Abstract
Atherosclerotic stenosis of cerebral arteries or intracranial large artery disease (ICLAD) is a major cause of stroke especially in Asians, Hispanics and Africans, but relatively little is known about gene expression changes in vessels at risk. This study compares comprehensive gene expression profiles in the middle cerebral artery (MCA) of New Zealand White rabbits exposed to two stroke risk factors i.e. hypertension and/or hypercholesterolemia, by the 2-Kidney-1-Clip method, or dietary supplementation with cholesterol. Microarray and Ingenuity Pathway Analyses of the MCA of the hypertensive rabbits showed up-regulated genes in networks containing the node molecules: UBC (ubiquitin), P38 MAPK, ERK, NFkB, SERPINB2, MMP1 and APP (amyloid precursor protein); and down-regulated genes related to MAPK, ERK 1/2, Akt, 26 s proteasome, histone H3 and UBC. The MCA of hypercholesterolemic rabbits showed differentially expressed genes that are surprisingly, linked to almost the same node molecules as the hypertensive rabbits, despite a relatively low percentage of ‘common genes’ (21 and 7%) between the two conditions. Up-regulated common genes were related to: UBC, SERPINB2, TNF, HNF4A (hepatocyte nuclear factor 4A) and APP, and down-regulated genes, related to UBC. Increased HNF4A message and protein were verified in the aorta. Together, these findings reveal similar nodal molecules and gene pathways in cerebral vessels affected by hypertension or hypercholesterolemia, which could be a basis for synergistic action of risk factors in the pathogenesis of ICLAD.
Introduction
Atherosclerotic stenosis of large arteries at the base of the brain or intracranial large artery disease (ICLAD) is a major cause of stroke especially in Asians, Hispanics and Africans [1], and is possibly the most common vascular lesion in the world [2]. It affects the middle cerebral artery (MCA), intracranial portion of the internal carotid artery, vertebrobasilar artery and the posterior and anterior cerebral arteries [1]. ICLAD carries a poor prognosis in terms of subsequent vascular event and death, and there is 25 - 30% incidence of recurrence in the 2 years after stroke [3], [4]. The disease is also prevalent among 53% of vascular dementia and 18% of Alzheimer’s disease patients of Asian ethnicity [1], [5].
The risk factors for ICLAD include hypertension, diabetes, hypercholesterolemia and cigarette smoking [6], and a strong association is found between asymptomatic ICLAD presenting as intracranial stenosis or calcification with large artery stiffness, and patients with untreated hypertension [7]. Arterial stiffness is a major determinant of increased systolic blood pressure, and is associated with lesions in intracranial arteries [8]. Prolonged elevation of blood pressure leads to reduction in vessel cross sectional area, increased wall thickness and accelerated plaque formation [9], [10]. Moreover, hypertension is thought to drive the atherosclerotic changes from larger to smaller vessels, and from extracranial- to intracranial vessels [11], [12]. Hypercholesterolemia is also a risk factor for ICLAD [6], and ischemic stroke from both extracranial and intracranial large-artery atherothromboembolism is associated with increased dietary intake of saturated fat, physical inactivity, obesity, and diabetes [13]. Reduction of cholesterol levels with statin treatment delays the progression of lesions in patients with ICLAD [14]. Increased lipoprotein is an independent biochemical risk factor for the development of ICLAD [15], and increased serum cholesterol is associated with elevated levels of oxidized low density lipoprotein [16]. The latter inhibits nitric oxide in endothelial cells to induce vasospasm [17] or increases tissue factor activity in these cells, to promote thrombosis [18]. Other factors that could contribute to ICLAD include increased oxidative stress in vessel walls [19]. A combination of hypercholesterolemia and hypertension may result in greater damage to vessels [9], [20].
Epidemiological studies indicate that there is increased risk of a second stroke especially in the first 1 or 2 years of post-stroke event [3], [4], [21], [22]. The reasons for this are not fully understood, but almost certainly involve gene expression changes at the vascular level that drive the atherothrombotic process. Thus far, however, there have been no studies to delineate global gene expression or gene network profiles in large intracerebral arteries at risk of atherothrombosis.
The present study was carried out to compare gene expression and morphological changes in intracranial vessels of rabbits, after exposure to hypertension and/or hypercholesterolemia. These conditions were induced by mostly non-genetically based methods, to reduce possible confounding effects during microarray analysis. The middle cerebral artery (MCA) was chosen for study, as this vessel is often affected in ICLAD [1], [23], [24], [25].
Materials and Methods
Animals
Male New Zealand White rabbits were used as it is the gold standard in atherosclerosis studies [26]. Although it is possible to produce hypertension in rats and mice, it is difficult to produce hypercholesterolemia in these animals [27]. The very small size of the MCA in rats and mice also hinders gene expression analyses of these vessels. Rabbits were approximately 8 weeks old (young adults) and weighed 2.0–2.5 kg each at the start of the experiments. Two sets of experiments were carried out: i) to determine gene expression changes in the MCA after hypertension, and ii) to determine gene expression changes in the MCA after hypercholesterolemia plus sham operation, and gene expression changes in the MCA after hypertension plus hypercholesterolemia. The first set of experiments were carried on 6 rabbits with the Goldblatt 2-Kidney 1-Clip (2K1C) method used to induce hypertension and fed with normal diet, vs. 6 sham operated controls on a normal diet. The second set of experiments were carried out on 6 rabbits on a high cholesterol diet with sham operation, 6 rabbits with 2K1C to induce hypertension plus a high cholesterol diet, and 6 rabbits on a normal diet.
The 2K1C procedure to induce hypertension was carried out as previous described [28]. In brief, animals were anesthetized with ketamine (75 mg/kg)/xylazine (10 mg/kg) cocktail followed by isoflurane maintenance, and the left renal artery exposed. The artery was partially occluded by attachment of a U-shaped silver ‘clip’ with a 0.6 mm slot. The clip was in left in place until the animals were sacrificed. Sham operated animals received the same surgical procedures as the 2K1C group, except that the renal artery was not partially occluded after its exposure. Animals that were subsequently treated with high cholesterol diet were allowed to recover from surgery for 1 week before treatment with diet containing cholesterol. Rabbits on this diet were fed with GPR diet +1% cholesterol (Glen Forrest Stockfeeders, Australia). Sham operated control rabbits were fed with GPR diet without cholesterol. All procedures including animals were approved by the Institutional Animal Care and Use Committee of the National University of Singapore, and carried out in accordance with guidelines of the National Advisory Committee for Laboratory Animal Research.
Measurement of Body Weight, Mean Arterial Pressure and Serum Total Cholesterol
Rabbits were anaesthetized by intramuscular injection of ketamine/xylazine cocktail, followed by mean arterial pressure measurements, and collection of blood. Mean arterial pressure was recorded from the ‘middle’ ear artery (Powerlab 4/30, AD Instruments, CO, USA), and blood samples obtained for cholesterol analysis, at 0, 4, 10 and 12 weeks. Approximately 3 mL of blood was withdrawn from the artery and collected in BD Vacutainer® Serum Tubes with Clot activator and silicone-coated interior (Becton Dickinson, NJ, USA). Whole blood was centrifuged at 1,000 g for 15 min, and the serum transferred to new vials and kept frozen at −80°C till analysis. Serum total cholesterol levels were measured by a fluorometric assay (Ex/Em 535/587 nm, BioVision Inc., CA, USA). Samples were analyzed in triplicates and read with a microplate reader (Infinite® i-control, Tecan Trading AG, Switzerland).
Tissue Harvesting and RNA Extraction
The 2K1C rabbits on normal diet, sham operated control rabbits on normal diet, or hypercholesterolemia plus sham operated rabbits and hypertension plus hypercholesterolemia rabbits were sacrificed 12 weeks after surgery. Sham operated control rabbits or untreated controls on a normal diet were sacrificed after a similar time. Animals were deeply anaesthetized by ketamine/xylazine cocktail and euthanized by intravenous injection of pentobarbital (250 mg/kg). The brains were removed and hemisected. The left half of the brain was immersed in 4% paraformaldehyde in 0.1M phosphate buffer in preparation for histology or electron microscopy (see below). The right MCA was identified and quickly stripped off the surface of the brain without any underlying cortical tissue, immersed in RNAlater® (Ambion, TX, USA), frozen in liquid nitrogen and stored in −80°C till further analysis. Total RNA was extracted using TRizol reagent (Invitrogen, CA, USA) according to the manufacturer’s protocol. Extracted RNA was purified using the RNeasy® Micro Kit (Qiagen, CA, USA). The cerebral neocortex, hippocampus, liver, kidney, aorta and other organs were also removed and snap frozen or stored in paraformaldehyde for future analyses.
DNA Microarray Analysis
Ten µL of total RNA from the MCA of four rabbits from each group were submitted to Genomax Technologies, Singapore. RNA quality was confirmed using an Agilent 2100 Bioanalyzer. cRNA was then generated and labeled using the one-cycle target labeling method, and hybridized to the 1-colour Agilent Rabbit Microarray (G2519F-020908; Agilent Technologies, CA, USA), according to the manufacturer’s protocol. Data was collected and exported into GeneSpring v11 software (Agilent Technologies) for analysis, using a parametric test based on the cross gene error model. Differentially expressed genes (DEGs) are those that show significantly increased or decreased expression compared to sham-operated controls using one-way ANOVA with Tukey HSD post-hoc test and corrected for multiple comparisons using Benjamini Hochberg FDR. P<0.01 was considered significant. In this study, to reduce false positives, only DEGs with greater than 4-fold change (or in the case of common genes between two data sets, greater than 4-fold change in at least one data set) were presented and used in IPA network analyses.
Network Analyses
The gene sets were analyzed using the Ingenuity Pathway Analysis (IPA) software (Ingenuity® Systems, www.ingenuity.com). Gene identifiers and corresponding expression values of up-regulated or down-regulated DEGS with more than 4-fold change was uploaded into IPA application. Each identifier mapped to its corresponding object in Ingenuity’s Knowledge Base, and was overlaid onto a global molecular network developed from information contained in the Ingenuity Knowledge Base. “Focus Genes” (Network Eligible genes) are defined as DEGs that have at least one other molecule in the Knowledge Base that interacts with it to form a “network”. The latter shows interactions between focus genes and ‘node molecules’ in the network, and how they work together at the molecular level.
Electron Microscopy
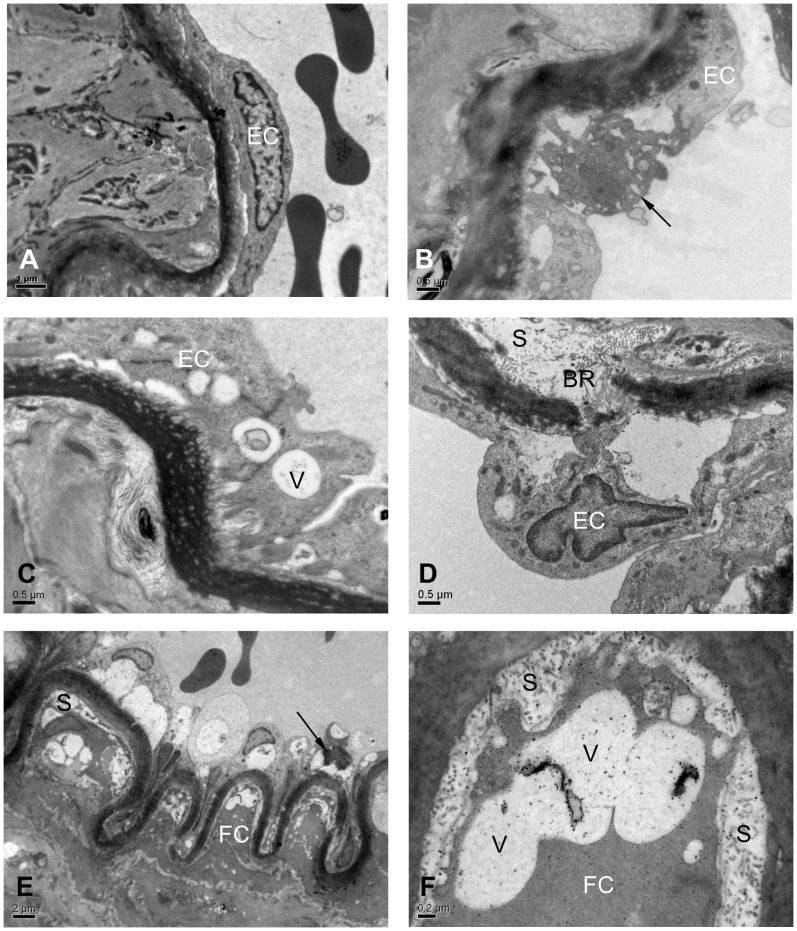
The left half of the brain was dissected out, fixed in 4% paraformaldehyde and 0.1 M phosphate buffer, and kept at 4°C. Blocks containing the MCA were osmicated, dehydrated in an ascending series of ethanol and acetone and embedded in Araldite. Thin sections were cut, mounted on Formvar coated copper grids and stained with lead citrate. They were viewed using a Jeol 1010 electron microscope (Jeol, Tokyo, Japan).
Quantitative Real-time PCR
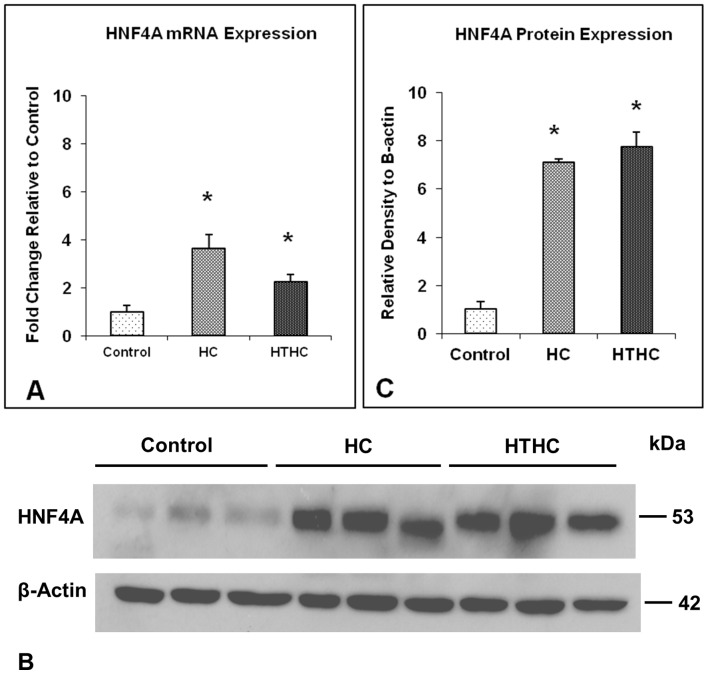
The mRNA of one of the node molecules identified by IPA, HNF4A, was further verified in the aorta by real-time RT-PCR. This was necessary, as only a small amount of RNA could be extracted from the rabbit MCA. Purified RNA from the descending aorta of 4 hypercholesterolemia plus sham, 4 hypertension plus hypercholesterolemia and 4 untreated control rabbits were reverse-transcribed with the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, CA, USA). Reaction conditions were 25°C for 10 min, 37°C for 120 min and 85°C for 5 sec. Real-time PCR amplification was performed using the 7500 Real time PCR system with TaqMan® Universal PCR Master Mix and probes. The PCR conditions were initial incubation of 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. Rabbit TaqMan® probe for HNF4A was purchased from Applied Biosystems. Rabbit ß-actin TaqMan® probe was used as an internal control. The fold change in expression was calculated using the 2-delta delta CT method as described previously [29]. Possible significant differences between the means were analyzed, using one-way ANOVA with Bonferroni’s multiple comparison post hoc test. p<0.05 was considered significant.
Western Blot Analysis
Aorta samples were homogenized in 10 volumes of ice-cold lysis buffer (150 mM sodium chloride, 50 mM Tris–hydrochloride, 0.25 mM EDTA, 1% Triton X-100, 0.1% sodium orthovanadate, and 0.1% protease inhibitor cocktail, pH 7.4), followed by centrifugation at 10,000 g for 10 min at 4°C. The supernatant was then collected, and protein concentrations measured using the Bio-Rad protein assay kit. The homogenates (20 µg) were resolved in 10% SDS-polyacrylamide gels under reducing conditions and electrotransferred to a polyvinylidene difluoride (PVDF) membrane. Non-specific binding sites on the PVDF membrane were blocked by incubation in 5% non-fat milk in tris-buffered saline-0.1% Tween 20 (TBST) for 1 h. The PVDF membrane was incubated overnight at 4°C with a mouse monoclonal anti-HNF4A antibody (K9218, Abcam, Cambridge, UK) diluted 1∶500 in 5% non-fat milk/TBST. After washing with TBST, the membrane was incubated with horseradish peroxidase-conjugated secondary anti-mouse IgG (Pierce, IL, USA) for 1 h at room temperature. Immunoreactivity was visualized using a chemiluminescence substrate (Millipore, MA, USA). Loading controls were carried out by incubating the blots at room temperature for 30 min with stripping buffer (100 mM 2-mercaptoethanol, 2% SDS, and 62.5 mM Tris-hydrochloride, pH 6.7), followed by reprobing with a mouse monoclonal antibody to ß-actin (Sigma, MO, USA; diluted 1∶10,000 in TBST). Exposed films containing blots were scanned and densities of the bands normalized to those of ß-actin. Possible significant differences between the values from treated and control rabbits were analyzed, using one-way ANOVA with Bonferroni’s multiple comparison post hoc test. p<0.05 was considered significant.
Histochemistry and Immunohistochemistry
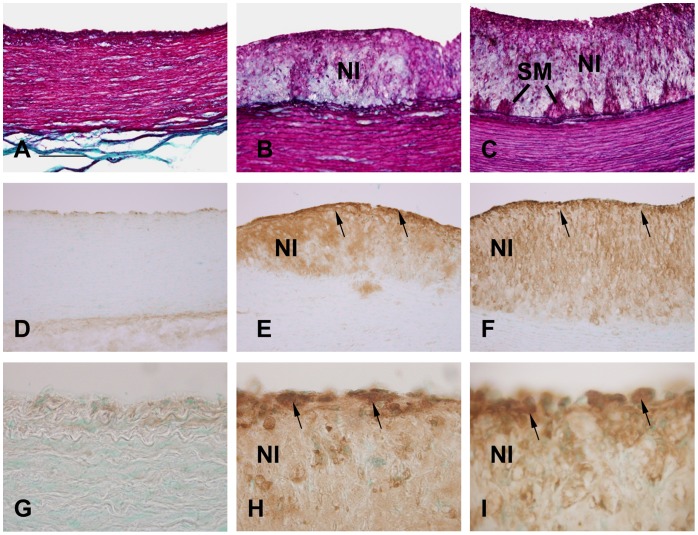
Aorta samples were sectioned at 40 µm using a freezing microtome. Sections were processed for histochemistry using Masson’s Trichrome histochemical stain, or immunoperoxidase staining. The latter sections were incubated in a blocking solution composed of 5% donkey serum (Vector) and 0.1% Triton X-100 for 1 h, followed by incubation with mouse monoclonal antibody to HNF4A (diluted 1∶100 in PBS) overnight. The sections were then washed three times in PBS and incubated with biotinylated anti-mouse secondary antibody. Immunoreaction product was visualized using an avidin-biotinylated horseradish peroxidase kit (Vector Laboratories, Burlingame, USA). Histochemically or immunohistochemically stained sections were mounted on glass slides and viewed using a light microscope (IX70, Olympus, Japan).
Results
1. Body Weight, Mean Arterial Pressure and Serum Total Cholesterol Levels
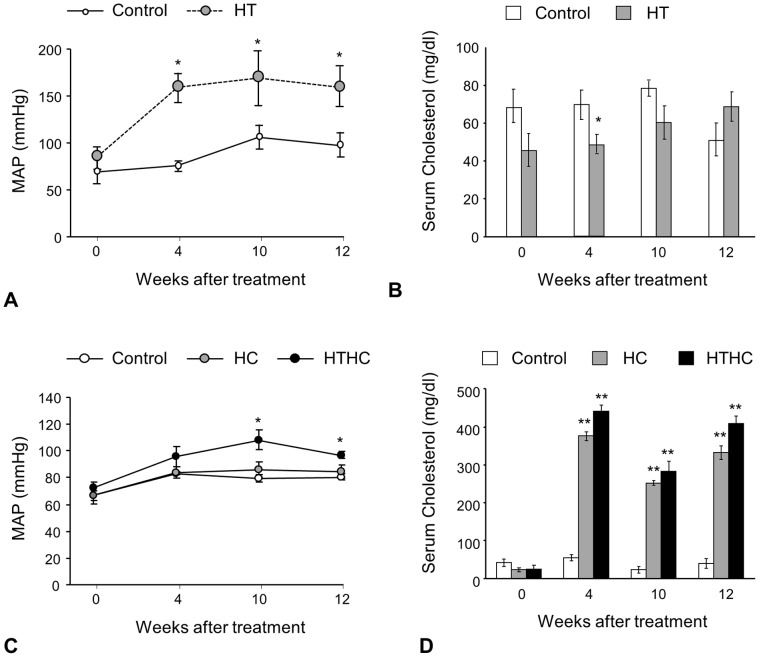
The average body weight was not significantly different between the 2K1C and sham operated groups (data not shown),but mean arterial pressure of 2K1C group was markedly higher than that of sham group at 4, 10, 12 weeks after the surgery (Figure 1A). The serum total cholesterol level remained at a low level (<100 mg/dl) for both groups throughout the experiment, and no difference was found between the groups, except for a slightly lower value in the 2K1C group on week 4 (Figure 1B). The average body weight among all groups was not significantly different (data not shown).
Figure 1. Mean arterial pressure and serum cholesterol levels in rabbits.
(A) Mean arterial pressure in hypertension only rabbits. (B) Serum cholesterol levels in hypertension only rabbits. (C) Mean arterial pressure in hypercholesterolemia plus sham- and hypertension plus hypercholesterolemia rabbits. (D) Serum cholesterol levels in hypercholesterolemia plus sham- and hypertension plus hypercholesterolemia rabbits. H: Hypertension only. HC: Hypercholesterolesterolemia plus sham operation. HTHC: Hypertension plus hypercholesterolemia. MAP: mean arterial pressure. Data are expressed as mean ± SEM. *p<0.05, **p<0.01 vs. control (Student’s t-test in A,B; repeated measure ANOVA followed by Tukey test in C,D).
Increased mean arterial pressure was found in the 2K1C plus cholesterol-fed rabbits (Figure 1C), and markedly elevated serum total cholesterol levels (>200mg/dl) were found in both the hypercholesterolemia plus sham- and 2K1C plus hypercholesterolememia groups at 4, 10 and 12 weeks, compared to control rabbits on a normal diet (Figure 1D).
2. Microarray Analyses
2.1. Microarray analyses of the hypertension only group
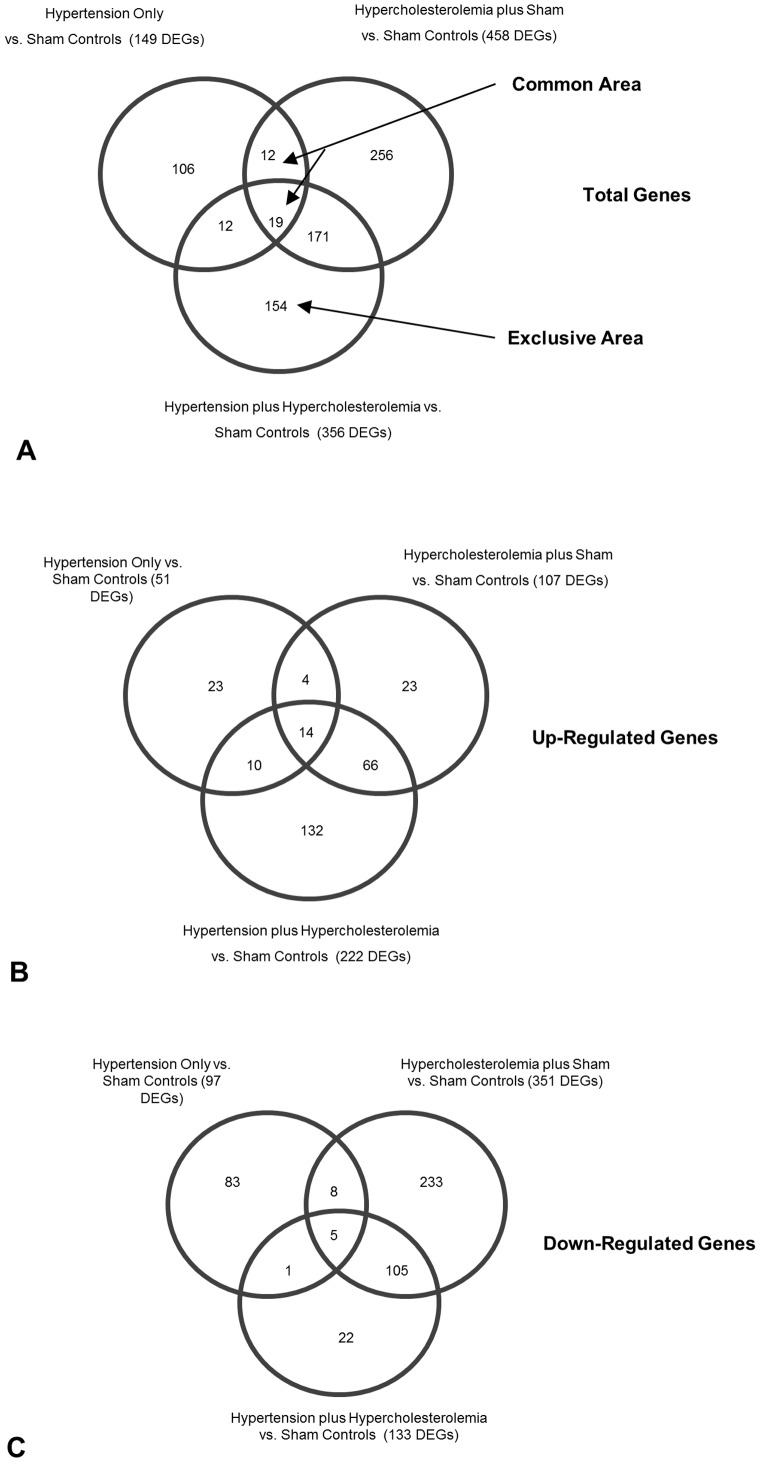
The gene expression profile in the MCA of the hypertension only group was compared with that of sham operated controls on a normal diet. After unknown genes and repeated probes of the same genes were omitted, 51 up-regulated and 97 down-regulated genes (greater than 4-fold change) were found in the MCA (Figure 2). Among the highly up-regulated genes in the MCA of the hypertension only group compared to sham controls were FAM167A, CERS3 and FAM53C (Table 1). Among the highly down-regulated genes were FOXN1, NSRP1 and THUMPD3 (Table 2). The panel of genes was imported into IPA to analyze network interactions.
Figure 2. Venn diagram of DEGs in the MCA of hypertension only rabbits; hypercholesterolemia plus sham operated rabbits; and hypertension plus hypercholesterolemia rabbits; all vs. sham operated control rabbits.
A: total number of genes, B: up-regulated genes C: down-regulated genes (One gene which is common between the Hypertension only- and Hypertension plus hypercholesterolemia group was both up- and down-regulated, and omitted).
Table 1. Up-regulated genes in the MCA of ‘hypertension only’ rabbits vs. sham controls with greater than 4-fold change.
| Gene | Gene Symbol | Corrected p-value | Hypertension only | Hyperchole-sterolemia plus sham | Hypertension plus hyperchole-sterolemia |
| Fold Change | Fold Change | Fold Change | |||
| family with sequence similarity 167, member A | FAM167A | 0.00017 | 14.04 | 2.44 | 5.11 |
| ceramide synthase 3 | CERS3 | 0.00486 | 11.44 | −1.50 | 3.68 |
| family with sequence similarity 53, member C | FAM53C | <0.00001 | 9.87 | 2.48 | 4.03 |
| tubulin tyrosine ligase-like family, member 5 | TTLL5 | 0.00005 | 8.16 | −1.18 | 1.86 |
| olfactory receptor, family 1, subfamily J, member 1 | OR1J1 | 0.00002 | 8.14 | 3.83 | −1.85 |
| Interstitial collagenase Precursor (EC 3.4.24.7)(Matrix metalloproteinase-1)(MMP-1) | MMP1 | 0.00149 | 8.12 | 2.20 | 6.33 |
| Nibrin-like | LOC100352398∧ | 0.00171 | 7.92 | 4.71 | 3.74 |
| TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa | TAF15∧, + | 0.00004 | 7.45 | 4.71 | 4.65 |
| zinc finger, DHHC-type containing 23 | ZDHHC23 | 0.00001 | 7.18 | 3.22 | 5.64 |
| ankyrin and armadillo repeat containing | ANKAR∧, + | 0.00006 | 7.05 | 7.99 | 5.23 |
| N(alpha)-acetyltransferase 25, NatB auxiliary subunit | NAA25∧, + | <0.00001 | 6.43 | 8.91 | 18.66 |
| corticotropin releasing hormone binding protein | CRHBP | 0.00005 | 6.38 | 2.23 | 4.02 |
| Cytochrome P450 1A2 (EC 1.14.14.1)(CYPIA2)(Cytochrome P450 isozyme 4)(Cytochrome P450 LM4)(Cytochrome P450-PM4) | CYP1A2 | 0.00004 | 6.15 | 1.73 | −1.90 |
| KIAA0232 | KIAA0232 | 0.00002 | 6.13 | 1.77 | 1.44 |
| Fanconi anemia, complementation group C | FANCC∧, + | <0.00001 | 6.10 | 4.25 | 6.39 |
| NEDD8 activating enzyme E1 subunit 1 | NAE1 | 0.00009 | 6.10 | 3.26 | −2.34 |
| Zinc finger protein 638 | LOC100345662 | 0.00005 | 6.01 | 1.02 | −2.87 |
| Uncharacterized protein C1orf50 | C1orf50 | 0.00006 | 5.81 | 3.80 | 8.01 |
| Mix paired-like homeobox | MIXL1 | 0.00093 | 5.80 | 3.54 | 4.64 |
| Serine proteinase inhibitor, clade B, member 2 (Predicted) | SERPINB2∧, + | 0.00005 | 5.63 | 6.99 | 9.10 |
| IDI1 protein-like | LOC100346274∧, + | 0.00042 | 5.51 | 4.87 | 6.67 |
| B-cell receptor-associated protein 31 | LOC100356247 | 0.00464 | 5.45 | 2.83 | 1.09 |
| sperm associated antigen 6 | SPAG6 | 0.00139 | 5.39 | 1.31 | 2.69 |
| meprin A, beta | MEP1B∧, + | 0.00291 | 5.37 | 5.12 | 5.85 |
| G protein-coupled receptor kinase interacting ArfGAP 1 | GIT1 | <0.00001 | 5.22 | 2.76 | 3.91 |
| ciliary rootlet coiled-coil, rootletin | CROCC | 0.00003 | 5.08 | 2.33 | 2.79 |
| SRY (sex determining region Y)-box 2-like | LOC100341629 | 0.00001 | 5.05 | 1.43 | −7.56 |
| Fibroblast growth factor binding protein 1-like | LOC100353835∧, + | 0.00122 | 5.01 | 4.01 | 4.63 |
| ankyrin repeat and SOCS box-containing 4 | ASB4∧ | 0.00027 | 4.98 | 4.06 | 3.51 |
| Double zinc ribbon and ankyrin repeat-containing protein 1 | DZANK1∧, + | <0.00001 | 4.87 | 5.04 | 7.37 |
| CD46 molecule, complement regulatory protein | CD46 | 0.00230 | 4.85 | 3.14 | −1.11 |
| programmed cell death 11 | PDCD11 | 0.00003 | 4.85 | −1.62 | −1.11 |
| signal-induced proliferation-associated 1 like 3 | SIPA1L3∧, + | 0.00001 | 4.80 | 8.45 | 6.50 |
| relaxin/insulin-like family peptide receptor 2 | RXFP2∧, + | 0.00029 | 4.79 | 5.62 | 6.96 |
| PHD and ring finger domains 1 | PHRF1 | 0.00002 | 4.69 | 1.13 | 1.17 |
| guanylate cyclase 2D, membrane (retina-specific) | GUCY2D∧, + | 0.00008 | 4.64 | 4.11 | 8.34 |
| EH domain binding protein 1 | EHBP1 | 0.00171 | 4.60 | 2.08 | −1.19 |
| testis-specific kinase 2 | TESK2∧ | 0.00571 | 4.50 | 5.11 | 2.41 |
| Golgin subfamily B member 1 | LOC100340420 | 0.00001 | 4.32 | −2.30 | −1.39 |
| coiled-coil domain containing 54 | CCDC54 | 0.00669 | 4.25 | 2.96 | 4.62 |
| solute carrier family 39 (zinc transporter), member 12 | SLC39A12 | 0.00006 | 4.23 | 2.48 | 4.96 |
| l(3)mbt-like (Drosophila) | L3MBTL1 | 0.00011 | 4.21 | 1.80 | 1.38 |
| chemokine (C-C motif) ligand 1 | CCL1 | <0.00001 | 4.18 | 1.13 | 1.37 |
| ribonuclease, RNase A family, 1 (pancreatic) | RNASE1∧, + | <0.00001 | 4.17 | 5.62 | 4.82 |
| DIS3 mitotic control homolog (S. cerevisiae)-like 2 | DIS3L2∧ | 0.00024 | 4.17 | 4.32 | 3.96 |
| Cold shock domain containing E1, RNA-binding | CSDE1 | 0.00002 | 4.16 | 1.89 | 2.99 |
| antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5 (MFI2) | MFI2 | 0.00386 | 4.04 | 2.47 | 1.09 |
| Glucose-fructose oxidoreductase domain containing 2-like | LOC100351150∧, + | 0.00001 | 4.01 | 6.34 | 12.42 |
| coiled-coil domain containing 89 | CCDC89 | 0.00238 | 4.01 | 1.64 | 3.00 |
∧Differentially expressed genes that are common between ‘hypertension only’ and ‘hypercholesterolemia plus sham’ groups (both vs. sham controls).+Differentially expressed genes that are common among ‘hypertension only’, ‘hypercholesterolemia plus sham’, and ‘hypertension plus hypercholesterolemia’ groups (all vs. sham controls).
Table 2. Down-regulated genes in the MCA of ‘hypertension only’ rabbits vs. sham controls with greater than 4-fold change.
| Gene | Gene Symbol | Corrected p-value | Hypertension only | Hyperchole-sterolemia plus sham | Hypertension plus hyperchole-sterolemia |
| Fold Change | Fold Change | Fold Change | |||
| forkhead box N1 | FOXN1∧, + | 0.00048 | −26.20 | −56.89 | −45.12 |
| nuclear speckle splicing regulatory protein 1 | NSRP1 | <0.00001 | −24.13 | −1.56 | −1.23 |
| THUMP domain containing 3 | THUMPD3 | <0.00001 | −17.18 | 1.04 | 1.31 |
| AT rich interactive domain 2 (ARID, RFX-like) | ARID2 | <0.00001 | −16.92 | −2.86 | −2.29 |
| Ribosomal protein S3a-like | LOC100354966∧ | <0.00001 | −16.54 | −5.74 | −1.83 |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 5 | NDUFAF5 | 0.00007 | −15.92 | −2.60 | −1.44 |
| ADAM metallopeptidase with thrombospondin type 1 motif, 17 | ADAMTS17∧ | 0.00001 | −15.85 | −7.25 | −3.06 |
| peptidylprolyl isomerase G (cyclophilin G) | PPIG∧ | <0.00001 | −12.61 | −6.20 | −1.81 |
| hematopoietic prostaglandin D synthase | HPGDS∧, + | 0.00043 | −12.38 | −21.44 | −13.63 |
| cyclin H | CCNH | <0.00001 | −10.89 | −3.52 | −1.34 |
| LYR motif containing 7 | LYRM7 | 0.00001 | −10.41 | −1.63 | −1.65 |
| male-specific lethal 2 homolog (Drosophila) | MSL2 | 0.00022 | −9.72 | −2.22 | −2.10 |
| secreted frizzled-related protein 4 | SFRP4 | <0.00001 | −9.32 | −1.07 | −1.11 |
| ankyrin 2, neuronal | ANK2 | 0.00002 | −9.32 | −1.39 | −1.31 |
| Ras-related protein Rab-7a | RAB7A | 0.00001 | −9.20 | −1.18 | −1.35 |
| NLR family, pyrin domain containing 5 | NLRP5 | <0.00001 | −8.60 | 1.51 | 2.02 |
| Integrin-linked kinase-associated serine/threonine phosphatase | ILKAP | 0.00001 | −8.57 | 1.49 | 1.39 |
| Histone H3.3B-like | LOC100341225 | 0.00209 | −8.43 | −1.36 | −2.17 |
| membrane-spanning 4-domains, subfamily A, member 1 | MS4A1 | 0.00073 | −7.86 | −3.98 | −2.53 |
| sorting nexin 9 | SNX9 | 0.00016 | −7.82 | −1.54 | −1.78 |
| protein tyrosine phosphatase, non-receptor type 23 | PTPN23 | 0.00003 | −7.80 | 1.69 | 1.30 |
| large subunit GTPase 1 homolog (S. cerevisiae) | LSG1∧ | <0.00001 | −7.69 | −20.38 | −1.93 |
| coiled-coil domain containing 59 | CCDC59 | <0.00001 | −7.66 | −2.29 | −1.40 |
| Protein SLX4IP | SLX4IP | 0.00008 | −7.64 | −1.90 | −1.79 |
| ribosomal protein S12 | RPS12 | 0.00079 | −7.57 | −1.56 | −1.28 |
| breast carcinoma amplified sequence 2 | BCAS2 | 0.00001 | −7.39 | −1.24 | −1.18 |
| Secreted protein, acidic, cysteine-rich (osteonectin) | SPARC | 0.00030 | −7.26 | −2.00 | −1.62 |
| mitochondrial ribosomal protein L15 | MRPL15∧, + | <0.00001 | −6.97 | −23.01 | −9.57 |
| Translocase of inner mitochondrial membrane domain-containing protein 1 | TIMMDC1 | 0.00021 | −6.72 | 1.06 | −1.07 |
| olfactory receptor, family 4, subfamily D, member 2 | OR4D2 | 0.00009 | −6.61 | −1.25 | 1.18 |
| tumor necrosis factor, alpha-induced protein 8 | TNFAIP8 | 0.00148 | −6.59 | −2.12 | −2.30 |
| LIM domains containing 1 | LIMD1 | 0.00007 | −6.10 | −2.35 | −1.61 |
| Neurolysin, mitochondrial Precursor | NLN | 0.00072 | −6.05 | −1.23 | −1.39 |
| Osteoglycin | OGN | 0.00004 | −5.96 | −1.58 | −1.49 |
| Ribosomal protein L35-like | LOC100339185 | 0.00176 | −5.85 | −3.09 | −1.69 |
| transient receptor potential cation channel, subfamily V, member 5 (TRPV5) | TRPV5 | 0.00002 | −5.80 | −1.90 | −1.49 |
| Glycine cleavage system protein H (aminomethyl carrier) | GCSH | <0.00001 | −5.68 | −1.84 | −1.61 |
| translocase of outer mitochondrial membrane 5 homolog (yeast) | TOMM5∧ | 0.00014 | −5.67 | −4.05 | −1.60 |
| SEC24 family, member A (S. cerevisiae) | SEC24A | 0.00205 | −5.62 | −2.86 | −2.06 |
| matrilin 2 | MATN2 | <0.00001 | −5.55 | 1.73 | 1.69 |
| ubiquitin specific peptidase 46 | USP46 | 0.00039 | −5.54 | −2.52 | −1.45 |
| purine-rich element binding protein A | PURA | 0.00034 | −5.54 | 1.21 | 1.12 |
| hydroxyprostaglandin dehydrogenase 15- (NAD) | HPGD | 0.00366 | −5.51 | −2.30 | −2.60 |
| parkinson protein 7 | PARK7 | <0.00001 | −5.44 | 1.27 | 1.07 |
| IL2-inducible T-cell kinase | ITK | 0.00003 | −5.37 | 1.07 | 1.25 |
| ribosomal protein L26 | RPL26∧ | 0.00016 | −5.27 | −14.60 | −2.81 |
| endothelial PAS domain protein 1 | EPAS1 | 0.00279 | −5.24 | −2.30 | −1.70 |
| ATP-sensitive inward rectifier potassium channel Kir6.2 (KIR62) | KIR62 | 0.00032 | −5.21 | −1.25 | 1.08 |
| Kruppel-like factor 12 | KLF12 | 0.00008 | −5.14 | −1.25 | −1.15 |
| TBC1 domain family, member 8B (with GRAM domain) | TBC1D8B | 0.00101 | −5.13 | −1.47 | −1.01 |
| Nucleophosmin 1 isoform 1 (Predicted) | NPM1 | 0.00001 | −5.12 | −1.40 | −1.40 |
| BUD31 homolog (S. cerevisiae) | BUD31 | 0.00343 | −5.08 | −1.20 | −1.30 |
| poly(A) polymerase alpha | PAPOLA | <0.00001 | −5.03 | −2.29 | −1.26 |
| ribosomal protein L37a | RPL37A | <0.00001 | −4.98 | 1.23 | 1.10 |
| splicing factor 3a, subunit 3, 60 kDa | SF3A3 | 0.00003 | −4.91 | −1.55 | −1.36 |
| C1q and tumor necrosis factor related protein 1 | C1QTNF1 | 0.00019 | −4.90 | −3.22 | −1.22 |
| family with sequence similarity 107, member B | FAM107B | 0.00140 | −4.88 | −3.41 | −2.58 |
| integral membrane protein 2C | ITM2C | 0.00015 | −4.87 | 1.64 | 1.30 |
| Alpha-1-B glycoprotein | A1BG | 0.00121 | −4.87 | 2.01 | 1.19 |
| Histone deacetylase complex subunit SAP18-like | LOC100356364 | 0.00003 | −4.86 | −1.85 | −1.65 |
| phosphorylase kinase, beta (PHKB | PHKB | 0.00033 | −4.83 | −1.23 | −1.12 |
| Vacuolar protein sorting-associated protein 51 homolog | VPS51 | 0.00004 | −4.74 | 2.15 | 1.65 |
| Uncharacterized protein C11orf57 | C11orf57 | 0.00365 | −4.65 | −1.71 | −1.25 |
| exostoses (multiple)-like 3 | EXTL3 | 0.00001 | −4.65 | −3.53 | −4.65 |
| Ferritin heavy chain Fragment (Ferritin H subunit) | FTH1 | 0.00003 | −4.64 | 1.06 | −1.09 |
| Uncharacterized protein C15orf62, mitochondrial Precursor | C15orf62 | 0.00018 | −4.58 | 1.21 | 1.48 |
| lysine (K)-specific demethylase 4A | KDM4A | <0.00001 | −4.57 | 2.48 | 2.70 |
| NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6, 17kDa | NDUFB6 | 0.00116 | −4.56 | −1.76 | −1.55 |
| p21 protein (Cdc42/Rac)-activated kinase 4 | PAK4 | 0.00002 | −4.56 | −1.16 | −1.05 |
| armadillo repeat containing, X-linked 3 | ARMCX3 | 0.00161 | −4.49 | −1.40 | −1.37 |
| Transmembrane tyrosine kinase receptor | c-kit | 0.00981 | −4.48 | −2.92 | −2.29 |
| leucine rich repeat (in FLII) interacting protein 2 | LRRFIP2 | 0.00001 | −4.47 | 1.23 | 1.14 |
| cardiac calcium channel beta-subunit (CAB2) | CAB2 | 0.00008 | −4.46 | 1.34 | 1.20 |
| HERPUD family member 2 | LOC100350332 | 0.00003 | −4.42 | −1.71 | −1.80 |
| family with sequence similarity 177, member A1 | FAM177A1∧ | 0.00016 | −4.41 | −10.75 | −1.95 |
| acyl-CoA synthetase long-chain family member 4 | ACSL4 | 0.00397 | −4.40 | −2.03 | −1.94 |
| interleukin-1 receptor-associated kinase 1 binding protein 1 | IRAK1BP1 | 0.00001 | −4.38 | −2.94 | −1.75 |
| polyamine modulated factor 1 binding protein 1 | PMFBP1 | <0.00001 | −4.34 | 2.06 | 2.81 |
| Proteasome (prosome, macropain) 26s subunit, non-ATPase, 4 | PSMD4∧, + | 0.00005 | −4.34 | −17.67 | −11.67 |
| dpy-19-like 1 (C. elegans) | DPY19L1 | <0.00001 | −4.32 | 1.14 | 1.10 |
| mastermind-like 2 (Drosophila) | MAML2 | 0.00134 | −4.29 | −1.57 | 1.05 |
| choline phosphotransferase 1 | CHPT1 | 0.00014 | −4.26 | −1.63 | −1.47 |
| Aggrecanase-2 | ADAMTS-11∧, + | 0.00005 | −4.26 | −9.31 | −6.51 |
| nuclear protein localization 4 homolog (S. cerevisiae) | NPLOC4 | 0.00342 | −4.25 | −2.86 | −1.62 |
| B-cell translocation gene 1, anti-proliferative | BTG1 | 0.00030 | −4.24 | −1.80 | −1.81 |
| galactokinase 2 | GALK2 | 0.00002 | −4.22 | 1.67 | 1.54 |
| N(alpha)-acetyltransferase 50, NatE catalytic subunit | NAA50 | 0.00132 | −4.21 | −1.38 | −1.99 |
| Tubulin polyglutamylase complex subunit 2-like | LOC100353679 | 0.00044 | −4.19 | 1.09 | −1.02 |
| Glycine N-methyltransferase Fragment (EC 2.1.1.20) | GNMT | 0.00006 | −4.17 | −2.35 | −1.18 |
| Protein FAM210B | FAM210B | 0.00130 | −4.16 | 1.36 | 1.38 |
| RNA pseudouridylate synthase domain containing 4 [ | RPUSD4 | 0.00011 | −4.11 | 1.05 | 1.04 |
| nucleotide-binding oligomerization domain containing 2 | NOD2 | 0.00111 | −4.09 | −1.39 | −1.44 |
| cysteine conjugate-beta lyase 2 | CCBL2 | 0.00688 | −4.07 | −1.10 | −1.03 |
| BRCA2 and CDKN1A interacting protein | BCCIP∧ | <0.00001 | −4.06 | −14.00 | −2.58 |
| major facilitator superfamily domain containing 9 | MFSD9 | 0.00383 | −4.04 | −1.26 | −1.52 |
| Olfactory receptor 572 (Predicted) | OLFR572 | 0.00007 | −4.03 | 1.30 | 2.27 |
| neuroplastin | NPTN | 0.00010 | −4.02 | −1.19 | −1.19 |
∧ Differentially expressed genes that are common between ‘hypertension only’ and ‘hypercholesterolemia plus sham’ groups (both vs. sham controls).+Differentally expressed genes that are common among ‘hypertension only’, ‘hypercholesterolemia plus sham’, and ‘hypertension plus hypercholesterolemia’ groups (all vs. sham controls).
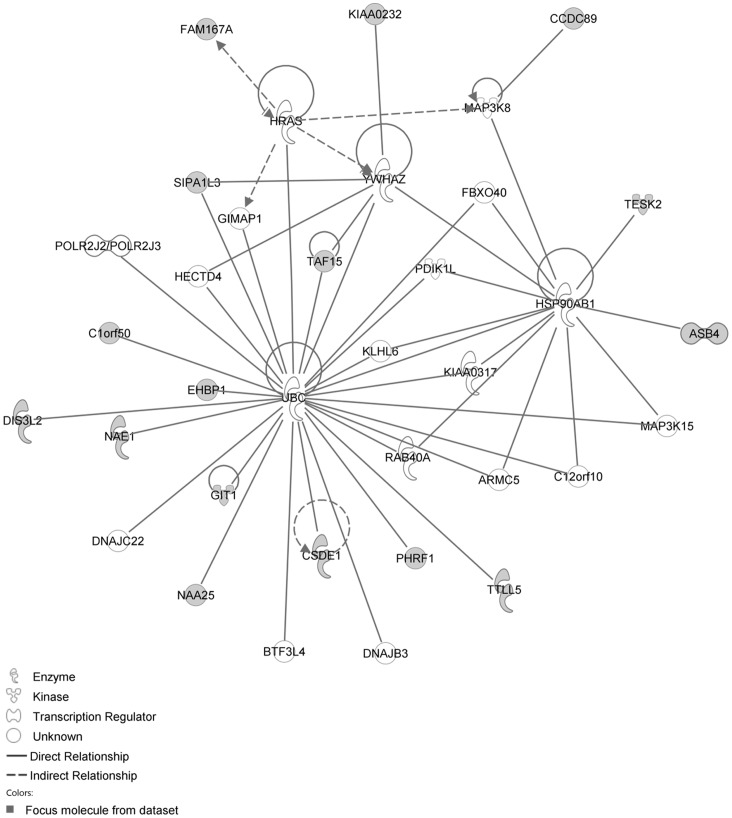
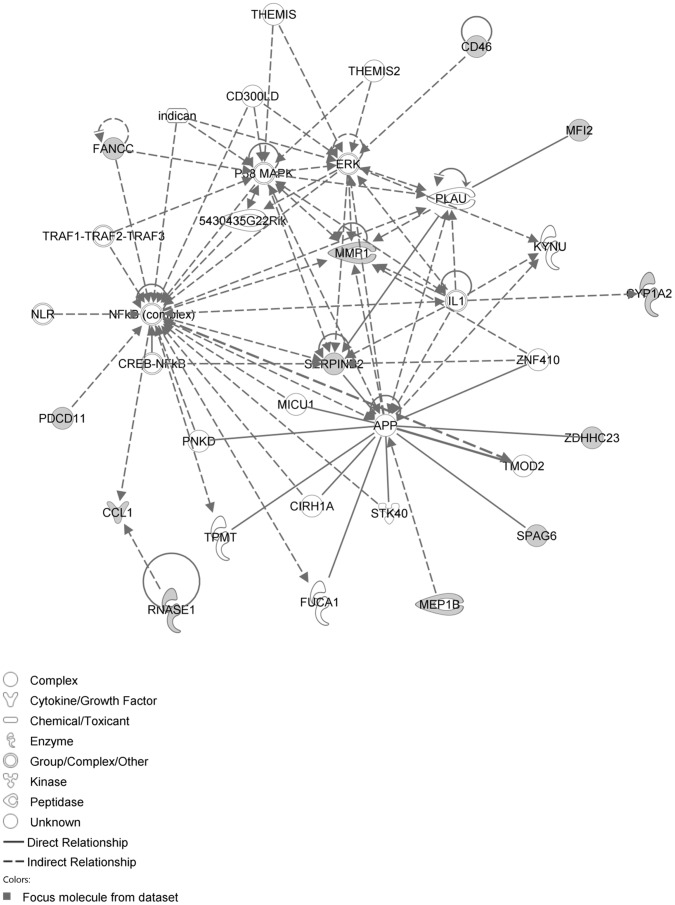
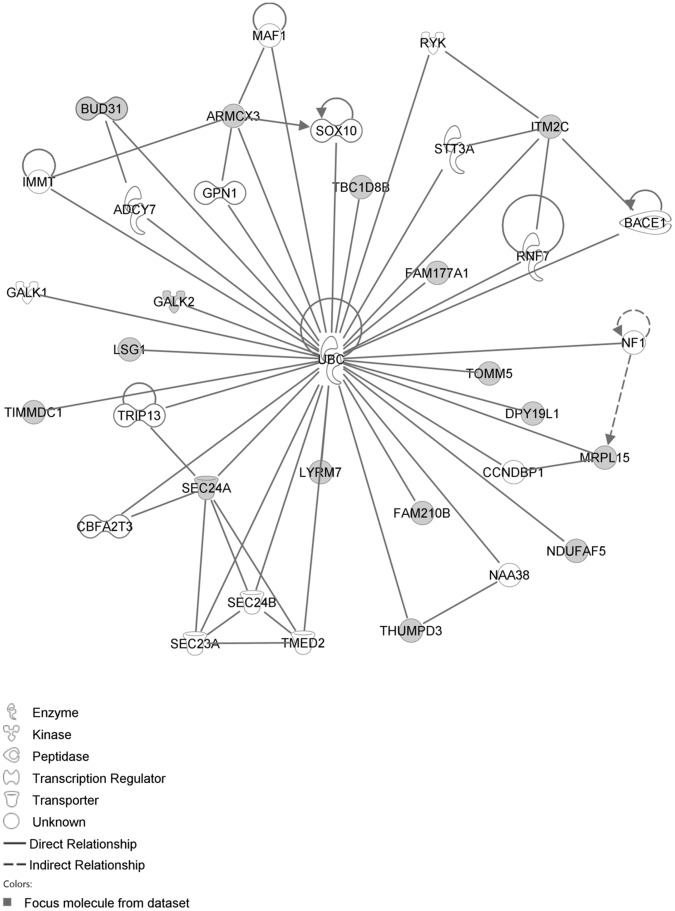
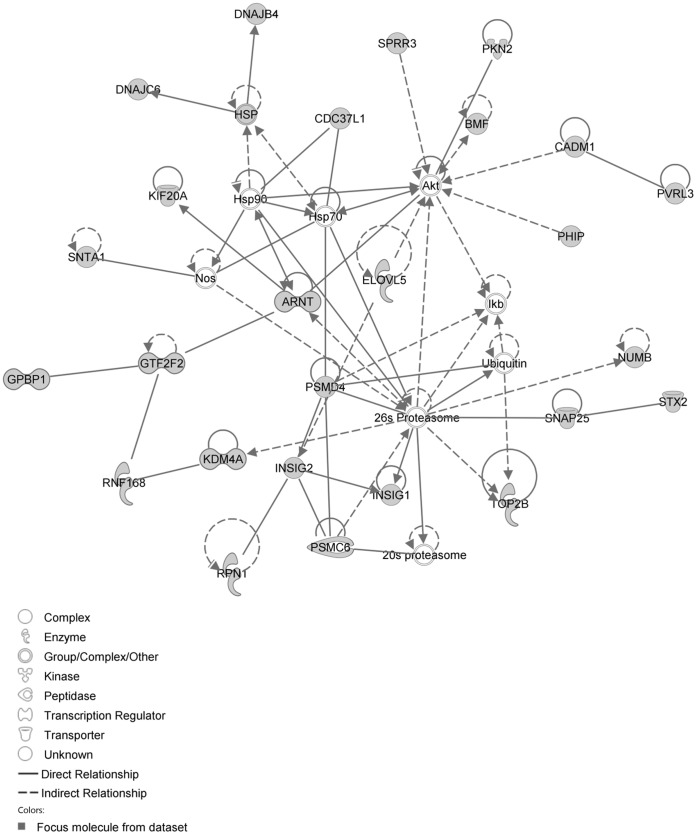
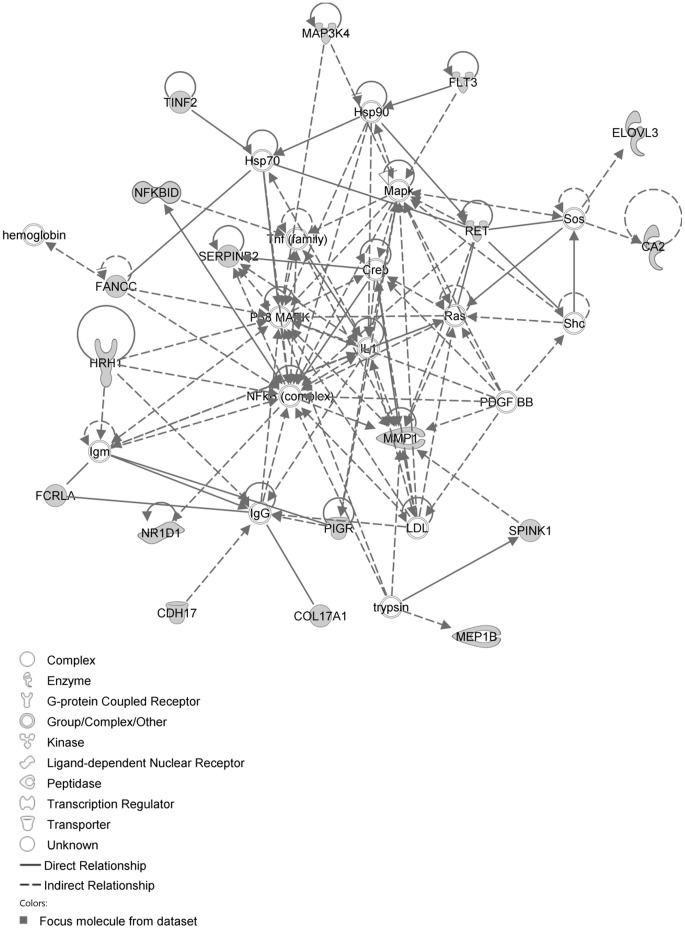
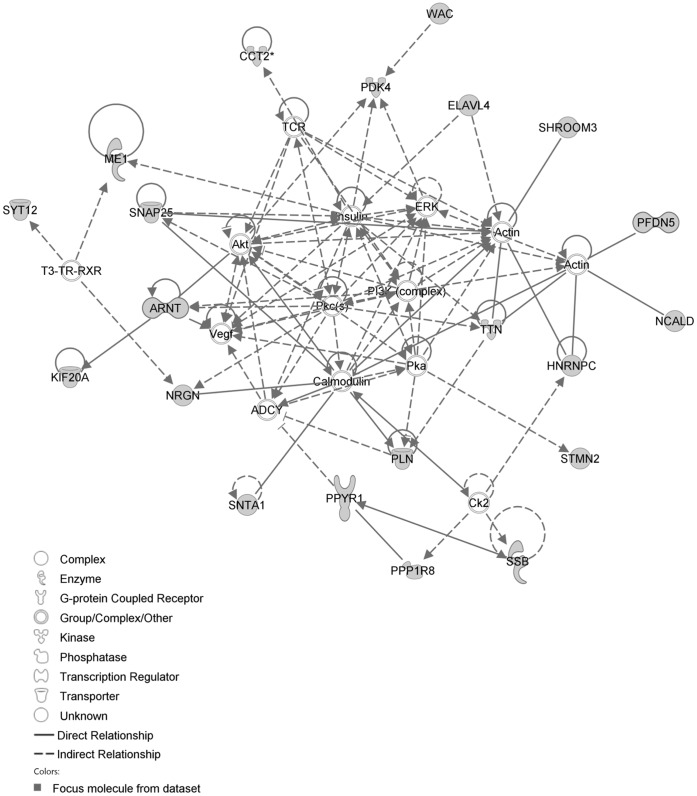
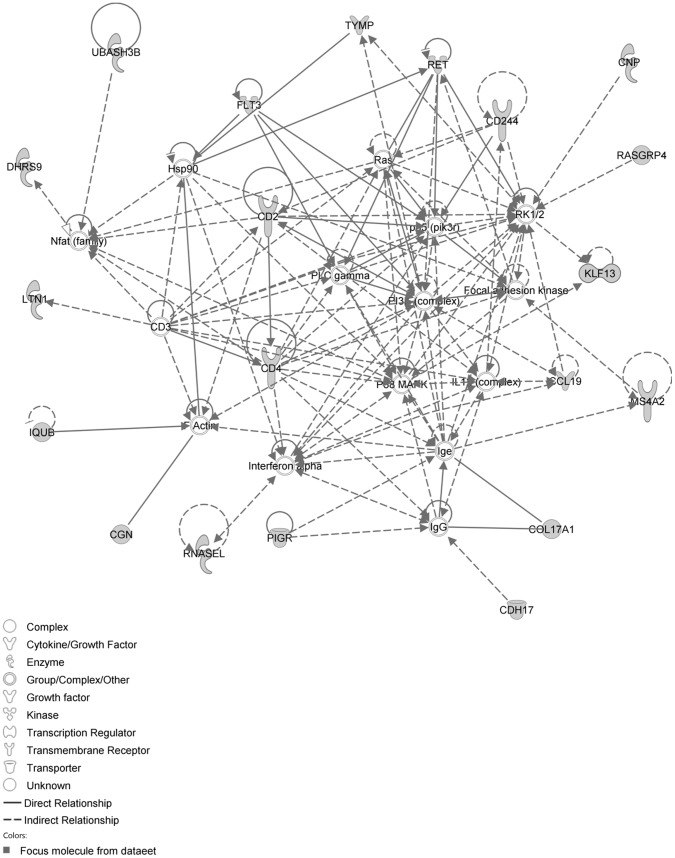
The IPA network with the ‘largest number of up-regulated focus genes’, contained 16 focus genes with functions in Cancer, Connective Tissue Disorders, Skeletal and Muscular Disorders. Focus genes in this network were ASB4, C1orf50, CCDC89, CSDE1, DIS3L2, EHBP1, FAM167A, GIT1, KIAA0232, NAA25, NAE1, PHRF1, SIPA1L3, TAF15, TESK2 and TTLL5. They were related to the ‘node molecule’, UBC (ubiquitin) (Figure 3, Table 1). The network with the second largest number of up-regulated focus genes had 12 focus genes, with functions in Cell-mediated Immune Response, Cellular Development, Cellular Function and Maintenance. Focus genes were CCL1, CD46, CYP1A2, FANCC, MEP1B, MFI2, MMP1, PDCD11, RNASE1, SERPINB2, SPAG6 and ZDHHC23; they were related to P38 MAPK, ERK, NFkB, SERPINB2, MMP1 and APP (Figure 4, Table 1).
Figure 3. IPA network showing the network with the largest number of up-regulated focus genes in the MCA of the hypertension only group, compared with sham operated controls.
Nodes are displayed using various shapes that represent functional classes of gene products. Focus genes in this network are indicated in grey nodes. Solid and dotted lines indicate direct and indirect interactions, respectively.
Figure 4. IPA network showing the network with the second largest number of up-regulated focus genes in the hypertension only group, compared with sham operated controls.
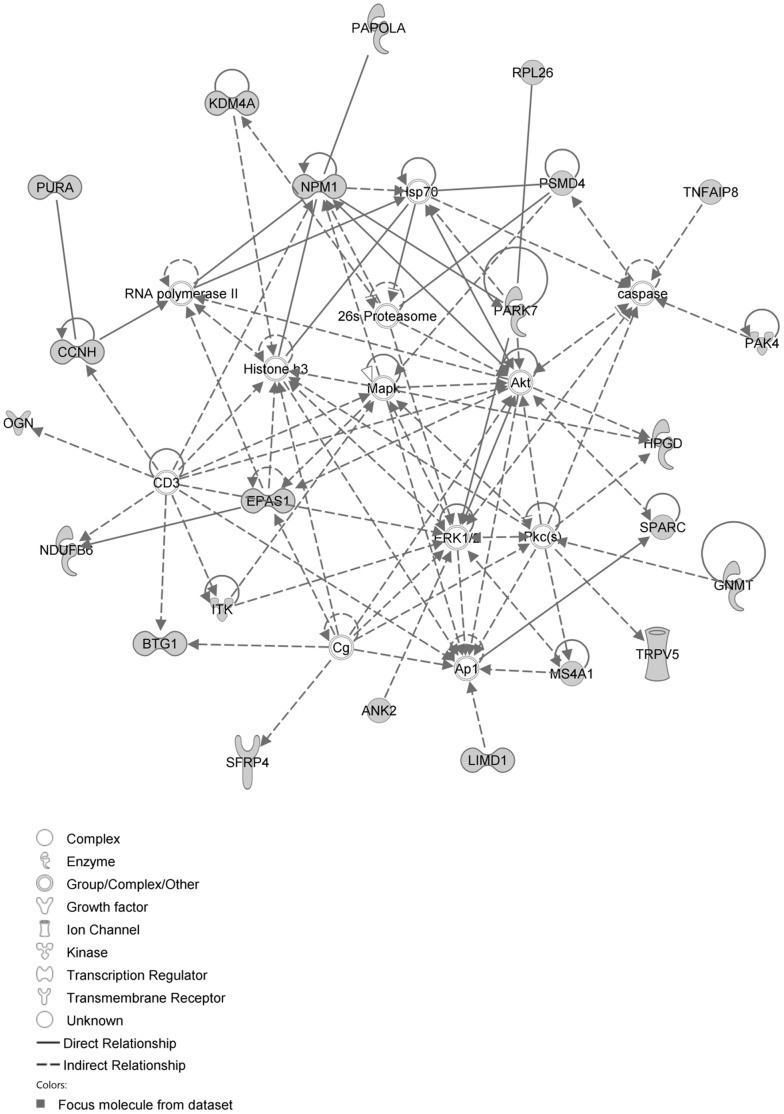
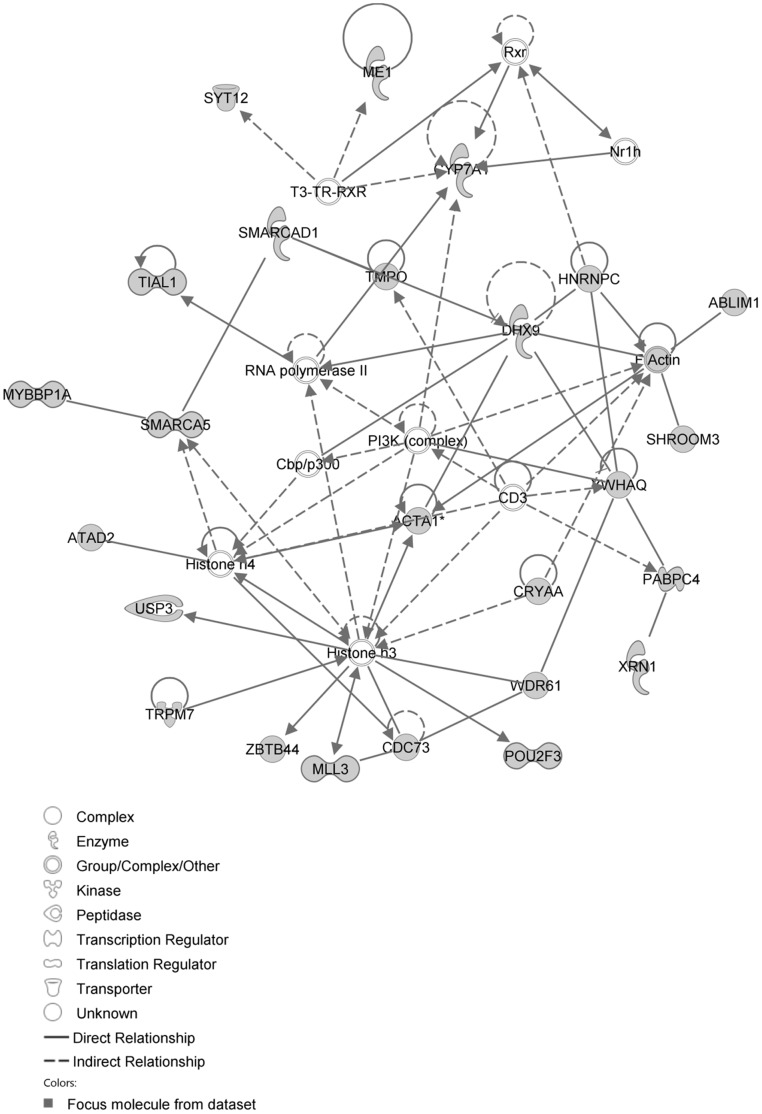
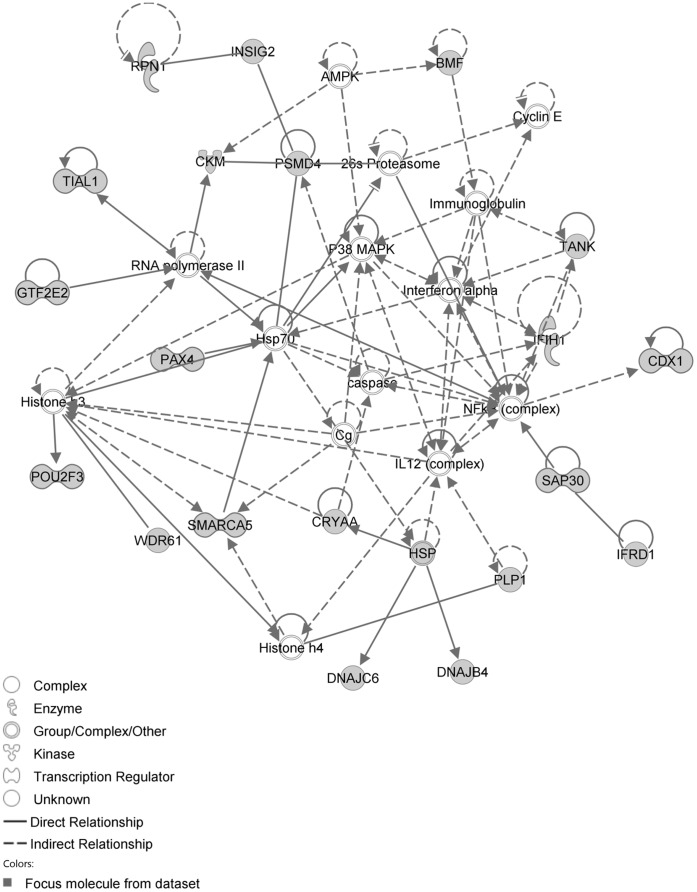
The network with the largest number of down-regulated focus genes contained 23 focus genes with functions in Cardiovascular System Development and Function, Organismal Development, Cell Morphology. Focus genes in this network were ANK2, BTG1, CCNH, EPAS1, GNMT, HPGD, ITK, KDM4A, LIMD1, MS4A1, NDUFB6, NPM1, OGN, PAK4, PAPOLA, PARK7, PSMD4, PURA, RPL26, SFRP4, SPARC, TNFAIP8 and TRPV5. They were related to MAPK, ERK1/2, Akt, 26s proteasome, histone H3 and PKC (Figure 5, Table 2). The network with the second largest number of down-regulated focus genes had 16 focus genes with functions in Cell Death and Survival, Embryonic Development, Cellular Development. Focus genes were ARMCX3, BUD31, DPY19L1, FAM177A1, FAM210B, GALK2, ITM2C, LSG1, LYRM7, MRPL15, NDUFAF5, SEC24A, TBC1D8B, THUMPD3, TIMMDC1 and TOMM5; they were related to UBC (ubiquitin)(Figure 6, Table 2).
Figure 5. IPA network showing the network with the largest number of down-regulated focus genes in the hypertension only group, compared with sham operated controls.
Figure 6. IPA network showing the network with the second largest number of down-regulated focus genes in the hypertension only group, compared with sham operated controls.
2.2. Microarray analyses of the hypercholesterolemia plus sham group
The gene expression profile in the MCA of the hypercholesterolemia plus sham group was compared with that of sham controls on a normal diet. After unknown and repeated genes were omitted, 107 up-regulated and 351 down-regulated genes (greater than 4-fold change) were found (Figure 2). Among the highly up-regulated genes in the MCA of the hypercholesterolemia plus sham group compared to sham controls were SLFN14, CA1, and LOC100357902 (Table 3). Among the highly down-regulated genes were LOC100125984, PFDN5 and CUL3 (Table 4).
Table 3. Up-regulated genes in the MCA of ‘hypercholesterolemia plus sham’ rabbits vs. sham controls with greater than 4-fold change.
| Gene | Gene Symbol | Corrected p-value | Hyperchole-sterolemia plus sham | Hypertension only | Hypertension plus hyperchole-sterolemia |
| Fold Change | Fold Change | Fold Change | |||
| schlafen family member 14 | SLFN14 | 0.00565 | 14.04 | 2.01 | 16.33 |
| carbonic anhydrase I | CA1 | 0.00531 | 10.02 | 2.38 | 10.07 |
| Gap junction alpha-3 protein-like | LOC100357902 | 0.00689 | 9.62 | 2.92 | 9.96 |
| EPH receptor A1 | EPHA1 | 0.00008 | 8.98 | 3.03 | 20.31 |
| N(alpha)-acetyltransferase 25, NatB auxiliary subunit | NAA25∧, + | <0.00001 | 8.91 | 6.43 | 18.66 |
| transmembrane protein 212 | TMEM212 | 0.00039 | 8.84 | 3.48 | 7.93 |
| family with sequence similarity 46, member C | FAM46C | 0.00016 | 8.51 | 1.72 | 6.92 |
| signal-induced proliferation-associated 1 like 3 | SIPA1L3∧, + | 0.00001 | 8.45 | 4.80 | 6.50 |
| SP110 nuclear body protein | SP110 | <0.00001 | 8.42 | 2.06 | 15.47 |
| Uncharacterized protein C11orf71 | C11orf71 | 0.00446 | 8.33 | 2.97 | 9.22 |
| ankyrin and armadillo repeat containing | ANKAR∧, + | 0.00006 | 7.99 | 7.05 | 5.23 |
| teneurin transmembrane protein 4 | TENM4 | 0.00004 | 7.56 | 3.08 | 3.55 |
| sterol O-acyltransferase 2 | SOAT2 | 0.00003 | 7.18 | 3.01 | 5.94 |
| Serine proteinase inhibitor, clade B, member 2 (Predicted) | SERPINB2∧, + | 0.00005 | 6.99 | 5.63 | 9.10 |
| Fc receptor-like A | FCRLA | 0.00083 | 6.76 | 1.94 | 5.14 |
| tetraspanin 33 | TSPAN33 | 0.00078 | 6.37 | 3.97 | 5.53 |
| Glucose-fructose oxidoreductase domain containing 2-like | LOC100351150∧, + | 0.00001 | 6.34 | 4.01 | 12.42 |
| CKLF-like MARVEL transmembrane domain containing 2 | CMTM2 | 0.00071 | 6.28 | 2.38 | 7.79 |
| solute carrier family 9 (sodium/hydrogen exchanger), member 8 | SLC9A8 | 0.00175 | 6.14 | 3.55 | 10.32 |
| spectrin, alpha, erythrocytic 1 (elliptocytosis 2) | SPTA1 | 0.00375 | 6.03 | 2.75 | 8.20 |
| kinesin family member 13B | KIF13B | 0.00002 | 6.02 | 2.35 | 9.02 |
| aminolevulinate, delta-, synthase 2 | ALAS2 | 0.00072 | 5.86 | 1.97 | 5.93 |
| protease, serine, 38 | PRSS38 | 0.00058 | 5.74 | 1.95 | 6.51 |
| UHRF1 binding protein 1 | UHRF1BP1 | 0.00074 | 5.70 | 2.63 | 4.69 |
| Bardet-Biedl syndrome 5 | LOC100342443 | 0.00004 | 5.62 | 2.76 | 8.15 |
| relaxin/insulin-like family peptide receptor 2 | RXFP2∧, + | 0.00029 | 5.62 | 4.79 | 6.96 |
| V-set and immunoglobulin domain containing 2 | VSIG2 | 0.00168 | 5.53 | 3.51 | 4.60 |
| REC8 homolog (yeast) | REC8 | <0.00001 | 5.52 | 1.93 | 4.31 |
| Gonadotropin-releasing hormone receptor | GnRHR | 0.00008 | 5.47 | 3.11 | 10.39 |
| transmembrane and coiled-coil domains 4 | TMCO4 | 0.00007 | 5.42 | 2.43 | 4.28 |
| Serine incorporator 1 | LOC100357075 | 0.00015 | 5.37 | 2.50 | 4.74 |
| transmembrane protein 173 | TMEM173 | 0.00141 | 5.36 | 2.88 | 4.70 |
| solute carrier family 9 (sodium/hydrogen exchanger), member 5 | SLC9A5 | 0.00057 | 5.36 | 2.33 | 4.19 |
| MLX interacting protein | MLXIP | 0.00069 | 5.36 | 3.02 | 4.09 |
| BCL2-like 10 (apoptosis facilitator) | BCL2L10 | 0.00018 | 5.34 | 1.67 | 4.64 |
| follicle stimulating hormone receptor | FSHR | 0.00023 | 5.31 | 1.24 | 8.27 |
| zinc finger, CW type with PWWP domain 2 | ZCWPW2 | 0.00011 | 5.29 | 2.31 | 5.21 |
| Mitogen-activated protein kinase kinase kinase 4 | MAP3K4 | 0.00196 | 5.19 | 3.17 | 4.93 |
| PSMC3 interacting protein | PSMC3IP | 0.00003 | 5.12 | 1.49 | 3.23 |
| meprin A, beta | MEP1B∧, + | 0.00291 | 5.12 | 5.37 | 5.85 |
| testis-specific kinase 2 | TESK2∧ | 0.00571 | 5.11 | 4.50 | 2.41 |
| Double zinc ribbon and ankyrin repeat-containing protein 1 | DZANK1∧, + | <0.00001 | 5.04 | 4.87 | 7.37 |
| nuclear receptor subfamily 1, group D, member 1 | NR1D1 | 0.00562 | 4.98 | 2.35 | 6.11 |
| corin, serine peptidase | CORIN | 0.00006 | 4.93 | 3.59 | 4.63 |
| deltex homolog 3 (Drosophila) | DTX3 | 0.00013 | 4.92 | 3.47 | 4.56 |
| IDI1 protein-like | LOC100346274∧, + | 0.00042 | 4.87 | 5.51 | 6.67 |
| RNA binding motif protein 6 | RBM6 | 0.00003 | 4.85 | 1.73 | 6.15 |
| TNF receptor-associated factor 3 interacting protein 1 | TRAF3IP1 | 0.00043 | 4.80 | 1.29 | 5.56 |
| TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68 kDa | TAF15∧, + | 0.00004 | 4.71 | 7.45 | 4.65 |
| Nibrin-like | LOC100352398∧ | 0.00171 | 4.71 | 7.92 | 3.74 |
| LY6/PLAUR domain containing 5 | LYPD5 | 0.00004 | 4.70 | 2.68 | 7.31 |
| Aquaporin 1 (Colton blood group) | AQP1 | 0.00372 | 4.69 | 1.44 | 2.33 |
| 4-hydroxyphenylpyruvate dioxygenase-like | HPDL | 0.00008 | 4.68 | 2.22 | 8.45 |
| Mast cell-expressed membrane protein 1 | MCEMP1 | 0.00003 | 4.67 | 1.74 | 8.27 |
| amyloid beta (A4) precursor protein-binding, family B, member 3 | APBB3 | <0.00001 | 4.66 | 1.82 | 3.33 |
| chymase like protein | LOC100008644 | 0.00009 | 4.58 | 1.84 | 8.36 |
| transmembrane protease, serine 11E | TMPRSS11E | <0.00001 | 4.57 | 1.26 | 6.76 |
| carboxypeptidase A5 | CPA5 | 0.00007 | 4.56 | 3.29 | 9.11 |
| Fc receptor-like 3 | FCRL3 | 0.00022 | 4.52 | 3.78 | -1.16 |
| Carbonic anhydrase 2 (EC 4.2.1.1) | CA2 | 0.00964 | 4.51 | 1.67 | 4.68 |
| pleckstrin homology domain containing, family G (with RhoGef domain) member 6 | PLEKHG6 | 0.00011 | 4.47 | 3.23 | 9.24 |
| BMS1-like, ribosome assembly protein | LOC100339194 | 0.00003 | 4.46 | 3.51 | 3.99 |
| potassium channel tetramerisation domain containing 19 | KCTD19 | 0.00004 | 4.40 | 1.51 | 6.56 |
| G protein-coupled receptor 52 | GPR52 | 0.00010 | 4.40 | 2.26 | 5.75 |
| cyclin B3 | CCNB3 | 0.00012 | 4.39 | 1.68 | 6.05 |
| Cytochrome P450, family 4, subfamily A, polypeptide 5 | CYP4A5 | 0.00044 | 4.39 | 3.27 | 6.74 |
| kelch-like 17 (Drosophila) | KLHL17 | <0.00001 | 4.38 | 2.72 | 3.65 |
| WD repeat domain 13 | WDR13 | 0.00063 | 4.37 | 2.31 | 3.14 |
| O-sialoglycoprotein endopeptidase-like | LOC100348675 | 0.00039 | 4.36 | 3.87 | 3.71 |
| Dmx-like 2 | DMXL2 | 0.00657 | 4.36 | 1.23 | 3.79 |
| CD2 molecule | CD2 | 0.00002 | 4.35 | 1.47 | 8.04 |
| NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75 kDa | LOC100341941 | 0.00022 | 4.34 | 2.69 | 5.46 |
| DIS3 mitotic control homolog (S. cerevisiae)-like 2 | DIS3L2∧ | 0.00024 | 4.32 | 4.17 | 3.96 |
| general transcription factor IIE, polypeptide 2, beta 34 kDa | GTF2E2 | 0.00360 | 4.29 | 2.33 | 3.63 |
| doublesex and mab-3 related transcription factor 2 | DMRT2 | 0.00002 | 4.28 | 1.81 | 7.32 |
| ribonuclease, RNase A family, 1 (pancreatic) | RNASE1∧, + | 0.00010 | 4.26 | 3.23 | 3.15 |
| Paralemmin 2 | PALM2 | 0.00116 | 4.26 | 2.43 | 3.57 |
| Fanconi anemia, complementation group C | FANCC∧, + | <0.00001 | 4.25 | 6.10 | 6.39 |
| collagen, type VII, alpha 1 | COL7A1 | 0.00600 | 4.25 | 2.00 | 2.98 |
| Serine/threonine kinase 23, muscle-specific serine kinase 1 70 (Predicted) | SRPK3 | 0.00027 | 4.23 | 1.35 | 8.51 |
| O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase | OGT | <0.00001 | 4.20 | 1.98 | 3.65 |
| SP100 nuclear antigen | SP100 | 0.00039 | 4.19 | 3.36 | 3.25 |
| Uncharacterized protein C1orf51 | C1orf51 | 0.00039 | 4.19 | 2.02 | 3.10 |
| Islet amyloid polypeptide Precursor; Fragment (Amylin) | IAPP | 0.00004 | 4.17 | 1.40 | 5.78 |
| family with sequence similarity 19 (chemokine (C-C motif)-like), member A4 | FAM19A4 | 0.00024 | 4.17 | 1.65 | 5.97 |
| MORN repeat containing 4 | MORN4 | <0.00001 | 4.14 | 1.45 | 4.53 |
| leucine-rich repeat-containing G protein-coupled receptor 5 | LGR5 | 0.00019 | 4.14 | 3.32 | 5.36 |
| spleen tyrosine kinase | SYK | 0.00072 | 4.13 | 1.52 | 4.88 |
| guanylate cyclase 2D, membrane (retina-specific) | GUCY2D∧, + | 0.00008 | 4.11 | 4.64 | 8.34 |
| Pregnancy-zone protein | PZP | 0.00099 | 4.11 | 1.20 | 4.60 |
| methyl-CpG binding domain protein 6 | MBD6 | <0.00001 | 4.11 | 2.26 | 3.11 |
| TERF1 (TRF1)-interacting nuclear factor 2 | TINF2 | 0.00011 | 4.10 | 3.47 | 6.41 |
| ankyrin repeat and SOCS box-containing 4 | ASB4∧ | 0.00027 | 4.06 | 4.98 | 3.51 |
| tripartite motif-containing 65 | TRIM65 | 0.00079 | 4.05 | 2.63 | 4.87 |
| inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon | IKBKE | 0.00003 | 4.04 | 1.93 | 3.39 |
| coiled-coil domain containing 54 | CCDC54 | 0.00196 | 4.04 | 3.11 | 4.57 |
| arylformamidase | AFMID | 0.00100 | 4.03 | 1.65 | 5.81 |
| 3-oxo-5-alpha-steroid 4-dehydrogenase 2-like | LOC100343882 | 0.00403 | 4.02 | 2.74 | 3.56 |
| Fibroblast growth factor binding protein 1-like | LOC100353835∧, + | 0.00122 | 4.01 | 5.01 | 4.63 |
| ATPase, Cu++ transporting, alpha polypeptide | ATP7A | 0.00404 | 4.01 | 1.59 | 2.58 |
| 2,3-bisphosphoglycerate mutase (BPGM) | BPGM | 0.00098 | 4.00 | 1.21 | 4.18 |
∧ Differentially expressed genes that are common between ‘hypertension only’ and ‘hypercholesterolemia plus sham’ groups (both vs. sham controls).+Differentially expressed genes that are common among ‘hypertension only’, ‘hypercholesterolemia plus sham’, and ‘hypertension plus hypercholesterolemia’ groups (all vs. sham controls).
Table 4. Down-regulated genes in the MCA of ‘hypercholesterolemia plus sham’ rabbits vs. sham controls with greater than 4-fold change.
| Gene | GeneSymbol | Corrected p-value | Hyperchole-sterolemia plus sham | Hypertension only | Hypertension plus hyperchole-sterolemia |
| Fold Change | Fold Change | Fold Change | |||
| beta tropomyosin (LOC100125984) | LOC100125984 | <0.00001 | −97.92 | −2.30 | −17.13 |
| prefoldin subunit 5 | PFDN5 | <0.00001 | −84.52 | −1.78 | −6.79 |
| cullin 3 | CUL3 | <0.00001 | −73.37 | 1.05 | −6.10 |
| family with sequence similarity 184, member B | FAM184B | <0.00001 | −68.05 | −1.32 | −25.72 |
| crystallin, alpha A (CRYAA) | CRYAA | <0.00001 | −60.93 | −1.50 | −10.81 |
| forkhead box N1 | FOXN1∧, + | 0.00048 | −56.89 | −26.20 | −45.12 |
| Tumor necrosis factor receptor superfamily, member 11b | TNFRSF11B | <0.00001 | −55.91 | 1.69 | −33.58 |
| Glutamate dehydrogenase 1-like | LOC100351029 | <0.00001 | −55.82 | 1.18 | −3.96 |
| 3′(2′), 5′-bisphosphate nucleotidase 1 | BPNT1 | <0.00001 | −52.27 | 1.05 | −25.01 |
| interferon-related developmental regulator 1 | IFRD1 | <0.00001 | −51.14 | 1.33 | −4.37 |
| manganese superoxide dismutase | SOD-2 | <0.00001 | −46.42 | −1.23 | −14.80 |
| GC-rich promoter binding protein 1 | GPBP1 | <0.00001 | −42.73 | −2.55 | −3.71 |
| microtubule associated tumor suppressor 1 | MTUS1 | <0.00001 | −41.71 | −1.88 | −20.40 |
| ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide-like 1 | UQCRFS1P1 | <0.00001 | −40.99 | 1.08 | −23.74 |
| Lupus La protein | SSB | <0.00001 | −40.45 | −1.63 | −10.68 |
| transmembrane protein 109 (TMEM109) | TMEM109 | <0.00001 | −39.14 | −1.07 | −4.26 |
| Progesterone receptor membrane component 1 | LOC100357097 | <0.00001 | −38.66 | 1.04 | −21.91 |
| Solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 | SLC9A2 | 0.00007 | −36.54 | −2.62 | −7.21 |
| Obg-like ATPase 1 | OLA1 | <0.00001 | −35.58 | 1.04 | −2.87 |
| Chaperonin containing TCP1, subunit 2 (beta) | CCT2 | <0.00001 | −35.56 | −1.91 | −30.96 |
| androgen-induced 1 | AIG1 | 0.00002 | −35.48 | −3.30 | −14.63 |
| endoplasmic reticulum lectin 1 | ERLEC1 | <0.00001 | −34.54 | 1.35 | −2.93 |
| glyceraldehyde-3-phosphate dehydrogenase, spermatogenic | GAPDHS | <0.00001 | −33.66 | 1.01 | −32.91 |
| transmembrane protein 38B | TMEM38B | <0.00001 | −31.53 | 1.05 | −18.80 |
| Alpha-tubulin N-acetyltransferase | ATAT1 | <0.00001 | −29.43 | 1.37 | −2.20 |
| OTU domain containing 6A | OTUD6A | 0.00011 | −28.99 | −3.59 | −5.70 |
| chaperonin containing TCP1, subunit 2 (beta) (CCT2) | CCT2 | <0.00001 | −28.67 | −1.23 | −4.57 |
| Lysosomal-associated membrane protein 2-like | LOC100350946 | <0.00001 | −26.85 | −1.78 | −15.88 |
| tumor protein, translationally-controlled 1 (TPT1) | TPT1 | 0.00008 | −26.77 | −1.45 | −1.80 |
| Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide-like | LOC100358299 | <0.00001 | −26.07 | −1.13 | −3.37 |
| AHA1, activator of heat shock 90kDa protein ATPase homolog 2 (yeast) | AHSA2 | 0.00081 | −23.77 | −3.66 | −5.22 |
| mitochondrial ribosomal protein L15 | MRPL15∧, + | <0.00001 | −23.01 | −6.97 | −9.57 |
| zinc finger protein 638 | ZNF638 | <0.00001 | −22.80 | −1.10 | −2.82 |
| proteasome (prosome, macropain) 26s subunit, ATPase, 6 | PSMC6 | <0.00001 | −22.71 | −1.36 | −3.80 |
| v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | KRAS | <0.00001 | −22.11 | −1.18 | −15.47 |
| myosin IE | MYO1E | <0.00001 | −21.50 | 1.35 | −2.84 |
| hematopoietic prostaglandin D synthase | HPGDS∧, + | 0.00043 | −21.44 | −12.38 | −13.63 |
| insulin induced gene 2 | INSIG2 | <0.00001 | −21.07 | −3.78 | −4.71 |
| Putative homeodomain transcription factor 2 | PHTF2 | <0.00001 | −20.84 | −1.69 | −3.07 |
| Cytoplasmic beta-actin | LOC100009506 | <0.00001 | −20.51 | −1.19 | −7.80 |
| TRAF family member-associated NFKB activator | TANK | 0.00001 | −20.39 | −1.28 | −5.66 |
| zinc finger protein 642 | ZFP69 | 0.00058 | −20.24 | −1.58 | −12.02 |
| SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 | SMARCA5 | <0.00001 | −19.50 | 1.08 | −8.08 |
| Transmembrane protein 14C-like | LOC100357487 | 0.00875 | −18.42 | 1.43 | −1.74 |
| zinc finger protein 326 | ZNF326 | <0.00001 | −18.36 | 1.09 | −6.29 |
| DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) | DCUN1D4 | <0.00001 | −18.27 | −1.65 | −15.90 |
| PHD finger protein 6 | PHF6 | 0.00001 | −17.91 | −1.84 | −3.80 |
| CD302 molecule | CD302 | <0.00001 | −17.51 | −1.14 | −4.14 |
| ORM1-like 3 (S. cerevisiae) | ORMDL3 | <0.00001 | −17.49 | −1.24 | −16.75 |
| large subunit GTPase 1 homolog (S. cerevisiae) | LSG1∧ | 0.00001 | −17.14 | 1.16 | −2.15 |
| FGGY carbohydrate kinase domain containing | FGGY | 0.00007 | −16.26 | −2.04 | −3.77 |
| Eukaryotic translation initiation factor 2 subunit 1 | EIF2S1 | 0.00001 | −16.08 | −2.04 | −14.40 |
| ribophorin I | RPN1 | 0.00002 | −15.81 | −1.30 | −4.51 |
| periphilin 1 | PPHLN1 | <0.00001 | −15.71 | 1.14 | −2.19 |
| mitochondrial ribosomal protein L1 | MRPL1 | <0.00001 | −15.71 | −1.06 | −12.00 |
| tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide (YWHAQ) | YWHAQ | <0.00001 | −15.13 | −1.44 | −2.31 |
| Sin3A-associated protein, 30 kDa | SAP30 | <0.00001 | −14.99 | −1.09 | −16.96 |
| ribosomal protein L26 | RPL26∧ | 0.00016 | −14.60 | −5.27 | −2.81 |
| amyloid beta precursor protein (cytoplasmic tail) binding protein 2 | APPBP2 | <0.00001 | −14.30 | −1.11 | −2.02 |
| nucleolar protein 10 | NOL10 | <0.00001 | −14.02 | 1.08 | −5.15 |
| TIA1 cytotoxic granule-associated RNA binding protein-like 1 | TIAL1 | 0.00001 | −13.80 | 2.34 | −4.88 |
| Titin | TTN | 0.00001 | −13.60 | −2.26 | −6.90 |
| asparagine-linked glycosylation 9, alpha-1,2-mannosyltransferase homolog (S. cerevisiae) | ALG9 | <0.00001 | −13.51 | −1.33 | −2.12 |
| Aryl hydrocarbon receptor nuclear translocator (ARNT protein)(Dioxin receptor, nuclear translocator)(Hypoxia-inducible factor 1 beta)(HIF-1 beta) | ARNT | <0.00001 | −13.43 | 1.04 | −5.12 |
| BRCA2 and CDKN1A interacting protein | BCCIP∧ | <0.00001 | −13.26 | −1.00 | −2.51 |
| WW domain containing adaptor with coiled-coil | WAC | 0.00001 | −13.20 | −3.12 | −11.91 |
| Uncharacterized LOC100347346 | LOC100347346 | 0.00002 | −13.05 | 1.33 | −2.30 |
| Tweety homolog 1 (Drosophila) | TTYH1 | 0.00108 | −12.82 | −5.16 | −27.02 |
| ubiquitin specific peptidase 16 | USP16 | <0.00001 | −12.55 | 1.16 | −1.82 |
| UDP-glucuronosyltransferase | UGT2C1 | <0.00001 | −12.36 | 2.01 | −2.07 |
| Heterogeneous nuclear ribonucleoprotein C (hnRNP C) | HNRNPC | <0.00001 | −12.22 | −1.33 | −7.49 |
| Nucleolar protein 11 | NOL11 | 0.00032 | −11.98 | 2.09 | −13.01 |
| guanylate cyclase 1, soluble, beta 3 | GUCY1B3 | 0.00010 | −11.97 | −2.52 | −6.79 |
| SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 | SMARCD3 | <0.00001 | −11.91 | −1.18 | −2.86 |
| up-regulated during skeletal muscle growth 5 homolog (mouse) | USMG5 | <0.00001 | −11.86 | −1.47 | −2.36 |
| endothelial differentiation gene 7 protein | LOC100009484 | 0.00562 | −11.73 | −1.86 | −7.39 |
| cysteine-rich protein 3 | CRIP3 | <0.00001 | −10.99 | 1.45 | −4.26 |
| interferon induced with helicase C domain 1 | IFIH1 | 0.00001 | −10.96 | 1.21 | −5.60 |
| Solute carrier family 2, facilitated glucose transporter member 3 Fragment (Glucose transporter type 3, brain)(GLUT-3) | SLC2A3 | 0.00019 | −10.95 | −1.81 | −11.60 |
| eukaryotic translation initiation factor 3, subunit M | EIF3M | <0.00001 | −10.81 | −1.38 | −2.09 |
| PDLIM1 interacting kinase 1 like | PDIK1L | 0.00010 | −10.76 | −2.49 | −2.61 |
| F-box and WD repeat domain containing 7 | FBXW7 | <0.00001 | −10.71 | 1.36 | −2.36 |
| IQ motif containing GTPase activating protein 3 | IQGAP3 | 0.00007 | −10.54 | 1.25 | −9.37 |
| caudal type homeobox 1 | CDX1 | 0.00001 | −10.42 | −1.18 | −6.08 |
| RUN and FYVE domain containing 1 | RUFY1 | 0.00004 | −10.38 | −1.02 | −2.46 |
| lysine (K)-specific demethylase 4A | KDM4A | <0.00001 | −10.29 | −2.03 | −3.47 |
| RWD domain containing 3 | RWDD3 | <0.00001 | −10.16 | 1.03 | −2.04 |
| syntrophin, alpha 1 (dystrophin-associated protein A1, 59 kDa, acidic component) (SNTA1) | SNTA1 | <0.00001 | −9.97 | −1.04 | −5.98 |
| secretion regulating guanine nucleotide exchange factor | SERGEF | 0.00011 | −9.83 | −1.08 | −1.71 |
| dehydrogenase/reductase (SDR family) member 13 | DHRS13 | <0.00001 | −9.82 | 1.02 | −4.23 |
| SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 | SMARCA1 | <0.00001 | −9.77 | 1.18 | −2.16 |
| RIO kinase 2 | LOC100347237 | <0.00001 | −9.56 | −1.53 | −3.14 |
| PDZD4 protein (Predicted) | PDZD4 | 0.00005 | −9.45 | −1.47 | −3.15 |
| adaptor-related protein complex 4, sigma 1 subunit | AP4S1 | <0.00001 | −9.37 | −1.09 | −2.92 |
| neurocalcin delta | NCALD | 0.00037 | −9.22 | −1.93 | −6.13 |
| Bcl2 modifying factor | BMF | <0.00001 | −9.18 | −1.04 | −4.55 |
| signal recognition particle 54 kDa | SRP54 | <0.00001 | −9.16 | 1.22 | −1.88 |
| Cytochrome P450, family 7, subfamily A, polypeptide 1 | CYP7A1 | <0.00001 | −9.16 | 1.93 | −3.27 |
| ring finger protein 38 | RNF38 | 0.00023 | −9.14 | −3.33 | −4.93 |
| Zinc finger protein 800 (Predicted) | ZNF800 | 0.00004 | −9.12 | −1.81 | −2.07 |
| DnaJ (Hsp40) homolog, subfamily B, member 4 | DNAJB4 | <0.00001 | −9.11 | −1.86 | −4.76 |
| claudin 16 | CLDN16 | 0.00014 | −9.09 | −1.23 | −3.61 |
| WW domain containing E3 ubiquitin protein ligase 1 | WWP1 | <0.00001 | −9.02 | −1.11 | −2.33 |
| Actin, alpha skeletal muscle (Alpha-actin-1) | ACTA1 | 0.00001 | −8.93 | −1.11 | −3.08 |
| TANK-binding kinase 1 | TBK1 | 0.00002 | −8.82 | −1.66 | −2.80 |
| IQ motif and Sec7 domain 3 | IQSEC3 | 0.00193 | −8.73 | −2.18 | −3.18 |
| karyopherin alpha 4 (importin alpha 3) | KPNA4 | <0.00001 | −8.63 | −1.27 | −1.92 |
| MACRO domain containing 2 | MACROD2 | 0.00003 | −8.51 | −1.57 | −2.19 |
| transmembrane protein 70 | TMEM70 | 0.00001 | −8.48 | −1.36 | −3.70 |
| SRY (sex determining region Y)-box 5 | SOX5 | 0.00036 | −8.38 | −1.20 | −2.75 |
| ring finger protein 222 | RNF222 | 0.00001 | −8.36 | −1.78 | −6.02 |
| Annexin A2 | LOC100357671 | 0.00008 | −8.32 | 1.11 | −3.60 |
| enabled homolog (Drosophila) | ENAH | 0.00036 | −8.30 | 1.27 | −2.87 |
| cell division cycle 37 homolog (S. cerevisiae)-like 1 | CDC37L1 | 0.00001 | −8.25 | −2.44 | −3.73 |
| zinc finger RNA binding protein | ZFR | <0.00001 | −8.22 | −1.06 | −2.45 |
| protein kinase C, beta (PRKCB), transcript variant II | PRKCB | 0.00026 | −8.21 | 3.41 | −3.02 |
| tetratricopeptide repeat domain 21B | TTC21B | <0.00001 | −8.19 | 1.01 | −3.64 |
| shroom family member 3 | SHROOM3 | <0.00001 | −7.95 | −3.82 | −4.60 |
| phospholamban (LOC100009299) | PLN | 0.00002 | −7.94 | −2.00 | −4.25 |
| Thioredoxin interacting protein | TXNIP | 0.00017 | −7.93 | −1.22 | −2.37 |
| Destrin | LOC100358081 | <0.00001 | −7.84 | −1.07 | −2.32 |
| Programmed cell death 6 interacting protein | LOC100341540 | 0.00018 | −7.83 | −1.88 | −3.78 |
| DEAH (Asp-Glu-Ala-His) box polypeptide 9 | DHX9 | <0.00001 | −7.81 | 1.34 | −2.39 |
| MAX dimerization protein 1 | MXD1 | 0.00035 | −7.73 | −1.51 | −3.29 |
| Acidic (leucine-rich) nuclear phosphoprotein 32 family, member E | LOC100349957 | 0.00032 | −7.65 | −1.50 | −2.24 |
| protein kinase N2 | PKN2 | 0.00001 | −7.59 | −1.47 | −3.08 |
| 5′-3′ exoribonuclease 1 | XRN1 | 0.00029 | −7.58 | −1.69 | −2.98 |
| NADP-dependent oxidoreductase domain-containing protein 1 | NOXRED1 | 0.00140 | −7.55 | −3.36 | −3.35 |
| adenosine deaminase, tRNA-specific 2, TAD2 homolog (S. cerevisiae) | ADAT2 | 0.00032 | −7.55 | 1.09 | −2.38 |
| NADH dehydrogenase-like | LOC100347823 | <0.00001 | −7.55 | 2.02 | −7.93 |
| Cold shock domain containing E1, RNA-binding | CSDE1 | 0.00001 | −7.53 | −2.74 | −3.37 |
| IQ motif containing E | IQCE | <0.00001 | −7.52 | 1.75 | −5.18 |
| POU class 2 homeobox 3 | POU2F3 | <0.00001 | −7.48 | 1.89 | −4.78 |
| H6 family homeobox 2 | HMX2 | 0.00121 | −7.46 | −1.58 | −1.86 |
| Kruppel-like factor 1 (erythroid) | KLF1 | <0.00001 | −7.43 | 1.07 | −3.10 |
| AP2 associated kinase 1 | AAK1 | 0.00571 | −7.42 | −2.56 | −3.14 |
| zinc finger protein 330 | ZNF330 | 0.00012 | −7.41 | −1.48 | −8.54 |
| NudC domain containing 2 | NUDCD2 | <0.00001 | −7.37 | −1.41 | −2.72 |
| spermatogenesis associated 24 | SPATA24 | 0.00015 | −7.37 | −1.43 | −3.75 |
| GC-rich sequence DNA-binding factor candidate isoform 1 (Predicted) | RA_m005_jsm426FEr | 0.00019 | −7.35 | 1.03 | −1.46 |
| gap junction protein, delta 3, 31.9 kDa | GJD3 | <0.00001 | −7.33 | 1.35 | −4.40 |
| zinc finger, MYM-type 1 | ZMYM1 | 0.00017 | −7.33 | −1.60 | −3.17 |
| cytochrome P450 IIC16 mRNA (CYP2C16) | CYP2C16 | <0.00001 | −7.31 | 2.43 | −3.15 |
| ASF1 anti-silencing function 1 homolog A (Predicted) | ASF1A | <0.00001 | −7.31 | −1.11 | −2.33 |
| protein phosphatase 1, regulatory (inhibitor) subunit 8 | PPP1R8 | <0.00001 | −7.27 | 1.09 | −4.24 |
| ADAM metallopeptidase with thrombospondin type 1 motif, 17 | ADAMTS17∧ | 0.00001 | −7.25 | −15.85 | −3.06 |
| myosin, light chain 6B, alkali, smooth muscle and non-muscle | MYL6B | 0.00002 | −7.23 | −1.58 | −4.66 |
| bone morphogenetic protein 7 (bmp7) | BMP7 | <0.00001 | −7.23 | −1.14 | −2.05 |
| clathrin, light chain A | CLTA | <0.00001 | −7.22 | −1.05 | −2.84 |
| pancreatic polypeptide receptor 1 (PPYR1) | PPYR1 | 0.00003 | −7.22 | −1.10 | −4.12 |
| sal-like 1 (Drosophila) | SALL1 | <0.00001 | −7.20 | −1.03 | −3.00 |
| Crystallin, zeta-like 1 (Predicted) | CRYZL1 | <0.00001 | −7.20 | −1.58 | −2.86 |
| junctional adhesion molecule 2 | JAM2 | 0.00121 | −7.11 | −2.54 | −3.40 |
| potassium channel tetramerisation domain containing 15 | KCTD15 | <0.00001 | −7.11 | −1.16 | −3.93 |
| WW domain binding protein 4 (formin binding protein 21) | WBP4 | <0.00001 | −7.08 | −1.63 | −2.77 |
| actin, alpha 1, skeletal muscle | ACTA1 | 0.00003 | −7.04 | −1.28 | −2.10 |
| host cell factor C1 regulator 1 (XPO1 dependent) | HCFC1R1 | 0.00001 | −6.97 | −1.52 | −3.01 |
| integrin, beta 8 (ITGB8) | ITGB8 | 0.00098 | −6.93 | −1.98 | −2.79 |
| OTU domain containing 7A | OTUD7A | <0.00001 | −6.87 | −1.30 | −2.70 |
| THAP domain containing 6 | THAP6 | 0.00024 | −6.86 | −2.75 | −2.73 |
| sialic acid binding Ig-like lectin 14 | SIGLEC14 | <0.00001 | −6.80 | 1.14 | −4.41 |
| nucleolar protein 8 | NOL8 | 0.00005 | −6.74 | −1.62 | −2.14 |
| epithelial cell transforming sequence 2 oncogene | ECT2 | 0.00003 | −6.73 | 1.12 | −2.57 |
| WD repeat domain 61 | WDR61 | 0.00029 | −6.72 | −1.07 | −1.47 |
| Ribosomal protein L7-like | LOC100339887 | 0.00001 | −6.70 | −1.23 | −5.10 |
| kinesin family member 20A | KIF20A | 0.00003 | −6.68 | −2.21 | −5.16 |
| ELOVL family member 5, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast) | ELOVL5 | <0.00001 | −6.66 | −1.34 | −2.32 |
| phospholipase A2, group IVC (cytosolic, calcium-independent) | PLA2G4C | 0.00006 | −6.55 | −1.01 | −3.36 |
| complement component 1, q subcomponent-like 3 | C1QL3 | 0.00293 | −6.49 | −2.42 | −6.91 |
| ribosomal protein L3-like | RPL3L | <0.00001 | −6.49 | 1.11 | −2.23 |
| actin, beta-like 2 | ACTBL2 | 0.00008 | −6.44 | −3.04 | −2.71 |
| proteolipid protein 1 (PLP1) | PLP1 | 0.00339 | −6.44 | −1.79 | −6.50 |
| DnaJ (Hsp40) homolog, subfamily C, member 6 | DNAJC6 | 0.00010 | −6.43 | −2.42 | −6.45 |
| zinc finger and BTB domain containing 44 | ZBTB44 | 0.00001 | −6.43 | −1.92 | −2.58 |
| poliovirus receptor-related 3 | PVRL3 | <0.00001 | −6.30 | 1.08 | −2.09 |
| protein phosphatase, EF-hand calcium binding domain 1 | PPEF1 | 0.00008 | −6.28 | −1.45 | −2.25 |
| Acidic ribosomal phosphoprotein P0 | 36B4 | 0.00003 | −6.27 | 1.43 | −4.94 |
| peptidylprolyl isomerase G (cyclophilin G) | PPIG∧ | <0.00001 | −6.20 | −12.61 | −1.81 |
| mediator complex subunit 19 | MED19 | 0.00002 | −6.16 | −1.27 | −2.30 |
| complement component 1, r subcomponent-like | C1RL | <0.00001 | −6.15 | 1.42 | −1.33 |
| homeobox C11 | HOXC11 | 0.00026 | −6.13 | −1.10 | −3.83 |
| Cell division cycle 37 protein | LOC100344117 | 0.00041 | −6.12 | 1.19 | −3.81 |
| glycine-N-acyltransferase-like 3 | GLYATL3 | 0.00001 | −6.10 | −1.23 | −2.57 |
| family with sequence similarity 177, member A1 | FAM177A1∧ | <0.00001 | −6.05 | −1.32 | −1.70 |
| poly(rC) binding protein 3 | PCBP3 | 0.00109 | −6.03 | −1.27 | −3.94 |
| KIAA0305-like | LOC100358165 | <0.00001 | −6.02 | −1.11 | −1.64 |
| dymeclin | DYM | <0.00001 | −5.97 | −1.57 | −3.07 |
| inositol polyphosphate phosphatase-like 1 | INPPL1 | 0.00005 | −5.96 | −1.14 | −4.87 |
| enolase 2 (gamma, neuronal) | ENO2 | <0.00001 | −5.92 | 1.21 | −2.24 |
| bactericidal/permeability-increasing protein-like 3 | BPIFB6 | 0.00001 | −5.92 | −1.79 | −6.83 |
| klotho (KL) | KL | 0.00379 | −5.89 | −2.06 | −2.74 |
| ELMO/CED-12 domain containing 2 | ELMOD2 | 0.00024 | −5.87 | −2.02 | −2.22 |
| Proteasome (prosome, macropain) 26s subunit, non-ATPase, 4 | PSMD4∧, + | <0.00001 | −5.84 | −1.49 | −9.66 |
| pleckstrin homology domain interacting protein | PHIP | 0.00039 | −5.84 | −2.72 | −3.73 |
| NCK-associated protein 1 | NCKAP1 | <0.00001 | −5.79 | −1.16 | −1.71 |
| ATPase family, AAA domain containing 2 | ATAD2 | 0.00002 | −5.78 | 1.13 | −2.53 |
| low density lipoprotein-related protein 1 (LRP1) | LRP1 | 0.00009 | −5.78 | −1.98 | −2.03 |
| ISL LIM homeobox 2 | ISL2 | 0.00007 | −5.75 | −1.36 | −4.24 |
| cell adhesion molecule 1 | CADM1 | 0.00002 | −5.75 | 1.10 | −3.03 |
| Ribosomal protein S3a-like | LOC100354966∧ | <0.00001 | −5.74 | −16.54 | −1.83 |
| Filamin A, alpha | FLNA | <0.00001 | −5.73 | 1.88 | −3.35 |
| integrin, alpha 10 | ITGA10 | 0.00014 | −5.71 | −1.72 | −2.44 |
| SERTA domain containing 4 | SERTAD4 | 0.00006 | −5.69 | 1.01 | −2.01 |
| Poly(A) binding protein, cytoplasmic 4 (inducible form) | PABPC4 | 0.00009 | −5.67 | 1.10 | −4.16 |
| immunoglobulin superfamily, member 9 | IGSF9 | 0.00223 | −5.66 | −2.79 | −3.20 |
| guanine nucleotide binding protein (G protein), gamma 3 | GNG3 | <0.00001 | −5.64 | 1.07 | −4.14 |
| nerve growth factor receptor | NGFR | <0.00001 | −5.62 | 2.10 | −3.05 |
| Intraflagellar transport 140-like | LOC100355837 | <0.00001 | −5.61 | 3.58 | −8.45 |
| Ribosomal protein L12-like | LOC100353722 | 0.00007 | −5.60 | −1.03 | −3.99 |
| ring finger and CCCH-type zinc finger domains 2 | RC3H2 | <0.00001 | −5.59 | 1.15 | −1.86 |
| Small proline-rich protein 3 | SPRR3 | <0.00001 | −5.59 | 2.41 | −1.85 |
| hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) (HIF1A) | HIF1A | 0.00001 | −5.59 | 1.16 | −2.44 |
| ATG4 autophagy related 4 homolog C (S. cerevisiae) | ATG4C | 0.00003 | −5.54 | −1.12 | −1.29 |
| XK, Kell blood group complex subunit-related, X-linked | XKRX | 0.00006 | −5.51 | −1.43 | −1.35 |
| ankyrin repeat and SOCS box-containing 10 | ASB10 | <0.00001 | −5.49 | −1.64 | −2.80 |
| aminoadipate aminotransferase | AADAT | 0.00005 | −5.48 | −1.41 | −1.78 |
| zinc finger, ZZ-type containing 3 | ZZZ3 | 0.00007 | −5.44 | −1.49 | −2.11 |
| transgelin 3 | TAGLN3 | 0.00003 | −5.41 | −1.12 | −1.93 |
| SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 | SMARCAD1 | <0.00001 | −5.41 | 1.04 | −2.04 |
| actinin, alpha 2 | ACTN2 | 0.00001 | −5.36 | 1.37 | −3.35 |
| synaptotagmin XII | SYT12 | 0.00001 | −5.36 | 1.14 | −6.91 |
| UDP glycosyltransferase 8 | UGT8 | 0.00127 | −5.36 | −1.08 | −2.71 |
| RasGEF domain family, member 1A | RASGEF1A | 0.00001 | −5.35 | 1.29 | −2.64 |
| hematopoietic SH2 domain containing | HSH2D | 0.00001 | −5.31 | −3.19 | −3.85 |
| 3-hydroxyisobutyrate dehydrogenase Fragment (HIBADH)(EC 1.1.1.31) | HIBADH | 0.00001 | −5.27 | 1.30 | −2.30 |
| Vacuolar protein sorting 4 homolog A (S. cerevisiae) | VPS4A | 0.00028 | −5.27 | −1.17 | −3.45 |
| Synaptosomal-associated protein 25 | LOC100340057 | 0.00007 | −5.26 | 1.05 | −2.54 |
| MAPK scaffold protein 1 | LAMTOR3 | 0.00002 | −5.24 | 1.03 | −2.10 |
| Ellis van Creveld syndrome | EVC | 0.00258 | −5.24 | −1.67 | −3.16 |
| topoisomerase (DNA) II beta 180 kDa | TOP2B | <0.00001 | −5.23 | −1.03 | −1.58 |
| RAB23, member RAS oncogene family | RAB23 | 0.00066 | −5.20 | −2.40 | −2.59 |
| N-myristoyltransferase 2 | NMT2 | 0.00005 | −5.19 | 1.14 | −1.51 |
| transmembrane emp24 protein transport domain containing 4 | TMED4 | 0.00077 | −5.19 | −1.21 | −3.23 |
| general transcription factor IIF, polypeptide 2, 30 kDa | GTF2F2 | 0.00007 | −5.17 | −1.26 | −1.92 |
| ribosomal protein L32 | RPL32 | 0.00002 | −5.17 | −1.92 | −1.46 |
| syntaxin 2 | STX2 | 0.00003 | −5.16 | −1.46 | −2.73 |
| succinate-CoA ligase, alpha subunit | SUCLG1 | <0.00001 | −5.14 | −1.02 | −1.93 |
| activating transcription factor 1 | ATF1 | 0.00020 | −5.14 | −2.13 | −3.43 |
| serum amyloid protein A (LOC100009259) | LOC100009259 | 0.00151 | −5.09 | 1.30 | −3.10 |
| peroxisomal biogenesis factor 1 | PEX1 | <0.00001 | −5.06 | −1.07 | −1.71 |
| interleukin-1 receptor-associated kinase 1 binding protein 1 | IRAK1BP1 | <0.00001 | −5.06 | −1.31 | −2.74 |
| ubiquitin specific peptidase 3 | USP3 | 0.00011 | −5.06 | −1.16 | −2.32 |
| BEN domain containing 6 | BEND6 | 0.00046 | −5.04 | −1.29 | −3.94 |
| Ribosomal protein S14-like | LOC100343884 | 0.00002 | −5.03 | 1.67 | −2.65 |
| HNF1 homeobox B | HNF1B | 0.00233 | −5.02 | −1.62 | −3.13 |
| Ribosomal protein S7-like | LOC100345715 | 0.00012 | −4.96 | −1.91 | −4.17 |
| GTPase activating protein (SH3 domain) binding protein 2 | G3BP2 | <0.00001 | −4.96 | −1.63 | −2.98 |
| insulin induced gene 1 | INSIG1 | 0.00148 | −4.93 | −1.34 | −3.58 |
| discs, large homolog 2 (Drosophila) | DLG2 | 0.00140 | −4.92 | −1.24 | −3.97 |
| syntrophin, gamma 1 | SNTG1 | 0.00002 | −4.90 | −1.42 | −3.33 |
| cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) | CDC73 | 0.00714 | −4.89 | −1.83 | −2.18 |
| family with sequence similarity 26, member F | FAM26F | 0.00573 | −4.88 | −1.47 | −2.67 |
| galactoside 2-L-fucosyltransferase (RFT-I) | RFT-I | 0.00014 | −4.85 | −1.47 | −2.96 |
| Ribosomal protein L36a-like | LOC100358069 | 0.00001 | −4.82 | −1.04 | −4.28 |
| microtubule associated monoxygenase, calponin and LIM domain containing 1 | MICAL1 | 0.00010 | −4.81 | −1.87 | −4.30 |
| Signal peptidase complex subunit 2 homolog | LOC100339691 | <0.00001 | −4.81 | 1.37 | −3.46 |
| Eukaryotic translation initiation factor 1 | LOC100345195 | 0.00015 | −4.81 | −1.22 | −4.44 |
| Poly(rC) binding protein 2-like | LOC100357382 | <0.00001 | −4.79 | 1.04 | −2.27 |
| kynureninase (L-kynurenine hydrolase) | KYNU | 0.00143 | −4.73 | 1.09 | −2.92 |
| glutamate receptor, ionotropic, N-methyl D-aspartate-like 1B | GCOM2 | 0.00001 | −4.72 | −1.28 | −1.74 |
| NADH dehydrogenase (ubiquinone) Fe-S protein 5, 15 kDa (NADH-coenzyme Q reductase) | LOC100342714 | 0.00018 | −4.70 | 1.30 | −3.33 |
| Signal sequence receptor, beta-like | LOC100342387 | 0.00006 | −4.66 | 1.96 | −3.48 |
| family with sequence similarity 107, member B | FAM107B | 0.00026 | −4.66 | −1.36 | −2.73 |
| Kinesin family member 5B-like | LOC100352693 | 0.00002 | −4.64 | 1.24 | −3.27 |
| tRNA aspartic acid methyltransferase 1 | TRDMT1 | 0.00300 | −4.63 | −1.58 | −2.39 |
| prostaglandin reductase 2 | PTGR2 | 0.00345 | −4.62 | −3.40 | −2.54 |
| zinc finger CCCH-type containing 7A | ZC3H7A | 0.00017 | −4.62 | −1.72 | −2.16 |
| guanine nucleotide binding protein (G protein), beta polypeptide 3 | GNB3 | 0.00016 | −4.62 | −1.21 | −2.80 |
| GS homeobox 2 | GSX2 | 0.00414 | −4.59 | −1.08 | −1.32 |
| CD164 molecule, sialomucin-like | LOC100346891 | 0.00024 | −4.58 | −2.13 | −3.13 |
| hect domain and RLD 5 | HERC5 | 0.00147 | −4.58 | 1.41 | −3.94 |
| non imprinted in Prader-Willi/Angelman syndrome 1 | NIPA1 | 0.00486 | −4.58 | −1.55 | −3.40 |
| numb homolog (Drosophila) | NUMB | <0.00001 | −4.58 | −1.11 | −3.22 |
| G protein-coupled receptor 68 | GPR68 | 0.00006 | −4.57 | 1.92 | −2.43 |
| listerin E3 ubiquitin protein ligase 1 | LTN1 | 0.00001 | −4.57 | −1.31 | −3.33 |
| methyltransferase like 10 | METTL10 | 0.00033 | −4.56 | 1.88 | −1.47 |
| CD83 molecule | CD83 | 0.00003 | −4.55 | 1.76 | −3.16 |
| Aldosterone synthase Fragment (EC 1.14.15.4) | CYP11B2 | 0.00001 | −4.53 | 2.52 | −3.03 |
| luteinizing hormone beta polypeptide (LHB) | LHB | 0.00001 | −4.52 | 1.46 | −2.78 |
| Two pore channel 3 | TPCN3 | 0.00008 | −4.52 | 1.47 | −3.83 |
| actin binding LIM protein 1 | ABLIM1 | 0.00010 | −4.52 | −1.61 | −2.20 |
| short coiled-coil protein | SCOC | 0.00010 | −4.52 | −1.12 | −2.70 |
| anti-Mullerian hormone receptor, type II (AMHR2) | AMHR2 | <0.00001 | −4.50 | 1.15 | −2.55 |
| family with sequence similarity 13, member C | FAM13C | 0.00108 | −4.49 | −1.91 | −3.43 |
| ring finger protein 168, E3 ubiquitin protein ligase | RNF168 | <0.00001 | −4.48 | −1.29 | −2.85 |
| MCG49427-like | LOC100343268 | <0.00001 | −4.46 | −1.65 | −1.99 |
| Aggrecanase-2 | ADAMTS-11∧, + | 0.00387 | −4.44 | −1.79 | −4.02 |
| NECAP endocytosis associated 2 | LOC100357803 | 0.00001 | −4.43 | 1.37 | −3.64 |
| DRAK1 (RDRAK1) | RDRAK1 | 0.00005 | −4.43 | −1.64 | −3.03 |
| thymopoietin | TMPO | 0.00001 | −4.42 | −1.05 | −1.77 |
| serine threonine kinase 39 (STE20/SPS1 homolog, yeast) | STK39 | 0.00026 | −4.41 | −2.06 | −5.10 |
| Crystallin, zeta | LOC100348912 | <0.00001 | −4.39 | 1.45 | −1.57 |
| Myeloid/lymphoid or mixed-lineage leukemia 3 | MLL3 | 0.00008 | −4.39 | −1.32 | −2.18 |
| discs, large homolog 3 (Drosophila) | DLG3 | 0.00008 | −4.39 | −1.32 | −3.99 |
| golgin, RAB6-interacting | GORAB | 0.00047 | −4.37 | −1.57 | −2.86 |
| Short coiled-coil protein-like | LOC100354452 | 0.00008 | −4.36 | −1.44 | −2.77 |
| glycerol kinase 5 (putative) | GK5 | 0.00019 | −4.35 | −1.74 | −2.42 |
| regulatory factor X, 2 (influences HLA class II expression) | RFX2 | 0.00003 | −4.34 | −1.12 | −2.22 |
| synaptosomal-associated protein, 25 kDa | SNAP25 | 0.00087 | −4.33 | 1.08 | −7.28 |
| MYB binding protein (P160) 1a | MYBBP1A | 0.00246 | −4.32 | −1.79 | −2.30 |
| chromatin modifying protein 2B | CHMP2B | 0.00002 | −4.31 | −1.18 | −2.23 |
| protein arginine methyltransferase 3 | PRMT3 | 0.00005 | −4.31 | −1.57 | −1.91 |
| Laminin, alpha 2 | LAMA2 | <0.00001 | −4.30 | 1.71 | −3.04 |
| family with sequence similarity 98, member B | FAM98B | 0.00294 | −4.29 | −2.05 | −2.96 |
| ankyrin repeat domain 44 | ANKRD44 | <0.00001 | −4.26 | 1.36 | −2.44 |
| family with sequence similarity 175, member A | FAM175A | 0.00028 | −4.26 | −1.59 | −2.22 |
| crumbs homolog 1 (Drosophila) | CRB1 | 0.00016 | −4.23 | 3.73 | −1.72 |
| TBC1 domain family, member 15 | TBC1D15 | 0.00297 | −4.23 | −1.87 | −6.40 |
| Zinc finger, FYVE domain containing 21-like | LOC100354231 | 0.00016 | −4.20 | 2.03 | −3.51 |
| Homeobox protein | HEX | 0.00003 | −4.19 | −1.18 | −2.79 |
| TPA-induced transmembrane protein | TTMP | 0.00179 | −4.18 | −1.69 | −2.76 |
| RAB3 GTPase activating protein subunit 2 (non-catalytic) | RAB3GAP2 | <0.00001 | −4.17 | −1.25 | −2.52 |
| sarcolemmal associated protein-3 (LOC100009199) | SLAP | 0.00065 | −4.17 | −1.68 | −2.65 |
| family with sequence similarity 175, member B | FAM175B | 0.00057 | −4.17 | −3.10 | −1.67 |
| guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 | GNAI1 | 0.00012 | −4.15 | −1.79 | −4.70 |
| Beta amyloid protein precursor | LOC100009546 | 0.00002 | −4.15 | 1.09 | −3.20 |
| golgi integral membrane protein 4 | GOLIM4 | 0.00004 | −4.15 | −1.69 | −2.86 |
| nuclear transport factor 2-like export factor 2 | NXT2 | 0.00009 | −4.15 | 1.04 | −2.86 |
| sodium channel, voltage-gated, type VII, alpha | SCN7A | 0.00103 | −4.14 | −2.44 | −3.92 |
| transmembrane protein 123 | TMEM123 | 0.00005 | −4.13 | −1.52 | −2.83 |
| protein tyrosine phosphatase domain containing 1 | PTPDC1 | 0.00286 | −4.11 | 1.13 | −3.15 |
| malic enzyme 1, NADP(+)-dependent, cytosolic | ME1 | 0.00007 | −4.11 | −1.52 | −4.16 |
| fibrillarin-like 1 | FBLL1 | 0.00229 | −4.11 | −1.58 | −4.00 |
| Nuclear casein kinase and cyclin-dependent kinase substrate 1-like | LOC100355717 | <0.00001 | −4.10 | 1.60 | −3.60 |
| Ligand-dependent nuclear receptor-interacting factor 1 | LRIF1 | 0.00001 | −4.07 | −1.07 | −1.82 |
| solute carrier family 44, member 1 | SLC44A1 | 0.00007 | −4.07 | −1.06 | −1.92 |
| Single-stranded DNA binding protein 2-like | LOC100345861 | 0.00001 | −4.07 | 1.01 | −2.98 |
| OMA1 homolog, zinc metallopeptidase (S. cerevisiae) | OMA1 | 0.00076 | −4.06 | −1.34 | −2.37 |
| glia maturation factor, beta | GMFB | 0.00106 | −4.05 | −1.74 | −2.63 |
| transient receptor potential cation channel, subfamily M, member 7 | TRPM7 | 0.00041 | −4.05 | −1.49 | −3.21 |
| translocase of outer mitochondrial membrane 5 homolog (yeast) | TOMM5∧ | 0.00014 | −4.05 | −5.67 | −1.60 |
| fibroblast growth factor 13 | FGF13 | 0.00088 | −4.00 | −2.20 | −4.12 |
| Heterogeneous nuclear ribonucleoprotein A1 | LOC100343627 | 0.00001 | −4.00 | 2.43 | −2.17 |
∧ Differentially expressed genes that are common between ‘hypertension only’ and ‘hypercholesterolemia plus sham’ groups (both vs. sham controls).+Differentially expressed genes that are common among ‘hypertension only’, ‘hypercholesterolemia plus sham’, and ‘hypertension plus hypercholesterolemia’ groups (all vs. sham controls).
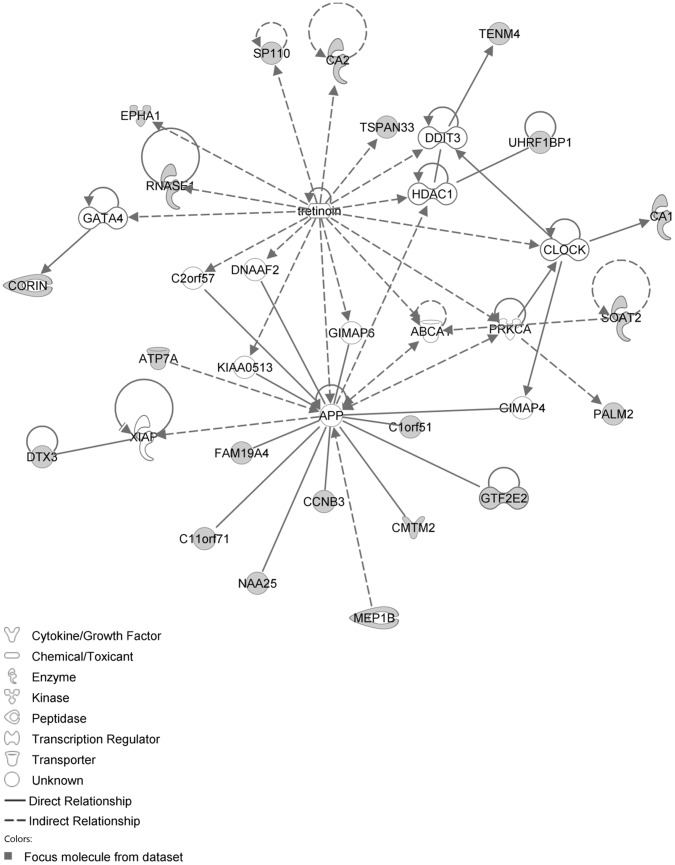
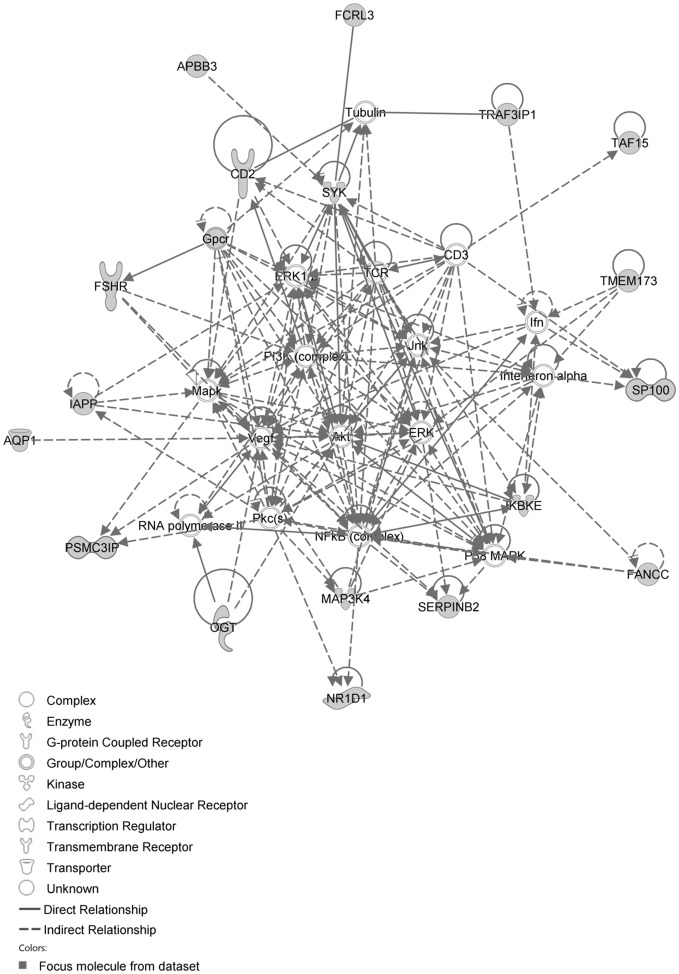
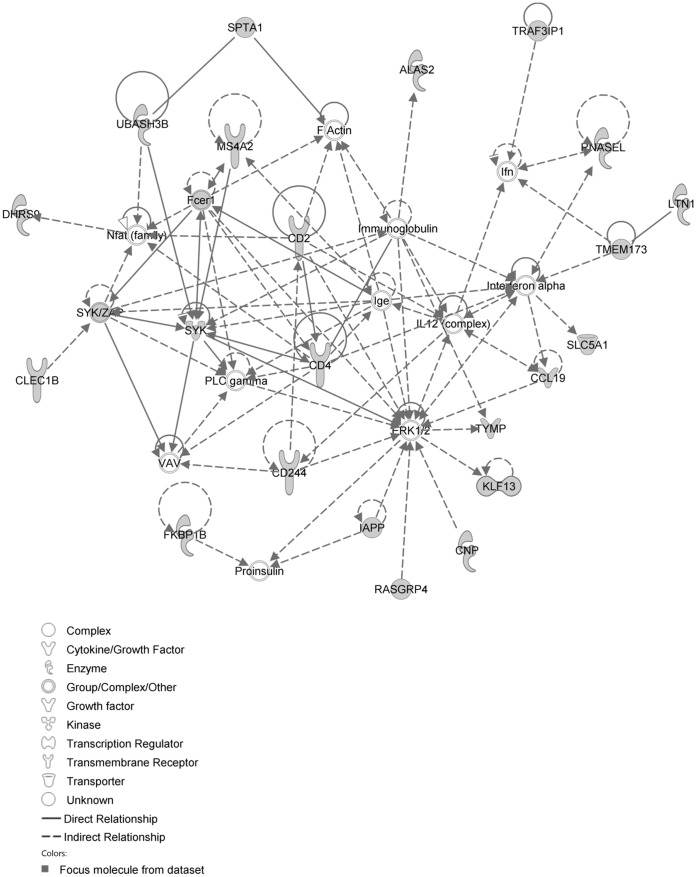
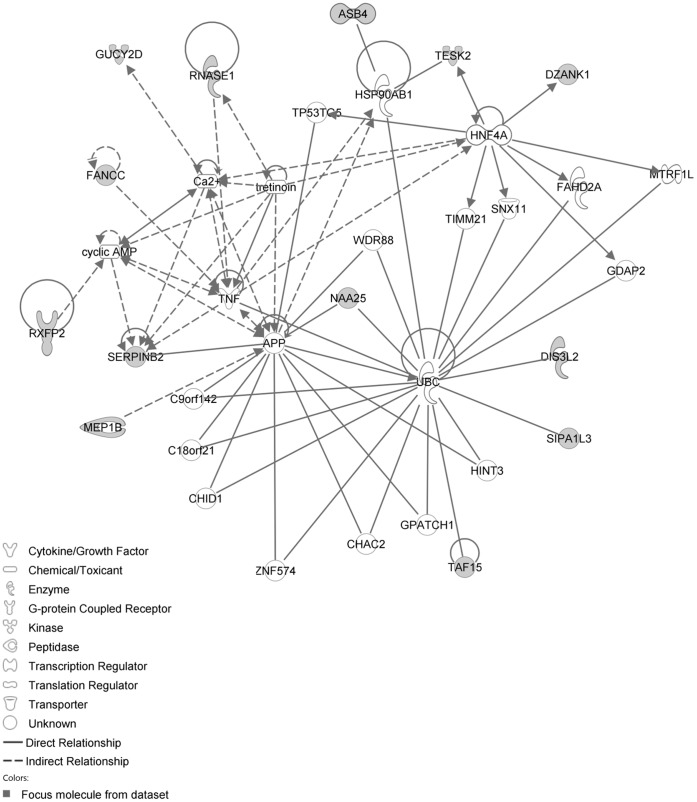
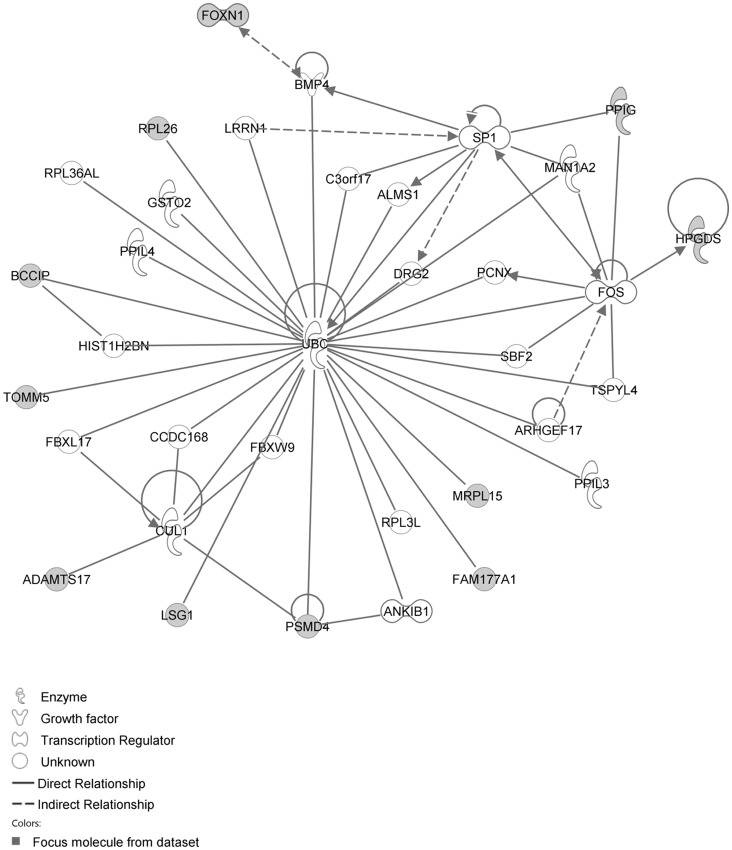
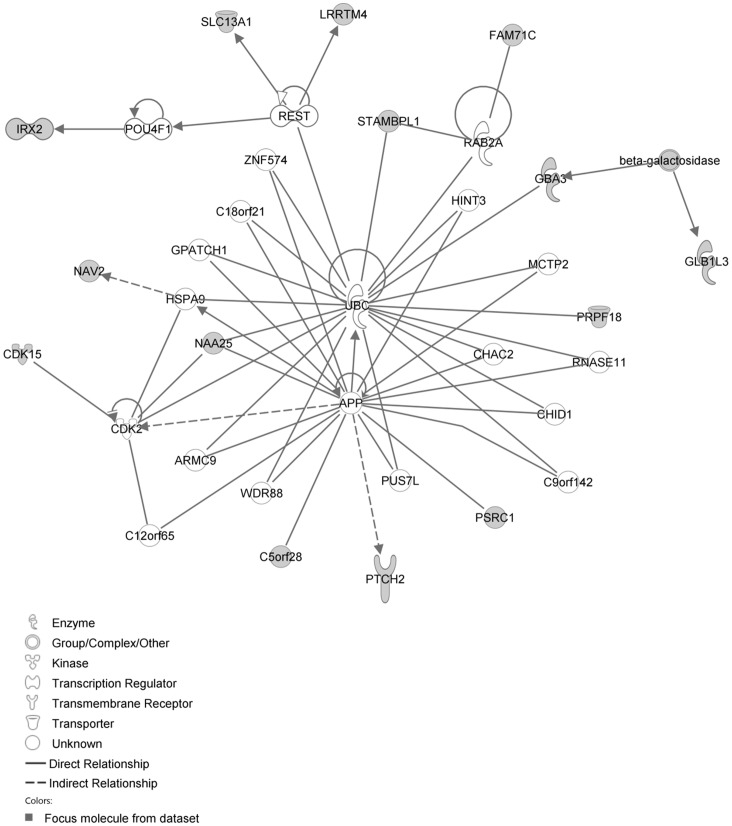
The IPA network with the ‘largest number of up-regulated focus genes’, contained 21 focus genes with functions in Cell Death and Survival, Lipid Metabolism, Small Molecule Biochemistry. Focus genes in this network were ATP7A, C11orf71, C1orf51, CA1, CA2, CCNB3, CMTM2, CORIN, DTX3, EPHA1, FAM19A4, GTF2E2, MEP1B, NAA25, PALM2, RNASE1, SOAT2, SP110, TENM4, TSPAN33 and UHRF1BP1. They were related to the ‘node molecules’, APP and tretinoin (Figure 7, Table 3). The next network of up-regulated focus genes had 18 focus genes, with functions in Organ Morphology, Reproductive System Development and Function, Developmental Disorder. Focus genes were APBB3, AQP1, CD2, FANCC, FCRL3, FSHR, IAPP, IKBKE, MAP3K4, NR1D1, OGT, PSMC3IP, SERPINB2, SP100, SYK, TAF15, TMEM173 and TRAF3IP1; they were related to P38 MAPK, ERK1/2, NFkB, SERPINB2, Akt, interferon alpha and VEGF (Figure 8, Table 3).
Figure 7. IPA network showing the network with the largest number of up-regulated focus genes in the hypercholesterolemia plus sham group, compared with sham operated controls.
Figure 8. IPA network showing the network with the second largest number of up-regulated focus genes in the hypercholesterolemia plus sham group, compared with sham operated controls.
The network with the largest number of down-regulated focus genes contained 26 focus genes with functions in Tissue Development, Connective Tissue Disorders, and Developmental Disorder. Focus genes in this network were ARNT, BMF, CADM1, CDC37L1, DNAJB4, DNAJC6, ELOVL5, GPBP1, GTF2F2, INSIG1, INSIG2, KDM4A, KIF20A, NUMB, PHIP, PKN2, PSMC6, PSMD4, PVRL3, RNF168, RPN1, SNAP25, SNTA1, SPRR3, STX2 and TOP2B. They were related to Ubiquitin, 26s Proteasome and Akt (Figure 9, Table 4). The next network of down-regulated focus genes had 25 focus genes with functions in Organ Morphology, Visual System Development and Function, Lipid Metabolism. Focus genes were ABLIM1, ACTA1, ATAD2, CDC73, CRYAA, CYP7A1, DHX9, HNRNPC, ME1, MLL3, MYBBP1A, PABPC4, POU2F3, SHROOM3, SMARCA5, SMARCAD1, SYT12, TIAL1, TMPO, TRPM7, USP3, WDR61, XRN1, YWHAQ and ZBTB44; they were related to histone H3 and F Actin (Figure 10, Table 4).
Figure 9. IPA network showing the network with the largest number of down-regulated focus genes in the hypercholesterolemia plus sham group, compared with sham operated controls.
Figure 10. IPA network showing the network with the second largest number of down-regulated focus genes in the hypercholesterolemia plus sham group, compared with sham operated controls.
2.3. Microarray analyses of the hypertension plus hypercholesterolemia group
The gene expression profile in the MCA of the hypertension plus hypercholesterolemia group was compared with that of sham controls on a normal diet. After unknown and repeated genes were omitted, 222 up-regulated and 133 down-regulated genes (greater than 4-fold change) were found (Figure 2). Among the highly up-regulated genes in the MCA of the hypertension plus hypercholesterolemia group compared to sham controls were EPHA1, SP110, SLFN14 (Table 5). Among the highly down-regulated genes were FOXN1, TNFRSF11B and GAPDHS (Table 6).
Table 5. Up-regulated genes in the MCA of ‘hypertension plus hypercholesterolemia’ rabbits vs. sham controls with greater than 4-fold change.
| Description | GeneSymbol | Corrected p-value | Hypertension plus hyperchole-sterolemia | Hypertension only | Hyperchole-sterolemia plus sham |
| Fold change | Fold change | Fold change | |||
| EPH receptor A1 | EPHA1 | 0.00008 | 20.31 | 3.03 | 8.98 |
| SP110 nuclear body protein | SP110 | <0.00001 | 15.47 | 2.06 | 8.42 |
| schlafen family member 14 | SLFN14 | 0.00614 | 13.64 | 1.33 | 10.65 |
| Glucose-fructose oxidoreductase domain containing 2-like | LOC100351150∧, + | 0.00001 | 12.42 | 4.01 | 6.34 |
| Gonadotropin-releasing hormone receptor | GnRHR | 0.00008 | 10.39 | 3.11 | 5.47 |
| solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 | SLC9A8 | 0.00175 | 10.32 | 3.55 | 6.14 |
| carbonic anhydrase I [ | CA1 | 0.00531 | 10.07 | 2.38 | 10.02 |
| Gap junction alpha-3 protein-like | LOC100357902 | 0.00689 | 9.96 | 2.92 | 9.62 |
| pleckstrin homology domain containing, family G (with RhoGef domain) member 6 | PLEKHG6 | 0.00011 | 9.24 | 3.23 | 4.47 |
| Uncharacterized protein C11orf71 | C11orf71 | 0.00446 | 9.22 | 2.97 | 8.33 |
| carboxypeptidase A5 | CPA5 | 0.00007 | 9.11 | 3.29 | 4.56 |
| Serine proteinase inhibitor, clade B, member 2 (Predicted) | SERPINB2∧, + | 0.00005 | 9.10 | 5.63 | 6.99 |
| kinesin family member 13B | KIF13B | 0.00002 | 9.02 | 2.35 | 6.02 |
| MRS2 magnesium homeostasis factor homolog (S. cerevisiae) | MRS2 | <0.00001 | 8.85 | 2.38 | 3.90 |
| Serine/threonine kinase 23, muscle-specific serine kinase 1 70 (Predicted) | SRPK3 | 0.00027 | 8.51 | 1.35 | 4.23 |
| 4-hydroxyphenylpyruvate dioxygenase-like | HPDL | 0.00008 | 8.45 | 2.22 | 4.68 |
| HEAT repeat containing 3 | LOC100357872 | 0.00100 | 8.34 | 2.38 | 3.77 |
| guanylate cyclase 2D, membrane (retina-specific) | GUCY2D∧, + | 0.00008 | 8.34 | 4.64 | 4.11 |
| membrane-spanning 4-domains, subfamily A, member 2 | MS4A2 | 0.00891 | 8.29 | 2.43 | 3.81 |
| Mast cell-expressed membrane protein 1 | MCEMP1 | 0.00003 | 8.27 | 1.74 | 4.67 |
| follicle stimulating hormone receptor | FSHR | 0.00023 | 8.27 | 1.24 | 5.31 |
| spectrin, alpha, erythrocytic 1 (elliptocytosis 2) | SPTA1 | 0.00375 | 8.20 | 2.75 | 6.03 |
| Bardet-Biedl syndrome 5 | LOC100342443 | 0.00004 | 8.15 | 2.76 | 5.62 |
| Uncharacterized protein C1orf50 | C1orf50 | 0.00006 | 8.01 | 5.81 | 3.80 |
| transmembrane protein 212 | TMEM212 | 0.00039 | 7.93 | 3.48 | 8.84 |
| CKLF-like MARVEL transmembrane domain containing 2 | CMTM2 | 0.00071 | 7.79 | 2.38 | 6.28 |
| Double zinc ribbon and ankyrin repeat-containing protein 1 | DZANK1∧, + | <0.00001 | 7.37 | 4.87 | 5.04 |
| doublesex and mab-3 related transcription factor 2 | DMRT2 | 0.00002 | 7.32 | 1.81 | 4.28 |
| LY6/PLAUR domain containing 5 | LYPD5 | 0.00004 | 7.31 | 2.68 | 4.70 |
| kinesin family member 21A | KIF21A | 0.00003 | 7.31 | 2.10 | 2.93 |
| N(alpha)-acetyltransferase 25, NatB auxiliary subunit | NAA25∧, + | 0.00099 | 7.23 | 1.04 | 3.68 |
| General transcription factor IIB | LOC100359058 | 0.00015 | 7.00 | 2.64 | 3.67 |
| relaxin/insulin-like family peptide receptor 2 | RXFP2∧, + | 0.00029 | 6.96 | 4.79 | 5.62 |
| interleukin 20 receptor, alpha | IL20RA | <0.00001 | 6.96 | 2.30 | 3.79 |
| family with sequence similarity 46, member C | FAM46C | 0.00016 | 6.92 | 1.72 | 8.51 |
| Fructose-1,6-bisphosphatase 1 | FBP1 | 0.00008 | 6.82 | 1.96 | 2.85 |
| transmembrane protease, serine 11E | TMPRSS11E | <0.00001 | 6.76 | 1.26 | 4.57 |
| IDI1 protein-like | LOC100346274∧, + | 0.00042 | 6.67 | 5.51 | 4.87 |
| solute carrier family 18 (vesicular monoamine), member 1 | SLC18A1 | 0.00074 | 6.64 | 1.42 | 3.45 |
| IQ motif and ubiquitin domain containing | IQUB | 0.00104 | 6.64 | 1.90 | 2.98 |
| family with sequence similarity 71, member C | FAM71C | 0.00026 | 6.63 | 2.04 | 3.24 |
| protease, serine, 38 | PRSS38 | 0.00058 | 6.51 | 1.95 | 5.74 |
| signal-induced proliferation-associated 1 like 3 | SIPA1L3∧, + | 0.00001 | 6.50 | 4.80 | 8.45 |
| secretoglobin, family 3A, member 1 | SCGB3A1 | 0.00010 | 6.42 | −1.18 | 3.56 |
| STAM binding protein-like 1 | STAMBPL1 | 0.00286 | 6.39 | 1.40 | 2.37 |
| Fanconi anemia, complementation group C | FANCC∧, + | <0.00001 | 6.39 | 6.10 | 4.25 |
| tripartite motif-containing 67 | TRIM67 | 0.00175 | 6.39 | 2.15 | 3.25 |
| Serum response factor binding protein 1-like | LOC100349313 | 0.00139 | 6.34 | 2.18 | 3.53 |
| Interstitial collagenase Precursor (EC 3.4.24.7)(Matrix metalloproteinase-1)(MMP-1) | MMP1 | 0.00149 | 6.33 | 8.12 | 2.20 |
| G protein-coupled receptor 119 | GPR119 | 0.00058 | 6.30 | 2.41 | 2.00 |
| RNA binding motif protein 6 | RBM6 | 0.00003 | 6.15 | 1.73 | 4.85 |
| 2′,3′-cyclic nucleotide 3′ phosphodiesterase | CNP | <0.00001 | 6.14 | 1.42 | 3.45 |
| nuclear receptor subfamily 1, group D, member 1 | NR1D1 | 0.00562 | 6.11 | 2.35 | 4.98 |
| cyclin B3 | CCNB3 | 0.00012 | 6.05 | 1.68 | 4.39 |
| steroidogenic acute regulatory protein | STAR | 0.00007 | 6.05 | 3.36 | 3.31 |
| CD4 molecule (CD4) | CD4 | 0.00004 | 6.02 | 1.17 | 2.77 |
| DOMON domain-containing protein FRRS1L | FRRS1L | 0.00003 | 5.99 | 1.37 | 3.57 |
| Tubulin tyrosine ligase-like family, member 5 | LOC100359240 | 0.00019 | 5.99 | 1.63 | 2.92 |
| family with sequence similarity 19 (chemokine (C-C motif)-like), member A4 | FAM19A4 | 0.00024 | 5.97 | 1.65 | 4.17 |
| sterol O-acyltransferase 2 | SOAT2 | 0.00003 | 5.94 | 3.01 | 7.18 |
| aminolevulinate, delta-, synthase 2 | ALAS2 | 0.00072 | 5.93 | 1.97 | 5.86 |
| thymidine phosphorylase | TYMP | 0.00005 | 5.87 | 1.17 | 3.87 |
| dynein heavy chain domain 1 | DNHD1 | 0.00134 | 5.87 | 2.10 | 3.49 |
| insulin-like 5 (INSL5), | INSL5 | 0.00018 | 5.85 | 1.39 | 3.03 |
| meprin A, beta | MEP1B∧, + | 0.00291 | 5.85 | 5.37 | 5.12 |
| arylformamidase | AFMID | 0.00100 | 5.81 | 1.65 | 4.03 |
| Islet amyloid polypeptide | IAPP | 0.00004 | 5.78 | 1.40 | 4.17 |
| Transcription elongation factor A (SII)-like 2-like | LOC100340955 | 0.00015 | 5.75 | 3.13 | 3.22 |
| G protein-coupled receptor 52 | GPR52 | 0.00010 | 5.75 | 2.26 | 4.40 |
| solute carrier family 5 (sodium/glucose cotransporter), member 9 (SLC5A9) | SLC5A9 | 0.00002 | 5.73 | 3.82 | 3.28 |
| Kruppel-like factor 13 | KLF13 | 0.00007 | 5.69 | 1.11 | 3.32 |
| Granulate-macrophage stimulating factor (Predicted) | GMCSF | 0.00004 | 5.68 | 2.22 | 3.39 |
| listerin E3 ubiquitin protein ligase 1 | LTN1 | 0.00009 | 5.66 | 3.05 | 2.75 |
| zinc finger, DHHC-type containing 23 | ZDHHC23 | 0.00001 | 5.64 | 7.18 | 3.22 |
| carboxylesterase 8 (putative) | CES4A | 0.00023 | 5.63 | 1.47 | 3.83 |
| C-type lectin domain family 1, member B | CLEC1B | 0.00584 | 5.60 | 1.50 | 3.65 |
| TNF receptor-associated factor 3 interacting protein 1 | TRAF3IP1 | 0.00043 | 5.56 | 1.29 | 4.80 |
| solute carrier family 15, member 5 | SLC15A5 | 0.00075 | 5.55 | 1.80 | 2.70 |
| glucosidase, beta, acid 3 (cytosolic) | GBA3 | 0.00012 | 5.55 | 2.27 | 2.77 |
| tetraspanin 33 | TSPAN33 | 0.00078 | 5.53 | 3.97 | 6.37 |
| Proline-rich protein 3 | PRR3 | 0.00129 | 5.50 | 1.24 | 3.46 |
| chemokine (C-C motif) ligand 19 | CCL19 | 0.00055 | 5.50 | 2.16 | 2.39 |
| NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa | LOC100341941 | 0.00022 | 5.46 | 2.69 | 4.34 |
| AchR epsilon subunit Fragment | CHRNE | 0.00010 | 5.43 | 1.33 | 3.22 |
| Transmembrane protein C5orf28 | C5orf28 | 0.00319 | 5.38 | 2.31 | 2.49 |
| dehydrogenase/reductase (SDR family) member 9 | DHRS9 | 0.00168 | 5.37 | 3.41 | 3.41 |
| FCH and double SH3 domains 1 | FCHSD1 | 0.00015 | 5.37 | 2.85 | 3.24 |
| leucine-rich repeat-containing G protein-coupled receptor 5 | LGR5 * | 0.00019 | 5.36 | 3.32 | 4.14 |
| Uncharacterized protein C7orf72 | C7orf72 | 0.00114 | 5.35 | -1.29 | 2.68 |
| family with sequence similarity 71, member F2 | FAM71F2 | 0.00643 | 5.34 | 1.21 | 2.13 |
| otopetrin 2 | OTOP2 | 0.00006 | 5.31 | 3.74 | 3.23 |
| ELOVL fatty acid elongase 3 | ELOVL3 | 0.00012 | 5.30 | 1.83 | 3.05 |
| solute carrier family 7 (anionic amino acid transporter), member 13 | SLC7A13 | 0.00134 | 5.30 | 1.82 | 2.81 |
| Uncharacterized protein C2orf71 | C2orf71 | 0.00177 | 5.30 | 1.36 | 3.60 |
| ankyrin and armadillo repeat containing | ANKAR∧, + | 0.00006 | 5.23 | 7.05 | 7.99 |
| solute carrier family 14 (urea transporter), member 1 (Kidd blood group) | SLC14A1 | 0.00027 | 5.21 | 1.61 | 3.38 |
| zinc finger, CW type with PWWP domain 2 | ZCWPW2 | 0.00011 | 5.21 | 2.31 | 5.29 |
| Fc receptor-like A | FCRLA | 0.00083 | 5.14 | 1.94 | 6.76 |
| RAS guanyl releasing protein 4 | RASGRP4 | 0.00058 | 5.11 | 2.61 | 3.68 |
| family with sequence similarity 167, member A | FAM167A | 0.00017 | 5.11 | 14.04 | 2.44 |
| ATPase, aminophospholipid transporter, class I, type 8B, member 2 | ATP8B2 | 0.00360 | 5.07 | 3.16 | 3.14 |
| Cytochrome P450, family 4, subfamily A, polypeptide 5 | CYP4A5 | 0.00061 | 5.05 | 1.03 | 3.12 |
| ATPase, H+/K+ exchanging, alpha polypeptide (ATP4A) | ATP4A | <0.00001 | 5.02 | 1.47 | 3.53 |
| CD2 molecule | CD2 | 0.00064 | 4.99 | 1.35 | 3.15 |
| 3-oxo-5-alpha-steroid 4-dehydrogenase 2-like | LOC100343882 | 0.00001 | 4.99 | 2.48 | 2.64 |
| ankyrin repeat and SOCS box-containing 4 | ASB4∧ | <0.00001 | 4.97 | 6.36 | 3.70 |
| phosphatidylcholine transfer protein | PCTP | 0.00001 | 4.96 | 2.13 | 3.26 |
| solute carrier family 39 (zinc transporter), member 12 | SLC39A12 | 0.00006 | 4.96 | 4.23 | 2.48 |
| transmembrane emp24 protein transport domain containing 6 | TMED6 | 0.00040 | 4.96 | 2.10 | 3.19 |
| actin-related protein T1 | ACTRT1 | 0.00005 | 4.94 | 2.95 | 3.11 |
| Wings apart-like homolog | LOC100348678 | 0.00041 | 4.94 | 2.27 | 2.92 |
| Fructose-1,6-bisphosphatase 1 | FBP1 | 0.00050 | 4.88 | 1.51 | 2.15 |
| spleen tyrosine kinase | SYK | 0.00072 | 4.88 | 1.52 | 4.13 |
| tripartite motif-containing 65 | TRIM65 | 0.00079 | 4.87 | 2.63 | 4.05 |
| neuron navigator 2 | NAV2 | 0.00080 | 4.87 | 1.80 | 2.69 |
| potassium channel tetramerisation domain containing 19 | KCTD19 | 0.00123 | 4.87 | 1.42 | 3.10 |
| spermatogenesis associated, serine-rich 1 | SPATS1 | 0.00201 | 4.83 | 2.77 | 2.57 |
| solute carrier family 5 (sodium/glucose cotransporter), member 1 | SLC5A1 | 0.00001 | 4.80 | 2.26 | 2.95 |
| tripartite motif-containing 35 | TRIM35 | 0.00084 | 4.80 | 1.38 | 3.16 |
| cytochrome P450, family 26, subfamily C, polypeptide 1 | CYP26C1 | 0.00224 | 4.79 | -1.58 | 2.30 |
| Polymeric immunoglobulin receptor | PIGR | 0.00025 | 4.75 | 1.40 | 2.82 |
| Serine incorporator 1 | LOC100357075 | 0.00015 | 4.74 | 2.50 | 5.37 |
| Ras and Rab interactor-like | RINL | 0.00135 | 4.71 | 1.67 | 3.40 |
| ubiquitin associated and SH3 domain containing B | UBASH3B | 0.00803 | 4.70 | 2.95 | 3.29 |
| UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 (GalNAc-T13) | GALNT13 | <0.00001 | 4.70 | 2.52 | 1.93 |
| transmembrane protein 173 | TMEM173 | 0.00141 | 4.70 | 2.88 | 5.36 |
| UHRF1 binding protein 1 | UHRF1BP1 | 0.00074 | 4.69 | 2.63 | 5.70 |
| PRP18 pre-mRNA processing factor 18 homolog (S. cerevisiae) | PRPF18 | 0.00144 | 4.68 | 1.10 | 2.28 |
| Carbonic anhydrase 2 (EC 4.2.1.1)(Carbonic anhydrase II)(CA-II)(Carbonate dehydratase II) | CA2 | 0.00964 | 4.68 | 1.67 | 4.51 |
| spinster homolog 3 (Drosophila) | SPNS3 | 0.00049 | 4.68 | 2.40 | 3.63 |
| galactosidase, beta 1-like 3 | GLB1L3 | 0.00003 | 4.67 | 1.24 | 3.03 |
| iroquois homeobox 2 | IRX2 | 0.00052 | 4.66 | -1.04 | 3.40 |
| collagen, type XVII, alpha 1 | COL17A1 | 0.00028 | 4.65 | 1.07 | 2.60 |
| patched homolog 2 (Drosophila) | PTCH2 | 0.00005 | 4.65 | 2.55 | 3.84 |
| TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68 kDa | TAF15∧, + | 0.00004 | 4.65 | 7.45 | 4.71 |
| BCL2-like 10 (apoptosis facilitator) | BCL2L10 | 0.00018 | 4.64 | 1.67 | 5.34 |
| Mix1 homeobox-like 1 (Xenopus laevis) | MIXL1 | 0.00093 | 4.64 | 5.80 | 3.54 |
| Fibroblast growth factor binding protein 1-like | LOC100353835∧, + | 0.00122 | 4.63 | 5.01 | 4.01 |
| corin, serine peptidase | CORIN | 0.00006 | 4.63 | 3.59 | 4.93 |
| V-set and immunoglobulin domain containing 2 | VSIG2 | 0.00168 | 4.60 | 3.51 | 5.53 |
| Pregnancy-zone protein | PZP | 0.00099 | 4.60 | 1.20 | 4.11 |
| DEAQ box RNA-dependent ATPase 1 | DQX1 | 0.00520 | 4.60 | 1.85 | 2.94 |
| Potassium channel, subfamily K, member 1 | KCNK1 | 0.00217 | 4.58 | 3.22 | 1.68 |
| purinergic receptor P2Y, G-protein coupled, 10 | P2RY10 | 0.00197 | 4.57 | 1.99 | 2.24 |
| coiled-coil domain containing 54 | CCDC54 | 0.00196 | 4.57 | 3.11 | 4.04 |
| somatostatin receptor 2 | SSTR2 | 0.00077 | 4.56 | 2.78 | 2.44 |
| deltex homolog 3 (Drosophila) | DTX3 | 0.00013 | 4.56 | 3.47 | 4.92 |
| zinc finger, DHHC-type containing 19 | ZDHHC19 | 0.00241 | 4.55 | 1.65 | 3.32 |
| ret proto-oncogene | RET | 0.00036 | 4.55 | 1.88 | 2.71 |
| MORN repeat containing 4 | MORN4 | <0.00001 | 4.53 | 1.45 | 4.14 |
| Oxytocin receptor | OTXR | 0.00517 | 4.52 | 1.10 | 2.58 |
| serine peptidase inhibitor, Kazal type 1 | SPINK1 | 0.00055 | 4.50 | 1.60 | 2.53 |
| membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) | MPP2 | 0.00409 | 4.50 | 2.36 | 2.89 |
| solute carrier family 13 (sodium/sulfate symporters), member 1 | SLC13A1 | 0.00325 | 4.49 | 1.59 | 1.58 |
| follistatin-like 5 | FSTL5 | 0.00071 | 4.49 | 3.17 | 2.31 |
| Mitochondrial ribosomal protein S15-like | LOC100345026 | 0.00064 | 4.49 | 2.82 | 2.65 |
| TERF1 (TRF1)-interacting nuclear factor 2 | TINF2 | 0.00325 | 4.49 | 2.19 | 3.29 |
| keratin associated protein 19-4 | KRTAP19-4 | 0.00044 | 4.48 | 1.36 | 2.78 |
| XK, Kell blood group complex subunit-related, X-linked | XKRX | 0.00051 | 4.47 | 2.36 | 2.54 |
| Chloride intracellular channel 2 (Predicted) | CLIC2 | 0.00279 | 4.46 | 1.60 | 5.43 |
| FK506 binding protein 1B, 12.6 kDa | FKBP1B | 0.00274 | 4.46 | −1.46 | 2.32 |
| phospholipase C, zeta 1 | PLCZ1 | 0.00002 | 4.45 | 1.01 | 2.92 |
| CD244 molecule, natural killer cell receptor 2B4 | CD244 | 0.00003 | 4.44 | −1.55 | 2.90 |
| carboxyl ester lipase (bile salt-stimulated lipase) (CEL) | CEL | 0.00016 | 4.44 | 1.98 | 2.68 |
| ADAM metallopeptidase with thrombospondin type 1 motif, 18 | ADAMTS18 | 0.00213 | 4.43 | 1.94 | 2.90 |
| UDP-glucuronosyltransferase | UGT2C1 | 0.00035 | 4.42 | 3.99 | 2.75 |
| fms-related tyrosine kinase 3 | FLT3 | 0.00270 | 4.41 | 1.80 | 3.63 |
| mitogen-activated protein kinase binding protein 1 | MAPKBP1 | 0.00003 | 4.41 | 3.08 | 2.68 |
| Ammonium transporter Rh type C (Rhesus blood group family type C glycoprotein)(Rh family type C glycoprotein)(Rh type C glycoprotein) | RHCG | 0.00032 | 4.41 | 2.20 | 3.29 |
| Kelch-like 1 (Drosophila) | KLHL1 | 0.00003 | 4.35 | −1.02 | 2.80 |
| leucine rich repeat transmembrane neuronal 4 | LRRTM4 | 0.00002 | 4.34 | 3.58 | 2.67 |
| proteoglycan 3 | PRG3 | 0.00254 | 4.33 | 1.60 | 3.79 |
| Ligand of numb-protein X 1-like | LOC100343259 | 0.00568 | 4.33 | 1.19 | 2.97 |
| chymase like protein | LOC100008644 | 0.00060 | 4.32 | −1.31 | 2.21 |
| sex comb on midleg-like 4 (Drosophila) | SCML4 | 0.00854 | 4.32 | 1.40 | 2.94 |
| BMS1-like, ribosome assembly protein | LOC100339194 | 0.00017 | 4.31 | 2.21 | 3.44 |
| Uncharacterized protein C2orf61 | C2orf61 | 0.00893 | 4.31 | 3.08 | 3.04 |
| REC8 homolog (yeast) | REC8 | <0.00001 | 4.31 | 1.93 | 5.52 |
| Protein FAM55A | FAM55A | 0.00326 | 4.30 | 1.16 | 1.70 |
| cyclin-dependent kinase 15 | CDK15 | 0.00129 | 4.28 | 1.93 | 3.69 |
| transmembrane and coiled-coil domains 4 | TMCO4 | 0.00007 | 4.28 | 2.43 | 5.42 |
| Rac GTPase activating protein 1 pseudogene | RACGAP1P | 0.00023 | 4.27 | 1.43 | 2.33 |
| family with sequence similarity 170, member A | FAM170A | 0.00008 | 4.23 | 2.43 | 2.99 |
| Mitogen-activated protein kinase kinase kinase 4 | MAP3K4 | 0.00788 | 4.22 | 2.40 | 3.16 |
| ATPase, H+ transporting, lysosomal 70 kDa, V1 subunit A | ATP6V1A | 0.00397 | 4.19 | 1.38 | 1.78 |
| solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 | SLC9A5 | 0.00057 | 4.19 | 2.33 | 5.36 |
| 2,3-bisphosphoglycerate mutase (BPGM) | BPGM | 0.00098 | 4.18 | 1.21 | 4.00 |
| FRY-like | FRYL | 0.00291 | 4.18 | 1.46 | 2.56 |
| clusterin-like 1 (retinal) | CLUL1 | 0.00072 | 4.17 | 2.09 | 2.68 |
| Superiorcervical ganglia, neural specific 10 | LOC100344464 | 0.00228 | 4.14 | 1.78 | 1.59 |
| Histamine receptor H1 | HRH1 | 0.00001 | 4.13 | 1.92 | 2.38 |
| Cingulin (Predicted) | CGN | 0.00236 | 4.12 | 1.43 | 3.44 |
| nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, delta | NFKBID | 0.00103 | 4.12 | 1.13 | 3.05 |
| ribonuclease, RNase A family, 1 (pancreatic)] | RNASE1∧, + | 0.00043 | 4.11 | 2.83 | 3.92 |
| MLX interacting protein | MLXIP | 0.00069 | 4.09 | 3.02 | 5.36 |
| Nuclear prelamin A recognition factor | LOC100344024 | 0.00002 | 4.09 | 2.47 | 2.46 |
| actin, beta-like 2 | ACTBL2 | 0.00023 | 4.09 | 1.32 | 2.55 |
| ghrelin receptor (LOC100101582) | LOC100101582 | 0.00062 | 4.08 | 1.40 | 2.17 |
| olfactomedin 2 | OLFM2 | 0.00385 | 4.07 | 3.66 | 2.50 |
| cytochrome c oxidase assembly homolog 10 (yeast) | COX10 | 0.00160 | 4.07 | 2.06 | 3.97 |
| ribonuclease L (2′,5′-oligoisoadenylate synthetase-dependent) | RNASEL | 0.00028 | 4.06 | −1.16 | 1.98 |
| keratin associated protein 11-1 | KRTAP11-1 | 0.00013 | 4.04 | 2.16 | 2.57 |
| family with sequence similarity 53, member C | FAM53C | <0.00001 | 4.03 | 9.87 | 2.48 |
| DnaJ (Hsp40) homolog, subfamily C, member 5 beta | DNAJC5B | 0.00016 | 4.03 | −1.01 | 2.35 |
| receptor (chemosensory) transporter protein 1 | RTP1 | 0.00012 | 4.03 | −1.25 | 1.76 |
| protocadherin 8 | PCDH8 | 0.00005 | 4.02 | 3.20 | 3.09 |
| Ubiquitin specific protease 39 | LOC100344046 | 0.00343 | 4.02 | 2.49 | 2.75 |
| corticotropin releasing hormone binding protein | CRHBP | 0.00005 | 4.02 | 6.38 | 2.23 |
| proline/serine-rich coiled-coil 1 | PSRC1 | 0.00571 | 4.01 | 1.63 | 2.68 |
| cadherin 17, LI cadherin (liver-intestine) | CDH17 | 0.00459 | 4.00 | 1.59 | 2.72 |
∧ Differentially expressed genes that are common between ‘hypertension only’ and ‘hypercholesterolemia plus sham’ groups (both vs. sham controls).+Differentially expressed genes that are common among ‘hypertension only’, ‘hypercholesterolemia plus sham’, and ‘hypertension plus hypercholesterolemia’ groups (all vs. sham controls).
Table 6. Down-regulated genes in the MCA of ‘hypertension plus hypercholesterolemia’ rabbits vs. sham controls with greater than 4-fold change.
| Description | GeneSymbol | Corrected p-value | Hypertension plus hyperchole-sterolemia | Hypertension only | Hyperchole-sterolemia plus sham |
| Fold Change | Fold Change | Fold Change | |||
| forkhead box N1 | FOXN1∧, + | 0.00048 | −45.12 | −26.20 | −56.89 |
| Tumor necrosis factor receptor superfamily, member 11b | TNFRSF11B | <0.00001 | −33.58 | 1.69 | −55.91 |
| glyceraldehyde-3-phosphate dehydrogenase, spermatogenic | GAPDHS | <0.00001 | −32.91 | 1.01 | −33.66 |
| Chaperonin containing TCP1, subunit 2 (beta) | CCT2 | <0.00001 | −30.96 | −1.91 | −35.56 |
| Tweety homolog 1 (Drosophila) | TTYH1 | 0.00108 | −27.02 | −5.16 | −12.82 |
| family with sequence similarity 184, member B | FAM184B | <0.00001 | −25.72 | −1.32 | −68.05 |
| 3′ (2′), 5′-bisphosphate nucleotidase 1 | BPNT1 | <0.00001 | −25.01 | 1.05 | −52.27 |
| ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 pseudogene 1 | UQCRFS1P1 | <0.00001 | −23.74 | 1.08 | −40.99 |
| Progesterone receptor membrane component 1 | LOC100357097 | <0.00001 | −21.91 | 1.04 | −38.66 |
| microtubule associated tumor suppressor 1 | MTUS1 | <0.00001 | −20.40 | −1.88 | −41.71 |
| transmembrane protein 38B | TMEM38B | <0.00001 | −18.80 | 1.05 | −31.53 |
| beta tropomyosin (LOC100125984) | LOC100125984 | <0.00001 | −17.13 | −2.30 | −97.92 |
| ORM1-like 3 (S. cerevisiae) | ORMDL3 | <0.00001 | −16.75 | −1.24 | −17.49 |
| DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) | DCUN1D4 | <0.00001 | −15.90 | −1.65 | −18.27 |
| Lysosomal-associated membrane protein 2-like | LOC100350946 | <0.00001 | −15.88 | −1.78 | −26.85 |
| v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | KRAS | <0.00001 | −15.47 | −1.18 | −22.11 |
| androgen-induced 1 | AIG1 | 0.00002 | −14.63 | −3.30 | −35.48 |
| Eukaryotic translation initiation factor 2 subunit 1 | EIF2S1 | 0.00001 | −14.40 | −2.04 | −16.08 |
| hematopoietic prostaglandin D synthase | HPGDS∧, + | 0.00043 | −13.63 | −12.38 | −21.44 |
| Nucleolar protein 11 | NOL11 | 0.00032 | −13.01 | 2.09 | −11.98 |
| zinc finger protein 642 | ZFP69 | 0.00058 | −12.02 | −1.58 | −20.24 |
| mitochondrial ribosomal protein L1 | MRPL1 | <0.00001 | −12.00 | −1.06 | −15.71 |
| WW domain containing adaptor with coiled-coil | WAC | 0.00001 | −11.91 | −3.12 | −13.20 |
| Solute carrier family 2, facilitated glucose transporter member 3 | SLC2A3 | 0.00019 | −11.60 | −1.81 | −10.95 |
| crystallin, alpha A (CRYAA) | CRYAA | <0.00001 | −10.81 | −1.50 | −60.93 |
| Lupus La protein | SSB | <0.00001 | −10.68 | −1.63 | −40.45 |
| somatostatin | SST | 0.00169 | −10.49 | 1.22 | −4.19 |
| Manganese superoxide dismutase | SOD-2 | <0.00001 | −9.93 | −1.46 | −51.78 |
| mitochondrial ribosomal protein L15 | MRPL15∧, + | <0.00001 | −9.57 | −6.97 | −23.01 |
| Proteasome (prosome, macropain) 26s subunit, non-ATPase, 4 | PSMD4∧, + | <0.00001 | −9.55 | −1.65 | −12.56 |
| IQ motif containing GTPase activating protein 3 | IQGAP3 | 0.00007 | −9.37 | 1.25 | −10.54 |
| cellular disintegrin ADAM 6d (ADAM6) | ADAM6 | 0.00009 | −9.13 | −1.03 | −3.07 |
| mRNA for prolactin receptor | PRLR | 0.00001 | −8.55 | 1.23 | −1.32 |
| zinc finger protein 330 | ZNF330 | 0.00012 | −8.54 | −1.48 | −7.41 |
| ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D) | ELAVL4 | 0.00033 | −8.49 | 1.79 | −3.26 |
| Intraflagellar transport 140-like | LOC100355837 | <0.00001 | −8.45 | 3.58 | −5.61 |
| SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 | SMARCA5 | <0.00001 | −8.08 | 1.08 | −19.50 |
| NADH dehydrogenase-like | LOC100347823 | <0.00001 | −7.93 | 2.02 | −7.55 |
| zinc finger RNA binding protein | ZFR | <0.00001 | −7.90 | −1.47 | −64.53 |
| Cytoplasmic beta-actin | LOC100009506 | <0.00001 | −7.80 | −1.19 | −20.51 |
| stathmin-like 2 | STMN2 | 0.00129 | −7.60 | 1.78 | −4.10 |
| SRY (sex determining region Y)-box 2-like | LOC100341629 | 0.00001 | −7.56 | 5.05 | 1.43 |
| synaptosomal-associated protein, 25kDa | SNAP25 | 0.00087 | −7.28 | 1.08 | −4.33 |
| general transcription factor IIE, polypeptide 2, beta 34 kDa | GTF2E2 | <0.00001 | −7.27 | 1.06 | −7.79 |
| Solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 | SLC9A2 | 0.00007 | −7.21 | −2.62 | −36.54 |
| olfactomedin 3 | OLFM3 | 0.00005 | −6.99 | −1.25 | −3.51 |
| pyruvate dehydrogenase kinase, isozyme 4 | PDK4 | 0.00711 | −6.98 | −2.51 | −3.47 |
| myelin-associated oligodendrocyte basic protein | MOBP | 0.00776 | −6.94 | −1.01 | −3.86 |
| synaptotagmin XII | SYT12 | 0.00001 | −6.91 | 1.14 | −5.36 |
| Titin | TTN | 0.00001 | −6.90 | −2.26 | −13.60 |
| BPI fold containing family B, member 6 | BPIFB6 | 0.00001 | −6.83 | −1.79 | −5.92 |
| guanylate cyclase 1, soluble, beta 3 | GUCY1B3 | 0.00010 | −6.79 | −2.52 | −11.97 |
| prefoldin subunit 5 | PFDN5 | <0.00001 | −6.79 | −1.78 | −84.52 |
| proteolipid protein 1 (PLP1) | PLP1 | 0.00339 | −6.50 | −1.79 | −6.44 |
| sodium channel, voltage-gated, type VII, alpha | SCN7A | 0.00021 | −6.43 | −1.77 | −12.29 |
| TBC1 domain family, member 15 | TBC1D15 | 0.00297 | −6.40 | −1.87 | −4.23 |
| DnaJ (Hsp40) homolog, subfamily C, member 6 | DNAJC6 | 0.00001 | −6.36 | −1.48 | −9.39 |
| zinc finger protein 326 | ZNF326 | <0.00001 | −6.29 | 1.09 | −18.36 |
| neurocalcin delta | NCALD | 0.00037 | −6.13 | −1.93 | −9.22 |
| cullin 3 | CUL3 | <0.00001 | −6.10 | 1.05 | −73.37 |
| caudal type homeobox 1 | CDX1 | 0.00001 | −6.08 | −1.18 | −10.42 |
| ring finger protein 222 | RNF222 | 0.00001 | −6.02 | −1.78 | −8.36 |
| syntrophin, alpha 1 | SNTA1 | <0.00001 | −5.98 | −1.04 | −9.97 |
| Creatine kinase, muscle | CKM | 0.00005 | −5.96 | 2.01 | −1.29 |
| Sin3A-associated protein, 30 kDa | SAP30 | <0.00001 | −5.90 | 1.30 | −43.67 |
| signal recognition particle 54 kDa | SRP54 | <0.00001 | −5.72 | 1.62 | −16.53 |
| OTU domain containing 6A | OTUD6A | 0.00011 | −5.70 | −3.59 | −28.99 |
| complement component 1, q subcomponent-like 3 | C1QL3 | 0.00232 | −5.67 | −2.89 | −9.59 |
| TRAF family member-associated NFKB activator | TANK | 0.00001 | −5.66 | −1.28 | −20.39 |
| interferon induced with helicase C domain 1 | IFIH1 | 0.00001 | −5.60 | 1.21 | −10.96 |
| Paired box protein Pax-4 (Predicted) | PAX4 | 0.00001 | −5.45 | 2.04 | −2.45 |
| IQ motif containing E | IQCE | <0.00001 | −5.18 | 1.75 | −7.52 |
| kinesin family member 20A | KIF20A | 0.00003 | −5.16 | −2.21 | −6.68 |
| nucleolar protein 10 | NOL10 | <0.00001 | −5.15 | 1.08 | −14.02 |
| Aryl hydrocarbon receptor nuclear translocator (ARNT protein)(Dioxin receptor, nuclear translocator)(Hypoxia-inducible factor 1 beta)(HIF-1 beta) | ARNT | <0.00001 | −5.12 | 1.04 | −13.43 |
| Ribosomal protein L7-like | LOC100339887 | 0.00001 | −5.10 | −1.23 | −6.70 |
| serine threonine kinase 39 | STK39 | 0.00026 | −5.10 | −2.06 | −4.41 |
| SPHK1 interactor, AKAP domain containing | SPHKAP | 0.00629 | −4.99 | −2.87 | −3.70 |
| NADP-dependent oxidoreductase domain-containing protein 1 EC = 1.-.-.- | NOXRED1 | 0.00004 | −4.98 | −1.28 | −11.39 |
| neurogranin (protein kinase C substrate, RC3) | NRGN | 0.00291 | −4.98 | −1.08 | −2.47 |
| Acidic ribosomal phosphoprotein P0 | 36B4 | 0.00003 | −4.94 | 1.43 | −6.27 |
| ring finger protein 38 | RNF38 | 0.00023 | −4.93 | −3.33 | −9.14 |
| TIA1 cytotoxic granule-associated RNA binding protein-like 1 | TIAL1 | 0.00001 | −4.88 | 2.34 | −13.80 |
| GDNF family receptor alpha 4 | GFRA4 | 0.00025 | −4.88 | 1.29 | −2.93 |
| inositol polyphosphate phosphatase-like 1 | INPPL1 | 0.00005 | −4.87 | −1.14 | −5.96 |
| POU class 2 homeobox 3 | POU2F3 | <0.00001 | −4.78 | 1.89 | −7.48 |
| DnaJ (Hsp40) homolog, subfamily B, member 4 | DNAJB4 | <0.00001 | −4.76 | −1.86 | −9.11 |
| sulfotransferase family 4A, member 1 (SULT4A1) | SULT4A1 | 0.00285 | −4.75 | −1.75 | −3.88 |
| poly(rC) binding protein 3 | PCBP3 | <0.00001 | −4.72 | 1.19 | −8.33 |
| insulin induced gene 2 | INSIG2 | <0.00001 | −4.71 | −3.78 | −21.07 |
| guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 | GNAI1 | 0.00012 | −4.70 | −1.79 | −4.15 |
| KIAA1549 | KIAA1549 | 0.00019 | −4.67 | −1.05 | −2.55 |
| myosin, light chain 6B, alkali, smooth muscle and non-muscle | MYL6B | 0.00002 | −4.66 | −1.58 | −7.23 |
| exostoses (multiple)−like 3 | EXTL3 | 0.00001 | −4.65 | −4.65 | −3.53 |
| Heterogeneous nuclear ribonucleoprotein C (hnRNP C) | HNRNPC | <0.00001 | −4.61 | −3.29 | −35.05 |
| shroom family member 3 | SHROOM3 | <0.00001 | −4.60 | −3.82 | −7.95 |
| chaperonin containing TCP1, subunit 2 (beta) (CCT2) | CCT2 | <0.00001 | −4.57 | −1.23 | −28.67 |
| enabled homolog (Drosophila) | ENAH | 0.00002 | −4.57 | −1.23 | −16.36 |
| Bcl2 modifying factor | BMF | <0.00001 | −4.55 | −1.04 | −9.18 |
| ribophorin I | RPN1 | 0.00002 | −4.51 | −1.30 | −15.81 |
| zinc finger CCCH-type containing 7B | ZC3H7B | 0.00176 | −4.48 | −3.56 | −3.09 |
| inhibin, beta E | INHBE | 0.00005 | −4.42 | 1.58 | −1.73 |
| sialic acid binding Ig-like lectin 14 | SIGLEC14 | <0.00001 | −4.41 | 1.14 | −6.80 |
| gap junction protein, delta 3, 31.9kDa | GJD3 | <0.00001 | −4.40 | 1.35 | −7.33 |
| interferon-related developmental regulator 1 | IFRD1 | <0.00001 | −4.37 | 1.33 | −51.14 |
| TPA-induced transmembrane protein | TTMP | 0.00014 | −4.35 | −1.23 | −10.35 |
| POU class 3 homeobox 4 | POU3F4 | 0.00030 | −4.33 | 1.01 | −3.55 |
| microtubule associated monoxygenase, calponin and LIM domain containing 1 | MICAL1 | 0.00010 | −4.30 | −1.87 | −4.81 |
| Ribosomal protein L36a-like | LOC100358069 | 0.00001 | −4.28 | −1.04 | −4.82 |
| cysteine-rich protein 3 | CRIP3 | <0.00001 | −4.26 | 1.45 | −10.99 |
| transmembrane protein 109 (TMEM109) | TMEM109 | <0.00001 | −4.26 | −1.07 | −39.14 |
| Phospholamban | PLN | 0.00002 | −4.25 | −2.00 | −7.94 |
| ISL LIM homeobox 2 | ISL2 | 0.00007 | −4.24 | −1.36 | −5.75 |
| protein phosphatase 1, regulatory (inhibitor) subunit 8 | PPP1R8 | <0.00001 | −4.24 | 1.09 | −7.27 |
| dehydrogenase/reductase (SDR family) member 13 | DHRS13 | <0.00001 | −4.23 | 1.02 | −9.82 |
| sarcolemmal associated protein-3 (LOC100009199) | SLAP | <0.00001 | −4.19 | −1.47 | −9.38 |
| Ribosomal protein S7-like | LOC100345715 | 0.00012 | −4.17 | −1.91 | −4.96 |
| Eukaryotic translation initiation factor 1 | LOC100345195 | 0.00004 | −4.16 | −1.06 | −5.61 |
| sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D | SEMA4D | <0.00001 | −4.16 | 1.19 | −3.24 |
| malic enzyme 1, NADP(+)-dependent, cytosolic | ME1 | 0.00007 | −4.16 | −1.52 | −4.11 |
| Poly(A) binding protein, cytoplasmic 4 (inducible form) | PABPC4 | 0.00009 | −4.16 | 1.10 | −5.67 |
| WD repeat domain 61 | WDR61 | <0.00001 | −4.14 | 1.14 | −30.33 |
| guanine nucleotide binding protein (G protein), gamma 3 | GNG3 | <0.00001 | −4.14 | 1.07 | −5.64 |
| CD302 molecule | CD302 | <0.00001 | −4.14 | −1.14 | −17.51 |
| fibroblast growth factor 13 | FGF13 | 0.00088 | −4.12 | −2.20 | −4.00 |
| pancreatic polypeptide receptor 1 (PPYR1) | PPYR1 | 0.00003 | −4.12 | −1.10 | −7.22 |
| membrane-associated ring finger (C3HC4) 6, E3 ubiquitin protein ligase | MARCH6 | 0.00091 | −4.11 | −3.08 | −3.82 |
| ADP-ribosylation factor 4-like | LOC100341862 | 0.00015 | −4.11 | −1.20 | −2.92 |
| bone morphogenetic protein 7 | BMP7 | 0.00001 | −4.08 | −1.21 | −11.02 |
| potassium channel, subfamily K, member 18 | KCNK18 | 0.00022 | −4.05 | 1.14 | −2.91 |
| Aggrecanase-2 | ADAMTS-11∧, + | 0.00387 | −4.02 | −1.79 | −4.44 |
∧ Differentially expressed genes that are common between ‘hypertension only’ and ‘hypercholesterolemia plus sham’ groups (both vs. sham controls).+Differentially expressed genes that are common among ‘hypertension only’, ‘hypercholesterolemia plus sham’, and ‘hypertension plus hypercholesterolemia’ groups (all vs. sham controls).
The IPA network with the ‘largest number of up-regulated focus genes’, contained 22 focus genes with functions in Tissue Development, Connective Tissue Disorders, Developmental Disorder. Focus genes in this network were ALAS2, CCL19, CD2, CD4, CD244, CLEC1B, CNP, DHRS9, FKBP1B, IAPP, KLF13, LTN1, MS4A2, RASGRP4, RNASEL, SLC5A1, SPTA1, SYK, TMEM173, TRAF3IP1, TYMP and UBASH3B. They were related to ERK1/2, Interferon alpha, IL12 complex and SYK (complex), CD2, CD4 and CD244 (Figure 11, Table 5). The next network of up-regulated focus genes had 18 focus genes, with functions in Organ Morphology, Visual System Development and Function, Lipid Metabolism. Focus genes were CA2, CDH17, COL17A1, ELOVL3, FANCC, FCRLA, FLT3, HRH1, MAP3K4, MEP1B, MMP1, NFkBID, NR1D1, PIGR, RET, SERPINB2, SPINK1 and TINF2; they were related to P38 MAPK, NFkB, SERPINB2, MMP1 and TNF (Figure 12, Table 5).
Figure 11. IPA network showing the network with the largest number of up-regulated focus genes in the hypertension plus hypercholesterolemia group, compared with sham operated controls.
Figure 12. IPA network showing the network with the second largest number of up-regulated focus genes in the hypertension plus hypercholesterolemia group, compared with sham operated controls.
The network with the largest number of down-regulated focus genes contained 21 focus genes with functions in Cardiovascular Disease, Cellular Assembly and Organization, Post-Translational Modification. Focus genes in this network were ARNT, CCT2, ELAVL4, HNRNPC, KIF20A, ME1, NCALD, NRGN, PDK4, PFDN5, PLN, PPP1R8, PPYR1, SHROOM3, SNAP25, SNTA1, SSB, STMN2, SYT12, TTN and WAC. They were related to ERK, Akt, PKC, Vegf, actin and insulin. (Figure 13, Table 6). The next network of down-regulated focus genes had 20 focus genes with functions in Endocrine System Disorders, Gastrointestinal Disease, and Hereditary Disorder. Focus genes were BMF, CDX1, CKM, CRYAA, DNAJB4, DNAJC6, GTF2E2, IFIH1, IFRD1, INSIG2, PAX4, PLP1, POU2F3, PSMD4, RPN1, SAP30, SMARCA5, TANK, TIAL1 and WDR61; they were related to P38 MAPK, NFkB, 26S proteasome, histone H3, interferon alpha and IL12 (Figure 14, Table 6).
Figure 13. IPA network showing the network with the largest number of down-regulated focus genes in the hypertension plus hypercholesterolemia group, compared with sham operated controls.
Figure 14. IPA network showing the network with the second largest number of down-regulated focus genes in the hypertension plus hypercholesterolemia group, compared with sham operated controls.
2.4. Microarray analyses of the common area between hypertension only- and hypercholesterolemia plus sham groups
The gene expression profile in the common area between the hypertension only- and hypercholesterolemia plus sham groups was also compared with that of sham controls on a normal diet (Fig. 2A). After unknown and repeated genes were omitted, 18 up-regulated and 13 down-regulated genes (greater than 4-fold change) were found (Figure 2). Among the highly up-regulated genes in the MCA of the hypertension only group compared to sham controls were Nibrin like (LOC100352398), TAF15 and ANKAR (Table 7). Among the highly down-regulated genes in the MCA of the hypertension only group were FOXN1, ribosomal protein S3a-like and ADAMTS17 (Table 8).
Table 7. Up-regulated genes in the MCA that are common between ‘hypertension only’ and ‘hypercholesterolemia plus sham’ rabbits (both vs. sham controls) with greater than 4-fold change (see Fig. 2).
| Gene | Gene Symbol | Corrected p-value | Hypertension only | Hyperchole-sterolemia plus sham | Hypertension plus hyperchole-sterolemia |
| Fold Change | Fold Change | Fold Change | |||
| Nibrin-like | LOC100352398 | 0.00171 | 7.92 | 4.71 | 3.74 |
| TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68 kDa | TAF15 | 0.00004 | 7.45 | 4.71 | 4.65 |
| ankyrin and armadillo repeat containing | ANKAR | 0.00006 | 7.05 | 7.99 | 5.23 |
| N(alpha)-acetyltransferase 25, NatB auxiliary subunit | NAA25 | <0.00001 | 6.43 | 8.91 | 18.66 |
| Fanconi anemia, complementation group C | FANCC | <0.00001 | 6.10 | 4.25 | 6.39 |
| Serine proteinase inhibitor, clade B, member 2 (Predicted) | SERPINB2 | 0.00005 | 5.63 | 6.99 | 9.10 |
| IDI1 protein-like | LOC100346274 | 0.00042 | 5.51 | 4.87 | 6.67 |
| meprin A, beta | MEP1B | 0.00291 | 5.37 | 5.12 | 5.85 |
| Fibroblast growth factor binding protein 1-like | LOC100353835 | 0.00122 | 5.01 | 4.01 | 4.63 |
| ankyrin repeat and SOCS box-containing 4 | ASB4 | 0.00027 | 4.98 | 4.06 | 3.51 |
| Ankyrin repeat-containing protein C20orf12 | DZANK1 | <0.00001 | 4.87 | 5.04 | 7.37 |
| signal-induced proliferation-associated 1 like 3 | SIPA1L3 | 0.00001 | 4.80 | 8.45 | 6.50 |
| relaxin/insulin-like family peptide receptor 2 | RXFP2 | 0.00029 | 4.79 | 5.62 | 6.96 |
| guanylate cyclase 2D, membrane (retina-specific) | GUCY2D | 0.00008 | 4.64 | 4.11 | 8.34 |
| testis-specific kinase 2 | TESK2 | 0.00571 | 4.50 | 5.11 | 2.41 |
| Ribonuclease, RNase A family, 1 (pancreatic) | RNASE1 | <0.00001 | 4.17 | 5.62 | 4.82 |
| DIS3 mitotic control homolog (S. cerevisiae)-like 2 | DIS3L2 | 0.00024 | 4.17 | 4.32 | 3.96 |
| Glucose-fructose oxidoreductase domain containing 2-like | LOC100351150 | 0.00001 | 4.01 | 6.34 | 12.42 |
Table 8. Down-regulated genes in the MCA that are common between ‘hypertension only’ and ‘hypercholesterolemia plus sham’ rabbits (both vs. sham controls) with greater than 4-fold change.
| Gene | Gene Symbol | Corrected p-value | Hypertension only | Hyperchole-sterolemia plus sham | Hypertension plus hyperchole-sterolemia |
| Fold Change | Fold Change | Fold Change | |||
| forkhead box N1 | FOXN1 | 0.00048 | −26.20 | −56.89 | −45.12 |
| Ribosomal protein S3a-like | LOC100354966 | <0.00001 | −16.54 | −5.74 | −1.83 |
| ADAM metallopeptidase with thrombospondin type 1 motif, 17 | ADAMTS17 | 0.00001 | −15.85 | −7.25 | −3.06 |
| peptidylprolyl isomerase G (cyclophilin G) | PPIG | <0.00001 | −12.61 | −6.20 | −1.81 |
| hematopoietic prostaglandin D synthase | HPGDS | 0.00043 | −12.38 | −21.44 | −13.63 |
| large subunit GTPase 1 homolog (S. cerevisiae) | LSG1 | <0.00001 | −7.69 | −20.38 | −1.93 |
| mitochondrial ribosomal protein L15 | MRPL15 | <0.00001 | −6.97 | −23.01 | −9.57 |
| translocase of outer mitochondrial membrane 5 homolog (yeast) | TOMM5 | 0.00014 | −5.67 | −4.05 | −1.60 |
| ribosomal protein L26 | RPL26 | 0.00016 | −5.27 | −14.60 | −2.81 |
| family with sequence similarity 177, member A1 | FAM177A1 | 0.00016 | −4.41 | −10.75 | −1.95 |
| Proteasome (prosome, macropain) 26s subunit, non-ATPase, 4 | PSMD4 | 0.00005 | −4.34 | −17.67 | −11.67 |
| Aggrecanase-2 | ADAMTS-11 | 0.00005 | −4.26 | −9.31 | −6.51 |
| BRCA2 and CDKN1A interacting protein | BCCIP | <0.00001 | −4.06 | −14.00 | −2.58 |
A single network of up-regulated focus genes was found, which contained 13 focus genes with functions in Drug Metabolism, Endocrine System Development and Function, Lipid Metabolism. Focus genes in this network were ASB4, DIS3L2, DZANK1, FANCC, GUCY2D, MEP1B, NAA25, RNASE1, RXFP2, SERPINB2, SIPA1L3, TAF15 and TESK2. They were related to UBC, APP, SERPINB2, TNF and HNF4A (Figure 15, Table 7).
Figure 15. IPA network showing the network with the largest number of up-regulated focus genes in the common area between the hypertension only and hypercholesterolemia plus sham group, compared with sham operated controls.
A single network of down-regulated focus genes was found, which contained 11 focus genes with functions in Cell Morphology, Embryonic Development, and Cellular Compromise. Focus genes were ADAMTS17, BCCIP, FAM177A1, FOXN1, HPGDS, LSG1, MRPL15, PPIG, PSMD4, RPL26 and TOMM5. They were related to UBC (Figure 16, Table 8).
Figure 16. IPA network showing the network with the largest number of down-regulated focus genes in the common area between the hypertension only and hypercholesterolemia plus sham group, compared with sham operated controls.
2.5. Microarray analyses of the ‘exclusive area’ in the hypertension plus hypercholesterolemia group
The gene expression profile in the exclusive area of the hypertension plus hypercholesterolemia group was compared with that of sham controls on a normal diet (Fig. 2A). After unknown and repeated genes were omitted, 132 up-regulated and 22 down-regulated genes (greater than 4-fold change) were found (Figure 2). Among the highly up-regulated genes in the MCA of the hypertension plus hypercholesterolemia compared to sham controls were SLFN, MRS2 and HEAT repeat containing (LOC100357872) (Table 9). Among the highly down-regulated genes in the MCA of the hypertension group were SST, ADAM6, and PRLR (Table 10). The panel of genes was analyzed by IPA.
Table 9. Up-regulated genes in the MCA that are exclusive to ‘hypertension plus hypercholesterolemia’ rabbits vs. sham controls with greater than 4-fold change.
| Gene | GeneSymbol | Corrected p-value | Hypertension plus hypercholesterolemia |
| Fold change | |||
| schlafen family member 14 | SLFN14 | 0.00614 | 13.64 |
| MRS2 magnesium homeostasis factor homolog (S. cerevisiae) | MRS2 | 0.00000 | 8.85 |
| HEAT repeat containing 3 | LOC100357872 | 0.00100 | 8.34 |
| membrane-spanning 4-domains, subfamily A, member 2 | MS4A2 | 0.00891 | 8.29 |
| kinesin family member 21A | KIF21A | 0.00003 | 7.31 |
| N(alpha)-acetyltransferase 25, NatB auxiliary subunit | NAA25 | 0.00099 | 7.23 |
| General transcription factor IIB | LOC100359058 | 0.00015 | 7.00 |
| interleukin 20 receptor, alpha | IL20RA | 0.00000 | 6.96 |
| solute carrier family 18 (vesicular monoamine), member 1 | SLC18A1 | 0.00074 | 6.64 |
| IQ motif and ubiquitin domain containing | IQUB | 0.00104 | 6.64 |
| family with sequence similarity 71, member C | FAM71C | 0.00026 | 6.63 |
| secretoglobin, family 3A, member 1 | SCGB3A1 | 0.00010 | 6.42 |
| STAM binding protein-like 1 | STAMBPL1 | 0.00286 | 6.39 |
| tripartite motif-containing 67 | TRIM67 | 0.00175 | 6.39 |
| Serum response factor binding protein 1-like | LOC100349313 | 0.00139 | 6.34 |
| G protein-coupled receptor 119 | GPR119 | 0.00058 | 6.30 |
| 2′,3′-cyclic nucleotide 3′ phosphodiesterase | CNP | 0.00000 | 6.14 |
| steroidogenic acute regulatory protein | STAR | 0.00007 | 6.05 |
| CD4 molecule (CD4) | CD4 | 0.00004 | 6.02 |
| DOMON domain-containing protein FRRS1L | FRRS1L | 0.00003 | 5.99 |
| Tubulin tyrosine ligase-like family, member 5 | LOC100359240 | 0.00019 | 5.99 |
| thymidine phosphorylase | TYMP | 0.00005 | 5.87 |
| dynein heavy chain domain 1 | DNHD1 | 0.00134 | 5.87 |
| Insulin-like 5 | INSL5 | 0.00018 | 5.85 |
| Transcription elongation factor A (SII)-like 2-like | LOC100340955 | 0.00015 | 5.75 |
| Solute carrier family 5 (sodium/glucose cotransporter), member 9 | SLC5A9 | 0.00002 | 5.73 |
| Kruppel-like factor 13 | KLF13 | 0.00007 | 5.69 |
| Granulate-macrophage stimulating factor (Predicted) | GMCSF | 0.00004 | 5.68 |
| listerin E3 ubiquitin protein ligase 1 | LTN1 | 0.00009 | 5.66 |
| carboxylesterase 8 (putative) | CES4A | 0.00023 | 5.63 |
| C-type lectin domain family 1, member B | CLEC1B | 0.00584 | 5.60 |
| solute carrier family 15, member 5 | SLC15A5 | 0.00075 | 5.55 |
| glucosidase, beta, acid 3 (cytosolic) | GBA3 | 0.00012 | 5.55 |
| Proline-rich protein 3 (MHC class I region proline-rich protein CAT56) | PRR3 | 0.00129 | 5.50 |
| chemokine (C-C motif) ligand 19 | CCL19 | 0.00055 | 5.50 |
| AchR epsilon subunit | CHRNE | 0.00010 | 5.43 |
| Transmembrane protein C5orf28 | C5orf28 | 0.00319 | 5.38 |
| dehydrogenase/reductase (SDR family) member 9 | DHRS9 | 0.00168 | 5.37 |
| FCH and double SH3 domains 1 | FCHSD1 | 0.00015 | 5.37 |
| Uncharacterized protein C7orf72 | C2orf72 | 0.00114 | 5.35 |
| family with sequence similarity 71, member F2 | FAM71F2 | 0.00643 | 5.34 |
| otopetrin 2 | OTOP2 | 0.00006 | 5.31 |
| elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3 | ELOVL3 | 0.00012 | 5.30 |
| solute carrier family 7, (cationic amino acid transporter, y+ system) member 13 | SLC7A13 | 0.00134 | 5.30 |
| Uncharacterized protein C2orf71 | C2orf71 | 0.00177 | 5.30 |
| solute carrier family 14 (urea transporter), member 1 (Kidd blood group) | SLC14A1 | 0.00027 | 5.21 |
| RAS guanyl releasing protein 4 | RASGRP4 | 0.00058 | 5.11 |
| ATPase, class I, type 8B, member 2 | ATP8B2 | 0.00360 | 5.07 |
| Cytochrome P450, family 4, subfamily A, polypeptide 5 | CYP4A5 | 0.00061 | 5.05 |
| Pregnancy-zone protein | PZP | 0.00652 | 5.02 |
| ATPase, H+/K+ exchanging, alpha polypeptide (ATP4A) | ATP4A | 0.00000 | 5.02 |
| CD2 molecule | CD2 | 0.00064 | 4.99 |
| 3-oxo-5-alpha-steroid 4-dehydrogenase 2-like | LOC100343882 | 0.00001 | 4.99 |
| phosphatidylcholine transfer protein | PCTP | 0.00001 | 4.96 |
| transmembrane emp24 protein transport domain containing 6 | TMED6 | 0.00040 | 4.96 |
| actin-related protein T1 | ACTRT1 | 0.00005 | 4.94 |
| Wings apart-like homolog | LOC100348678 | 0.00041 | 4.94 |
| Fructose-1,6-bisphosphatase 1 | FBP1 | 0.00050 | 4.88 |
| neuron navigator 2 | NAV2 | 0.00080 | 4.87 |
| potassium channel tetramerisation domain containing 19 | KCTD19 | 0.00123 | 4.87 |
| spermatogenesis associated, serine-rich 1 | SPATS1 | 0.00201 | 4.83 |
| Solute carrier family 5 (sodium/glucose cotransporter), member 1 | LOC100009262 | 0.00001 | 4.80 |
| tripartite motif-containing 35 | TRIM35 | 0.00084 | 4.80 |
| cytochrome P450, family 26, subfamily C, polypeptide 1 | CYP26C1 | 0.00224 | 4.79 |
| Polymeric immunoglobulin receptor Precursor (Poly-Ig receptor)(PIGR) | PIGR | 0.00025 | 4.75 |
| Ras and Rab interactor-like | RINL | 0.00135 | 4.71 |
| ubiquitin associated and SH3 domain containing B | UBASH3B | 0.00803 | 4.70 |
| UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 (GalNAc-T13) | GALNT13 | 0.00000 | 4.70 |
| PRP18 pre-mRNA processing factor 18 homolog (S. cerevisiae) | PRPF18 | 0.00144 | 4.68 |
| spinster homolog 3 (Drosophila) | SPNS3 | 0.00049 | 4.68 |
| galactosidase, beta 1-like 3 | GLB1L3 | 0.00003 | 4.67 |
| iroquois homeobox 2 | IRX2 | 0.00052 | 4.66 |
| collagen, type XVII, alpha 1 | COL17A1 | 0.00028 | 4.65 |
| patched 2 | PTCH2 | 0.00005 | 4.65 |
| DEAQ box RNA-dependent ATPase 1 | DQX1 | 0.00520 | 4.60 |
| Potassium channel, subfamily K, member 1 | KCNK1 | 0.00217 | 4.58 |
| purinergic receptor P2Y, G-protein coupled, 10 | P2RY10 | 0.00197 | 4.57 |
| somatostatin receptor 2 | SSTR2 | 0.00077 | 4.56 |
| zinc finger, DHHC-type containing 19 | ZDHHC19 | 0.00241 | 4.55 |
| ret proto-oncogene | RET | 0.00036 | 4.55 |
| Oxytocin receptor | OXTR | 0.00517 | 4.52 |
| serine peptidase inhibitor, Kazal type 1 | SPINK1 | 0.00055 | 4.50 |
| membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) | MPP2 | 0.00409 | 4.50 |
| solute carrier family 13 (sodium/sulfate symporters), member 1 | SLC13A1 | 0.00325 | 4.49 |
| follistatin-like 5 | FSTL5 | 0.00071 | 4.49 |
| Mitochondrial ribosomal protein S15-like | LOC100345026 | 0.00064 | 4.49 |
| TERF1 (TRF1)-interacting nuclear factor 2 | TINF2 | 0.00325 | 4.49 |
| keratin associated protein 19-4 | KRTAP19-4 | 0.00044 | 4.48 |
| XK, Kell blood group complex subunit-related, X-linked | XKRX | 0.00051 | 4.47 |
| FK506 binding protein 1B, 12.6 kDa | FKBP1B | 0.00274 | 4.46 |
| phospholipase C, zeta 1 | PLCZ1 | 0.00002 | 4.45 |
| CD244 molecule, natural killer cell receptor 2B4 | CD244 | 0.00003 | 4.44 |
| carboxyl ester lipase (bile salt-stimulated lipase) (CEL) | CEL | 0.00016 | 4.44 |
| ADAM metallopeptidase with thrombospondin type 1 motif, 18 | ADAMTS18 | 0.00213 | 4.43 |
| UDP-glucuronosyltransferase | UGT2C1 | 0.00035 | 4.42 |
| fms-related tyrosine kinase 3 | FLT3 | 0.00270 | 4.41 |
| mitogen-activated protein kinase binding protein 1 | MAPKBP1 | 0.00003 | 4.41 |
| Ammonium transporter Rh type C (Rhesus blood group family type C glycoprotein)(Rh family type C glycoprotein)(Rh type C glycoprotein) | RHCG | 0.00032 | 4.41 |
| Kelch-like 1 (Drosophila) | KLHL1 | 0.00003 | 4.35 |
| leucine rich repeat transmembrane neuronal 4 | LRRTM4 | 0.00002 | 4.34 |
| proteoglycan 3 | PRG3 | 0.00254 | 4.33 |
| Ligand of numb-protein X 1-like | LOC100343259 | 0.00568 | 4.33 |
| Chymase like protein | LOC100008644 | 0.00060 | 4.32 |
| sex comb on midleg-like 4 (Drosophila) | SCML4 | 0.00854 | 4.32 |
| BMS1-like, ribosome assembly protein | LOC100339194 | 0.00017 | 4.31 |
| Uncharacterized protein C2orf61 | C2orf61 | 0.00893 | 4.31 |
| Protein FAM55A | LOC100009342 | 0.00326 | 4.30 |
| cyclin-dependent kinase 15 | CDK15 | 0.00129 | 4.28 |
| Rac GTPase activating protein 1 pseudogene | RACGAP1P | 0.00023 | 4.27 |
| family with sequence similarity 170, member A | FAM170A | 0.00008 | 4.23 |
| Mitogen-activated protein kinase kinase kinase 4 | MAP3K4 | 0.00788 | 4.22 |
| ATPase, H+ transporting, lysosomal 70 kDa, V1 subunit A | ATP6V1A | 0.00397 | 4.19 |
| FRY-like | FRYL | 0.00291 | 4.18 |
| clusterin-like 1 (retinal) | CLUL1 | 0.00072 | 4.17 |
| Superiorcervical ganglia, neural specific 10 | LOC100344464 | 0.00228 | 4.14 |
| histamine receptor H1 | HRH1 | 0.00001 | 4.13 |
| Cingulin (Predicted) | Cgn | 0.00236 | 4.12 |
| nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, delta | NFKBID | 0.00103 | 4.12 |
| ribonuclease, RNase A family, 1 (pancreatic) | RNASE1 | 0.00043 | 4.11 |
| Nuclear prelamin A recognition factor | LOC100344024 | 0.00002 | 4.09 |
| actin, beta-like 2 | ACTBL2 | 0.00023 | 4.09 |
| Ghrelin receptor | LOC100101582 | 0.00062 | 4.08 |
| olfactomedin 2 | OLFM2 | 0.00385 | 4.07 |
| COX10 homolog, cytochrome c oxidase assembly protein, heme A: farnesyltransferase (yeast) | COX10 | 0.00160 | 4.07 |
| ribonuclease L (2′,5′-oligoisoadenylate synthetase-dependent) | RNASEL | 0.00028 | 4.06 |
| keratin associated protein 11-1 | KRTAP11-1 | 0.00013 | 4.04 |
| DnaJ (Hsp40) homolog, subfamily C, member 5 beta | DNAJC5B | 0.00016 | 4.03 |
| receptor (chemosensory) transporter protein 1 | RTP1 | 0.00012 | 4.03 |
| protocadherin 8 | PCDH8 | 0.00005 | 4.02 |
| Ubiquitin specific protease 39 | LOC100344046 | 0.00343 | 4.02 |
| proline/serine-rich coiled-coil 1 | PSRC1 | 0.00571 | 4.01 |
| cadherin 17, LI cadherin (liver-intestine) | CDH17 | 0.00459 | 4.00 |
Table 10. Down-regulated genes in the MCA that are exclusive to ‘hypertension plus hypercholesterolemia’ rabbits vs. sham controls with greater than 4-fold change.
| Gene | GeneSymbol | Corrected p-value | Hypertension plus hypercholesterolemia |
| Fold change | |||
| somatostatin | SST | 0.00169 | −10.49 |
| cellular disintegrin ADAM 6d (ADAM6) | ADAM6 | 0.00009 | −9.13 |
| prolactin receptor | PRLR | 0.00001 | −8.55 |
| ELAV (embryonic lethal, abnormal vision, Drosophila)−like 4 (Hu antigen D) | ELAVL4 | 0.00033 | −8.49 |
| Proteolipid protein 1 | PLP1 | 0.00578 | −8.28 |
| stathmin-like 2 | STMN2 | 0.00129 | −7.60 |
| olfactomedin 3 | OLFM3 | 0.00005 | −6.99 |
| pyruvate dehydrogenase kinase, isozyme 4 | PDK4 | 0.00711 | −6.98 |
| myelin-associated oligodendrocyte basic protein | MOBP | 0.00776 | −6.94 |
| Paired box protein Pax-4 (Predicted) | PAX4 | 0.00001 | −5.45 |
| SPHK1 interactor, AKAP domain containing | SPHKAP | 0.00629 | −4.99 |
| neurogranin (protein kinase C substrate, RC3) | NRGN | 0.00291 | −4.98 |
| GDNF family receptor alpha 4 | GFRA4 | 0.00025 | −4.88 |
| Sulfotransferase family 4A, member 1 | SULT4A1 | 0.00285 | −4.75 |
| KIAA1549 | KIAA1549 | 0.00019 | −4.67 |
| zinc finger CCCH-type containing 7B | ZC3H7B | 0.00176 | −4.48 |
| inhibin, beta E | INHBE | 0.00005 | −4.42 |
| POU class 3 homeobox 4 | POU3F4 | 0.00030 | −4.33 |
| sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D | SEMA4D | 0.00000 | −4.16 |
| membrane-associated ring finger (C3HC4) 6, E3 ubiquitin protein ligase | MARCH6 | 0.00091 | −4.11 |
| ADP-ribosylation factor 4-like | LOC100341862 | 0.00015 | −4.11 |
| potassium channel, subfamily K, member 18 | KCNK18 | 0.00022 | −4.05 |
The IPA network with the largest number of up-regulated focus genes contained 20 focus genes with functions in Cell Death and Survival, Cellular Compromise, Inflammatory Response. Focus genes in this network were CCL19, CD2, CD4, CD244, CDH17, CGN, CNP, COL17A1, DHRS9, FLT3, IQUB, KLF13, LTN1, MS4A2, PIGR, RASGRP4, RET, RNASEL, TYMP and UBASH3B. They were related to P38 MAPK, ERK 1/2, interferon alpha and CD2, CD4 and CD244 (Figure 17, Table 9). The next network of up-regulated focus genes had 14 focus genes with functions in Cell Cycle, Nervous System Development and Function, Cell Death and Survival. Focus genes were C5orf28, CDK15, FAM71C, GBA3, GLB1L3, IRX2, LRRTM4, NAA25, NAV2, PRPF18, PSRC1, PTCH2, SLC13A1 and STAMBPL1; they were related to UBC and APP (Figure 18, Table 9).
Figure 17. IPA network showing the network with the largest number of up-regulated focus genes in the hypertension plus hypercholesterolemia group (exclusive area), compared with sham operated controls.
Figure 18. IPA network showing the network with the second largest number of up-regulated focus genes in the hypertension plus hypercholesterolemia group (exclusive area), compared with sham operated controls.
The network with the largest number of down-regulated focus genes contained 10 focus genes with functions in Digestive System Development and Function, Hepatic System Development and Function, Organ Morphology. Focus genes in this network were ELAVL4, GFRA4, KCNK18, PAX4, PDK4, POU3F4, PRLR, SPHKAP, SST and STMN2. They were related to MAPK, ERK and insulin (Figure 19, Table 10). The next network of down-regulated focus genes also had 10 focus genes with functions in Metabolic Disease, Gene Expression, and Cellular Compromise. Focus genes were INHBE, KIAA1549, MARCH6, MOBP, NRGN, OLFM3, PLP1, SEMA4D, SULT4A1 and ZC3H7B; they were related to UBC and APP (Figure 20, Table 10).
Figure 19. IPA network showing the network with the largest number of down-regulated focus genes in the hypertension plus hypercholesterolemia group (exclusive area), compared with sham operated controls.
Figure 20. IPA network showing the network with the second largest number of down-regulated focus genes in the hypertension plus hypercholesterolemia group (exclusive area), compared with sham group.
3. Electron Microscopy of the MCA
The MCA of sham operated rabbits on a normal diet showed continuous healthy appearing endothelial cells (Figure 21A). In comparison, the MCA of hypertension only rabbits contained pyknotic endothelial cells (Figure 21B), while that of hypercholesterolemia plus sham rabbits showed large intracellular vacuoles in endothelial cells (Figure 21C). The above changes were exacerbated in the hypertension plus hypercholesterolemia rabbits, and pyknotic endothelial cells, breaks in the basement membrane, and large extracellular spaces were present between the basement membrane and underlying smooth muscle cells (Figure 21D, E). In addition, subendothelial foam cells were observed (Figure 21E, F) consistent with early atherosclerotic changes. The tunica media and tunica adventitia had a normal appearance.
Figure 21. Electron micrographs of the MCA.
(A) Sham operated rabbit on a normal diet showing continuous endothelial cells (EC) (B) Hypertension only rabbit, showing a pyknotic cell (arrow) in the endothelial layer (EC). (C) Hypercholesterolemia plus sham rabbit, showing large intracellular vacuoles (V) in endothelial cells. (D) Hypertension plus hypercholesterolemia rabbit, showing breaks in the basement membrane (BR), and a large extracellular space (S) between the basement membrane and the underlying smooth muscle cells. (E) Hypertension plus hypercholesterolemia rabbit, showing a pyknotic cell among the endothelial layer (arrow), and presence subendothelial foam cells (FC). (F) Higher magnification of a foam cell in E, showing intracellular vacuoles, and extracellular spaces (S) containing collagen fibrils. Scale: A = 1 µm, B–D = 0.5 µm, E = 2 µm, F = 0.2 µm.
4. Vascular Changes in the Aorta
4.1. RT-PCR
HNF4A mRNA expression was increased in the aorta of the hypercholesterolemia plus sham-operated group- and hypertension plus hypercholesterolemia group (3.64 and 2.25-fold change respectively), compared to controls on a normal diet (Figure 22A).
Figure 22. HNF4A expression in the aorta.
(A) Real-time RT-PCR analyses of HNF4A in the aorta of control, hypercholesterolemia plus sham, and hypertension plus hypercholesterolemia rabbits. The mean and standard error are shown. *p<0.05 vs. controls by one-way ANOVA with Bonferroni’s multiple comparison post-hoc test (n = 4 in each group). (B) Western blot analyses of HNF4A in the aorta of control, hypercholesterolemia plus sham and hypertension plus hypercholesterolemia rabbits. (C) Densitometric analyses of HNF4A protein, normalized to ß-actin. The mean and standard error are shown. *p<0.05 vs. control by one-way ANOVA with Bonferroni’s multiple comparison post-hoc test (n = 3 in each group). Abbreviations as in Fig. 1.
4.2. Western blot analyses
The antibody to HNF4A detected a 53 kDa band in homogenates of the aorta consistent with the expected molecular weight of the protein (Figure 22B). Increased density of the HNF4A band relative to beta actin was found in homogenates from hypercholesterolemia plus sham- and hypertension plus hypercholesterolemia group compared to controls, indicating up-regulation of HNF4A protein expression after exposure to hypertension and/or hypercholesterolemia (Figure 22C).
4.3. Histochemistry and immunohistochemistry
The general structure of the aorta was examined by Masson’s Trichrome staining (Figure 23A–C). Hypercholesterolemia plus sham rabbits as well as the hypertension plus hypercholesterolemia rabbits showed neointimal formation along part of the circumference of the vessel. This was associated with migration of red-staining, smooth muscle cells from the tunica media into the neointima (Figure 23C). The changes were more pronounced in the hypertension plus hypercholesterolemia than the hypercholesterolemia plus sham rabbits (Figure 23A–C). The tunica media and tunica adventitia had a normal appearance.
Figure 23. Histochemical and immunohistochemical staining of the aorta from rabbits exposed to stroke risk factors.
A, D, G: Rabbits on normal diet. B, E, H: Hypercholesterolemia plus sham group. C, F, I: Hypertension plus hypercholesterolemia group. A-C: Aorta of rabbits stained by Masson’s Trichrome. Increased thickness of the neointima (NI) is seen in the hypercholesterolemia plus sham group (B, arrow). The changes are exacerbated in the hypertension plus hypercholesterolemia group (C, arrow). SM = smooth muscle cells in the neointima. D, E, F: Aorta of rabbits immunostained with a mouse monoclonal antibody to HNF-4A. Very little or no labeling is present in normal rabbits, but dense staining is observed in endothelial cells in the hypercholesterolemia plus sham, and hypertension plus hypercholesterolemia rabbits (arrows). G, H,I: Higher magnification of the aorta of rabbits immunostained with mouse monoclonal antibody to HNF-4A, showing dense staining in endothelial cells of hypercholesterolemia plus sham, and hypertension plus hypercholesterolemia rabbits (arrows). Scale: A-F = 200 µm, G-I = 70 µm.
Immunostaining of the aorta with HNF4A antibody showed that the endothelial layer of hypercholesterolemia plus sham group and the hypertension plus hypercholesterolemia group were densely stained for HNF4A, compared to controls (Figure 23D–I). Immunolabel was observed in the nucleus and cytoplasm of endothelial cells and other cells near the endothelial layer. No staining was observed in the tunica media or adventitia (Figure 23D-I). These results indicate that increased HNF4A gene expression in the aorta occurred mainly in endothelial cells.
Discussion
The present study was carried out to elucidate differential gene expression changes in the MCA of rabbits exposed to two stroke risk factors, i.e. hypertension and/or hypercholesterolemia. Of the DEGs in the MCA that were altered by a single risk factor, hypertension alone vs. sham controls on a normal diet, FAM167A had the highest fold change, followed by CERS3 and FAM53C. FAM167A encodes a ubiquitously expressed gene, the function of which remains unknown. CERS3 (ceramide synthase 3) catalyzes the condensation of sphinganine and fatty acyl-coenzyme A to form dihydroceramide, which is oxidized to ceramide [30]. Among the down-regulated DEGs in the MCA of the hypertension only group were FOXN1, NSRP1 and THUMPD3. Forkhead transcription factor is essential for thymus development [31] and keratinocyte differentiation [32].
Of the DEGs in the MCA that were up-regulated in the hypercholesterolemia plus sham group vs. sham controls on a normal diet, SLFN14 had the largest fold change, followed by CA1 and Gap protein alpha-3 protein-like (LOC100357902). SLFN14 is part of the Schlafen family of proteins which have growth regulatory properties [33]. CA1 (carbonic anhydrase I) is a member of the carbonic anhydrase family that catalyzes the hydration and dehydration of CO2/H2CO3 [34] and gene mutation is associated with rheumatoid arthritis [35].
Among the DEGs that were down-regulated in the hypercholesterolemia plus sham group vs. sham controls were beta tropomyosin (LOC100125984), PFDN5 and CUL3. The latter is a member of the cullin protein family [36], [37] involved in ubiquitination [38], and gene polymorphism is associated with hypertension [39].
Of the DEGs in the MCA that were up-regulated in the hypertension plus hypercholesterolemia group vs. sham controls on a normal diet, EPHA1 had the largest fold change, followed by SP110, and SLFN14. EPHA1 (Ephrin receptor A1) has recently been identified in large-scale genome-wide association studies to be one of the risk genes for late onset Alzheimer’s disease (AD) [40], [41]. SLFN14 has been mentioned in the hypertension only group.
Among the DEGs that were down-regulated in the hypertension plus hypercholesterolemia group were FOXN1, TNFRSF11B, and GAPDHS. TNFRSF11B (tumor necrosis factor receptor superfamily, member 11b) is the gene encoding osteoprotegerin, a member of the tumor necrosis factor receptor superfamily of cytokines [42] involved in bone resorption [43] and vascular diseases [44], [45]. Gene polymorphism of TNFRSF11B is a risk factor for ischemic stroke [46]. FOXN1 has been mentioned in the hypertension only group.
Of the DEGs in the that were up-regulated in common between the hypertension only- and hypercholesterolemia plus sham groups (both vs. sham controls), Nibrin like (LOC100352398) showed the largest fold change, followed by TAF15, and ANKAR. TAF15 (TATA box binding protein associated factor 15) is a member of the FET family of RNA-binding proteins [47]. ANKAR (ankyrin and armadillo repeat containing) is one of the genes affected in aortic dilatation/dissection [48].
Among the DEGs that were down-regulated in common, between the hypertension only, and hypercholesterolemia plus sham groups (both vs. sham controls) were FOXN1, Ribosomal protein S3-like (LOC100354966) and ADAMTS17 (ADAM metallopeptidase with thrombospondin type 1 motif, 17). The ADAMTS family of genes is involved in cancer, arthritis and coagulation [49], and variants of ADAMTS17 are associated with pediatric stroke [50].
The network in the MCA with largest number of up-regulated focus genes affected by hypertension showed many focus genes related to the ‘node molecule’, ubiquitin, a regulatory protein that directs other proteins to the proteasome [51]. Apart from chronic neurodegenerative diseases, the ubiquitin-proteasome system is implicated in brain ischemia by inducing cell damage or leukocyte infiltration into the brain [51]. The network with the second largest number of up-regulated molecules was related to P38 MAPK and ERK. P38 MAPK (mitogen-activated protein kinase) is a member of the MAPK family involved in stress-related signal transductions [52], and sustained activation can result in apoptosis in various cell types [53], [54], [55]. Inhibition of P38 activity is reported to reduce infarct volume and neurological deficits [52], [56] as well as cytokine expression after stroke [52]. ERK1/2 (extracellular signal-regulated kinase 1/2) is a well-characterized member of the MAPK family that is activated by mitogens or stressors, and plays an important role in cell differentiation and proliferation [57], [58]. Phosphorylated ERK1/2 is increased after cerebral ischemia/reperfusion, and the ERK pathway is involved in both neuroprotection and cell death [58]. Other focus genes in this network were related to NF-κB, SERPINB2, MMP1 and APP. NF-κB (nuclear factor-kappa B) is a central regulator of inflammation and apoptosis [59] and is active in many chronic inflammatory diseases including atherosclerosis [60]. It could have damaging effects in cerebral ischemia [61], [62], and inhibition of NF-κB decreases neointimal formation [63], [64], [65] and reduces infarct volume and neurological deficits after stroke [66]. On the other hand, NF-κB activation could also be neuroprotective [67], [68], as it participates in cell death/survival pathways through regulation of pro- and anti-apoptotic genes [69], [70]. SERPINB2 (serpin peptidase inhibitor, clade B (ovalbumin), member 2), also known as plasminogen activator inhibitor (PAI) type 2 is a physiological inhibitor of urokinase plasminogen activator (uPA) [71]. Increased SERPINB2 expression is found in the AD brain [72], and after brain ischemia or trauma, particularly in the basement membrane and endothelial cells of vessels adjacent to the lesion [73]. MMP1 (matrix metallopeptidase 1) belongs to a family of protein-digesting enzymes that degrades the extracellular matrix in both physiological and pathological conditions including stroke [74]. MMP1 is increased in atherosclerotic plaques [75] and gene polymorphism is suggested to influence the risk of coronary heart disease [76]. APP (β-amyloid precursor protein) can be processed by an amyloidogenic pathway to form A-beta. The latter and vascular risk factors [77], [78] play important roles in the pathogenesis of AD [79], [80], and endothelial dysfunction in APP overexpressing mice increases the susceptibility of the brain to AD pathology [81] and cerebral ischemia [82].
The network in the MCA with the largest number of down-regulated focus genes affected by hypertension was related to MAPK, ERK 1/2, Akt, 26s proteasome, histone and PKC, while the network with the second largest number of down-regulated focus genes was related to UBC. Akt is a serine/threonine kinase that is activated by PI3K in various growth factors-mediated signaling cascades [83]. The PI3K/Akt signaling pathway is important in mediating cell survival [83], [84] and Akt activity is shown to confer neuroprotection after ischemic brain injury [85], [86]. PKC (protein kinase C) is a serine-threonine kinase family that is important in regulating cellular functions, and several isozymes of PKC such as PKC<$>\raster="rg1"<$>, PKCδ and PKCγ, are associated with cerebral ischemic and reperfusion injury [87], [88]. The use of PKCδ peptide inhibitors is reported to alleviate reperfusion injury and reduce stroke infarct size [87], [88], [89]. 26s Proteasome is an essential component of the ubiquitin-proteasome system which functions to degrade cellular proteins [90]. The exact role of the ubiquitin-proteasome system in cerebral ischemia is at present unclear, and deleterious effects of proteasome malfunction, as well as beneficial effects of proteasome inhibition on cerebral ischemia have been reported [51], [90]. Histones affect gene transcription by binding to DNA; hence changes in histone H3 may affect the expression of downstream molecules in vessels during hypercholesterolemia. Interestingly, promising outcomes from the use of histone deacetylase (HDACs) inhibitors have been reported in preclinical stroke models [91].
Gene network analysis of the MCA after hypercholesterolemia (plus sham operation), surprisingly, showed very similar networks as that the hypertension only group. This was despite a relatively low percentage of genes in the common area between these two conditions (20.8% of hypertension genes and 6.8% of hypercholesterolemia genes were in the common area, respectively). The results suggest recruitment of different focus genes that are related to similar ‘node molecules’, as in the following example: the network in the MCA with largest number of up-regulated focus genes affected by hypercholesterolemia showed many molecules related to APP. This is similar to hypertension only rabbits, and could indicate synergism of the two risk factors in affecting the expression of molecules related to APP. Other focus genes were related to tretinoin or all-trans retinoic acid (ATRA) [92], a molecule known to modulate A-beta associated memory deficits and neuropathological alterations in animal models of AD [92], [93].
The network with the second largest number of up-regulated focus genes also had many similarities with the hypertension only group, with P38 MAPK, ERK 1/2, NFkB, SERPINB2 and Akt being central players, together with focus genes related to interferon alpha and VEGF. Interferon alpha is a member of a family of nonspecific antiviral agents with immunomodulatory and cytostatic properties [94]. Although other isoforms of the interferon family such as interferon beta and gamma are associated with atherosclerosis [94], a possible role of interferon alpha in this condition is yet unknown. Vegf (vascular endothelial growth factor) is involved in conditions such as atherosclerosis, cerebral edema, brain and vascular repair following ischemia [95], and plasma vegf values are increased immediately after stroke [96].
The network in the MCA with largest number of down-regulated focus genes affected by hypercholesterolemia is related to ubiquitin, 26s proteasome and Akt, and the network with the second largest number of down-regulated focus genes is related to histone H3 and F actin. These changes in ubiquitin, proteasome, and histone H3 are very similar to that of hypertension only animals. Down-regulation of genes related to actin may affect process outgrowth or motility of vascular cells, and actin cytoskeleton signaling is one of the functional pathways that are related to male-specific ischemic stroke genes [97].
The network in the MCA with largest number of up-regulated focus genes affected by hypertension plus hypercholesterolemia showed many molecules related to ERK 1/2, interferon alpha, IL12, SYK, CD2, CD4, and CD244. IL12 (interleukin 12) is a proinflammatory and immunomodulatory cytokine [98] released in response to tissue injury [99]. Elevated serum levels of IL12 are observed in patients with acute myocardial infarction [100], traumatic brain injury [101] and ischemic stroke [102]; in addition, IL 12 signaling is related to female-specific ischemic stroke genes [97]. SYK (spleen tyrosine kinase) is a non-receptor tyrosine kinase [103] involved in signaling cascades in platelets [104]. The use of inhibitors of SYK is a potential treatment for occlusive vascular disease, due to its effect in modulating platelet aggregation and thrombus formation [104]. SYK also phosphorylates tau [105] and α-synuclein [106]. CD2 (CD2 molecule) is a member of the immunoglobulin superfamily and mediates the activation of T and natural killer cells [107]. CD4 (CD4 molecule) is expressed on the surface of T cells [108] and CD4+ T cells are regulators of humoral and cellular immune response [109]. Lack of CD4+ T cells is associated with decreased lesion size after stroke [110]. CD244 (CD244 molecule, natural killer cell receptor 2B4) is part of the ‘signaling lymphocyte activation molecule’ (SLAM) family of receptors [111] and is present on natural killer cells, activated CD8+ T cells, and monocytes [112], [113]. It is postulated that CD244 may be involved in a pathway that promotes inflammatory neurological disease [114]. The network with the second largest number of up-regulated focus genes was related to P38 MAPK, NFkB, SERPINB2, MMP1 and Tnf (family). TNF (tumor necrosis factor) plays a key role in increasing the expression of inflammation related genes in atherosclerosis [115], and expression is increased in brain during ischemia [116] or in patients who suffer intracerebral hemorrhage [117]. Antagonism of the TNF-α receptor modulates neurovascular injury and improves neurobehavioral outcomes, after intracerebral hemorrhage in mice [118].
The network in the MCA with largest number of down-regulated focus genes affected by hypertension plus hypercholesterolemia showed many molecules related to ERK, Akt, PKC, Vegf, actin, and insulin. Diabetes mellitus and insulin resistance increase the risk of ICLAD and stroke [119], [120], [121], [122], [123]. Moreover, patients with diabetes show poorer post-stroke functional outcomes in terms of mortality [124], [125] and cognition [126]. These findings may be related to the effect of insulin on activation of NFkB and generation of pro-inflammatory factors involved in atherogenesis [127]. The network with the second largest number of down-regulated focus genes was related to P38 MAPK, NFkB, 26s proteasome, histone H3, interferon alpha and IL12.
The common area between the hypertension only- and hypercholesterolemia plus sham groups showed up-regulated focus genes related to UBC, APP, SERPINB2, TNF and HNF4A, and down-regulated focus genes related to UBC. HNF4A (hepatocyte nuclear factor 4 alpha) is a ligand activated nuclear transcription factor that regulates the expression of many genes involved in lipid transport and glucose metabolism and are associated with cell cycle, immunity, apoptosis, stress response and cancer [128]. Increased expression of HNF4A was shown by RT-PCR and Western blot, and the protein immunolocalized to endothelial cells in the aorta. Since HNF4A suppresses hepatocyte proliferation in adult mice [129], increased expression may likewise affect the turnover of endothelial cells. The effect of this on atherosclerosis is unclear, although excess proliferation of vascular smooth muscle cells is known to have an atherogenic effect [130].
The hypertension plus hypercholesterolemia ‘exclusive’ area showed up-regulated and down-regulated pathways, related to many of the node molecules mentioned above. This area is tentatively interpreted as containing genes that are exacerbated by two risk factors, and the effects tends towards recruitment of additional molecules into existing networks rather than initiation of new networks.
An ischemic cerebrovascular event or transient ischemic attack is a risk factor for a subsequent event [21], [22], [131]. The reasons for this are multifactorial [132] and may be partly due to presence of existing atherosclerotic lesions. The present findings extend these concepts to the gene network level, and delineates pathways related to NF-κB and TNF that are involved in inflammation and atherosclerosis [60], [133], as well as focus genes related to ubiquitin, proteasome, histone, HNF4A, insulin and APP. It is hoped that these could provide a framework for better understanding of pathophysiological mechanisms, and development of new therapies for ICLAD.
Funding Statement
This research was supported by the National Research Foundation Singapore under its Competitive Research Programme (CRP Award NRF-CRP 3-2008-1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Wong LK (2006) Global burden of intracranial atherosclerosis. Int J Stroke 1: 158–159. [DOI] [PubMed] [Google Scholar]
- 2. Gorelick PB, Wong KS, Bae HJ, Pandey DK (2008) Large artery intracranial occlusive disease: a large worldwide burden but a relatively neglected frontier. Stroke 39: 2396–2399. [DOI] [PubMed] [Google Scholar]
- 3. Wong KS, Li H (2003) Long-term mortality and recurrent stroke risk among Chinese stroke patients with predominant intracranial atherosclerosis. Stroke 34: 2361–2366. [DOI] [PubMed] [Google Scholar]
- 4. Mazighi M, Tanasescu R, Ducrocq X, Vicaut E, Bracard S, et al. (2006) Prospective study of symptomatic atherothrombotic intracranial stenoses: the GESICA study. Neurology 66: 1187–1191. [DOI] [PubMed] [Google Scholar]
- 5. De Silva DA, Ancalan M, Doshi K, Chang HM, Wong MC, et al. (2009) Intracranial large artery disease in Alzheimer’s disease and vascular dementia among ethnic Asians. Eur J Neurol 16: 643–645. [DOI] [PubMed] [Google Scholar]
- 6. Ingall TJ, Homer D, Baker HL Jr, Kottke BA, O’Fallon WM, et al. (1991) Predictors of intracranial carotid artery atherosclerosis. Duration of cigarette smoking and hypertension are more powerful than serum lipid levels. Arch Neurol 48: 687–691. [DOI] [PubMed] [Google Scholar]
- 7. Zhang J, Li Y, Wang Y, Niu W, Zhang Y, et al. (2011) Arterial stiffness and asymptomatic intracranial large arterial stenosis and calcification in hypertensive chinese. Am J Hypertens 24: 304–309. [DOI] [PubMed] [Google Scholar]
- 8. O’Rourke MF, Safar ME (2005) Relationship between aortic stiffening and microvascular disease in brain and kidney: cause and logic of therapy. Hypertension 46: 200–204. [DOI] [PubMed] [Google Scholar]
- 9. Xu CP, Glagov S, Zatina MA, Zarins CK (1991) Hypertension sustains plaque progression despite reduction of hypercholesterolemia. Hypertension 18: 123–129. [DOI] [PubMed] [Google Scholar]
- 10.Hollander W, Prusty S, Kemper T, Rosene DL, Moss MB (1993) The effects of hypertension on cerebral atherosclerosis in the cynomolgus monkey. Stroke 24: 1218–1226; discussion 1226–1217. [DOI] [PubMed]
- 11.Ropper AH, Adams RD, Victor M, Samuels MA (2009) Adams and Victor’s principles of neurology. New York: McGraw-Hill Medical. x, 1572 p. p.
- 12. Mills S, Cain J, Purandare N, Jackson A (2007) Biomarkers of cerebrovascular disease in dementia. Br J Radiol 80 Spec No 2: S128–145. [DOI] [PubMed] [Google Scholar]
- 13. Hankey GJ, Wong KS, Chankrachang S, Chen C, Crimmins D, et al. (2010) Management of cholesterol to reduce the burden of stroke in Asia: consensus statement. Int J Stroke 5: 209–216. [DOI] [PubMed] [Google Scholar]
- 14. Mok VC, Lam WW, Fan YH, Wong A, Ng PW, et al. (2009) Effects of statins on the progression of cerebral white matter lesion: Post hoc analysis of the ROCAS (Regression of Cerebral Artery Stenosis) study. J Neurol 256: 750–757. [DOI] [PubMed] [Google Scholar]
- 15. Vrhovski-Hebrang D, Flegar-Mestric Z, Preden-Kerekovic V, Perkov S, Hebrang A, et al. (2002) Biochemical risk factors in angiographically established stenosis of cerebral arteries. Croat Med J 43: 696–701. [PubMed] [Google Scholar]
- 16. Chen CH, Jiang T, Yang JH, Jiang W, Lu J, et al. (2003) Low-density lipoprotein in hypercholesterolemic human plasma induces vascular endothelial cell apoptosis by inhibiting fibroblast growth factor 2 transcription. Circulation 107: 2102–2108. [DOI] [PubMed] [Google Scholar]
- 17. Giardina JB, Tanner DJ, Khalil RA (2001) Oxidized-LDL enhances coronary vasoconstriction by increasing the activity of protein kinase C isoforms alpha and epsilon. Hypertension 37: 561–568. [DOI] [PubMed] [Google Scholar]
- 18. Weis JR, Pitas RE, Wilson BD, Rodgers GM (1991) Oxidized low-density lipoprotein increases cultured human endothelial cell tissue factor activity and reduces protein C activation. FASEB J 5: 2459–2465. [DOI] [PubMed] [Google Scholar]
- 19. Steinberg D (2009) The LDL modification hypothesis of atherogenesis: an update. J Lipid Res 50 Suppl: S376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alexander RW (1995) Theodore Cooper Memorial Lecture. Hypertension and the pathogenesis of atherosclerosis. Oxidative stress and the mediation of arterial inflammatory response: a new perspective. Hypertension 25: 155–161. [DOI] [PubMed] [Google Scholar]
- 21. Burn J, Dennis M, Bamford J, Sandercock P, Wade D, et al. (1994) Long-term risk of recurrent stroke after a first-ever stroke. The Oxfordshire Community Stroke Project. Stroke 25: 333–337. [DOI] [PubMed] [Google Scholar]
- 22. Viitanen M, Eriksson S, Asplund K (1988) Risk of recurrent stroke, myocardial infarction and epilepsy during long-term follow-up after stroke. Eur Neurol 28: 227–231. [DOI] [PubMed] [Google Scholar]
- 23. Ritman EL, Lerman A (2007) The dynamic vasa vasorum. Cardiovasc Res 75: 649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Olsen TS, Skriver EB, Herning M (1985) Cause of cerebral infarction in the carotid territory. Its relation to the size and the location of the infarct and to the underlying vascular lesion. Stroke 16: 459–466. [DOI] [PubMed] [Google Scholar]
- 25. Brown AT, Skinner RD, Flores R, Hennings L, Borrelli MJ, et al. (2010) Stroke location and brain function in an embolic rabbit stroke model. J Vasc Interv Radiol 21: 903–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yanni AE (2004) The laboratory rabbit: an animal model of atherosclerosis research. Lab Anim 38: 246–256. [DOI] [PubMed] [Google Scholar]
- 27. Russell JC, Proctor SD (2006) Small animal models of cardiovascular disease: tools for the study of the roles of metabolic syndrome, dyslipidemia, and atherosclerosis. Cardiovasc Pathol 15: 318–330. [DOI] [PubMed] [Google Scholar]
- 28. Akabane S, Natsume T, Matsushima Y, Deguchi F, Kuramochi M, et al. (1985) Alterations in renal Na+K+ATPase activity and [3H]ouabain binding in Goldblatt hypertensive rabbits. J Hypertens 3: 469–474. [PubMed] [Google Scholar]
- 29. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 30. Merrill AH Jr, Jones DD (1990) An update of the enzymology and regulation of sphingomyelin metabolism. Biochim Biophys Acta 1044: 1–12. [DOI] [PubMed] [Google Scholar]
- 31. Nowell CS, Bredenkamp N, Tetelin S, Jin X, Tischner C, et al. (2011) Foxn1 regulates lineage progression in cortical and medullary thymic epithelial cells but is dispensable for medullary sublineage divergence. PLoS Genet 7: e1002348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phillips MA, Qin Q, Hu Q, Zhao B, Rice RH (2013) Arsenite Suppression of BMP Signaling in Human Keratinocytes. Toxicol Appl Pharmacol. [DOI] [PMC free article] [PubMed]
- 33. Schwarz DA, Katayama CD, Hedrick SM (1998) Schlafen, a new family of growth regulatory genes that affect thymocyte development. Immunity 9: 657–668. [DOI] [PubMed] [Google Scholar]
- 34. Supuran CT (2008) Carbonic anhydrases–an overview. Curr Pharm Des 14: 603–614. [DOI] [PubMed] [Google Scholar]
- 35. Chang X, Zheng Y, Yang Q, Wang L, Pan J, et al. (2012) Carbonic anhydrase I (CA1) is involved in the process of bone formation and is susceptible to ankylosing spondylitis. Arthritis Res Ther 14: R176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Du M, Sansores-Garcia L, Zu Z, Wu KK (1998) Cloning and expression analysis of a novel salicylate suppressible gene, Hs-CUL-3, a member of cullin/Cdc53 family. J Biol Chem 273: 24289–24292. [DOI] [PubMed] [Google Scholar]
- 37. Furukawa M, He YJ, Borchers C, Xiong Y (2003) Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat Cell Biol 5: 1001–1007. [DOI] [PubMed] [Google Scholar]
- 38. Petroski MD, Deshaies RJ (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6: 9–20. [DOI] [PubMed] [Google Scholar]
- 39. Boyden LM, Choi M, Choate KA, Nelson-Williams CJ, Farhi A, et al. (2012) Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature 482: 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, et al. (2011) Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet 43: 429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, et al. (2011) Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet 43: 436–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Simonet WS, Lacey DL, Dunstan CR, Kelley M, Chang MS, et al. (1997) Osteoprotegerin: a novel secreted protein involved in the regulation of bone density. Cell 89: 309–319. [DOI] [PubMed] [Google Scholar]
- 43.Fu YX, Gu JH, Zhang YR, Tong XS, Zhao HY, et al.. (2013) Osteoprotegerin influences the bone resorption activity of osteoclasts. Int J Mol Med. [DOI] [PubMed]
- 44. Ueland T, Yndestad A, Oie E, Florholmen G, Halvorsen B, et al. (2005) Dysregulated osteoprotegerin/RANK ligand/RANK axis in clinical and experimental heart failure. Circulation 111: 2461–2468. [DOI] [PubMed] [Google Scholar]
- 45. Ziegler S, Kudlacek S, Luger A, Minar E (2005) Osteoprotegerin plasma concentrations correlate with severity of peripheral artery disease. Atherosclerosis 182: 175–180. [DOI] [PubMed] [Google Scholar]
- 46. Biscetti F, Straface G, Giovannini S, Santoliquido A, Angelini F, et al. (2013) Association between TNFRSF11B gene polymorphisms and history of ischemic stroke in Italian diabetic patients. Hum Genet 132: 49–55. [DOI] [PubMed] [Google Scholar]
- 47. Kovar H (2011) Dr. Jekyll and Mr. Hyde: The Two Faces of the FUS/EWS/TAF15 Protein Family. Sarcoma 2011: 837474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Meienberg J, Rohrbach M, Neuenschwander S, Spanaus K, Giunta C, et al. (2010) Hemizygous deletion of COL3A1, COL5A2, and MSTN causes a complex phenotype with aortic dissection: a lesson for and from true haploinsufficiency. Eur J Hum Genet 18: 1315–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Le Goff C, Cormier-Daire V (2011) The ADAMTS(L) family and human genetic disorders. Hum Mol Genet 20: R163–167. [DOI] [PubMed] [Google Scholar]
- 50. Arning A, Hiersche M, Witten A, Kurlemann G, Kurnik K, et al. (2012) A genome-wide association study identifies a gene network of ADAMTS genes in the predisposition to pediatric stroke. Blood 120: 5231–5236. [DOI] [PubMed] [Google Scholar]
- 51. Meller R (2009) The role of the ubiquitin proteasome system in ischemia and ischemic tolerance. Neuroscientist 15: 243–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Barone FC, Irving EA, Ray AM, Lee JC, Kassis S, et al. (2001) Inhibition of p38 mitogen-activated protein kinase provides neuroprotection in cerebral focal ischemia. Med Res Rev 21: 129–145. [DOI] [PubMed] [Google Scholar]
- 53. Frasch SC, Nick JA, Fadok VA, Bratton DL, Worthen GS, et al. (1998) p38 mitogen-activated protein kinase-dependent and -independent intracellular signal transduction pathways leading to apoptosis in human neutrophils. J Biol Chem 273: 8389–8397. [DOI] [PubMed] [Google Scholar]
- 54. Aoshiba K, Yasui S, Hayashi M, Tamaoki J, Nagai A (1999) Role of p38-mitogen-activated protein kinase in spontaneous apoptosis of human neutrophils. J Immunol 162: 1692–1700. [PubMed] [Google Scholar]
- 55. Xia Z, Dickens M, Raingeaud J, Davis RJ, Greenberg ME (1995) Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science 270: 1326–1331. [DOI] [PubMed] [Google Scholar]
- 56. Barone FC, Irving EA, Ray AM, Lee JC, Kassis S, et al. (2001) SB 239063, a second-generation p38 mitogen-activated protein kinase inhibitor, reduces brain injury and neurological deficits in cerebral focal ischemia. J Pharmacol Exp Ther 296: 312–321. [PubMed] [Google Scholar]
- 57. Roux PP, Blenis J (2004) ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev 68: 320–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sawe N, Steinberg G, Zhao H (2008) Dual roles of the MAPK/ERK1/2 cell signaling pathway after stroke. J Neurosci Res 86: 1659–1669. [DOI] [PubMed] [Google Scholar]
- 59. Hoffmann A, Baltimore D (2006) Circuitry of nuclear factor kappaB signaling. Immunol Rev 210: 171–186. [DOI] [PubMed] [Google Scholar]
- 60. Monaco C, Paleolog E (2004) Nuclear factor kappaB: a potential therapeutic target in atherosclerosis and thrombosis. Cardiovasc Res 61: 671–682. [DOI] [PubMed] [Google Scholar]
- 61. Nurmi A, Lindsberg PJ, Koistinaho M, Zhang W, Juettler E, et al. (2004) Nuclear factor-kappaB contributes to infarction after permanent focal ischemia. Stroke 35: 987–991. [DOI] [PubMed] [Google Scholar]
- 62. Schneider A, Martin-Villalba A, Weih F, Vogel J, Wirth T, et al. (1999) NF-kappaB is activated and promotes cell death in focal cerebral ischemia. Nat Med 5: 554–559. [DOI] [PubMed] [Google Scholar]
- 63. Breuss JM, Cejna M, Bergmeister H, Kadl A, Baumgartl G, et al. (2002) Activation of nuclear factor-kappa B significantly contributes to lumen loss in a rabbit iliac artery balloon angioplasty model. Circulation 105: 633–638. [DOI] [PubMed] [Google Scholar]
- 64. Yoshimura S, Morishita R, Hayashi K, Yamamoto K, Nakagami H, et al. (2001) Inhibition of intimal hyperplasia after balloon injury in rat carotid artery model using cis-element ‘decoy’ of nuclear factor-kappaB binding site as a novel molecular strategy. Gene Ther 8: 1635–1642. [DOI] [PubMed] [Google Scholar]
- 65. Zuckerbraun BS, McCloskey CA, Mahidhara RS, Kim PK, Taylor BS, et al. (2003) Overexpression of mutated IkappaBalpha inhibits vascular smooth muscle cell proliferation and intimal hyperplasia formation. J Vasc Surg 38: 812–819. [DOI] [PubMed] [Google Scholar]
- 66. Xu L, Zhan Y, Wang Y, Feuerstein GZ, Wang X (2002) Recombinant adenoviral expression of dominant negative IkappaBalpha protects brain from cerebral ischemic injury. Biochem Biophys Res Commun 299: 14–17. [DOI] [PubMed] [Google Scholar]
- 67. van den Tweel ER, Kavelaars A, Lombardi MS, Groenendaal F, May M, et al. (2006) Selective inhibition of nuclear factor-kappaB activation after hypoxia/ischemia in neonatal rats is not neuroprotective. Pediatr Res 59: 232–236. [DOI] [PubMed] [Google Scholar]
- 68. Blondeau N, Widmann C, Lazdunski M, Heurteaux C (2001) Activation of the nuclear factor-kappaB is a key event in brain tolerance. J Neurosci 21: 4668–4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dutta J, Fan Y, Gupta N, Fan G, Gelinas C (2006) Current insights into the regulation of programmed cell death by NF-kappaB. Oncogene 25: 6800–6816. [DOI] [PubMed] [Google Scholar]
- 70. Barkett M, Gilmore TD (1999) Control of apoptosis by Rel/NF-kappaB transcription factors. Oncogene 18: 6910–6924. [DOI] [PubMed] [Google Scholar]
- 71. Medcalf RL, Stasinopoulos SJ (2005) The undecided serpin. The ins and outs of plasminogen activator inhibitor type 2. FEBS J 272: 4858–4867. [DOI] [PubMed] [Google Scholar]
- 72. Akiyama H, Ikeda K, Kondo H, Kato M, McGeer PL (1993) Microglia express the type 2 plasminogen activator inhibitor in the brain of control subjects and patients with Alzheimer’s disease. Neurosci Lett 164: 233–235. [DOI] [PubMed] [Google Scholar]
- 73. Dietzmann K, von Bossanyi P, Krause D, Wittig H, Mawrin C, et al. (2000) Expression of the plasminogen activator system and the inhibitors PAI-1 and PAI-2 in posttraumatic lesions of the CNS and brain injuries following dramatic circulatory arrests: an immunohistochemical study. Pathol Res Pract 196: 15–21. [DOI] [PubMed] [Google Scholar]
- 74. Horstmann S, Kalb P, Koziol J, Gardner H, Wagner S (2003) Profiles of matrix metalloproteinases, their inhibitors, and laminin in stroke patients: influence of different therapies. Stroke 34: 2165–2170. [DOI] [PubMed] [Google Scholar]
- 75. Morgan AR, Rerkasem K, Gallagher PJ, Zhang B, Morris GE, et al. (2004) Differences in matrix metalloproteinase-1 and matrix metalloproteinase-12 transcript levels among carotid atherosclerotic plaques with different histopathological characteristics. Stroke 35: 1310–1315. [DOI] [PubMed] [Google Scholar]
- 76. Ye S, Gale CR, Martyn CN (2003) Variation in the matrix metalloproteinase-1 gene and risk of coronary heart disease. Eur Heart J 24: 1668–1671. [DOI] [PubMed] [Google Scholar]
- 77. Skoog I, Gustafson D (2002) Hypertension and related factors in the etiology of Alzheimer’s disease. Ann N Y Acad Sci 977: 29–36. [DOI] [PubMed] [Google Scholar]
- 78. Meyer JS, Rauch GM, Rauch RA, Haque A, Crawford K (2000) Cardiovascular and other risk factors for Alzheimer’s disease and vascular dementia. Ann N Y Acad Sci 903: 411–423. [DOI] [PubMed] [Google Scholar]
- 79. Altman R, Rutledge JC (2010) The vascular contribution to Alzheimer’s disease. Clin Sci (Lond) 119: 407–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zlokovic BV (2011) Neurovascular pathways to neurodegeneration in Alzheimer’s disease and other disorders. Nat Rev Neurosci 12: 723–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Iadecola C, Zhang F, Niwa K, Eckman C, Turner SK, et al. (1999) SOD1 rescues cerebral endothelial dysfunction in mice overexpressing amyloid precursor protein. Nat Neurosci 2: 157–161. [DOI] [PubMed] [Google Scholar]
- 82. Zhang F, Eckman C, Younkin S, Hsiao KK, Iadecola C (1997) Increased susceptibility to ischemic brain damage in transgenic mice overexpressing the amyloid precursor protein. J Neurosci 17: 7655–7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Dudek H, Datta SR, Franke TF, Birnbaum MJ, Yao R, et al. (1997) Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science 275: 661–665. [DOI] [PubMed] [Google Scholar]
- 84. Brunet A, Datta SR, Greenberg ME (2001) Transcription-dependent and -independent control of neuronal survival by the PI3K-Akt signaling pathway. Curr Opin Neurobiol 11: 297–305. [DOI] [PubMed] [Google Scholar]
- 85. Wu J, Li Q, Wang X, Yu S, Li L, et al. (2013) Neuroprotection by curcumin in ischemic brain injury involves the akt/nrf2 pathway. PLoS One 8: e59843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhao H, Shimohata T, Wang JQ, Sun G, Schaal DW, et al. (2005) Akt contributes to neuroprotection by hypothermia against cerebral ischemia in rats. J Neurosci 25: 9794–9806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Chou WH, Messing RO (2005) Protein kinase C isozymes in stroke. Trends Cardiovasc Med 15: 47–51. [DOI] [PubMed] [Google Scholar]
- 88. Bright R, Mochly-Rosen D (2005) The role of protein kinase C in cerebral ischemic and reperfusion injury. Stroke 36: 2781–2790. [DOI] [PubMed] [Google Scholar]
- 89. Bright R, Raval AP, Dembner JM, Perez-Pinzon MA, Steinberg GK, et al. (2004) Protein kinase C delta mediates cerebral reperfusion injury in vivo. J Neurosci 24: 6880–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wojcik C, Di Napoli M (2004) Ubiquitin-proteasome system and proteasome inhibition: new strategies in stroke therapy. Stroke 35: 1506–1518. [DOI] [PubMed] [Google Scholar]
- 91. Langley B, Brochier C, Rivieccio MA (2009) Targeting histone deacetylases as a multifaceted approach to treat the diverse outcomes of stroke. Stroke 40: 2899–2905. [DOI] [PubMed] [Google Scholar]
- 92. Sodhi RK, Singh N (2013) All-trans retinoic acid rescues memory deficits and neuropathological changes in mouse model of streptozotocin-induced dementia of Alzheimer’s type. Prog Neuropsychopharmacol Biol Psychiatry 40: 38–46. [DOI] [PubMed] [Google Scholar]
- 93. Ding Y, Qiao A, Wang Z, Goodwin JS, Lee ES, et al. (2008) Retinoic acid attenuates beta-amyloid deposition and rescues memory deficits in an Alzheimer’s disease transgenic mouse model. J Neurosci 28: 11622–11634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Young JL, Libby P, Schonbeck U (2002) Cytokines in the pathogenesis of atherosclerosis. Thromb Haemost 88: 554–567. [PubMed] [Google Scholar]
- 95.Greenberg DA, Jin K (2013) Vascular endothelial growth factors (VEGFs) and stroke. Cell Mol Life Sci. [DOI] [PMC free article] [PubMed]
- 96. Matsuo R, Ago T, Kamouchi M, Kuroda J, Kuwashiro T, et al. (2013) Clinical significance of plasma VEGF value in ischemic stroke - research for biomarkers in ischemic stroke (REBIOS) study. BMC Neurol 13: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Tian Y, Stamova B, Jickling GC, Liu D, Ander BP, et al. (2012) Effects of gender on gene expression in the blood of ischemic stroke patients. J Cereb Blood Flow Metab 32: 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Trinchieri G (1998) Interleukin-12: a cytokine at the interface of inflammation and immunity. Adv Immunol 70: 83–243. [DOI] [PubMed] [Google Scholar]
- 99. DeGraba TJ (1998) The role of inflammation after acute stroke: utility of pursuing anti-adhesion molecule therapy. Neurology 51: S62–68. [DOI] [PubMed] [Google Scholar]
- 100. Zhou RH, Shi Q, Gao HQ, Shen BJ (2001) Changes in serum interleukin-8 and interleukin-12 levels in patients with ischemic heart disease in a Chinese population. J Atheroscler Thromb 8: 30–32. [DOI] [PubMed] [Google Scholar]
- 101. Arand M, Melzner H, Kinzl L, Bruckner UB, Gebhard F (2001) Early inflammatory mediator response following isolated traumatic brain injury and other major trauma in humans. Langenbecks Arch Surg 386: 241–248. [DOI] [PubMed] [Google Scholar]
- 102. Zaremba J, Losy J (2006) Interleukin-12 in acute ischemic stroke patients. Folia Neuropathol 44: 59–66. [PubMed] [Google Scholar]
- 103. Sada K, Takano T, Yanagi S, Yamamura H (2001) Structure and function of Syk protein-tyrosine kinase. J Biochem 130: 177–186. [DOI] [PubMed] [Google Scholar]
- 104. Speich HE, Grgurevich S, Kueter TJ, Earhart AD, Slack SM, et al. (2008) Platelets undergo phosphorylation of Syk at Y525/526 and Y352 in response to pathophysiological shear stress. Am J Physiol Cell Physiol 295: C1045–1054. [DOI] [PubMed] [Google Scholar]
- 105. Lebouvier T, Scales TM, Hanger DP, Geahlen RL, Lardeux B, et al. (2008) The microtubule-associated protein tau is phosphorylated by Syk. Biochim Biophys Acta 1783: 188–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Negro A, Brunati AM, Donella-Deana A, Massimino ML, Pinna LA (2002) Multiple phosphorylation of alpha-synuclein by protein tyrosine kinase Syk prevents eosin-induced aggregation. FASEB J 16: 210–212. [DOI] [PubMed] [Google Scholar]
- 107. Crawford K, Stark A, Kitchens B, Sternheim K, Pantazopoulos V, et al. (2003) CD2 engagement induces dendritic cell activation: implications for immune surveillance and T-cell activation. Blood 102: 1745–1752. [DOI] [PubMed] [Google Scholar]
- 108. Hedrick SM (2002) T cell development: bottoms-up. Immunity 16: 619–622. [DOI] [PubMed] [Google Scholar]
- 109. Stockinger B, Bourgeois C, Kassiotis G (2006) CD4+ memory T cells: functional differentiation and homeostasis. Immunol Rev 211: 39–48. [DOI] [PubMed] [Google Scholar]
- 110. Gu L, Xiong X, Zhang H, Xu B, Steinberg GK, et al. (2012) Distinctive effects of T cell subsets in neuronal injury induced by cocultured splenocytes in vitro and by in vivo stroke in mice. Stroke 43: 1941–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Veillette A, Latour S (2003) The SLAM family of immune-cell receptors. Curr Opin Immunol 15: 277–285. [DOI] [PubMed] [Google Scholar]
- 112. Speiser DE, Colonna M, Ayyoub M, Cella M, Pittet MJ, et al. (2001) The activatory receptor 2B4 is expressed in vivo by human CD8+ effector alpha beta T cells. J Immunol 167: 6165–6170. [DOI] [PubMed] [Google Scholar]
- 113. Tangye SG, Phillips JH, Lanier LL, Nichols KE (2000) Functional requirement for SAP in 2B4-mediated activation of human natural killer cells as revealed by the X-linked lymphoproliferative syndrome. J Immunol 165: 2932–2936. [DOI] [PubMed] [Google Scholar]
- 114. Enose-Akahata Y, Matsuura E, Oh U, Jacobson S (2009) High expression of CD244 and SAP regulated CD8 T cell responses of patients with HTLV-I associated neurologic disease. PLoS Pathog 5: e1000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. McKellar GE, McCarey DW, Sattar N, McInnes IB (2009) Role for TNF in atherosclerosis? Lessons from autoimmune disease. Nat Rev Cardiol 6: 410–417. [DOI] [PubMed] [Google Scholar]
- 116.Perez-de-Puig I, Martin A, Gorina R, Rosa XD, Martinez E, et al.. (2013) Induction of hemeoxygenase (HO)-1 expression after inhibition of HO activity promotes inflammation and worsens ischemic brain damage in mice. Neuroscience. [DOI] [PubMed]
- 117. Fang HY, Ko WJ, Lin CY (2007) Inducible heat shock protein 70, interleukin-18, and tumor necrosis factor alpha correlate with outcomes in spontaneous intracerebral hemorrhage. J Clin Neurosci 14: 435–441. [DOI] [PubMed] [Google Scholar]
- 118.King MD, Alleyne CH, Jr., Dhandapani KM (2013) TNF-alpha receptor antagonist, R-7050, improves neurological outcomes following intracerebral hemorrhage in mice. Neurosci Lett. [DOI] [PMC free article] [PubMed]
- 119. Park JH, Kwon HM (2008) Association between metabolic syndrome and previous ischemic lesions in patients with intracranial atherosclerotic stroke. Clin Neurol Neurosurg 110: 215–221. [DOI] [PubMed] [Google Scholar]
- 120. De Silva DA, Woon FP, Lee MP, Chen CL, Chang HM, et al. (2009) Metabolic syndrome is associated with intracranial large artery disease among ethnic Chinese patients with stroke. J Stroke Cerebrovasc Dis 18: 424–427. [DOI] [PubMed] [Google Scholar]
- 121. Rundek T, Gardener H, Xu Q, Goldberg RB, Wright CB, et al. (2010) Insulin resistance and risk of ischemic stroke among nondiabetic individuals from the northern Manhattan study. Arch Neurol 67: 1195–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Wannamethee SG, Perry IJ, Shaper AG (1999) Nonfasting serum glucose and insulin concentrations and the risk of stroke. Stroke 30: 1780–1786. [DOI] [PubMed] [Google Scholar]
- 123.Kaneda Y, Ishikawa S, Sadakane A, Gotoh T, Kayaba K, et al.. (2013) Insulin Resistance and Risk of Cerebral Infarction in a Japanese General Population: The Jichi Medical School Cohort Study. Asia Pac J Public Health. [DOI] [PubMed]
- 124. Tuomilehto J, Rastenyte D, Jousilahti P, Sarti C, Vartiainen E (1996) Diabetes mellitus as a risk factor for death from stroke. Prospective study of the middle-aged Finnish population. Stroke 27: 210–215. [DOI] [PubMed] [Google Scholar]
- 125. Prosser J, MacGregor L, Lees KR, Diener HC, Hacke W, et al. (2007) Predictors of early cardiac morbidity and mortality after ischemic stroke. Stroke 38: 2295–2302. [DOI] [PubMed] [Google Scholar]
- 126. Luchsinger JA, Tang MX, Stern Y, Shea S, Mayeux R (2001) Diabetes mellitus and risk of Alzheimer’s disease and dementia with stroke in a multiethnic cohort. Am J Epidemiol 154: 635–641. [DOI] [PubMed] [Google Scholar]
- 127. Bienek R, Marek B, Kajdaniuk D, Borgiel-Marek H, Piecha T, et al. (2012) Adiponectin, leptin, resistin and insulin blood concentrations in patients with ischaemic cerebral stroke. Endokrynol Pol 63: 338–345. [PubMed] [Google Scholar]
- 128. Bolotin E, Liao H, Ta TC, Yang C, Hwang-Verslues W, et al. (2010) Integrated approach for the identification of human hepatocyte nuclear factor 4alpha target genes using protein binding microarrays. Hepatology 51: 642–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Bonzo JA, Ferry CH, Matsubara T, Kim JH, Gonzalez FJ (2012) Suppression of hepatocyte proliferation by hepatocyte nuclear factor 4alpha in adult mice. J Biol Chem 287: 7345–7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Rivard A, Andres V (2000) Vascular smooth muscle cell proliferation in the pathogenesis of atherosclerotic cardiovascular diseases. Histol Histopathol 15: 557–571. [DOI] [PubMed] [Google Scholar]
- 131. Bergman D (2011) Preventing recurrent cerebrovascular events in patients with stroke or transient ischemic attack: the current data. J Am Acad Nurse Pract 23: 659–666. [DOI] [PubMed] [Google Scholar]
- 132. Hillen T, Coshall C, Tilling K, Rudd AG, McGovern R, et al. (2003) Cause of stroke recurrence is multifactorial: patterns, risk factors, and outcomes of stroke recurrence in the South London Stroke Register. Stroke 34: 1457–1463. [DOI] [PubMed] [Google Scholar]
- 133. Csiszar A, Wang M, Lakatta EG, Ungvari Z (2008) Inflammation and endothelial dysfunction during aging: role of NF-kappaB. J Appl Physiol 105: 1333–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]