar Omega luminometer (BMG Labtech) for 1 second/well. Unpaired t tests were run using GraphPad Software to determine significance as mentioned in text.
Western Blots
BHK21 cells were infected at an MOI specified in text. At times indicated, cells were lysed in RIPA buffer with protease and phosphatase inhibitors (1mM PMSF, 1mM benzamidine, 100nM okadaic acid, 100nM microcysin and 100nM sodium fluoride). 20μg of total lysate were separated on a 10% SDS-PAGE gel and transferred to PVDF (Bio-Rad 162-0177). Blots were probed with polyclonal antibodies specific to viral M and G proteins and actin (Santa Cruz, SC-47778).
Immunofluorescence
VSV. A549 cells were seeded in 6 well plates (Corning) the previous day and infected with VSV-eGFP at MOI 5 for 6 hours (Whitlow et al., 2006). Cells were fixed with 4% formaldehyde and stained for VSV-G using 8G5 and DAPI to delineate the nuclei. EBOV. All pathogenic Ebola virus work was completed in the BSL-4 laboratory at the United States Army Medical Research Institute of Infectious Diseases (USAMRIID) following approved Standard Operating Procedures (SOP). A549 cells were seeded in chamber slides the previous day and infected with EBOV-eGFP at MOI 1.0 (Towner et al., 2005). Cells were fixed at 65hpi with 10% neutral buffered formalin for 24 hours. Cells were mounted with coverslips using ProLong Gold antifade reagent with DAPI (Invitrogen) and imaged on Axiovert 200M microscope (Zeiss). Rift Valley fever virus. All RVFV work was completed in the BSL-3+ laboratory at USAMRIID following approved SOPs. A549 cells were seeded in a 96 well plate (Corning) the previous day and infected with RVFV-GFP at MOI 0.1 or MOI 5. Infection was monitored by GFP expression using a Nikon Eclipse TE2000-S fluorescence microscope, and imaged at 49 hpi. The fluorescence was measured using a Modulus Fluorometer (Turner Biosystems) at 49 hpi.
VSV Minigenome Assay
BSR-T7/5 cells at approximately 90% confluence were infected with VACV-T7 at an MOI of 1 in serum-free Opti-MEM (Life Technologies 31985-062). One hour later cells were transfected with 3μg WN, 1μg WP, 0.6μg WL and 3μg VSV(-)GFP using 10μL Lipfectamine 2000 (Life Technologies 11668-019). Six hours after transfection, media was removed and replaced with DMEM + 7% FBS. At 24 hpi, resuspended cells were placed in a 96 well plate (Corning) and well fluorescence was measured on a Tecan Infinity M1000 fluorescence plate reader (Tecan Group Ltd.), using 488 excitation and 507 emission wavelengths.
RNA labeling
BHK21 cells at approximately 70% confluence were infected with VSV-Indiana San Juan at an MOI of 10. Cells were treated with 40μM 3402E or the equivalent volume of DMSO as described in the text. At 3hpi, 2μg/mL actinomycin-D was added to cells to inhibit cellular transcription. From 4-6hpi, 20μCi/mL 3H-uridine (Perkin Elmer NET367001MC) was added to cells to label ongoing viral RNA synthesis. At 6hpi, cells were lysed in Buffer RLT, and total RNA was purified using the Qiagen RNeasy mini kit (74106) according to the manufacturer’s instructions. Purified RNA was separated on a 1% denaturing agarose-formaldehyde gel. The gel was dehydrated and fixed with methanol and incubated overnight in scintillation fluid to amplify the radioactive signal. The gel was then dehydrated and exposed to film to detect labeled RNA (Kodak BioMax MS).
Ebolavirus Minigenome Assay
BSR-T7/5 cells at approximately 90% confluence were transfected with the Ebola virus plasmids (Muhlberger et al., 1999) at the following ratios: Ebo-RLuc 1ug: pTNP-Ebo 0.167ug:pTVP35-Ebo 0.08ug: pTL-Ebo 0.08:pVP30-Ebo 0.1ug using Lipofectamine 2000 (Life Technologies 11668-019). At 22 hours post transfection, cells were lysed and luciferase read according to manufacturer’s instructions using Renilla-Glo Luciferase Assay system (Promega) on a LUMIstar Omega luminometer (BMG Labtech) for 1 second/well.
Ebola Fluorescence Assay
All pathogenic Ebola virus experiments were completed in the BSL-4 laboratory at USAMRIID following approved SOPs. A549 cells were seeded in 96-well plates the previous day and infected with EBOV-eGFP at MOI 1.0 with DMSO, or 40μM CMLDBU3402, CMLDBU3279, or CMLDBU3466. Plates were read on SpectraMax M5 (Molecular Devices) using GFP settings: excitation 485 nm, emission 515 nm with 495 nm cutoff.
Supplementary Material
Highlights.
Identified indoline alkaloid-type compounds as antivirals
Compounds show capacity to block multiple viruses
Mechanism of action is inhibiting viral RNA synthesis
New chemical scaffold for targeting a broad range of human pathogens
Acknowledgements
We would like to thank Darci Smith for assistance with the RVFV experiment, Aaron Beeler for his help in coordinating CMLD compound screening, and Sean Whelan for sharing the VSV minigenome system. We thank Elke Muhlberger for use of the EBOV minigenome system. CMF was supported by the Postgraduate Research Participation Program and the U.S. Army Research and Medical Command administered by the Oak Ridge Institute for Science and Education (ORISE). This work was supported by in part by P41 086180. Also in part by NIH RO1AI1096159-01 and K22AI-064606 to JHC. We thank NSF for supporting the purchase of the NMR (CHE 0619339) and HRMS (CHE 0443618) spectrometers.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Benson SC, Lee L, Wei W, Ni F, Olmos JJ, Strom KR, Beeler AB, Cheng KC, Inglese J, Kota S, et al. Truncated Aspidosperma Alkaloid-Like Scaffolds: Unique Structures For The Discovery Of New, Bioactive Compounds. Heterocycles. 2012;84:135–155. [Google Scholar]
- Brown LE, Chih-Chien Cheng K, Wei WG, Yuan P, Dai P, Trilles R, Ni F, Yuan J, MacArthur R, Guha R, et al. Discovery of new antimalarial chemotypes through chemical methodology and library development. Proc Natl Acad Sci U S A. 2011;108:6775–6780. doi: 10.1073/pnas.1017666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron CE, Castro C. The mechanism of action of ribavirin: lethal mutagenesis of RNA virus genomes mediated by the viral RNA-dependent RNA polymerase. Current opinion in infectious diseases. 2001;14:757–764. doi: 10.1097/00001432-200112000-00015. [DOI] [PubMed] [Google Scholar]
- Connor JH, McKenzie MO, Parks GD, Lyles DS. Antiviral activity and RNA polymerase degradation following Hsp90 inhibition in a range of negative strand viruses. Virology. 2007;362:109–119. doi: 10.1016/j.virol.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S, Maag D, Arnold JJ, Zhong W, Lau JY, Hong Z, Andino R, Cameron CE. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nature medicine. 2000;6:1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- Furuta Y, Takahashi K, Shiraki K, Sakamoto K, Smee DF, Barnard DL, Gowen BB, Julander JG, Morrey JD. T-705 (favipiravir) and related compounds: Novel broad-spectrum inhibitors of RNA viral infections. Antiviral Res. 2009;82:95–102. doi: 10.1016/j.antiviral.2009.02.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowen BB, Wong MH, Jung KH, Sanders AB, Mendenhall M, Bailey KW, Furuta Y, Sidwell RW. In vitro and in vivo activities of T-705 against arenavirus and bunyavirus infections. Antimicrobial agents and chemotherapy. 2007;51:3168–3176. doi: 10.1128/AAC.00356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann HH, Kunz A, Simon VA, Palese P, Shaw ML. Broad-spectrum antiviral that interferes with de novo pyrimidine biosynthesis. Proc Natl Acad Sci U S A. 2011;108:5777–5782. doi: 10.1073/pnas.1101143108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggins JW. Prospects for treatment of viral hemorrhagic fevers with ribavirin, a broad-spectrum antiviral drug. Reviews of infectious diseases. 1989;11(Suppl 4):S750–761. doi: 10.1093/clinids/11.supplement_4.s750. [DOI] [PubMed] [Google Scholar]
- Lefrancois L, Lyles DS. The interaction of antibody with the major surface glycoprotein of vesicular stomatitis virus. I. Analysis of neutralizing epitopes with monoclonal antibodies. Virology. 1982a;121:157–167. [PubMed] [Google Scholar]
- Lefrancois L, Lyles DS. The interaction of antibody with the major surface glycoprotein of vesicular stomatitis virus. II. Monoclonal antibodies of nonneutralizing and cross-reactive epitopes of Indiana and New Jersey serotypes. Virology. 1982b;121:168–174. doi: 10.1016/0042-6822(82)90126-x. [DOI] [PubMed] [Google Scholar]
- Li J, Chorba JS, Whelan SP. Vesicular stomatitis viruses resistant to the methylase inhibitor sinefungin upregulate RNA synthesis and reveal mutations that affect mRNA cap methylation. Journal of virology. 2007;81:4104–4115. doi: 10.1128/JVI.02681-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle G. Toxicity of antiretroviral nucleoside and nucleotide analogues: is mitochondrial toxicity the only mechanism? Drug safety : an international journal of medical toxicology and drug experience. 2000;23:467–481. doi: 10.2165/00002018-200023060-00001. [DOI] [PubMed] [Google Scholar]
- Muhlberger E, Weik M, Volchkov VE, Klenk HD, Becker S. Comparison of the transcription and replication strategies of marburg virus and Ebola virus by using artificial replication systems. J Virol. 1999;73:2333–2342. doi: 10.1128/jvi.73.3.2333-2342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni F, Kota S, Takahashi V, Strosberg AD, Snyder JK. Potent inhibitors of hepatitis C core dimerization as new leads for anti-hepatitis C agents. Bioorg Med Chem Lett. 2011;21:2198–2202. doi: 10.1016/j.bmcl.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng LF, Kim SS, Matchacheep S, Lei X, Su S, Lin W, Runguphan W, Choe WH, Sakamoto N, Ikeda M, et al. Identification of novel epoxide inhibitors of hepatitis C virus replication using a high-throughput screen. Antimicrobial agents and chemotherapy. 2007;51:3756–3759. doi: 10.1128/AAC.00233-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidel LM, Stollar V. Mutations that confer resistance to mycophenolic acid and ribavirin on Sindbis virus map to the nonstructural protein nsP1. Virology. 1991;181:490–499. doi: 10.1016/0042-6822(91)90881-b. [DOI] [PubMed] [Google Scholar]
- Schwegmann A, Brombacher F. Host-directed drug targeting of factors hijacked by pathogens. Science signaling. 2008;1:re8. doi: 10.1126/scisignal.129re8. [DOI] [PubMed] [Google Scholar]
- Smith DR, McCarthy S, Chrovian A, Olinger G, Stossel A, Geisbert TW, Hensley LE, Connor JH. Inhibition of heat-shock protein 90 reduces Ebola virus replication. Antiviral Res. 2010;87:187–194. doi: 10.1016/j.antiviral.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeter DG, Witkowski JT, Khare GP, Sidwell RW, Bauer RJ, Robins RK, Simon LN. Mechanism of action of 1- -D-ribofuranosyl-1,2,4-triazole-3-carboxamide (Virazole), a new broad-spectrum antiviral agent. Proc Natl Acad Sci U S A. 1973;70:1174–1178. doi: 10.1073/pnas.70.4.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Furuta Y, Fukuda Y, Kuno M, Kamiyama T, Kozaki K, Nomura N, Egawa H, Minami S, Shiraki K. In vitro and in vivo activities of T-705 and oseltamivir against influenza virus. Antiviral chemistry & chemotherapy. 2003;14:235–241. doi: 10.1177/095632020301400502. [DOI] [PubMed] [Google Scholar]
- Tan DS. Diversity-oriented synthesis: exploring the intersections between chemistry and biology. Nat Chem Biol. 2005;1:74–84. doi: 10.1038/nchembio0705-74. [DOI] [PubMed] [Google Scholar]
- Thomas GL, Spandl RJ, Glansdorp FG, Welch M, Bender A, Cockfield J, Lindsay JA, Bryant C, Brown DF, Loiseleur O, et al. Anti-MRSA agent discovery using diversity-oriented synthesis. Angewandte Chemie (International ed. 2008;47:2808–2812. doi: 10.1002/anie.200705415. [DOI] [PubMed] [Google Scholar]
- Towner JS, Paragas J, Dover JE, Gupta M, Goldsmith CS, Huggins JW, Nichol ST. Generation of eGFP expressing recombinant Zaire ebolavirus for analysis of early pathogenesis events and high-throughput antiviral drug screening. Virology. 2005;332:20–27. doi: 10.1016/j.virol.2004.10.048. [DOI] [PubMed] [Google Scholar]
- van der Vries E, Stelma FF, Boucher CA. Emergence of a multidrug-resistant pandemic influenza A (H1N1) virus. N Engl J Med. 2010;363:1381–1382. doi: 10.1056/NEJMc1003749. [DOI] [PubMed] [Google Scholar]
- Wei W, Cai C, Kota S, Takahashi V, Ni F, Strosberg AD, Snyder JK. New small molecule inhibitors of hepatitis C virus. Bioorg Med Chem Lett. 2009;19:6926–6930. doi: 10.1016/j.bmcl.2009.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan SP, Barr JN, Wertz GW. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr Top Microbiol Immunol. 2004;283:61–119. doi: 10.1007/978-3-662-06099-5_3. [DOI] [PubMed] [Google Scholar]
- Whitlow ZW, Connor JH, Lyles DS. Preferential translation of vesicular stomatitis virus mRNAs is conferred by transcription from the viral genome. J Virol. 2006;80:11733–11742. doi: 10.1128/JVI.00971-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf MC, Freiberg AN, Zhang T, Akyol-Ataman Z, Grock A, Hong PW, Li J, Watson NF, Fang AQ, Aguilar HC, et al. A broad-spectrum antiviral targeting entry of enveloped viruses. Proc Natl Acad Sci U S A. 2010;107:3157–3162. doi: 10.1073/pnas.0909587107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Patel NK, McAuliffe JM, Zhu W, Wachter L, McCarthy MP, Suzich JA. Natural Polymorphisms and Resistance-Associated Mutations in the Fusion Protein of Respiratory Syncytial Virus (RSV): Effects on RSV Susceptibility to Palivizumab. Journal of Infectious Diseases. 2012;205:635–638. doi: 10.1093/infdis/jir790. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.