Summary
Most pandemics—eg, HIV/AIDS, severe acute respiratory syndrome, pandemic influenza—originate in animals, are caused by viruses, and are driven to emerge by ecological, behavioural, or socioeconomic changes. Despite their substantial effects on global public health and growing understanding of the process by which they emerge, no pandemic has been predicted before infecting human beings. We review what is known about the pathogens that emerge, the hosts that they originate in, and the factors that drive their emergence. We discuss challenges to their control and new efforts to predict pandemics, target surveillance to the most crucial interfaces, and identify prevention strategies. New mathematical modelling, diagnostic, communications, and informatics technologies can identify and report hitherto unknown microbes in other species, and thus new risk assessment approaches are needed to identify microbes most likely to cause human disease. We lay out a series of research and surveillance opportunities and goals that could help to overcome these challenges and move the global pandemic strategy from response to pre-emption.
This is the third in a Series of three papers about zoonoses
Introduction
The emergence of novel infectious diseases such as HIV/AIDS, severe acute respiratory syndrome (SARS), and pandemic influenza has shown the vulnerability of human beings to new zoonotic health threats. The mortality and morbidity associated with HIV/AIDS have devastated communities in some countries and led to global changes in public health. The rapid pandemic spread of SARS coronavirus in 2003 and a new triple-reassortant H1N1 influenza in 2009 both resulted in substantial economic loss as the pathogens exploited—and in some instances closed down—global travel and trade networks.1 These vulnerabilities emphasise the need for a systematic, pre-emptive approach that aims to prevent the spread, or even the initial emergence, of pandemics.
The emergence of novel pandemic agents often seems to be inherently unpredictable.2 Indeed, no pathogens have been predicted before their first appearance. However, patterns in the origins and spread of new pathogens can be noted and are an intrinsic, albeit ad-hoc, part of surveillance strategy. For example, more than 60% of the roughly 400 emerging infectious diseases that have been identified since 1940 are zoonotic,3 and these pathogens are the focus of particular public health interest.4, 5 Similarly, specific geographical regions or interfaces between people, wildlife, livestock, and the environment have been identified as the origins of recent emerging infectious diseases, and thus are targets for intense surveillance.3, 5, 6, 7 Analysis of previous emergence events has led to a better understanding of the causes (so-called drivers) of emergence.6, 8 These advances, coupled with a better understanding of the dynamics of pathogen transmission, ecology, and evolution as they emerge and spread, promise the possibility to predict pandemics. Here we review these findings and the most promising strategies to improve anticipation, prediction, and pre-emption of the next pandemic zoonosis at the source.
Key messages.
-
•
Most recent pandemics, such as HIV/AIDS, severe acute respiratory syndrome, and pandemic influenza, are caused by zoonotic pathogens (ie, pathogens harboured by non-human animals), are viral diseases, and originated in wildlife
-
•
Such infections are usually driven to emerge by ecological, behavioural, or socioeconomic changes
-
•
Technological advances in mathematical modelling, diagnostics, communication, and informatics enable targeted global surveillance of emerging and previously unknown infections in both human beings and other species
-
•
New risk-assessment approaches show promise for the use of these capabilities to predict and pre-empt potential pandemics at their source (eg, in wildlife or other animals), and need to be further developed
Origins and dynamics of pandemic zoonoses
When the origins of emerging infectious diseases are traced back to first emergence in the human population, some distinctive patterns are revealed that could be used in disease control.3, 6 First, the frequency with which new pathogens emerge is increasing, even when the increased surveillance globally is taken into account,3 suggesting that efforts to coordinate the global strategy to fight pandemics are timely and of growing importance.9, 10 Second, the emergence of all major groups of emerging infectious diseases correlates strongly with human population density, supporting the hypothesis that disease emergence is driven by largely anthropogenic changes, such as the expansion of agriculture, travel routes, and trade, and changes in land use.11 Finally, the emergence of zoonotic pathogens of wildlife origin (which have dominated the pandemics of the past 100 years) correlates strongly with both human density and the global distribution of wildlife biodiversity.3, 8 Spatially explicit models can be used to identify the regions most likely to produce the next emerging zoonoses (so-called hotspots of emerging infectious disease).3 These hotspots are regions where human activities take place against a background of high wildlife biodiversity, with concomitant microbial biodiversity (figure 1 ). Targeting of surveillance to such regions provides a rationale for better allocation of global resources to prevent emerging infectious disease or rapidly deal with outbreaks.3
Figure 1.
Global hotspots for emerging infectious diseases that originate in wildlife
A database of all known emerging infectious diseases3 since 1940 was used to identify the most likely origins of each separate emergence event. Presence or absence of infections emerging from wildlife was analysed with logistic regression against a series of known drivers, including human population density, change in human population density, and wildlife diversity (mammalian species richness), gridded at 1 km2 resolution. The global distribution of model outputs gives a measure of the likelihood of a region to generate a new zoonotic emerging infectious disease that originates in wildlife. Because previous pandemics have mainly originated in wildlife, these maps identify hotspots where the next pandemic is most likely to originate.
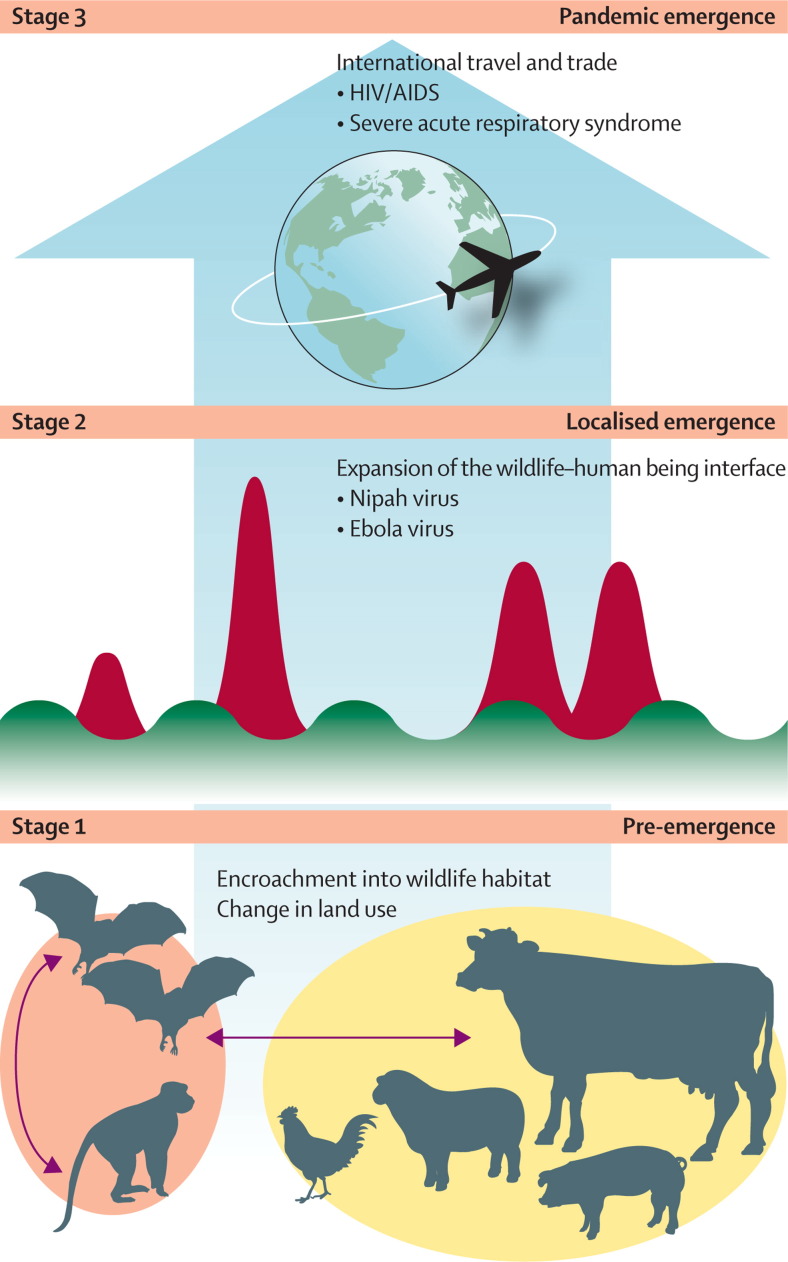
The process through which pandemic zoonoses emerge can be analysed to identify the crucial control points and specific research challenges (panel 1 , figure 2 ). The model we adopt here (developed by Daszak) to assess pandemic potential has three stages—no human infection (stage 1), localised human infection (spillover; stage 2), and widespread transmission and global dissemination (stage 3). The frequency at which stages 1 and 2 occur is unknown but probably high. That human populations are continually exposed to a wide variety of non-human-animal pathogens, many of which infect human beings, seems reasonable to assume. To assess the role of pathogen biology in emergence, investigators need to know how many pathogens people are exposed to and how many successfully cross the species barrier. A complete inventory of spillovers is not available, but researchers have a good working knowledge of pathogens of domestic livestock and pets (species in frequent contact with human beings). Almost 50% of the roughly 1000 species of pathogens that are noted in livestock and pets are zoonotic,17 implying that any barriers between these hosts and human beings are routinely breached by many different pathogens. More than 50% of the recognised pathogens of human beings can infect other vertebrate hosts.18 Many non-human pathogens can infect several hosts, and clear examples of viruses transferring between different animal hosts to cause outbreaks in other species have been reported.17, 19 Pathogens can transfer from human beings to animals and between different animal species before being transferred back to people, allowing remixing and evolution with spill-back and potentially enhanced pathogenicity (eg, influenza).20, 21
Panel 1. Stages in disease emergence.
Several researchers have separated the process of emergence into distinct steps or stages. Morse originally divided the process into two steps, introduction (into a new species, such as human beings) and establishment/dissemination.6, 12 Wolfe and colleagues13 subsequently developed a model with five stages—a so-called pathogen pyramid—to represent successively greater pathogen adaptation to successful human infection and transmission, from inability to infect human beings (stage 1) to exclusively human disease (stage 5).
Figure 2 shows an alternative depiction of the stages of emergence, which emphasises the dynamics of infection rather than pathogen properties. The frequency of these events probably diminishes from stage 1 to stage 2, and the final stage, true pandemic emergence (ie, intercontinental spread of a pathogen in human beings), is far rarer than the initial localised emergence events.
Stage 1 (pre-emergence): the putative pandemic pathogen is still in its natural reservoir. Ecological, social, or socioeconomic changes (eg, change in land use) alter the dynamics of pathogen transmission within the host or between hosts and allow the pathogen to expand within its host population, spread to a new region, or be transmitted to another non-human host population or species. Each of these changes increases the likelihood of the pathogen making contact with and spilling over into human beings (and thus progressing to stage 2).1, 6, 7 The drivers that cause stage 1 emergence tend to be large-scale environmental, agricultural, or demographic shifts—eg, moving of livestock to a region for the first time, or transportation of wildlife from a region for food. Such events happened before the emergence of Nipah virus in Malaysia in 1997, after intensively managed pig farms and fruit orchards were built in a region frequented by fruit bats (the natural reservoir). These bats began to feed on fruit trees around pigsties, enabling viral transmission to pigs and stage 1 emergence.14 Similarly, change in land use in Africa has brought livestock, people, and wildlife into the same habitat, leading to multidirectional pathogen transmission between livestock and non-human primates (stage 1), and some spillover into people (stage 2).15
Stage 2 (localised emergence): initial spillover of a wildlife or livestock pathogen to people. Causes range from handling of butchered wildlife to exposure to fomites in wildlife markets or livestock farms, or in the wild. Outcomes vary widely, from small clusters of human cases (eg, Menangle virus)16 to large outbreaks, some with limited person-to-person transmission (eg, Ebola virus) and some without (eg, Hendra virus).
Stage 3 (full pandemic emergence): sustained person-to-person transmission and large-scale spread, often aided by global air travel (eg, HIV/AIDS, severe acute respiratory syndrome) or the international movement of reservoir hosts or vectors through trade (eg, West Nile virus). Stage 3 pandemics are rare because even pathogens capable of some person-to-person transmission might not be able to maintain long enough chains of transmission to spread (eg, Nipah virus in Bangladesh).
Figure 2.
Emergence of pandemic zoonotic disease
Stage 1 is a pre-emergence state, in which naturally occurring microbes are transmitted between their animal reservoirs. Disturbances to the ecology of these populations (eg, due to changes in land use) change the dynamics of microbial transmission and can lead to a heightened risk of pathogen spillover to other non-human wildlife or livestock hosts (but not people). Stage 2 is localised emergence, either through self-limiting spillover events (green peaks and troughs, representing the rise and fall in numbers of infected people with time) or large- scale spillover (red peaks, representing spikes in the number of infected people with time), that leads to person-to-person transmission for a few pathogen generations. In stage 3, some spillover events might lead to indefinitely sustained person-to-person outbreaks, international or global spread, and the emergence of a true pandemic. The size, spread, and potential effect of events increase from stage 1 to stage 3, but the frequency falls so that full stage 3 pandemics are quite rare. By dissecting this process and analysing the interactions of the underlying drivers with the risk of spillover and spread, development of a more structured approach to pandemic prevention is possible. The ultimate goal of successful pandemic prevention is to move the control point to stage 1.
Human pathogens from all taxa contain zoonotic species. Roughly 80% of viruses, 50% of bacteria, 40% of fungi, 70% of protozoa, and 95% of helminths that infect human beings are zoonotic.18 Most of the identified reservoirs are mammalian (roughly 80%) or, to a lesser extent, avian,18, 22 although people share some pathogens with invertebrates, which act as vectors23 or intermediate hosts. Identification of the key taxonomic groups that are sources for the emergence of zoonotic disease could help to improve targeting of surveillance and interventions. Ungulates are the mammalian taxa with which human beings share the most pathogens17—perhaps not surprising because, as major food sources, these animals are often in close proximity to people, and their diseases have been studied extensively. Rodents, carnivores, and primates are also well represented. Although pathogens deemed emerging or re-emerging are disproportionately likely to be zoonotic,18 their reservoirs are much the same as those of non-emerging zoonoses.22
The situation, however, is substantially different for pathogens that have reached stage 3. A disproportionate number of these pathogens are viruses, suggesting that viruses have the potential to evolve more rapidly than do other kinds of pathogen. However, this finding could be due to ascertainment bias. Less complete knowledge is available about the diversity of viruses than that of other pathogens, and researchers might still be missing many at stages 1 and 2.24 Although a lot of stage 3 pathogens that infect only people don't strictly have reservoirs (eg, HIV-1), many are thought to have had zoonotic origins, including in non-human primates, which contrasts strongly with the minor contribution of primates as reservoirs for zoonoses in general. If risk is a function of contact frequency and probability of successful adaptation to human beings, pathogens acquired from other primates might already be better adapted to successful transmission than those from other mammals (exposure from rodents, bats,25 and other common vertebrates are the most frequent).
Many pandemic zoonoses (ie, those that reach stage 3), such as HIV/AIDS,26 have achieved sustained person-to-person transmission without the need for a non-human reservoir. For example, SARS, which originated from the SARS-like coronaviruses of bats, emerged in China in 2003 and was due to hunting and trading of bats for food.27 In the wildlife markets of southern China these bat viruses seemed to become stage 1 pathogens, which spilled over to civets before being transmitted to people and achieving stage 2.28 SARS coronavirus then underwent repeated cycles of transmission in people, and spread nationally and then globally (ie, reached stage 3), including 251 cases as far away as Toronto. Non-human hosts can have a role in the maintenance and transmission of some stage 3 pathogens (eg, influenza A virus).20, 21 Of the 400 or so known emerging pathogens, roughly 100 occur only as human pathogens—ie, such specialisation is rare.29 Some of the seemingly specialist human pathogens are thought to have had zoonotic origins much further back in evolutionary time (eg, Plasmodium falciparum).30 This pattern could be the norm, but the origins of most specialist human pathogens are unknown.13 Much more is known about the dynamics of pandemic pathogens once they can be transmitted among people. Transmission rates are often higher in dense than in sparse populations, and spread is often greatly enhanced by air travel or human migration. The mathematics of these spreading events is well known, and a sophisticated array of computational models have been used to back-predict such events accurately—eg, the first case clusters of SARS31 and the subsequent global spread, including the country-by-country distribution of human cases.32 Models based on air travel, realistic human contact networks, and individual countries' capacities to deal with outbreaks can now produce reasonably accurate predictions of future pandemic spread (particularly for influenza).33, 34, 35 However, less is known about the factors that enable pathogens to move from stage 2 to stage 3 and ultimately achieve pandemic status. During this process, stuttering chains of novel human transmission (as with human monkeypox in Africa, for example36, 37) can become self-sustaining (which monkeypox has still not achieved in people36). The distribution and behaviour of human beings, reservoir hosts, and vectors are crucial for emergence, and human demographics and behaviour often establish capacity for person-to-person transmission.
Differential host physiology, especially age and immunocompetence, affect susceptibility to infection.38, 39 The surge in immunocompromised patients during the HIV/AIDS pandemic was associated with an upsurge in other emerging pathogens.7, 9 Host factors often receive less research focus than does pathogen virulence, but improved knowledge of host responses is essential to understand the species barrier and why some zoonotic pathogens are benign in their natural hosts but lethal when introduced to other species.38, 39
In view of the importance of human activities in emergence, integration of social sciences research—particularly that focused on human behaviour and demographics—into models of infectious disease emergence and evolution is essential to understand pandemics.40, 41 The importance of human exposure throughout the emergence process also suggests that simple behavioural precautions could greatly reduce risk. Risks to hunters, food handlers, and livestock workers from occupational exposure could be reduced in hotspots of emerging infectious diseases though routine sanitation and biosafety precautions, as has been tried with H5N1 in agricultural settings.42 Nosocomial spread could be prevented by stringent adherence to infection control practices—eg, use of sterile injection equipment.
A successful pathogen needs to possess the molecular machinery to infect people, invade human cells or tissues, replicate, and access tissues from which it can exit the host and transmit (directly or indirectly) to other people.38, 43 Historical examples are few, and thus to quantify the relative contribution of exposure frequency versus phylogenetic relatedness to the risk of successful infection is difficult. Analysis of host–pathogen databases of primates shows that closely related sympatric host species are more likely to share parasite species than are distantly related sympatric host species.44 Although any pathogen from a mammalian (or avian) host is a potential threat, that exposure of people to novel pathogens from our closest animal relatives poses the greater risk to public health is at least plausible.
Microbes often evolve substantially during emergence (eg, SIVcpz evolved into HIV-1), and the role of evolutionary adaptation in enabling pathogen establishment in human populations is a subject of active research.45, 46 Alternatively, some pathogens can spread between human beings without evolutionary change from the genotypes present in the wildlife host (eg, Ebola virus), and thus can enter the human population at stage 2. Phylodynamics, which combines a modelling framework for host, epidemiological, and molecular data,47, 48 especially for RNA viruses, shows particular promise for understanding of patterns of viral evolution during epidemics.48, 49
Testing of the relative importance of exposure and host relatedness in stage 2 emergence is an important challenge. For plant pathogens, experimental infection of closely or distantly related plants with a range of fungal pathogens has shown that host relatedness is a key factor in the ability of a pathogen to infect a novel host.50 Such a study would be logistically and ethically challenging in mammals, but experimental work with several model systems and various other organisms is underway, and systematic studies of host–parasite databases might provide an alternative. These studies might provide new surveillance opportunities by allowing improved targeting of the key wildlife species most likely to harbour the next emerging zoonosis.
Pathogen discovery
Identification of zoonotic pathogens has been substantially improved by advances in molecular diagnostics, which make possible the identification of hitherto unknown pathogens in nature (so-called pathogen discovery).51, 52 This process also includes differential diagnosis of infectious disease and surveillance for known or novel microbes that are normally benign in natural hosts but have the potential to become pathogenic in other hosts. Traditionally, microbe hunters used many techniques, including in-vitro and in-vivo culture, immunohistochemistry, electron microscopy, and serology. Immunohistochemistry of clinical samples was used successfully in several cases. Molecular techniques were used to first identify sin nombre virus, Nipah virus, West Nile virus, SARS coronavirus, and Lujo virus, among others. With the advent of unbiased molecular methods, the use of visualisation techniques has already begun to shift from discovery to provision of evidence of causal relations between an infectious agent and disease, by locating evidence of the agent at sites of pathological changes, including in both chronic and acute disease.51
Rapid, cost-effective molecular techniques allow broad-scale screening of samples, and pathogen discovery could be a strategy in wildlife to pre-empt pandemic zoonotic disease emergence through early identification. This strategy might be particularly effective when enhanced by hotspot modelling, which allows targeting of specific regions with the highest spillover risks. Historically, most surveillance efforts have been designed to detect individual pathogens in natural reservoirs, vectors, or at-risk human or animal populations through use of individual (so-called singleplex) PCR assays or serology. However, the costs for multiplex PCR, microarrays, and high-throughput sequencing continue to fall, and thus these assays are increasingly used as primary techniques for syndromic surveillance and studies of microbial diversity and discovery.51, 52 The challenge with any of these technologies is determination of the relevance of a microbial signature. 5000 species of mammals and 10 000 species of birds are known, each of which probably has at least several unique endemic viruses. Therefore the total number of new pathogens awaiting discovery is probably far higher than the 2000 to 3000 characterised. When other vertebrate-associated microbes are also considered, clearly the unfocused application of high-throughput screening could rapidly exhaust resources without substantively reducing pandemic risk. Therefore, discovery efforts need to be directed toward reservoirs and vectors where human being–animal interfaces6 and hotspot modelling3 or other information suggest an increased risk of transmission. In the past few years, the abundance and availability of molecular sequence data have made possible the identification of unknown microbes in nature based on similarities to the sequences of known organisms, even in crude nucleic acid extracts (so-called metagenomics).51, 52 This approach has clear value during the early stages of emergence. However, sequence data became available while an epidemic was still in progress for the first time only during the West Nile outbreak in 1999, and then again during the 2003 SARS outbreak and the 2009 influenza H1N1 pandemic. This information was invaluable for rapid development of diagnostics, identification of the source of the infection, and pathogen discovery. However, the rapid sequencing and identification of new pathogens before large-scale spread remain a substantial challenge.
Prediction of pandemic potential in novel microbes
Advances in meta-genomic technology have led to efforts in which molecular data are prospectively used to identify potential human pathogens in other species. The goal of such studies is to characterise novel microbes and assess probable virulence and transmissibility in people. However, crucial challenges exist in prediction of the effects of these new microbes, especially for viruses.53 In some cases (eg, coronaviruses54), host range can at least partially be predicted by receptor specificity and other factors.24, 54 When virulence factors are known (as for many bacteria38), meta-genomic analysis for these factors can be very useful.52 Such analysis is more challenging with viruses than with other microbes,19, 43, 55 although the experimental development of mutations in H5N1 that increase transmissibility in a ferret model has allowed targeted surveillance for isolates that are most likely to become efficient human pathogens.56 However, even when putative human pathogens can infect human-cell cultures, they are often unable to infect people. Genetically humanised murine models have been developed for SARS57 and hepatitis C virus (which usually can infect only human beings and chimpanzees).58 Humanised mice,59 or other model systems, could perhaps be adapted to screen for the ability of newly discovered microbes from wildlife reservoirs to infect people. Workers at the Rockefeller Foundation Virus Program in the mid-20th century used a low-tech version of the strategy—intracerebral inoculation—on suckling mice, which are susceptible to many infections by this route.60 However, screening in humanised mice is not yet widely available. In most cases, prediction of human virulence or transmissibility from molecular or phylogenetic data alone is still not feasible (another crucial unmet need), although promising work has been done with in-vitro tests to assess ability to induce a hyperinflammatory cytokine response in human cells as a predictor of virulence.61, 62 Molecular data will be of increasing predictive value as the global genomic database expands, thereby allowing wider comparisons and identification of commonalities. The predictive value will increase further if these molecular data are correlated with epidemiological and clinical data, and as our understanding of host requisites for transmission and barriers to cross-species transfer improves. As programmes for pathogen discovery in wildlife expand rapidly, the development of risk assessment criteria for the importance of these microbes is crucial. In our opinion, crucial questions remain and further research could substantially improve our understanding and our capacity to prevent pandemics before they emerge (panel 2 ).
Panel 2. Prediction of pandemic potential.
Programmes for pathogen discovery (including within wildlife populations) are expanding rapidly, and development of risk assessment and prioritisation criteria for these microbes is crucial. Several lines of inquiry could substantially increase capacity to prevent pandemics before emergence.
-
•
Research about the relative importance of host relatedness versus contact frequency: evidence shows that pathogens from closely related host species might be better able to spill over from one to the other.63 Emerging pathogens in people have originated in both closely related host species (eg, HIV-1 from chimpanzees26) and distantly related hosts with which human beings have had close recent contact (eg, severe acute respiratory syndrome from bats and civets,27 Nipah virus from bats and pigs,14 influenza from pigs and birds20, 64).
-
•
Analysis of viral relatedness as a predictor of emergence: wildlife viruses that are more closely related to known human pathogens are often assumed to be more likely to infect people than are those not similar to human pathogens. However, with the possible exception of the orthomyxoviruses and paramyxoviruses, the viral families that caused the most important recent zoonoses in human beings (ie, lentiviruses and coronaviruses) were recognised to have the potential for emergence only after they had already done so. Analysis of viral traits and phylogenetic relations, and how these correlate with pathogenicity after a virus spills over, will be crucial.65, 66
-
•
Host range and plasticity: emerging pathogens often have a broad host range. Pathogens that are more able to undergo successful transmission between different host taxa are also more likely to infect human beings than are those less able to transmit between different host taxa.22 These mechanisms should be elucidated and predictive correlations made.
-
•
Estimates of a virus's ability to evolve: the factors that allow a pathogen to successfully jump species are poorly known, but might include high mutability and an absence of proofreading to correct mutations, which are thought to explain the high proportion of negative-stranded RNA viruses in emerging pathogens.19, 29 So-called evolvability67 might help to explain why some pathogens have a high propensity for host jumps.19
-
•
Research about host–receptor interactions: receptor binding is an essential first step in cell infection and has been associated with changes in host and tissue tropism.19, 24, 45 Prediction of receptor binding necessitates understanding of the interactions for commonly expressed receptors (eg, sialic acids or heparan sulfate proteoglycans) or ease of adaptation of the virus to a new host receptor.24 However, the cell receptor is still unknown for more than 50% of human viruses. Highly host-specific receptor structures are a barrier to infection of a new host. Receptor binding is a necessary (but not sufficient) condition to enable viral entry to a cell and successful replication. Changing of receptor affinity (from avian-type to mammalian-type) was the first step in the adaption of H5N1 avian influenza virus to mammalian transmission, but other mutations were also needed.68, 69 Intracellular barriers and host resistance factors also exist for many viruses.43, 55, 70, 71 Viral genes that interfere with these functions might be identified on the basis of signatures or similarity to known genes.
-
•
Capacity to exploit new routes of transmission: widespread outbreaks are often correlated with high transmission efficiency, which might be partly related to the route or frequency of spread.72 Human behaviour is an important component that should be integrated into any predictive model,41 but the aspects of behaviour that matter are established by the pathogen's mode of transmission.
-
•
Prediction of virulence in human beings: along with prediction of human transmissibility, perhaps the biggest challenge is to assess the likelihood that a wildlife or livestock virus will cause noteworthy disease if the virus does infect people. True risk to human beings is related to both the transmissibility of a pathogen and the severity of the disease that results, and many zoonotic viruses that infect people cause no disease (eg, simian foamy viruses73) or mild symptoms (eg, Menangle virus16). Why some viruses that are benign in their natural hosts induce a severe or lethal hyperinflammatory response in a new host (eg, Ebola virus, sin nombre virus) is not well understood, but the causative pathogen components or structures should be identified and the mechanisms of species specificity further elucidated.
-
•
Patterns of host–virus coevolution: coevolutionary relationships in a group of related viruses and their wildlife hosts can be assessed easily by analysing genetic sequences.74 Strong patterns of coevolution during recent evolutionary time suggest stable long-term interactions with little host-switching, but pathogens that have frequently moved from one host to another have poorly aligned coevolutionary trees. Improved understanding of the pathogen's opportunities for transfer are needed for prediction.
These research questions are part of a much-needed basic research programme to investigate prediction of pandemics. However, previous evidence of pathogen spillover is a simple predictor of possible future emergence, and an active interface between human beings and other species is an obvious sine qua non. The capacity of a novel virus to transmit to people, and especially to cause illness, is a clue that the virus can replicate well in human beings, and thus could gain the properties that allow onward transmission. Spillover infections just below the threshold for self-sustainment in people (ie, infections with a basic reproduction number slightly <1) have been suggested as the prime epidemics-in-waiting,75, 76 although they are difficult to identify in advance. Thus, spillover events of any magnitude should be carefully monitored. For H5N1, intensive monitoring of each spillover case in endemic countries is crucial for prediction of if and when this pathogen moves from stage 2 to 3. Additionally, because pandemic zoonoses are such a large threat to global public health, to understand the ecological, virological, and social rules governing their emergence seems important. A concerted, interdisciplinary research focus on the process of disease emergence is urgently needed, and should include all sectors of the medical sciences, ecologists, social scientists, and wildlife biologists, among others.
Global strategy for surveillance and prevention
For emerging infections, strengthening of public health surveillance worldwide to provide early warning has been the primary recommendation of expert groups for the past two decades.4, 5, 10, 77, 78 Although greatly improved, public health surveillance capabilities remain restricted and fragmented, and have uneven global coverage.20, 64, 79 ProMED and the ProMED-mail e-mail LISTSERV and website were developed in the 1990s to improve early-warning capacity.77 This notion has expanded substantially, with a series of new initiatives such as WHO's Global Outbreak Alert and Response Network;80 a tripartite One Health initiative involving WHO, the Food and Agriculture Organization of the UN, and the World Organisation for Animal Health; HealthMap.org (and similarly Canada's Global Public Health Intelligence Network), a system that does high-capacity searching and filtering of internet feeds to identify novel pathogen outbreaks as rapidly as possible;81 and, in the USA, the Centers for Disease Control and Prevention's Global Disease Detection programme, the Department of Defense's Global Emerging Infections Surveillance and Response System, and other initiatives. These initiatives have been assisted by regional coordination and the formation of, for example, the European Centre for Disease Prevention and Control, and subregional networks such as the Mekong Basin Disease Surveillance Network.82, 83 The adoption of the revised International Health Regulations, which have a target of establishment of minimum core capacities, or specific plans, by mid-2012, by the 194 member states of WHO was a noteworthy advance.84 The revised Regulations incorporate a broad definition of outbreaks of public health concern, which include outbreaks in non-human animals, and provides an incentive for low-income countries to build capacity for pandemic surveillance and prevention. Although implementation will probably be delayed because many countries have so far been unable to achieve minimum core capacity goals, the adoption of the Regulations should greatly improve standards for surveillance, reporting, and response.82
Efforts to reduce or prevent pandemic zoonoses before they emerge in people have also begun, and might form the template for a new, globally coordinated pandemic prevention strategy. For influenza, these efforts have been focused on animal surveillance as a strategy to identify periods when the risk of spillover to people is high.64 Wider surveillance in wildlife to include targeted pathogen discovery has been called for.3, 5, 7, 64 However, few coordinated efforts have been implemented to pre-empt zoonotic disease emergence with wildlife surveillance. The Emerging Pandemic Threats programme is a noteworthy project initiated in 2009 by the US Agency for International Development. The programme is a suite of capacity-building investments designed to rapidly identify (and eventually predict) the emergence of new public health threats and increase country-level capacities to mitigate the potential effects of these threats.85 It draws heavily from efforts to address the H5N1 threat, and emphasises a strategic approach that builds on the understanding that the health and wellbeing of people, animals, and the environment are inextricably linked; promotes a One Health approach that spans the animal health, public health, environmental, and conservation communities;86 targets promotion of policies and capacities to identify and minimise the risk of emergence of new disease; and uses a risk-based approach to target investments where the likelihood of disease emergence is greatest.
The Emerging Pandemic Threats programme, through its PREDICT component, has developed an approach in which predictive modelling is used to identify the regions, wildlife hosts, and human being–animal interfaces most likely to propagate the next emerging zoonosis. The approach brings together experts from specialties, including wildlife ecology, epidemiology, genetics, virology, informatics, and veterinary medicine, all focused on building of a global early warning system for emerging diseases that move between wildlife and people. The programme's first goal is to obtain timely and reliable data for zoonotic threats, through internet surveillance of reports of unusual events in hotspot countries, analyses of the capacity of pathogens to emerge and then spread under different social systems in hotspots, and in-depth sample collection from the wildlife hosts most likely to harbour zoonoses. Samples are analysed to identify known zoonotic pathogens and new, closely related viruses, and then those deemed most likely to infect and cause illness in people are more fully characterised.
PREDICT is active in 20 developing countries in emerging infectious disease hotspots and focuses on surveillance at human being–animal interfaces where cross-species transmission is most likely. Infection in the natural or reservoir hosts is often asymptomatic,20 and thus testing of seemingly healthy animals is essential, but might be resource intensive, and unproductive if done randomly. To avoid this issue, PREDICT uses a combination of risk modelling to target locations, interfaces, and host taxa, and computerised data collection and analysis and active wildlife field sampling at high-risk sites to collect and identify viruses that might transfer from wildlife and cause disease. The programme partners with national and local governments, in-country scientists, and other local specialists who are active in outbreak reporting, microbial characterisation, and pathogen discovery, in collaboration with other US Agency for International Development Emerging Pandemic Threats projects as appropriate. These partnerships are intended to ensure the longevity of the programme by building capacity in the most in-need regions. The programme's scale gives some indication of how feasible this type of approach is for identification of the many thousands of novel pathogens that are probably in wildlife globally. In the first two years of the programme, around 100 000 samples from 20 000 animals (mainly bats, rodents, and non-human primates) in 20 countries have yielded 150 novel viruses from families known to harbour zoonoses. These data will be used to further refine global hotspot mapping and modelling strategies and test hypotheses about zoonotic transmission. They will also be shared as open source information, after the appropriate country clearances, through an interactive application at HealthMap.org. In selected high-risk regions, the associations between viral diversity, biological diversity, patterns of human contact with wildlife and livestock, and changes in land use are being explored to decipher the rules that govern disease emergence.
The ultimate goal of the Emerging Pandemic Threats programme is to develop a strategy to prevent future pandemics at the source before they infect human beings—an ambitious goal that requires more than building of health-care capacity or surveillance and diagnostic programmes. The challenge to true pandemic prevention (and pre-emption) is how to address the underlying drivers that are essentially ecological (eg, juxtaposition of livestock production and wildlife populations) or occur on large spatial scales because of economic activity (eg, change in land use related to development of tropical forests).1 In the case of both Nipah virus14 and H5N1,87, 88 economic development resulted in changes to animal production that led to the opening of a new niche for a pathogen. Could the seemingly opposing forces of economic development and public health have been reconciled before rather than after these outbreaks occurred? Expansion of so-called health impact assessments is a possible approach.12 Incentives for industries with roles in activities that propagate pandemics could be linked to development initiatives. For example, concessions in development of logging or mining could include better food supply chains as an alternative to bushmeat hunting, better clinics for migrant workers than are available, and more intensive surveillance of livestock at these crucial interfaces. Similarly, efforts to curtail the wildlife trade for food and pets in hotspot and other countries could include creation of incentives for consumers that lead to certification of industries promoting healthy practices.
Conclusion
The basis on which to build a global pandemic prevention strategy has changed substantially over the past few decades. A newly revised global reporting system of outbreaks and new molecular methods for pathogen identification and discovery are available, and advances in communications technology—eg, access to mobile phones and the Internet, even in remote areas—have enabled improved reporting. The early identification of a new human coronavirus respiratory disease from Saudi Arabia89 and a novel haemorrhagic fever virus in central Africa90 are examples of the great improvement in surveillance capabilities.
Understanding of the process of emergence and spread has moved from anecdotal through analytical to potentially predictive. Researchers are positioned to move from a paucity of data to a wealth of information on potential pathogens in nature. The challenge is to develop the basic research agenda to allow potential pathogens to be distinguished from harmless microbes by use of molecular sequence data only (the most commonly collected information), or information that can be deduced from these data—eg, structures of key proteins.
Political will for countries to act together to strengthen global networks against pandemic emergence also seems to have become positive. This new approach to pandemic prevention is shown by the handling of the discovery, in 1997, of a highly pathogenic influenza A (H5N1) of avian origin that infects people (precursor of the now widespread H5N1 avian influenza).91 Since then, and despite the virus's continued inability to transmit effectively between people, the public health community has recognised that a lethal virus circulating only in wildlife and domestic animals creates extraordinary opportunities to mitigate future risk.64 The response to H5N1 has implicated not only clinical, diagnostic, and therapeutic advances, but also better understanding of avian ecology, the economics of poultry production in low-income countries, and the ecology of the virus across the virus's broad host range. For perhaps the first time, the response to a zoonotic pandemic has included development agencies improving individual countries' abilities to identify new zoonoses early and mitigate quickly any new health threats arising within their borders.
The challenge is to establish whether and how researchers can intervene before a pathogen reaches the human population and develop appropriate triggers for action. Zoonotic diseases, by definition, should be a key mission of human-health agencies, agricultural authorities and producers, and natural resource managers, all working cooperatively. Substantial investments in each of these challenges are essential because the ecological and social changes worldwide that allow the emergence of infectious diseases are increasing at an unprecedented rate. Integration of efforts and coordination of budgetary resources for prevention and control is clearly a challenge that governments, both local and national, need to confront, and building of capacity to sustain these efforts might be the greatest challenge of all.
Search strategy and selection criteria
We searched PubMed and ISI Web of Knowledge with the terms “emerging infectio*”, “zoonos*”, or “pathogen discovery” in combination with the terms “modeling”, “prediction”, “surveillance”, “evolution”, “ecology”, or “methodology” for papers published in any language before Sept 25, 2012. We did our searches when we began to develop and write the paper and again before submission of the revised, final version. Some coauthors provided references that they deemed of particular importance. We largely selected publications from the past 5 years, but did not exclude commonly referenced and highly regarded older publications. We also searched the reference lists of articles identified by our searches and selected those judged relevant. Reviews and book chapters are cited to provide readers with more detailed information and references than is possible in the space allowed. Our reference list was modified on the basis of comments from peer reviewers.
Acknowledgments
Acknowledgments
This research was funded by the US Agency for International Development (USAID) Emerging Pandemic Threats PREDICT project (USAID cooperative agreement GHN-A-009-00010-00). WIL is supported by grants from the National Institutes of Health (AI079231, AI57158), and the Defense Threat Reduction Agency. PD is supported by an Ecology and Evolution of Infectious Diseases award from the Fogarty International Center of the National Institutes of Health (2R01-TW005869) and the National Science Foundation Human and Social Dynamics programme (BCS-0826779). MW receives support from the Wellcome-Trust-funded VIZIONS project, and SSM is supported by the Arts and Letters Foundation. We thank Murray Trostle for his invaluable insights and Aleksei A Chmura (EcoHealth Alliance) for figure 2. The contents are the responsibility of the authors and do not necessarily reflect the views of USAID or the United States Government. This paper is dedicated to the memory of Prof Joshua Lederberg (1925–2008).
Contributors
SSM and PD selected the topics for review, wrote portions of the paper, and coordinated and edited the contributions of all other authors. SSM did the searches of scientific literature, with contributions from the other authors. CZ-T analysed the underlying data and produced the hotspot map; all other authors contributed sections of the Series paper, provided references, and contributed to revisions. All authors reviewed the paper.
Conflicts of interest
We declare that we have no conflicts of interest.
References
- 1.Karesh WB, Dobson A, Lloyd-Smith JO. Ecology of zoonoses: natural and unnatural histories. Lancet. 2012;380:1936–1945. doi: 10.1016/S0140-6736(12)61678-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy FA. Emerging zoonoses. Emerg Infect Dis. 1998;4:429–435. doi: 10.3201/eid0403.980324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones KE, Patel N, Levy M. Global trends in emerging infectious diseases. Nature. 2008;451:990–994. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lederberg J, Shope RE, Oaks SC. Emerging infections: microbial threats to health in the United States. National Academies Press; Washington, DC: 1992. [PubMed] [Google Scholar]
- 5.Keusch GT, Pappaioanou M, Gonzalez MC, Scott KA, Tsai P. Sustaining global surveillance and response to emerging zoonotic diseases. National Academies Press; Washington, DC: 2009. [PubMed] [Google Scholar]
- 6.Morse SS. Factors in the emergence of infectious disease. Emerg Infect Dis. 1995;1:7–15. doi: 10.3201/eid0101.950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daszak P, Cunningham AA, Hyatt AD. Emerging infectious diseases of wildlife—threats to biodiversity and human health. Science. 2000;287:443–449. doi: 10.1126/science.287.5452.443. [DOI] [PubMed] [Google Scholar]
- 8.Keesing F, Belden LK, Daszak P. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature. 2010;468:647–652. doi: 10.1038/nature09575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morens DM, Folkers GK, Fauci AS. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430:242–249. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smolinski MS, Hamburg MA, Lederberg J. Microbial threats to health: emergence, detection, and response. National Academies Press; Washington, DC: 2003. [PubMed] [Google Scholar]
- 11.Weiss RA, McMichael AJ. Social and environmental risk factors in the emergence of infectious diseases. Nat Med. 2004;10:S70–S76. doi: 10.1038/nm1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morse SS. Regulating viral traffic. Issues Sci Technol. 1990;7:81–84. [Google Scholar]
- 13.Wolfe ND, Dunavan CP, Diamond J. Origins of major human infectious diseases. Nature. 2007;447:279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pulliam JRC, Epstein JH, Dushoff J. Agricultural intensification, priming for persistence and the emergence of Nipah virus: a lethal bat-borne zoonosis. J R Soc Interface. 2012;9:89–101. doi: 10.1098/rsif.2011.0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldberg TL, Gillespie TR, Rwego IB, Estoff EL, Chapman CA. Forest fragmentation and bacterial transmission among nonhuman primates, humans and livestock, Uganda. Emerg Infect Dis. 2008;14:1375–1382. doi: 10.3201/eid1409.071196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mackenzie JS. Emerging viral diseases: an Australian perspective. Emerg Infect Dis. 1999;5:1–8. doi: 10.3201/eid0501.990101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cleaveland S, Laurenson MK, Taylor LH. Diseases of humans and their domestic mammals: pathogen characteristics, host range and the risk of emergence. Phil Trans R Soc Lond B. 2001;356:991–999. doi: 10.1098/rstb.2001.0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor LH, Latham SM, Woolhouse MEJ. Risk factors for human disease emergence. Phil Trans R Soc Lond B. 2001;356:983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parrish CR, Holmes EC, Morens DM. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol Molec Biol Rev. 2008;72:457–470. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barber MR, Guan Y, Magor KE, Peiris JS, Webster RG. The role of animal surveillance in influenza preparedness: the consequence of inapparent infection in ducks and pigs. Influenza Other Respir Viruses. 2011;5(suppl 1):8–11. [PubMed] [Google Scholar]
- 21.Dawood FS, Jain S, Finelli L. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360:2605–2615. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- 22.Woolhouse MEJ, Gowtage-Sequeria S. Host range and emerging and re-emerging pathogens. Emerg Infect Dis. 2005;11:1842–1847. doi: 10.3201/eid1112.050997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kilpatrick AM, Randolph SE. Drivers, dynamics, and control of emerging vector-borne zoonotic diseases. Lancet. 2012;380:1946–1955. doi: 10.1016/S0140-6736(12)61151-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woolhouse MEJ, Scott F, Hudson Z, Howey R, Chase-Topping M. Human viruses: discovery and emergence. Phil Trans R Soc B. 2012;367:2864–2871. doi: 10.1098/rstb.2011.0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev. 2006;19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keele BF, Van Heuverswyn F, Li YY. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li WD, Shi ZL, Yu M. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 28.Guan Y, Zheng BJ, He YQ. Isolation and characterization of viruses related to the SARS coronavirus from animals in Southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 29.Woolhouse MEJ, Taylor LH, Haydon DT. Population biology of multihost pathogens. Science. 2001;292:1109–1112. doi: 10.1126/science.1059026. [DOI] [PubMed] [Google Scholar]
- 30.Liu WM, Li YY, Learn GH. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature. 2010;467:420–425. doi: 10.1038/nature09442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson RM, Fraser C, Ghani AC. Epidemiology, transmission dynamics and control of SARS: the 2002–2003 epidemic. Phil Trans R Soc Lond B. 2004;359:1091–1105. doi: 10.1098/rstb.2004.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hufnagel L, Brockmann D, Geisel T. Forecast and control of epidemics in a globalized world. Proc Natl Acad Sci USA. 2004;101:15124–15129. doi: 10.1073/pnas.0308344101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hosseini P, Sokolow SH, Vandegrift KJ, Kilpatrick AM, Daszak P. Predictive power of air travel and socio-economic data for early pandemic spread. PLoS One. 2010;5:e12763. doi: 10.1371/journal.pone.0012763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferguson NM, Cummings DAT, Cauchemez S. Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature. 2005;437:209–214. doi: 10.1038/nature04017. [DOI] [PubMed] [Google Scholar]
- 35.Chan J, Holmes A, Rabadan R. Network analysis of global influenza spread. PLoS Comp Biol. 2010;6:e10010005. doi: 10.1371/journal.pcbi.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Damon I. Status of human monkeypox: clinical disease, epidemiology and research. Vaccine. 2011;29(suppl 4):D54–D59. doi: 10.1016/j.vaccine.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 37.Fenner F. Human monkeypox, a newly discovered human virus disease. In: Morse SS, editor. Emerging viruses. Oxford University Press; Oxford: 1993. pp. 176–183. [Google Scholar]
- 38.Finlay BB, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Molec Biol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fukuyama S, Kawaoka Y. The pathogenesis of influenza virus infections: the contributions of virus and host factors. Curr Opin Immunol. 2011;23:481–486. doi: 10.1016/j.coi.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simini F, Gonzalez MC, Maritan A, Barabasi AL. A universal model for mobility and migration patterns. Nature. 2012;484:96–100. doi: 10.1038/nature10856. [DOI] [PubMed] [Google Scholar]
- 41.Janes CR, Corbett KK, Jones JH, Trostle J. Emerging infectious diseases: the role of social sciences. Lancet. 2012;380:1885–1887. doi: 10.1016/S0140-6736(12)61725-5. [DOI] [PubMed] [Google Scholar]
- 42.Ssematimba A, Hagenaars TJ, de Wit JJ. Avian influenza transmission risks: analysis of biosecurity measures and contact structure in Dutch poultry farming. Prev Vet Med. 2012 doi: 10.1016/j.prevetmed.2012.09.001. published online Sept 19. [DOI] [PubMed] [Google Scholar]
- 43.Webby R, Hoffmann E, Webster R. Molecular constraints to interspecies transmission of viral pathogens. Nat Med. 2004;10:S77–S81. doi: 10.1038/nm1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davies TJ, Pedersen AB. Phylogeny and geography predict pathogen community similarity in wild primates and humans. Proc Roy Soc B. 2008;275:1695–1701. doi: 10.1098/rspb.2008.0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pepin KM, Lass S, Pulliam JRC, Read AF, Lloyd-Smith JO. Identifying genetic markers of adaptation for surveillance of viral host jumps. Nat Rev Microbiol. 2010;8:802–813. doi: 10.1038/nrmicro2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lloyd-Smith JO, George D, Pepin KM. Epidemic dynamics at the human-animal interface. Science. 2009;326:1362–1367. doi: 10.1126/science.1177345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grenfell BT, Pybus OG, Gog JR. Unifying the epidemiological and evolutionary dynamics of pathogens. Science. 2004;303:327–332. doi: 10.1126/science.1090727. [DOI] [PubMed] [Google Scholar]
- 48.Rambaut A, Pybus OG, Nelson MI, Viboud C, Taubenberger JK, Holmes EC. The genomic and epidemiological dynamics of human influenza A virus. Nature. 2008;453:615–619. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holmes EC. The evolution and emergence of RNA viruses. Oxford University Press; Oxford: 2009. [Google Scholar]
- 50.Gilbert GS, Webb CO. Phylogenetic signal in plant pathogen-host range. Proc Natl Acad Sci USA. 2007;104:4979–4983. doi: 10.1073/pnas.0607968104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lipkin WI. Microbe hunting. Microbiol Molec Biol Revs. 2010;74:363–377. doi: 10.1128/MMBR.00007-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Relman DA. Microbial genomics and infectious diseases. N Engl J Med. 2011;365:347–357. doi: 10.1056/NEJMra1003071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morse SS. Toward an evolutionary biology of viruses. In: Morse SS, editor. The evolutionary biology of viruses. Wolters Kluwer/Raven Press; New York: 1994. pp. 1–28. [Google Scholar]
- 54.Graham RL, Baric RS. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J Virol. 2010;84:3134–3146. doi: 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuiken T, Holmes EC, McCauley J, Rimmelzwaan GF, Williams CS, Grenfell BT. Host species barriers to influenza virus infections. Science. 2006;312:394–397. doi: 10.1126/science.1122818. [DOI] [PubMed] [Google Scholar]
- 56.Russell CA, Fonville JM, Brown AE. The potential for respiratory droplet–transmissible A/H5N1 influenza virus to evolve in a mammalian host. Science. 2012;336:1541–1547. doi: 10.1126/science.1222526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baric RS, Sims AC. Humanized mice develop coronavirus respiratory disease. Proc Natl Acad Sci USA. 2005;102:8073–8074. doi: 10.1073/pnas.0503091102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dorner M, Horwitz JA, Robbins JB. A genetically humanized mouse model for hepatitis C virus infection. Nature. 2011;474:208–211. doi: 10.1038/nature10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kalscheuer H, Danzl N, Onoe T. A model for personalized in vivo analysis of human immune responsiveness. Sci Transl Med. 2012;4:125ra30. doi: 10.1126/scitranslmed.3003481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Downs WG. The Rockefeller Foundation Virus Program: 1951–1971 with update to 1981. Annu Rev Med. 1982;33:1–29. doi: 10.1146/annurev.me.33.020182.000245. [DOI] [PubMed] [Google Scholar]
- 61.Wolf B, Morgan H, Krieg J. A whole blood in vitro cytokine release assay with aqueous monoclonal antibody presentation for the prediction of therapeutic protein induced cytokine release syndrome in humans. Cytokine. 2012;60:828–837. doi: 10.1016/j.cyto.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 62.Vidal JM, Kawabata TT, Thorpe R. In vitro cytokine release assays for predicting cytokine release syndrome: the current state-of-the-science. Report of a European Medicines Agency Workshop. Cytokine. 2010;51:213–215. doi: 10.1016/j.cyto.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 63.Streicker DG, Turmelle AS, Vonhof MJ, Kuzmin IV, McCracken GF, Rupprecht CE. Host phylogeny constrains cross-species emergence and establishment of rabies virus in bats. Science. 2010;329:676–679. doi: 10.1126/science.1188836. [DOI] [PubMed] [Google Scholar]
- 64.Peiris JSM, Poon LLM, Guan Y. Surveillance of animal influenza for pandemic preparedness. Science. 2012;335:1173–1174. doi: 10.1126/science.1219936. [DOI] [PubMed] [Google Scholar]
- 65.Pulliam JRC. Viral host jumps: moving toward a predictive framework. Ecohealth. 2008;5:80–91. doi: 10.1007/s10393-007-0149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pulliam JRC, Dushoff J. Ability to replicate in the cytoplasm predicts zoonotic transmission of livestock viruses. J Infect Dis. 2009;199:565–568. doi: 10.1086/596510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Burke DS. The evolvability of emerging viruses. In: Nelson AM, Horsburgh CR, editors. Pathology of emerging infections. American Society for Microbiology; Washington, DC: 1998. pp. 1–12. [Google Scholar]
- 68.Imai M, Watanabe T, Hatta M. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Herfst S, Schrauwen EJA, Linster M. Airborne transmission of Influenza A/H5N1 virus between ferrets. Science. 2012;336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moresco EM, LaVine D, Beutler B. Toll-like receptors. Curr Biol. 2011;21:R488–R493. doi: 10.1016/j.cub.2011.05.039. [DOI] [PubMed] [Google Scholar]
- 71.Moresco EM, Beutler B. Resisting viral infection: the gene by gene approach. Curr Opin Virol. 2011;1:513–518. doi: 10.1016/j.coviro.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ewald PW. Evolution of infectious disease. Oxford University Press; New York: 1996. [Google Scholar]
- 73.Wolfe ND, Switzer WM, Carr JK. Naturally acquired simian retrovirus infections in central African hunters. Lancet. 2004;363:932–937. doi: 10.1016/S0140-6736(04)15787-5. [DOI] [PubMed] [Google Scholar]
- 74.Cui J, Han NIJ, Streicker D. Evolutionary relationships between bat coronaviruses and their hosts. Emerg Infect Dis. 2007;13:1526–1532. doi: 10.3201/eid1310.070448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Antia R, Regoes RR, Koella JC, Bergstrom CT. The role of evolution in the emergence of infectious diseases. Nature. 2003;426:658–661. doi: 10.1038/nature02104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bull J, Dykhuizen D. Epidemics-in-waiting. Nature. 2003;426:609–610. doi: 10.1038/426609a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morse SS, Rosenberg BH, Woodall J. ProMED global monitoring of emerging diseases: design for a demonstration program. Health Policy. 1996;38:135–153. doi: 10.1016/0168-8510(96)00863-9. [DOI] [PubMed] [Google Scholar]
- 78.National Biosurveillance Advisory Subcommittee Improving the nation's ability to detect and respond to 21st century urgent health threats: second report of the National Biosurveillance Advisory Subcommittee. http://www.cdc.gov/about/advisory/pdf/NBASFinalReport_April2011.pdf (accessed Nov 12, 2012).
- 79.Morse SS. Public health surveillance and infectious disease detection. Biosec Bioterr. 2012;10:6–16. doi: 10.1089/bsp.2011.0088. [DOI] [PubMed] [Google Scholar]
- 80.WHO Global Outbreak Alert & Response Network. http://www.who.int/csr/outbreaknetwork/en/ (accessed Sept 25, 2012).
- 81.Chan EH, Brewer TF, Madoff LC. Global capacity for emerging infectious disease detection. Proc Natl Acad Sci USA. 2010;107:21701–21706. doi: 10.1073/pnas.1006219107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Long WJ. Pandemics and peace: public health cooperation in zones of conflict. United States Institute of Peace Press; Washington, DC: 2011. [Google Scholar]
- 83.Kimball AM, Moore M, French HM. Regional infectious disease surveillance networks and their potential to facilitate the implementation of the international health regulations. Med Clin North Am. 2008;92:1459–1471. doi: 10.1016/j.mcna.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.WHO . International Health Regulations (2005) 2nd ed. World Health Organization; Geneva: 2008. [Google Scholar]
- 85.University of California, Davis PREDICT. http://www.vetmed.ucdavis.edu/ohi/predict/index.cfm (accessed Nov 12, 2012).
- 86.Karesh WB, Cook RA. One world—one health. Clin Med. 2009;9:259–260. doi: 10.7861/clinmedicine.9-3-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Beigel JH, Farrar J, Han AM. Avian influenza A (H5N1) infection in humans. N Engl J Med. 2005;53:1374–1385. doi: 10.1056/NEJMra052211. [DOI] [PubMed] [Google Scholar]
- 88.Writing committee of the second WHO consultation on clinical aspects of human infection with avian influenza A (H5N1) virus Update on avian influenza A (H5N1) virus infection in humans. N Engl J Med. 2008;358:261–273. doi: 10.1056/NEJMra0707279. [DOI] [PubMed] [Google Scholar]
- 89.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus ADME, Fouchier RAM. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 90.Grard G, Fair JN, Lee D. A novel rhabdovirus associated with acute hemorrhagic fever in central Africa. PLoS Pathog. 2012;8:e1002924. doi: 10.1371/journal.ppat.1002924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Claas ECJ, Osterhaus ADME, van Beek R. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]