Abstract
HMGNs are nucleosome-binding proteins that alter the pattern of histone modifications and modulate the binding of linker histones to chromatin. The HMGN3 family member exists as two splice forms, HMGN3a which is full-length and HMGN3b which lacks the C-terminal RD (regulatory domain). In the present study, we have used the Glyt1 (glycine transporter 1) gene as a model system to investigate where HMGN proteins are bound across the locus in vivo, and to study how the two HMGN3 splice variants affect histone modifications and gene expression. We demonstrate that HMGN1, HMGN2, HMGN3a and HMGN3b are bound across the Glyt1 gene locus and surrounding regions, and are not enriched more highly at the promoter or putative enhancer. We conclude that the peaks of H3K4me3 (trimethylated Lys4 of histone H3) and H3K9ac (acetylated Lys9 of histone H3) at the active Glyt1a promoter do not play a major role in recruiting HMGN proteins. HMGN3a/b binding leads to increased H3K14 (Lys14 of histone H3) acetylation and stimulates Glyt1a expression, but does not alter the levels of H3K4me3 or H3K9ac enrichment. Acetylation assays show that HMGN3a stimulates the ability of PCAF [p300/CREB (cAMP-response-element-binding protein)-binding protein-associated factor] to acetylate nucleosomal H3 in vitro, whereas HMGN3b does not. We propose a model where HMGN3a/b-stimulated H3K14 acetylation across the bodies of large genes such as Glyt1 can lead to more efficient transcription elongation and increased mRNA production.
Keywords: acetylation, chromatin, elongation, epigenetics, HMGN, p300/CREB (cAMP-response-element-binding protein)-binding protein-associated factor (PCAF)
INTRODUCTION
HMGN (high-mobility group nucleosome-binding) family members are nuclear proteins that bind to nucleosomes and alter chromatin structure and function (reviewed in [1]). They modulate transcription and thus regulate gene expression patterns in vivo [2,3], as well as being important for DNA repair [4] and replication [5]. Studies using knockout mice and cultured cells have revealed roles for HMGNs in early embryogenesis, in differentiation and in the response to various stresses (reviewed in [6]). There are four canonical members of the family, HMGN1–4, and they share a highly conserved NBD (nucleosome-binding domain), a bipartite NLS (nuclear localization sequence/signal) 1 and 2, and an RD (regulatory domain) [7]. A related protein named HMGN5 also contains the conserved NBD, but has a large C-terminal acidic RD [8]. In the present study, we focus on the role of HMGN3, which is the only HMGN family member to exist as two splice variants, HMGN3a and HMGN3b [3,9]. The shorter HMGN3b variant lacks the C-terminal RD, but it has not been shown whether the two variants play distinct roles in vivo.
Many studies have investigated the mechanisms used by HMGNs to influence transcription. Experiments using chromatin templates assembled in vitro revealed a role for HMGNs in unfolding chromatin and modulating transcription [1,10–12]. Both in vitro and in vivo studies have demonstrated that HMGNs can alter chromatin structure in a variety of ways, including counteracting linker histone H1 [13], inhibiting chromatin remodelling complexes [14] and altering the level of histone modifications. In particular, HMGN2, and to a lesser extent HMGN1, stimulate acetylation of nucleosomal H3K14 (Lys14 of histone H3) by PCAF [p300/CREB (cAMP-response-element-binding protein)-binding protein-associated factor] in vitro [15,16]. HMGNs can also modulate the MSK1 (mitogenand stress-activated kinase 1)- and RSK2 (ribosomal S6 kinase 2)-mediated phosphorylation of H2AS1 (Ser1 of histone H2A), H3S10 (Ser10 of histone H3) and H3S28 (Ser28 of histone H3) in nucleosomal substrates [16–19]. Analyses of domain-swap and deletion mutations have revealed that the RD of HMGN2 is responsible for stimulating H3K14 acetylation by PCAF, whereas the NLS2 region of HMGN1 is responsible for inhibiting H3S10 phosphorylation by MSK1 [16].
There are several reports of functional and/or physical interactions between HMGNs and transcription factors, including TR/RXR (retinoid X receptor) [20], PITX2 (pituitary homeobox 2)–β-catenin [21], ERα (oestrogen receptor α) [22], SRF (serum-response factor) [22] and PDX-1 (pancreatic and duodenal homeobox-1) [23]. In most of these cases, the transcription factor is responsive to extracellular signals [e.g. thyroid hormone (TR/RXR), oestrogen (ERα and SRF) and Wnt signalling (PITX2–β-catenin)]. The extracellular signal thus acts via the transcription factor to regulate HMGN binding [21,22]. Furthermore, in the examples of TR/RXR, PITX2–β-catenin and PDX-1, the HMGN protein also seems to promote the DNA binding of the transcription factor [20,21,23]. Thus HMGNs could influence transcription by modulating the DNA binding of specific transcription factors, and in some cases this appears to be independent of the effects of HMGNs on chromatin structure [20,21].
The ability of HMGNs to affect transcription will also depend on where HMGNs are bound with respect to individual genes. Few studies have combined functional analysis of how HMGNs regulate a gene with detailed analysis of where HMGNs bind to the gene in question. In the examples mentioned above, HMGN binding at a gene promoter is stimulated by certain transcription factors, and results in either activation or repression of the gene. In the case of Sox9, however, HMGN1 is enriched at other regions across the locus rather than the promoter, and is thought to repress gene expression in mouse limb bud cells [24]. At the Hsp70 (heat-shock protein 70) gene, HMGN1 is bound evenly across the entire gene locus but promotes histone acetylation at the nucleosome near the promoter, and stimulates heat-shock-induced transcription at early time points [25]. A recent genome-wide study by Zhao and co-workers found that HMGN1 is preferentially bound to the promoters of active genes and at DHSs (DNase I-hypersensitive sites) [26]. However, there is not yet any functional data to show whether all of these genes are actually regulated by HMGN1.
In the present study, we focused on a known HMGN3 target gene, Glyt1 (glycine transporter 1, also known as Slc6a9)[3] to investigate where the two HMGN3 splice variants bind to this locus and how they modulate gene expression. Glyt1 plays an essential role at glycinergic and glutamatergic synapses in the brain and CNS (central nervous system) (reviewed in [27]), but is also expressed in several other tissues including the liver, lung and pancreas [28]. We have previously shown that Glyt1 is up-regulated by overexpression of HMGN3 in murine Hepa-1 cells [3], and we wanted to investigate which regions of the gene are preferentially bound by HMGN3a/b. We were particularly interested in whether modifications of the histone H3 N-terminal tail might play a role in HMGN3 targeting, as there is evidence that HMGNs interact with the N-terminal tail of histone H3, and that the presence of histone tails stabilizes the interaction of HMGNs with nucleosome core particles [29,30]. We also aimed to investigate whether the binding of either full-length HMGN3a or the shorter HMGN3b alters the pattern of histone modifications at Glyt1 and how this might be related to the activation of gene expression.
EXPERIMENTAL
Plasmid construction
Insulator sequences from plasmid pBAW3 [a gift from G. Felsenfeld, NIH (National Institutes of Health)] were inserted on either side of the expression cassette of plasmid pTRE (Clontech) to minimize chromatin position effects after random integration into the genome. Tandem copies of the core insulator from HS4 of the chicken β-globin locus were digested with BglII/SpeI, blunt-ended and either had XhoI linkers ligated for insertion into the XhoI site or were ligated into the blunted SapI site of pTRE to create pTRE-INS2. Both insulator insertions are in the same direction. The open reading frame of mouse HMGN3a or HMGN3b was amplified by PCR and inserted into the MluI and SalI sites of pTRE-INS2 to create pTRE-INS2-N3a and pTREINS2-N3b.
Generation and culture of HMGN-expressing cell lines
The `control' cell line (clone 2–9) was derived from the mouse hepatoma line Hepa-1. Cells were transfected with pTet-ON and pTet-tTS (Clontech) and selected in DMEM (Dulbecco's modified Eagle's medium) with 10% FBS (fetal bovine serum) and 400 μg/ml G418. H-N3b (clone 293-73) and H-N3a (clone 291-10) cell lines were derived from clone 2–9 following transfection with pTK-hyg and either pTRE-INS-N3b or pTRE-INS2-N3a respectively, and selection with 400 μg/ml hygromycin. For HDAC (histone deacetylase) inhibitor treatments, control cells were incubated with 2 mM or 5 mM sodium butyrate, or 165 nM trichostatin A for 3 h. These concentration ranges have been used previously to inhibit deacetylases and induce gene expression in Hepa-1 and human hepatoma cell lines [31–33].
RT (reverse transcription)–PCR
RNA was extracted using RNeasy kits (Qiagen), and reverse transcribed using Superscript III reverse transcriptase (Invitrogen) and oligo(dT)16 according to the manufacturer's instructions. An aliquot of the cDNA was used in real-time PCR using SYBR Green (Roche) using a Stratagene Mx3000P Q-PCR machine according to the manufacturer's instructions. The PCR primer sequences are listed in the Supplementary Tables S1 and S2 (available at http://www.BiochemJ.org/bj/442/bj4420495add.htm). Primer sequences for Gcn5 (general control of amino acid synthesis 5), Pcaf and p300 are courtesy of OriGene. For each sample, the mean Ct from three replicates was taken. Expression levels were normalized to Gapdh (glyceraldehyde-3-phosphate dehydrogenase) and the control sample using the comparative ΔΔCT method.
ChIP (chromatin immunoprecipitation) and Western blotting
ChIP was carried out as described by Duan et al. [34] with minor modifications. This protocol results in significantly higher immunoprecipitation efficiency and reproducibility for HMGN3 ChIPs, compared with that described previously [3]. Cells were removed from culture plates by scraping into growth medium. Protein-DNA cross-linking was performed by incubating cell suspensions with formaldehyde at a final concentration of 0.5% for 5 min at room temperature (23 °C) with gentle agitation. Glycine was added to 0.125 M and incubated for 5 min to quench the reaction. Chromatin was sonicated to an average size of 500 bp. Aliquots were pre-cleared with normal IgG for 1 h followed by incubation with Protein G–agarose (Millipore) overnight. Pre-cleared chromatin was then incubated with 4–10 μg of specific antibody for 2 h at 4 °C. Normal IgG (10 μg) control immunoprecipitations were always performed in parallel to experimental IPs for each batch of chromatin. Immune complexes were collected by incubation with 30 μl of Protein G–agarose for 2 h at 4 °C. Complexes were washed and eluted, cross-links were reversed, and DNA was purified by Qiagen MinElute columns. For Western blotting, cross-links in chromatin samples were reversed by heating to 95 °C for 20 min in SDS/PAGE loading buffer. Perchloric acid extracts for HMGN3 Western blots were prepared as described previously [35] and concentrations assayedbymeasuringthe A280 using an ND1000 Nanodrop spectrophotometer. Samples were run on SDS/15% PAGE gels and blotted on to PVDF, and membranes were probed with the antibodies listed above. Blots were imaged using a Fujifilm LAS3000 imager and quantified using Aida Image Data Analyzer software.
Antibodies
The N3-Cter antibody was raised in rabbit against the peptide VEEAQRTESIEKEGE and affinity purified (Eurogentec). It is specific to HMGN3, and does not recognize HMGN1 or HMGN2 (Figure 1d). Antibodies against HMGN1 and HMGN2 were raised against a 15-amino-acid peptide from the C-terminus of each protein and affinity purified (Eurogentec). They are specific to their respective HMGN proteins (results not shown). Antibodies 2752 and 2751 raised against HMGN3 internal peptides have been described previously [9]. Anti-H3K14ac (acetylated Lys14 of histone H3) (07–353), anti-H3K9ac (acetylated Lys9 of histone H3) (07–352) were from Upstate Biotechnology. Anti-H3K4me3 (trimethylated Lys4 of histone H3) was from Abcam (ab8580, Figure 3) or from Millipore (07–473, Figure 5), and normal IgG (I5006) was from Sigma.
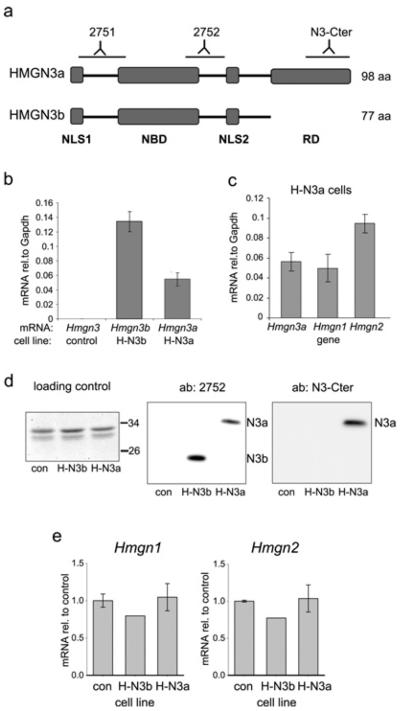
Figure 1. Overexpression of Hmgn3b and Hmgn3a in Hepa-1 cells.
(a) Schematic diagram of HMGN3a and its shorter splice form, HMGN3b. The two NLS, the NBD and the RD are indicated. The locations of peptides used to raise antibodies 2751, 2752 and N3-Cter are indicated. (b) Hmgn3a and Hmgn3b were overexpressed in H-N3b and H-N3a cells respectively, and were barely detectable in control cells. Hmgn3 expression was quantified by real-time quantitative PCR and is plotted as a percentage of Gapdh expression. (c) Hmgn3a mRNA was expressed at levels comparable with those of Hmgn1 and Hmgn2 in H-N3a cells. (d) HMGN3b and HMGN3a proteins were produced at similar levels in H-N3b and H-N3a cells respectively. Acid-soluble proteins from the three cell lines were run on three parallel SDS/PAGE gels. Left-hand panel: a strip from the Coomassie Blue-stained protein gel showing the relative protein levels in the extracts. The bands shown are the most prominent on the gel, and correspond to the linker histone variants. The positions of the 26 and 34 kDa size markers are indicated. Middle and right-hand panels: Western blots were probed with antibodies 2752 and N3-Cter. No other bands were visible on the blots. Quantification of the bands in the 2752 blot revealed relative intensities of 1.73:1 for HMGN3b/HMGN3a. (e) Hmgn1 (left-hand histogram) and Hmgn2 (right-hand histogram) mRNA levels are unchanged in H-N3b and H-N3a cell lines compared with the control. Hmgn mRNA was normalized to Gapdh and is plotted relative to expression in the control cells (con).
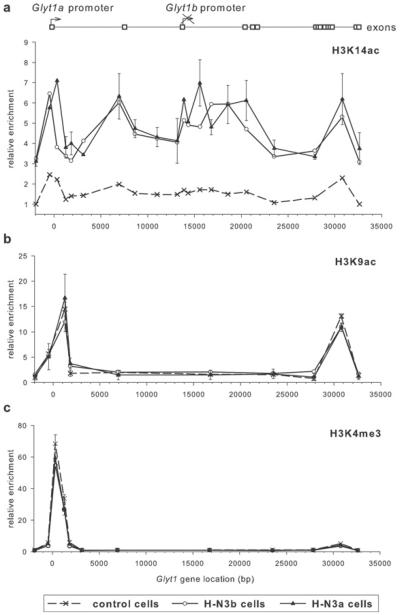
Figure 3. H3K14 acetylation across the Glyt1 locus is increased in cells expressing Hmgn3a or Hmgn3b.
ChIP profiles for (a) H3K14ac, (b) H3K9ac and (c) H3K4me3 across the Glyt1 locus in control, H-N3b and H-N3a cells (means±S.E.M., n =2 ChIP replicates).
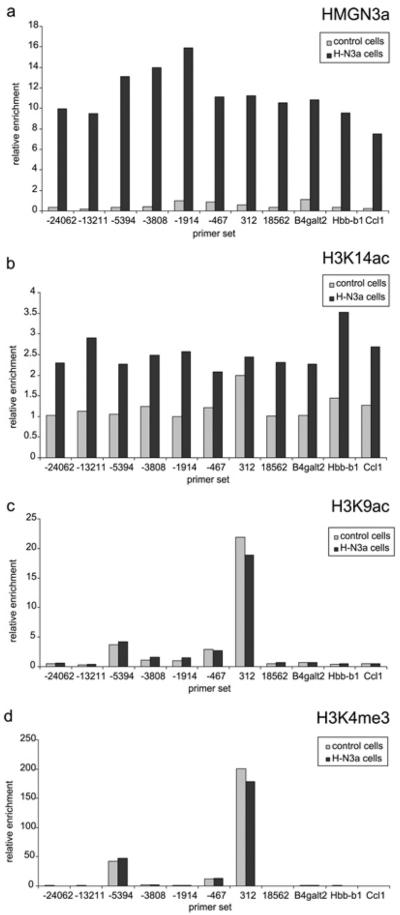
Figure 5. HMGN3a binding is not limited to the Glyt1 gene body.
Relative enrichment for (a) HMGN3a (b) H3K14ac (c) H3K9ac and (d) H3K4me3 at various sites outwith the Glyt1 gene body in control cells and H-N3a cells. Locations of numbered primer sets near the Glyt1 locus are shown in Figure 4. B4galt2 is a gene downstream of Glyt1 as shown in Figure 4. Hbb-b1 is the β-globin adult major gene and Ccl1 is a chemokine gene, both of which are repressed in Hepa-1 cells. HMGN3 represses the tumour necrosis factor α (TNFα)-induced expression of Ccl1 in mouse embryonic fibroblasts (K.L. West, unpublished work). ChIP enrichments were normalized to H3 enrichment for each primer set.
Quantitative PCR
Real-time quantitative PCR was performed on the immuno-precipitated samples using the Stratagene Mx3000P machine and SYBR Green master mix (Roche). Primer pairs were designed across non-repetitive regions of the Glyt1 gene, and checked for specificity and amplification efficiency. Primer sequences are listed in the Supplementary Online Data. PCRs contained either 0.19% of each immunoprecipitate or 0.5 ng of input DNA, and were carried out in duplicate. Enrichments were calculated as a ratio of the PCR readings in immunoprecipitate and input DNAs (ΔCT). Log2 ΔCT values were converted into fold enrichments, taking into account the PCR efficiency of each primer pair. In Figures 2 and 3, fold enrichments for each experimental immunoprecipitation were normalized to the mean of the control IgG enrichments (mean calculated from all Glyt1 primer pairs), to control for variation in immunoprecipitation efficiency between chromatin preparations. In Figures 5 and 6, fold enrichments were normalized to those for H3 at each primer set. For each histogram shown (i.e. for each antibody), the data are scaled so that the enrichment in control cells at primer set −1914 is set to 1.
Figure 2. HMGN3a and HMGN3b are bound across the Glyt1 locus.
(a) ChIP profiles comparing HMGN3a binding across the Glyt1 locus in H-N3a cells and control cells. ChIPs were carried out using the N3-Cter antibody (means±S.E.M., n =2 ChIP replicates). Numbering starts at the first transcribed base pair. The relative positions of the TSS and 14 exons for Glyt1a (active) are shown in the schematic diagram above the graph. The first exon for Glyt1b, which is not expressed in Hepa cells and is not included in the Glyt1a transcript, is also shown. (b) ChIP profiles using internal HMGN3 antibodies (2751 and 2752 combined) in H-N3a, H-N3b and control cells. Points in the control cell profile represent the mean and S.E.M. from three ChIP replicates.
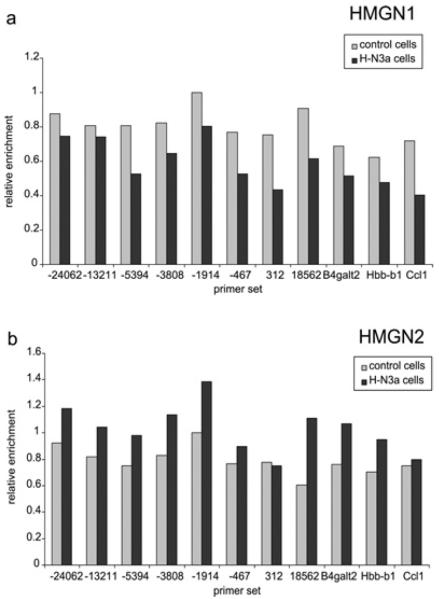
Figure 6. HMGN1 and HMGN2 are enriched to similar levels at all genomic locations tested.
Relative enrichment for HMGN1 (a) and HMGN2 (b) in control cells and H-N3a cells at the same genomic locations tested in Figure 5. Enrichments were normalized to H3 for each primer set.
Gel-retardation and in vitro acetylation assays
Gel-retardation assays were performed with nucleosome core particles and recombinant HMGN3 protein as described previously [9]. In vitro acetylation assays were performed in HAT buffer [50 mM Tris/HCl, pH 8.0, 10% glycerol (v/v), 1 mM DTT (dithiothreitol), 0.1 mM EDTA and 5 mM butyric acid]. Each 10 μl reaction contained: 0.1 mg/ml nucleosome cores, 0.2 mM [1-14C]acetyl-CoA (20 nCi), 100 ng of recombinant PCAF (Upstate Cell Signalling Solutions, 14–309), and various amounts of HMGN (added at specific molar ratio to core particles varied from 0.2 to 3.2). The assay was performed at 37 °C for 30 min. The reactions were stopped by the addition of an equal volume of an SDS-gel sample buffer [100 mM Tris/HCl (pH 6.8), 200 mM DTT, 2% SDS, 0.1% Bromophenol Blue and 20% glycerol], denatured for 5 min at 95 °C, and the proteins were resolved by SDS/PAGE (15% gel). The gels were stained with Coomassie Blue for estimation of protein quantities, soaked in enlightening enhancer solution (Dupont) for 30 min and vacuum dried. The radioactivity incorporated into the protein bands was visualized by a PhosphorImager (Molecular Dynamics) and quantified with ImageQuant software.
RESULTS
The relationship between HMGN3 binding and histone modifications across the Glyt1 gene
In order to study the relationship between HMGN3 and histone modifications at the Glyt1 gene, we generated Hepa-1 cell lines in which either HMGN3a, or its C-terminally truncated splice variant, HMGN3b (Figure 1a), is ectopically expressed. The new cell lines are named H-N3a and H-N3b respectively. Real-time RT-PCR and Western blotting showed that the control (parent) cell line had undetectable levels of HMGN3 (Figures 1b and 1d). Western blotting using the antibody 2752, which recognizes an internal peptide common to both HMGN3 splice forms (Figure 1a) [9], revealed similar levels of HMGN3b and HMGN3a proteins in the H-N3b and H-N3a cell lines respectively (Figure 1d). The N3-Cter antibody was raised against a peptide in the C-terminal RD of HMGN3a, and so did not detect HMGN3b protein (Figure 1d). Real-time RT–PCR indicated that the Hmgn3b and Hmgn3a transgenes are expressed in H-N3b and H-N3a cells respectively, at levels comparable with those of other Hmgn family members (Figures 1b and 1c). The levels of endogenous Hmgn1 and Hmgn2 mRNA were altered by less than 20% by the ectopic expression of Hmgn3a or Hmgn3b (Figure 1e).
We used ChIP assays to investigate the profile of HMGN3 binding across the Glyt1 locus. The locus is approximately 35 kb long, and Glyt1 expression can be driven from the Glyt1a or Glyt1b promoters, resulting in two splice forms with different 5′ exons [36]. In Hepa-1 cells, the Glyt1a promoter is active, but the promoter for the alternative splice form, Glyt1b, is inactive (Figure 2a). ChIP assays using the N3-Cter antibody revealed HMGN3a binding across the entire Glyt1 gene body in H-N3a cells (Figure 2a). The average relative enrichment for HMGN3a across the locus in H-N3a cells is 17-fold, compared with an average enrichment of 0.5-fold in the control cell line (Figure 2a), and 0.56-fold for non-immune IgG (results not shown). ChIPs with antibodies against internal HMGN3 peptides (2751 and 2752) generated a similar binding profile (Figure 2b), although the average enrichment across the locus was lower, at 4.7-fold. This may reflect the reduced accessibility of the internal epitopes compared with the C-terminal epitope [37].
The binding profile of HMGN3b in H-N3b cells was assayed using antibodies 2751 and 2752. The data show that HMGN3b is bound across the Glyt1 gene body with an average enrichment of 5.5-fold, which is comparable with the enrichment of HMGN3a in H-N3a cells (Figure 2b). The precise pattern of peaks and troughs varied between the HMGN3a and HMGN3b profiles. Both profiles had small peaks at the TSS (transcription start site) and the inactive Glyt1b start site, but peaks approximately 7 and 21 kb into the gene were more variable. Despite these differences, it is clear that ectopic HMGN3a/b expression in H-N3a/b cells leads to HMGN3a/b binding across the Glyt1 gene body, and there is no major peak at the promoter or TSS.
In order to investigate whether histone modifications may be involved in recruiting HMGN3, or whether HMGN3 affects the levels of certain modifications, we profiled H3K14ac, H3K9ac and H3K4me3 across the Glyt1 locus in the different cell lines. The level of H3K14ac across Glyt1 was significantly increased in cells overexpressing HMGN3a or HMGN3b (Figure 3a). In control cells, small peaks of H3K14ac were found at the TSS and the 3′ end of the gene, with an average enrichment of 1.6-fold across all points. In H-N3a and H-N3b cells, H3K14 acetylation was increased across the gene, resulting in average enrichments of 4.9- and 4.5-fold respectively. The peaks of H3K14 acetylation at the 5′ and 3′ ends of the gene were increased, and additional peaks within the gene body are also apparent. Interestingly, the shape of the H3K14ac binding profile in H-N3a cells is similar to that of HMGN3a (C-ter antibody), which is reflected in a moderate correlation coefficient of 0.66 between the two data sets. For comparison, the correlation between the IgG profile (results not shown) and H3K14ac in these cells is negligible, at −0.18. These data suggest that binding of HMGN3a or HMGN3b to chromatin in vivo leads to increased H3K14 acetylation either on the same nucleosome or adjacent nucleosomes.
H3K9ac and H3K4me3 were both highly enriched in tight peaks at the TSS, and H3K9ac was also present in a clear peak near the 3′ end of the gene (Figures 3b and 3c). These profiles are distinct from that of HMGN3, suggesting that H3K9ac and H4K4me3 do not have a significant role in HMGN3 recruitment. Furthermore, the peaks do not change in cells overexpressing HMGN3a or HMGN3b, indicating that HMGN3 binding does not alter the levels of H3K9ac or H3K4me3.
HMGN protein binding at multiple genomic locations
The recent study by Cuddapah et al. [26] revealed HMGN1 peaks at the enhancers and/or promoters of many active genes. Enhancers have been defined as DHSs enriched in H3K4me1 (monomethylated Lys4 of histone H3) and p300 [38–40]. In order to identify any putative enhancers of the Glyt1 gene, we used the UCSC genome browser [41] to examine the genomic context of the Glyt1 gene and to compare it with publicly available ChIP-seq data from the ENCODE project [42,43]. As can be seen in Figure 4, there are several genes immediately downstream of Glyt1, whereas the upstream region is intergenic. Notably, at just over 5 kb upstream of the Glyt1 TSS there is a putative enhancer region that is DNase I hypersensitive and enriched for H3K4me1, p300 and CTCF (CCCTC-binding factor) in most of the tissues studied.
Figure 4. ENCODE data for the Glyt1 locus and surrounding regions.
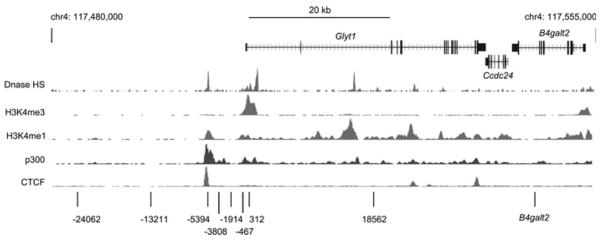
Screenshot from the UCSC genome browser [41] of data generated by the Ren laboratory (Ludwig Institute for Cancer Research, USCD) as part of the ENCODE consortium (genome build NCBI37/mm9) (http://genome.ucsc.edu/) [42,43]. The locations of Glyt1 and the two downstream genes, Ccdc24 and B4galt2 are indicated. ChIP-seq data from mouse cerebellum for DNase I hypersensitivity and for H3K4me1, H3K4me3 and CTCF are shown below, as is data for p300 in mouse heart tissue. The locations of the additional primer sets used in Figure 5 are indicated at the bottom. Primer set −5394 is located at a putative enhancer, as indicated by the peaks of H3K4me1 and p300 and the DHS [38–40].
We used ChIP to investigate whether HMGN3a is bound at the putative Glyt1 enhancer and at other regions outwith the Glyt1 gene. As shown in Figure 5(a), HMGN3a binding in H-N3a cells was comparable at all genomic locations studied: the upstream intergenic region, the putative enhancer, the Glyt1 promoter, the Glyt1 gene body, the downstream B4galt2 gene and two unrelated genes Hbb-b1 and CCl1. This reveals that HMGN3a is not specifically targeted to the Glyt1 gene, and is not enriched at its promoter or putative enhancer.
H3K14ac was enriched at the Glyt1 TSS in control cells, but not at the putative enhancer. Expression of HMGN3a led to an increase in H3K14ac levels at all locations tested, showing that the effect of HMGN3 on H3K14 acetylation is not limited to the Glyt1 gene body. Peaks of H3K9ac and H3K4me3 were observed at the putative enhancer (primer set –5394) as well as at the TSS (primer set 312) (Figures 5b and 5c). The heights of these peaks were the same in the control cells as in the H-N3a cells, providing additional evidence that HMGN3a does not alter the levels of these modifications.
HMGN1 and HMGN2 are the most predominant HMGN isoforms in the parent Hepa-1 cell line, and their mRNA levels are not altered by the ectopic expression of HMGN3 (Figure 1). ChIP was used to determine whether these isoforms are found in peaks at the Glyt1 enhancer or promoter, and to investigate whether their binding profiles are altered by HMGN3 expression. Figures 6(a) and 6(b) show that HMGN1 and HMGN2 binding is comparable at all genomic locations tested, with no peaks at the enhancer, promoter or TSS of Glyt1. The enrichment of HMGN1 decreased by an average of 25% (± 10%) in the H-N3a cells compared with the control cells. Conversely, HMGN2 enrichment increased by an average of 30% (± 20%) in H-N3a cells. These data show that all three HMGN isoforms can bind at the Glyt1 locus, but that none of them are specifically enriched at the regulatory elements.
HMGN3 and histone acetylation
To investigate how HMGN3 expression and histone acetylation correlates with Glyt1a transcription, we quantified the levels of Glyt1a mRNA in the different cell lines (Figure 7a). Glyt1a expression is increased by 2.3- and 2.6-fold in H-N3b and H-N3a cells respectively, suggesting that either the HMGN3 binding and/or the increased H3K14ac levels have contributed to increased transcription from the Glyt1 locus. To investigate whether histone acetylation alone can affect Glyt1a expression, control cells were treated with the HDAC inhibitors sodium butyrate or trichostatin A for 3 h (Figure 7b). These treatments induced Glyt1a expression by 2.3- and 1.9-fold respectively, indicating that histone acetylation can stimulate Glyt1a expression in the absence of HMGN3.
Figure 7. Glyt1a expression is increased in cells expressing Hmgn3a or Hmgn3b, and in cells treated with HDAC inhibitors.
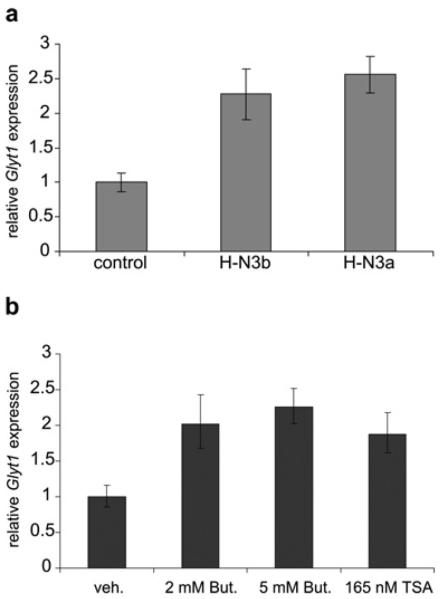
(a) Expression of Glyt1a mRNA in control, H-N3b and H-N3a cells. Expression was normalized to Gapdh and is plotted relative to that in control cells (means±S.E.M., n =2–4 samples). Data using primers specific to the Glyt1a isoform is presented. Primers specific to exon 8 (further downstream) gave similar results (results not shown). Glyt1b expression was barely detectable above background and is not shown. (b) Expression of Glyt1a mRNA in control cells treated with either sodium butyrate (2 mM or 5 mM in water) or trichostatin A (165 nM in ethanol) for 3 h. Expression was normalized to Gapdh and is plotted relative to expression in cells treated with vehicle alone (water or ethanol).
Western blotting was performed to determine whether the increased H3K14 acetylation detected at the Glyt1 locus in H-N3b and H-N3a cells (Figure 2) was also observed in bulk chromatin. The results show that global levels of H3K14ac are actually decreased in H-N3b and H-N3a cells relative to the control cells (Figures 8a and 8b). H3K9ac levels were not significantly altered, in agreement with the ChIP data. Furthermore, levels of the HATs (histone acetyltransferases) (Gcn5, p300 and Pcaf) are unchanged or reduced in H-N3b and H-N3a cells compared with control cells (Figure 8c), showing that HMGN3 does not increase the expression of these chromatin modifiers. These results indicate that the increase in H3K14 acetylation over the Glyt1 locus is unlikely to be due to indirect effects of HMGN3 on HAT or HAT co-factor expression.
Figure 8. Global acetylation levels are not altered in cells expressing Hmgn3a or Hmgn3b.
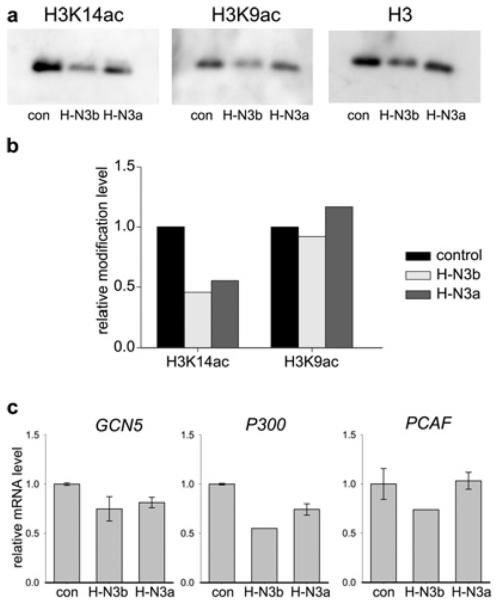
(a) Western blots of chromatin from control, H-N3b and H-N3a cells, probed with antibodies against H3K14ac, H3K9ac or histone H3. (b) Band intensities from the blots in (a) were normalized to those of H3, and modification levels are plotted relative to those in control cells. (c) Expression of the HATs Gcn5, p300 and Pcaf in the different cell lines was quantified by real-time quantitative PCR. Expression was normalized to Gapdh and is plotted relative to that in control cells.
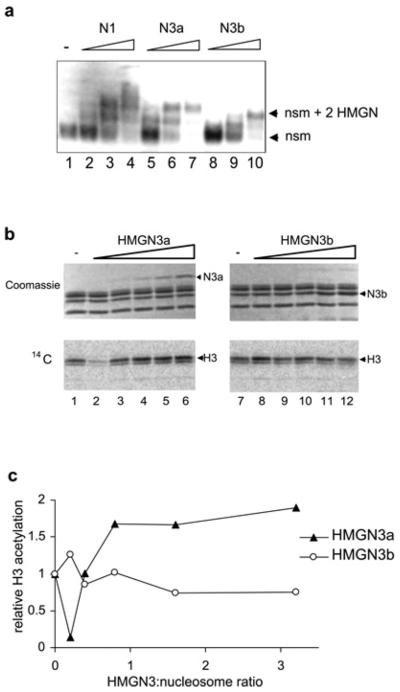
One of the main HATs that acetylates H3K14 is PCAF, and previous reports have shown that other HMGN family members can stimulate histone H3 acetylation by PCAF in vitro [15,16]. To investigate whether HMGN3a and HMGN3b can also stimulate histone acetylation in vitro, HMGN3a and HMGN3b were first titrated against nucleosome core particles and the binding efficiency monitored by gel-retardation assays (Figure 9a). The results indicated that both HMGN3a and HMGN3b bind to nucleosomes with efficiency similar to HMGN1, as has been previously reported [9]. Pre-bound HMGN3–nucleosome complexes were then incubated with purified PCAF and radiolabelled acetyl-CoA. It is known that the main residue acetylated by PCAF is H3K14 [44]. After SDS/PAGE, 14C incorporation into histones was quantified as a measure of acetylation activity (Figures 9b and 9c). These data show that HMGN3a can stimulate acetylation of nucleosomal H3 by up to 2-fold, whereas HMGN3b, which lacks the RD, does not have any effect in this assay. This result is in agreement with previous findings that the RD of HMGNs can enhance PCAF-mediated acetylation of H3K14 [16].
Figure 9. HMGN3a stimulates the ability of PCAF to acetylate nucleosomal histone H3 in vitro.
(a) Gel-retardation assays showing the extent of HMGN3 binding to nucleosome core particles. Nucleosome core particles were incubated with increasing amounts of purified recombinant HMGN1 (lanes 2–4), HMGN3a (lanes 5–7) or HMGN3b (lanes 8–10) in 2×TBE prior to electrophoresis on a 5% polyacrylamide gel. The molar ratios of HMGN to nucleosome core particle were 0 (lane 1), 0.8 (lanes 2, 5 and 8), 1.6 (lanes 3, 6 and 9) and 3.2 (lanes 4, 7 and 10). The expected stoichiometry of HMGN/nucleosome binding is 2:1. (b) In vitro acetylation of nucleosomal H3 by PCAF. Nucleosome core particles were incubated with various amounts of HMGN protein, radiolabelled acetyl-CoA and PCAF. After electrophoresis, the gels were stained with Coomassie Blue for the estimation of protein quantities (upper panels) and exposed to a phosphorimaging plate for quantification of 14C incorporation. Arrowheads indicate the positions of HMGN3a and HMGN3b. HMGN3b co-migrated with H2A/H2B. (c) Quantification of the H3 acetylation assayed in (b).
DISCUSSION
In the present study, we used the Glyt1 locus as a model system to study the relationship between HMGN proteins, active histone modifications and gene expression. We confirmed that Glyt1 is atarget of HMGN3, as Glyt1a expression is increased when HMGN3 is ectopically expressed. However, HMGN3 is not enriched in specific peaks at the gene promoter, TSS or putative enhancer. Instead, it is bound throughout the gene locus, and is found at similar levels at other unrelated genomic locations. HMGN1 and HMGN2 were also bound at fairly constant levels at all the genomic regions tested. We found that ectopic expression of HMGN3a or HMGN3 leads to increased H3K14ac levels at all locations. In contrast, levels of H3K9ac and H3K4me3 enrichment at the Glyt1a promoter and putative enhancer are unaffected. In vitro experiments showed that HMGN3a, but not HMGN3b, is able to stimulate the ability of PCAF, one of the main H3K14 acetyltransferases, to acetylate nucleosomal H3. The results indicate that HMGN3a binding leads to a direct increase in H3K14 acetylation, and a concomitant increase in Glyt1a transcription. A role for histone acetylation in Glyt1a regulation is supported by the demonstration that treatment with HDAC inhibitors also leads to an increase in Glyt1a expression in vivo.
HMGN binding across the Glyt1 locus
A recent study profiled genome-wide HMGN1 binding in human CD4+ T-cells [26], and found that HMGN1 is preferentially bound at the promoters of active genes. In addition, many of the HMGN1 peaks overlapped with DHSs and were enriched in active chromatin marks such as H3K4me3 and H2A.Z [26]. In contrast with these genome-wide data, we found that HMGN1, HMGN2 and HMGN3 are bound fairly evenly across the 35 kb Glyt1 locus, the putative enhancer, upstream intergenic regions and at other unrelated genes.
We observed no correlation between HMGN binding and the H3K4me3 and H3K9ac modifications that mark the promoter and putative enhancer. These results indicate that HMGN proteins are not specifically recruited by the histone modifications usually found at active gene promoters and enhancers. This is consistent with previous studies on individual genes, which have observed a variety of HMGN-binding profiles. In the case of Glut2 (glucose transporter 2), HMGN3 is enriched in a 3 kb region at the promoter, and this enrichment requires the transcription factor Pdx1 [23]. Knockdown of HMGN3 reduces the binding of several key transcription factors to the Glut2 promoter, reduces H3K14 acetylation levels, and reduces Glut2 expression by approximately 80% [23]. Peaks of HMGN1 binding are also found at three regions of Sox9, although not including the promoter, and HMGN1 is important for repressing Sox9 expression in mouse limb bud cells [24]. In contrast, an even distribution of HMGN1 binding was observed across the Hsp70 locus, and HMGN1 was shown to be important for the heat shock-induced nucleosome remodelling and acetylation at the Hsp70 promoter [25]. Our data indicate that the transcription factors and/or chromatin structural elements required for enhanced HMGN recruitment are absent from the Glyt1 locus in Hepa-1-derived cells. It will be interesting to investigate whether the HMGN peaks observed at regulatory elements on a genome-wide scale [26] are due to recruitment by specific transcription factors, or by some other characteristic of open/actively transcribed chromatin.
There does appear to be a relationship between the peaks of HMGN3a/b and H3K14ac and the Glyt1 exons, although there are not enough primer sets to draw any firm conclusions. This relationship would be consistent with the genome-wide analysis [26], which revealed that HMGN1 is enriched at the intron–exon boundaries of expressed genes. It is well established that nucleosomes are well positioned relative to intron–exon boundaries, and that nucleosomes within exons are enriched in active histone modifications [45]. It is thought that chromatin structure might influence the rate of RNA polymerase II movement and exon usage during co-transcriptional splicing [45].
Stimulation of H3K14 acetylation by HMGN3a and HMGN3b
The ability of HMGN3a to stimulate H3K14 acetylation in vitro is consistent with previous data showing that HMGN2 and HMGN1 can stimulate the ability of PCAF to acetylate nucleosomal H3 by 3.5-fold and 2-fold respectively [15,16]. Kinetic data indicated that HMGN1 promotes H3K14 acetylation by making the H3 tail a better enzymatic substrate, rather than by increasing the binding affinity of PCAF or by decreasing the action of a specific deacetylase [15]. The RD is required for both HMGN1 and HMGN2 to stimulate acetylation by PCAF [15,16]. The RD is absent from HMGN3b, which explains why we could not detect HMGN3b-mediated stimulation of acetylation of mononucleosomes in vitro. It is notable that while H3K14 acetylation is increased by HMGN3a, acetylation of the nearby residue, H3K9, is unaffected in vivo. Similar results were observed for HMGN1, which stimulates H3K14 acetylation, yet inhibits H3S10 phosphorylation and H3K9 acetylation [15–17,22].
Given that HMGN3b does not stimulate nucleosomal acetylation by PCAF in vitro, it is unclear as to why it should promote H3K14 acetylation across the Glyt1 locus in vivo. It is possible that HMGN3b can alter the structure of the H3 tail in chromatin to promote acetylation by PCAF, whereas in mononucleosomes this is not possible. Indeed, it has previously been shown that the acetylation activity of recombinant PCAF increases in proportion to the length of the nucleosomal array substrate [46]. It is also possible that the recombinant PCAF used these in vitro experiments has a different response to HMGN3b compared with the various PCAF, GCN5 and Elp3 complexes that can acetylate H3K14ac in vivo (see below). Alternatively, HMGN3a and/or HMGN3b may alter the chromatin structure of the Glyt1 locus by another mechanism, for example by competing with the linker histone H1 [11,47], which could increase accessibility to HATs and thus promote transcription [11,47].
Acetylation of H3K14 across the Glyt1 gene body increases transcription
The relationship between histone acetylation and transcription is well documented. H3K14 acetylation is regulated by a balance of acetylation and deacetylation activities [48]. In yeast, H3K14 acetylation on gene bodies is deposited by GCN5 within the SAGA complex [49–51], and by Elp3 within the elongator complex [52], whereas Clr3 is the main H3K14 deacetylase [51]. In metazoans, GCN5 and its close homologue, PCAF, are found in several highly related 2 MDa complexes (the TFTC, STAGA and PCAF/GCN5 complexes) as well as the 700 kDa ATAC complex (reviewed in [53]). These complexes have been shown to acetylate nucleosomal H3K14, in addition to other residues, although it is clear that subunit composition influences the substrate specificity of the HAT complex [53,54].
On highly transcribed genes, H3 acetylation by SAGA is linked to nucleosome eviction and thus increased elongation efficiency [50]. However, it has also been shown that genes transcribed at lower levels do not lose nucleosomes during transcription, and that the level of acetylation across these genes is closely correlated with the rate of transcription [50]. This is consistent with the results of the present study, where the level of H3 acetylation is related to the level of Glyt1 transcription. The nucleosome remodelling complex RSC is known to be recruited by H3K14 acetylation via its bromodomains [55], and has been shown to increase elongation efficiency by RNA polymerase II in vitro [56]. Furthermore, prior acetylation of chromatin by SAGA increases the ability of RSC to stimulate transcription elongation [56,57].
We suggest that H3K14 acetylation across the Glyt1 gene body may improve elongation efficiency, possibly by promoting the recruitment of the nucleosome remodelling complex RSC. HMGN1 has previously been shown to stimulate RNA polymerase II elongation on isolated SV40 minichromosomes [58], but a link between HMGNs and transcription elongation in vivo has not been demonstrated. The Glyt1a primary transcript is relatively long (35 kb), and straddles the silent Glyt1b promoter, so it may be particularly sensitive to improvements in elongation efficiency.
In summary, our analysis has revealed that although HMGN3 expression stimulates Glyt1 transcription, its binding is not specifically enriched at the gene body or its regulatory elements. HMGN3 expression does not alter the level of the histone modifications associated with the TSS, but does increase the level of H3K14ac across the locus. We propose a model whereby HMGN3-stimulated H3K14 acetylation across the bodies of genes such as Glyt1 can lead to more efficient transcription elongation and increased mRNA production.
Supplementary Material
ACKNOWLEDGEMENT
We thank Dr Adam West (University of Glasgow) for scientific advice and comments on the paper before submission.
FUNDING This work was supported by the Biotechnology and Biological Sciences Research Council [grant number BB/C006496/1]; the University of Malaya, Kuala Lumpur, Malaysia (Ph.D. funding to G.M.); the Association for International Cancer Research [grant number 07-0127]; a European Community Marie Curie International Reintegration Grant [contract number 006652]; and the Centre for Cancer Research intramural program of the National Cancer Institute and National Institutes of Health.
Abbreviations used
- ChIP
chromatin immunoprecipitation
- CTCF
CCCTC-binding factor
- DHS
DNase I-hypersensitive site
- DTT
dithiothreitol
- ERα
oestrogen receptor α
- Gapdh
glyceraldehyde-3-phosphate dehydrogenase
- GCN5
general control of amino acid synthesis 5
- Glut2
glucose transpoter 2
- Glyt1
glycine transporter 1
- H3K4me1
monomethylated Lys4 of histone H3
- H3K4me3
trimethylated Lys4 of histone H3
- H3K9ac
acetylated Lys9 of histone H3
- H3K14
Lys14 of histone H3
- H3K14ac
acetylated Lys14 of histone H3
- H3S10
Ser10 of histone H3
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- HMGN
high-mobility group nucleosome-binding
- Hsp70
heat-shock protein 70
- MSK1
mitogen- and stress-activated kinase 1
- NBD
nucleosome-binding domain
- NLS
nuclear localization sequence/signal
- PCAF
p300/CREB (cAMP-response-element-binding protein)-binding protein-associated factor
- PDX-1
pancreatic and duodenal homeobox-1
- PITX2
pituitary homeobox 2
- RD
regulatory domain
- RT
reverse transcription
- RXR
retinoid X receptor
- SRF
serum-response factor
- TSS
transcription start site
Footnotes
AUTHOR CONTRIBUTION Gráinne Barkess, Yuri Postnikov, Chrisanne Campos, Shivam Mishra, Gokula Mohan, Sakshi Verma and Katherine West carried out the experiments and analysed the data. Michael Bustin and Katherine West supervised the work. Katherine West wrote the paper, with help from Grainne Barkess, Yuri Postnikov and Michael Bustin. All authors reviewed and commented on the paper.
REFERENCES
- 1.Postnikov U, Bustin M. Regulation of chromatin structure and function by HMGN proteins. Biochim. Biophys. Acta. 2010;1799:62–68. doi: 10.1016/j.bbagrm.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rochman M, Taher L, Kurahashi T, Cherukuri S, Uversky VN, Landsman D, Ovcharenko I, Bustin M. Effects of HMGN variants on the cellular transcription profile. Nucleic Acids Res. 2011;39:1–12. doi: 10.1093/nar/gkq1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.West KL, Castellini MA, Duncan MK, Bustin M. Chromosomal proteins HMGN3a and HMGN3b regulate the expression of glycine transporter 1. Mol. Cell. Biol. 2004;24:3747–3756. doi: 10.1128/MCB.24.9.3747-3756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birger Y, West KL, Postnikov YV, Lim JH, Furusawa T, Wagner JP, Laufer CS, Kraemer KH, Bustin M. Chromosomal protein HMGN1 enhances the rate of DNA repair in chromatin. EMBO J. 2003;22:1665–1675. doi: 10.1093/emboj/cdg142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vestner B, Bustin M, Gruss C. Stimulation of replication efficiency of a chromatin template by chromosomal protein HMG-17. J. Biol. Chem. 1998;273:9409–9414. doi: 10.1074/jbc.273.16.9409. [DOI] [PubMed] [Google Scholar]
- 6.Furusawa TCS. Developmental function of HMGN proteins. Biochim. Biophys. Acta. 2010;1799:69–73. doi: 10.1016/j.bbagrm.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bustin M. Chromatin unfolding and activation by HMGN(*) chromosomal proteins. Trends Biochem. Sci. 2001;26:431–437. doi: 10.1016/s0968-0004(01)01855-2. [DOI] [PubMed] [Google Scholar]
- 8.Rochman M, Malicet C, Bustin M. HMGN5/NSBP1: a new member of the HMGN protein family that affects chromatin structure and function. Biochim. Biophys. Acta. 2010;1799:86–92. doi: 10.1016/j.bbagrm.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.West KL, Ito Y, Birger Y, Postnikov Y, Shirakawa H, Bustin M. HMGN3a and HMGN3b, two protein isoforms with a tissue-specific expression pattern, expand the cellular repertoire of nucleosome-binding proteins. J. Biol. Chem. 2001;276:25959–25969. doi: 10.1074/jbc.M101692200. [DOI] [PubMed] [Google Scholar]
- 10.Trieschmann L, Alfonso PJ, Crippa MP, Wolffe AP, Bustin M. Incorporation of chromosomal proteins HMG-14/HMG-17 into nascent nucleosomes induces an extended chromatin conformation and enhances the utilization of active transcription complexes. EMBO J. 1995;14:1478–1489. doi: 10.1002/j.1460-2075.1995.tb07134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding HF, Bustin M, Hansen U. Alleviation of histone H1-mediated transcriptional repression and chromatin compaction by the acidic activation region in chromosomal protein HMG-14. Mol. Cell. Biol. 1997;17:5843–5855. doi: 10.1128/mcb.17.10.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paranjape SM, Krumm A, Kadonaga JT. HMG17 is a chromatin-specific transcriptional coactivator that increases the efficiency of transcription initiation. Genes Dev. 1995;9:1978–1991. doi: 10.1101/gad.9.16.1978. [DOI] [PubMed] [Google Scholar]
- 13.Catez F, Brown DT, Misteli T, Bustin M. Competition between histone H1 and HMGN proteins for chromatin binding sites. EMBO Rep. 2002;3:760–766. doi: 10.1093/embo-reports/kvf156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rattner BP, Yusufzai T, Kadonaga JT. HMGN proteins act in opposition to ATP-dependent chromatin remodeling factors to restrict nucleosome mobility. Mol. Cell. 2009;34:620–626. doi: 10.1016/j.molcel.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim JH, West KL, Rubinstein Y, Bergel M, Postnikov YV, Bustin M. Chromosomal protein HMGN1 enhances the acetylation of lysine 14 in histone H3. EMBO J. 2005;24:3038–3048. doi: 10.1038/sj.emboj.7600768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ueda T, Postnikov YV, Bustin M. Distinct domains in high mobility group N variants modulate specific chromatin modifications. J. Biol. Chem. 2006;281:10182–10187. doi: 10.1074/jbc.M600821200. [DOI] [PubMed] [Google Scholar]
- 17.Lim JH, Catez F, Birger Y, West KL, Prymakowska-Bosak M, Postnikov YV, Bustin M. Chromosomal protein HMGN1 modulates histone H3 phosphorylation. Mol. Cell. 2004;15:573–584. doi: 10.1016/j.molcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 18.Shimada M, Nakadai T, Fukuda A, Hisatake K. cAMP-response element-binding protein (CREB) controls MSK1-mediated phosphorylation of histone H3 at the c-fos promoter in vitro. J. Biol. Chem. 2010;285:9390–9401. doi: 10.1074/jbc.M109.057745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Postnikov YV, Belova GI, Lim JH, Bustin M. Chromosomal protein HMGN1 modulates the phosphorylation of serine 1 in histone H2A. Biochemistry. 2006;45:15092–15099. doi: 10.1021/bi0613271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amano T, Leu K, Yoshizato K, Shi YB. Thyroid hormone regulation of a transcriptional coactivator in Xenopus laevis: implication for a role in postembryonic tissue remodeling. Dev. Dyn. 2002;223:526–535. doi: 10.1002/dvdy.10075. [DOI] [PubMed] [Google Scholar]
- 21.Amen M, Espinoza HM, Cox C, Liang X, Wang J, Link TM, Brennan RG, Martin JF, Amendt BA. Chromatin-associated HMG-17 is a major regulator of homeodomain transcription factor activity modulated by Wnt/β-catenin signaling. Nucleic Acids Res. 2008;36:462–476. doi: 10.1093/nar/gkm1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu N, Hansen U. HMGN1 modulates estrogen-mediated transcriptional activation through interactions with specific DNA-binding transcription factors. Mol. Cell. Biol. 2007;27:8859–8873. doi: 10.1128/MCB.01724-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ueda T, Furusawa T, Kurahashi T, Tessarollo L, Bustin M. The nucleosome binding protein HMGN3 modulates the transcription profile of pancreatic β cells and affects insulin secretion. Mol. Cell. Biol. 2009;29:5264–5276. doi: 10.1128/MCB.00526-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furusawa T, Lim JH, Catez F, Birger Y, Mackem S, Bustin M. Down-regulation of nucleosomal binding protein HMGN1 expression during embryogenesis modulates Sox9 expression in chondrocytes. Mol. Cell. Biol. 2006;26:592–604. doi: 10.1128/MCB.26.2.592-604.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belova GI, Postnikov YV, Furusawa T, Birger Y, Bustin M. Chromosomal protein HMGN1 enhances the heat shock-induced remodeling of Hsp70 chromatin. J. Biol. Chem. 2008;283:8080–8088. doi: 10.1074/jbc.M709782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cuddapah S, Schones DE, Cui K, Roh TY, Barski A, Wei G, Rochman M, Bustin M, Zhao K. Genomic profiling of HMGN1 reveals an association with chromatin at regulatory regions. Mol. Cell. Biol. 2011;31:700–709. doi: 10.1128/MCB.00740-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zafra F, Gimenez C. Glycine transporters and synaptic function. IUBMB Life. 2008;60:810–817. doi: 10.1002/iub.128. [DOI] [PubMed] [Google Scholar]
- 28.Kim KM, Kingsmore SF, Han H, Yang-Feng TL, Godinot N, Seldin MF, Caron MG, Giros B. Cloning of the human glycine transporter type 1: molecular and pharmacological characterization of novel isoform variants and chromosomal localization of the gene in the human and mouse genomes. Mol. Pharmacol. 1994;45:608–617. [PubMed] [Google Scholar]
- 29.Trieschmann L, Martin B, Bustin M. The chromatin unfolding domain of chromosomal protein HMG-14 targets the N-terminal tail of histone H3 in nucleosomes. Proc. Natl. Acad. Sci. U.S.A. 1998;95:5468–5473. doi: 10.1073/pnas.95.10.5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crippa MP, Alfonso PJ, Bustin M. Nucleosome core binding region of chromosomal protein HMG-17 acts as an independent functional domain. J. Mol. Biol. 1992;228:442–449. doi: 10.1016/0022-2836(92)90833-6. [DOI] [PubMed] [Google Scholar]
- 31.Rada-Iglesias A, Enroth S, Ameur A, Koch CM, Clelland GK, Respuela-Alonso P, Wilcox S, Dovey OM, Ellis PD, Langford CF, et al. Butyrate mediates decrease of histone acetylation centered on transcription start sites and down-regulation of associated genes. Genome Res. 2007;17:708–719. doi: 10.1101/gr.5540007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beedanagari SR, Taylor RT, Hankinson O. Differential regulation of the dioxin-induced Cyp1a1 and Cyp1b1 genes in mouse hepatoma and fibroblast cell lines. Toxicol. Lett. 2010;194:26–33. doi: 10.1016/j.toxlet.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zelko IN, Folz RJ. Sp1 and Sp3 transcription factors mediate trichostatin A-induced and basal expression of extracellular superoxide dismutase. Free Radical Biol. Med. 2004;37:1256–1271. doi: 10.1016/j.freeradbiomed.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 34.Duan Z, Stamatoyannopoulos G, Li Q. Role of NF-Y in in vivo regulation of the γ -globin gene. Mol. Cell. Biol. 2001;21:3083–3095. doi: 10.1128/MCB.21.9.3083-3095.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Postnikov YV, Lehn DA, Robinson RC, Friedman FK, Shiloach J, Bustin M. The cooperative binding of chromosomal protein HMG-14 to nucleosome cores is reduced by single point mutations in the nucleosomal binding domain. Nucleic Acids Res. 1994;22:4520–4526. doi: 10.1093/nar/22.21.4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borowsky B, Hoffman BJ. Analysis of a gene encoding two glycine transporter variants reveals alternative promoter usage and a novel gene structure. J. Biol. Chem. 1998;273:29077–29085. doi: 10.1074/jbc.273.44.29077. [DOI] [PubMed] [Google Scholar]
- 37.Bustin M, Crippa MP, Pash JM. Immunochemical analysis of the exposure of high mobility group protein 14 and 17 surfaces in chromatin. J. Biol. Chem. 1990;265:20077–20080. [PubMed] [Google Scholar]
- 38.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 41.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenbloom KR, Dreszer TR, Pheasant M, Barber GP, Meyer LR, Pohl A, Raney BJ, Wang T, Hinrichs AS, Zweig AS, et al. ENCODE whole-genome data in the UCSC Genome Browser: update 2012. Nucleic Acids Res. 2012;38:D620–D625. doi: 10.1093/nar/gkp961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schiltz RL, Mizzen CA, Vassilev A, Cook RG, Allis CD, Nakatani Y. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem. 1999;274:1189–1192. doi: 10.1074/jbc.274.3.1189. [DOI] [PubMed] [Google Scholar]
- 45.Dhami P, Saffrey P, Bruce AW, Dillon SC, Chiang K, Bonhoure N, Koch CM, Bye J, James K, Foad NS, et al. Complex exon-intron marking by histone modifications is not determined solely by nucleosome distribution. PLoS ONE. 2010;5:e12339. doi: 10.1371/journal.pone.0012339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herrera JE, Schiltz RL, Bustin M. The accessibility of histone H3 tails in chromatin modulates their acetylation by P300/CBP-associated factor. J. Biol. Chem. 2000;275:12994–12999. doi: 10.1074/jbc.275.17.12994. [DOI] [PubMed] [Google Scholar]
- 47.Rochman M, Postnikov Y, Correll S, Malicet C, Wincovitch S, Karpova TS, McNally JG, Wu X, Bubunenko NA, Grigoryev S, Bustin M. The interaction of NSBP1/HMGN5 with nucleosomes in euchromatin counteracts linker histone-mediated chromatin compaction and modulates transcription. Mol. Cell. 2009;35:642–656. doi: 10.1016/j.molcel.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wittschieben BO, Fellows J, Du W, Stillman DJ, Svejstrup JQ. Overlapping roles for the histone acetyltransferase activities of SAGA and elongator in vivo. EMBOJ. 2000;19:3060–3068. doi: 10.1093/emboj/19.12.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Govind CK, Zhang F, Qiu H, Hofmeyer K, Hinnebusch AG. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol. Cell. 2007;25:31–42. doi: 10.1016/j.molcel.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 50.Kristjuhan A, Svejstrup JQ. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBOJ. 2004;23:4243–4252. doi: 10.1038/sj.emboj.7600433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnsson A, Durand-Dubief M, Xue-Franzen Y, Ronnerblad M, Ekwall K, Wright A. HAT-HDAC interplay modulates global histone H3K14 acetylation in gene-coding regions during stress. EMBO Rep. 2009;10:1009–1014. doi: 10.1038/embor.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winkler GS, Kristjuhan A, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3517–3522. doi: 10.1073/pnas.022042899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagy Z, Riss A, Fujiyama S, Krebs A, Orpinell M, Jansen P, Cohen A, Stunnenberg HG, Kato S, Tora L. The metazoan ATAC and SAGA coactivator HAT complexes regulate different sets of inducible target genes. Cell. Mol. Life Sci. 2007;67:611–628. doi: 10.1007/s00018-009-0199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schiltz RL, Mizzen CA, Vassilev A, Cook RG, Allis CD, Nakatani Y. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem. 1999;274:1189–1192. doi: 10.1074/jbc.274.3.1189. [DOI] [PubMed] [Google Scholar]
- 55.Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- 56.Carey M, Li B, Workman JL. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol. Cell. 2006;24:481–487. doi: 10.1016/j.molcel.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kasten M, Szerlong H, Erdjument-Bromage H, Tempst P, Werner M, Cairns BR. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ding HF, Rimsky S, Batson SC, Bustin M, Hansen U. Stimulation of RNA polymerase II elongation by chromosomal protein HMG- 14. Science. 1994;265:796–799. doi: 10.1126/science.8047885. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.