Abstract
Typically, hepatitis B virus (HBV) decays in patients under therapy in a biphasic manner. However, more complex decay profiles of HBV DNA (e.g. flat partial response, triphasic, stepwise) have also been observed in some treated patients with no clear understanding of their origin. We recently introduced the notion of a critical drug efficacy, εc, such that if overall drug efficacy, εtot, is higher than the critical drug efficacy, i.e., εtot > εc, then viral levels will continually decline on therapy, while if εtot < εc then viral loads will initially decline but ultimately stabilize at a new set point, as seen in flat partial responders. Using the idea of critical efficacy and including hepatocyte proliferation in a viral kinetic model we can account for these complex HBV DNA decays. The model predicts that complex profiles such as those exhibiting a plateau or shoulder phase as well as a class of stepwise declines occur only in patients in whom the majority of hepatocytes are infected before therapy. In conclusion, we show via kinetic modeling how a variety of HBV DNA decay profiles can arise in treated patients.
Keywords: mathematical modeling, hepatitis B virus, multiphase viral decay, critical drug efficacy
INTRODUCTION
Infection with hepatitis B virus (HBV) affects hundreds of millions of people worldwide and is the major cause of hepatocellular carcinoma (1). Moreover, 10% of HIV-infected people are co-infected with HBV (~ 4 million people worldwide). This important health burden has led to the development of several approved therapies for chronic HBV infection (1), including interferon-α (IFN) and nucleot(s)ide analogues (NA) that interfere with the reverse transcription step of the HBV lifecycle.
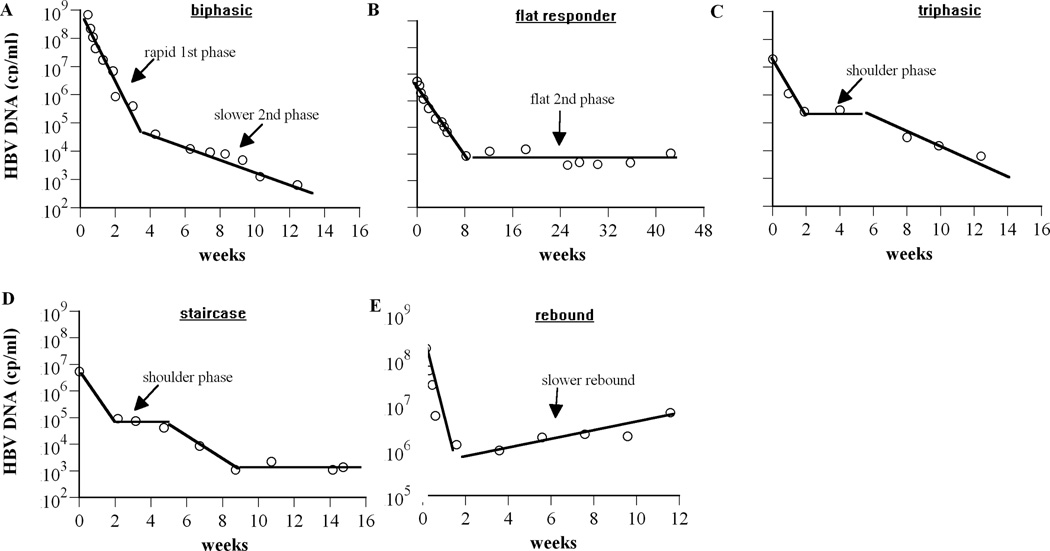
While in most patients HBV DNA levels fall during therapy, the pattern of fall is varied. Often HBV DNA falls in a biphasic manner, with a fast first phase decrease of several logs over a few weeks, followed by a slower but continued decrease over many weeks (Fig. 1A). In others, HBV DNA decay under therapy seems to proceed in triphasic-like (Fig. 1C) or staircase-like (Fig. 1D) fashions (2), with periods of viral decrease interspersed with more or less long periods of stable viral load. However, in many patients primary treatment failure is observed, with less than 1 log decay over the first 6 months of treatment (3); in others, secondary treatment failure occurs, when in spite of an initial viral response, HBV DNA remains flat (Fig. 1B) or increases (Fig. 1E) more than 1 log over nadir in two successive treatment visits over more than a month apart (3).
Figure 1.
Complex HBV DNA decline profiles observed in patients during drug therapy. (A) Biphasic decay (patient 16 from Lewin et al. (2), treated with lamivudine and famciclovir), (B) flat 2nd phase (patient 8472 digitized from Colombatto et al. (19); patient was treated with pegylated intereferon-α2a), (C) triphasic decay (Figure 2B digitized from Tsiang et al. (11); patient was treated with adefovir dipivoxil), (D) staircase decay (patient 6 from Lewin et al. (2), treated with lamivudine and famciclovir), and (E) slow rebound (patient 10 from Lewin et al. (2), treated with lamivudine). Experimental data are represented by open circles. Solid lines were used to emphasize plausible phases of viral decay.
A better understanding of the origins of these varied profiles of HBV DNA decay under therapy could lead to improved management of HBV treatment. While host factors, such as genetic background, drug metabolism, and strength of the immune response, and viral factors, such as genotype, quasi-species diversity, and drug resistance could affect the HBV DNA decay profile, here we show how viral dynamic factors alone can explain many of the observed patterns. To provide this explanation we introduce the concept of a critical drug effectiveness, which has previously been used to explain HIV (4–6) and hepatitis C (7) infection dynamics under therapy. We show how the effectiveness of therapy in a given patient, relative to the patient’s critical drug effectiveness, and the fraction of hepatocytes infected at treatment baseline dynamically influence the pattern of HBV DNA decay under therapy.
METHODS
Model Description
Recently, we extended (7, 8) a model of HCV infection (9) to account for the proliferation of hepatocytes. Along analogous lines, here we generalize a previous model of HBV infection and treatment (2) by including proliferation of uninfected hepatocytes, T, and infected hepatocytes, I. Our previous model for HCV infection (7, 8) does not include the possibility of noncytolytic cure of infected cells, which we include in the HBV model. The models also differ in that HCV treatment involves ribavirin whose effect was modeled by the production of noninfectious virions (7), an effect not needed when modeling HBV treatment (2, 10–12). The equations describing our new model of HBV infection and treatment are
| (1) |
where V represents virus, HBV. The model assumes that uninfected hepatocytes susceptible to HBV infection (target cells), T, are produced at a constant rate s, die at rate, dT, per cell and are infected by virus, V, at constant rate β. Uninfected and infected hepatocytes can proliferate with maximum proliferation rates rT and rI, respectively. However, as the total hepatocyte number, T+I, approaches its natural level, Tmax, we assume proliferation slows down according to a homeostasis process in which only the total number of hepatocytes matter and not whether they are infected or not (13). In order to have a physiologically realistic model (i.e., T before infection less than Tmax) the hepatocyte influx rate s, needs to be less than dTTmax. Here the influx represents both uninfected hepatocytes that become susceptible to infection and de novo generation of hepatocytes from stem cells (14). Due to the burden of supporting HBV replication, we allow infected cells to proliferate slower than uninfected cells, i.e., rI ≤ rT. Infected hepatocytes die at a constant rate δ per cell and are possibly “cured” by noncytolytic processes at a constant rate ρ per cell. As hepatocytes die the total number of hepatocytes will be reduced from Tmax, and proliferation will resume. In the absence of treatment, each productively infected cell releases new infectious virions at rate p. Virus particles are assumed to be cleared at rate c per particle. IFN and NA therapies are assumed to lower p by a factor (1-ε) – where ε is the effectiveness of drug therapy in blocking virion production – and may also lower β by a fraction (1-η) – where η is the effectiveness of therapy in blocking infection. For simplicity, we define in our model an overall drug effectiveness, εtot, where 1 − εtot = (1 − η)(1 − ε). Before treatment, a patient with chronic HBV infection is assumed to be in an infected steady state with εtot=0.
We can find a critical drug efficacy, εc, such that when εtot is higher than εc, viral levels are predicted to continually decline on therapy (4–8). One can calculate εc from our model in a manner analogous to the methods we previously used for analyzing HCV models (7, 8). As shown in the online suppplemental material, we find
| (2) |
where T̅0 is the steady state number of hepatocytes in an uninfected individual (see online supplemental material).
RESULTS
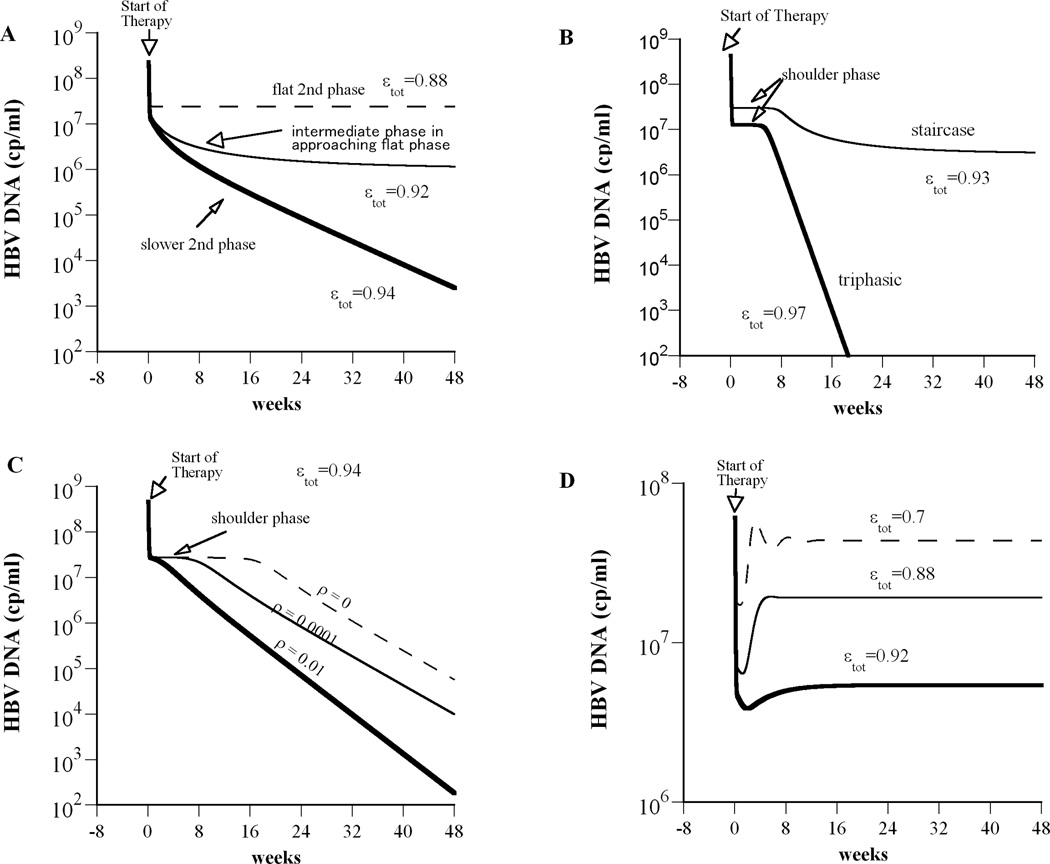
We introduced a model of treatment of HBV infection, Eq. (1), that includes proliferation of uninfected and infected hepatocytes. In the context of this model, we define the concept of a critical drug efficacy, εc (see Methods), such that treatments with efficacy higher than the critical efficacy are predicted to be successful in reducing HBV viral load. Moreover, in the case of successful drug therapy, i.e., εtot > ε c, HBV DNA is predicted to decline in either the typical biphasic manner (Fig. 2A; thick solid line) or in a triphasic manner (Fig. 2B; thick solid line). When the efficacy of treatment is lower than the critical drug efficacy, i.e., εtot < ε c, therapy is predicted to be unsuccessful with HBV DNA initially falling, but then achieving a new steady state, i.e. plateau level. In this case, the predicted patterns of HBV DNA decay include a flat second phase after an initial fast first phase decline (Fig. 2A, dashed line); a slow approach to the new steady-state (Fig. 2A, thin line); a stepwise decline pattern, with an intermediate period of constant viral load (Fig. 2B).
Figure 2.
Model simulations predict complex HBV DNA decline profiles during drug therapy. We solved our model (Eq. 1) numerically using Berkeley-Madonna (version 7.0.2; http://www.berkeleymadonna.com). (A) A flat second phase occurs when εtot<εc (dashed line; here εtot=0.88; εc=0.934). When εtot is close to εc (but still εtot < εc), then after the rapid viral decay phase an intermediate slower phase is predicted that approaches a flat final phase (thin solid line; εtot=0.92). When εtot > εc we obtain a biphasic viral decline (thick line; εtot=0.94). (B) If we decrease the ratio of the target cell to the infected cell replication rate from rT/rI = 8.5 (used in (A)) to rT/rI= 2, a shoulder phase emerges. Staircase and triphasic viral decay profiles are represented by thin and thick solid lines, respectively. For the staircase profile total drug efficacy, εtot=0.93, is lower than the critical drug efficacy, εc=0.934 and therefore after the third phase of viral decay the viral load stabilizes in a steady state until end of treatment. For the triphasic profile εtot> εc (εtot=0.97) and thus viral will continue to decline under therapy. (C) If we increase the rate constant for cure of infected cells, ρ, from 0 (i.e., no cure) to 0.01 day−1, the shoulder phase of the triphasic decline disappears and we obtain a biphasic viral decline. Here εtot=0.94 and rT/rI = 1.25. (D) Viral rebound during treatment occurs when the hepatocyte replication rate is low, rT=0.51 day−1, rT/rI = 2 and εtot< εc (η=0). Except as noted, model parameters used were: Tmax = 1.9×107 cells ml−1, dT = 0.004 day−1, p = 92.3 virions cell−1 day−1, β= 7.1×10−9 ml day−1 virions−1, c = 3.5 day−1, δ= 0.25 day−1, ρ=0 day−1, rT= 3.0 day−1, rT/rI = 8.5, η=0.5 and s= 1 cell ml−1 day−1.
Which pattern is actually observed within a patient depends on the relationship between the critical drug efficacy and the overall treatment efficacy for that individual (Table 1). When the total drug effectiveness is higher than εc, the viral load should decline to below the limit of detection. The model predicts that this decline may be biphasic (Fig. 2A) or triphasic (Fig. 2B), depending on whether or not a shoulder phase exists. During the shoulder phase, not only the virus but also the number of infected cells must remain constant. Under potent therapy, few new infections occur (15), and thus proliferation of infected cells is the major source of new infected cells. Thus, proliferation helps compensate for the loss of infected cells, a feature that was not possible in earlier models (2, 11, 12). This proliferation can keep the number of infected cells constant for some period of time, but because uninfected cells are assumed to proliferate faster than infected ones their relative number increases, and due to competition for space and other resources this ultimately slows down the proliferation of infected cells to the point that infected cell numbers decline and the shoulder phase ends. We recently showed (7) that the shoulder phase ends approximately when the number of uninfected hepatocytes reaches the same order of magnitude as the infected ones. This demarks the transition from the shoulder phase to the third phase of viral decline. When the ratio of uninfected cells to infected cells at the start of therapy is close to or greater than one, the shoulder phase does not exist. Thus, our model predicts that triphasic responses will only be seen in patients in which a majority of hepatocytes are infected before therapy is begun.
Table 1.
HBV DNA kinetic patterns under therapy
| Drug effectiveness, εtot, and patient’s critical drug efficacy, εc |
Ratio of HBV- infected cells to uninfected cells at the start of therapy [I:T] |
Ratio of (T+I) to Tmax at start of therapy |
HBV DNA decay pattern |
Example |
|---|---|---|---|---|
| εtot<εc | I:T<1 | (T+I):Tmax~1 | flat 2nd phase | Fig. 2A; Fig. 3C |
| I:T>1 | (T+I):Tmax~1 | complex decay in approaching flat phase | Fig. 2B; Lewin et al. (2) | |
| any I:T | (T+I):Tmax<<1 | rebound | Fig. 2D; Fig. 3D | |
| εtot~ εc(εtot< εc) | I:T<1 | 0<(T+I):Tmax≤1 | intermediate slope phase that approaches a final flat phase | Fig. 2A; Heathcote et al. (30) |
| εtot>εc | I:T<1 | 0<(T+I):Tmax≤1 | biphasic | Fig. 2A; Fig. 3A |
| I:T>1 | 0<(T+I):Tmax≤1 | triphasic | Fig. 2B; Fig. 3B |
Interestingly, the model predicts that the length of the shoulder phase is dependent on how fast the number of uninfected hepatocytes, T, reaches the same order of magnitude as the infected ones, I, assuming that T<I at baseline. An illustration of this notion is shown in Fig. 2 in which the time needed for T to reach I is governed by the ratio of the target cell to the infected cell replication rate, rT/rI (Fig. 2B). It may be possible that some patients exhibiting biphasic decline also have T<I at baseline but a shoulder phase will not be observed if fast replication of uninfected hepatocytes causes T to increase to approximately I during the first phase of viral decline.
Clearance of HBV infected hepatocytes has been shown to occur through non-cytolytic mechanisms in HBV transgenic mice (16) and strongly suggested to occur during acute HBV infection in chimpanzees (17) and possibly humans. Analysis of our model shows that “cure” of infected cells, ρ, can shorten the shoulder phase of the triphasic decline or even abolish it completely, depending on its magnitude (Fig. 2C).
For reasons that are not fully understood, therapy is not equally effective in all patients. For patients in which the overall efficacy of drug therapy is lower than εc, the viral load will decline to a viral plateau lower than baseline, and if the patient is compliant with therapy the virus will remain at the plateau until the end of therapy, until therapy is changed (18), or until drug resistance develops (3). Again, the specific profile of decay depends on the ratio of uninfected cells to infected cells at baseline as well as viral kinetic parameters. According to the model, a flat second phase can be obtained with large death rates of infected cells, δ, a feature that was not possible in earlier models in which target cells were held constant (11, 12).
Lewin et al. (2) observed that some HBV treated patients exhibited a staircase or stepwise decline in HBV DNA under treatment. Our model can explain a class of stepwise decays in which there is a flat second phase followed by a second drop in HBV DNA and a final flat phase (Fig. 2B). Note that in our model all parameters are held constant and thus the stepwise decreases in viral load are not caused by parameter changes. The “two-step declines” that our model generates are a special case of triphasic decays, that are characterized by having a shoulder phase, but in which εtot < εc so that therapy is not potent enough to continually eliminate virus and hence leads to a final state in which virus is still present. However, simulating the model with over 275,000 choices of sets of parameters spanning the biologically feasible ranges of all the parameters showed that the model could only generate modest sized second drops, i.e., less than 1 log (Fig. 2B). For patients in whom this second drop is very large (e.g., Patients 6 and 11 in Fig. 5 from (2)), other effects such as changes in the immune response may be playing a role.
In some treated patients a viral rebound is observed during therapy (15). Several explanations for this viral rebound have been suggested, such as the development of antiviral drug resistance or/and poor adherence to therapy (3). Here we offer another explanation. Our model shows that viral rebound can occur during suboptimal therapy, i.e., when εtot < εc, in the absence of resistance if the total number of hepatocytes susceptible to HBV infection increases during therapy (Fig. 2D; Table 1). Moreover, depending of the total drug efficacy, HBV DNA levels may rebound to baseline levels or rebound and stabilize at a lower therapy-induced set point (Fig. 2D).
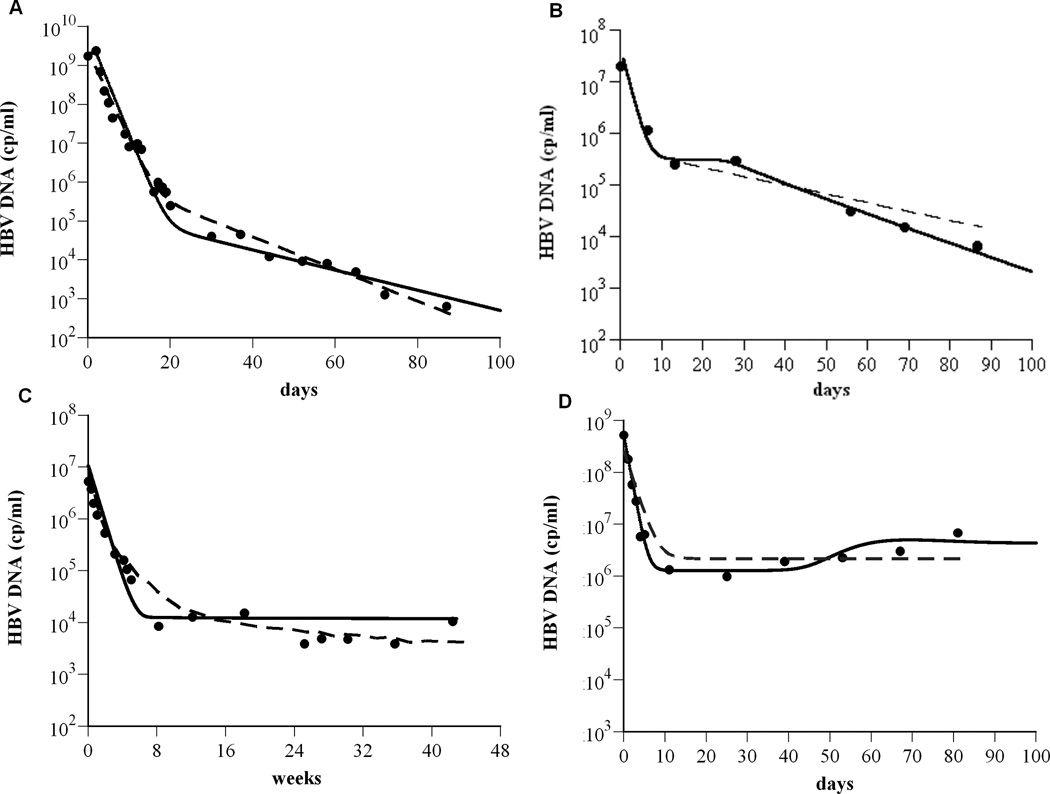
We compared our model with experimental HBV DNA data. By varying patient specific parameters the kinetics of HBV decline predicted by our model can be shown to quantitatively agree with: (i) a biphasic decline (Fig. 3A) observed in a HBeAg-positive patient (Patient 16 from (2)) treated with lamivudine and famciclovir), (ii) a triphasic decline (Fig. 2B) observed in an HBeAg-positive patient (Patient F in Fig. 5 from (11)) treated with adefovir dipivoxil (ADV), (iii) a flat second phase decline (Fig. 3C) observed in HBeAg-negative patients (e.g., Patient 8472 from (19)) treated with pegylated intereferon-α2a, and (iv) a slow viral rebound (Fig. 3D) observed in an HBeAg-positive patient (Patient 10 from (2)) treated with lamivudine.
Figure 3.
The model is consistent with experimental data. To show that the model (Eq. 1) agrees with experimental data (as shown in Fig. 1), exhibiting biphasic (A), triphasic (B), flat 2nd phase (C) viral decays, and slow viral rebound (D), the solution for V(t) from Eq. (1) was compared (solid lines) to the HBV DNA (●) using Berkeley-Madonna (version 7.0.2; http://www.berkeleymadonna.com/). For the theoretical curves, we fixed Tmax = 1.9×107 cells ml−1; ρ=0 day−1; s = 1 cell day−1 ml−1. The other parameter values used to generate the theoretical curves in panels (A) to (D), respectively, are: dT = 0.2×10−3, 2.7×10−3, 5.3×10−3, and 9.0×10−4 day−1; δ= 0.07, 0.22, 0.25, and 0.06 day−1; β= 1×10−10, 2.5×10−7, 1.9×10−6, and 6.6×10−8 ml virions−1 day−1; rT = 3.0, 0.9, 0.8, and 0.5 day−1; rT : rI = 5.0, 3.3, 2.9, and 7.2; c = 0.6, 0.7, 0.18, and 1.0 day−1; p = 87, 5, 1.4, and 164 virions cell−1 day−1; ε= 0.999934, 0.991, 0.9988, and 0.997; η=0.5, 0.5, 0.2, and 0.5; ε c = 0.75, 0.994, 0.99908, and 0.9987. Previous model predictions (dashed lines) for (A) and (D) were calculated from (2) and for (B) and (C) were digitized from (11) and (19), respectively.
Discussion
The field of viral dynamic modeling is predicated on the assumption that the kinetic pattern of viral decay under therapy can be informative. In the case of HIV, viral dynamic analyses revealed that very rapid viral replication and clearance were occurring despite the relatively constant level of viremia observed in untreated patients. Similar analyses then were performed for HCV and HBV (see review (20)). Here we have extended viral kinetic analyses of HBV infection in order to shed light on the origin of some complex HBV DNA decay patterns. In many patients on treatment, the now classic biphasic decay of HBV DNA is observed (2, 10, 11, 19, 21). However, Lewin et al. (2) pointed out that in some patients more complex decay profiles occur. Here we have shown, as in the case of HCV infection (7), that patients can exhibit a triphasic decay or a fast first phase followed by a gradual approach to a steady state HBV level – patterns that are not a classic second phase. We also showed that a class of stepwise declines in which there is < 1 log drop in HBV DNA after a flat second phase (Fig. 2B) as well as viral rebounds (Fig. 2D and Fig. 3D) could be also be explained by this new model.
Including hepatocyte proliferation (of both infected and uninfected cells) in an HBV kinetic model and introducing the notion of critical drug efficacy allows us to explain not only the typical biphasic HBV decay but also more complex viral kinetic patterns that have been observed under therapy. Four main features, according to the model, define the HBV DNA decay patterns under therapy: (i) the ratio of HBV-infected cells to uninfected cells at the start of therapy, (ii) the ratio at the start of therapy of the total number of cells (T+I) to the total number of hepatocytes in a healthy liver, Tmax, (iii) the overall drug effectiveness, εtot, and (iv) the patient’s critical drug efficacy, εc. The relationship between these model features and outcome of HBV DNA kinetic patterns are summarized in Table 1. Unfortunately, the model presented here has additional parameters that characterize uninfected and infected cell kinetics, which cannot be estimated from HBV DNA kinetics. Thus, a future challenge will be to collect additional data from liver biopsies about hepatocyte population kinetics both before and during treatment and correlate it with HBV DNA decay profiles under treatment.
Viral rebound can occur, according to the model, when εtot < εc and T+I<<Tmax at the start of therapy. If Tmax represents total hepatocyte number in an uninfected healthy liver then T+I<<Tmax may not be realistic as it indicates a drastic decrease in hepatocyte number. However, if one assumes that our model is only concerned with a subset of hepatocytes that are susceptible to infection due to their state of differentiation or other host factors, then Tmax would represent this number of susceptible hepatocytes in a healthy liver and T and I the subsets of these cells that are uninfected and infected during chronic infection. In this case, having I+T<<Tmax (pretreatment) may be reasonable as this does not necessarily imply a huge decrease in total hepatocyte number. Experimental results during acute HBV infection (22, 23) or woodchuck hepatitis (24), show that the majority of hepatocytes are HBV infected before the specific immune response against HBV is induced. However, once a cell-mediated immune response is present, HBV DNA is cleared from serum or reaches a viral set point. Recently, using a mathematical model of acute HBV infection in humans, we predicted the appearance of a population of cells refractory to infection or in which viral production is inhibited (25). If, in patients that go on to chronicity, this “antiviral state” is maintained, Tmax may represent only a small fraction of susceptible cells to HBV infection. This possibility is suggested by studies involving chronically infected patients (26–28) and animals (29) showing that only a small fraction of hepatocytes appear to be infected.
Drug resistance develops over long periods (months to years) of treatment with nucleos(t)ide analogues (NAs) against HBV (1). Virological breakthrough, during this period of time, is defined as a ≥1 log10 cp/ml increase in HBV DNA level from nadir in two consecutive samples 1 month apart. However, as shown here, in cases of viral rebound within days or weeks post initiation of therapy and especially under IFN monotherapy or in combination with NAs, in which drug resistance is less likely to develop, an increase of susceptible cells may lead to an increase in HBV replication. In these cases, careful analysis for the nature of viral breakthrough (i.e., drug resistance vs. an increase of susceptible cells) needs to be preformed in order to distinguish between the two processes.
While our new model can successfully explain a large number of observed HBV decay patterns, there are patterns that the model does not explain such as the step-wise viral declines seen with lamivudine alone or in combination with famciclovir (patients 6 and 11 from Lewin et al. (2), respectively) in which there is >1 log decline in viral load from the shoulder phase to the final steady-state level that lasts until end of therapy. The model also suggests that from a viral dynamic viewpoint when εtot > εc, HBV DNA should ultimately be cleared. Thus, the model does not properly account for the low rate of treatment success (<8%); i.e., loss of HBsAg, seroconversion to anti-HBs and undetectable level of HBV DNA six month after stopping therapy (1). However, the model does not consider covalently closed circular HBV DNA, HbsAg, nor a dynamic immune response to HBV. Future models will need to address these issues.
In summary, we show that by including proliferation of both uninfected and infected cells, a viral kinetic model can account for complex HBV DNA decays observed in treated patients. In addition, we introduced the notion of critical drug efficacy that implies that each patient has a particular viral-host state in which a certain drug effectiveness needs to be imposed in order to reduce HBV viral load below detection. In addition, we predict that viral breakthrough early on treatment can be explained by an increase of susceptible cells without drug resistance development and/or poor treatment compliance. Further studies of viral and host dynamics, based on these results, may be able to better identify the factors giving rise to successful therapy.
Supplementary Material
Acknowledgments
Financial support: This research was performed under the auspices of the U.S. Department of Energy under contract DE-AC52-06NA25396 and supported by National Institutes of Health (NIH) grant RR06555 (to A.S.P). R.M.R and H.D were supported by grant P20-RR18754 from the National Center for Research Resources (NCRR), a component of the NIH. H.D is supported by the University of Illinois Gastrointestinal and Liver Disease (UIC GILD) Association. This study is solely the responsibility of the authors and does not necessarily represent the official views of NCRR or NIH.
Abbreviations
- HBV
Hepatitis B virus
References
- 1.Hoofnagle JH, Doo E, Liang TJ, Fleischer R, Lok AS. Management of hepatitis B: summary of a clinical research workshop. Hepatology. 2007;45:1056–1075. doi: 10.1002/hep.21627. [DOI] [PubMed] [Google Scholar]
- 2.Lewin SR, Ribeiro RM, Walters T, Lau GK, Bowden S, Locarnini S, Perelson AS. Analysis of hepatitis B viral load decline under potent therapy: complex decay profiles observed. Hepatology. 2001;34:1012–1020. doi: 10.1053/jhep.2001.28509. [DOI] [PubMed] [Google Scholar]
- 3.Lok AS, Zoulim F, Locarnini S, Bartholomeusz A, Ghany MG, Pawlotsky JM, Liaw YF, et al. Antiviral drug-resistant HBV: standardization of nomenclature and assays and recommendations for management. Hepatology. 2007;46:254–265. doi: 10.1002/hep.21698. [DOI] [PubMed] [Google Scholar]
- 4.Wein LM, D'Amato RM, Perelson AS. Mathematical analysis of antiretroviral therapy aimed at HIV-1 eradication or maintenance of low viral loads. J Theor Biol. 1998;192:81–98. doi: 10.1006/jtbi.1997.0622. [DOI] [PubMed] [Google Scholar]
- 5.Callaway DS, Perelson AS. HIV-1 infection and low steady state viral loads. Bull Math Biol. 2002;64:29–64. doi: 10.1006/bulm.2001.0266. [DOI] [PubMed] [Google Scholar]
- 6.Huang Y, Rosenkranz SL, Wu H. Modeling HIV dynamics and antiviral response with consideration of time-varying drug exposures, adherence and phenotypic sensitivity. Math Biosci. 2003;184:165–186. doi: 10.1016/s0025-5564(03)00058-0. [DOI] [PubMed] [Google Scholar]
- 7.Dahari H, Ribeiro RM, Perelson AS. Triphasic decline of hepatitis C virus RNA during antiviral therapy. Hepatology. 2007;46:16–21. doi: 10.1002/hep.21657. [DOI] [PubMed] [Google Scholar]
- 8.Dahari H, Lo A, Ribeiro RM, Perelson AS. Modeling hepatitis C virus dynamics: Liver regeneration and critical drug efficacy. J Theor Biol. 2007;247:371–381. doi: 10.1016/j.jtbi.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neumann AU, Lam NP, Dahari H, Gretch DR, Wiley TE, Layden TJ, Perelson AS. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998;282:103–107. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 10.Lau GK, Tsiang M, Hou J, Yuen S, Carman WF, Zhang L, Gibbs CS, et al. Combination therapy with lamivudine and famciclovir for chronic hepatitis B-infected Chinese patients: a viral dynamics study. Hepatology. 2000;32:394–399. doi: 10.1053/jhep.2000.9143. [DOI] [PubMed] [Google Scholar]
- 11.Tsiang M, Rooney JF, Toole JJ, Gibbs CS. Biphasic clearance kinetics of hepatitis B virus from patients during adefovir dipivoxil therapy. Hepatology. 1999;29:1863–1869. doi: 10.1002/hep.510290626. [DOI] [PubMed] [Google Scholar]
- 12.Nowak MA, Bonhoeffer S, Hill AM, Boehme R, Thomas HC, McDade H. Viral dynamics in hepatitis B virus infection. Proc.Natl.Acad.Sci.U.S.A. 1996;93:4398–4402. doi: 10.1073/pnas.93.9.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahari H, Major M, Zhang X, Mihalik K, Rice CM, Perelson AS, Feinstone SM, et al. Mathematical modeling of primary hepatitis C infection: Noncytolytic clearance and early blockage of virion production. Gastroenterology. 2005;128:1056–1066. doi: 10.1053/j.gastro.2005.01.049. [DOI] [PubMed] [Google Scholar]
- 14.Theise ND, Nimmakayalu M, Gardner R, Illei PB, Morgan G, Teperman L, Henegariu O, et al. Liver from bone marrow in humans. Hepatology. 2000;32:11–16. doi: 10.1053/jhep.2000.9124. [DOI] [PubMed] [Google Scholar]
- 15.Neumann AU. Hepatitis B viral kinetics: a dynamic puzzle still to be resolved. Hepatology. 2005;42:249–254. doi: 10.1002/hep.20831. [DOI] [PubMed] [Google Scholar]
- 16.Guidotti LG, Chisari FV. Noncytolytic control of viral infections by the innate and adaptive immune response. Annu.Rev.Immunol. 2001;19:65–91. doi: 10.1146/annurev.immunol.19.1.65. 65–91. [DOI] [PubMed] [Google Scholar]
- 17.Guidotti LG, Rochford R, Chung J, Shapiro M, Purcell R, Chisari FV. Viral clearance without destruction of infected cells during acute HBV infection. Science. 1999;284:825–829. doi: 10.1126/science.284.5415.825. [DOI] [PubMed] [Google Scholar]
- 18.Hezode C, Chevaliez S, Bouvier-Alias M, Roudot-Thoraval F, Brillet R, Zafrani ES, Dhumeaux D, et al. Efficacy and safety of adefovir dipivoxil 20 mg daily in HBeAg-positive patients with lamivudine-resistant hepatitis B virus and a suboptimal virological response to adefovir dipivoxil 10 mg daily. J Hepatol. 2007;46:791–796. doi: 10.1016/j.jhep.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 19.Colombatto P, Civitano L, Bizzarri R, Oliveri F, Choudhury S, Gieschke R, Bonino F, et al. A multiphase model of the dynamics of HBV infection in HBeAg-negative patients during pegylated interferon-alpha2a, lamivudine and combination therapy. Antivir Ther. 2006;11:197–212. [PubMed] [Google Scholar]
- 20.Perelson AS. Modelling viral and immune system dynamics. Nat Rev Immunol. 2002;2:28–36. doi: 10.1038/nri700. [DOI] [PubMed] [Google Scholar]
- 21.Zeuzem S, de Man RA, Honkoop P, Roth WK, Schalm SW, Schmidt JM. Dynamics of hepatitis B virus infection in vivo. J Hepatol. 1997;27:431–436. doi: 10.1016/s0168-8278(97)80345-5. [DOI] [PubMed] [Google Scholar]
- 22.Thimme R, Wieland S, Steiger C, Ghrayeb J, Reimann KA, Purcell RH, Chisari FV. CD8(+) T cells mediate viral clearance and disease pathogenesis during acute hepatitis B virus infection. J.Virol. 2003;77:68–76. doi: 10.1128/JVI.77.1.68-76.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertoletti A, Ferrari C. Kinetics of the immune response during HBV and HCV infection. Hepatology. 2003;38:4–13. doi: 10.1053/jhep.2003.50310. [DOI] [PubMed] [Google Scholar]
- 24.Summers J, Jilbert AR, Yang W, Aldrich CE, Saputelli J, Litwin S, Toll E, et al. Hepatocyte turnover during resolution of a transient hepadnaviral infection. Proc.Natl.Acad.Sci U.S.A. 2003;100:11652–11659. doi: 10.1073/pnas.1635109100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ciupe SM, Ribeiro RM, Nelson PW, Dusheiko G, Perelson AS. The role of cells refractory to productive infection in acute hepatitis B viral dynamics. Proc Natl Acad Sci U S A. 2007;104:5050–5055. doi: 10.1073/pnas.0603626104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gowans EJ, Burrell CJ, Jilbert AR, Marmion BP. Detection of hepatitis B virus DNA sequences in infected hepatocytes by in situ cytohybridisation. J Med Virol. 1981;8:67–78. doi: 10.1002/jmv.1890080106. [DOI] [PubMed] [Google Scholar]
- 27.Govindarajan S, Conrad A, Lim B, Valinluck B, Kim AM, Schmid P. Study of preneoplastic changes of liver cells by immunohistochemical and molecular hybridization techniques. Arch Pathol Lab Med. 1990;114:1042–1045. [PubMed] [Google Scholar]
- 28.Bannasch P, Haertel T, Su Q. Significance of hepatic preneoplasia in risk identification and early detection of neoplasia. Toxicol Pathol. 2003;31:134–139. doi: 10.1080/01926230390173923. [DOI] [PubMed] [Google Scholar]
- 29.Mason WS, Jilbert AR, Summers J. Clonal expansion of hepatocytes during chronic woodchuck hepatitis virus infection. Proc Natl Acad Sci U S A. 2005;102:1139–1144. doi: 10.1073/pnas.0409332102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heathcote E, Gane E, DeMan R, Lee S, Flisiak R, Manns MP, Tchernev K, et al. A Randomized, Double - Blind, Comparison of TenofovirDF ( TDF ) versus Adefovir Dipivoxil ( ADV ) for the Treatment of HBeAg Positive Chronic Hepatitis B ( CHB ):Study GS - US - 174 - 0103. Hepatology. 2007;46:861A. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.