Abstract
Fusarium graminearum, the causal agent of Fusarium head blight in cereal crops, produces mycotoxins such as trichothecenes and zearalenone in infected plants. Here, we focused on the function of FgLaeA in F. graminearum, a homolog of Aspergillus nidulans LaeA encoding the global regulator for both secondary metabolism and sexual development. Prior to gene analysis, we constructed a novel luciferase reporter system consisting of a transgenic F. graminearum strain expressing a firefly luciferase gene under control of the promoter for either TRI6 or ZEB2 controlling the biosynthesis of these mycotoxins. Targeted deletion of FgLaeA led to a dramatic reduction of luminescence in reporter strains, indicating that FgLaeA controls the expression of these transcription factors in F. graminearum; reduced toxin accumulation was further confirmed by GC-MS analysis. Overexpression of FgLaeA caused the increased production of trichothecenes and additional metabolites. RNA seq-analysis revealed that gene member(s) belonging to ∼70% of total tentative gene clusters, which were previously proposed, were differentially expressed in the ΔFgLaeA strain. In addition, ΔFgLaeA strains exhibited an earlier induction of sexual fruiting body (perithecia) formation and drastically reduced disease symptoms in wheat, indicating that FgLaeA seems to negatively control perithecial induction, but positively control virulence toward the host plant. FgLaeA was constitutively expressed under both mycotoxin production and sexual development conditions. Overexpression of a GFP-FgLaeA fusion construct in the ΔFgLaeA strain restored all phenotypic changes to wild-type levels and led to constitutive expression of GFP in both nuclei and cytoplasm at different developmental stages. A split luciferase assay demonstrated that FgLaeA was able to interact with FgVeA, a homolog of A. nidulans veA. Taken together, these results demonstrate that FgLaeA, a member of putative FgVeA complex, controls secondary metabolism, sexual development, and virulence in F. graminearum, although the specific regulation pattern differs from that of LaeA in A. nidulans.
Introduction
Fusarium graminearum (teleomorph: Gibberella zeae), a homothallic ascomycetous fungus, is an important pathogen of major cereal plants that causes diseases such as head blight (scab), stalk rot, and ear rot [1]–[3]. Previously classified as a single panmictic species, the F. graminearum species complex consists of over 16 phylogenetically distinct species or lineages found throughout the world [4]–[9]. The ability of F. graminearum to produce sexual progeny (ascospores) on overwintering cereal debris is essential for completion of the recurrent cycle of the plant diseases [10]. In addition to direct yield losses, F. graminearum produces mycotoxins such as trichothecenes and zearalenone in host plants, threatening human and animal health [2]. Trichothecenes, potent inhibitors of eukaryotic protein biosynthesis, are associated with feed refusal, vomiting, diarrhea, dermatitis, and hemorrhages in farm animals [2], [11]. Trichothecenes also appear to contribute to the virulence of F. graminearum on host plants [12]. Zearalenone causes estrogenic disorders in laboratory rats, mice, and farm-raised swine [2].
Genes involved in the biosynthesis of mycotoxins and other secondary metabolites (e.g., those encoding metabolic enzymes and transporters) usually reside close to one another on a chromosome, forming a gene cluster. Expression of these gene clusters is controlled at multiple levels. A pathway-specific transcription factor located within a gene cluster can regulate the expression of other members of the same cluster (e.g., TRI6 and ZEB2 for the regulations of trichothecenes and zearalenone biosynthesis, respectively, in F. graminearum) [13], [14]. Expression of these pathway-specific transcription factors is affected by environmental conditions such as ambient pH and availability of nitrogen, which are in turn regulated by other types of transcription factors, often located outside of the gene cluster (e.g., AreA and/or PacC regulate the gene clusters for gibberellic acid, fumonisin, and trichothecenes) [15]–[19].
In addition to specific transcription factors, the discovery of global regulators provides new insight into the regulatory hierarchy for fungal secondary metabolites. These master regulators operate in a relatively nonspecific manner by acting at the level of chromatin remodeling. One of the most intensively studied global regulators is LaeA, a nuclear protein containing a methyltransferase domain. It was first identified as a positive regulator of various secondary metabolites in Aspergillus nidulans [20]. It has been shown to modulate the function of the so-called velvet (VeA) complex consisting of at least 10 different proteins including VeA, VeA-like, VelB, and VosA. This complex coordinates both secondary metabolism and morphological development in response to light [21], [22]. Under dark conditions, LaeA interacts with the VeA–VelB heterodimeric complex in the nucleus, forming the heterotrimeric VeA complex (VelB–VeA–LaeA), which is responsible for activating secondary metabolism and regulating asexual/sexual development [21], [22]. Additionally, LaeA has been shown to control both the protein levels of other VeA members (VosA, VelB, and VeA), as well as physical interactions between them, thereby indirectly regulating fungal development [22].
The functional roles of LaeA have been demonstrated in several fungal species including Aspergillus and Penicillium (Eurotiomycetes) [20], [23]–[26], Cochliobolus heterostrophus (Dothideomycetes) [27], and F. fujikuroi [28], F. verticillioides [29], F. oxysporum [30], and Trichoderma reesei (Sordariomycetes) [31], [32]. In F. graminearum, homologs of three VeA complex members have been identified, two of which, FgVeA and FgVelB, have been shown to regulate mycotoxin production and fungal development, similar to those in Aspergillus spp. [33]–[35]. However, function of the F. graminearum LaeA homolog (designated FgLaeA) and the existence of the nuclear heterotrimeric FgVeA complex FgVelB–FgVeA–FgVelB have yet to be established.
In this study, we provide a detailed functional characterization of FgLaeA in F. graminearum using newly developed firefly luciferase reporter systems for toxin production and protein–protein interaction, respectively, and other strategies. Targeted gene deletion, complementation, and overexpression demonstrated that FgLaeA functions as a positive regulator in the production of various mycotoxins, asexual development (only in the dark), and virulence on host plant as well as in gene expression for the corresponding secondary metabolites, while acting as a negative regulator in sexual development (albeit in a light-independent manner). In addition, in vivo protein interaction between FgVeA and FgLaeA was clearly demonstrated. In contrast to other fungal LaeA homologs, the FgLaeA protein was constitutively localized in both the cytoplasm and nucleus under both dark and light conditions.
Results
Targeted Deletion, Complementation, and Overexpression of FgLaeA
A genome-wide search for homologs of the functionally characterized fungal LaeA-like genes, such as A. nidulans LaeA (GenBank accession no. AAQ95166.1) and F. fujikuroi FfLae1 (FN548141), identified two open reading frames (ORFs) as possible LaeA homologs based on sequence homology, both carrying a conserved methyltransferase domain, in F. graminearum. The first ORF (annotated as FGSG_00657.3 in the F. graminearum genome database), encodes for 317 amino acids, interrupted by 6 introns, which was confirmed by reverse transcription-polymerase chain reaction (RT-PCR), but different from the previous annotation in the F. graminearum database. The intron positions are as follows: 25–87, 191–243, 393–445, 503–552, 703–749, and 1176–1222 (nucleotide numbering starts with 1 at the first nucleotide of the FgLaeA ORF as annotated in the genome database). Its coding region showed the highest similarity to FfLae1 (with 70% identity over 282 residues), but a lower similarity to LaeA (27.8% over 180 residues). We designated this ORF FgLaeA, as previously described [33], due to its highest sequence homology to FfLae1 from a closely related Fusarium species. The second ORF (FGSG_07660, previously designated FgVIP1 [33]) shared 35.8% identity to LaeA, and 30.6% identity to FfLae1 over 296 residues. Targeted deletion of both genes was performed in WT strain Z3643, resulting in the ΔFgLaeA and ΔFgVIP1 strains, respectively. Integration of gene deletion constructs into the fungal genome via double crossover was verified by Southern blot (Fig. 1, data not shown for ΔFgVIP1). Because the ΔFgVIP1 strains showed no dramatic changes in major fungal traits, further gene deletions using the FLTRI6 and FLZEB2 strains (the luciferase reporter strains for biosynthesis of trichothecenes and zearalenone, respectively; for details, see the subheading “Development of a firefly luciferase reporter system for mycotoxin production” in the result section) were done for only FgLaeA, generating the FLTRI6ΔFgLaeA and FLZEB2ΔFgLaeA strains, respectively (Fig. 1). Genetic complementation of a ΔFgLaeA strain derived from Z3643 was achieved by introducing an intact copy of FgLaeA (including ∼1 kb of its native promoter region) into the genome of the ΔFgLaeA strain, generating the so-called add-back strains (Fig. 1). For generating transgenic F. graminearum strains overexpressing FgLaeA, we complemented the ΔFgLaeA strain with an intact copy of the FgLaeA ORF fused to a green fluorescence protein gene (GFP) under control of a strong promoter from the Cryphonectria parasitica Crp gene [36], generating the OE::GFP-FgLaeA (OE) strain.
Figure 1. Targeted deletion of FgLaeA from the genome of the F. graminearum Z3643 (WT), FLTRI6, or FLZEB2 strains.
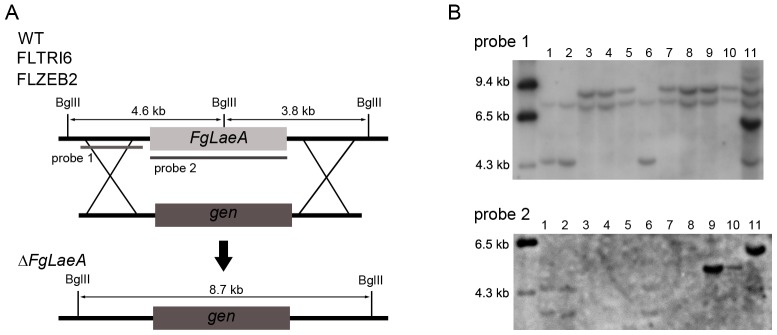
(A) Deletion scheme. (B) BglII-digested genomic DNA gel blot hybridized with probe 1 (upper panel) and probe 2 (lower), respectively; positions are indicated in A. Lanes 1, 2, and 6: the Z3643, FLTRI6, and FLZEB2 strains; lanes 3 and 4: the FgLaeA-deletion strains derived from Z3643 (designated ΔFgLaeA); lanes 5 and 7: the FgLaeA-deletion strains derived from FLTRI6 (FLTRI6ΔFgLaeA); lane 7: the FLZEB2ΔFgLaeA strain; lanes 9 and 10: the FgLaeA add-back strains derived from the ΔFgLaeA strain; lane 11: the FgLaeA-overexpression strain derived from the ΔFgLaeA strain. DNA size markers are indicated on the left side of the gel. Note that a ∼7.5-kb nonspecific hybridizing band was observed in all of the strains examined when hybridized with probe 1.
Hyphal Growth, Pigmentation, Conidiation, and Sexual Development in the ΔFgLaeA and OE::FgLaeA Strains
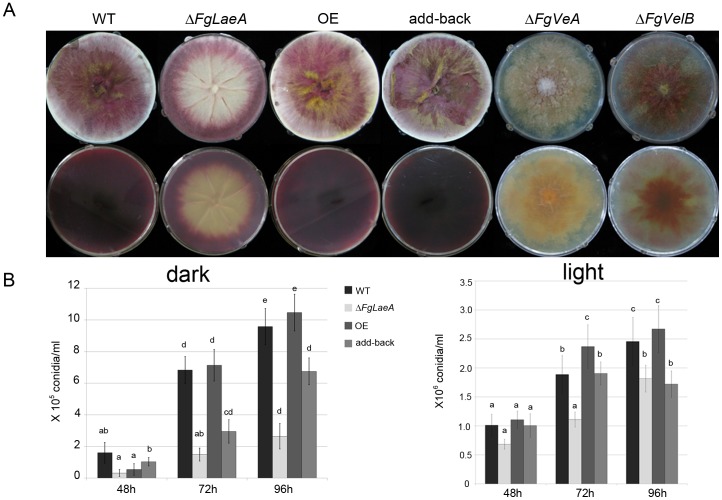
Compared to wild-type (WT) progenitors Z3643, FLTRI6, and FLZEB2, all ΔFgLaeA strains examined exhibited a slight reduction in radial growth, but a significant reduction in red hyphal pigmentation on potato dextrose agar (PDA; Difco Laboratories, Detroit, MI, USA), CM [3], and carrot agar (Fig. 2). However, this reduction was relatively modest compared with the phenotypes caused by the deletion of FgVeA [33] or FgVelB [34], which showed fewer aerial hyphae with reduced or abolished pigmentation (Fig. 2). The reduced pigmentation phenotype caused by ΔFgLaeA was less robust than a typical albino mutant of F. graminearum (e.g., that generated by a deletion of PKS12, the gene responsible for biosynthesis of the red pigment aurofusarin [37]). Aurofusarin production was still observed in young marginal hyphae of mutant colonies, but older regions of the colony were albino, resulting in ring-shaped pigmentation (Fig. 2). This pigmentation pattern was also seen in the marginal regions between different growing colonies (Fig. S1). Fewer aerial mycelia, with lower hydrophobicity were observed only on the albino region in the ΔFgLaeA strains; however, the formation of thick aerial hyphae was retained in the pigmented margin areas, similar to those from WT strains. This reduced pigmentation phenotype was restored to WT levels in both the FgLaeA-add-back and OE strains (Fig. 2). However, this phenotype is dependent upon growth conditions, as the same ΔFgLaeA strain was completely albino when grown in complete liquid medium (Fig. S1).
Figure 2. Hyphal growth, pigmentation, and conidiation.
(A) Colony morphology of the WT (Z3643), ΔFgLaeA, OE (FgLaeA-overexpression), add-back, ΔFgVeA [32], and ΔFgVelB [33] strains on CM plates. Upper and lower panels show the morphology in surface and undersurface of plates, respectively. (B) The number of conidia formed in CMC liquid medium under dark and light conditions. The y-axis represents the number of conidia (×105 or ×106/mL). Data shown are the mean values obtained from three independent samples. Statistical analysis was performed with ANOVA and Duncan’s multiple range test. The same letter above bars represents no significant difference.
Conidiation of WT strains grown in carboxymethyl cellulose (CMC) liquid medium was strongly favored (∼10–15-fold increase) under 24-h light condition compared to those maintained in the dark. ΔFgLaeA strains showed no dramatic changes under light condition, while showed reduced levels of conidiation under dark condition (Fig. 2). No significant changes were detected among the WT, OE, and add-back strains (Fig. 2).
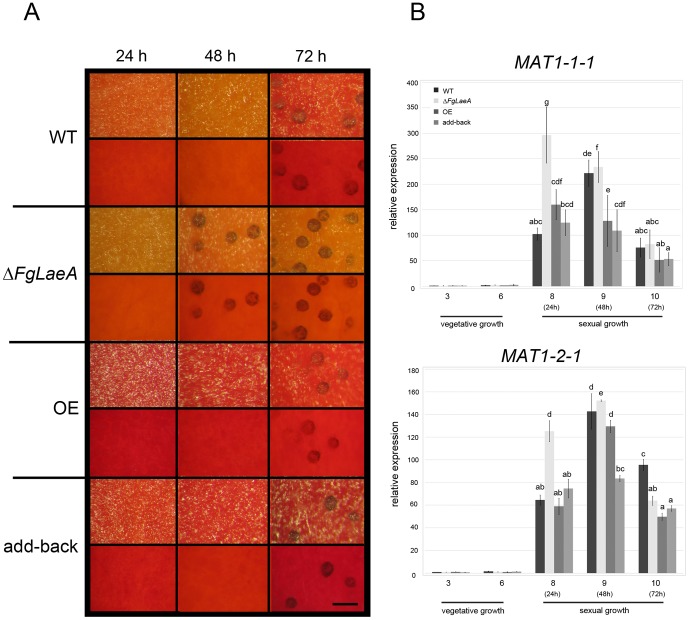
Sexual development (self-fertility) of ΔFgLaeA strains was determined based on perithecia production on carrot agar. The WT Z3643 strain began to produce protoperithecia approximately 72 h after sexual induction on carrot agar under both 24-h light and 12-h dark/light cycle conditions (Fig. 3). After an additional 3- or 4-day incubation, mature perithecia containing asci, each with eight ascospores formed. In contrast, five independent experiments confirmed that all ΔFgLaeA strains derived from the Z3643, FLTRI6, and FLZEB2 strains began producing protoperithecia approximately 24 h earlier than the WT strain under the same culture conditions (Fig. 3A). However, neither the WT nor ΔFgLaeA strains produced protoperithecia on carrot agar under 24-h dark conditions (data not shown). This early induction of protoperithecia formation in the ΔFgLaeA strains was restored to WT levels in both the add-back and OE strains (Fig. 3A). When a conidial suspension of the ΔFgLaeA strain was spermatized for the self-sterile ΔMAT1-2 strain, fertile perithecia successfully formed on carrot agar, indicating that the ΔFgLaeA strain has no defect in male fertility (data not shown).
Figure 3. Protoperithecia formation and the expression of MAT genes.
(A) Protoperithecia formation on carrot agar in the WT (Z3643), ΔFgLaeA, OE (FgLaeA-overexpression), and add-back strains. Time since perithecia induction (i.e., the removal of aerial mycelia, which had previously grown for 7 days in the dark) is indicated above the pictures. Upper and lower panels for each strain show the surface and undersurface, respectively, of carrot agar plates. The size bar indicates 200 µm. (B) Relative transcript levels for two MAT genes (x-axis) accumulated in the fungal strains shown in (A) at each of the growth time points (days, y-axis) for total RNA extraction. 3 and 6: days 3 and 6 under vegetative growth, respectively; 8, 9, and 10: days 8, 9, and 10 following perithecial induction, respectively (i.e. 24 h, 48 h, and 72 h after perithecia induction, respectively). GzRPS16 (FGSG_09438.3) was used as an endogenous control for data normalization [52]. MAT1-1-1 and MAT1-2-1 transcript levels from a 3-day-old vegetative sample of a F. graminearum WT strain were used as references. Statistical analysis was performed with ANOVA and Duncan’s multiple range test. The same letter above bars represents no significant difference.
Mycotoxin Production and Virulence of the ΔFgLaeA and OE::GFP-FgLaeA Strains
The effect of FgLaeA deletion and overexpression on the production of mycotoxins [trichothecenes (e.g. deoxynivalenol, DON; 15-acetyl DON, 15ADON) and zearalenone (ZEA)] in F. graminearum was determined using the luminescence reporter system, chemical analysis, and qRT-PCR. In all tests, the WT strain produced both DON and ZEA with concomitant gene expression levels under appropriate conditions, whereas toxin production in the ΔFgLaeA strains significantly decreased. Luminescence signal intensity from cell lysates extracted from FLTRI6ΔFgLaeA strains grown in AG medium for 6 days were at least 10-fold lower than those from the FLTRI6 strain, demonstrating that the Tri6 transcript level significantly decreased due to ΔFgLaeA; no significant difference in luminescence was observed between FLTRI6 strains grown under dark and light conditions (Fig. 4A). Similarly, luminescence intensities for ZEB2 expression in the FLZEB2ΔFgLaeA strain decreased ∼2-fold compared to FLZEB2 when grown in AG medium (Fig. 4B) and ∼6-fold when grown on a rice substrate for 2 weeks (data not shown). GC-MS analysis using extracts from the fungal rice culture confirmed that 15ADON and ZEA were detected in the Z3643 and add-back strains, whereas no detectable amount of 15ADON or a ∼30-fold-reduced level of ZEA was detected in the ΔFgLaeA strain derived from Z3643 (Fig. 5, Table S1). However, a FgLaeA-overexpression strain (OE8) produced ∼11-fold-increased amounts of 15ADON along with additional metabolites, such as butenolide and culmorin, which were not detected in Z3643; no ZEA levels were altered (Fig. 5, Table S1). The same effect of ΔFgLaeA on the DON and ZEA production was also confirmed using the other wild-type strain (9F1) of F. graminearum that produces a significant amount of culmorin [38] (Fig S2). In addition, qRT-PCR confirmed that the gene transcript levels for theses metabolites, Tri6 (FGSG_03536) required for 15ADON and CLM1 (FGSG_10397) for culmorin were drastically reduced in the ΔFgLaeA strain, but increased in the OE strain when grown in AG medium (Fig. 4C).
Figure 4. Mycotoxin production determined by luminescence signals and gene transcript levels.
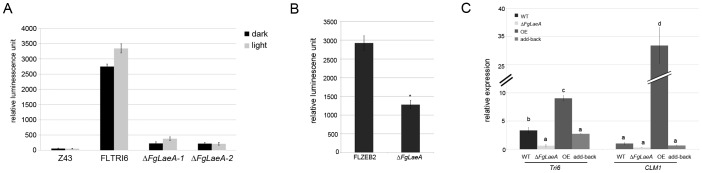
(A) Luminescence signals in the cell lysates from the Z3643 (Z43), FLTRI6, and two independent FLTRI6ΔFgLaeA (ΔFgLaeA-1 and ΔFgLaeA-2) strains grown in AG liquid medium for 6 days. (B) Luminescence signals in cell lysates from the FLZEB2 and FLZEB2ΔFgLaeA (ΔFgLaeA) strains grown in SG liquid medium for 8 days. (C) Relative transcript levels for secondary metabolites accumulated in the WT, ΔFgLaeA (derived from Z3643), FgLaeA-overexpression (OE), and add-back strains grown in AG liquid medium for 6 days, as determined by qRT-PCR. The amount of CLM1 transcript in the WT strain was used as a reference.//: discontinuity. Statistical analysis was performed with ANOVA and Duncan’s multiple range test. The same letter above bars represents no significant difference.
Figure 5. Reconstructed ion chromatogram of fungal rice culture extracts.
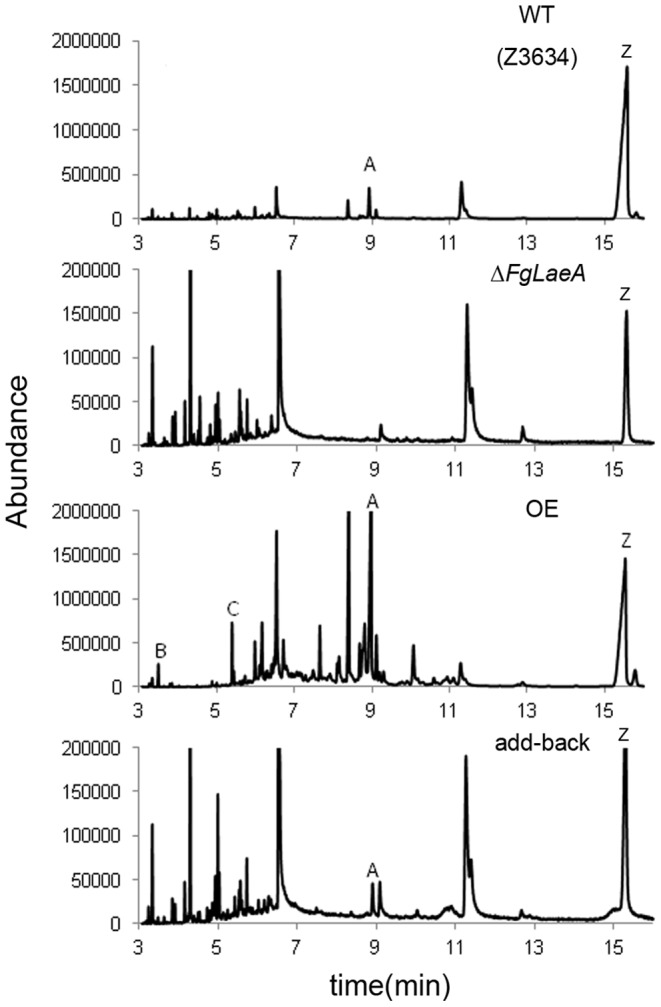
WT (Z3643), the wild-type Z3643 strain; ΔFgLaeA, a Z3643ΔFgLaeA strain derived from Z3643; OE, a FgLaeA-overexpression strain derived from the Z3643ΔFgLaeA strain; add-back, a FgLaeA-add-back strain derived from the Z3643ΔFgLaeA strain. A, 15ADON; B, butenolide; C, culmorin; Z, zearalenone.
In plant inoculation tests, the WT, OE, and add-back strains examined caused typical head blight leading to complete bleaching on wheat. The ΔFgLaeA strains were able to colonized inoculated spikelets, but rarely spread to the adjacent ones (Fig. 6).
Figure 6. Virulence of the F. graminearum strains on wheat heads.

The photographs were taken 14 days after inoculation. WT, wild-type strain Z3643; ΔFgLaeA, the ΔFgLaeA strain derived from Z3643; OE, the FgLaeA-overexpression strain derived from Z3643; add-back, the FgLaeA-add-back strain derived from the ΔFgLaeA strain.
Expression Pattern of FgLaeA, FgVeA and FgVelB in the Fungal Strains
qRT-PCR analysis revealed that the FgLaeA transcript was constitutively but weakly expressed in WT strain Z3643 during vegetative growth, perithecial formation on carrot agar, and trichothecene production in AG medium (Fig. 7). Except during conidiation in CMC medium, and 6 days after perithecia induction on carrot agar, FgLaeA transcript levels remained stable across all time points examined, staying within 1.5-fold relative to a reference gene (either EF1A or FGSG_09438) (Fig. 7). The constitutive low expression of FgLaeA under these conditions was clearly confirmed by RNA-seq analysis (unpublished data). The RPKM (reads per kilobase per million mapped reads) values of the FgLaeA transcripts accumulated under conditions for sexual development, trichothecene production, and vegetative hyphal growth were 4.3, 5.5, and 7.6, respectively. However, two other genes, FgVeA (FGSG_11955.3) and FgVelB (FGSG_01362.3), which may represent a FgVeA complex, showed sexual developmental-specific expression patterns and higher expression levels under all three conditions compared to FgLaeA. RPKMs for FgVeA and FgVelB were 325.9, 100.9, 126.4, and 630.9, 56.8, 33.0, respectively. Note that the FgVelB transcript, which significantly accumulated only on carrot agar for sexual development, could be clearly detected in a Northern blot hybridization [34]. In contrast, the FgLaeA transcript levels in two independent OE strains (OE7 and OE8) dramatically increased (up to ∼1,200-fold) compared to those in Z3643 during the entire growth conditions examined; those in the add-back strains were not significantly different from those in Z3643 (Fig. S3A).
Figure 7. Expression of FgLaeA under various culture conditions.
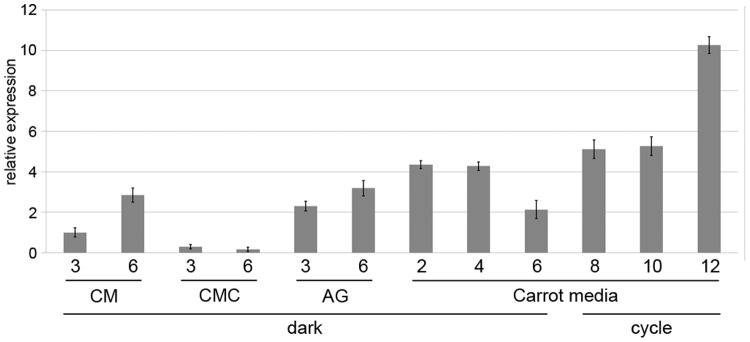
Relative FgLaeA transcript levels in the WT Z3643 strain grown in complete liquid medium for vegetative growth, CMC liquid medium for conidiation, AG liquid medium (AG) for trichothecene production, and on carrot agar for sexual development under dark or dark/light cycle conditions (cycle). Days of incubation following inoculation in each medium is shown in on the x-axis. GzRPS16 (FGSG_09438.3) was used as an endogenous control for data normalization [51]. The amount of FgLaeA transcript from a 3-day-old sample in complete liquid medium was used as a reference.
Expressions of two genes (FgVeA and FgVelB) for putative members of the FgVeA complex were dramatically elevated in the ΔFgLaeA strain on carrot agar (under the sexual developmental stage); only the FgVeA transcript level was increased in CM medium (Fig. S3B).
Expression of Mating-type (MAT) Genes in the ΔFgLaeA Strain under Sexual Development
To investigate a possible genetic cause for the early sexual induction in the ΔFgLaeA strain, we compared gene expression profiles at the mating-type (MAT) locus, the master regulator for sexual development, among the WT, ΔFgLaeA, OE, and add-back strains. qRT-PCR analysis confirmed that the transcript levels of two MAT genes (MAT1-1-1 and MAT1-2-1, both of which are required for early sexual stage development in F. graminearum [39]), were elevated ∼2–3-fold in the ΔFgLaeA strain compared to the WT strain 24 h after perithecial induction on carrot agar (Fig. 3B); no changes were observed in the OE and add-back strains during the same time points for perithecial induction compared to those in WT (Fig. 3B). However, the other three MAT genes (MAT1-1-2, MAT1-1-3, and MAT1-2-3) showed no dramatic differences in transcript levels among the strains examined (data not shown).
Differentially Expressed Genes (DEGs) in the ΔFgLaeA Strain under Trichothecene Production Conditions
To identify genes regulated under control of FgLaeA during trichothecene production, we performed RNA-seq analysis using total RNA extracted from mycelia of ΔFgLaeA and Z3643 strains grown in AG liquid medium for 60 h, an early induction stage for trichothecene production. Analysis of transcriptional profiles revealed a total of 799 genes differentially regulated ≥2-fold in the FLTRI6ΔFgLaeA strain compared to its WT progenitor FLTRI6; 444 and 355 genes were significantly down- and upregulated, respectively (Tables S2, S3, S4). Gene ontology (GO) analysis using blast2go (http://www.blast2go.com) revealed enrichment of DEGs associated with catalytic activity among GO molecular functions, and enrichment of DEGs associated with metabolic and cellular processes among GO biological processes (Fig. S4).
Searching against the conserved domain database (CDD) revealed that 332 of the DEGs (63.4%) found within the database contained enzymatic domains or were related to enzyme functions, including 72 genes (13.7%) involved in metabolism. In contrast, only 16 genes (3.0%) were identified as those probably involved in development and signal transduction (Table S5). In addition, 8 and 17 genes for transcription factors (TFs) were identified as down- and upregulated, respectively (Tables S2, S3, S4); however, none of the TFs differentially regulated in the FLTRI6ΔFgLaeA strain were associated with distinct functions based on gene deletion assays [40]. Unlike in the A. nidulans ΔlaeA strain [21], three major members of a putative FgVeA complex, FgVeA, FgVelB, and FgVosA (FGSG_06774.3), were not significantly differentially regulated in the FLTRI6ΔFgLaeA strain in AG medium; FgVeA and FgVosA showed only 1.48- and 1.43-fold increases, respectively, compared to FLTRI6 (note that the transcript levels of both FgVeA and FgVelB were dramatically increased in the ΔFgLaeA strain under sexual development condition, confirmed by qRT-PCR).
To identify genes for secondary metabolites, we compared the DEGs against members of the 77 tentative functional gene clusters (TFCs) in F. graminearum, previously identified using five types of functional descriptions [41]. Gene member(s) belonging to 37 TFCs and 19 TFCs were identified in downregulated and upregulated genes in the FLTRI6ΔFgLaeA strain, respectively (Tables S2, S3, S4). Moreover, members of nine TFCs were found in both down- and upregulated genes without overlapping.
Among the downregulated genes, seven genes (Tri3–Tri14) in the Tri gene cluster for trichothecene biosynthesis were identified; Tri101 (FGSG_07896.3), located outside the gene cluster, was not differentially expressed. Three polyketide synthase (PKS) genes including PKS10, responsible for biosynthesis of fusarin C, and two PKS genes (PKS2 and PKS14) whose chemical products have not yet been identified, a non-ribosomal peptide synthetase gene (NPS7), and CLM1 (FGSG_10397) for culmorin biosynthesis were also identified. However, all of these genes except PKS10 showed very low RPKM values (<0.5) in the WT FLTRI6 strain; eight members of a putative PKS10 cluster (FGSG_07798–07807) showed higher RPKM values ranging from 5.4 to 90.0 in FLTRI6. In addition to these key enzyme genes, three additional genes in the PKS cluster (PKS5, PKS11, and PKS12 for aurofusarin), one NPS gene (NPS12), and one butenolide cluster gene were downregulated. Six genes with contiguous gene numbers (FGSG_10608–10614), which were not deduced as a TFC in the previous study [41], were also downregulated (Tables S2, S3, S4). In contrast, no key enzyme genes in any TFCs were identified among the upregulated genes, with the exception of PKS7 (FGSG_08795.3).
Mapping of the DEGs onto the F. graminearum genome revealed the DEGs to be dispersed throughout the four chromosomes; however, they were not evenly distributed on each chromosome. The genomic positions of some DEGs seemed to be enriched in specific regions such as subtelomeres, or genome-wide locations of histone H3-lysine methylations (H3K7me3 or H3Kme2) (Freitag, personal communication) (Fig. S5).
Comparison of DEGs in the ΔFgLaeA and ΔFgVelB Strains
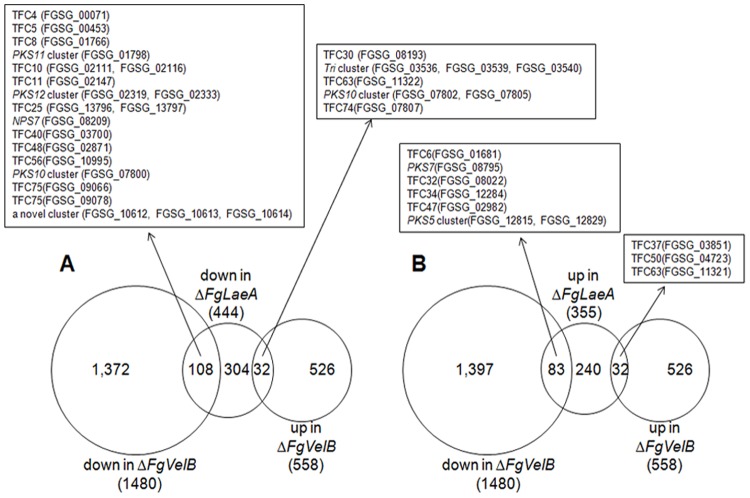
We compared DEGs in the ΔFgLaeA strain under toxin production conditions with those obtained from the ΔFgVelB strain grown under sexual development conditions [34]. Although growth conditions for the gene deletion strains were different from each other, more than 30% of DEGs in the ΔFgLaeA strain overlapped with those from the ΔFgVelB strain (Fig. 8). Among 444 genes downregulated in the ΔFgLaeA strain, 108 (24.3%) and 32 (7.2%) genes were similarly downregulated and upregulated, respectively, in the ΔFgVelB strain; genes belonging to 16 TFCs were downregulated in both strains. Gene members of five TFCs including the Tri gene cluster were differentially expressed between strains, exhibiting downregulation in the ΔFgLaeA strain and upregulation in the ΔFgVelB strain. Similarly, 83 (23.3%) and 32 (9.0%) genes from the sets of down- and upregulated genes in the ΔFgVelB strain overlapped with those upregulated in the ΔFgLaeA strain, respectively (Fig. 8). Gene members of three TFCs were upregulated in both strains, while six other TFCs also showed differential expression between strains (Fig. 8).
Figure 8. Number of genes expressed differentially in the F. graminearum ΔFgLaeA and ΔFgVelB strains.
(A) Downregulated genes; (B) upregulated genes. The numbers of DEGs for each category is indicated in parentheses below the fungal strains. TFC, tentative functional cluster according to Lee (2010) [40].
Cellular Localization of FgLaeA
To determine the cellular localization of the protein product of FgLaeA under various developmental stages, we constructed two additional transgenic fungal strains derived from the ΔFgLaeA strain, as well as the OE::GFP-FgLaeA strain described above. The ΔFgLaeA::nGFP-FgLaeA strain was generated by introducing an intact copy of FgLaeA fused to GFP under its native promoter into the ΔFgLaeA strain. The OE::RFP-His::GFP-FgLaeA strain was constructed by introducing the Pcrp-GFP-FgLaeA construct into the genome of a F. graminearum strain (H4::RFP) carrying a red fluorescence protein (RFP) fused to a histone gene [42]. The red pigmentation and perithecia formation pattern in both ΔFgLaeA::nGFP-FgLaeA and H4::RFP-OE::GFP-FgLaeA strains were restored to WT levels (Fig. 2). However, GFP expression in the former strains was not visualized by fluorescence microscopy.
In both OE::GFP-FgLaeA and H4::RFP-OE::GFP-FgLaeA strains, GFP-FgLaeA commonly localized to both the nucleus and cytoplasm in fungal cells grown under all of stages examined, including newly formed conidia, germinating conidia, mature hyphae, and ascospores under both dark and light conditions (Fig. 9). Occasionally, nuclear enrichment of GFP signals was not obvious; instead, GFP expression was clearly seen in cellular compartments other than the nucleus (Fig. 9B).
Figure 9. Expression of GFP in the F. graminearum OE::GFP-FgLaeA strain grown on CM plates.
(A) Vegetative hyphae grown under dark/light cycle conditions; (B) those grown under constant dark conditions; (C) ascospore in ascus; (D) conidia; (E) conidium 3 h after germination; (F) apical tip of the germ tube after 7 h germination. Each picture consists of DIC, H4::RFP, GFP::FgLaeA and a merged picture of the RFP and GFP images, respectively. White arrows indicate the nuclei in each image. Size bar is 10 µm.
Protein Interactions between FgLaeA and FgVeA, FgLaeA and FgVelB, and FgVeA and FgVelB
To determine if the FgLaeA protein is a member of a possible FgVeA complex (i.e., a direct interaction occurred between FgLaeA and FgVeA), we employed the split luciferase complementation assay, a sensitive and efficient method of monitoring in vivo protein–protein interactions in filamentous ascomycetes [43]. For this assay, we generated plasmid vectors carrying the entire coding region of FgLaeA fused to a DNA region encoding an N-terminal fragment of luciferase (FgLaeA–nLuc) and the FgVeA fused to a C-terminal fragment (cLuc–FgVeA), respectively, and introduced either or both of them to the genome of Z3643. All cell lysates from negative controls grown in complete liquid medium, which were transgenic F. graminearum strains carrying a single empty vector expressing only nLuc or cLuc (pFNLucG or pFCLucH), a single-fused gene (FNLuc–FgLaeA or FCLuc–FgVeA), or the single fused gene along with the corresponding singly empty vector, exhibited only background levels of luminescence similar to WT strains (Table 1). In contrast, all fungal transformants co-expressing both of the fused proteins, FNLuc–FgLaeA and FCLuc–FgVeA, exhibited ∼1,000–2,300-fold increased luminescence relative to negative controls (Table 1). Similarly, we examined whether other F. graminearum LaeA-like proteins, including FgVIP1 described above, had the capacity to interact with FgVeA. No significant increase in luminescence intensity was found in the fungal strains co-expressing the FNLuc–FgVIP1 and FCLuc–FgVeA compared to the negative controls (Table 1). In addition, we examined the interactions between FgLaeA and FgVelB as well as between FgVeA and FgVelB. Transgenic F. graminearum strains co-expressing the FNLuc-FgVelB and FCluc-FgVeA constructs showed higher levels of luminescence compared to WT strains (Table 1), demonstrating the FgVeA-FgVelB interaction. Interestingly, the luminescence levels in the cell lysates from the fungal strains carrying both FNLuc-VelB and FCLuc-FgLaeA were also higher than those from the WT strains and negative controls although the luminescence levels for the FgLaeA-FgVelB interaction were much lower than those from the other fungal strains showing positive signals (Table 1).
Table 1. Luminescence activity1 in fungal cell lysates.
| Strain | Interaction | Plasmids inserted | RLU2 |
| Z3643 | no | None | 15-±6 |
| cLUCnLUC | no | pFNLucG & pFCLucH | 23±13 |
| GzFNCS-1 | FBP1-SKP1 | pFNLuc-FBP1G &pFCLuc-SKP1H | 19,815±7,268 |
| VeALaeA-5 | FgLaeA-FgVeA | pFNLuc-FgLaeAG &pFCLuc-FgVeAH | 67,555±15,605 |
| VeALaeA-8 | FgLaeA-FgVeA | pFNLuc-FgLaeAG &pFCLuc-FgVeAH | 28,849±5,341 |
| VeAVelB-4 | FgVeA-FgVelB | pFNLuc-FgVelBG &pFCLuc-FgVeAH | 2,596±1,790 |
| VeAVelB-5 | FgVeA-FgVelB | pFNLuc-FgVelBG &pFCLuc-FgVeAH | 2,786±1,543 |
| VelBLaeA-1 | FgLaeA-FgVelB | pFNLuc-FgVelBG &pFCLuc-FgLaeAH | 249±94 |
| VelBLaeA-4 | FgLaeA-FgVelB | pFNLuc-FgVelBG &pFCLuc-FgLaeAH | 505±294 |
| VeAVIP1-6 | FgVIP1-FgVeA | pFNLuc-FgVIP1G &pFCLuc-FgVeAH | 29±6 |
| VeAVIP1-9 | FgVIP1-FgVeA | pFNLuc-FgVIP1G &pFCLuc-FgVeAH | 39±4 |
| LaeAN-1 | FgLaeA-FN3 | pFNLuc-FgLaeAG & pFCLucH | 48±2 |
| LaeAN-3 | FgLaeA-FN | pFNLuc-FgLaeAG & pFCLucH | 35±4 |
| LaeAC-1 | FC3-FgLaeA | pFNLucG & pFCLuc-FgLaeAH | 28±3 |
| VeAC-2 | FC-FgVeA | pFNLucG & pFCLuc-FgVeAH | 22±4 |
| VeAC-3 | FC-FgVeA | pFNLucG & pFCLuc-FgVeAH | 28±8 |
| VelBC-3 | FC-FgVelB | pFNLucG & pFCLuc-FgVelBH | 30±5 |
| VIP1C-2 | FgVIP1-FC | pFNLuc-FgVIP1G & pFCLucH | 11±7 |
| VIP1C-3 | FgVIP1-FC | pFNLuc-FgVIP1G & pFCLucH | 15±2 |
obtained from three replicates.
relative light units per microgram of protein per min.
FC: a C-terminal fragment of the firefly luciferase, FN: a N-terminal fragment.
Development of a Firefly Luciferase Reporter System for Mycotoxin Production
To circumvent several problems in the chemical analysis of mycotoxin production, we developed a new reporter system for mycotoxin production in F. graminearum using firefly luciferase. A DNA construct carrying the firefly Photinus pyralis luciferase gene (designated Fluc) under control of a putative promoter region (∼1,000 bp) from either TRI6 or ZEB2 (transcription factors controlling the production of trichothecenes and zearalenone, respectively) was inserted into its native genomic region in F. graminearum strain Z3643 via homologous recombination (Fig. S6, S7). The single crossover integration events, confirmed by Southern hybridizations, resulted in fungal strains designated FLTRI6 and FLZEB2, respectively (Figs. S6, 7). No differences were seen between strains FLTRI6 and FLZEB2 and their WT progenitor (Z3643) for hyphal growth, pigmentation, conidiation, sexual development, and toxin production (data not shown). Strain FLTRI6 was grown in AG liquid medium, which favors trichothecene production [15], for 7 days. Luminescence signal intensities from cell lysates peaked at day 6, whereas only basal levels were detected in complete medium (CM; Fig S6C). Gas chromatography-mass spectrometry (GC-MS) analysis revealed that the FLTRI6 strain began producing both DON and 15ADON at day 4, and showed maximum levels of toxin production at day 7; no detectable level of trichothecenes were produced in complete liquid medium during the entire incubation periods (data not shown). Similarly, the luminescence signals from the FLZEB2 strain grown in SG liquid medium, which favors zearalenone production [14], increased up to day 8, but showed only basal level when grown in CM (Fig. S7C). Higher luminescence signals and toxin accumulations were detected on 2-week-old rice cultures of the FLZEB2 strain, a more favorable condition for the production of both toxins (data not shown). A similar pattern in time-course increases between luminescence signals and toxin accumulations from these strains demonstrated that both the FLTRI6 and FLZEB2 strains could be used as luminescence reporter systems for qualitative and quantitative determination of mycotoxin production in F. graminearum.
Discussion
The results presented here clearly demonstrate that a LaeA-like gene (FgLaeA) of F. graminearum is involved in controlling developmental processes, secondary metabolism, and virulence on host plant. Reduced pigmentation of aerial mycelia in the ΔFgLaeA strains is one of the most common growth-related phenotype in the LaeA-deletion strains of the previously characterized filamentous fungi except C. heterostrophus [27], in which pigmentation was increased, even though the biosynthetic pathways for each polyketide pigment were different (Table 2). This phenotypic change could have been attributable to the role of FgLaeA in controlling secondary metabolism described below. However, the leaky pigmentation phenotype on agar plates indicates that PKS12-mediated aurofusarin production is not tightly regulated by FgLaeA, especially when hyphal elongation was physically blocked (e.g., by Petri dish wall or neighbor colonies on agar plates). The relatively minor effect of ΔFgLaeA on light-dependent asexual development (conidiation) under dark condition was also similar to those of LaeA homologs in other fungi, in which the conidiation mostly decreased (Table 2). However, no disturbance of light-dependent conidiation by either the absence or overproduction of FgLaeA implies that FgLaeA is not involved in a light-sensing pathway for conidiation in F. graminearum.
Table 2. Comparison of phenotypic changesa caused by the LaeA-like gene deletion or overexpression in filamentous fungi.
| Species | Hyphal growth & morphology | Pigmentation | Conidiation | Sexual development | Secondary metabolismb | Virulence | References |
| A. nidulans | similar to WT | loss | reduced | increased smaller, lowerfertile cleisothecia | reduced for ST and PN; increased for PN, LOV, & monocolin J | NAc | [20], [22], [25] |
| A. fumigatus | similar to WT; more hyphal mass | loss | reduced | NDd | reduced for gliotoxin | impaired | [20], [25] |
| A. flavus | similar to WT | loss | reduced; increased on peanut | no sclerotia; increased | reduced for AF, cyclopiazonic acid, &kojic acid; increased for AF & additional metabolites | NAc | [24] |
| P. chrysogenum | curly & hyperbranching hyphae | NDd | reduced, light-dependent | NDd | reduced for PN | NAc | [43] |
| C. heterostrophus | reduced growth; reducedtolerance to H2O2 | increased; reduced | increased under vegetative | female-sterile;female-sterile | reduced for T-toxin in the dark;increased for T-toxin in both | reduced | [27] |
| T. reesei | similar to WT | loss | reduced; increased; light-dependent | NDd | loss of cellulases; increased for cellulases | NAc | [31], [32] |
| F. fujikuroi | similar to WT | slightly reduced;increased | reduced | NDd | reduced for GA; increased for bikaverin | reduced | [28] |
| F. verticillioides | NDd | NDd | NDd | NDd | reduced for bikaverin, fusaric acid,& fusarin C | NDd | [29] |
| F. oxysporum | increased growth rate; no aerialmycelium; reducedhydrophobicity | NDd | reduced | NDd | reduced for siderophores,beauvericin, & fusaric acidin the dark | reduced | [30] |
| F. graminearum | fewer aerial mycelium; reducedhydrophobicity; similar to WT | reduced or loss;similar to WT | reduced only in the dark;light-dependent;similar to WT | slightly increased;light-dependentsimilar to WT | reduced for DON, ZEA, CM, &fusarin C; light-independent;increased for DON, BT, & CM | reduced | this study |
phenotypes caused by FgLaeA-overexpression were shown in bold in the same column for each description.
abbreviations for metabolites: PN, penicillin; LOV, lovastatin; AF, aflatoxins; GA, gibberellic acids; DON, deoxynivalenol; ZEA, zearalenone; BT, butenolide; CM, culmorin.
not applicable;
not determined.
In contrast to these conserved functions, an earlier induction of perithecia formation in the ΔFgLaeA strain (and recovery to WT levels in both add-back and OE strains) provided genetic evidence demonstrating a distinct role of FgLaeA during sexual development in F. graminearum. This phenotype could be comparable to the elevated, constitutive formation of fruiting bodies (cleistothecia) in the A. nidulans ΔlaeA strain (Table 2), implying a conserved role of LaeA as a negative regulator for initiating sexual development in both fungi. However, the effect of gene deletions on completion of sexual development were different between the two species. The A. nidulans ΔlaeA strain was not able to form Hülle cells, which nurse the young cleistothecia, leading to the production of smaller, less fertile cleistothecia [22]; no obvious impairments in maturation of fertile perithecia were observed in F. graminearum.
The other striking difference between F. graminearum LaeA and LaeA from other species is the effect of light on sexual development. In A. nidulans, cleistothecia formation occurs preferentially in the dark and is inhibited by light. This responsiveness, required for proper sexual development, is disrupted in ΔlaeA strains, as elevated cleistothecia formation is observed even in the presence of light [22]. In contrast, the light effect on inducing sexual development in F. graminearum is completely opposite to that of A. nidulans; perithecia formation occurs exclusively under either light or dark/light cycle conditions, and is completely inhibited under dark condition. Even though the absence of FgLaeA enhanced sexual development, it did not affect light-dependent perithecia formation, suggesting that FgLaeA may play a role in repressing sexual developmental processes, especially during early stages. However, unlike A. nidulans laeA, FgLaeA is not involved in sensing light signals for initiating sexual development, which is similar to its role in regulating conidiation, as described above. The effect of LaeA deletion in F. graminearum was opposite that of C. heterostrophus, when ΔChlae1 caused female sterility, suggesting a role in positive regulation of sexual reproduction by ChLAE in this species [27]. Even though this phenotypic change in the ΔFgLaeA strain seems relatively subtle, the distinct role of FgLaeA in sexual development becomes more readily apparent when compared to phenotypic changes caused by the deletion of other velvet complex member genes in F. graminearum (e.g., ΔFgVeA and ΔFgVelB) [33]-[35]. Loss of FgVeA and FgVelB completely abolished the capability of F. graminearum to form perithecia, which is opposite to effect of ΔFgLaeA. Downregulation of three MAT genes in the ΔFgVelB strain, confirmed by a microarray analysis, is consistent with FgVelB as a positive regulator of MAT gene expression [34]. In contrast, increased MAT transcript levels in the ΔFgLaeA strain during early stage of perithecia formation, confirmed by qRT-PCR, could be attributable to the enhanced sexual development phenotype, indicating that FgLaeA controls MAT gene expression in a manner different from that of the other FgVeA complex members (i.e., independently of the FgVeA complex). In addition, upregulation of FgVeA and FgVelB in the ΔFgLaeA strain under perithecial induction stage, but not under toxin production stage also suggests a specific role for FgLaeA (as a negative regulator) in modulating FgVeA complex for sexual development, as proposed in A. nidulans [21], [22].
In contrast to developmental processes, secondary metabolism was significantly affected by ΔFgLaeA in F. graminearum, consistent with the role of LaeA homologs in many filamentous fungi (Table 2). Not only reduced production of several mycotoxins and their corresponding gene expression levels in the ΔFgLaeA strain, but increased production of these metabolites by FgLaeA overexpression clearly confirms FgLaeA as a positive regulator of secondary metabolite biosynthesis. In addition, RNA-seq analysis of a ΔFgLaeA strain (FLTRI6ΔFgLaeA) grown under conditions favoring trichothecene production supports the role of FgLaeA as a global regulator for secondary metabolite production, including these mycotoxins. Among the DEGs in the ΔFgLaeA strain were genes for key enzymes (e.g., PKS, NRPS, terpenoid synthase, transporters, and cytochrome P450) and other members F. graminearum TFCs for secondary metabolites [41]. These genes were identified at a high frequency (56 of 77), indicating that these TFCs may be globally regulated by FgLaeA, although in many TFCs, only a small number of genes were found to be differentially expressed. Further RNA-seq analyses using an FgLaeA-overexpressing strain, as well as the ΔFgLaeA strain under different conditions favoring other metabolites, would be necessary to confirm a global role for FgLaeA in controlling secondary metabolite production in F. graminearum. Localization of some DEGs in subtelomeric or H3K7 methylation regions of each of the F. graminearum chromosomes would support a role for FgLaeA in chromatin-based regulation of gene expression for secondary metabolites, as proposed in Aspergillus species [44]. Taken together, these data provide conclusive evidence that FgLaeA has a conserved role in secondary metabolism at the global level in F. graminearum, as demonstrated in other species of Fusarium (F. fujikuroi, F. verticillioides, and F. oxysporum), as well as distantly related species (Table 2).
Fewer symptoms on wheat displayed in the ΔFgLaeA strain could be attributed to impaired ability of this strain to produce a secondary metabolite required for disease development, such as trichothecenes [12], which is comparable to the effect of other fungal LaeA genes on the production of secondary metabolites required for virulence in A. fumigatus [25], C. heterostrophus [27], and F. fujikuroi [28]. However, more attenuated symptoms caused by ΔFgLaeA than those by the gene disruption for trichothecene biosynthesis [12] suggests that FgLaeA controls additional secondary metabolisms and/or regulatory pathways essential for disease development in F. graminearum.
The next question we wanted to ask was whether the regulatory roles of FgLaeA described above are associated with those of other members of the putative FgVeA complex, as in A. nidulans and other species [21], [23], [27], [28], [30], [45]. To address this question, we would need to demonstrate a direct interaction between FgLaeA and FgVeA in F. graminearum, similar to that seen in A. nidulans, Penicillium chrysogenum, and F. fujikuroi [21], [28], [45], in which protein–protein interactions were confirmed by either yeast two-hybrid or bimolecular fluorescence complementation (BiFC) analysis. A previous study showed no interaction between FgLaeA and FgVeA using a yeast two-hybrid method [33]. Evidence provided in this study directly contradicts this finding; we demonstrated a clear in vivo protein–protein interaction between FgLaeA and FgVeA in F. graminearum using a newly developed split-luciferase complementation assay [43], implying that FgLaeA plays a role as a member of the FgVeA complex. In addition, the interaction between FgVeA and FgVelB, which was also confirmed by split luciferase complementaion, clearly supports the existence of the heterotrimeric FgLaeA–FgVeA–FgVelB complex in F. graminearum. Interestingly, positive (but lower) luminescence signals from the interaction between FgLaeA and FgVelB, which has not been intensively investigated in other fungi except A. nidulans [21], [22], may suggest a possibility for the direct interaction between these two protein in F. graminearum. However, it is also possible that FgVeA acts as a bridge to bring the FNLuc-FgVelB and FCLuc-FgLaeA into close proximity, as shown in A. nidulans [22], which leads to a partial complementation of split luciferase domains. Furthermore, more than 30% of DEGs in the ΔFgLaeA strain overlapped with those in the ΔFgVelB strain despite the difference in culture conditions for each gene deletion strain (Fig. 8). These overlapping DEGs may be common targets of both FgLaeA and FgVelB, or the FgLaeA–FgVeA–FgVelB complex. The presence of DEGs displaying opposite gene expression patterns between two strains (i.e., 32 genes downregulated the ΔFgLaeA strain and upregulated in the ΔFgVelB strain, and 83 genes upregulated in the ΔFglaeA strain and downregulated the ΔFgVelB strain) suggests their expression may be differentially controlled by the FgVeA complex, which is growth condition-dependent. For example, the trichothecene gene cluster is positively controlled by the FgVeA complex in agmatine liquid medium, while negatively controlled during the sexual development, as previously suggested [34].
In addition to protein–protein interactions, constitutive nuclear localization of LaeA homologs would be important evidence regarding the regulatory role of LaeA. Nuclear localization of the OE::GFP-FgLaeA product during various developmental stages, such as mature hyphae, conidia, germinating conidia, and ascospores, regardless of light signals, was clearly demonstrated in this study. However, exclusive nuclear enrichment of a LaeA–GFP signal in A. nidulans, as evidenced based on the nuclear/cytoplasmic GFP signal ratio [21], was not evident in F. graminearum (Fig. 9), in which significant levels of GFP expression were visualized in the cytoplasm as well as in the nucleus. This localization could be inferred based on nonspecific accumulation of the excessively producing GFP–FgLaeA in the cytoplasm. However, considering that the LaeA–GFP constructs used in cellular localization analyses in A. nidulans [20], [21] and F. fujikuroi [28] were expressed under the control of strong promoters such as those for alcA, niiA, and gpd in A. nidulans, the cytoplasmic distribution of GFP–FgLaeA may be not an artifact, but reflect a distinct function of FgLaeA compared to other fungi.
In conclusion, FgLaeA plays important roles in controlling secondary metabolism, various developmental processes, and virulence in F. graminearum, not only through functions conserved across several filamentous fungi (e.g., as a member of the FgVeA complex), but also through distinct additional functions (e.g., FgVeA complex-independent), which may reflect the unique life style of F. graminearum. Looking beyond FgLaeA, the efficiency and usefulness of the firefly luciferase reporter system for mycotoxin production developed here was clearly demonstrated in this study. This system has a potential for application in other investigations, e.g., using a recipient strain for an insertional mutagenesis to identify a novel gene controlling the expression of Tri6 or ZEB2.
Materials and Methods
Fungal Strains and Culture Conditions
The F. graminearum WT strains Z3643, provided by Dr. Robert L. Bowden (USDA-ARS Plant Science and Entomology Research Unit, Manhattan, KS, USA), and 9F1 [38] belongs to lineage 7 of the F. graminearum species complex [7], [46]. T43ΔM2-2, acting as a female in an outcross, is a MAT1-2-1-deleted strain derived from Z3643 [47]. A F. graminearum transgenic strain carrying histone H4 fused with RFP [42] was used for the nuclear localization of FgLaeA. The WT and transgenic strains derived from Z3643 were stored in 20% glycerol at –70°C. Conidiation was induced in CMC [48] liquid medium. For sexual development, strains were inoculated and incubated on carrot agar as previously described [3], [47]. For genomic DNA extraction, each strain was grown in 50 mL CM [3] at 25°C for 72 h on a rotary shaker (150 rpm). For trichothecene production and total RNA extraction, 1 mL conidial suspension (105/mL) of each strain was inoculated into the AG-amended liquid medium [49], as previously described. For zearalenone production and total RNA extraction, fungal strains were grown in SG liquid medium or on rice substrate, as previously described [14].
Nucleic Acid Manipulations and Primers
Fungal genomic DNA was prepared as described previously [3], [50], and total RNA was extracted from mycelia using an Easy-Spin Total RNA Extraction kit (Intron Biotech, Seongnam, Korea) according to the manufacturer’s instructions. Other general procedures for nucleic acid manipulations were performed as described previously [51]. DNA gel blots were hybridized with biotinylated DNA probes prepared using the BioPrime DNA labeling system (Invitrogen, Carlsbad, CA, USA) and developed using the BrightStar® BioDetect™ Kit (Ambion, Austin, TX, USA). All PCR primers used in this study (Table S5) were synthesized by the Bioneer Corporation (Chungwon, Korea). qRT-PCR was performed with SYBR Green Super Mix (Bio-Rad, Hercules, CA, USA) using first-strand cDNA synthesized from total RNA; amplification efficiencies of all genes were determined as previously described [52]. Gene expression was measured in three biological replicates from each time point. GzRPS16 (FGSG_09438.3) and EF1A (FGSG_08811.3) were used as endogenous controls for data normalization [52].
Mycotoxin Analysis, Virulence Test, and Luciferase Assay
F. graminearum strains were grown in 20 mL AG liquid medium for 6 days for trichothecene production and extracted as described previously [53]. For production of both trichothecenes and zearalenone, and other metabolites, fungal strains were grown in triplicate on 40 g of rice substrate inoculated with spores washed from V8 juice plate for 7 days in the dark at 25°C. The fungal cultures were extracted on a rotary shaker with 100 ml ethyl acetate, dried on a rotoevaporator, and re-dissolved in 20 ml acetonitrile-water (86∶14). Five milliliter of this solution was cleaned with a Romer MycoSep225® Trich column, dried under a stream of nitrogen, and re-dissolved in 1 ml methanol. GC-MS analysis was performed on a HP 6890 has chromatograph fitted with HP-5MS (30 m×0.25 mm film thickness) and a 5973 mass detector. The carrier gas was helium with a 20∶1 split ratio and a 20 ml/min slot flow. The column was held at 120°C at injection, heated to 260°C at 25°C/min and held at 260°C for 14 min. Compounds were identified by comparison of GC retention time and mass spectral fragmentation with those of standard compounds.
The virulence of fungal strains was determined on wheat heads. Briefly, conidia of each strain were harvested from CMC culture, suspended in sterile water (1×106 conidia/ml), and injected into a floret in the basal spikelet of the wheat (cv. Eunpamil) head at mid-anthesis. The plants were placed in a greenhouse for two weeks after a 3-day incubation in a humidity chamber.
Luciferase activity in cell lysates from fungal strains was measured using GloMax® 96 Microplate Luminometer (Promega) as previously described [43].
RNA-seq Analysis
PolyA tailed transcript RNA (mRNA) was enriched from total RNAs prescreened for quality using Oligo dT beads (TruSeq RNA Sample Prep. Kit, FC-122-1001,1002; Illumina, San Diego, CA, USA) and fragmented for cDNA synthesis by reverse transcriptase (SuperScript II Reverse Transcriptase, part no. 18064-014; Invitrogen). The synthesized cDNA underwent end-repair and subsequent 3′ A tailing for adaptor ligation (TruSeq RNA Sample Prep. Kit, FC-122-1001,1002; Illumina). After amplification of cDNA with adaptors, 300–400-bp cDNA fragments were finally selected. Paired-end sequencing of the amplified cDNA library was performed using the HiSeq2000 platform [TruSeq SBS Kit v3–HS (200 cycles), ILFC-401-3001; Illumina). RNA-seq reads were aligned to the F. graminearum genome sequences available from the Broad Institute (http://www.broadinstitute.org/annotation/genome/fusarium_group/MultiDownloads.html) and normalized to RPKM values for genes by using the CLC Genomics Workbench RNA-seq data analysis pipeline according to the developer’s instructions. RPKM values from each strain were used for identification of DEG sets. The RNA-seq data have been deposited in the NCBI Short Read Archive (http://www.ncbi.nlm.nih.gov/sra; SRP022085).
Constructions of Vectors and Fusion PCR Products for Fungal Transformation
The DNA construct for deletion of FgLaeA from the genomes of F. graminearum strains Z3643, FLTRI6, and FLZEB2 was created using a split-marker recombination procedure as previously described [39], [54]. The 5′ and 3′ flanking regions of the FgLaeA ORF were amplified with the primer pairs 657for5/657revtail5 and 657fortail3/657rev3, respectively, fused to the geneticin resistance gene (gen) cassette, which was amplified from pII99 [55] with the primers Gen-for and Gen-rev, in the second round of PCR, and used as a template to generate split markers with the new nested primer sets, 657nest5/Gen-revN and 657nest3/Gen-forN, respectively (Table S6).
DNA plasmids carrying FgLaeA fused to GFP under control of the cryparin gene (Crp) promoter from C. parasitica [36] (PCrp-GFP-FgLaeA), and the same construct under control of the native promoter region of FgLaeA (Pnative-GFP-FgLaeA), were generated using a double-joint PCR [56]. For the PCrp–GFP–FgLaeA construct, three amplicons were amplified from pCHPH1 [36], pIGPAPA [47], and Z3643 with the primer pairs Pcrp_for/Pcrp-GFP tai/rev, GFP_for/GFP_rev, and LaeA-GFP tail/LaeA rev, respectively, mixed in a 1∶1:3 molar ratio, and finally fused together using the nested primer set Pcrp_for and LaeA 3rd fusion. For Pnative-GFP-FgLaeA, the 1.0-kb native promoter region of FgLaeA was amplified from Z3643 with primers pLaeA for and pLaeA rev. The final fusion product was amplified with the nested primer set pLaeA for and LaeA 3rd fusion. Each of these two fusion PCR products was cloned into pGEMT (Promega), creating pPCrp–GFP–FgLaeA and pPnative–GFP–FgLaeA, respectively. The DNA plasmid (pPnative–FgLaeA) used in generating the add-back strain was constructed by amplification of a 3.2-kb FgLaeA region including the FgLaeA ORF with its 5′ (0.9 kb) and 3′ (1.0 kb) flanking regions with primers LaeA5R and LaeA3F, followed by cloning into pGEMT.
For insertion of the firefly luciferase gene (FLuc) under control of the putative promoter region of Tri6, a 1.8-kb fragment upstream of the 5′ end of Tri ORF was fused to the FLuc ORF (1.6 kb) using a single-joint PCR method [56]. The promoter region and FLuc were amplified from the genomic DNA of Z3643 and the plasmid DNA pSPluc+NF fusion vector (Promega, Madison, WI, USA) with primer pairs Tri6SPL5/Tri6Luc3 and SPluc5/SPluc3 (Table S6), respectively. The two amplicons were mixed in a 1∶1 molar ratio and fused in a second round of PCR, followed by a third round of PCR using the nested primer set SPLuc5nest and TSPluc3nest (Table S6). The amplified product was cloned into pGEMT (Promega), creating pPTri6-FLuc. A SalI-digested hygB cassette, derived from pBCATPH, was cloned into pPTri6-FLuc, resulting in a 8.9-kb plasmid pPTri6-FLuc-H. Similarly, the plasmid DNA carrying the FLuc gene fused to a promoter of ZEB2 was constructed as described above. The ZEB2 promoter region (1.8 kb) was amplified from Z3643 with the primers ZEBSPLuc5 and ZEBSPLuc3 for the first round of PCR and fused to the FLuc PCR product in the second round, followed by a third round with primers ZSLuc5nest and SPLuc3nest. Using the same cloning procedure described above, the final vector pPZEB2–Fluc-H was constructed.
The amplified fusion products or constructed DNA plasmids were added into the protoplasts of WT F. graminearum strains for transformation [39], [57]; pPCrp–GFP–FgLaeA, pPnative–GFP–FgLaeA, and pPnative–FgLaeA were added into the ΔFgLaeA strain along with pII99 carrying gen, respectively, for co-transformation.
Split Luciferase Complementation Assay
For split luciferase complementation, the coding regions of the FgLaeA and FgVIP1 genes, which were amplified from total RNA of Z3643 with the primer pairs LaeAnLuc-for2/LaeA-inrev3 and VIP1-forNT/VIP1-revNT, respectively, were introduced into the SalI site of the DNA plasmid pFNLucG [43] carrying an N-terminal fragment of FLuc and gen using the In-Fusion® HD Cloning Kit (Clontech, Mountain View, CA, USA) as previously described [43], creating pFNLuc-FgLaeAG and pFNLuc-FgVIP1G (Table 1), respectively. Similarly, the cDNA of FgVeA (FGSG_11956.3) was cloned into pFCLucH [43] carrying a C-terminal fragment of FLuc and hygB, generating pFCLuc–FgVeAH (Table 1). The DNA plasmid pFCLuc–FgVeAH was added into the protoplasts of Z3643 along with either pFNLuc–FgLaeAG or pFNLuc–FgVIP1G for selection of fungal transformants carrying both plasmids. Luciferase activity was measured from the cell lysates of the transformants grown in CM liquid medium for 3 days, as previously described [43]. As a positive control, we included the transgenic F. graminearum GzFNCS-1 strain showing high luciferase activity driven by an in vivo protein–protein interaction between FBP1 and SKP1, fused to NLuc and CLuc, respectively [43]. The experiment was repeated three times.
Microscopic Observations
Microscopic observations were performed using an image analysis system consisting of a microscope (Leica DM 2000) with attached digital camera (Leica DFC 550) and a computer.
Supporting Information
(A) Pigmentation of the ΔFgLaeA strain in the marginal regions between different growing colonies grown on CM agar plate, and (B) when grown in CM liquid medium. Left and right, WT and ΔFgLaeA strain, respectively.
(TIF)
Reconstructed ion chromatograms of rice culture extracts from the fungal strains derived from 9F1. WT (9F1), the wild-type 9F1 strain; ΔFgLaeA, a 9F1 ΔFgLaeA strain derived from 9F1; add-back, a 9F1 FgLaeA-add-back strain derived from the 9F1ΔFgLaeA strain. A, 15ADON; C, culmorin; Z, zearalenone.
(TIF)
Expression of FgLaeA, FgVeA and FgVelB in various F. gramineaum strains. (A) Relative transcript levels of FgLaeA in the FgLaeA-overexpression and add-back strains, and (B) those of FgVeA and FgVelB in the ΔFgLaeA strain derived from Z3643, which were grown in CM liquid medium for vegetative growth, CMC liquid medium for conidiation, AG liquid medium for trichothecene production, and on carrot agar for sexual development. Days of incubation following inoculation in each medium are shown in on the x-axis. cycle, 12 h-dark/12 h-light cycle. The amounts of FgVeA and FgVelB transcripts from a 6-day-old sample in AG liquid medium were used as references in (A) and (B), respectively.
(TIF)
Gene ontology analysis of DEGs in the Δ FgLaeA strain grown in AG liquid medium for 60 h.
(TIF)
Distribution of DEGs in the Δ FgLaeA strain on each chromosome of the F. graminearum PH-1 strain. The genomic locations of histone 3 lysine methylations (H3K7me3 and H3K4me2) were indicated by thin black and red bars, respectively, below the positions of DEGs.
(TIF)
Generation of a firefly luciferase reporter system for trichothecene production. (A) Scheme for the insertion of the FLuc gene under control of a promoter region of Tri6 into the genome of the F. graminearum Z3643 (WT) strain by homologous recombination. (B) SalI-digested genomic DNA gel blot probed with the entire vector. Lane 1, WT strain; lane 2, the FLTRI6 strain. DNA size markers are indicated on the left side of the gel. (C) Luminescence signals in the cell lysate from FLTRI6 grown in AG and complete liquid media, respectively. Days postinoculation (DPI) are indicated below the days of incubation.
(TIF)
Generation of firefly luciferase reporter system for zearalenone production. (A) Scheme for the insertion of the FLuc gene under control of a ZEB2 promoter into the genome of the F. graminearum Z3643 (WT) strain by homologous recombination. (B) SalI-digested genomic DNA gel blot probed with the entire vector. Lane 1, WT strain; lanes 2 and 3, the FLZEB2 strains. DNA size markers are indicated on the left side of the gel. (C) Luminescence signals in the cell lysate from a FLZEB2 strain grown in SG and complete liquid media, respectively. Days postinoculation (DPI) are indicated below the days of incubation. Asterisks above the bars indicate a significant difference between two culture conditions for each DPI according to Tukey’s test.
(TIF)
Quantification of 15ADON and ZEA levels in fungal rice culture extracts.
(XLSX)
Genes down-regulated in the Δ FgLaeA strain grown in agmatine-amended liquid medium for 60 h.
(XLSX)
Genes up-regulated in the Δ FgLaeA strain grown in agmatine-amended liquid medium.
(XLSX)
DEGs belonging to tentative functional gene clusters from the Δ FgLaeA strain.
(XLSX)
Grouping of differentially expressed genes in the Δ FgLaeA strain by functional domains.
(XLSX)
Primers used in this study.
(DOCX)
Acknowledgments
We are grateful to Crystal Probyn for her technical assistance, and Stephanie Folmar and Jennifer Teresi for conducting the virulence test.
Funding Statement
This research was supported by the R&D Convergence Center Support Program, Ministry for Food, Agriculture, Forestry and Fisheries, Republic of Korea, and by a grant from the Next-Generation Bio Green21 Program (no. PJ008210), the Rural Development Administration, Republic of Korea. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. McMullen M, Jones R, Gallenberg D (1997) Scab of wheat and barley: a re-emerging disease of devastating impact. Plant Dis 81: 1340–1348. [DOI] [PubMed] [Google Scholar]
- 2.Desjardins AE (2006) Fusarium mycotoxin: chemistry, genetics and biology. St. Paul, MN, U.S.A.: APS Press.
- 3.Leslie JF, Summerell BA (2006) The Fusarium lab manual. Ames, IA, U.S.A.: Blackwell.
- 4. Starkey DE, Ward TJ, Aoki T, Gale LR, Kistler HC, et al. (2007) Global molecular surveillance reveals novel Fusarium head blight species and trichothecene toxin diversity. Fungal Genet Biol 44: 1191–1204. [DOI] [PubMed] [Google Scholar]
- 5. Sarver BA, Ward TJ, Gale LR, Broz K, Kistler HC, et al. (2011) Novel Fusarium head blight pathogens from Nepal and Louisiana revealed by multilocus genealogical concordance. Fungal Genet Biol 48: 1096–1107. [DOI] [PubMed] [Google Scholar]
- 6. O’Donnell K, Ward TJ, Aberra D, Kistler HC, Aoki T, et al. (2008) Multilocus genotyping and molecular phylogenetics resolve a novel head blight pathogen within the Fusarium graminearum species complex from Ethiopia. Fungal Genet Biol 45: 1514–1522. [DOI] [PubMed] [Google Scholar]
- 7. O’Donnell K, Kistler HC, Tacke BK, Casper HH (2000) Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc Natl Acad Sci U S A 97: 7905–7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. O’Donnell K, Ward TJ, Geiser DM, Corby Kistler H, Aoki T (2004) Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genet Biol 41: 600–623. [DOI] [PubMed] [Google Scholar]
- 9. Yli-Mattila T, Gagkaeva T, Ward TJ, Aoki T, Kistler HC, et al. (2009) A novel Asian clade within the Fusarium graminearum species complex includes a newly discovered cereal head blight pathogen from the Russian Far East. Mycologia 101: 841–852. [DOI] [PubMed] [Google Scholar]
- 10. Trail F, Xu H, Loranger R, Gadoury D (2002) Physiological and environmental aspects of ascospore discharge in Gibberella zeae (anamorph Fusarium graminearum). Mycologia 94: 181–189. [PubMed] [Google Scholar]
- 11.Marasas WFO, Nelson PE, Toussoun TA (1984) Toxigenic Fusarium species: identity and mycotoxicology. University Park: The Pennsylvania State University Press.
- 12. Proctor RH, Hohn TM, McCormick SP (1995) Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol Plant Microbe Interact 8: 593–601. [DOI] [PubMed] [Google Scholar]
- 13. Hohn TM, Krishna R, Proctor RH (1999) Characterization of a transcriptional activator controlling trichothecene toxin biosynthesis. Fungal Genet Biol 26: 224–235. [DOI] [PubMed] [Google Scholar]
- 14. Kim YT, Lee YR, Jin J, Han KH, Kim H, et al. (2005) Two different polyketide synthase genes are required for synthesis of zearalenone in Gibberella zeae . Mol Microbiol 58: 1102–1113. [DOI] [PubMed] [Google Scholar]
- 15. Gardiner DM, Osborne S, Kazan K, Manners JM (2009) Low pH regulates the production of deoxynivalenol by Fusarium graminearum . Microbiology 155: 3149–3156. [DOI] [PubMed] [Google Scholar]
- 16. Tudzynski B, Homann V, Feng B, Marzluf GA (1999) Isolation, characterization and disruption of the areA nitrogen regulatory gene of Gibberella fujikuroi . Mol Gen Genet 261: 106–114. [DOI] [PubMed] [Google Scholar]
- 17. Kim H, Woloshuk CP (2008) Role of AREA, a regulator of nitrogen metabolism, during colonization of maize kernels and fumonisin biosynthesis in Fusarium verticillioides . Fungal Genet Biol 45: 947–953. [DOI] [PubMed] [Google Scholar]
- 18. Flaherty JE, Pirttila AM, Bluhm BH, Woloshuk CP (2003) PAC1, a pH-regulatory gene from Fusarium verticillioides . Appl Environ Microbiol 69: 5222–5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Min K, Shin Y, Son H, Lee J, Kim JC, et al. (2012) Functional analyses of the nitrogen regulatory gene areA in Gibberella zeae . FEMS Microbiol Lett 334: 66–73. [DOI] [PubMed] [Google Scholar]
- 20. Bok JW, Keller NP (2004) LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell 3: 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bayram O, Krappmann S, Ni M, Bok JW, Helmstaedt K, et al. (2008) VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320: 1504–1506. [DOI] [PubMed] [Google Scholar]
- 22. Bayram OS, Bayram O, Valerius O, Park HS, Irniger S, et al. (2010) LaeA control of velvet family regulatory proteins for light-dependent development and fungal cell-type specificity. PLoS Genet 6: e1001226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Amaike S, Keller NP (2009) Distinct roles for VeA and LaeA in development and pathogenesis of Aspergillus flavus . Eukaryot Cell 8: 1051–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kale SP, Milde L, Trapp MK, Frisvad JC, Keller NP, et al. (2008) Requirement of LaeA for secondary metabolism and sclerotial production in Aspergillus flavus . Fungal Genet Biol 45: 1422–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bok JW, Balajee SA, Marr KA, Andes D, Nielsen KF, et al. (2005) LaeA, a regulator of morphogenetic fungal virulence factor. Eukaryot Cell 4: 1574–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Perrin RM, Fedorova ND, Bok JW, Cramer RA, Wortman JR, et al. (2007) Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog 3: e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu D, Oide S, Zhang N, Choi MY, Turgeon BG (2012) ChLae1 and ChVel1 regulate T-toxin production, virulence, oxidative stress response, and development of the maize pathogen Cochliobolus heterostrophus . PLoS Pathog 8: e1002542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiemann P, Brown DW, Kleigrewe K, Bok JW, Keller NP, et al. (2010) FfVel1 and FfLae1, components of a velvet-like complex in Fusarium fujikuroi, affect differentiation, secondary metabolism and virulence. Mol Microbiol. [DOI] [PMC free article] [PubMed]
- 29. Butchko RA, Brown DW, Busman M, Tudzynski B, Wiemann P (2012) Lae1 regulates expression of multiple secondary metabolite gene clusters in Fusarium verticillioides . Fungal Genet Biol 49: 602–612. [DOI] [PubMed] [Google Scholar]
- 30. Lopez-Berges MS, Hera C, Sulyok M, Schafer K, Capilla J, et al. (2013) The velvet complex governs mycotoxin production and virulence of Fusarium oxysporum on plant and mammalian hosts. Mol Microbiol 87: 49–65. [DOI] [PubMed] [Google Scholar]
- 31. Seiboth B, Karimi RA, Phatale PA, Linke R, Hartl L, et al. (2012) The putative protein methyltransferase LAE1 controls cellulase gene expression in Trichoderma reesei . Mol Microbiol 84: 1150–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Karimi-Aghcheh R, Bok JW, Phatale PA, Smith KM, Baker SE, et al. (2013) Functional analyses of Trichoderma reesei LAE1 reveal conserved and contrasting roles of this regulator. G3 (Bethesda) 3: 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jiang J, Liu X, Yin Y, Ma Z (2011) Involvement of a velvet protein FgVeA in the regulation of asexual development, lipid and secondary metabolisms and virulence in Fusarium graminearum . PLoS One 6: e28291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee J, Myong K, Kim JE, Kim HK, Yun SH, et al. (2012) FgVelB globally regulates sexual reproduction, mycotoxin production and pathogenicity in the cereal pathogen Fusarium graminearum . Microbiology 158: 1723–1733. [DOI] [PubMed] [Google Scholar]
- 35. Jiang J, Yun Y, Liu Y, Ma Z (2012) FgVELB is associated with vegetative differentiation, secondary metabolism and virulence in Fusarium graminearum . Fungal Genet Biol 49: 653–662. [DOI] [PubMed] [Google Scholar]
- 36. Kwon BR, Kim MJ, Park JA, Chung HJ, Kim JM, et al. (2009) Assessment of the core cryparin promoter from Cryphonectria parasitica for heterologous expression in filamentous fungi. Appl Microbiol Biotechnol 83: 339–348. [DOI] [PubMed] [Google Scholar]
- 37. Kim JE, Han KH, Jin J, Kim H, Kim JC, et al. (2005) Putative polyketide synthase and laccase genes for biosynthesis of aurofusarin in Gibberella zeae . Appl Environ Microbiol 71: 1701–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McCormick SP, Alexander NJ, Harris LJ (2010) CLM1 of Fusarium graminearum encodes a longiborneol synthase required for culmorin production. Appl Environ Microbiol 76: 136–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kim HK, Cho EJ, Lee S, Lee YS, Yun SH (2012) Functional analyses of individual mating-type transcripts at MAT loci in Fusarium graminearum and Fusarium asiaticum . FEMS Microbiol Lett 337: 89–96. [DOI] [PubMed] [Google Scholar]
- 40. Son H, Seo YS, Min K, Park AR, Lee J, et al. (2011) A phenome-based functional analysis of transcription factors in the cereal head blight fungus, Fusarium graminearum . PLoS Pathog 7: e1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee W (2010) Comprehensive discovery of fungal gene clusters: unexpected co-work reflected at the genomic level. München, Germany: Technische Universität München.
- 42. Lee S, Son H, Lee J, Min K, Choi GJ, et al. (2011) Functional analyses of two acetyl coenzyme A synthetases in the ascomycete Gibberella zeae . Eukaryot Cell 10: 1043–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim HK, Cho EJ, Jo S, Sung BR, Lee S, et al. (2012) A split luciferase complementation assay for studying in vivo protein-protein interactions in filamentous ascomycetes. Curr Genet 58: 179–189. [DOI] [PubMed] [Google Scholar]
- 44. Shwab EK, Bok JW, Tribus M, Galehr J, Graessle S, et al. (2007) Histone deacetylase activity regulates chemical diversity in Aspergillus . Eukaryot Cell 6: 1656–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hoff B, Kamerewerd J, Sigl C, Mitterbauer R, Zadra I, et al. (2010) Two components of a velvet-like complex control hyphal morphogenesis, conidiophore development, and penicillin biosynthesis in Penicillium chrysogenum . Eukaryot Cell 9: 1236–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee SH, Lee S, Choi D, Lee YW, Yun SH (2006) Identification of the down-regulated genes in a mat1–2-deleted strain of Gibberella zeae, using cDNA subtraction and microarray analysis. Fungal Genet Biol 43: 295–310. [DOI] [PubMed] [Google Scholar]
- 47. Lee J, Lee T, Lee YW, Yun SH, Turgeon BG (2003) Shifting fungal reproductive mode by manipulation of mating type genes: obligatory heterothallism of Gibberella zeae . Mol Microbiol 50: 145–152. [DOI] [PubMed] [Google Scholar]
- 48. Capellini RA, Peterson JL (1965) Macroconidium formation in submerged cultures by a nonsporulating strain of Gibberella zeae . Mycologia 57: 962–966. [Google Scholar]
- 49. Gardiner DM, Kazan K, Manners JM (2009) Nutrient profiling reveals potent inducers of trichothecene biosynthesis in Fusarium graminearum . Fungal Genet Biol 46: 604–613. [DOI] [PubMed] [Google Scholar]
- 50. Chi MH, Park SY, Lee YH (2009) A quick and safe method for fungal DNA extraction. Plant Pathology J 25: 108–111. [Google Scholar]
- 51.Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Plainview, U.S.A.: Cold Spring Harbor Laboratory Press.
- 52. Kim HK, Yun SH (2011) Evaluation of potential reference genes for quantitative RT-PCR analysis in Fusarium graminearum under different culture conditions. Plant Pathol J 27: 301–309. [Google Scholar]
- 53. Seo JA, Kim JC, Lee DH, Lee YW (1996) Variation in 8-ketotrichothecenes and zearalenone production by Fusarium graminearum isolates from corn and barley in Korea. Mycopathologia 134: 31–37. [DOI] [PubMed] [Google Scholar]
- 54. Catlett NL, Lee BN, Yoder OC, Turgeon BG (2003) Split-marker recombination for efficient targeted deletion of fungal genes. Fungal Genet Newsl 50: 9–11. [Google Scholar]
- 55. Namiki F, Matsunaga M, Okuda M, Inoue I, Nishi K, et al. (2001) Mutation of an arginine biosynthesis gene causes reduced pathogenicity in Fusarium oxysporum f. sp. melonis . Mol Plant Microbe Interact 14: 580–584. [DOI] [PubMed] [Google Scholar]
- 56. Yu JH, Hamari Z, Han KH, Seo JA, Reyes-Dominguez Y, et al. (2004) Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41: 973–981. [DOI] [PubMed] [Google Scholar]
- 57. Lee J, Son H, Lee YW (2011) Estrogenic compounds compatible with a conditional gene expression system for the phytopathogenic fungus Fusarium graminearum . Plant Pathol J 27: 349–353. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Pigmentation of the ΔFgLaeA strain in the marginal regions between different growing colonies grown on CM agar plate, and (B) when grown in CM liquid medium. Left and right, WT and ΔFgLaeA strain, respectively.
(TIF)
Reconstructed ion chromatograms of rice culture extracts from the fungal strains derived from 9F1. WT (9F1), the wild-type 9F1 strain; ΔFgLaeA, a 9F1 ΔFgLaeA strain derived from 9F1; add-back, a 9F1 FgLaeA-add-back strain derived from the 9F1ΔFgLaeA strain. A, 15ADON; C, culmorin; Z, zearalenone.
(TIF)
Expression of FgLaeA, FgVeA and FgVelB in various F. gramineaum strains. (A) Relative transcript levels of FgLaeA in the FgLaeA-overexpression and add-back strains, and (B) those of FgVeA and FgVelB in the ΔFgLaeA strain derived from Z3643, which were grown in CM liquid medium for vegetative growth, CMC liquid medium for conidiation, AG liquid medium for trichothecene production, and on carrot agar for sexual development. Days of incubation following inoculation in each medium are shown in on the x-axis. cycle, 12 h-dark/12 h-light cycle. The amounts of FgVeA and FgVelB transcripts from a 6-day-old sample in AG liquid medium were used as references in (A) and (B), respectively.
(TIF)
Gene ontology analysis of DEGs in the Δ FgLaeA strain grown in AG liquid medium for 60 h.
(TIF)
Distribution of DEGs in the Δ FgLaeA strain on each chromosome of the F. graminearum PH-1 strain. The genomic locations of histone 3 lysine methylations (H3K7me3 and H3K4me2) were indicated by thin black and red bars, respectively, below the positions of DEGs.
(TIF)
Generation of a firefly luciferase reporter system for trichothecene production. (A) Scheme for the insertion of the FLuc gene under control of a promoter region of Tri6 into the genome of the F. graminearum Z3643 (WT) strain by homologous recombination. (B) SalI-digested genomic DNA gel blot probed with the entire vector. Lane 1, WT strain; lane 2, the FLTRI6 strain. DNA size markers are indicated on the left side of the gel. (C) Luminescence signals in the cell lysate from FLTRI6 grown in AG and complete liquid media, respectively. Days postinoculation (DPI) are indicated below the days of incubation.
(TIF)
Generation of firefly luciferase reporter system for zearalenone production. (A) Scheme for the insertion of the FLuc gene under control of a ZEB2 promoter into the genome of the F. graminearum Z3643 (WT) strain by homologous recombination. (B) SalI-digested genomic DNA gel blot probed with the entire vector. Lane 1, WT strain; lanes 2 and 3, the FLZEB2 strains. DNA size markers are indicated on the left side of the gel. (C) Luminescence signals in the cell lysate from a FLZEB2 strain grown in SG and complete liquid media, respectively. Days postinoculation (DPI) are indicated below the days of incubation. Asterisks above the bars indicate a significant difference between two culture conditions for each DPI according to Tukey’s test.
(TIF)
Quantification of 15ADON and ZEA levels in fungal rice culture extracts.
(XLSX)
Genes down-regulated in the Δ FgLaeA strain grown in agmatine-amended liquid medium for 60 h.
(XLSX)
Genes up-regulated in the Δ FgLaeA strain grown in agmatine-amended liquid medium.
(XLSX)
DEGs belonging to tentative functional gene clusters from the Δ FgLaeA strain.
(XLSX)
Grouping of differentially expressed genes in the Δ FgLaeA strain by functional domains.
(XLSX)
Primers used in this study.
(DOCX)